Abstract
Cultivated grapevines are commonly grafted on closely related species to cope with specific biotic and abiotic stress conditions. The three North American Vitis species V. riparia, V. rupestris, and V. berlandieri, are the main species used for breeding grape rootstocks. Here, we report the diploid chromosome-scale assembly of three widely used rootstocks derived from these species: Richter 110 (110R), Kober 5BB, and 101–14 Millardet et de Grasset (Mgt). Draft genomes of the three hybrids were assembled using PacBio HiFi sequences at an average coverage of 53.1 X-fold. Using the tool suite HaploSync, we reconstructed the two sets of nineteen chromosome-scale pseudomolecules for each genome with an average haploid genome size of 494.5 Mbp. Residual haplotype switches were resolved using shared-haplotype information. These three reference genomes represent a valuable resource for studying the genetic basis of grape adaption to biotic and abiotic stresses, and designing trait-associated markers for rootstock breeding programs.
Subject terms: Plant genetics, Plant breeding
| Measurement(s) | Genome Assembly Sequence |
| Technology Type(s) | PacBio Sequel System |
| Sample Characteristic - Organism | Vitis cinerea var. helleri x Vitis rupestris • Vitis riparia x Vitis rupestris • Vitis cinerea var. helleri x Vitis riparia |
| Sample Characteristic - Location | State of California |
Background & Summary
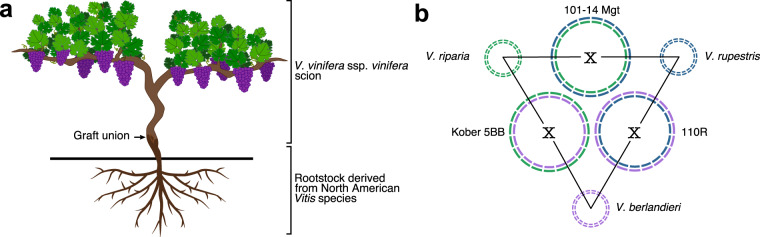
Cultivated grapevines (Vitis vinifera ssp. vinifera) are usually grafted onto rootstocks derived from North American Vitis species (Fig. 1a). This practice was established during the 19th century in response to the near devastation of European vineyards by the grape root aphid phylloxera (Daktulosphaira vitifoliae Fitch)1. Grape phylloxera was introduced into Europe in the 1850s through the movement of plant material from North America2. Most North American Vitis species are resistant to phylloxera, likely as a result of co-evolution with the insect in their native environment. Vitis riparia and Vitis rupestris were the first wild grape species used as rootstock because they root easily from hardwood cuttings and have good grafting compatibility with the berry-producing scions3. However, these two species were not suitable for calcareous soils, which are common in Europe. Vitis berlandieri, another North American grape species, was then found to be resistant to phylloxera and lime-tolerant, although it poorly roots from dormant cuttings4. To introduce the lime-tolerance of V. berlandieri and improve its rootability, new rootstocks were bred crossing V. berlandieri with either V. riparia or V. rupestris. Today, commercialized rootstocks are mainly hybrids of these three grape species5. Among these, Richter 110 (110R; V. berlandieri x V. rupestris), Kober 5BB (V. berlandieri x V. riparia), and 101–14 Millardet et de Grasset (Mgt; V. riparia x V. rupestris) are the most commonly used worldwide (Fig. 1b). In addition to their resistance to phylloxera, grape rootstocks are chosen based on tolerance to biotic (e.g. nematodes) and abiotic stresses (e.g. drought), preference of soil physicochemical properties, and the vigor level they confer to the scion6. For instance, 101–14 Mgt generally triggers the precocity of the vegetative growth despite a moderate vigor, whereas 110R and Kober 5BB confer high vigor and delay plant maturity7. 110R is known for its drought tolerance and excess soil moisture has negative impacts on its development6. In contrast, 101–14 Mgt and Kober 5BB are not considered drought-tolerant and grow well in moist soils6. The three rootstocks also have different levels of tolerance to nematodes depending on the nematode species6,8.
Fig. 1.
Description of the three grape rootstocks 101–14 Mgt, 110R, and Kober 5BB. (a) Wine grapevine scion (Vitis vinifera spp. vinifera) grafted onto a rootstock from another Vitis species. (b) Schematic representation of haplotype composition of 101–14 Mgt, 110R, and Kober 5BB. Each pair of rootstocks shares a set of chromosomes from the same parental Vitis species. Shared haplotypes are represented with the same color.
