Abstract
Exploring individual hallmarks of brain ageing is important. Here, we propose the age-related glucose metabolism pattern (ARGMP) as a potential index to characterize brain ageing in cognitively normal (CN) elderly people. We collected 18F-fluorodeoxyglucose (18F-FDG) PET brain images from two independent cohorts: the Alzheimer’s Disease Neuroimaging Initiative (ADNI, N = 127) and the Xuanwu Hospital of Capital Medical University, Beijing, China (N = 84). During follow-up (mean 80.60 months), 23 participants in the ADNI cohort converted to cognitive impairment. ARGMPs were identified using the scaled subprofile model/principal component analysis method, and cross-validations were conducted in both independent cohorts. A survival analysis was further conducted to calculate the predictive effect of conversion risk by using ARGMPs. The results showed that ARGMPs were characterized by hypometabolism with increasing age primarily in the bilateral medial superior frontal gyrus, anterior cingulate and paracingulate gyri, caudate nucleus, and left supplementary motor area and hypermetabolism in part of the left inferior cerebellum. The expression network scores of ARGMPs were significantly associated with chronological age (R = 0.808, p < 0.001), which was validated in both the ADNI and Xuanwu cohorts. Individuals with higher network scores exhibited a better predictive effect (HR: 0.30, 95% CI: 0.1340 ~ 0.6904, p = 0.0068). These findings indicate that ARGMPs derived from CN participants may represent a novel index for characterizing brain ageing and predicting high conversion risk into cognitive impairment.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11357-022-00588-2.
Keywords: Pattern, Brain ageing, Positron emission tomography, Glucose metabolism
Introduction
Ageing is a complex process occurring in both normal ageing and pathological conditions. Exploring hallmarks of brain ageing in cognitively normal (CN) elderly people is important for understanding the normal ageing process [1, 2]. Currently, existing indices used to characterize individual brain ageing focus on the cellular and molecular levels, such as mitochondrial dysfunction, accumulation of oxidatively damaged molecules, aberrant neuronal network activity, and dysregulated energy metabolism [3–5]. However, the complexity of measurement methods limits the wide application of these indices in clinical practice. In addition, the myriad of indices introduced by previous studies cannot be universally used in all conditions due to different mechanisms and their variable patterns. For instance, these indices may be influenced by both healthy brain ageing and pathological disruptions, e.g. amyloid-β (Aβ) and tau pathologies in Alzheimer’s disease (AD) [3]. Therefore, an alternative in vivo index is needed.
Glucose metabolism alterations in the brain may represent an alternative index of brain ageing. Ageing appears to deteriorate the systemic control of glucose utilization, which, in turn, may increase the risk of reducing glucose uptake in brain regions [6]. Studies have reported that brain glucose metabolism abnormalities occur with ageing and reflect cognitive decline with high sensitivity [7, 8]. Thus, mapping age-related regional glucose metabolism changes may represent a promising avenue for understanding the neurobiological foundation of normal brain ageing. 18F-fluorodeoxyglucose (18F-FDG) positron emission tomography (PET) is a well-established technique to visualize and quantify the resting-state cerebral glucose metabolic rate in vivo and has been used to elucidate the normal ageing process of human brains [9–11]. Using 18F-FDG PET, previous studies have shown that normal brain ageing is associated with specific patterns of regional cerebral glucose metabolism, such as the anterior cingulate/medial prefrontal cortex [1, 7, 12]. Therefore, it is possible to estimate brain ageing by generating a glucose metabolism pattern using 18F-FDG PET.
Currently, data-driven methods are usually applied to estimate brain ageing using chronological age as the training label [13–15]. In particular, the scaled subprofile model/principal component analysis (SSM/PCA) method is frequently used to characterize brain glucose metabolism. SSM/PCA is a multivariate statistical method that drives a covariance pattern of voxel-based differences in brain metabolism [16]. This feature extraction method can enhance the identification of significant patterns in multivariate imaging data and mirror the underlying relationships between brain regions that are not captured by univariate techniques [17, 18]. Additionally, it enables the assessment of network-level alterations [19]. Therefore, this method is an optimal tool to evaluate abnormalities in functional brain organization in neurodegenerative disorders. Currently, the SSM/PCA method has been widely used to investigate brain metabolic patterns in several disorders, such as AD, Parkinson’s disease (PD), and multiple system atrophy (MSA) [20–23]. Thus, we hypothesized that the SSM/PCA method could also be used to assess age-related glucose metabolism patterns (ARGMPs) in CN older adults.
In the present study, we extracted ARGMPs from 18F-FDG PET images and generated the ARGMP expression score in each participant as a potential index to characterize brain ageing. The primary purposes of this study were (1) to validate the robustness of ARGMPs in both Western and Chinese cohorts; (2) to assess whether ARGMPs in normal ageing are independent from the AD pathological process; and (3) to investigate whether individuals with high ARGMPs have a high risk of developing cognitive impairment during follow-up.
Materials and methods
Overall study procedures
A total of 211 CN elderly people were collected from two independent cohorts: the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort (Cohort A, N = 127) from the ADNI database (http://adni.loni.usc.edu/) and the Xuanwu cohort (Cohort B, N = 84) from the Department of Neurology, Xuanwu Hospital of Capital Medical University, Beijing, China. All participants underwent 18F-FDG PET scans. Using the SSM/PCA algorithm, ARGMPs were identified based on the combined cohort. Then, network expression scores were used to quantify ARGMPs, and the association of individual network scores with chronological age in the combined cohort was assessed. To validate the robustness of the ARGMPs, metabolic brain network analysis was conducted in the ADNI and Xuanwu cohorts. We separately assessed the correlation between individual network scores and chronological age in both independent cohorts. In addition, according to brain Aβ and the standardized network score, the entire ADNI cohort was divided into four stages (stage 0, Aβ-Network score-; stage 1, Aβ-Network score + ; stage 2, Aβ + Network score-; and stage 3, Aβ + Network score +), and the conversion rate of CN participants to cognitive impairment in each stage was calculated. Survival analysis was further conducted to compare the predictive effect of conversion risk among different stages. The procedures of this study are shown in Fig. 1.
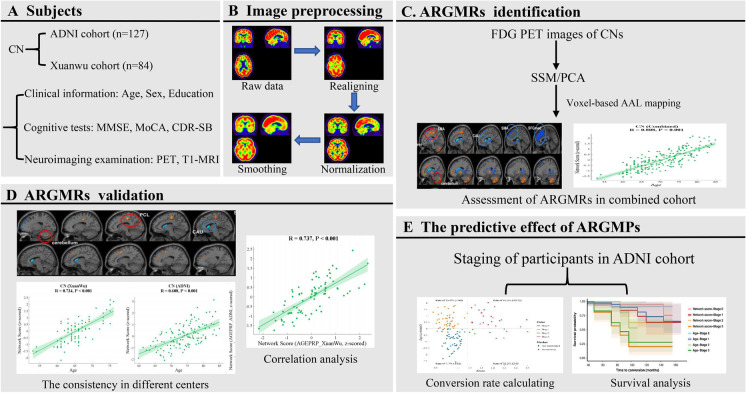
Fig. 1.
Whole procedure of the present study. (A) Clinical and neuroimaging date collected from two cohorts: the ADNI cohort and the Xuanwu cohort. (B) The image preprocessing procedure. (C) Extracting ARGMPs using the SSM/PCA method. (D) Cross-validations of ARGMPs in both the ADNI cohort and the Xuanwu cohort. (E) Assessing the predictive effect of ARGMPs
Participants
For all CN participants of the two cohorts, clinical (sex, age, education, and Mini-Mental State Examination (MMSE)) and neuroimaging (18F-FDG PET and T1-MRI image) data were collected. Each participant in the ADNI cohort underwent 18F-florbetapir (AV45) PET scans at baseline. Participants in the ADNI cohort completed the Montreal Cognitive Assessment (MoCA), and those in the Xuanwu cohort completed the MoCA-Basic Version (MoCA-B). In addition, the clinical severity was also evaluated using the Clinical Dementia Rating Sum of Boxes (CDR-SB) for participants in the ADNI cohort. The combination of the two cohorts was defined as the combined cohort. The inclusion criteria for CN participants were as follows: (1) no cognitive complaints; (2) no history of stroke, hypertension, central nervous system diseases, or mental illness; and (3) an MMSE score above or equal to 27.
Specifically, according to whether the cerebral-to-cerebellar AV45 standardized uptake value ratio (SUVR) was below 1.18 [24], CN participants in the ADNI cohort were classified into Aβ-negative CN (CN-, N = 96) and Aβ-positive CN (CN + , N = 31) groups. Two experienced neuroradiologists at Xuanwu Hospital, who were blinded to the classification, also visually evaluated the amyloid-PET images. The detailed selection process for participants in the ADNI cohort is shown in Supplementary Fig. S1. However, due to the lack of 18F-florbetapir (AV45) PET scans, the CN participants from the Xuanwu cohort were not divided into CN- and CN + subgroups.
Participants with CDR-SB scores ≥ 1 at the follow-up visit were considered as the transformers. Two experienced neurologists (CS and YH) evaluated the clinical diagnosis of conversion to cognitive impairment. In our study, during the follow-up (follow-up range: 6 ~ 168 months, mean follow-up period: 80.60 months), 23 participants in the ADNI cohort developed cognitive impairment (conversion rate: 18.11%).
This study was approved by the Institutional Review Board of the ADNI and the Research Ethics Committee of Xuanwu Hospital, Beijing, China (ClinicalTrials.gov Identifier: NCT03370744). All participants voluntarily participated in this study and provided written informed consent.
Image acquisition protocol
In the ADNI cohort, all participants were invited to undergo optional 18F-FDG PET and 18F-AV45 PET scans in three-dimensional acquisition mode. The process of data acquisition in the ADNI cohort is described in detail in the imaging protocol column of the ADNI database (http://adni.loni.usc.edu/). In the Xuanwu cohort, PET and T1-MRI data were simultaneously obtained on an integrated 3.0 T time-of-flight (TOF) PET/MR scanner (SIGNA PET/MR, GE Healthcare, Milwaukee, Wisconsin, USA) for each participant. For FDG-PET, each participant was instructed to fast for at least 6 h and was required to have a confirmed serum glucose level below 8 mmol/L. The images were acquired approximately 40 min after an intravenous injection of 3.7 MBq/kg of 18F-FDG, and the data were recorded by using a TOF ordered subset expectation maximization algorithm with the following parameters: scan duration = 35 min, eight iterations, 32 subsets matrix = 192 × 192, field of view (FOV) = 350 × 350, and half-width height = 3. Three-dimensional T1-weighted magnetization-prepared rapid gradient echo scans were performed with the following parameters: spoiled gradient-recalled echo (SPGR) sequence, FOV = 256 × 256 mm2, matrix = 256 × 256, slice thickness = 1 mm, gap = 0, slice number = 192, repetition time (TR) = 6.9 ms, echo time (TE) = 2.98 ms, inversion time (TI) = 450 ms, flip angle = 12°, and voxel size = 1 × 1 × 1 mm3.
