Abstract
Many plants associate with arbuscular mycorrhizal fungi for nutrient acquisition, while legumes also associate with nitrogen-fixing rhizobial bacteria. Both associations rely on symbiosis signaling and here we show that cereals can perceive lipochitooligosaccharides (LCOs) for activation of symbiosis signaling, surprisingly including Nod factors produced by nitrogen-fixing bacteria. However, legumes show stringent perception of specifically decorated LCOs, that is absent in cereals. LCO perception in plants is activated by nutrient starvation, through transcriptional regulation of Nodulation Signaling Pathway (NSP)1 and NSP2. These transcription factors induce expression of an LCO receptor and act through the control of strigolactone biosynthesis and the karrikin-like receptor DWARF14-LIKE. We conclude that LCO production and perception is coordinately regulated by nutrient starvation to promote engagement with mycorrhizal fungi. Our work has implications for the use of both mycorrhizal and rhizobial associations for sustainable productivity in cereals.
Subject terms: Arbuscular mycorrhiza, Strigolactone, Plant signalling
Lipochitooligosaccharide (LCO) perception by legumes is required to establish symbiotic relationships with nitrogen fixing bacteria. Here the authors show that nutrient starvation can activate LCO perception in cereals to promote symbiotic association with arbuscular mycorrhizal fungi.
Introduction
Plant growth and development largely depends on the absorption of mineral nutrients, with nitrogen (N) and phosphorus (P) being the principal limitations to plant productivity1. These nutrients are widely applied in agriculture as inorganic fertilizers to maximize crop yields. Fertilizer over-use pollutes terrestrial and aquatic environments, significantly impacts biodiversity, and contributes to a sizable proportion of agricultural greenhouse gas emissions2–4.
It is estimated that most land plants associate with beneficial arbuscular mycorrhizal fungi (AMF) that capture mineral nutrients and water from the soil5–7. During this association the fungus makes contact with the plant root following recognition of plant-derived strigolactones that promote fungal growth and the production of fungal signals, short-chain chitooligosaccharides (COs), and lipochitooligosaccharides (LCOs)8–10. In legumes, the recognition of COs and LCOs activates symbiosis signaling in epidermal cells of the plant root9–11, which facilitates the processes necessary to accommodate AMF12. In contrast, reports in rice demonstrate recognition of primarily COs11,13,14, with very limited perception of LCOs14, for establishment of the AMF association. Initial intracellular infection of root atrichoblast cells allows entry to the root, followed by intercellular fungal growth through the root tissue. Within inner root cortical cells, the fungus produces extensively branched arbuscular intrusions, surrounded by plant membrane, creating a large surface interface for nutrient exchange. In parallel extensive fungal colonization of the surrounding soil provides greater access to mineral nutrient capture than the plant root alone can achieve. Sources of N and P captured by the fungus are exchanged for carbon derived from photosynthesis, the sole carbon source that AMF can acquire15–18.
Plant species within the N-fixing clade have augmented their symbiotic associations with the addition of interactions with N-fixing bacteria, that allow the plant access to the unlimited supply of atmospheric N through conversion to ammonium. The evolution of this association recruited preexisting processes associated with the AMF association, including symbiosis signaling19,20. In AMF-host species, detection of LCOs and/or COs leads to the activation of the common symbiosis signaling pathway, which is essential for accommodating the fungal symbiont11,21 and in legumes the same signaling pathway drives the establishment of the interaction with N-fixing bacteria leading to the production of nodules22–24. Consequently, it is feasible that manipulation of these pre-existing mechanisms in cereals may enable the engineering of N-fixing symbioses.
Maintaining AMF and rhizobial symbioses are expensive to the plant in terms of supplying sources of carbon from photosynthesis, and as such the symbioses are tightly regulated by the availability of nutrients, particularly N-availability in the regulation of nodulation and P-availability in the regulation of AMF symbioses. Plants preferentially enter these associations when these nutrients are too limiting to be satisfied by direct root capture25,26, and it has long been known that in nutrient-replete conditions, the symbioses are strongly inhibited27,28. In the case of nodulation, CEP-peptides that act as N-starvation signals, promote the production of miR2111, which inhibits a negative regulator of nodulation in the root29,30. P-regulation of the AMF symbiosis is in part controlled by PHR231,32, a transcription factor that controls many aspects of the P-starvation response, as well as NSP1 and NSP2 that regulate the production of strigolactones33. PHR2 has been demonstrated to directly regulate many genes associated with arbuscule function, in particular nutrient exchange between the plant and the fungus31, but to also control the expression of genes associated with CO and LCO perception and the symbiosis signaling pathway32, implicating roles both in the early recognition of AMF fungi at the root surface, as well as regulating arbuscule function in the root cortex. Here, we demonstrate the mechanism by which LCO recognition in the plant is coordinately controlled with LCO production by AMF, through the action of NSP1 and NSP2, facilitating nutrient control of AMF colonization.
Results
Cereals show non-stringent perception of LCOs
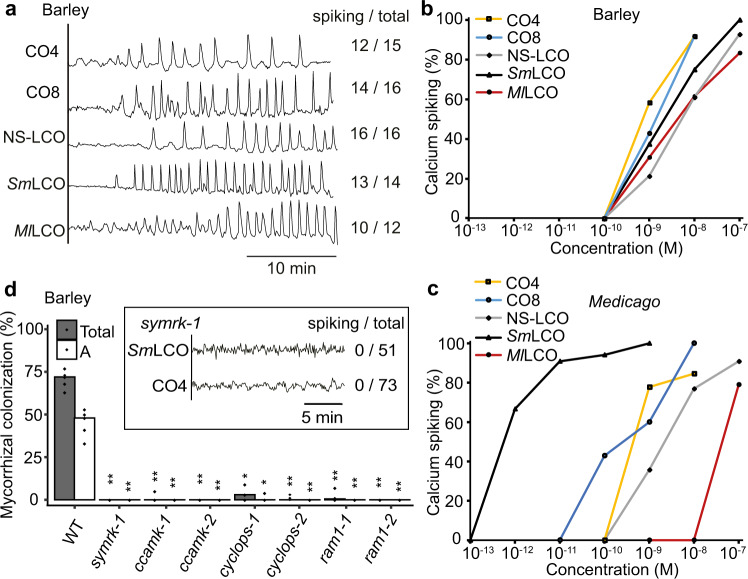
Whereas legumes show robust induction of symbiosis signaling in response to LCOs and COs12, results from rice are inconsistent, with reports of either no LCO responses11 or occasional non-periodic calcium oscillations14, and inconsistent results on the impact of mutations in the LCO receptor homolog NFR514,34. To rationalize the discrepancy of CO and LCO perception in legumes and cereals, we generated an array of cereal lines carrying the calcium reporter cameleon YC3.6, allowing measurement of symbiotic calcium oscillations. At early developmental stages (up to 5-day-old plants) in barley, we only observed CO-induced oscillations, but no responses to LCOs (Supplementary Fig. 1a), consistent with what we previously reported in rice11. However, two-week-old barley roots grown under nutrient depletion showed nuclear calcium oscillations in atrichoblasts in response to COs and LCOs (Fig. 1a). Barley perceived LCOs produced by AMF (non-sulfated LCO: NS-LCO), as well as those produced by the N-fixing rhizobial bacteria Sinorhizobium meliloti (SmLCO) and Mezorhizobium loti (MlLCO). These calcium oscillations are comparable to those induced by LCOs in Medicago truncatula and Lotus japonicus12. Dose response curves reveal that barley shows responses at equivalent concentrations for all molecules tested (Fig. 1b), whereas M. truncatula strongly discriminates between LCOs produced by its bacterial symbiont S. meliloti versus its non-symbiont M. loti (Fig. 1c). Symbiotic calcium oscillations are a function of symbiosis signaling and consistently barley lines mutated in multiple components of this signaling pathway (Supplementary Fig. 1b) abolish associations with the AMF Rhizophagus irregularis (Fig. 1d and Supplementary Fig. 1c). SYMRK, a co-receptor upstream of calcium oscillations in symbiosis signaling, is essential for both LCO and CO-induced calcium oscillations (Fig. 1d, inset).
Fig. 1. LCOs and COs activate calcium oscillations in barley.
a Representative calcium traces of atrichoblasts of barley (H. vulgare) lateral roots grown in nutrient depletion, responding to 10−8 M CO4, 10−8 M CO8, 10−7 M NS-LCO (non-sulphated LCO produced by R. irregularis), 10−7 M SmLCO (LCO produced by S. meliloti), and 10−7 M MlLCO (LCO produced by Mesorhizobium loti). Numbers indicate cells responding compared to total cells analyzed. Dose-response curves for LCO and CO induction of calcium oscillations in barley (b) and M. truncatula (c) lateral roots. Calcium spiking (%) indicates the ratio of cells responding with calcium oscillations. d AMF colonization measured at 7 weeks post-inoculation of barley wild type (WT), symrk, ccamk, cyclops, and ram1 mutants, with the inset showing representative traces of barley symrk mutant atrichoblasts responding to 10−7 M SmLCO and 10−7 M CO4. Numbers indicate cells responding compared to total cells analyzed. n = 3-5 biologically independent plants. Total: total colonization; A: arbuscules. For statistical analysis a one-sided Wilcoxon test was performed. **p < 0.01.
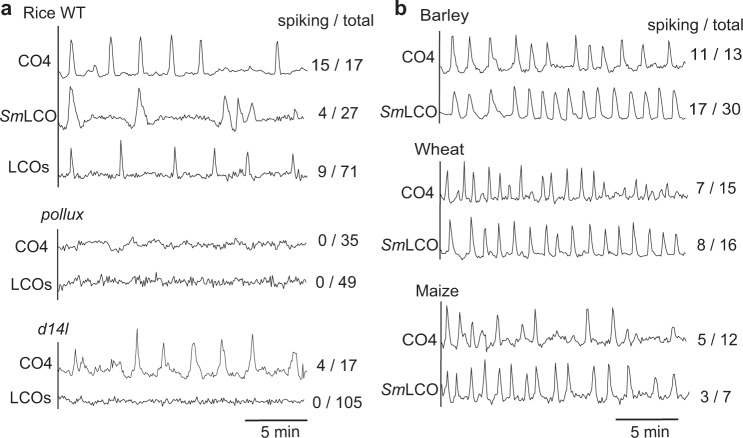
We previously reported that rice activates calcium oscillations in response to COs, not LCOs11, while others have shown non-periodic responses to LCOs14. Our previous work was undertaken in young rice seedlings, the only stage when YC3.6 was detectable in the available rice line and a stage when barley shows only CO responses (Supplementary Fig. 1a). A new rice YC3.6 line allowed analysis at later stages of plants grown under nutrient depletion, and now both LCO and CO responses could be observed, in a manner dependent on POLLUX, a component of symbiosis signaling required for calcium oscillations21,35 (Fig. 2a). The karrikin receptor Dwarf14-Like (D14L), has been shown to be essential for the establishment of AMF colonization in rice and acts through the inhibition of the SMAX1 suppressor36,37. d14l mutants of rice show the complete absence of a transcriptional response to AMF, suggesting a very early role in allowing AMF recognition36. Consistent with these earlier observations, we found that LCO responsiveness was abolished in d14l and CO responsiveness was attenuated (Fig. 2a).