In addition to their commercial importance, rootstocks are valuable to study the genetic bases of grape adaptation to biotic and abiotic stresses9. However, to date only two genomes of V. riparia have been published10,11 and no reference genome is available for any of the commonly used rootstocks. This article describes the chromosome-scale assemblies of 110R, Kober 5BB, and 101–14 Mgt. Genomes were sequenced using highly accurate long-read sequencing (HiFi, Pacific Biosciences) and assembled with Hifiasm12. Each diploid draft genome was then scaffolded into two sets of pseudomolecules using the tool suite HaploSync13, and haplotypes were assigned to each Vitis parent based on sequence similarity between the haplotypes derived from the same species. These genomes represent an important resource for investigating the genetic basis of resistance to environmental factors and designing markers to accelerate rootstock breeding programs.
Methods
Library preparation and sequencing
Young leaves (1–2 cm-wide) were collected from 110R (FPS 01), Kober 5BB (FPS 06), and 101–14 Mgt (FPS 01) at Foundation Plant Services (University of California Davis, Davis, CA) and immediately frozen and ground to powder in liquid nitrogen. High molecular weight genomic DNA was extracted from 1 g of ground leaf tissue as described in Chin et al.14, and 12 µg of high molecular weight gDNA was sheared to a size distribution between 15 and 20 kbp using the Megaruptor® 2 (Diagenode, Denville, NJ, USA). For each accession, one HiFi sequencing library was prepared using the SMRTbell® Express Template Prep Kit 2.0 followed by immediate treatment with the Enzyme Clean Up Kit (Pacific Biosciences, Menlo Park, CA, USA). Libraries were size-selected using a BluePippin (Sage Sciences, Beverly, MA, USA) and HiFi SMRTbell® templates longer than 15 kbp were collected. Size-selected library fractions were cleaned using AMPure PB beads (Pacific Biosciences, Menlo Park, CA, USA). Concentration and final size distribution of the libraries were evaluated using a Qubit™ 1X dsDNA HS Assay Kit (Thermo Fisher, Waltham, MA, USA) and Femto Pulse System (Agilent, Santa Clara, CA, USA), respectively. HiFi libraries of 110R and Kober 5BB were sequenced using a PacBio Sequel II system (Pacific Biosciences, CA, USA) at the DNA Technology Core Facility, University of California, Davis (Davis, CA, USA). For 101–14 Mgt, sequencing was performed by Corteva Agriscience (Johnston, IA, USA) as an award from Pacific Biosciences to Dr. Noé Cochetel. An average of 26.5 ± 3.8 Gbp sequences were generated for each genome, corresponding to 53.1 ± 7.7 X-fold coverage of a 500 Mbp haploid genome (Table 1).
Table 1.
Genome assembly statistics of the three rootstocks.
| 101–14Mgt | 110R | Kober 5BB | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sequencing | ||||||||||||||||||
| Sequencing Depth (Gbp) X-Fold coverage* | 26.3 | 30.5 | 22.8 | |||||||||||||||
| 53x | 61x | 46x | ||||||||||||||||
| Draft Assembly | ||||||||||||||||||
| Cumulative length (bp) | 1,021,000,930 | 1,006,052,903 | 1,018,035,111 | |||||||||||||||
| Number of sequences | 656 | 348 | 214 | |||||||||||||||
| Average sequence length (bp) | 1,556,404 | 2,890,957 | 4,757,173 | |||||||||||||||
| Maximum sequence length (bp) | 38,550,893 | 40,445,604 | 32,011,292 | |||||||||||||||
| N50 Length (bp) | 14,459,101 | 13,727,353 | 14,854,816 | |||||||||||||||
| N50 Index | 24 | 28 | 26 | |||||||||||||||
| Count | % | Count | % | Count | % | |||||||||||||
| Complete BUSCOs (Total 2,326) | 2,290 | 98.5% | 2,295 | 98.7% | 2,294 | 98.6% | ||||||||||||
| Complete BUSCOs Single | 18 | 0.8% | 27 | 1.2% | 74 | 3.2% | ||||||||||||
| Complete BUSCOs Duplicated | 2,272 | 97.7% | 2,268 | 97.5% | 2,220 | 95.4% | ||||||||||||
| Pseudomolecules | ||||||||||||||||||
| HaplotypeV. riparia | HaplotypeV. rupestris | Unplaced | HaplotypeV. berlandieri | HaplotypeV. rupestris | Unplaced | HaplotypeV. berlandieri | HaplotypeV. riparia | Unplaced | ||||||||||