Image preprocessing
All PET images and T1-MRI images were preprocessed using statistical parametric mapping software (SPM12; https://www.fil.ion.ucl.ac.uk/spm/software/spm12/) in MATLAB (Version R2014a; MathWorks, Natick, MA, USA). We first used the realigning method to ensure that all frames in the dynamic PET scans were motion-corrected to the first frame, processed the output single average functional image, and reduced system or head motion errors. Then, the average functional image was normalized to the standard Montreal Neurological Institute (MNI) brain space using the deformation field from the MRI image to the MNI space. Finally, normalized functional images were smoothed to reduce noise and improve image quality using an isotropic Gaussian smoothing kernel with a Gaussian filter of 8 mm full-width at half-maximum.
SSM/PCA
To explore the effect of ageing on glucose metabolism in the brains of CN participants, metabolic brain network analysis was conducted on their FDG PET images to identify ARGMPs using the SSM/PCA voxel-based spatial covariance mapping algorithm. The SSM/PCA method was implemented using ScAnVp (Scan Analysis and Visualization Processor) software, version 7.0w (available at http://www.feinsteinneuroscience.org at the Centre for Neuroscience, the Feinstein Institute for Medical Research, Manhasset, NY). FDG PET images were first multiplied using a binary mask (determined by the standardized automated anatomical labelling (AAL) template) to eliminate artefacts associated with noise and areas unrelated to brain activity, including the ventricle and white matter, from subsequent analysis. Then, the SSM/PCA method was used to derive the group invariant subprofile (GIS) and the participant score of each principal component (PC) in the spatial covariance model of the combined CN group. Specifically, the logarithmically transformed training set image data matrix is double centred to obtain the subject residual profile (SPR), which reveals a covariance structure that has biological significance for the group. The SPR is expressed as follows:
where is the row mean of the log values and GMP (group mean profile) is the mean row image. Next, PCA is applied to the SRP covariance matrix to derive orthogonal eigenvectors, associated eigenvalues, and participant scores to obtain subject scaling factors (SSFs) and GIS using Eq. 2.
We selected the series of PCs with the maximal correlation to chronological age in the multiple linear regression analysis, with the participant scores of the first series of PCs (cumulative variance accounted for 60%) and age as the independent and dependent variables, respectively. ARGMPs were identified based on the linear combination of the regression coefficients and GISs of these selected PCs. The stability of the ARGMPs is based on a bootstrapping scheme. The voxels were obtained by bootstrapping 1000 times on ARGMPs at p < 0.001 with an absolute weight threshold of 1.4 and a cluster threshold of 100 voxels (800 mm3). They were then mapped to the AAL atlas to determine anatomical brain regions that exhibited age-related changes in glucose metabolism.
Using the voxel-based topographic profile rating algorithm, network expression scores were used to quantify ARGMPs in all participants. The correlation between network scores and chronological age of CN participants in the combined cohort was used to assess the accuracy and reliability of the ARGMPs. To validate the robustness of the ARGMPs, metabolic brain network analysis was also conducted in the ADNI and Xuanwu cohorts. In addition, to validate the independence of ARGMPs in the normal ageing process and AD pathological process, the correlation between network scores and chronological age was assessed separately in the CN- and CN + subgroups of the ADNI cohort.
Staging the transformers in the ADNI cohort
All CN participants in the ADNI cohort underwent amyloid-PET scans. The Aβ positivity threshold was calculated as an SUVR of 1.18. Then, we calculated the standardized chronological age and standardized network score, with a score of 0 as the threshold. According to whether the Aβ was greater than 1.18 and the standardized chronological age was greater than 0, the entire ADNI cohort was divided as follows: stage 0, Aβ-Age-; stage 1, Aβ-Age + ; stage 2, Aβ + Age-; and stage 3, Aβ + Age + . According to whether the standardized network score was greater than 0, participants in the ADNI cohort were also classified into four stages: stage 0, Aβ-Network score-; stage 1, Aβ-Network score + ; stage 2, Aβ + Network score-; and stage 3, Aβ + Network score + . In each stage, the conversion rate was calculated. The standardized values (z scores) of individual chronological age and network score were calculated using the following formula:
where Mage denotes mean chronological age, SDage denotes the standard deviation of the chronological age, MNS denotes the mean network score, and SDNS denotes the standard deviation of the network score.
Statistical analysis
A two-sample t-test was performed to compare differences in continuous variables, and a Chi-square test was conducted to assess categorical variables. A multiple linear regression model was used, controlling for sex and years of education as covariates. Pearson’s correlation coefficient was then used to describe the correlation between the network scores and chronological age of the CN individuals in each cohort. To validate the independence of ARGMPs from AD-related pathology, we further evaluated the correlation between the network scores and chronological age in the CN- and CN + subgroups. The Gramm toolbox (available at https://github.com/piermorel/gramm) in MATLAB was used to plot and visualize all of the statistical data presented in this paper.
Survival was estimated using the Kaplan–Meier method, and any differences in survival were evaluated using a log-rank test in R version 3.6.3. To assess the validity of ARGMPs in predicting high conversion risk to cognitive impairment, we first compared the survival probability between the standardized network score > 0 (Network score +) group and the standardized network score < 0 (Network score-) group. Next, the survival probability between the standardized chronological age > 0 (Age +) group and standardized chronological age < 0 (Age-) group was also compared. Moreover, four subgroups, the Aβ-Network score-, Aβ-Network score + , Aβ + Network score-, and Aβ + Network score + , were compared to evaluate the risk of conversion. Hazard ratios were used to indicate the risk of conversion among different groups. The p-value was calculated using the log-rank test. P < 0.05 was considered statistically significant.
Results
Demographic characteristics of participants at baseline
There were a total of 211 CN participants (age range 53.0 ~ 84.8 years) in the present study, including 127 CN participants (age range 59.8 ~ 84.8 years) from the ADNI cohort and 84 CN participants (age range 53.0 ~ 76.0 years) from the Xuanwu cohort. Table 1 shows the detailed demographic and clinical details of all participants at baseline. Significant differences were observed in age (p < 0.001), sex (p < 0. 001), and educational years (p < 0.001) between the ADNI and Xuanwu CN cohorts, while no significant differences in the MMSE (p = 0.933) or MoCA (p = 0.061) were observed. In addition, the CN + subgroup was older than the CN- subgroup (p < 0.001). The follow-up period of the ADNI cohort was 80.60 ± 36.2 months, with a total of 23 participants (18.11%) converting to cognitive impairment.
Table 1.
Demographic and clinical characteristics of CN participants at baseline
| The combined cohort | ADNI cohort | Xuanwu cohort | |||
|---|---|---|---|---|---|
| Total | CN- | CN + | |||
| N | 211 | 127 | 96 | 31 | 84 |
| Age (y) | 70.5 ± 6.6 | 73.9 ± 5.4 | 72.88 ± 5.5 | 76.88 ± 4.1## | 65.3 ± 4.7** |
| Sex (F/M) | 134/77 | 68/59 | 47/49 | 21/10 | 66/18** |
| Education (y) | 15.1 ± 3.2 | 16.5 ± 2.6 | 16.60 ± 2.6 | 16.00 ± 2.5 | 13.1 ± 2.9** |
| MMSE | 29.3 ± 0.8 | 29.3 ± 0.8 | 29.32 ± 0.8 | 29.16 ± 1.0 | 29.3 ± 0.8 |
| CDR-SB | / | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | / |
| MoCA/MoCA-Ba | 26.2 ± 2.1 | 26.0 ± 2.3 | 26.12 ± 2.3 | 25.67 ± 2.1 | 26.6 ± 1.9 |
| Follow-up (m) | / | 80.60 ± 36.2 | 84.06 ± 35.9 | 69.87 ± 34.9 | / |
| Conversion rate | / | 23 (18.11%) | 11 (11.46%) | 12 (38.71%) | / |
Abbreviations: MMSE, Mini-Mental State Examination; MoCA, Montreal Cognitive Assessment; MoCA-B, Montreal Cognitive Assessment-Basic Version; CDR-SB, Clinical Dementia Rating Sum of Boxes; CN, cognitively normal; CN-, Aβ negative cognitively normal; CN + , Aβ positive cognitively normal; y, year; F, female; M, male; m, months; a, MoCA used in the ADNI cohort, and MoCA-B used in the Xuanwu cohort
* means p < 0.05 compared to the CN group in the ADNI cohort
** means p < 0.001 compared to the CN group in the ADNI cohort
## means p < 0.001 compared to the CN- group in the ADNI cohort
Supplementary Table S1 shows the demographic and clinical information for transformers (n = 23) and nontransformers (n = 104) in the ADNI cohort at baseline. There were significant differences in age (p = 0.008) and MMSE scores (p = 0.040) between transformers and nontransformers, while no differences in sex, education, or MoCA scores were observed.
SSM/PCA based on the combined cohort
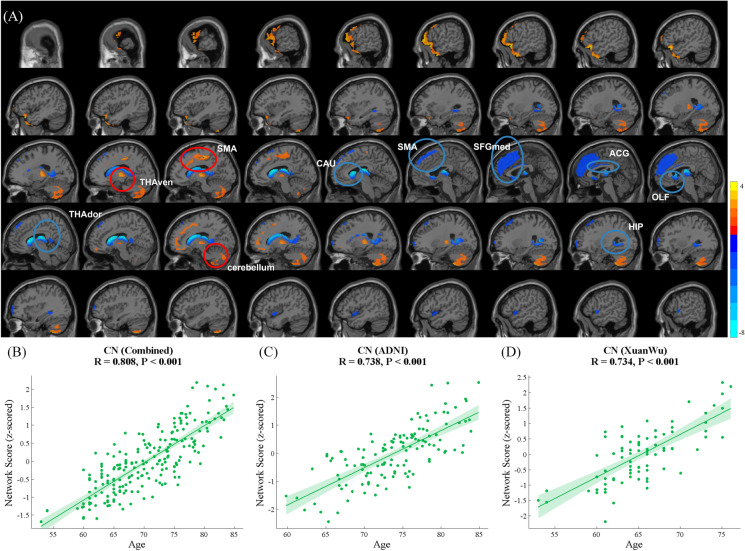
Network analysis for CN participants in the combined cohort evaluated the first eight PCs, including 51.5% participant × voxel variance. ARGMPs were identified by linear combination with the first six PCs (variance accounted for 22.9%, 9.2%, 6.0%, 3.9%, 3.2%, and 2.4%, and cumulative variance accounted for 49.7%) whose participant scores maximally correlated with chronological age. Brain regions that exhibited reduced glucose metabolism with ageing were primarily distributed in the left supplementary motor area (SMA.L), bilateral olfactory cortex (OLF), medial superior frontal gyrus (SFGmed), anterior cingulate and paracingulate gyri (ACG), caudate nucleus (CAU), and dorsal thalamus (THAdor). Brain regions manifesting increased glucose metabolism with ageing included the bilateral ventral thalamus (THAven) and part of the cerebellum (Cerebellum_Crus II) (Fig. 2A, Table 2). There were significant positive correlations between network scores and chronological age for the CN participants in the combined cohort (R = 0.808, p < 0.001, Fig. 2B), ADNI cohort (R = 0.785, p < 0.001, Fig. 2C), and Xuanwu cohort (R = 0.668, p < 0.001, Fig. 2D).