Fig. 2. COs and LCOs activate calcium oscillations in cereals.
a Representative traces of atrichoblasts from lateral roots in rice wild type (WT), pollux and d14l mutants responding to 10−5 M CO4, 10−7 M SmLCO and LCOs (1:1 mix of 10−7 M sulfated-LCO and 10−7 M NS-LCO). b Representative calcium traces of barley, wheat, and maize atrichoblasts responding to 10−7 M CO4 and 10−7 M SmLCO. All plants were grown under nutrient deficient conditions. Numbers indicate cells responding compared to total cells analyzed.
The observation of LCO induction of symbiosis signaling in both barley and rice suggests that LCO recognition may be a common feature of cereals generally. To assess this, we analysed wheat and maize lines carrying YC3.6 and tested for LCO and CO-induced symbiotic calcium oscillations. We observed responses comparable to that observed in barley in both wheat and maize (Fig. 2b). We conclude that CO and LCO induction of symbiosis signaling are a common feature of legumes and cereals, with legumes augmenting the stringency of LCO recognition to differentiate rhizobial species.
Nutrient regulation of LCO recognition involves NSP1 and NSP2
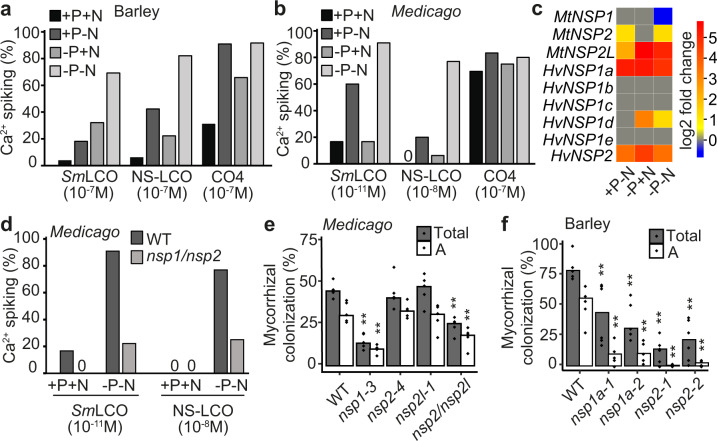
We only observed LCO recognition in barley when the plants were depleted for nutrients and the onset of LCO responsiveness was associated with developmental markers of nutrient starvation (Supplementary Fig. 1d). We, therefore, further assessed the regulation of symbiosis signaling under different nutrient regimes. Barley and M. truncatula only show activation of LCO perception under N- and/or P-starvation (Fig. 3a, b), but the oscillation patterns are not equivalent among each nutrient regime, with the combination of N- and P-starvation displaying more frequent and robust oscillations (Supplementary Fig. 2a, b). The principal difference between the barley and M. truncatula responses to nutrient starvation is a strong preference for N-starvation in LCO perception in M. truncatula (Fig. 3b), reflecting the additional importance of LCO regulation in the control of the association with N-fixing bacteria. We found no impact of nutrient starvation for CO induction of calcium oscillations in M. truncatula, but some nutrient regulation of the CO response in barley (Fig. 3a, b). However, the degree of nutrient regulation of CO-induced calcium oscillations in barley was considerably less than that observed for LCO responses (Fig. 3a).
Fig. 3. NSP1 and NSP2 control nutrient regulation of symbiosis signaling and AMF colonization.
Nutrient regulation of symbiosis signaling in 16-day-old barley (a) and M. truncatula (b) lateral roots grown under different nutrient regimes, responding to SmLCO, NS-LCO, or CO4. Calcium spiking (%) indicates the number of cells responding with calcium oscillations, with 0 indicating no calcium oscillations observed. c A heatmap showing the expression of M. truncatula and barley NSP1 and NSP2 homologs upon N- and/or P- starvation. +P-N represents the comparison of gene expression under +P−N to +P + N ( + P−N/ + P + N), similarly −P + N and −P-N representing −P + N/ + P + N and −P−N/ + P + N. d The percentage of M. truncatula atrichoblasts on lateral roots showing calcium oscillations in wild type and the nsp1-1/nsp2-2 double mutant, grown under nutrient replete or deplete conditions. e AMF colonization of M. truncatula mutants grown under P-limited conditions, measured at 3 weeks post inoculation. n = 5 biologically independent samples. f Root-length colonization of barley wild type, nsp1a, and nsp2 mutants grown under P-limited conditions, measured at 7 weeks post inoculation. n = 5–6 biologically independent plants. Total: total colonization; A: arbuscules. p-values for colonization levels were determined by a one-sided Wilcoxon test. **p < 0.01; *0.01 < p < 0.05.
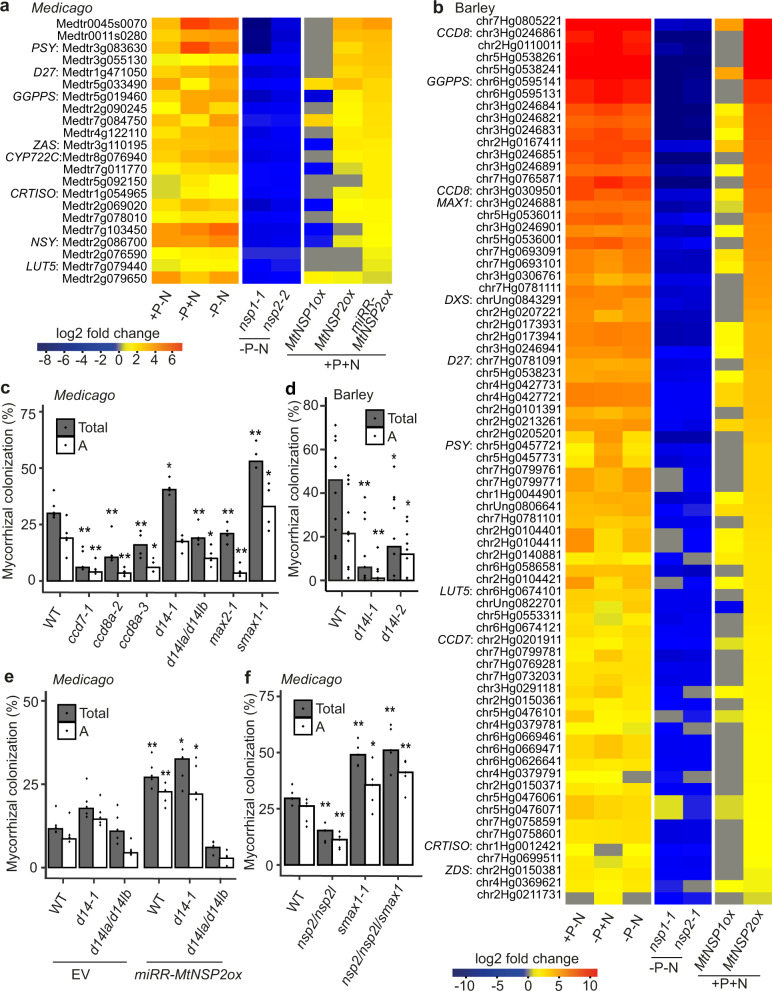
Nodulation Signaling Pathway (NSP)1 and NSP2 were discovered because of their major roles in establishing root nodule symbiosis in M. truncatula38,39, but were secondarily shown to also be necessary for appropriate AMF colonization9,40,41. Among the NSP1 and NSP2 gene families in M. truncatula and barley are a number of genes regulated by the nutrient status (Fig. 3c and Supplementary Fig. 3a, b), suggesting a possible role in the nutrient regulation of symbiosis. The promotion of LCO responsiveness by nutrient starvation, measured by activation of calcium oscillations, was greatly attenuated in M. truncatula nsp1/nsp2, compared to wild type (Fig. 3d). That this is relevant to symbiosis is supported by the observation that M. truncatula nsp1 shows defects in AMF colonization (Fig. 3e and Supplementary Fig. 4a–b), as previously reported41. However, no colonization defects were observed in single mutants of M. truncatula NSP2 or its homolog NSP2L (Fig. 3e and Supplementary Fig. 4a–c), but a double mutant between these two genes, nsp2/nsp2l, displayed reduced AMF colonization (Fig. 3e). Barley possesses only a single ortholog of NSP2 (Supplementary Fig. 3b), that is transcriptionally responsive to nutrient starvation (Fig. 3c), but five potential orthologs of NSP1 (Supplementary Fig. 3a), one of which NSP1a shows a very strong transcriptional response to nutrient starvation (Fig. 3c). Mutation of HvNSP1a and HvNSP2 revealed major functions in AMF colonization, with reduced total colonization and arbuscule formation (Fig. 3f and Supplementary Fig. 5a). The nsp mutant phenotypes in barley are more severe than any of the M. truncatula nsp mutants (Fig. 3e, f), revealing a major function for these genes during AMF associations in cereals.
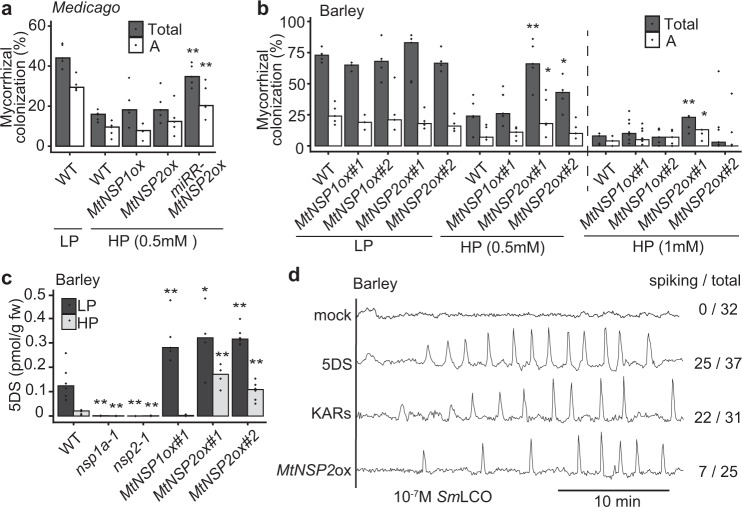
NSP2 in M. truncatula is controlled by miR171h and, consistent with previous work that overexpression of a miR171h-resistant version of NSP2 (miRR-NSP2), but not native NSP2, increased AMF colonization under P-limiting condition40, we observed that only miRR-NSP2 overexpression promoted AMF colonization in M. truncatula under repressive P concentrations (Fig. 4a), while native NSP1 or NSP2 overexpression showed no effect (Fig. 4a). In wild-type barley, AMF colonization is suppressed with increased P concentrations (Supplementary Fig. 6a), and overexpression of M. truncatula (Mt)NSP2, but not MtNSP1 (Supplementary Fig. 6b–c), in barley also partially blocked P-suppression of mycorrhization (Fig. 4b and Supplementary Fig. 6d). We conclude that the mis-regulation of NSP2 is sufficient to partially overcome P-suppression of mycorrhization in both M. truncatula and barley, pointing towards a conserved dominance of NSP2 in the control of the AMF symbiosis.
Fig. 4. Overexpression of NSP2 promotes AMF colonization and strigolactone biosynthesis.
a AMF colonization measured at 5 weeks post-inoculation in M. truncatula roots overexpressing MtNSP1, MtNSP2, and a miR171h-resistant version of NSP2 (miRR-MtNSP2), grown under high P (HP) conditions, with wild-type (WT) under low P (LP) and HP conditions as controls. WT indicates roots transformed with an empty vector. n = 5 biologically independent samples. b Root-length colonization at 7 weeks post-inoculation in barley roots overexpressing MtNSP1 and MtNSP2 grown under different P levels. n = 3–5 biologically independent plants. c Strigolactone levels in barley nsp mutants and NSP overexpression lines under both LP and HP conditions. n = 4–6 biologically independent samples. d Representative calcium traces of atrichoblasts on lateral roots of barley responding to SmLCO. Wild-type plants were grown under repressive P-conditions and pretreated for 16 h with buffer (mock), 0.1 μM 5-deoxystrigol (5DS), or a mixture of 0.1 μM karrikin 1 and karrikin 2 (KARs), as well as transgenic plants overexpressing MtNSP2 grown under repressive P-conditions. Numbers indicate cells responding with calcium oscillations compared to total cells analyzed. Total: total colonization; A: arbuscules. p-values were determined by a one-sided Wilcoxon test. **p < 0.01; * 0.01 < p < 0.05.