| Cumulative length (bp) | 492,356,428 | 492,600,706 | 36,133,067 | 495,178,401 | 491,477,282 | 19,903,444 | 505,179,188 | 489,908,332 | 23,334,789 | |||||||||
| GC percentage | 35.0% | 34.8% | 43.6% | 34.6% | 34.7% | 47.0% | 34.7% | 35.0% | 42.3% | |||||||||
| Number of sequences | 19 | 19 | 527 | 19 | 19 | 215 | 19 | 19 | 92 | |||||||||
| Average sequence length (bp) | 25,913,496 | 25,926,352 | 68,564 | 26,062,021 | 25,867,225 | 92,574 | 26,588,378 | 25,784,649 | 253,639 | |||||||||
| N50 Length (bp) | 25,475,941 | 25,378,183 | 69,079 | 26,414,266 | 25,747,756 | 143,097 | 26,431,197 | 25,800,664 | 2,015,173 | |||||||||
| N50 Index | 9 | 9 | 116 | 9 | 9 | 29 | 9 | 9 | 5 | |||||||||
| Count | % | Count | % | Count | % | Count | % | Count | % | Count | % | Count | % | Count | % | Count | % | |
| Complete BUSCOs (Total 2,326) | 2,284 | 98.2% | 2,284 | 98.2% | 9 | 0.4% | 2,282 | 98.1% | 2,276 | 97.9% | 2 | 0.1% | 2,286 | 98.3% | 2,283 | 98.2% | 37 | 1.6% |
| Complete BUSCOs Single | 2,240 | 96.3% | 2,240 | 96.3% | 9 | 0.4% | 2,239 | 96.3% | 2,230 | 95.9% | 2 | 0.1% | 2,239 | 96.3% | 2,237 | 96.2% | 34 | 1.5% |
| Complete BUSCOs Duplicated | 44 | 1.9% | 44 | 1.9% | 0 | 0.0% | 43 | 1.8% | 46 | 2.0% | 0 | 0.0% | 47 | 2.0% | 46 | 2.0% | 3 | 0.1% |
| PN40024 unique genes (Total 28,243) | 26,868 | 95.1% | 26,802 | 94.9% | 352 | 1.3% | 26,854 | 95.1% | 26,836 | 95.0% | 402 | 1.4% | 26,866 | 95.1% | 26,791 | 94.9% | 761 | 2.7% |
| Annotated Genes | 33,147 | 33,611 | 6,000 | 28,110 | 27,678 | 980 | 29,620 | 28,927 | 1,260 | |||||||||
| Annotated Proteins | 83,091 | 83,455 | 6,217 | 50,909 | 49,418 | 1,048 | 54,187 | 53,912 | 1,588 | |||||||||
| Repeat content | 50.4% | 50.0% | 53.9% | 49.3% | 49.6% | 75.2% | 49.9% | 50.3% | 75.0% | |||||||||
*based on 500Mbp genome size
Summary statistics of the genome sequencing, draft genome assembly, chromosome-scale genome assembly, and gene annotation of 101–14 Mgt, 110R, and Kober 5BB rootstocks.
Total RNA from V. berlandieri 9031, V. rupestris B38, and V. riparia HP-1 (PI588271) leaves was isolated using a Cetyltrimethyl Ammonium Bromide (CTAB)-based extraction protocol as described in Blanco-Ulate et al.15. RNA purity was evaluated with a Nanodrop 2000 spectrophotometer (Thermo Scientific, Hanover Park, IL, USA), and RNA integrity by electrophoresis and an Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA). RNA quantity was assessed with a Qubit 2.0 Fluorometer and a broad range RNA kit (Life Technologies, Carlsbad, CA, USA). Total RNA (300 ng, RNA Integrity Number >8.0) were used for library construction. Short-read cDNA libraries were prepared using the Illumina TruSeq RNA sample preparation kit v.2 (Illumina, CA, USA) following Illumina™ low-throughput protocol. Libraries were evaluated for quantity and quality with the High Sensitivity chip and an Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA). One library per species was sequenced using an Illumina HiSeq 4000 sequencer with a 2x100bp protocol (DNA Technology Core Facility, University of California, Davis, CA, USA). Long-read cDNA SMRTbell libraries were prepared for V. berlandieri and V. riparia. First-strand synthesis and cDNA amplification were accomplished using the NEB Next Single Cell/Low Input cDNA Synthesis & Amplification Module (New England, Ipswich, MA, USA). The cDNAs were subsequently purified with ProNex magnetic beads (Promega, WI, USA) following the instructions in the Iso-Seq Express Template Preparation for Sequel and Sequel II Systems protocol (Pacific Biosciences, Menlo Park, CA, USA). ProNex magnetic beads (86 µL) were used to select amplified cDNA (≥2 kbp). At least 80 ng of the size-selected amplified cDNA were used to prepare the cDNA SMRTbell library. DNA damage repair and SMRTbell ligation was performed with SMRTbell Express Template Prep Kit 2.0 (Pacific Biosciences, Menlo Park, CA, USA) following the manufacturer’s protocol. One SMRT cell was sequenced for each species on the PacBio Sequel I platform (DNA Technology Core Facility, University of California, Davis, CA, USA).