Fig. 2.
ARGMPs in CN adults in the combined cohort during normal ageing. The red areas indicate a significant increase in glucose metabolism with ageing, and the blue areas indicate a significant decrease (A). Scatter plot and fit results, with 95% confidence limits, between network scores and CN participant age in the combined cohort (B), ADNI cohort (C), and Xuanwu cohort (D); SMA, supplementary motor area; OLF, olfactory cortex; SFGmed, medial superior frontal gyrus; ACG, anterior cingulate and paracingulate gyri; CAU, caudate nucleus; THAdor, dorsal thalamus; THAven, bilateral ventral thalamus and part of Cerebellum_Crus II
Table 2.
Brain regions loading by the voxels survived from 1000 bootstrapping on ARGMP at p < 0.001, with absolute weight threshold of 1.4 and cluster threshold of 100 voxels (800 mm3)
| Brain region | AAL number | Laterality | MNI Coordinates | |||
|---|---|---|---|---|---|---|
| X | Y | Z | ||||
| Decreased Metabolism | ||||||
| Supplementary motor area | Frontal | 19 | L | -5.3 | 4.8 | 61.4 |
| Olfactory cortex | Prefrontal | 21 | L | -8.1 | 15.1 | -11.5 |
| Olfactory cortex | Prefrontal | 22 | R | 10.4 | 15.9 | -11.3 |
| Superior frontal gyrus, medial | Prefrontal | 23 | L | -4.8 | 49.2 | 30.9 |
| Superior frontal gyrus, medial | Prefrontal | 24 | R | 9.1 | 50.8 | 30.2 |
| Anterior cingulate and paracingulate gyri | Prefrontal | 31 | L | -4.0 | 35.4 | 14.0 |
| Anterior cingulate and paracingulate gyri | Prefrontal | 32 | R | 8.5 | 37.0 | 15.8 |
| Caudate nucleus | Subcortical | 71 | L | -11.5 | 11.0 | 9.2 |
| Caudate nucleus | Subcortical | 72 | R | 14.8 | 12.1 | 9.4 |
| Dorsal thalamus | Subcortical | 77 | L | -10.8 | -17.6 | 8.0 |
| Dorsal thalamus | Subcortical | 78 | R | 13.0 | -17.6 | 8.1 |
| Increased Metabolism | ||||||
| Ventral thalamus | Subcortical | 77 | L | -10.8 | -17.6 | 8.0 |
| Ventral thalamus | Subcortical | 78 | R | 13.0 | -17.6 | 8.1 |
| Cerebellum_Crus II | Cerebellum | 101 | L | - | - | - |
| Cerebellum_Crus II | Cerebellum | 103 | L | - | - | - |
| Cerebellum_Crus II | Cerebellum | 104 | R | - | - | - |
The consistency of ARGMPs between different centres
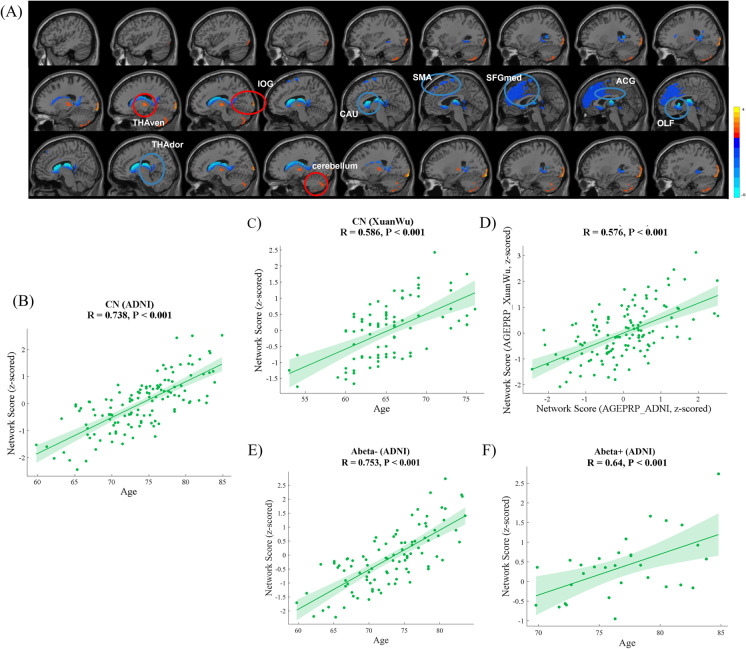
Network analysis for CN participants in the ADNI cohort evaluated the first six PCs, including 50.0% participant × voxel variance. ARGMPADNI was identified by linear combination with PC2, PC3, PC4, and PC5 (variance accounted for 9.0%, 4.8%, 4.4%, and 3.4%, respectively, and cumulative variance accounted for 21.6%), whose participant scores maximally correlated with chronological age. Brain regions that exhibited reduced glucose metabolism with ageing were distributed in the SMA.L, bilateral OLF, SFGmed, ACG, CAU, THAdor, and part of the vermis (Vermis_3). Brain regions that manifested increased glucose metabolism with ageing included the right inferior occipital gyrus (IOG.R), THAven, and part of the cerebellum (Cerebellum_Crus II) (Fig. 3A, Table 3). Significant correlations between the network scores on the ARGMPADNI and chronological age of CN participants were observed in both the ADNI (R = 0.738, p < 0.001, Fig. 3B) and Xuanwu cohorts (R = 0.586, p < 0.001, Fig. 3C). In addition, the network scores of the ARGMPADNI and the ARGMPXuanwu exhibited highly similar topographies (R = 0.576, p < 0.001, Fig. 3D).
Fig. 3.
ARGMPADNI in CN adults in the ADNI cohort during normal ageing. The red areas indicate a significant increase in glucose metabolism with ageing, and the blue areas indicate a significant decrease (A). Scatter plot and the fit results, with 95% confidence limits, between network scores and the age of the CN participants in the ADNI (B) and Xuanwu cohorts (C). Scatter plot and fit results with 95% confidence limits and between network scores on the ARGMPADNI and ARGMPXuanwu of the CN participants in the ADNI cohort (D). Correlations between the network scores and chronological age in the CN- (E) and CN + (F) subgroups in the ADNI cohort. SMA, supplementary motor area; OLF, olfactory cortex; SFGmed, medial superior frontal gyrus; ACG, anterior cingulate and paracingulate gyri; CAU, caudate nucleus; THAdor, dorsal thalamus; THAven, bilateral ventral thalamus; and part of Cerebellum_CrusII
Table 3.
Brain regions loading by the voxels survived from 1000 bootstrapping on ARGMPADNI at p < 0.001, with absolute weight threshold of 1.4 and cluster threshold of 100 voxels (800 mm3)
| Brain region | AAL number | Laterality | MNI Coordinates | |||
|---|---|---|---|---|---|---|
| X | Y | Z | ||||
| Decreased Metabolism | ||||||
| Supplementary motor area | Frontal | 19 | L | -5.3 | 4.8 | 61.4 |
| Olfactory cortex | Prefrontal | 21 | L | -8.1 | 15.1 | -11.5 |
| Olfactory cortex | Prefrontal | 22 | R | 10.4 | 15.9 | -11.3 |
| Superior frontal gyrus, medial | Prefrontal | 23 | L | -4.8 | 49.2 | 30.9 |
| Superior frontal gyrus, medial | Prefrontal | 24 | R | 9.1 | 50.8 | 30.2 |
| Anterior cingulate and paracingulate gyri | Prefrontal | 31 | L | -4.0 | 35.4 | 14.0 |
| Anterior cingulate and paracingulate gyri | Prefrontal | 32 | R | 8.5 | 37.0 | 15.8 |
| Caudate nucleus | Subcortical | 71 | L | -11.5 | 11.0 | 9.2 |
| Caudate nucleus | Subcortical | 72 | R | 14.8 | 12.1 | 9.4 |
| Dorsal thalamus | Subcortical | 77 | L | -10.8 | -17.6 | 8.0 |
| Dorsal thalamus | Subcortical | 78 | R | 13.0 | -17.6 | 8.1 |
| Vermis | Vermis | 110 | R | - | - | - |
| Increased Metabolism | ||||||
| Inferior occipital gyrus | Occipital | 54 | R | 38.2 | -82 | -7.6 |
| Ventral thalamus | Subcortical | 77 | L | -10.8 | -17.6 | 8.0 |
| Ventral thalamus | Subcortical | 78 | R | 13.0 | -17.6 | 8.1 |
| Cerebellum_Crus II | Cerebellum | 101 | L | - | - | - |
| Cerebellum_Crus II | Cerebellum | 103 | L | - | - | - |
| Cerebellum_Crus II | Cerebellum | 104 | R | - | - | - |
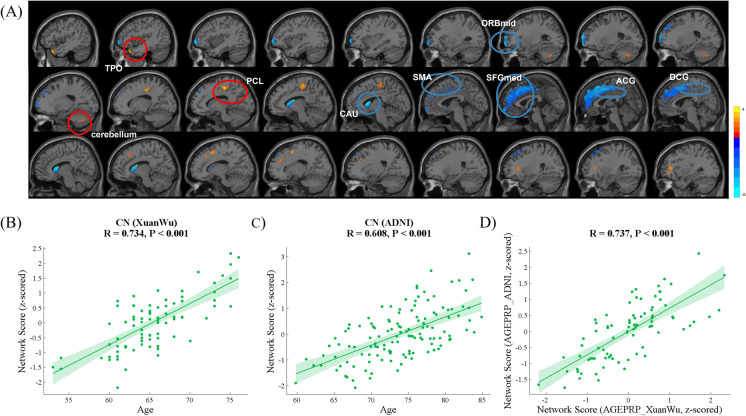
Network analysis for the CN participants in the Xuanwu cohort included the evaluation of the first nine PCs, including 50.1% participant × voxel variance. ARGMPXuanwu was identified by the linear combination with PC1, PC2, PC3, PC6, and PC7 (variance accounted for 23.9%, 6.2%, 4.9%, 2.4%, and 2.3%, respectively, and cumulative variance accounted for 39.7%), whose participant scores maximally correlated with chronological age. Brain regions characterized by reduced glucose metabolism with ageing included the left orbital part of the middle frontal gyrus (ORBmid.L), SMA.L, left medial orbital of the superior frontal gyrus (ORBsupmed.L), bilateral SFGmed, ACG, median cingulate and paracingulate gyri (DCG), and CAU. Brain regions displaying increased glucose metabolism with ageing were distributed in the left paracentral lobule (PCL.L), left temporal pole: superior and middle temporal gyrus (TPOsup.L, TPOmid.L), and part of the left inferior cerebellum (Fig. 4A, Table 4). Significant correlations were observed between the ARGMPXuanwu network scores and the chronological age of the CN participants in both the Xuanwu (R = 0.734, p < 0.001, Fig. 4B) and ADNI cohorts (R = 0.608, p < 0.001, Fig. 4C). The correlation between network scores of the ARGMPADNI and the ARGMPXuanwu was significantly positive (R = 0.737, p < 0.001, Fig. 4D).