NSP1 and NSP2 control strigolactone biosynthesis in response to nutrient starvation
P-starvation is associated with the biosynthesis of strigolactones42–44, in part controlled by NSP1 and NSP233,45 and this is necessary for mycorrhizal and rhizobial colonization46,47. Consistently, we found barley nsp1 and nsp2 mutants lost all P starvation-induced strigolactone production (Fig. 4c). Overexpression of both MtNSP1 and MtNSP2 in barley led to higher levels of strigolactone at low P, but only MtNSP2 overexpression led to higher levels of strigolactone at high P (Fig. 4c). To assess whether such strigolactone induction could explain the promotion of LCO recognition at repressive P, we grew barley, M. truncatula and wheat at high P and treated roots overnight with either 5-deoxystrigol (5DS) or a combination of karrikin 1 and karrikin 2 (KARs). Either treatment was sufficient to strongly promote LCO recognition at a suppressive P concentration (Fig. 4d and Supplementary Fig. 2c, d), with a combination of karrikins showing a greater effect than single karrikin treatment (Supplementary Fig. 2e). A minimum of 8 h of treatment was required to see an effect (Supplementary Table 1). Moreover, overexpression of MtNSP2 in barley also allows LCO perception under repressive P conditions (Fig. 4d). We conclude that NSP1 and NSP2 are necessary and sufficient for P-regulation of strigolactone biosynthesis and the promotion of LCO recognition.
To assess the contributions made by NSP1 and NSP2 to nutrient starvation, we analyzed the N- and P-starvation transcriptome in barley and M. truncatula nsp mutants. NSP1 is responsible for 12% of the N- and P-starvation transcriptome in M. truncatula and 15% in barley, while NSP2 is responsible for 18% in M. truncatula and 23% in barley (Supplementary Fig. 7 and Supplementary Table 2). In order to focus on those genes most directly regulated by these transcription factors, we selected the nutrient responsive genes across all starvation conditions that were nsp1 or nsp2-dependent, as well as induced by NSP2 overexpression (Fig. 5a, b). This focused group of NSP-regulated genes includes many enzymes involved in apocarotenoid biosynthesis (Supplementary Fig. 8a), including most of the strigolactone biosynthesis pathway, but also many enzymes involved in other apocarotenoids associated with mycorrhization: mycorradicins, blumenols, and zaxinones48. In M. truncatula, nsp1 mutants loose root expression of the strigolactone biosynthetic gene D27 (Supplementary Fig. 8b), as previously reported33,45, but overexpression of NSP1 alone is not sufficient to activate D27 or other strigolactone biosynthetic genes (Fig. 5a and Supplementary Fig. 8c). Rather overexpression of NSP2 alone has some gene induction ability, but a combination of NSP1 and NSP2 overexpression in M. truncatula maximally activates these genes (Supplementary Fig. 8c), revealing the combinatorial function of NSP1 and NSP2 in M. truncatula49. Of the focused group of NSP-regulated genes, 73% and 44% could function in apocarotenoid or small molecule biosynthesis in M. truncatula and barley, respectively (Fig. 5a, b). The apocarotenoid and associated pathways appear a principal target of NSP action, and thus we propose that component(s) of this pathway may explain NSP action in nutrient regulation of symbiotic perception.
Fig. 5. NSP2 overrides P-suppression via strigolactone production and D14L/SMAX1 signaling.
Heatmaps showing N- and/or P-starvation induced genes regulated by NSP1 and NSP2 and constitutively activated by miRNA-resistant MtNSP2 overexpression in M. truncatula (a) and MtNSP2 overexpression in barley (b). Genes involved in strigolactone biosynthesis are annotated. +P–N, −P + N, and −P−N represent the expression of these starvation-induced genes in wild-type plants by comparing −N or/and −P conditions to +P + N. The nsp mutants show gene expression in nsp mutants compared to wild-type plants under nutrient depletion, while NSPox shows NSP overexpression roots compared to wild type under nutrient-replete condition. c AMF colonization at 3 weeks post-inoculation of M. truncatula mutants defective in strigolactone biosynthesis and signaling, as well as karrikin signaling. Plants were grown under low P conditions. n = 4–5 biologically independent samples. d Root-length colonization at 7 weeks post-inoculation in barley wild type and d14l mutant roots under P deficient conditions. n = 10 biologically independent plants. e AMF colonization of wild type, d14, and the d14l double mutant with or without overexpression of miRR-MtNSP2. Plants were grown under high-P (0.5 mM PO43−) conditions for 5 weeks. EV: empty vector. n = 5 biologically independent samples. f AMF colonization at 3 weeks post inoculation of M. truncatula nsp2/nsp2l, smax1, and nsp2/nsp2l/smax1 triple mutants. Plants were grown under low-P conditions. n = 5 biologically independent samples. Total: total colonization; A: arbuscules. For statistical analysis, a one-sided Wilcoxon test was performed. **p < 0.01; *0.01 < p < 0.05.
NSP2 promotes AMF colonization through karrikin signaling
Both karrikin and strigolactone treatment overcame P-suppression of LCO recognition (Fig. 4d and Supplementary Fig. 2c, d), and these two molecules are perceived by different, but homologous receptors: D14L and D14, respectively. Mutations in the strigolactone biosynthesis genes CCD7 and CCD8a (two CCD8 homologs in M. truncatula, Supplementary Figs. 3c, 7b and 4b) show strong reductions in R. irregularis colonization (Fig. 5c), while mutants in D14 (Supplementary Figs. 3d, 4a and 4b) show enhanced R. irregularis colonization, although the significance of this effect was variable between experiments (Fig. 5c and Supplementary Fig. 4d). This suggests strigolactone biosynthesis is important for the AMF symbiosis, but perception by D14 is not required and may even have negative connotations for AMF colonization. In contrast, mutants in the karrikin receptor D14L (two copies in M. truncatula and a single ortholog in barley, Supplementary Fig. 3d) display decreased colonization by R. irregularis in both M. truncatula and barley (Fig. 5c, d and Supplementary Fig. 5b). MAX2/D3, encoding an F-box protein crucial for both strigolatone and karrikin-like signaling, is a positive regulator of AMF colonization, with its M. truncatula mutant (Supplementary Fig. 4a, b) showing reduced R. irregularis colonization (Fig. 5c). Consistently, mutants in the negative regulator SMAX1 (Supplementary Figs. 3e, 4a and 4b), that is targeted for degradation by D14L37, show enhanced R. irregularis colonization (Fig. 5c). While not equivalent in genetic penetrance, the mycorrhizal phenotypes of these biosynthesis and signaling genes in Medicago are comparable to what has been observed in rice and other species36,37,47,50,51. We conclude that strigolactones are required as secreted signals to AMF in the rhizosphere, but plant recognition of strigolactones through D14 has no role in this symbiosis. In contrast, the D14L receptor, that is essential to remove the SMAX1 suppressor, shows comparable positive regulation of AMF establishment, with SMAX1 negatively regulating this association.
D14L is required in rice to promote LCO recognition for activation of symbiotic calcium oscillations (Fig. 2a), suggesting a role for this signaling component in the establishment of AMF perception at the root surface. Considering that NSPs regulate enzymes involved in apocarotenoid biosynthesis (Fig. 5a, b) and also control nutrient-starvation induced promotion of LCO perception (Fig. 3d), we hypothesized that NSP1 and NSP2 may function in nutrient starvation through the production of the D14L ligand. To test this, we assessed NSP2 promotion of AMF colonization in d14 and d14l mutants. We found that the promotion of AMF colonization at restrictive P-concentrations by NSP2 overexpression was dependent on D14L, but not D14 (Fig. 5e). Consistently, AMF colonization of an nsp2/nsp2l/smax1 triple mutant resembled the higher colonization of smax1, rather than the low colonization observed in the nsp2/nsp2l double mutant (Fig. 5f). From this we conclude that NSP2 functions upstream of the D14L/SMAX1 signaling module.
Strigolactones have been reported to have negative and positive effects in the regulation of nodulation in pea, dependent on whether mutants in biosynthetic enzymes or the receptor were analyzed47,52. In M. truncatula we found that both ccd7 and ccd8a mutants develop more nodules (Supplementary Fig. 4e), reflecting a negative role for strigolactones in nodulation of M. truncatula. Consistently, strigolactone signaling, through D14, also represses nodulation, while karrikin-like signaling, through D14L has no impact (Supplementary Fig. 4e). We conclude that the positive effects of strigolactone biosynthesis and karrikin-like signaling are specific to the AMF symbiosis and do not have equivalent functions during the association with N-fixing bacteria.
NSP1 and NSP2 control nutrient starvation-induction of an LCO receptor in barley
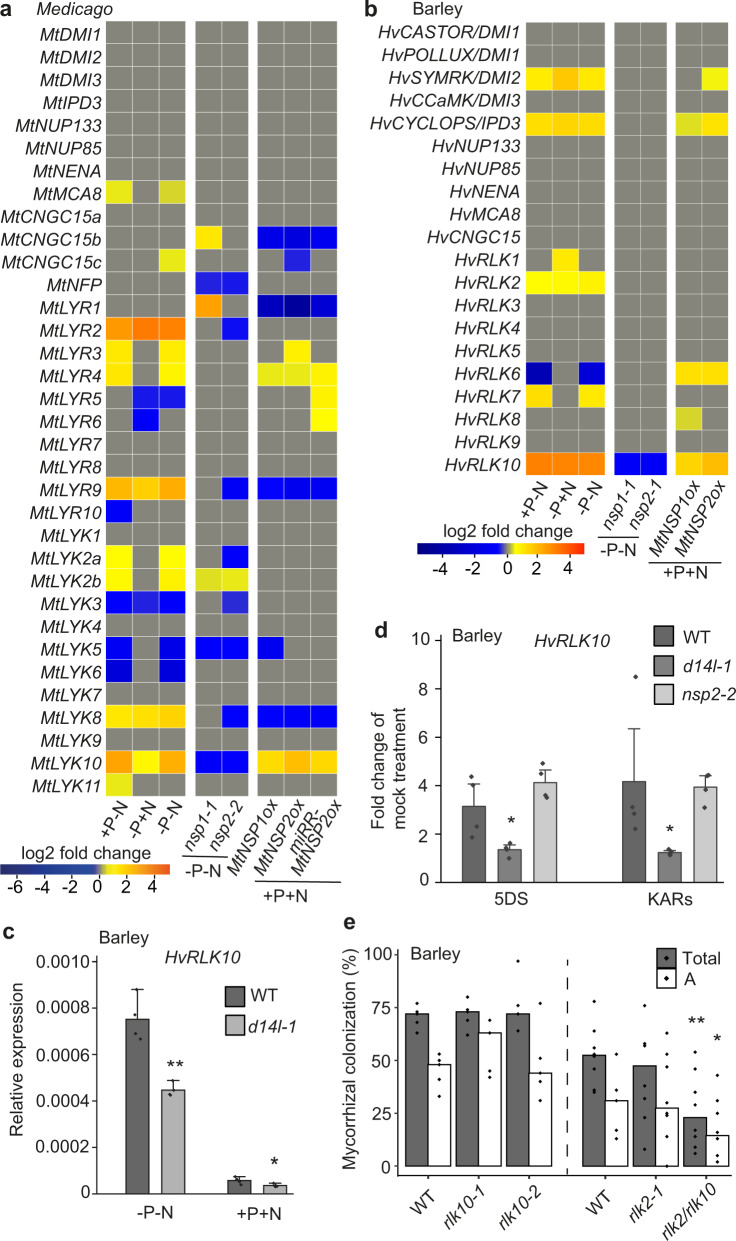
Previous work has reported that mutation of SMAX1 upregulates genes involved in the symbiosis signaling pathway, including the candidate LCO receptor NFR5, as well as SYMRK, CCaMK, and CYCLOPS37. Our finding that NSP2 functions through activation of D14L signaling and thus likely removal of SMAX1, implicates a potential role of NSP2 in regulating symbiosis signaling. Overexpression of MtNSP2 in barley significantly promotes the expression of SYMRK and CYCLOPS, as well as RLK10 (the NFR5 homolog in barley, Supplementary Fig. 3f), under nutrient replete conditions (Fig. 6b). Notably, while these genes are regulated by nutrient starvation, only RLK10 shows nutrient-starvation activation in a manner dependent on NSP1 and NSP2 (Fig. 6b), as well as D14L (Fig. 6c). 5DS and KARs treatments allow LCO perception under repressive P conditions (Fig. 4d) and consistently also promote expression of RLK10 (Fig. 6d and Supplementary Fig. 9b), in a manner dependent on D14L, but independent of NSP2 (Fig. 6d). These data reveal that the action of 5DS and KARs are both dependent on D14L, showing, at least in this experimental set-up, that D14L signaling can be promoted by 5DS, as well as KARs.