Genome assembly and pseudomolecule construction
HiFi reads were assembled using Hifiasm v.0.16.1-r37412. Multiple combinations of several assembly parameters were tested. A total of 1,939 assemblies were generated. The least fragmented assembly of each genotype was selected. The selected draft assemblies consisted of 406 ± 226 contigs with a N50 = 14.3 ± 0.6 Mbp (Table 1). Compared to other grape genomes previously generated with PacBio CLR technology, the PacBio HiFi reads greatly improves the contiguity of the draft assembly (PacBio CLR 1.2 ± 0.3 Mbp, Fig. 2a). Gene space completeness was assessed using BUSCO V.5.1 with the Viridiplantae and Embryophyta ODB10 datasets16 and by mapping PN40024 (V1 annotation17) single-copy genes using GMAP v.2019-09-12 (alignments with at least 80% coverage and 80% identity were considered). For each rootstock, the draft genome assembly underwent quality control and scaffolding into a diploid set of chromosome-scale pseudomolecules using HaploSync13 and the Vitis consensus genetic map developed by Zou et al.18. One cycle of HaploFill was used for each genotype. The use of PacBio HiFi reads reduced significantly the fragmentation of the draft assembly compared to recently published grape genomes sequenced using PacBio CLR technology (Fig. 2b)13,14,19. The lower fragmentation resulted in a 15 times smaller number of contigs necessary to scaffold a pseudomolecule (3.6 ± 2.0 HiFi contigs/pseudomolecule vs. 43.0 ± 20.6 CLR contigs/pseudomolecule) (Fig. 2b). Remarkably, in total across the three genomes, fifteen pseudomolecules were reconstructed from a single contig. Haplotype switches were identified based on sequence similarity of protein-coding sequences. Gene loci sequences of each rootstock were aligned against each others using minimap2 v.2.17-r94120 and the parameter “-x map-hifi”. Alignments with the highest coverage and identity were used to assign common species parentage and to detect haplotype switches along pseudomolecules (Fig. 3a). After manual correction of the haplotype switches, a second cycle of HaploFill13 was performed using the pseudomolecules derived from the same Vitis species as alternative haplotypes to help closing gaps with draft sequences.
Fig. 2.
PacBio HiFi sequencing technology substantially improves the contiguity of Vitis draft genome assembly. (a) Draft assembly fragmentation of 101–14 Mgt, 110R, Kober 5BB represented as distribution of contig NG(x) values. Muscadidia rotundifolia cv. Trayshed and V. vinifera cv. Cabernet Sauvignon, produced with CLR reads, were included as comparison. The NG(x) value is defined as the sequence length of the shortest contig necessary to achieve, cumulatively, a given fraction (x) of the expected diploid genome length (1 Gbp) when sequences are sorted from the longest to the shortest. Diploid assemblies produced with PacBio HiFi reads (101–14 Mgt, 110R, and Kober 5BB) resulted in a much more contiguous draft genome assembly compared to other grape genomes assembled with older long-read sequencing technologies despite a lower X-Fold coverage employed (PacBio Sequel CLR reads for Trayshed 140x X-Fold coverage;19,21 PacBio RSII CLR reads for Cabernet Sauvignon, 115X X-Fold coverage14). (b) Distribution of the number of contig scaffolded into complete pseudomolecules. The substantially lower fragmentation of the draft assemblies generated using PacBio HiFi reads (101–14 Mgt, 110R, and Kober 5BB) resulted on average in a 15x smaller number of contigs necessary to build a pseudomolecule.
Fig. 3.
Haplotyping based on intraspecific sequence similarity. Shared parental species information was used to assign each haplotype to either V. riparia, V. rupestris, or V. berlandieri based on sequence similarity. This allowed to resolved assembly errors (i.e. haplotype switches). (a) Example of an haplotype switch found on chromosome 8 of Kober 5BB (left panel). After scaffolding of the pseudomolecules, an haplotype switch was observed at the end of chromosome 8 of Kober 5BB. The genes in the contig Kober5BB_h1tg000016l on haplotype 1 were highly similar to the genes located in 101–14 Mgt haplotype 2 (red), suggesting that Kober5BB_h1tg000016l derived from V. riparia, whereas the genes of Kober5BB_h1tg000038l corresponded to genes in haplotype 2 of chromosome 8 of 110R (light green), suggesting that Kober5BB_h1tg000038l derived from V. berlandieri. An opposite pattern was observed on haplotype 2, with the genes of the first 18.9 Mbp of the pseudomolecule similar to the genes of the haplotype 2 of 110R (light green) and the genes from the last 4.2 Mbp similar to the genes of 101–14 Mgt haplotype 2 (red). The haplotype switch was corrected by interchanging the contig Kober5BB_h1tg000038l with the corresponding region in the alternative haplotype, consisting of Kober5BB_h2tg000109l and 4.2 Mbp of Kober5BB_h2tg000017l (right panel). (b) Sequence similarity between haplotypes represented as the average percentage of the haploid chromosome set length not affected by structural variants (>50 bp), SNPs or InDels when compared with another haplotype. (c) Distribution of the percentage of sequence similarity (as defined in B) between haplotypes derived from the same species and haplotypes derived from different species (Statistical testing was performed with pairwise Wilcoxon rank sum test, density plot was produced with adjust = 1, n = 4096, kernel = “cosine” parameters).