Fig. 4.
ARGMPXuanwu in CN adults in the Xuanwu cohort during normal ageing. The red areas indicate a significant increase in glucose metabolism associated with advancing age, and the blue areas indicate a significant decrease (A). Scatter plot and fit results, with 95% confidence limits, between network scores and the age of the CN participants in the Xuanwu cohort (B) and ADNI cohort (C). Scatter plot and the fit results, with 95% confidence limits and between network scores in the ARGMPXuanwu and ARGMPADNI of the CN participants in the ADNI cohort (D). SMA, supplementary motor area; SFGmed, medial superior frontal gyrus; ACG, anterior cingulate and paracingulate gyri; CAU, caudate nucleus; DCG, median cingulate and paracingulate gyri; ORBmid, orbital part of the middle frontal gyrus; TPO, temporal pole and part of Cerebellum_Crus II
Table 4.
Brain regions loading by the voxels survived from 1000 bootstrapping on ARGMPXuanwu at P < 0.001, with absolute weight threshold of 1.4 and cluster threshold of 100 voxels (800 mm3)
| Brain region | AAL number | Laterality | MNI Coordinates | |||
|---|---|---|---|---|---|---|
| X | Y | Z | ||||
| Decreased Metabolism | ||||||
| Middle frontal gyrus, orbital part | Prefrontal | 9 | L | -30.6 | 50.4 | -9.6 |
| Supplementary motor area | Frontal | 19 | L | -5.3 | 4.8 | 61.4 |
| Superior frontal gyrus, medial | Prefrontal | 23 | L | -4.8 | 49.2 | 30.9 |
| Superior frontal gyrus, medial | Prefrontal | 24 | R | 9.1 | 50.8 | 30.2 |
| Superior frontal gyrus, medial orbital | Prefrontal | 25 | L | -5.2 | 54.1 | -7.4 |
| Anterior cingulate and paracingulate gyri | Prefrontal | 31 | L | -4.0 | 35.4 | 14.0 |
| Anterior cingulate and paracingulate gyri | Prefrontal | 32 | R | 8.5 | 37.0 | 15.8 |
| Median cingulate and paracingulate gyri | Frontal | 33 | L | -5.5 | -14.9 | 41.6 |
| Median cingulate and paracingulate gyri | Frontal | 34 | R | 8.0 | -8.8 | 39.8 |
| Caudate nucleus | Subcortical | 71 | L | -11.5 | 11.0 | 9.2 |
| Caudate nucleus | Subcortical | 72 | R | 14.8 | 12.1 | 9.4 |
| Increased Metabolism | ||||||
| Paracentral lobule | Parietal | 69 | L | -7.6 | -25.4 | 70.1 |
| Temporal pole: superior temporal gyrus | Temporal | 83 | L | -39.9 | 15.1 | -20.2 |
| Temporal pole: middle temporal gyrus | Temporal | 87 | L | -36.3 | 14.6 | -34.1 |
| Cerebellum_Crus II | Cerebellum | 103 | L | - | - | - |
The correlation between network scores and chronological age in CN subgroups
Network analysis for the CN- and CN + subgroups in the ADNI cohort was also performed. For CN- participants, there was a significantly positive correlation between network scores and chronological age (R = 0.753, p < 0.001, Fig. 3E). Meanwhile, a significant association between network scores and chronological age was also observed in CN + participants (R = 0.640, p < 0.001, Fig. 3F).
The predictive effect of ARGMPs on the development of cognitive impairment at follow-up
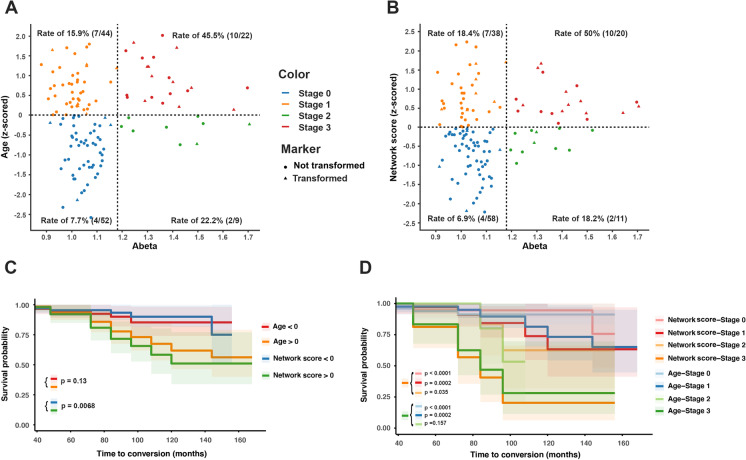
As shown in Fig. 5A, conversion stages (stage 0 ~ stage 3) were defined by thresholds of the Aβ and standardized chronological age. The rates of conversion in the four stages were 7.7%, 15.9%, 22.2%, and 45.5%, respectively. In Fig. 5B, four stages were classified by thresholds of the Aβ and standardized network score. The results showed that the rates of conversion in the four stages were 6.9%, 18.4%, 18.2%, and 50%. The conversion rate in stage 1 was lower in Aβ-age + individuals (Fig. 5A) than in Aβ-Network scores + individuals (Fig. 5B) (15.9% vs. 18.4%), while the conversion rate in stage 2 is higher in Fig. 5A than in Fig. 5B (22.2% vs. 18.2%). In stage 3, participants with Aβ + Network scores + exhibited a higher conversion rate than those with Aβ + Age + in the prediction models (45.5% vs. 50%).
Fig. 5.
Staging the transformers and survival analysis estimated by the Kaplan–Meier method. According to the threshold of Aβ (1.18) and the standardized chronological age (0), CN participants in the ADNI cohort were classified into four stages (stage 0 to 3). In each stage, the conversion rate of CN participants to cognitive impairment was calculated (A). According to the threshold of Aβ (1.18) and the standardized network score (0), CN participants in the ADNI cohort were also classified into four stages (stage 0 to 3). In each stage, the conversion rate of CN participants to cognitive impairment was calculated (B). In the survival analysis, we compared different predictive effects of conversion risk between higher network scores (standardized network score > 0, green line) and lower network scores (standardized network score > 0, blue line) (C) and between the older group (standardized chronological age > 0, orange line) and the younger group (standardized chronological age < 0, red line) (C). Furthermore, we compared different predictive effects of conversion risk among the standardized network score subgroup and the standardized chronological age subgroup: the Aβ + Network score + (stage 3), Aβ-Network score- (stage 0), Aβ-Network score + (stage 1), and Aβ + Network score- (stage 2); the Aβ + Age + (stage 3), Aβ-Age- (stage 0), Aβ-Age + (stage 1), and Aβ + Age- (stage 2) (D)
In the survival analysis, higher network scores (standardized network score > 0, green line) indicated a better predictive effect for developing a clinical diagnosis of cognitive impairment than lower network scores (standardized network score < 0, blue line) (HR: 0.30, 95% CI: 0.1340 ~ 0.6904, p = 0.0068, Fig. 5C). However, there was no significant difference between the older group (standardized chronological age > 0, orange line) and the younger group (standardized chronological age < 0, red line) (HR: 0.50, 95% CI: 0.2193 ~ 1.154, p = 0.1250, Fig. 5C). Furthermore, the predictive effect of the conversion risk to cognitive impairment in the Aβ + Network score + subgroup compared to that in the other three subgroups was as follows (Fig. 5D): the Aβ-Network score- subgroup (HR: 0.10, 95% CI: 0.0270 ~ 0.3704, p < 0.0001), Aβ-Network score + subgroup (HR: 0.21, 95% CI: 0.0668 ~ 0.6429, p = 0.0002), and Aβ + Network score- subgroups (HR: 0.25, 95% CI: 0.0810 ~ 0.7906, p = 0.035). However, compared with the other three subgroups, the predictive effect of the conversion risk in the Aβ + Age + subgroup was as follows (Fig. 5D): the Aβ-Age- subgroup (HR: 0.14, 95% CI: 0.0428 ~ 0.4650, p < 0.0001), Aβ-Age + subgroup (HR: 0.20, 95% CI: 0.0628 ~ 0.6227, p = 0.0002), and Aβ + Age- subgroups (HR: 0.38, 95% CI: 0.1162 ~ 1.260, p = 0.157).
Discussion
This study proposes ARGMPs as a novel index to characterize individual brain ageing. The primary findings were that (1) ARGMPs appear to be a stable and sensitive index for characterizing brain ageing; (2) this index is independent of AD pathological biomarkers; and (3) ARGMPs display better potential for predicting the risk of cognitive impairment than chronological age in CN people.
Compared to chronological age, the novel index appeared to be more reasonable in assessing normal brain ageing. Using the SSM/PCA method, brain ageing-specific spatial metabolic profiles were generated from linear combinations of several principal components [25]. This multivariate analysis method reduces the interindividual variations in FDG-PET images and provides complementary, clinically relevant information that may be superior to univariate measures [26]. In our study, ARGMPs derived from the ADNI and Xuanwu cohorts exhibited similar topographies, validating the robustness and repeatability of our findings. According to the biological definition of AD proposed in 2018 [27], individuals with evidence of Aβ deposition are categorized into the Alzheimer’s continuum. Thus, to eliminate the latent influence of AD pathology, we further analysed ARGMPs from the CN- and CN + subgroups. Notably, significantly positive correlations between network score and chronological age were also observed in these two subgroups. Our findings validated that the novel index was independent of AD pathological biomarkers. Moreover, given the potential impact of cognitive reserve on age-related brain changes [28–30], other risk factors (e.g. educational level, sex) were also regressed as covariates in our study, which further ensured that the novel index was stable.
