Abstract
Elevated serum urate (hyperuricemia) promotes crystalline monosodium urate tissue deposits and gout, with associated inflammation and increased mortality. To identify modifiers of uric acid pathologies, we performed a fly Genome-Wide Association Study (GWAS) on purine metabolites using the Drosophila Genetic Reference Panel strains. We tested the candidate genes using the Drosophila melanogaster model of hyperuricemia and uric acid crystallization (“concretion formation”) in the kidney-like Malpighian tubule. Medusa (mda) activity increased urate levels and inflammatory response programming. Conversely, whole-body mda knockdown decreased purine synthesis precursor phosphoribosyl pyrophosphate, uric acid, and guanosine levels; limited formation of aggregated uric acid concretions; and was sufficient to rescue lifespan reduction in the fly hyperuricemia and gout model. Levels of mda homolog FAM214A were elevated in inflammatory M1- and reduced in anti-inflammatory M2-differentiated mouse bone marrow macrophages, and influenced intracellular uric acid levels in human HepG2 transformed hepatocytes. In conclusion, mda/FAM214A acts in a conserved manner to regulate purine metabolism, promotes disease driven by hyperuricemia and associated tissue inflammation, and provides a potential novel target for uric acid–driven pathologies.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11357-022-00557-9.
Keywords: GWAS, Drosophila, Purines, Lifespan, Hyperuricemia, Inflammation
Introduction
An increase in uric acid body burden, reflected in elevated circulating levels of urate (termed hyperuricemia), promotes the development of tissue monosodium urate crystallization and uric acid urolithiasis in gout [1]. Gout and hyperuricemia prevalence are ~ 4% and ~ 21%, respectively, in the USA adult population, and increase with aging, and gout is a substantial global public health problem [2–4]. Moreover, with or without gout, hyperuricemia is associated with chronic kidney disease and several other disorders associated with aging, including hypertension, metabolic syndrome, type 2 diabetes, and coronary artery disease [5]. In addition, gout and hyperuricemia are positively correlated with and have been linked, as independent risk factors, with premature death and higher cardiovascular and all-cause mortality in clinical cohort studies [6–17].
Aging-related decline and hyperuricemia affect similar processes, including the interconnected generation of reactive oxygen species (ROS) and inflammation [18–22]. Xanthine oxidase, which is responsible for the direct enzymatic generation of uric acid, contributes to aging by promoting oxidative stress [23, 24]. However, the mechanisms by which hyperuricemia contributes to aging are incompletely defined. Since uric acid is the end product of purine degradation in humans, specifically defining the effects and regulation of uric acid in aging and aging-related diseases could provide novel targets for specific diseases of aging and healthspan.
We sought to identify novel genes which could impact uric acid metabolism and determine their role in purine metabolism and aging. Drosophila melanogaster has been a useful model for this type of analysis, as we have developed an in-house hyperuricemia fly model that takes advantage of the highly conserved purine metabolic pathway that includes multiple enzymes involved in de novo synthesis, salvage, and degradation of purine intermediates [25]. This model inhibits the conserved enzyme, urate oxidase (Uro), which catalyzes one of the final steps in purine degradation, the conversion of uric acid to allantoin, and demonstrates that Drosophila melanogaster is ideally suited to study both hyperuricemia and crystallopathies. Crystallopathies and elevated uric acid can be achieved through either dietary supplementation or inactivation of the gene for the interconvertible enzymatic forms of xanthine oxidase/xanthine dehydrogenase [25–32]. Yet, there is currently no study utilizing the power of Drosophila genetic tools to identify genes regulating the pathological phenotypes caused by these changes to purine metabolism.
Here, we tested the hypothesis that specific natural genetic variants are responsible for variation in levels of purine metabolites including uric acid. To do so, we used lines from the Drosophila Genetic Reference Panel (DGRP) [33]. Applying a Genome-Wide Association Study (GWAS) approach, we assessed purine metabolite levels from DGRP lines raised under conditions under which purine metabolism was differentially regulated by two different dietary conditions. We identified 5 genes that are associated with at least one purine metabolite [34]. We showed that the candidate gene CG9005, which we rename as a medusa (mda), is involved in purine regulation, the inflammatory process, uric acid crystallization-induced concretion formation in the Malpighian tubule, and shortened fly lifespan. Little is known about mda, beyond it being previously identified as regulating the lifespan response to tunicamycin-induced ER stress in flies [35]. mda was also identified via ChIP-Seq of 3rd-instar fly larvae as a target of the known ER stress response transcription factor xbp1 [36–38]. mda shares two domains, a domain of unknown function DUF4210 and the C-terminal of protein FAM214/SPAC3H8.04, with its human ortholog, Family With Sequence Similarity 214 Member A (FAM214A); functions have not yet been defined for both domains and the ortholog [39–41].