Fig. 6. NSP1 and NSP2 promote the expression of LysM-type receptors allowing LCO recognition.
Heatmaps representing the expression of genes encoding the common symbiosis signaling pathway and LysM-type receptor-like kinases in wild-type plants under N- and/or P-starvation, as well as in nsp mutants under nutrient deficient conditions and in NSP overexpression lines under nutrient-replete conditions in M. truncatula (a) and barley (b). +P−N, −P + N and -P-N represent the expression of these genes in wild-type plants under −N or/and −P conditions in comparison to +P + N conditions. nsp mutant columns show the expression of these genes in nsp mutants compared to wild type under nutrient-deficient conditions, while NSPox columns show NSP overexpression roots compared to wild type under nutrient-replete conditions. c Expression of barley RLK10 in wild type and d14l mutant roots. Plants were grown in nutrient deficient or sufficient conditions for 3 weeks. n = 4 biologically independent samples, ±s.e.m.; **indicates p < 0.01, *indicates 0.01 < p < 0.05, measured using a Student’s t-test (one-tailed, two-sample equal variance). d Induction of barley RLK10 by 5DS and KARs. Wild type, d14l-1 and nsp2-2 mutant plants were grown under repressive P-conditions and pretreated for 2 days on solid media containing 1 μM 5-deoxystrigol (5DS) or a mixture of 1 μM karrikin 1 and karrikin 2 (KARs). n = 4 biologically independent samples, ±s.e.m. *indicates 0.01 < p < 0.05, measured using a Student’s t-test (one-tailed, two-sample equal variance). e Root-length colonization of barley rlk10, rlk2, and rlk2/rlk10 double mutants grown under P-limited conditions, measured at 7 weeks post inoculation. The RLK10 mutation in the rlk2/rlk10 double mutant is equivalent to rlk10-1. n = 5–8 biologically independent plants. Total: total colonization; A: arbuscules. p-values for colonization levels were determined by a one-sided Wilcoxon test. **p < 0.01; *0.01 < p < 0.05.
Our data suggest that NSP and D14L control of AMF colonization, as a function of nutrient status, may principally be through the regulation of RLK10. To test this we generated mutants in RLK10 and its close homolog RLK2 (Supplementary Fig. 3f), which is also upregulated under nutrient starvation (Fig. 6b and Supplementary Fig. 9a). No significant defect of AMF colonization was observed in the rlk10 or rlk2 single mutants (Fig. 6e and Supplementary Fig. 9c, d), but a combination rlk2/rlk10 double mutant was significantly impaired in AMF colonization (Fig. 6e and Supplementary Fig. 9d). These two receptors contribute to LCO and CO perception with rlk2 attenuated in both LCO and CO-induced calcium oscillations and the rlk2/rlk10 double mutant completely insensitive to LCO perception (Table 1). We conclude that RLK10 acts as a LCO receptor and its nutrient regulation, in combination with that of RLK2, allows LCO recognition under nutrient starvation conditions.
Table 1.
RLK2 and RLK10 contribute to LCO and CO perception for induction of calcium spiking (cells spiking/total cells analysed)
| 10−7 M SmLCO (spiking/total) | 10-7 M CO4 (spiking/total) | |
|---|---|---|
| WT | 54 / 72 | 52 / 69 |
| rlk2 | 12 / 110 | 14 / 88 |
| rlk10 | 38 / 50 | 30 / 42 |
| rlk2/rlk10 | 0 / 91 | 13 / 80 |
A parallel assessment of nutrient-status and NSP-dependence on the regulation of symbiosis signaling genes in M. truncatula showed very little nutrient regulation of these signal transduction components (Fig. 6a), however, a number of genes encoding LysM-type receptor-like kinases showed nutrient-responsiveness, some of which were dependent on NSP1 and/or NSP2 (Fig. 6a). However, the known LCO receptors, NFP and LYK3, showed no upregulation in response to nutrient-starvation (Fig. 6a). We propose that other receptors, beyond those characterized, must explain nutrient responsive LCO perception in M. truncatula.
Discussion
Plants regulate associations with beneficial microorganisms as a function of the levels of N and P in their surrounding environment53–55, likely due to the higher energetic costs associated with a symbiotic route for nutrient capture16–18. The mechanisms that allow this nutrient regulation of microbial engagement are only just beginning to be elucidated. We demonstrate that one mechanism for nutrient control of AMF associations is through the regulation of LCO perception and its ability to activate symbiosis signaling22. This signaling pathway is essential for AMF colonization21 and is particularly associated with the initial recognition of AMF at the root surface11,56. We show that the perception of LCOs in both M. truncatula and barley is strongly regulated by the availability of both N and P, with a combination of N- and P-starvation maximally activating LCO recognition. The regulation of CO perception and its ability to activate symbiosis signaling is much less responsive to nutrient availability, suggesting that a mechanism specific to LCO perception is likely the principle target for regulation. We propose transcriptional regulation of the LCO receptors, RLK2 and RLK10 in barley, provides such a mechanism. However, we also observe nutrient control of SYMRK and CYCLOPS expression and their upregulation by NSP2 overexpression, pointing at additional targets for the nutrient regulation of this pathway. This may explain the qualitative differences we observe in the patterns of the calcium oscillations in plants grown under different nutrient regimes (Supplementary Fig. 2a, b). LCOs have recently been shown to be molecules produced by a range of fungal species, not only beneficial fungi57. However, LCO production is enhanced in AMF following the perception of plant-derived signals, such as strigolactones8–10, and this enhancement of LCO production has not been demonstrated in other species of fungi. We propose that even with LCOs produced by many fungi, there remain possible mechanisms for these molecules to act as symbiotic signals, either through the suite of decorated LCOs generated by specific fungi or by their enhanced production and delivery at the root surface. However, it is surprising that the rlk2/rlk10 mutant that shows no LCO induction of symbiosis signaling and a greatly attenuated response to CO4, still shows 50% colonization by AMF, in contrast to the lack of colonization in symbiosis signaling mutants. This suggests that additional signals from AMF, yet to be defined, may contribute to the activation of symbiosis signaling in the host plant.
We demonstrate that nutrient regulation of LCO perception is a function of the transcription factors NSP1 and NSP2. These genes regulate many components involved in the biosynthesis of apocarotenoids, in both M. truncatula and barley, suggesting that regulation of apocarotenoid biosynthesis is an important and conserved function of NSPs in plants. Numerous apocarotenoids have been associated with the AMF symbiosis, such as strigolactones, mycorradicins, blumenols, and zaxinones48. We demonstrate that NSPs are necessary and sufficient to activate strigolactone production, but the range of enzymes controlled by NSPs that are associated with apocarotenoid biosynthesis implies that these transcription factors are likely affecting a much broader range of apocarotenoid-derived small molecules. Our genetic analysis in M. truncatula is consistent with previous work in rice36,37,47,50,51 and demonstrates that strigolactones are primarily a signal exuded to the rhizosphere to promote interactions with the fungus, but their recognition by the plant via the receptor D14 is irrelevant to AMF associations and may even be detrimental. In contrast, the karrikin-like receptor D14L, that is essential for AMF colonization36, is essential for both appropriate and NSP2-driven enhancement of AMF infection. In our work, we found that D14L is essential for responses to karrikins, but surprisingly was also essential for the 5DS induction of RLK10 expression and resultant LCO perception. Villaécija-Aguilar et al.58 reported that root hair development in d14 is still responsive to 5DS58, suggesting a D14-independent strigolactone receptor. D14L paralogs in pea are able to bind 5DS in vitro and hydrolyse strigolactone-like molecules59, raising the possibility that strigolactones or strigolactone-derived molecules60 may bind and activate D14L.
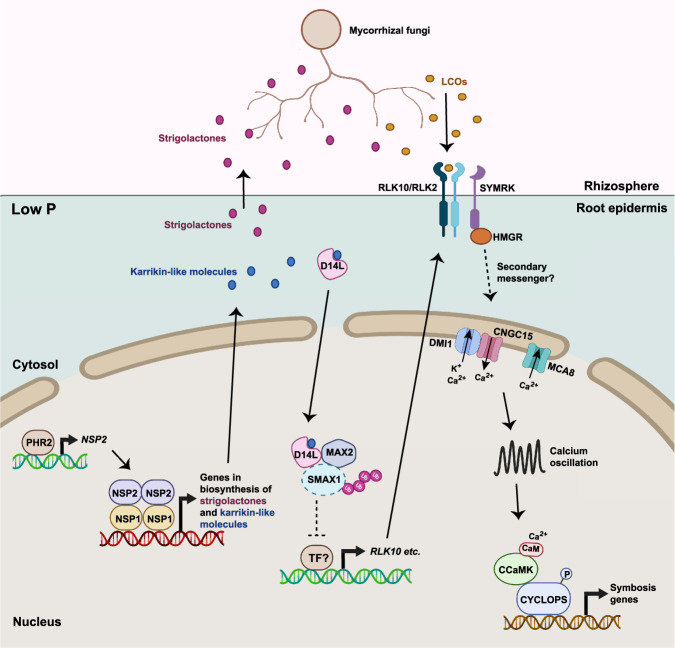
We propose that NSPs likely control the production of small molecules required for the activation of D14L and subsequent suppression of SMAX1 (Fig. 7). Consistent with this hypothesis, we observe that NSP2 overexpression in barley activates RLK10/NFR5, SYMRK, and CYCLOPS, genes that are also constitutively activated in the rice smax1 mutant37. Thus NSP regulation during nutrient starvation coordinates external rhizopheric signaling to AMF via the production of strigolactones, with internal plant signaling promoting the expression of the LCO receptor and some components of the symbiosis signaling pathway. As such, NSPs can coordinate multiple processes preparing both AMF and the host plant for symbiosis (Fig. 7).