The novel index revealed that the overlapping brain regions with age-related hypometabolism focused on the bilateral SFGmed, ACG, CAU, and left SMA based on the three cohorts (the combined cohort, the ADNI cohort, and the Xuanwu cohort), whereas age-related hypermetabolism was primarily distributed in part of the left Cerebellum_Crus II. Thus, age-related regional hypometabolism was primarily observed in the frontal lobe and striatum, suggesting that these regions may be more sensitive to normal brain ageing. The involvement of the prefrontal cortex (e.g. SFGmed, ACG) in normal brain ageing has been supported by previous studies [7, 31, 32]. In contrast with AD, the prefrontal cortex is regarded as the first structure affected by normal ageing. For instance, Moeller [33] provided evidence of decreased glucose metabolism in the medial frontal region of 130 healthy volunteers aged 21–90 years. Age-related differences in working memory load-related activity have also been observed in the SFGmed [32]. The prefrontal cortex has multiple physiological functions, especially in working memory, attention, and mood regulation [31, 34, 35]. The decline in glucose metabolism in the prefrontal cortex with increasing age may be attributed to several potential mechanisms, such as decreased neuronal function [36] and altered synaptic efficiency [7].
Our study also demonstrated decreased glucose metabolism in the left SMA with normal brain ageing. The SMA, which is a critical component of the cortical motor network, plays a major role in planning and executing rhythmic bilateral movements [37]. Growing evidence indicates that age-related decline in motor control is due to special brain structural and functional changes, such as grey matter atrophy in the pre- and postcentral gyri [38], reduced white matter integrity in the corpus callosum [39], abnormal functional connectivity, or hypometabolism within the cortical motor network. Current studies have reported decreased cerebral glucose metabolism in the left SMA in healthy older adults [40] and reduced SMA-primary motor cortex (M1) interactions [41] with ageing. In contrast with normal brain ageing, AD pathology initially affects the entorhinal cortex, while motor areas are generally damaged during the late stage. Therefore, age-related hypometabolism in the SMA may represent a distinct biomarker for identifying normal brain ageing. In our study, the subcortical CAU was another region manifesting significant metabolic decline with ageing. The CAU, a key component of the striatum, is associated with navigational skills [42, 43]. Previous work has revealed age-related reductions in perfusion in the CAU [44, 45].
Finally, we investigated the ability of ARGMPs to predict the conversion risk of CN individuals to cognitive impairment. Regional hypometabolism in AD patients has been reported in many previous studies and may represent a predictor of cognitive decline from mild cognitive impairment (MCI) to AD dementia [46, 47]. Decreased glucose metabolism is considered a biomarker of neurodegeneration, which is associated with the clinical progression of AD [48]. In this study, participants with evidence of Aβ deposition and high network scores exhibited higher conversion rates than other three subgroups, indicating that the combination of Aβ burden and ARGMPs has the potential to identify individuals at risk of developing cognitive impairment. We also assessed the prediction effect of chronological age. To the best of our knowledge, the prevalence of AD and the accumulation of Aβ increase with ageing [49]. We also found that older people with Aβ positivity evidence (stage 3) presented a relatively higher conversion rate than those at other stages, consistent with previous studies [50]. However, compared to chronological age, individuals with higher network scores had a higher risk of conversion than those with lower network scores.
It should be noted that this study has some limitations. First, age differences existed between the ADNI and Xuanwu cohorts, with the average age of the participants from Xuanwu Hospital being 8.6 years younger than those in the ADNI cohort. Correlations between ARGMP expression network scores and chronological age were identified in both cohorts, but whether this correlation exists in CN populations with wider age ranges needs to be validated in the future. Second, the underlying mechanism of the age-related metabolic pattern in older CN adults was not elucidated in the present study. Future studies are needed to investigate the development of ARGMPs through molecular pathways, such as neurotransmitter and endocrine signalling, etc. Third, the effect of other factors, especially biological sex, on the correlation between brain regional glucose metabolism dysfunction and chronological ageing was not considered in this study. Notably, sex is a key risk factor that contributes to variability in AD manifestation [51]. Previous studies have reported that females are at greater risk of developing AD, as well as exhibit more pathological changes [52–54]. However, the effect of biological sex on brain glucose metabolism remains unclear. Thus, the impact of sex differences on ARGMPs needs to be evaluated in future studies. Finally, the sample size in this study was relatively small, and only data in the ADNI cohort were longitudinal. Future multicentre studies with a larger sample size are essential to provide more accurate evidence. In addition, the similarities and differences between AD-related ageing and chronological ageing patterns warrant further study.
Conclusions
This study revealed ARGMP topographies in the spatial covariance brain network and the distribution of regional metabolism alterations that occur during normal brain ageing using 18F-FDG PET images. Cross-validation between the ADNI and Xuanwu cohorts confirmed highly reproducible ARGMPs across different CN populations and imaging instrumentation/protocols. The expression score of ARGMPs in individual participants objectively mirrored normal brain ageing, such as chronological age, and better predicted the conversion of CN participants to cognitive impairment. This study indicates that the glucose metabolism pattern in CN older adults may represent an effective biomarker of normal brain ageing and has the potential for future application in routine clinical practice.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Data collection and dissemination for this project were funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI): the National Institutes of Health (grant number U01 AG024904), and the Department of Defense (award numberW81XWH-12-2-0012). ADNI is funded by the National Institute of Aging and the National Institute of Biomedical Imaging and Bioengineering as well as through generous contributions from the following organizations: AbbVie, Alzheimer’s Association, Alzheimer’s Drug Discovery Foundation, Araclon Biotech, BioClinica Inc., Biogen, Bristol-Myers Squibb Company, CereSpir Inc., Eisai Inc., Elan Pharmaceuticals Inc., Eli Lilly and Company, EuroImmun, F. Hoffmann-La Roche Ltd. and its affiliated company Genentech Inc., Fujirebio, GE Healthcare, IXICO Ltd., Janssen Alzheimer Immunotherapy Research & Development LLC., Johnson & Johnson Pharmaceutical Research &Development LLC., Lumosity, Lundbeck, Merck & Co. Inc., Meso Scale Diagnostics LLC., NeuroRx Research, Neurotrack Technologies, Novartis Pharmaceuticals Corporation, Pfizer Inc., Piramal Imaging, Servier, Takeda Pharmaceutical Company, and Transition Therapeutics. The Canadian Institutes of Health Research are providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego, CA, USA. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California, CA, USA.
Michael W. Weiner7, Paul Aisen8, Ronald Petersen9, Clifford R. Jack9, William Jagust10, John Q. Trojanowski11, Arthur W. Toga12, Laurel Beckett13, Robert C. Green14, Andrew J. Saykin15, John Morris16, Leslie M. Shaw11, Zaven Khachaturian13,17, Greg Sorensen18, Lew Kuller19, Marcus Raichle16, Steven Paul20, Peter Davies21, Howard Fillit22, Franz Hefti23, David Holtzman16, Marek M. Mesulam24, William Potter25, Peter Snyder26, Adam Schwartz27, Tom Montine28, Ronald G. Thomas28, Michael Donohue28, Sarah Walter28, Devon Gessert28, Tamie Sather28, Gus Jiminez28, Danielle Harvey13, Matthew Bernstein9, Paul Thompson29, Norbert Schuff7,13, Bret Borowski9, Jeff Gunter9, Matt Senjem9, Prashanthi Vemuri9, David Jones9, Kejal Kantarci9, Chad Ward9, Robert A. Koeppe30, Norm Foster31, Eric M. Reiman32, Kewei Chen32, Chet Mathis19, Susan Landau10, Nigel J. Cairns16, Erin Householder16, Lisa Taylor-Reinwald16, Virginia Lee11, Magdalena Korecka11, Michal Figurski11, Karen Crawford12, Scott Neu12, Tatiana M. Foroud15, Steven G. Potkin33, Li Shen15, Kelley Faber15, Sungeun Kim15, Kwangsik Nho15, Leon Thal8, Neil Buckholtz34, Marylyn Albert35, Richard Frank36, John Hsiao34, Jeffrey Kaye37, Joseph Quinn37, Betty Lind37, Raina Carter37, Sara Dolen37, Lon S. Schneider12, Sonia Pawluczyk12, Mauricio Beccera12, Liberty Teodoro12, Bryan M. Spann12, James Brewer8, Helen Vanderswag8, Adam Fleisher8,32, Judith L. Heidebrink30, Joanne L. Lord30, Sara