Gene prediction and repeat annotation
Gene structural annotations were predicted using the procedures described in https://github.com/andreaminio/AnnotationPipeline-EVM_based-DClab21. For each rootstock, Iso-Seq data from the corresponding parental species were concatenated with the de novo assembled transcripts from RNA-seq reads before generating the gene models. Iso-Seq libraries underwent extraction, demultiplexing and error correction using IsoSeq3 v.3.3.0 protocol (https://github.com/PacificBiosciences/IsoSeq). Low-quality and single isoforms dataset were further polished using LSC v2.022. RNA-seq reads were quality-filtered and adapters were trimmed with Trimmomatic v.0.36 and the options “ILLUMINACLIP:2:30:10 LEADING:7 TRAILING:7 SLIDINGWINDOW:10:20 MINLEN:36”23. High-quality RNA-seq reads from each Vitis species were assembled with three different protocols: (i) Trinity v.2.6.524 with the “de novo” protocol, (ii) Trinity v.2.6.524 using the “On-genome” protocol, (iii) Stringtie v.1.3.4d25 using the reads found to align on the genome sequences with HISAT2 v.2.0.5 and the parameter “--very-sensitive”26. Transcript sequences common to the three assembly methods were then pooled with the Iso-Seq reads. Sequence redundancy was reduced using CD-HIT v4.627 with the parameters “cd-hit-est -c 0.99 -g 0 -r 0 -s 0.70 -aS 0.99”. Non-redundant transcripts were processed with PASA v.2.3.328 to obtain the final training model sets. Combined with data from public databases, the derived transcript and protein evidences were aligned on the genome assembly using a multi-aligner pipeline including Exonerate v.2.2.029 and Pasa v.2.3.328. To produce the final set of consensus gene models with EvidenceModeler v.1.1.130, ab initio predictions were also generated using Augustus v.3.0.331, BUSCO v.3.0.232, GeneMark v.3.4733, and SNAP v.2006-07-2834. For the repeat annotation, RepeatMasker v.open-4.0.635 was used. To assign a functional annotation to each of these gene models, results from diamond v2.0.13.15136,37 blastp matches on the Refseq plant protein database (https://ftp.ncbi.nlm.nih.gov/refseq/, retrieved January 17th, 2019) and from InterProScan v.5.28–67.038 were parsed through Blast2GO v.4.1.939. A total of 56,768 protein-coding gene loci were annotated in the genome assembly of 110R, 59,807 in Kober 5BB and 72,758 in 101–14 Mgt. On average, 124,991 ± 36,197 protein-coding alternative splicing variants were identified per haplotype. The unplaced sequences were composed of 2,747 ± 2,821 gene loci (Table 1).
Analysis of colinearity between haplotypes
Colinear gene loci were identified using MCScanX v.11.Nov.201340. Annotated protein-coding sequences of the three rootstocks were aligned against each other using GMAP v.2019-09-1241 with the parameters “-B 4 -x 30–split-output”. Alignments with both identity and coverage greater than 80% were retained. Alignments corresponding to annotated mRNA regions were identified using mapBed from Bedtools v2.29.242 with the parameters “-F 0.75 -f 0.5 -e”. Colinear blocks were then detected with MCScanx_h (MCScanX v.11.Nov.201340) tool using the following parameters “-s 10 -m 5 -w 5”.
Identification of sequence polymorphisms and structural variants between haplotypes
Pseudomolecule sequences were aligned against each other using nucmer tool from MUMmer4 v.4.0.0.beta543. SNPs and short indels between haplotypes were identified from alignments with show-snps tool (MUMmer4 v.4.0.0.beta543) with parameters “-Clr -x” and longer structural variants with show-diff tool (MUMmer4 v.4.0.0.beta543) with default parameters.
Data Records
Sequencing data were deposited at NCBI under BioProject number PRJNA858084, SRA accessions SRR2081042144, SRR2081042245, SRR2081042346, SRR2081042447, SRR2081042548, SRR2081042649, and SRR2081042750. Genome assemblies are available at EMBL-EBI under BioProject number PRJEB5501351. Genome assemblies, gene annotation and repeat annotation files are at Zenodo under the 10.5281/zenodo.682432352, and at http://www.grapegenomics.com53. A genome browser and a blast tool are available for each rootstock at http://www.grapegenomics.com53.