Fig. 7. P-regulation of LCO production and perception, controlled by NSP1/NSP2.
Under P-starvation conditions, NSP2 expression is promoted by PHR232, that functions with NSP1 to promote gene expression associated with the biosynthesis of strigolactones and unknown karrikin-like molecules. Strigolactones are exuded to the rhizosphere to activate hyphal branching of mycorrhizal fungi and stimulate the production of fungal signals, including LCOs8–10. Karrikin-like signaling via D14L removes SMAX1 suppression, allowing the expression of the LCO receptor, RLK10 in barley and NFR5 in rice, allowing LCO recognition and activation of symbiosis signaling, triggering nuclear calcium oscillations and symbiotic gene expression. Whether strigolactones or strigolactone-derived molecules act as endogenous karrikin-like molecules needs to be further assessed. Taken together, NSP1 and NSP2 coordinate fungal and plant processes associated with LCO production and perception, to facilitate the AMF symbiosis. Created with BioRender.com.
The transcription factors PHR1 and PHR2 are master regulators of the P-starvation response that have been demonstrated to also control associations with a range of microorganisms1,31,32,61,62. Of particular relevance is the very recent demonstrations that PHRs control the engagement with AMF, appearing to function at multiple levels during this association31,32. PHR2 directly binds the promoters of a range of genes in rice associated with cortical cells harboring arbuscules, with the regulation of proteins associated with nutrient exchange across the periarbuscular membrane31. In addition, PHR2 also directly regulates the expression of CERK1 and SYMRK in rice, that are associated with the perception of COs and the symbiosis signaling pathway, suggesting an additional role for PHR2 at the root surface for initial perception of AMF32. While the expression of the LCO receptor NFR514,34 is affected by the phr2 mutant, this gene is not directly regulated by PHR232. Instead, NSP2 is a direct target of PHR2 and we propose that the regulation of the LCO receptor by PHR2 is through the regulation of NSP2, as demonstrated here. This implies that PHR2 can regulate the symbiosis signaling pathway via two mechanisms: direct control of CERK1 and SYMRK and indirect control of NFR5, SYMRK, and CYCLOPS through the promotion of NSP2 expression. Overexpression of both NSP2 (this study) and PHR231,32 is sufficient to partially override P-suppression of AMF colonization, with the relative levels of impact comparable. This further supports a linear relationship between PHR2 and NSP2 function, at least for the regulation of AMF colonization at the root surface. NSP2 overexpression does not induce any of the PHR2-induced genes associated with the arbuscule31, making it difficult to judge whether NSPs also function downstream of PHR2 at this late stage of the AMF association. However, nsp mutants of barley show strong reductions in numbers of arbuscules (Fig. 3f), implying a possible role for NSPs also during arbuscule development.
Nutrient regulation of LCO recognition by NSPs is conserved between M. truncatula and barley, implying a process that probably evolved early in the plant kingdom, likely alongside the evolution of symbiosis signaling63, to support AMF symbioses. Within the last 100 million years, legumes augmented their symbiotic repertoire with the addition of associations with N-fixing bacteria. During this evolutionary process, legumes utilized the preexisting symbiosis signaling pathway to allow recognition of N-fixing bacteria64. Our work reveals that whereas both M. truncatula and barley can activate symbiosis signaling via equivalent signaling molecules, including LCO Nod factors produced by N-fixing bacteria, legumes appear to have expanded their specificity of LCO recognition, showing exquisite sensitivity for appropriately decorated LCOs generated by their N-fixing symbiont. This is consistent with the specific decorations on LCOs providing host legume specificity for N-fixing symbionts65. NSP1 and NSP2 have essential roles in nodulation38,39, and this function appears to be during nodule establishment, downstream of both symbiosis signaling and cytokinin signaling23,66. This suggests additional functions for NSP1 and NSP2 during nodulation, that may be unrelated to their functions in preparation of the plant for AMF colonization. An expansion for NSP2 function may have been facilitated by the duplication of this gene in legumes, with genetic redundancy between NSP2 and NSP2L for AMF associations in M. truncatula, but no such genetic redundancy for NSP2 during nodulation67. It remains unclear whether NSP2 function during nodulation is associated with apocarotenoid biosynthesis, however, the fact that neither strigolactones or D14L have positive functions during nodule establishment, points at quite different targets and modes of action for NSPs during nodule organogenesis.
Achieving sustainable productivity in agriculture necessitates a reduction in the global dependence on inorganic fertilizers and this in part could be achieved through better use of beneficial microbial associations. Our work demonstrates a mechanism to enhance colonization of AMF, in particular under suppressive P-conditions, using genes that have symbiotic specific functions, that is preferable to using genes such as PHR2 that have very broad effects on plant function. Enhancing AMF associations in agriculture has the potential to improve the efficiency of nutrient capture, reducing loss of these nutrients to the environment68. Furthermore, we have also demonstrated major differences between legumes and cereals in the stringency of LCO recognition, a process already well understood69. This is likely an important engineering target for generating N-fixing cereals. Agricultural processes that rely more heavily on beneficial microbial nutrient delivery as oppose to the application of inorganic fertilizers, have the potential to deliver more sustainable and equitable global food production.
Methods
Plant materials
Barley (Hordeum vulgare L. cv. Golden Promise), wheat (Triticum aestivum L. cv Fielder), maize (Zea mays L.), and rice (Oryza sativa cv. Nipponbare) were transformed with the calcium reporter cameleon YC3.6 using Agrobacterium tumefaciens-mediated transformation70–72. Barley symrk, ccamk, cyclops, ram1, nsp1, nsp2, and d14l mutants were generated with CRISPR/Cas973. YC3.6 was introduced into the barley symrk-1 and rice pollux36 and d14l37 through Agrobacterium-mediated transformation70. Individual T2 lines showing strong expression of YC3.6 were selected for calcium analysis. Medicago truncatula A17 was used as the wild type of nsp1-1, nsp2-2, and nsp1/nsp2 mutants. YC3.6 was incorporated into nsp1/nsp2 mutant by crossing with an existing stable YC3.6 line in A17 background. Medicago truncatula R108 was used as the control for the Tnt1 insertion mutants nsp1-3 (NF9220), nsp2-4 (NF10950), nsp2l-1 (NF17492), ccd7-1 (NF1485), ccd8a-2 (NF18323), ccd8a-3 (NF111036), ccd8a-3 (NF111036), d14-1 (NF18262), d14la-1 (NF13623), d14lb-1 (NF5873), max2-1 (NF18521), and smax1-1 (NF4373). All Tnt1 lines were ordered from the Samuel Roberts Noble Foundation collection74. The primers for genotyping of these Tnt1 mutants are listed in Table S4.
Plant growth conditions
Barley, wheat, and maize seeds were treated with 70% ethanol for 2 minutes, followed with three times wash in sterile water. The seeds were then surfaced sterilized by 5% sodium hypochlorite solution for 4 minutes and rinsed with sterile water 4–5 times. The sterilized seeds were plated on 1% water agar plates and imbibed at 4 °C for 3 days and then germinated in the dark at 22 °C for 2–3 days. Seedlings were cultivated in a 1:1 mixture of sterilized terragreen (Oil‐dry UK ltd) and sharp sand (BB Minerals) in growth chambers at 20 °C under 16 h light/8 h dark conditions with 80% relative humidity.
Rice seeds were surface-sterilized briefly in 70% (v/v) ethanol, then for 20 min in 3% (v/v) sodium hypochlorite, and finally rinsed three times in sterile water. Imbibed seeds were germinated on 0.6% (w/v) bactoagar in the dark at 30 °C for 3-7 days. Seedlings were transferred into cones containing sterile quartz sand and cultivated in growth chambers at 28 °C/20 °C, 12 h light/12 h dark cycle with 60% relative humidity.
M. truncatula seeds were scarified with sandpaper and surface-sterilized in 10% sodium hypochlorite solution for 6 mins and rinsed 4–5 times in sterile water. The seeds planted on 1% water agar were imbibed at 4 °C for 3 days and germinated overnight in the dark at 22 °C. Seedlings were cultivated in growth chambers at 20 °C under 16 h light/8 h dark conditions with 80% relative humidity.
Nuclear calcium imaging
For barley, wheat and maize calcium analyses in Figs. 1a and 2b, plants were grown in 1:1 mixture of sterilized Terragreen and sharp sand mix and watered with sterile water only for 25 days. For rice calcium analyses in Fig. 2a, plants were grown in cones containing sterile quartz sand and watered three times weekly for the first week, and subsequently watered once a week and fertilized twice a week with half Hoagland solution (25 μM PO43-) until plants were five weeks old. These growth conditions were previously demonstrated to promote efficient mycorrhizal colonization21. For calcium analyses in Fig. 1b, c, M. truncatula and barley seedlings were grown for 10 days on BNM75 plates without aminoethoxyvinylglycine (AVG, Sigma-Aldrich) until lateral roots emerged.
For nutrient treatments on M. truncatula and barley, seedlings were grown for 16 days on modified BNM plates with the concentrations of N and P as: -N-P, no NO3− and no PO43−; −N + P, no NO3− and 0.5 mM PO43−; + N-P, 5 mM NO3- and no PO43−; + N + P, 5 mM NO3− and 0.5 mM PO43−. The concentration of P used reflects that which was sufficient to significantly suppress AMF colonization in M. truncatula and barley (Fig. 4a, b).
For the pretreatment of strigolactone and karrikin on barley, M. truncatula and wheat, seedlings were grown on BNM plates (−N + P, no NO3− and 0.5 mM PO43−) for 5 days (barley and wheat) or 10 days (M. truncatula) until lateral roots emerged, and then were treated in BNM liquid medium with 0.1 μM or 1 μM (±)−5-deoxystrigol (Chiralix), a 0.1 μM or 1 μM mixture of karrikin 1 and karrikin 2 (Chiralix) or 1 μM GR24 (Chiralix), using BNM liquid adding the same volume of acetone (Fisher Chemical) as a control. Following the treatment of lateral roots with different concentrations of COs and LCOs, the measurement of calcium oscillations was performed using an inverted epifluorescence microscope (model TE2000; Nikon). Calcium recordings were collected and analyzed as described previously11.
Generation of barley CRISPR mutants and barley transgenic lines
The methods for creating barley CRISPR mutants are as described73. The specific target sequences of the guide RNAs, and the mutations present in each mutant plant, are listed in Supplementary Table 5. Hordeum vulgare cv. Golden Promise wild type or YC3.6 lines were transformed as previously described70 with the constructs (L2 plasmids) listed in Supplementary Table 3. Briefly, the constructs were transformed into Agrobacterium tumefaciens strain AGL1. Immature barley seeds (with embryo size around 1.5 mm in diameter) were harvested and sterilized with 70% ethanol for 2 minutes, followed by 3 washes in sterile water. The seeds were then sterilized with 5% sodium hypochlorite solution for 4 minutes and rinsed with sterile water 4–5 times. Immature embryos were isolated using fine forceps in a laminar flow hood and the embryonic axis was removed from the scutellum and discarded. They were then cultured on Callus induction (CI) plates70 at 23–24 °C in the dark overnight. The embryos were treated with Agrobacterium inoculum and transferred to new CI plates for co-cultivation. After 3 days, the embryos were transferred to CI plates with antibiotics: Timentin, to remove the Agrobacterium and hygromycin, for selection of transgenic cells. The following culture at 23–24 °C in the dark for 2 weeks, embryos were transferred to fresh CI plates and cultured under the same conditions for another 4 weeks to induce callus formation, with subculture to fresh plates after 2 weeks. Calli were then transferred to Transition medium under low light for 2 weeks. Calli that developed shoots were transferred to Regeneration plates70 until small plantlets formed. Individual plantlets were separated and transferred to rooting tubes. Once plantlets reached the top of the tubes and had good root systems, they were transferred to soil for growth to maturity.