S. Mason9, Colleen S. Albers9, David Knopman9, Kris Johnson9, Rachelle S. Doody38, Javier Villanueva-Meyer38, Munir Chowdhury38, Susan Rountree38, Mimi Dang38, Yaakov Stern38, Lawrence S. Honig38, Karen L. Bell38, Beau Ances16, Maria Carroll16, Sue Leon16, Mark A. Mintun16, Stacy Schneider16, Angela Oliver16, Daniel Marson39, Randall Griffith39, David Clark39, David Geldmacher39, John Brockington39, Erik Roberson39, Hillel Grossman40, Effie Mitsis40, Leyla de Toledo-Morrell41, Raj C. Shah41, Ranjan Duara42, Daniel Varon42, Maria T. Greig42, Peggy Roberts42, Chiadi Onyike35, Daniel D’Agostino35, Stephanie Kielb35, James E. Galvin43, Brittany Cerbone43, Christina A. Michel43, Henry Rusinek43, Mony J. de Leon43, Lidia Glodzik43, Susan De Santi43, P Murali Doraiswamy44, Jeffrey R. Petrella44, Terence Z. Wong44, Steven E. Arnold11, Jason H. Karlawish11, David Wolk11, Charles D. Smith45, Greg Jicha45, Peter Hardy45, Partha Sinha45, Elizabeth Oates45, Gary Conrad45, Oscar L. Lopez19, MaryAnn Oakley19, Donna M. Simpson35, Anton P. Porsteinsson46, Bonnie S. Goldstein46, Kim Martin46, Kelly M. Makino46, M Saleem Ismail46, Connie Brand46, Ruth A. Mulnard33, Gaby Thai33, Catherine McAdams-Ortiz33, Kyle Womack47, Dana Mathews47, Mary Quiceno47, Ramon Diaz-Arrastia47, Richard King47, Myron Weiner47, Kristen Martin-Cook47, Michael DeVous47, Allan I Levey48, James J. Lah48, Janet S. Cellar48, Jeffrey M. Burns48, Heather S. Anderson49, Russell H. Swerdlow49, Liana Apostolova29, Kathleen Tingus29, Ellen Woo29, Daniel H.S. Silverman29, Po H. Lu29, George Bartzokis29, Neill R. Graff-Radford50, Francine Parfitt50, Tracy Kendall50, Heather Johnson50, Martin R. Farlow15, Ann Marie Hake15, Brandy R. Matthews15, Scott Herring15, Cynthia Hunt15, Christopher H. van Dyck51, Richard E. Carson51, Martha G. MacAvoy51, Howard Chertkow52, Howard Bergman52, Chris Hosein52, Ging-Yuek Robin Hsiung53, Howard Feldman53, Benita Mudge53, Michele Assaly53, Charles Bernick54, Donna Munic54, Andrew Kertesz55, John Rogers55, Dick Trost55, Diana Kerwin24, Kristine Lipowski24, Chuang-Kuo Wu24, Nancy Johnson24, Carl Sadowsky56, Walter Martinez56, Teresa Villena56, Raymond Scott Turner57, Kathleen Johnson57, Brigid Reynolds57, Reisa A. Sperling14, Keith A. Johnson14, Gad Marshall14, Meghan Frey14, Barton Lane14, Allyson Rosen14, Jared Tinklenberg14, Marwan N. Sabbagh58, Christine M. Belden58, Sandra A. Jacobson58, Sherye A. Sirrel58, Neil Kowall58, Ronald Killiany59, Andrew E. Budson59, Alexander Norbash59, Patricia Lynn Johnson59, Joanne Allard59, Alan Lerner61, Paula Ogrocki61, Leon Hudson61, Evan Fletcher13, Owen Carmichae23, John Olichney13, Charles DeCarli13, Smita Kittur62, Michael Borrie63, T-Y. Lee63, Rob Bartha63, Sterling Johnson64, Sanjay Asthana64, Cynthia M. Carlsson64, Adrian Preda29, Dana Nguyen29, Pierre Tariot31, Stephanie Reeder31, Vernice Bates65, Horacio Capote65, Michelle Rainka65, Douglas W. Scharre66, Maria Kataki66, Anahita Adeli66, Earl A. Zimmerman67, Dzintra Celmins67, Alice D. Brown67, Godfrey D. Pearlson68, Karen Blank68, Karen Anderson68, Robert B. Santulli69, Tamar J. Kitzmiller69, Eben S. Schwartz69, Kaycee M. Sink70, Jeff D. Williamson70, Pradeep Garg70, Franklin Watkins70, Brian R. Ott71, Henry Querfurth71, Geoffrey Tremont71, Stephen Salloway72, Paul Malloy72, Stephen Correia72, Howard J. Rosen7, Bruce L. Miller7, Jacobo Mintzer73, Kenneth Spicer73, David Bachman73, Stephen Pasternak55, Irina Rachinsky55, Dick Drost55, Nunzio Pomara74, Raymundo Hernando74, Antero Sarrael74, Susan K. Schultz75, Laura L. Boles Ponto75, Hyungsub Shim75, Karen Elizabeth Smith75, Norman Relkin20, Gloria Chaing20, Lisa Raudin17,20, Amanda Smith76, Kristin Fargher76, Balebail Ashok Raj76, Thomas Neylan7, Jordan Grafman24, Melissa Davis8, Rosemary Morrison8, Jacqueline Hayes7, Shannon Finley7, Karl Friedl77, Debra Fleischman41, Konstantinos Arfanakis41, Olga James44, Dino Massoglia73, J Jay Fruehling64, Sandra Harding64, Elaine R. Peskind28, Eric C. Petrie66, Gail Li66, Jerome A. Yesavage78, Joy L. Taylor78 & Ansgar J. Furst78
7UC San Francisco, San Francisco, CA 94143, USA. 8UC San Diego, San Diego, CA 92093, USA. 9Mayo Clinic, Rochester, NY 14603, USA. 10UC Berkeley, Berkeley, CA 94720, USA. 11UPenn, Philadelphia, PA 9104, USA. 12USC, Los Angeles, CA 90089, USA. 13UC Davis, Davis, CA 95616, USA. 14Brigham and Women’s Hospital/Harvard Medical School, Boston, MA 02115, USA. 15Indiana University, Bloomington, IN 47405, USA. 16Washington University in St Louis, St Louis, MI 63130, USA. 17Prevent Alzheimer’s Disease 2020, Rockville, MD 20850, USA. 18Siemens, Munich 80333, Germany. 19University of Pittsburgh, Pittsburgh, PA 15260, USA. 20Weill Cornell Medical College, Cornell University, New York City, NY 10065, USA. 21Albert Einstein College of Medicine of Yeshiva University, Bronx, NY 10461, USA. 22AD Drug Discovery Foundation, New York City, NY 10019, USA. 23Acumen Pharmaceuticals, Livermore, CA 94551, USA. 24Northwestern University, Evanston and Chicago, IL 60208, USA. 25National Institute of Mental Health, Rockville, MD 20852, USA. 26Brown University, Providence, RI 02912, USA. 27Eli Lilly, Indianapolis, IN 46225, USA. 28University of Washington, Seattle, WA 98195, USA. 29UCLA, Los Angeles, CA 90095, USA. 30University of Michigan, Ann Arbor, MI 48109, USA. 31University of Utah, Salt Lake City, UT 84112, USA. 32Banner Alzheimer’s Institute, Phoenix, AZ 85006, USA. 33UC Irvine, Irvine, CA 92697, USA. 34National Institute on Aging, Bethesda, MD 20892, USA. 35Johns Hopkins University, Baltimore, MD 21218, USA. 36Richard Frank Consulting, Washington, DC 20001, USA. 37Oregon Health and Science University, Portland, OR 97239, USA. 38Baylor College of Medicine, Houston, TX 77030, USA. 39University of Alabama, Birmingham, AL 35233, USA. 40Mount Sinai School of Medicine, New York City, NY 10029, USA. 41Rush University Medical Center, Chicago, IL 60612, USA. 42Wien Center, Miami, FL 33140, USA. 43New York University, New York City, NY 10003, USA. 44Duke University Medical Center, Durham, NC 27710, USA. 45University of Kentucky, Lexington, KY 0506, USA. 46University of Rochester Medical Center, Rochester, NY 14642, USA. 47University of Texas Southwestern Medical School, Dallas, TX 75390, USA. 48Emory University, Atlanta, GA 30322, USA. 49Medical Center, University of Kansas, Kansas City, KS 66103, USA. 50Mayo Clinic, Jacksonville, FL 32224, USA. 51Yale University School of Medicine, New Haven, CT 06510, USA. 52McGill University/Montreal-Jewish General Hospital, Montreal, QC H3T 1E2, Canada. 53University of British Columbia Clinic for AD & Related Disorders, Vancouver, BC V6T 1Z3, Canada. 54Cleveland Clinic Lou Ruvo Center for Brain Health, Las Vegas, NV 89106, USA. 55St Joseph’s Health Care, London, ON N6A 4V2, Canada. 56Premiere Research Institute, Palm Beach Neurology, Miami, FL 33407, USA. 57Georgetown University Medical Center, Washington, DC 20007, USA. 58Banner Sun Health Research Institute, Sun City, AZ 85351, USA. 59Boston University, Boston, MA 02215, USA. 60Howard University, Washington, DC 20059, USA. 61Case Western Reserve University, Cleveland, OH 20002, USA. 62Neurological Care of CNY, Liverpool, NY 13088, USA. 63Parkwood Hospital, London, ON N6C 0A7, Canada. 64University of Wisconsin, Madison, WI 53706, USA. 65Dent Neurologic Institute, Amherst, NY 14226, USA. 66Ohio State University, Columbus, OH 43210, USA. 67Albany Medical College, Albany, NY 12208, USA. 68Hartford Hospital, Olin Neuropsychiatry Research Center, Hartford, CT 06114, USA. 69Dartmouth- Hitchcock Medical Center, Lebanon, NH 03766, USA. 70Wake Forest University Health Sciences, Winston-Salem, NC 27157, USA. 71Rhode Island Hospital, Providence, RI 02903, USA. 72Butler Hospital, Providence, RI 02906, USA. 73Medical University South Carolina, Charleston, SC 29425, USA. 74Nathan Kline Institute, Orangeburg, NY 10962, USA. 75University of Iowa College of Medicine, Iowa City, IA 52242, USA. 76University of South Florida: USF Health Byrd Alzheimer’s Institute, Tampa, FL 33613, USA. 77Department of Defense, Arlington, VA 22350, USA. 78Stanford University, Stanford, CA 94305, USA
Author contributions
JJ and YH conceived of the study. CL and XJ performed the statistical analysis. CS, GC, SJ drafted the initial manuscript. XJ and LL drew the pictures. All authors contributed to revision and editing of the manuscript.
Funding
This study was supported by grants received from the National Natural Science Foundation of China (grant numbers 61633018, 82020108013, 61603236, 81830059, and 81801052); the National Key Research and Development Program of China (grant numbers 2016YFC1306300, 2018YFC1312000, and 2018YFC1707704); the 111 Project (grant number D20031); the Shanghai Municipal Science and Technology Major Project (grant number 2017SHZDZX01); and the Beijing Municipal Commission of Health and Family Planning (grant number PXM2020_026283_000002).
Availability of data and materials
All data are available upon request from the authors.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jiehui Jiang, Can Sheng, and Guanqun Chen contributed equally to this work.
Contributor Information
Jiehui Jiang, Email: jiangjiehui@shu.edu.cn.