Technical Validation
The genome assemblies were evaluated for completeness of the diploid sequence and gene content, and for correct haplotype phasing. The average size of each set of 19 pseudomolecules was 494.5 ± 5.5 Mbp (diploid genome size: 1,015.0 ± 7.9 Mbp, Supplemental figure 1), which is close to the length of the parental haploid genome size estimated by flow cytometry (499.3 ± 37.3 Mbp54) suggesting that the three genomes were entirely assembled. Only 36.1 Mbp (3.5%), 19.9 Mbp (2.0%), and 23.3 Mbp (2.3%) of the draft sequences could not be placed into any pseudomolecules of 101–14 Mgt, 110R, and Kober 5BB genomes, respectively. The unplaced sequences were mostly composed of repeats (68.0% ± 12.3%). These results are comparable with the latest release of the V. vinifera PN40024 reference haploid genome assembly, for which the location of 27.4 Mbp (5.6%) remains undetermined55.
Each set of 19 pseudomolecules was evaluated for gene space completeness using both conserved single-copy orthologs of plant genes (BUSCOs) and the single-copy gene content of V. vinifera PN40024. Complete copies of 98.1 ± 0.14% of the BUSCO models were found in each set of pseudomolecules (Supplemental Table 1). Similarly, almost all of the single-copy genes of PN40024 aligned to each set of pseudomolecules (95.01% ± 0.3%). The gene space present in the unplaced sequences was limited to 0.69 ± 0.8% of the BUSCO models and 1.79 ± 0.8% of the PN40024 genes. The completeness of the gene space is another strong evidence that the assemblies are a complete representation of the diploid genomes of the three rootstocks. On both haplotypes of 101–14 Mgt we found more gene loci (33,379 ± 328) than in 110R and Kober 5BB (28,584 ± 863). Further genome-wide gene expression analyses are required to determine if the larger number of gene loci identified in 101–14 Mgt corresponds to a larger number of expressed transcripts than in the other rootstocks.
Using the pedigree information of each rootstock (Fig. 1b), we assigned each pseudomolecule to its parental Vitis species, i.e. either V. riparia, V. rupestris, or V. berlandieri. For each pseudomolecule, we identified the three pairs of haplotypes having the highest gene sequence similarity and assigned them to the shared parental Vitis species. This allowed us to manually detect and correct the phasing errors (i.e. haplotype switches) introduced during the assembly of the draft sequences or the scaffolding of the pseudomolecules (Fig. 3a). Whole-sequence comparison of the six haplotypes of each pseudomolecule showed that the haplotypes assigned to the same Vitis species were more similar (80.5% ± 1.4% identity) than those that do not share the same species (74.0% ± 3.3% identity; p value = 0.0003, W = 142, n = 30 unpaired Wilcoxon rank sum test; Fig. 3b,c). These results suggest that the haplotypes of the three rootstock genomes were correctly phased. Despite the variable levels of sequence polymorphism, pseudomolecules of the three rootstock genomes were highly colinear regardless of their species of origin. When considering both gene sequence similarity, gene order, and physical location, 73.1% ± 3.5% of the protein-coding loci were found in at least one colinear block when comparing haplotypes with shared parental origin, and 71.5% ± 3.5% between haplotypes of different species (Supplemental figure 2). Overall, an average of 82.4% ± 2.6% of the genomic sequences are covered by colinear blocks (Supplemental figure 3), which reflects a remarkable conservation of chromosome structure among these Vitis species.
Supplementary information
Acknowledgements
The RNAseq data of V. rupestris were kindly provided by Dr. Jason Londo, Cornell University. This work was funded by the NSF grant #1741627 and partially supported by funds to D.C. from the Louis P. Martini Endowment in Viticulture.
Author contributions
A.M., N.C. and D.C. conceived the work. A.M. conducted the bioinformatic analyses. R.F.-B. performed all the wet-lab activities associated with the project. A.M., N.C., M.M., D.C. wrote the manuscript.
Code availability
The pipeline used for gene structural and functional annotation is available in details at https://github.com/andreaminio/AnnotationPipeline-EVM_based-DClab.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41597-022-01753-0.
References
- 1.Millardet, A. Histoire des principales variétés et espéces de vignes d’origine américaine qui résistent au phylloxera (G. Masson, Paris, 1885).
- 2.Dodson Peterson, J. C. et al. Grape Rootstock Breeding and Their Performance Based on the Wolpert Trials in California. In Cantu, D. & Walker, M. A. (eds.) The Grape Genome, 301–318, 10.1007/978-3-030-18601-2_14 (Springer International Publishing, Cham, 2019).
- 3.Pongracz, D. P. Rootstocks for grape-vines. Publisher: Cape Town (South Africa) David Philip (1983).
- 4.Ravaz, L. Les vignes américaines: porte-greffes et producteurs-directs: caractéres, aptitudes (Goulet, 1902).