Mycorrhizal inoculation
For mycorrhization assays, the barley seedlings were inoculated with 5% or 10% (w/w) crude inoculum (produced on Tagetes multiflora) of R. irregularis and grown in a cone system at 28 °C/20 °C,12 h light/12 h dark cycle with 60% humidity. Plants were watered with water only for the first 1–2 weeks, then were nurtured with half-strength Hoagland solution containing 10 μM PO43− twice a week. For nutrient treatments on barley NSP overexpression lines, half-strength Hoagland solutions containing a range of P concentrations (10 μM, 500 μM, and 1 mM PO43−) were given twice a week repeatedly. The inoculated roots were harvested and stained by Trypan blue76. Stained roots were mounted on slides (ten root pieces per slide), and fungal colonization was quantified at 10 representative random points per root piece.
M. truncatula plants were grown in 4 x 4 x 4.5 cm3 pots containing sharp sand and 200 R. irregularis spores. R. irregularis spores were ordered from Premier Tech (Québec, Canada). Plants were fertilized twice per week with modified BNM liquid medium (5 mM NO3− and 10 μM PO43−) for 3 weeks. For nutrient treatments on hairy-root transformed NSP-overexpression plants, modified BNM solutions containing low and high P (10 μM and 500 μM PO43−, 5 mM NO3−) were given twice a week until harvesting at 5 weeks post inoculation. The harvest roots were stained with an ink-vinegar solution77. The roots were cut into 1 cm segments and pre‐cleared by incubation in 10% (w/v) KOH at 95 °C for 10 min. Roots were rinsed 3 times with distilled water and stained with a solution containing 5% ink and 5% acetic acid at 95 °C for 3–5 min. Roots were de‐stained in water for 1–2 days, and fungal structures were quantified using the gridline intersect method78. Fungal colonization was visualized using a Keyence digital microscope (Keyence VHX-5000).
Plasmid construction
The plasmids were all constructed using the Golden Gate cloning system79,80. The gene sequences were domesticated and synthesized, and then cloned into level 0 vector pMS or pMK (GeneArt, Thermo Fisher Scientific). The assemblies of level 1 and level 2 plasmids are listed in Table S3. All the plasmids used in this study are held for distribution in the ENSA project collection (https://www.ensa.ac.uk/). pMtD27::GUS transcriptional reporter was generated using 1045 bp of the MtD27 promotor and cloned in modified pCAMBIA2200 binary vector81.
Hairy root transformation
The hairy root transformation assay was performed as described82. For Fig. 4a, M. truncatula seedlings were transformed via A. rhizogenes strain ARqua1 mediated hairy root transformation with either an empty vector carrying a RUBY reporter83 or vectors with overexpression of NSP genes (Supplementary Table 3). Three weeks after transformation, plants with RUBY-expressing roots were selected for further assays.
For Supplementary Fig. 8b, c, M. truncatula seedlings were transformed via A. rhizogenes strain AR1193 mediated hairy root transformation with the vectors containing a DsRed reporter. The plants were grown on modFP plates for four weeks after hairy-root transformation. The transformed roots were screened using an Axiozoom V16 (Zeiss) or DMR/MZFLIII (Leica) microscope to visualize the transformation marker DsRed. The positive roots were harvested for GUS staining or gene expression analyses.
RNA extraction and RT-qPCR
Barley seedlings were grown in sterilized sharp sand for 21 days, fertilized twice a week with modified BNM liquid with the concentrations of N and P as: −N−P, no NO3− and no PO43−; −N + P, no NO3− and 0.5 mM PO43−; + N-P, 5 mM NO3− and no PO43−; + N + P, 5 mM NO3− and 0.5 mM PO43−. For the pretreatment of strigolactone and karrikin on barley, seedlings were grown on modified BNM plates (+N + P, 3 mM NO3- and 0.5 mM PO43−) for 3 days, and then were treated on modified BNM solid medium containing 1 μM (±)-5-deoxystrigol (Chiralix) and a 1 μM mixture of karrikin 1 and karrikin 2 (Chiralix) for 2 days.
M. truncatula seedlings of A17, nsp1-1, and nsp2-2 were grown on modified BNM plates for 15 days with the concentrations of N and P as: −N−P, no NO3− and no PO43−; −N + P, no NO3− and 3.75 mM PO43−; + N−P, 5 mM NO3− and no PO43−; + N + P, 5 mM NO3− and 3.75 mM PO43. The concentrations of P used reflect those calculated as being necessary for suppression of AMF colonization in M. truncatula55. The A. rhizogenes transformed M. truncatula roots with NSP overexpression were transferred into sterilized sharp sand after the selection by a RUBY reporter and watered with modified BNM liquid (+N + P, 5 mM NO3− and 0.5 mM PO43−) twice a week for three weeks. The roots were harvested and stored at −80 °C for RNA extraction.
Barley total RNA was extracted using the Spectrum™ Plant Total RNA kit (Sigma-Aldrich) and M. truncatula total RNA was isolated with the RNeasy PlantMini Kit (Qiagen), following the manufacturer’s instructions. RNA concentration was determined by NanoDrop One (Thermo Scientific). 1μg of total RNA was used for cDNA synthesis with the SensiFAST cDNA Synthesis Kit (Bioline). Real-time quantitative PCR (RT-qPCR) was performed using SensiFAST SYBR No-ROX Kit and semi-quantitative RT-PCR using GoTaq Green Master Mix (Promega). The primers used for gene expression analysis are listed in Supplementary Table 4.
RNA-seq and data analysis
Large scale transcriptomic RNA sequencing (RNA-seq) was performed by GeneWiz (Leipzig, Germany) and Novogene (Cambridge, United Kingdom). RNA samples were sent for library preparation and sequencing with Illumina NovaSeq Next Generation Sequencing system as paired-end 2 × 150 bp reads. Raw fastq files generated from sequencing were further subjected to quality controls with FastQC software84 and then mapped to the Medicago truncatula reference genome 4.0 (Mt4.0v1)85 from Phytozome database86 or the Hordeum vulgare cv. Golden Promise reference genome63,87, using STAR software88. The counts and FPKM (Fragments per kilobase per million mapped reads) values were calculated with featureCounts in R package Rsubread89. Non-metric multidimensional scaling was exploited to account for outliers90. Genes that showed low expression throughout all samples were removed by measuring CPM (counts per million) values using R package edgeR91 to filter out genes with CPM values corresponding to fewer than 10 raw counts. TMM expression values were obtained using R package edgeR91 to conduct TMM normalization of the library sizes and output CPM values normalized by the resultant effective library sizes. Differentially expressed genes (DEGs) were identified by pairwise comparisons of raw counts of nutrient replete versus depleted treatments or wildtype versus mutant/transgenic lines for nutrient and genotypic effects, respectively, using the R package DESeq292 with a false discovery rate (FDR) corrected p-value more significant than 0.05. The heatmaps of differential expression and hierarchical clustering were plotted with Python package Seaborn93, with hierarchical clustering applying the “correlation” metric and displaying DEGs with a threshold of absolute fold change over 1.5. Synonyms and descriptions of Medicago truncatula DEGs were obtained as described previously94. Putative synonyms and descriptions of Hordeum vulgare DEGs were assigned by employing the Orthofinder tool95,96 to infer orthology relationships to genes from other annotated genomes, followed by manual curation of descriptions. For a more robust inference, OrthoFinder analysis was run with default parameters, including four proteomes: Oryza sativa v7.097, Arabidopsis thaliana TAIR1098, Medicago truncatula Mt4.0v185, and Hordeum vulgare cv. Golden Promise v1r163.
Generation of phylogenetic trees
Homologs of genes of interest were searched against the genomes of Arabidopsis thaliana Araport1199, Fragaria vesca v4.015, Medicago truncatula Mt4.0v185, Lotus japonicus Gifu v1.2100, Oryza sativa v7.097, Zea mays PH207 v1.1101, and Hordeum vulgare63. Searches were performed with the tBLASTn algorithm v2.10.1 + 102 with the protein sequences of M. truncatula NSP1 and NSP2, and Arabidopsis CCD8, KAI2, and SMAX1 as query. LysM-RLK homologs were found by using previously identified Medicago genes from the family103 as query, combined with barley genes identified as containing both LysM (PF01476) and protein kinase (PF00069) domains from the Pfam database104 using HMMR 3.3.2105. Coding sequences of obtained hits were aligned with MAFFT v7106 and phylogenetic tree construction by maximum likelihood was performed using IQ-TREE with 1000 replicates of ultrafast bootstrap107. Trees were visualized and annotated on the Interactive Tree Of Life (iTOL) platform, with clades that included the gene(s) of interest pruned from the tree and bootstrap information displayed as circles108.
Measurement of strigolactones
Barley seedlings were grown in sterilized sand for six weeks, watered twice a week with a modified half-strength Hoaglands nutrient solution [2.4 mM KNO3, 1.6 mM Ca(NO3)2, 0.5 mM KH2PO4, 0.8 mM MgSO4, 0.18 mM FeSO4, 0.1 mM Na2EDTA, 4.5 µM MnCl2, 23 µM H3BO3, 0.3 µM CuSO4, 1.5 µM ZnCl2, and 0.1 µM Na2MoO4] (high P, HP). P starvation stress was induced by reducing the amount available PO4- to 0.02 mM KH2PO4 (low P, LP). After six weeks, all pots were rinsed with 1 L half-strength Hoaglands nutrient solution (either HP or LP), and grown for an additional week under these nutrient conditions before harvesting the roots. Strigolactones were extracted as previously described109,110, with some modifications. Extracts were evaporated to dryness, taken up in 100% hexane, and loaded on pre-equilibrated Silica gel SPE (Grace PureTM SPE Silica 100 mg/1 mL) columns. Samples were then eluted with 1 ml of hexane:ethyl acetate (1:9) and evaporated to dryness. The residue was dissolved in 200 μl of 20% acetonitrile/water (v/v) spiked with 10−6 M GR24. Samples were filtered using a 0.2 µm Mini spin column (BGB Analytik Benelux B.V.). and stored for maximum 24 h at 4 ˚C before UPLC-MS/MS analysis. SLs were analyzed by comparing retention times and mass transitions with a 5-deoxystrigol standard using a Waters Xevo TQ mass spectrometer equipped with an electrospray-ionization source and coupled to a Waters Acquity Ultraperformance LC system as previously described43 with some modifications111–113. GR24 served as an internal standard that was added before analysis. Detection and quantification of SLs were performed with six biological replicates.
Nodulation assay
M. truncatula plants were grown in 4 x 4 x 4.5 cm3 pots containing 1:1 mix of sterile terragreen/sharp sand for seven days and inoculated with 5 mL S. meliloti 2011 (OD600: 0.02) in each pot. Nodules were harvested for quantification at two weeks post inoculation.