Ying Han, Email: hanying@xwh.ccmu.edu.cn.
for the Alzheimer’s Disease Neuroimaging Initiative:
Michael W. Weiner, Paul Aisen, Ronald Petersen, Clifford R. Jack, William Jagust, John Q. Trojanowski, Arthur W. Toga, Laurel Beckett, Robert C. Green, Andrew J. Saykin, John Morris, Leslie M. Shaw, Zaven Khachaturian, Greg Sorensen, Lew Kuller, Marcus Raichle, Steven Paul, Peter Davies, Howard Fillit, Franz Hefti, David Holtzman, Marek M. Mesulam, William Potter, Peter Snyder, Adam Schwartz, Tom Montine, Ronald G. Thomas, Michael Donohue, Sarah Walter, Devon Gessert, Tamie Sather, Gus Jiminez, Danielle Harvey, Matthew Bernstein, Paul Thompson, Norbert Schuff, Bret Borowski, Jeff Gunter, Matt Senjem, Prashanthi Vemuri, David Jones, Kejal Kantarci, Chad Ward, Robert A. Koeppe, Norm Foster, Eric M. Reiman, Kewei Chen, Chet Mathis, Susan Landau, Nigel J. Cairns, Erin Householder, Lisa Taylor-Reinwald, Virginia Lee, Magdalena Korecka, Michal Figurski, Karen Crawford, Scott Neu, Tatiana M. Foroud, Steven G. Potkin, Li Shen, Kelley Faber, Sungeun Kim, Kwangsik Nho, Leon Thal, Neil Buckholtz, Marylyn Albert, Richard Frank, John Hsiao, Jeffrey Kaye, Joseph Quinn, Betty Lind, Raina Carter, Sara Dolen, Lon S. Schneider, Sonia Pawluczyk, Mauricio Beccera, Liberty Teodoro, Bryan M. Spann, James Brewer, Helen Vanderswag, Adam Fleisher, Judith L. Heidebrink, Joanne L. Lord, Sara S. Mason, Colleen S. Albers, David Knopman, Kris Johnson, Rachelle S. Doody, Javier Villanueva-Meyer, Munir Chowdhury, Susan Rountree, Mimi Dang, Yaakov Stern, Lawrence S. Honig, Karen L. Bell, Beau Ances, Maria Carroll, Sue Leon, Mark A. Mintun, Stacy Schneider, Angela Oliver, Daniel Marson, Randall Griffith, David Clark, David Geldmacher, John Brockington, Erik Roberson, Hillel Grossman, Effie Mitsis, Leyla de Toledo-Morrell, Raj C. Shah, Ranjan Duara, Daniel Varon, Maria T. Greig, Peggy Roberts, Chiadi Onyike, Daniel D’Agostino, Stephanie Kielb, James E. Galvin, Brittany Cerbone, Christina A. Michel, Henry Rusinek, Mony J. de Leon, Lidia Glodzik, Susan De Santi, P. Murali Doraiswamy, Jeffrey R. Petrella, Terence Z. Wong, Steven E. Arnold, Jason H. Karlawish, David Wolk, Charles D. Smith, Greg Jicha, Peter Hardy, Partha Sinha, Elizabeth Oates, Gary Conrad, Oscar L. Lopez, MaryAnn Oakley, Donna M. Simpson, Anton P. Porsteinsson, Bonnie S. Goldstein, Kim Martin, Kelly M. Makino, M. Saleem Ismail, Connie Brand, Ruth A. Mulnard, Gaby Thai, Catherine McAdams-Ortiz, Kyle Womack, Dana Mathews, Mary Quiceno, Ramon Diaz-Arrastia, Richard King, Myron Weiner, Kristen Martin-Cook, Michael DeVous, Allan I Levey, James J. Lah, Janet S. Cellar, Jeffrey M. Burns, Heather S. Anderson, Russell H. Swerdlow, Liana Apostolova, Kathleen Tingus, Ellen Woo, Daniel H. S. Silverman, Po H. Lu, George Bartzokis, Neill R. Graff-Radford, Francine Parfitt, Tracy Kendall, Heather Johnson, Martin R. Farlow, Ann Marie Hake, Brandy R. Matthews, Scott Herring, Cynthia Hunt, Christopher H. van Dyck, Richard E. Carson, Martha G. MacAvoy, Howard Chertkow, Howard Bergman, Chris Hosein, Ging-Yuek Robin Hsiung, Howard Feldman, Benita Mudge, Michele Assaly, Charles Bernick, Donna Munic, Andrew Kertesz, John Rogers, Dick Trost, Diana Kerwin, Kristine Lipowski, Chuang-Kuo Wu, Nancy Johnson, Carl Sadowsky, Walter Martinez, Teresa Villena, Raymond Scott Turner, Kathleen Johnson, Brigid Reynolds, Reisa A. Sperling, Keith A. Johnson, Gad Marshall, Meghan Frey, Barton Lane, Allyson Rosen, Jared Tinklenberg, Marwan N. Sabbagh, Christine M. Belden, Sandra A. Jacobson, Sherye A. Sirrel, Neil Kowall, Ronald Killiany, Andrew E. Budson, Alexander Norbash, Patricia Lynn Johnson, Joanne Allard, Alan Lerner, Paula Ogrocki, Leon Hudson, Evan Fletcher, Owen Carmichae, John Olichney, Charles DeCarli, Smita Kittur, Michael Borrie, T.-Y. Lee, Rob Bartha, Sterling Johnson, Sanjay Asthana, Cynthia M. Carlsson, Adrian Preda, Dana Nguyen, Pierre Tariot, Stephanie Reeder, Vernice Bates, Horacio Capote, Michelle Rainka, Douglas W. Scharre, Maria Kataki, Anahita Adeli, Earl A. Zimmerman, Dzintra Celmins, Alice D. Brown, Godfrey D. Pearlson, Karen Blank, Karen Anderson, Robert B. Santulli, Tamar J. Kitzmiller, Eben S. Schwartz, Kaycee M. Sink, Jeff D. Williamson, Pradeep Garg, Franklin Watkins, Brian R. Ott, Henry Querfurth, Geoffrey Tremont, Stephen Salloway, Paul Malloy, Stephen Correia, Howard J. Rosen, Bruce L. Miller, Jacobo Mintzer, Kenneth Spicer, David Bachman, Stephen Pasternak, Irina Rachinsky, Dick Drost, Nunzio Pomara, Raymundo Hernando, Antero Sarrael, Susan K. Schultz, Laura L. Boles Ponto, Hyungsub Shim, Karen Elizabeth Smith, Norman Relkin, Gloria Chaing, Lisa Raudin, Amanda Smith, Kristin Fargher, Balebail Ashok Raj, Thomas Neylan, Jordan Grafman, Melissa Davis, Rosemary Morrison, Jacqueline Hayes, Shannon Finley, Karl Friedl, Debra Fleischman, Konstantinos Arfanakis, Olga James, Dino Massoglia, J. Jay Fruehling, Sandra Harding, Elaine R. Peskind, Eric C. Petrie, Gail Li, Jerome A. Yesavage, Joy L. Taylor, and Ansgar J. Furst
References
- 1.Fjell AM, McEvoy L, Holland D, Dale AM, Walhovd KB. What is normal in normal aging? Effects of aging, amyloid and Alzheimer’s disease on the cerebral cortex and the hippocampus. Prog Neurobiol. 2014;117:20–40. doi: 10.1016/j.pneurobio.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toepper M. Dissociating Normal Aging from Alzheimer’s Disease: A View from Cognitive Neuroscience. J Alzheimer’s Dis. 2017;57:331–352. doi: 10.3233/JAD-161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mattson MP, Arumugam TV. Hallmarks of Brain Aging: Adaptive and Pathological Modification by Metabolic States. Cell Metab. 2018;27:1176–1199. doi: 10.1016/j.cmet.2018.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grimm A, Friedland K, Eckert A. Mitochondrial dysfunction: the missing link between aging and sporadic Alzheimer’s disease. Biogerontology. 2016;17:281–296. doi: 10.1007/s10522-015-9618-4. [DOI] [PubMed] [Google Scholar]
- 5.Błaszczyk JW. Energy metabolism decline in the aging brain-pathogenesis of neurodegenerative disorders. Metabolites. 2020;10(11):450. [DOI] [PMC free article] [PubMed]
- 6.Cunnane S, Nugent S, Roy M, Courchesne-Loyer A, Croteau E, Tremblay S, et al. Brain fuel metabolism, aging, and Alzheimer’s disease. Nutrition. 2011;27:3–20. doi: 10.1016/j.nut.2010.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pardo JV, Lee JT, Sheikh SA, Surerus-Johnson C, Shah H, Munch KR, et al. Where the brain grows old: decline in anterior cingulate and medial prefrontal function with normal aging. Neuroimage. 2007;35:1231–1237. doi: 10.1016/j.neuroimage.2006.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bi Q, Wang W, Niu N, Li H, Wang Y, Huang W, et al. Relationship between the disrupted topological efficiency of the structural brain connectome and glucose hypometabolism in normal aging. Neuroimage. 2021;226:117591. doi: 10.1016/j.neuroimage.2020.117591. [DOI] [PubMed] [Google Scholar]
- 9.Habeck C, Risacher S, Lee G, et al. Relationship between baseline brain metabolism measured using [18F]FDG PET and memory and executive function in prodromal and early Alzheimer’s disease. Brain Imaging Behav. 2012;6(4):568–83. [DOI] [PMC free article] [PubMed]
- 10.Chételat G, Landeau B, Salmon E, Yakushev I, Bahri MA, Mézenge F, et al. Relationships between brain metabolism decrease in normal aging and changes in structural and functional connectivity. Neuroimage. 2013;76:167–177. doi: 10.1016/j.neuroimage.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 11.Adams JN, Lockhart SN, Li L, Jagust WJ. Relationships Between Tau and Glucose Metabolism Reflect Alzheimer’s Disease Pathology in Cognitively Normal Older Adults. Cereb Cortex. 2019;29:1997–2009. doi: 10.1093/cercor/bhy078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inoue K, Ito H, Uchida S, Taki Y, Kinomura S, Tsuji I, et al. Decrease in glucose metabolism in frontal cortex associated with deterioration of microstructure of corpus callosum measured by diffusion tensor imaging in healthy elderly. Hum Brain Mapp. 2008;29:375–384. doi: 10.1002/hbm.20394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole JH, Poudel RPK, Tsagkrasoulis D, Caan MWA, Steves C, Spector TD, et al. Predicting brain age with deep learning from raw imaging data results in a reliable and heritable biomarker. Neuroimage. 2017;163:115–124. doi: 10.1016/j.neuroimage.2017.07.059. [DOI] [PubMed] [Google Scholar]
- 14.de Lange AG, Anatürk M, Suri S, Kaufmann T, Cole JH, Griffanti L, et al. Multimodal brain-age prediction and cardiovascular risk: The Whitehall II MRI sub-study. Neuroimage. 2020;222:117292. doi: 10.1016/j.neuroimage.2020.117292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beheshti I, Mishra S, Sone D, Khanna P, Matsuda H. T1-weighted MRI-driven Brain Age Estimation in Alzheimer’s Disease and Parkinson’s Disease. Aging Dis. 2020;11:618–628. doi: 10.14336/AD.2019.0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bergfield KL, Hanson KD, Chen K, Teipel SJ, Hampel H, Rapoport SI, et al. Age-related networks of regional covariance in MRI gray matter: Reproducible multivariate patterns in healthy aging. Neuroimage. 2010;49:1750–1759. doi: 10.1016/j.neuroimage.2009.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li TR, Dong QY, Jiang XY, Kang GX, Li X, Xie YY, et al. Exploring brain glucose metabolic patterns in cognitively normal adults at risk of Alzheimer’s disease: A cross-validation study with Chinese and ADNI cohorts. Neuroimage Clin. 2022;33:102900. doi: 10.1016/j.nicl.2021.102900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spetsieris PG, Ma Y, Dhawan V, Eidelberg D. Differential diagnosis of parkinsonian syndromes using PCA-based functional imaging features. Neuroimage. 2009;45:1241–1252. doi: 10.1016/j.neuroimage.2008.12.063. [DOI] [PubMed] [Google Scholar]