- 5.Riaz S, et al. Genetic diversity and parentage analysis of grape rootstocks. Theoretical and Applied Genetics. 2019;132:1847–1860. doi: 10.1007/s00122-019-03320-5. [DOI] [PubMed] [Google Scholar]
- 6.Christensen, L. P. Rootstock selection. Wine grape varieties in California. University of California, Oakland, CA, USA 12–15 (2003).
- 7.Dodson Peterson JC, Andrew Walker M. Influence of Grapevine Rootstock on Scion Development and Initiation of Senescence. Catalyst: Discovery into Practice. 2017;1:48. doi: 10.5344/catalyst.2017.16006. [DOI] [Google Scholar]
- 8.Ferris H, Zheng L, Walker MA. Resistance of Grape Rootstocks to Plant-parasitic Nematodes. Journal of nematology. 2012;44:377–386. [PMC free article] [PubMed] [Google Scholar]
- 9.Rahemi, A., Dodson Peterson, J. C. & Lund, K. T. Grape Rootstocks and Related Species (Springer International Publishing, Cham, 2022).
- 10.Girollet, N. et al. De novo phased assembly of the Vitis riparia grape genome. Scientific Data6, 1–8, 10/ghdrm3 (2019). [DOI] [PMC free article] [PubMed]
- 11.Patel, S. et al. Draft genome of the Native American cold hardy grapevine Vitis riparia Michx. ‘Manitoba 37’. Horticulture Research7, 10/gg53d4. ISBN: 4143802003162 Publisher: Springer US (2020). [DOI] [PMC free article] [PubMed]
- 12.Cheng, H., Concepcion, G. T., Feng, X., Zhang, H. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nature Methods18, 170–175, 10/ghz4s5 (2021). [DOI] [PMC free article] [PubMed]
- 13.Minio, A., Cochetel, N., Vondras, A. M., Massonnet, M. & Cantu, D. Assembly of complete diploid-phased chromosomes from draft genome sequences. G3 Genes|Genomes|Genetics jkac143, 10.1093/g3journal/jkac143 (2022). [DOI] [PMC free article] [PubMed]
- 14.Chin CS, et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nature Methods. 2016;13:1050–1054. doi: 10.1038/nmeth.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blanco-Ulate, B., Vincenti, E., Powell, A. L. & Cantu, D. Tomato transcriptome and mutant analyses suggest a role for plant stress hormones in the interaction between fruit and Botrytis cinerea. Frontiers in Plant Science4, 1–16, 10/gkzg3v (2013). [DOI] [PMC free article] [PubMed]
- 16.Manni M, Berkeley MR, Seppey M, Simão FA, Zdobnov EM. BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Molecular Biology and Evolution. 2021;38:4647–4654. doi: 10.1093/molbev/msab199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaillon, O. et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature449, 463–467, 10/ckfnh2 (2007). [DOI] [PubMed]
- 18.Zou, C. et al. Haplotyping the Vitis collinear core genome with rhAmpSeq improves marker transferability in a diverse genus. Nature Communications11, 413, 10/ghdrnk. Publisher: Springer US (2020). [DOI] [PMC free article] [PubMed]
- 19.Massonnet, M. et al. The genetic basis of sex determination in grapes. Nature communications11, 2902, 10/gjxrfm. Publisher: Springer US (2020). [DOI] [PMC free article] [PubMed]
- 20.Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics34, 3094–3100, 10/gdhbqt. _eprint: 1708.01492 (2018). [DOI] [PMC free article] [PubMed]
- 21.Cochetel N, et al. Diploid chromosome-scale assembly of the Muscadinia rotundifolia genome supports chromosome fusion and disease resistance gene expansion during Vitis and Muscadinia divergence. G3 Genes|Genomes|Genetics. 2021;11:jkab033. doi: 10.1093/g3journal/jkab033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Au, K. F., Underwood, J. G., Lee, L. & Wong, W. H. Improving PacBio Long Read Accuracy by Short Read Alignment. PLoS ONE7, 1–8, 10/f383xz (2012). [DOI] [PMC free article] [PubMed]
- 23.Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics30, 2114–2120, 10/f6cj5w (2014). [DOI] [PMC free article] [PubMed]
- 24.Haas, B. J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nature Protocols8, 1494–1512, 10/f22qdv (2013). [DOI] [PMC free article] [PubMed]
- 25.Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature Biotechnology33, 290–295, 10/f64s85 (2015). [DOI] [PMC free article] [PubMed]
- 26.Kim D, Langmead B, Salzberg SL. HISAT: A fast spliced aligner with low memory requirements. Nature Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, W. & Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics22, 1658–1659, 10/ct8g72 (2006). [DOI] [PubMed]
- 28.Haas, B. J. et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Research31, 5654–5666, 10/cgkkwd (2003). [DOI] [PMC free article] [PubMed]
- 29.Slater GSC, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:1–11. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haas, B. J. et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biology9, 10.1186/gb-2008-9-1-r7 (2008). [DOI] [PMC free article] [PubMed]
- 31.Stanke M, Tzvetkova A, Morgenstern B. AUGUSTUS at EGASP: using EST, protein and genomic alignments for improved gene prediction in the human genome. Genome biology. 2006;7(Suppl 1):1–8. doi: 10.1186/gb-2006-7-s1-s11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seppey, M., Manni, M. & Zdobnov, E. M. Gene Prediction: Methods and Protocols, vol. 1962 of Methods in Molecular Biology (Springer New York, New York, NY, 2019).