GUS staining
For histochemical GUS analysis, root tissues of pMtD27::GUS hairy roots were soaked in GUS staining solution (100 mM sodium phosphate buffer, pH 7.2, 10 mM EDTA, 0.1 % Triton X-100, 0.3 mg/ml X-Gluc) under vacuum for 15 min. Root tissues were then incubated 2–3 h in GUS staining solution at 37 °C. Staining is stopped at the same time for WT and nsp1-1 mutant by washing the roots with water, then roots are fixed in 50% ethanol for long-term storage and imagining using Axiozoom V16 (Zeiss) under brightfield.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank Jiangqi Wen for providing M. truncatula mutants from his core collection. We thank Roza Bilas, Georgia Hassell and Eirini Vlachaki for the quantification of AMF colonization in barley, as well as Jean Keller, Cyril Libourel, and Pierre-Marc Delaux for help with the phylogenetic analysis of the GRAS genes. We also thank Carolien Franken and Amber van Seters for strigolactone measurement, Melanie Craze and Sarah Bowden for wheat and rice transformation, and Tom Brutnell for oversight of maize transformation. This work was supported by the Bill and Melinda Gates Foundation and the Foreign, Commonwealth, and Development Office as OPP1172165, Engineering the Nitrogen Symbiosis for Africa, the Biotechnology and Biological Sciences Research Council as BB/K003712/1 and BB/J019356/, and the Gatsby Foundation as GAT3395/GLH.
Source data
Author contributions
X-R.L., J.S., D.A., J.C., U.P., and G.E.D.O. designed the experiments. X-R.L., J.S., D.A., R.H., W.K., L.H.L., C.H.C., A.J., and B.G. performed the assays. T.L. (Tom Lawrensen) and E.S. developed the crispr mutants in barley, H.L. and J.M. generated some of the M. truncatula mutants. W.H. and E.W. managed barley and wheat transformation. D.Z., T.L. (Tak Lee), E.J-B., and A.A. analyzed and interpreted RNA-seq data. X-R.L., E.J-B., D.Z., and G.E.D.O. wrote the paper, with input from all the authors.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Data availability
The raw data of RNA-sequencing have been deposited in the National Center for Biotechnology Information, Gene Expression Omnibus (GEO), with accession code GSE214698. Any additional information required to reanalyze the data reported in this study is available from the lead contact upon request. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xin-Ran Li, Jongho Sun, Doris Albinsky.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-022-33908-3.
References
- 1.Oldroyd, G. E. D. & Leyser, O. A plant’s diet, surviving in a variable nutrient environment. Science368, eaba0196 (2020). [DOI] [PubMed]
- 2.Azam HM, et al. Phosphorous in the environment: characteristics with distribution and effects, removal mechanisms, treatment technologies, and factors affecting recovery as minerals in natural and engineered systems. Environ. Sci. Pollut. Res. Int. 2019;26:20183–20207. doi: 10.1007/s11356-019-04732-y. [DOI] [PubMed] [Google Scholar]
- 3.Stevens CJ. Nitrogen in the environment. Science. 2019;363:578–580. doi: 10.1126/science.aav8215. [DOI] [PubMed] [Google Scholar]
- 4.Tian H, et al. A comprehensive quantification of global nitrous oxide sources and sinks. Nature. 2020;586:248–256. doi: 10.1038/s41586-020-2780-0. [DOI] [PubMed] [Google Scholar]
- 5.Wang B, Qiu YL. Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza. 2006;16:299–363. doi: 10.1007/s00572-005-0033-6. [DOI] [PubMed] [Google Scholar]
- 6.Brundrett MC, Tedersoo L. Evolutionary history of mycorrhizal symbioses and global host plant diversity. N. Phytol. 2018;220:1108–1115. doi: 10.1111/nph.14976. [DOI] [PubMed] [Google Scholar]
- 7.Püschel D, Bitterlich M, Rydlová J, Jansa J. Facilitation of plant water uptake by an arbuscular mycorrhizal fungus: a Gordian knot of roots and hyphae. Mycorrhiza. 2020;30:299–313. doi: 10.1007/s00572-020-00949-9. [DOI] [PubMed] [Google Scholar]
- 8.Besserer A, et al. Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol. 2006;4:e226. doi: 10.1371/journal.pbio.0040226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maillet F, et al. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature. 2011;469:58–63. doi: 10.1038/nature09622. [DOI] [PubMed] [Google Scholar]
- 10.Genre A, et al. Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. N. Phytol. 2013;198:190–202. doi: 10.1111/nph.12146. [DOI] [PubMed] [Google Scholar]
- 11.Sun J, et al. Activation of symbiosis signaling by arbuscular mycorrhizal fungi in legumes and rice. Plant Cell. 2015;27:823–838. doi: 10.1105/tpc.114.131326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng F, et al. A combination of chitooligosaccharide and lipochitooligosaccharide recognition promotes arbuscular mycorrhizal associations in Medicago truncatula. Nat. Commun. 2019;10:5047. doi: 10.1038/s41467-019-12999-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carotenuto G, et al. The rice LysM receptor-like kinase OsCERK1 is required for the perception of short-chain chitin oligomers in arbuscular mycorrhizal signaling. N. Phytol. 2017;214:1440–1446. doi: 10.1111/nph.14539. [DOI] [PubMed] [Google Scholar]
- 14.He J, et al. A LysM receptor heteromer mediates perception of arbuscular mycorrhizal symbiotic signal in rice. Mol. Plant. 2019;12:1561–1576. doi: 10.1016/j.molp.2019.10.015. [DOI] [PubMed] [Google Scholar]
- 15.An J, et al. A Medicago truncatula SWEET transporter implicated in arbuscule maintenance during arbuscular mycorrhizal symbiosis. N. Phytol. 2019;224:396–408. doi: 10.1111/nph.15975. [DOI] [PubMed] [Google Scholar]
- 16.Luginbuehl LH, et al. Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science. 2017;356:1175–1178. doi: 10.1126/science.aan0081. [DOI] [PubMed] [Google Scholar]
- 17.Jiang Y, et al. Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science. 2017;356:1172–1175. doi: 10.1126/science.aam9970. [DOI] [PubMed] [Google Scholar]
- 18.Keymer A, et al. Lipid transfer from plants to arbuscular mycorrhiza fungi. Elife. 2017;6:e29107. doi: 10.7554/eLife.29107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parniske M. Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat. Rev. Microbiol. 2008;6:763–775. doi: 10.1038/nrmicro1987. [DOI] [PubMed] [Google Scholar]
- 20.Oldroyd GED. Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 2013;11:252–263. doi: 10.1038/nrmicro2990. [DOI] [PubMed] [Google Scholar]
- 21.Gutjahr C, et al. Arbuscular mycorrhiza-specific signaling in rice transcends the common symbiosis signaling pathway. Plant Cell. 2008;20:2989–3005. doi: 10.1105/tpc.108.062414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Catoira R, et al. Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell. 2000;12:1647–1666. doi: 10.1105/tpc.12.9.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gleason C, et al. Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature. 2006;441:1149–1152. doi: 10.1038/nature04812. [DOI] [PubMed] [Google Scholar]
- 24.Tirichine L, et al. Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature. 2006;441:1153–1156. doi: 10.1038/nature04862. [DOI] [PubMed] [Google Scholar]
- 25.Chiu, C. H. & Paszkowski, U. Mechanisms and impact of symbiotic phosphate acquisition. Cold Spring Harb. Perspect. Biol. 11, a034603 (2019). [DOI] [PMC free article] [PubMed]
- 26.Nishida H, Suzaki T. Nitrate-mediated control of root nodule symbiosis. Curr. Opin. Plant Biol. 2018;44:129–136. doi: 10.1016/j.pbi.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 27.Hepper CM. The effect of nitrate and phosphate on the vasicular-arbuscular mycorrhizal infection of lettuce. N. Phytol. 1983;93:389–399. [Google Scholar]
- 28.Streeter J, Wong PP. Inhibition of legume nodule formation and N2 fixation by nitrate. Crit. Rev. Plant Sci. 1988;7:1–23. [Google Scholar]
- 29.Tsikou D, et al. Systemic control of legume susceptibility to rhizobial infection by a mobile microRNA. Science. 2018;362:233–236. doi: 10.1126/science.aat6907. [DOI] [PubMed] [Google Scholar]
- 30.Gautrat P, Laffont C, Frugier F. Compact Root Architecture 2 promotes root competence for nodulation through the miR2111 systemic effector. Curr. Biol. 2020;30:1339–1345.e1333. doi: 10.1016/j.cub.2020.01.084. [DOI] [PubMed] [Google Scholar]
- 31.Shi J, et al. A phosphate starvation response-centered network regulates mycorrhizal symbiosis. Cell. 2021;184:5527–5540.. doi: 10.1016/j.cell.2021.09.030. [DOI] [PubMed] [Google Scholar]
- 32.Das D, et al. PHOSPHATE STARVATION RESPONSE transcription factors enable arbuscular mycorrhiza symbiosis. Nat. Commun. 2022;13:477. doi: 10.1038/s41467-022-27976-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu W, et al. Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2. Plant Cell. 2011;23:3853–3865. doi: 10.1105/tpc.111.089771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyata K, et al. Evaluation of the role of the LysM receptor-like kinase, OsNFR5/OsRLK2 for AM symbiosis in rice. Plant Cell Physiol. 2016;57:2283–2290. doi: 10.1093/pcp/pcw144. [DOI] [PubMed] [Google Scholar]
- 35.Imaizumi-Anraku H, et al. Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature. 2005;433:527–531. doi: 10.1038/nature03237. [DOI] [PubMed] [Google Scholar]
- 36.Gutjahr C, et al. Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex. Science. 2015;350:1521–1524. doi: 10.1126/science.aac9715. [DOI] [PubMed] [Google Scholar]
- 37.Choi J, et al. The negative regulator SMAX1 controls mycorrhizal symbiosis and strigolactone biosynthesis in rice. Nat. Commun. 2020;11:2114. doi: 10.1038/s41467-020-16021-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaló P, et al. Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science. 2005;308:1786–1789. doi: 10.1126/science.1110951. [DOI] [PubMed] [Google Scholar]
- 39.Smit P, et al. NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science. 2005;308:1789–1791. doi: 10.1126/science.1111025. [DOI] [PubMed] [Google Scholar]
- 40.Lauressergues D, et al. The microRNA miR171h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting. NSP2. Plant J. 2012;72:512–522. doi: 10.1111/j.1365-313X.2012.05099.x. [DOI] [PubMed] [Google Scholar]
- 41.Delaux PM, Becard G, Combier JP. NSP1 is a component of the Myc signaling pathway. New Phytol. 2013;199:59–65. doi: 10.1111/nph.12340. [DOI] [PubMed] [Google Scholar]
- 42.Yoneyama K, Yoneyama K, Takeuchi Y, Sekimoto H. Phosphorus deficiency in red clover promotes exudation of orobanchol, the signal for mycorrhizal symbionts and germination stimulant for root parasites. Planta. 2007;225:1031–1038. doi: 10.1007/s00425-006-0410-1. [DOI] [PubMed] [Google Scholar]
- 43.López-Ráez JA, et al. Tomato strigolactones are derived from carotenoids and their biosynthesis is promoted by phosphate starvation. New Phytol. 2008;178:863–874. doi: 10.1111/j.1469-8137.2008.02406.x. [DOI] [PubMed] [Google Scholar]
- 44.Yoneyama K, et al. How do nitrogen and phosphorus deficiencies affect strigolactone production and exudation? Planta. 2012;235:1197–1207. doi: 10.1007/s00425-011-1568-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Zeijl A, et al. The strigolactone biosynthesis gene DWARF27 is co-opted in rhizobium symbiosis. BMC Plant Biol. 2015;15:260. doi: 10.1186/s12870-015-0651-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gomez-Roldan V, et al. Strigolactone inhibition of shoot branching. Nature. 2008;455:189–194. doi: 10.1038/nature07271. [DOI] [PubMed] [Google Scholar]
- 47.Foo E, Yoneyama K, Hugill CJ, Quittenden LJ, Reid JB. Strigolactones and the regulation of pea symbioses in response to nitrate and phosphate deficiency. Mol. Plant. 2013;6:76–87. doi: 10.1093/mp/sss115. [DOI] [PubMed] [Google Scholar]