- 19.Eidelberg D. Metabolic brain networks in neurodegenerative disorders: a functional imaging approach. Trends Neurosci. 2009;32:548–557. doi: 10.1016/j.tins.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nan Z, Gordon ML, Yilong M, Bradley C, Gomar JJ, Shichun P, et al. The Age-Related Perfusion Pattern Measured With Arterial Spin Labeling MRI in Healthy Subjects. Front Aging Neurosci. 2018;10:214. doi: 10.3389/fnagi.2018.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teune LK, Strijkert F, Renken RJ, Izaks GJ, de Vries JJ, Segbers M, et al. The Alzheimer’s disease-related glucose metabolic brain pattern. Curr Alzheimer Res. 2014;11:725–732. doi: 10.2174/156720501108140910114230. [DOI] [PubMed] [Google Scholar]
- 22.Wu T, Ma Y, Zheng Z, Peng S, Wu X, Eidelberg D, et al. Parkinson’s disease-related spatial covariance pattern identified with resting-state functional MRI. J Cereb Blood Flow Metab. 2015;35:1764–1770. doi: 10.1038/jcbfm.2015.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mudali D, Teune LK, Renken RJ, Leenders KL, Roerdink JB. Classification of Parkinsonian syndromes from FDG-PET brain data using decision trees with SSM/PCA features. Comput Math Methods Med. 2015;2015:136921. doi: 10.1155/2015/136921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen K, Roontiva A, Thiyyagura P, Lee W, Liu X, Ayutyanont N, et al. Improved power for characterizing longitudinal amyloid-β PET changes and evaluating amyloid-modifying treatments with a cerebral white matter reference region. J Nucl Med. 2015;56:560. doi: 10.2967/jnumed.114.149732. [DOI] [PubMed] [Google Scholar]
- 25.Iizuka T, Kameyama M. Spatial metabolic profiles to discriminate dementia with Lewy bodies from Alzheimer disease. J Neurol. 2020;267:1960–1969. doi: 10.1007/s00415-020-09790-8. [DOI] [PubMed] [Google Scholar]
- 26.Habeck C, Foster NL, Perneczky R, Kurz A, Alexopoulos P, Koeppe RA, et al. Multivariate and univariate neuroimaging biomarkers of Alzheimer’s disease. Neuroimage. 2008;40:1503–1515. doi: 10.1016/j.neuroimage.2008.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jack CR, Jr, Bennett DA, Blennow K, Carrillo MC, Dunn B, Haeberlein SB, et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimer’s Dementia. 2018;14:535–562. doi: 10.1016/j.jalz.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson RS, Yu L, Lamar M, Schneider JA, Boyle PA, Bennett DA. Education and cognitive reserve in old age. Neurology. 2019;92:e1041–e1050. doi: 10.1212/WNL.0000000000007036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stern Y, Arenaza-Urquijo EM, Bartrés-Faz D, Belleville S, Cantilon M, Chetelat G, et al. Whitepaper: Defining and investigating cognitive reserve, brain reserve, and brain maintenance. Alzheimer’s Dementia. 2020;16:1305–1311. doi: 10.1016/j.jalz.2018.07.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pettigrew C, Soldan A, Zhu Y, Cai Q, Wang MC, Moghekar A, et al. Cognitive reserve and rate of change in Alzheimer’s and cerebrovascular disease biomarkers among cognitively normal individuals. Neurobiol Aging. 2020;88:33–41. doi: 10.1016/j.neurobiolaging.2019.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sotoudeh N, Namavar MR, Zarifkar A, Heidarzadegan AR. Age-dependent changes in the medial prefrontal cortex and medial amygdala structure, and elevated plus-maze performance in the healthy male Wistar rats. IBRO Rep. 2020;9:183–194. doi: 10.1016/j.ibror.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bennett IJ, Rivera HG, Rypma B. Isolating age-group differences in working memory load-related neural activity: assessing the contribution of working memory capacity using a partial-trial fMRI method. Neuroimage. 2013;72:20–32. doi: 10.1016/j.neuroimage.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moeller JR, Ishikawa T, Dhawan V, Spetsieris P, Mandel F, Alexander GE, et al. The metabolic topography of normal aging. J Cereb Blood Flow Metab. 1996;16:385–398. doi: 10.1097/00004647-199605000-00005. [DOI] [PubMed] [Google Scholar]
- 34.Liu D, Gu X, Zhu J, Zhang X, Han Z, Yan W, et al. Medial prefrontal activity during delay period contributes to learning of a working memory task. Science. 2014;346:458–463. doi: 10.1126/science.1256573. [DOI] [PubMed] [Google Scholar]
- 35.Günseli E, Aly M. Preparation for upcoming attentional states in the hippocampus and medial prefrontal cortex. Elife. 2020;9:e53191. [DOI] [PMC free article] [PubMed]
- 36.Kakimoto A, Ito S, Okada H, Nishizawa S, Minoshima S, Ouchi Y. Age-Related Sex-Specific Changes in Brain Metabolism and Morphology. J Nucl Med. 2016;57:221–225. doi: 10.2967/jnumed.115.166439. [DOI] [PubMed] [Google Scholar]
- 37.Rojkova K, Volle E, Urbanski M, Humbert F, Dell’Acqua F, Thiebaut de Schotten M. Atlasing the frontal lobe connections and their variability due to age and education: a spherical deconvolution tractography study. Brain Struct Funct. 2016;221:1751–66. [DOI] [PubMed]
- 38.Salat DH, Buckner RL, Snyder AZ, Greve DN, Desikan RS, Busa E, et al. Thinning of the cerebral cortex in aging. Cereb Cortex. 2004;14:721–730. doi: 10.1093/cercor/bhh032. [DOI] [PubMed] [Google Scholar]
- 39.Sullivan EV, Rohlfing T, Pfefferbaum A. Quantitative fiber tracking of lateral and interhemispheric white matter systems in normal aging: relations to timed performance. Neurobiol Aging. 2010;31:464–481. doi: 10.1016/j.neurobiolaging.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakurai R, Fujiwara Y, Yasunaga M, Suzuki H, Kanosue K, Montero-Odasso M, et al. Association between Hypometabolism in the Supplementary Motor Area and Fear of Falling in Older Adults. Front Aging Neurosci. 2017;9:251. doi: 10.3389/fnagi.2017.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Green PE, Ridding MC, Hill KD, Semmler JG, Drummond PD, Vallence AM. Supplementary motor area-primary motor cortex facilitation in younger but not older adults. Neurobiol Aging. 2018;64:85–91. doi: 10.1016/j.neurobiolaging.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 42.Sodums DJ, Bohbot VD. Negative correlation between grey matter in the hippocampus and caudate nucleus in healthy aging. Hippocampus. 2020;30:892–908. doi: 10.1002/hipo.23210. [DOI] [PubMed] [Google Scholar]
- 43.Lithfous S, Dufour A, Després O. Spatial navigation in normal aging and the prodromal stage of Alzheimer’s disease: insights from imaging and behavioral studies. Ageing Res Rev. 2013;12:201–213. doi: 10.1016/j.arr.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 44.Chen JJ, Rosas HD, Salat DH. Age-associated reductions in cerebral blood flow are independent from regional atrophy. Neuroimage. 2011;55:468–478. doi: 10.1016/j.neuroimage.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galiano A, Mengual E, García de Eulate R, Galdeano I, Vidorreta M, Recio M, et al. Coupling of cerebral blood flow and functional connectivity is decreased in healthy aging. Brain Imaging Behav. 2020;14:436–50. doi: 10.1007/s11682-019-00157-w. [DOI] [PubMed] [Google Scholar]
- 46.An Y, Varma VR, Varma S, Casanova R, Dammer E, Pletnikova O, et al. Evidence for brain glucose dysregulation in Alzheimer’s disease. Alzheimer’s Dementia. 2018;14:318–329. doi: 10.1016/j.jalz.2017.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chételat G, Arbizu J, Barthel H, Garibotto V, Law I, Morbelli S, et al. Amyloid-PET and (18)F-FDG-PET in the diagnostic investigation of Alzheimer’s disease and other dementias. Lancet Neurol. 2020;19:951–962. doi: 10.1016/S1474-4422(20)30314-8. [DOI] [PubMed] [Google Scholar]
- 48.Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, et al. Toward defining the preclinical stages of Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dementia. 2011;7:280–292. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scheltens P, De Strooper B, Kivipelto M, Holstege H, Chételat G, Teunissen CE, et al. Alzheimer’s disease. Lancet. 2021;397(10284):1577–90. [DOI] [PMC free article] [PubMed]
- 50.Guo T, Landau SM, Jagust WJ. Detecting earlier stages of amyloid deposition using PET in cognitively normal elderly adults. Neurology. 2020;94:e1512–e1524. doi: 10.1212/WNL.0000000000009216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vila-Castelar C, Tariot PN, Sink KM, Clayton D, Langbaum JB, Thomas RG, et al. Sex differences in cognitive resilience in preclinical autosomal-dominant Alzheimer’s disease carriers and non-carriers: Baseline findings from the API ADAD Colombia Trial. Alzheimer’s Dement. 2022;10.1002/alz.12552. [DOI] [PMC free article] [PubMed]
- 52.Buckley RF, Mormino EC, Rabin JS, Hohman TJ, Landau S, Hanseeuw BJ, et al. Sex Differences in the Association of Global Amyloid and Regional Tau Deposition Measured by Positron Emission Tomography in Clinically Normal Older Adults. JAMA Neurol. 2019;76:542–551. doi: 10.1001/jamaneurol.2018.4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hohman TJ, Dumitrescu L, Barnes LL, Thambisetty M, Beecham G, Kunkle B, et al. Sex-Specific Association of Apolipoprotein E With Cerebrospinal Fluid Levels of Tau. JAMA Neurol. 2018;75:989–998. doi: 10.1001/jamaneurol.2018.0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oveisgharan S, Arvanitakis Z, Yu L, Farfel J, Schneider JA, Bennett DA. Sex differences in Alzheimer’s disease and common neuropathologies of aging. Acta Neuropathol. 2018;136:887–900. doi: 10.1007/s00401-018-1920-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available upon request from the authors.