- 33.Lomsadze A, Ter-Hovhannisyan V, Chernoff YO, Borodovsky M. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Research. 2005;33:6494–6506. doi: 10.1093/nar/gki937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korf, I. Gene finding in novel genomes. BMC Bioinformatics5, 59, 10/cdvb5x. ISBN: 1471-2105 (Electronic) (2004). [DOI] [PMC free article] [PubMed]
- 35.Smit, A. F. A., Hubley, R. & Green, P. RepeatMasker Open-4.0. Pages: 2013–2015 Publication Title: http://www.repeatmasker.org (2013).
- 36.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nature Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 37.Buchfink B, Reuter K, Drost H-G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nature Methods. 2021;18:366–368. doi: 10.1038/s41592-021-01101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones, P. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics (Oxford, England)30, 1236–40, 10/f53532 (2014). [DOI] [PMC free article] [PubMed]
- 39.Conesa A, et al. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 40.Wang, Y. et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Research40, 1–14, 10/fzn3xm (2012). [DOI] [PMC free article] [PubMed]
- 41.Wu, T. D. & Watanabe, C. K. GMAP: A genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics21, 1859–1875, 10/cjb8q8 (2005). [DOI] [PubMed]
- 42.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marçais, G. et al. MUMmer4: A fast and versatile genome alignment system. PLoS Computational Biology14, e1005944, 10/gcw64s (2018). [DOI] [PMC free article] [PubMed]
- 44.2022. NCBI Sequence Read Archive. SRR20810421
- 45.2022. NCBI Sequence Read Archive. SRR20810422
- 46.2022. NCBI Sequence Read Archive. SRR20810423
- 47.2022. NCBI Sequence Read Archive. SRR20810424
- 48.2022. NCBI Sequence Read Archive. SRR20810425
- 49.2022. NCBI Sequence Read Archive. SRR20810426
- 50.2022. NCBI Sequence Read Archive. SRR20810427
- 51.2022. ENA European Nucleotide Archive. PRJEB55013
- 52.Minio A, Cantu D, Cochetel N, Massonnet M, Figueroa-Balderas R. 2022. Supporting data: HiFi chromosome-scale diploid assemblies of the grape rootstocks 110R, Kober 5BB, and 101–14 Mgt. Zenodo. [DOI] [PMC free article] [PubMed]
- 53.Minio A, Cantu D. 2022. Grapegenomics.com: a web portal with genomic data and analysis tools for wild and cultivated grapevines. Zenodo. [DOI]
- 54.Lodhi, M. A. & Reisch, B. I. Nuclear DNA content of Vitis species, cultivars, and other genera of the Vitaceae. Theoretical and Applied Genetics90, 11–16, 10/cgwkss (1995). [DOI] [PubMed]
- 55.Canaguier A, et al. A new version of the grapevine reference genome assembly (12X.v2) and of its annotation (VCost.v3) Genomics Data. 2017;14:56–62. doi: 10.1016/j.gdata.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- 2022. NCBI Sequence Read Archive. SRR20810421
- 2022. NCBI Sequence Read Archive. SRR20810422
- 2022. NCBI Sequence Read Archive. SRR20810423
- 2022. NCBI Sequence Read Archive. SRR20810424
- 2022. NCBI Sequence Read Archive. SRR20810425
- 2022. NCBI Sequence Read Archive. SRR20810426
- 2022. NCBI Sequence Read Archive. SRR20810427
- 2022. ENA European Nucleotide Archive. PRJEB55013
- Minio A, Cantu D, Cochetel N, Massonnet M, Figueroa-Balderas R. 2022. Supporting data: HiFi chromosome-scale diploid assemblies of the grape rootstocks 110R, Kober 5BB, and 101–14 Mgt. Zenodo. [DOI] [PMC free article] [PubMed]
- Minio A, Cantu D. 2022. Grapegenomics.com: a web portal with genomic data and analysis tools for wild and cultivated grapevines. Zenodo. [DOI]
Supplementary Materials
Data Availability Statement
The pipeline used for gene structural and functional annotation is available in details at https://github.com/andreaminio/AnnotationPipeline-EVM_based-DClab.