- 48.Fiorilli V, Wang JY, Bonfante P, Lanfranco L, Al-Babili S. Apocarotenoids: old and new mediators of the arbuscular mycorrhizal symbiosis. Front. Plant Sci. 2019;10:1186. doi: 10.3389/fpls.2019.01186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hirsch S, et al. GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell. 2009;21:545–557. doi: 10.1105/tpc.108.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshida S, et al. The D3 F-box protein is a key component in host strigolactone responses essential for arbuscular mycorrhizal symbiosis. N. Phytol. 2012;196:1208–1216. doi: 10.1111/j.1469-8137.2012.04339.x. [DOI] [PubMed] [Google Scholar]
- 51.Liu G, et al. Strigolactones play an important role in shaping exodermal morphology via a KAI2-dependent pathway. iScience. 2019;17:144–154. doi: 10.1016/j.isci.2019.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Foo E, Davies NW. Strigolactones promote nodulation in pea. Planta. 2011;234:1073–1081. doi: 10.1007/s00425-011-1516-7. [DOI] [PubMed] [Google Scholar]
- 53.Bouwmeester HJ, Roux C, Lopez-Raez JA, Bécard G. Rhizosphere communication of plants, parasitic plants and AM fungi. Trends Plant Sci. 2007;12:224–230. doi: 10.1016/j.tplants.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 54.Balzergue C, Puech-Pages V, Becard G, Rochange SF. The regulation of arbuscular mycorrhizal symbiosis by phosphate in pea involves early and systemic signalling events. J. Exp. Bot. 2011;62:1049–1060. doi: 10.1093/jxb/erq335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Balzergue C, Chabaud M, Barker DG, Bécard G, Rochange SF. High phosphate reduces host ability to develop arbuscular mycorrhizal symbiosis without affecting root calcium spiking responses to the fungus. Front. Plant Sci. 2013;4:426–426. doi: 10.3389/fpls.2013.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kosuta S, et al. Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proc. Natl Acad. Sci. USA. 2008;105:9823–9828. doi: 10.1073/pnas.0803499105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rush TA, et al. Lipo-chitooligosaccharides as regulatory signals of fungal growth and development. Nat. Commun. 2020;11:3897. doi: 10.1038/s41467-020-17615-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Villaécija-Aguilar JA, et al. SMAX1/SMXL2 regulate root and root hair development downstream of KAI2-mediated signalling in Arabidopsis. PLoS Genet. 2019;15:e1008327. doi: 10.1371/journal.pgen.1008327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guercio AM, et al. Structural and functional analyses explain Pea KAI2 receptor diversity and reveal stereoselective catalysis during signal perception. Commun. Biol. 2022;5:126. doi: 10.1038/s42003-022-03085-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu E, et al. Catabolism of strigolactones by a carboxylesterase. Nat. Plants. 2021;7:1495–1504. doi: 10.1038/s41477-021-01011-y. [DOI] [PubMed] [Google Scholar]
- 61.Hiruma K, et al. Root endophyte Colletotrichum tofieldiae confers plant fitness benefits that are phosphate status dependent. Cell. 2016;165:464–474. doi: 10.1016/j.cell.2016.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Castrillo G, et al. Root microbiota drive direct integration of phosphate stress and immunity. Nature. 2017;543:513–518. doi: 10.1038/nature21417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Radhakrishnan GV, et al. An ancestral signalling pathway is conserved in intracellular symbioses-forming plant lineages. Nat. Plants. 2020;6:280–289. doi: 10.1038/s41477-020-0613-7. [DOI] [PubMed] [Google Scholar]
- 64.Markmann K, Parniske M. Evolution of root endosymbiosis with bacteria: How novel are nodules? Trends Plant Sci. 2009;14:77–86. doi: 10.1016/j.tplants.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 65.Dénarié J, Debellé F, Rosenberg C. Signaling and host range variation in nodulation. Annu. Rev. Microbiol. 1992;46:497–531. doi: 10.1146/annurev.mi.46.100192.002433. [DOI] [PubMed] [Google Scholar]
- 66.Madsen LH, et al. The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat. Commun. 2010;1:10. doi: 10.1038/ncomms1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oldroyd GE, Long SR. Identification and characterization of nodulation-signaling pathway 2, a gene of Medicago truncatula involved in Nod actor signaling. Plant Physiol. 2003;131:1027–1032. doi: 10.1104/pp.102.010710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bailey-Serres J, Parker JE, Ainsworth EA, Oldroyd GED, Schroeder JI. Genetic strategies for improving crop yields. Nature. 2019;575:109–118. doi: 10.1038/s41586-019-1679-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bozsoki Z, et al. Ligand-recognizing motifs in plant LysM receptors are major determinants of specificity. Science. 2020;369:663–670. doi: 10.1126/science.abb3377. [DOI] [PubMed] [Google Scholar]
- 70.Bartlett JG, Snape JW, Harwood WA. Intron-mediated enhancement as a method for increasing transgene expression levels in barley. Plant Biotechnol. J. 2009;7:856–866. doi: 10.1111/j.1467-7652.2009.00448.x. [DOI] [PubMed] [Google Scholar]
- 71.Milner MJ, et al. A PSTOL-like gene, TaPSTOL, controls a number of agronomically important traits in wheat. BMC Plant Biol. 2018;18:115. doi: 10.1186/s12870-018-1331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roth R, et al. A rice Serine/Threonine receptor-like kinase regulates arbuscular mycorrhizal symbiosis at the peri-arbuscular membrane. Nat. Commun. 2018;9:4677. doi: 10.1038/s41467-018-06865-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lawrenson, T. & Harwood, W. A. In Barley: Methods and Protocols (ed Wendy A. Harwood) 217–232 (Springer New York, 2019).
- 74.Tadege M, et al. Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 2008;54:335–347. doi: 10.1111/j.1365-313X.2008.03418.x. [DOI] [PubMed] [Google Scholar]
- 75.Ehrhardt DW, Atkinson EM, Long SR. Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti Nod factors. Science. 1992;256:998–1000. doi: 10.1126/science.10744524. [DOI] [PubMed] [Google Scholar]
- 76.Paszkowski U, Jakovleva L, Boller T. Maize mutants affected at distinct stages of the arbuscular mycorrhizal symbiosis. Plant J. 2006;47:165–173. doi: 10.1111/j.1365-313X.2006.02785.x. [DOI] [PubMed] [Google Scholar]
- 77.Vierheilig H, Coughlan AP, Wyss U, Piche Y. Ink and vinegar, a simple staining technique for arbuscular-mycorrhizal fungi. Appl. Environ. Microbiol. 1998;64:5004–5007. doi: 10.1128/aem.64.12.5004-5007.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Giovannetti M, Mosse B. An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. N. Phytol. 1980;84:489–500. [Google Scholar]
- 79.Patron NJ, et al. Standards for plant synthetic biology: a common syntax for exchange of DNA parts. New Phytol. 2015;208:13–19. doi: 10.1111/nph.13532. [DOI] [PubMed] [Google Scholar]
- 80.Feike D, et al. Characterizing standard genetic parts and establishing common principles for engineering legume and cereal roots. Plant Biotechnol. J. 2019;17:2234–2245. doi: 10.1111/pbi.13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Engler C, Kandzia R, Marillonnet S. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 2008;3:e3647. doi: 10.1371/journal.pone.0003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Boisson-Dernier A, et al. Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 2001;14:695–700. doi: 10.1094/MPMI.2001.14.6.695. [DOI] [PubMed] [Google Scholar]
- 83.He Y, Zhang T, Sun H, Zhan H, Zhao Y. A reporter for noninvasively monitoring gene expression and plant transformation. Hortic. Res. 2020;7:152. doi: 10.1038/s41438-020-00390-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data [Online]. (2010).
- 85.Tang H, et al. An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genom. 2014;15:312. doi: 10.1186/1471-2164-15-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Goodstein DM, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schreiber M, et al. A genome assembly of the barley ‘transformation reference’ cultivar Golden Promise. G3 (Bethesda) 2020;10:1823–1827. doi: 10.1534/g3.119.401010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 90.Kruskal J. Nonmetric multidimensional scaling: a numerical method. Psychometrika. 1964;29:115–129. [Google Scholar]
- 91.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Waskom, M. seaborn: statistical data visualization. J. Open Source Softw. 6, https://seaborn.pydata.org/ (2021).
- 94.Schiessl K, et al. NODULE INCEPTION recruits the lateral root developmental program for symbiotic nodule organogenesis in Medicago truncatula. Curr. Biol. 2019;29:3657–3668.e3655. doi: 10.1016/j.cub.2019.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Emms DM, Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ouyang S, et al. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res. 2006;35:D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lamesch P, et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2011;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cheng C-Y, et al. Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017;89:789–804. doi: 10.1111/tpj.13415. [DOI] [PubMed] [Google Scholar]
- 100.Mun T, Bachmann A, Gupta V, Stougaard J, Andersen SU. Lotus Base: an integrated information portal for the model legume Lotus japonicus. Sci. Rep. 2016;6:39447. doi: 10.1038/srep39447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hirsch CN, et al. Draft assembly of elite inbred line ph207 provides insights into genomic and transcriptome diversity in maize. Plant Cell. 2016;28:2700–2714. doi: 10.1105/tpc.16.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Camacho C, et al. BLAST+: architecture and applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gough, C., Cottret, L., Lefebvre, B. & Bono, J.-J. Evolutionary history of plant LysM receptor proteins related to root endosymbiosis. Front. Plant Sci. 9, PMC6039847 (2018). [DOI] [PMC free article] [PubMed]
- 104.Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2011;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–w259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kohlen W, et al. Strigolactones are transported through the xylem and play a key role in shoot architectural response to phosphate deficiency in nonarbuscular mycorrhizal host Arabidopsis. Plant Physiol. 2010;155:974–987. doi: 10.1104/pp.110.164640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.López-Ráez JA, et al. Does abscisic acid affect strigolactone biosynthesis? New Phytol. 2010;187:343–354. doi: 10.1111/j.1469-8137.2010.03291.x. [DOI] [PubMed] [Google Scholar]
- 111.Cardoso C, et al. Natural variation of rice strigolactone biosynthesis is associated with the deletion of two MAX1 orthologs. Proc. Natl Acad. Sci. USA. 2014;111:2379–2384. doi: 10.1073/pnas.1317360111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Song X, et al. IPA1 functions as a downstream transcription factor repressed by D53 in strigolactone signaling in rice. Cell Res. 2017;27:1128–1141. doi: 10.1038/cr.2017.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang L, et al. Strigolactone and karrikin signaling pathways elicit ubiquitination and proteolysis of SMXL2 to regulate hypocotyl elongation in Arabidopsis. Plant Cell. 2020;32:2251–2270. doi: 10.1105/tpc.20.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data of RNA-sequencing have been deposited in the National Center for Biotechnology Information, Gene Expression Omnibus (GEO), with accession code GSE214698. Any additional information required to reanalyze the data reported in this study is available from the lead contact upon request. Source data are provided with this paper.