Investigating how mda regulates lifespan in our hyperuricemia model, we identified its role as a regulator of the uric acid–induced innate immune inflammatory response downstream of xbp1. Moreover, we demonstrate the regulation of FAM214A in M1-differentiated mouse inflammatory macrophages and its capacity to regulate intracellular urate in transformed human HepG2 hepatocytes. The work raises new questions about the roles of altered purine metabolism and hyperuricemia in aging and aging-related pathologies. The results presented in this study suggest mda/FAM214A is a novel and major component of the conserved uric acid pathway that is relevant for metabolic diseases and aging. Importantly, mda/FAM214A is a potential target to ameliorate the negative side effects of elevated uric acid burden, without the detrimental consequences of targeting upstream genes with pleiotropic effects.
Methods
Fly lines, husbandry, and diet composition
We generated a hyperuricemia fly model by recombining the RU486-inducible ubiquitous Gal4 driver daughterless-GeneSwitch-Gal4 with a uricase RNAi transgenic line (Da-GS, Uro), and this fly line was crossed with either RNA inhibition (RNAi) or UAS overexpression of the gene of interest for various experiments. All fly lines were maintained on a standard fly yeast extract medium containing 1.55% yeast, 5% sucrose, 0.46% agar, 8.5% cornmeal, and 1% acid mix (a 1:1 mix of 10% propionic acid and 83.6% orthophosphoric acid) prepared in distilled water. To prepare the media, cornmeal (85 g), sucrose (50 g), active dry yeast (16 g, “Saf-instant”), and agar (4.6 g) were mixed in a liter of water and brought to boil under constant stirring. Once cooled down to 60 °C, 10 ml of the acid mix was added to the media. The media were then poured in vials (~ 10 ml/ vial) or bottles (50 ml/bottle) and allowed to cool down before storing at 4 °C for later usage. These vials or bottles were then seeded with live yeast just before the flies are transferred and used to maintain lab stocks, collect virgins, or set up crosses.
For each cross, 12–15 virgin females of the Da-GS, Uro RNAi, or another driver line were mated with 3–5 males of the altered expression line in bottles containing an intermediate diet with 1.55% yeast as a protein source. Flies mated for 5 days, and then were removed. Nine days later, non-virgin female progenies were sorted onto an AL diet. Four to 8 vials of 25 mated female flies per vial are collected for each diet, maintained at 25 °C and 65% relative humidity, and were on a 12-h light/dark cycle. For gene knockdown experiments, we used RNAi lines from FlyORF, Vienna Drosophila Resource Center, and Bloomington Stock Center55 (Table S1). To investigate effects, we employed the Gal4-UAS driver system for RNAi in specific tissues, as well as the drug-inducible GeneSwitch system25. Crosses utilizing the GeneSwitch system were sorted onto AL + (standard diet with 5% yeast + RU486 in 95% ethanol for a final RU486 concentration of 200 μM) or AL − (5% yeast + 95% ethanol) diets. Flies were maintained under these conditions and concretion formation and lifespan were tested. Except strains created by Bloomington in the valium background, all other fly strains were routinely outcrossed 6 times into the w1118 background to achieve genetic background homogeneity among different strains and provide isogenization. Due to the difficulty in backcrossing appropriate TRiP control lines, BL35785 and 36,303 empty vector strains were used as background controls for the Bloomington lines in these backgrounds.
Concretion assay
Flies were maintained on an AL + or AL − diet for 14 days. They were transferred to fresh media bi-daily, at which point dead flies were removed. The following transfer on day 14, living flies were frozen. Frozen flies were then placed on an agar plate with abdomens and anal plates within droplets of water, and guts were dissected using surgical pliers to pull the anal plate. Flies were marked as having a concretion if at least one concretion of any size was observed [25].
Lifespan analysis
Flies developed on standard fly 1.5% yeast extract medium were transferred to the necessary diet within 72 h after eclosion. For survivorship analysis, vials with 25 mated females were transferred to fresh food every other day, and fly survival was scored by counting the number of dead flies. Each lifespan was repeated at least once to generate independent biological replicates [34, 42–45]. We used Cox proportional hazards analysis implemented in the R package “survival” to analyze the significance of the interaction between two variables in several of the survival outcomes. We report the probability that B1,2 = 0, from fitting the formula phenotype = B1 × variable1 + B2 × variable2 + B1,2 × (variable1 × variable2). The respective p values can be found in Table S2.
Genome-wide association mapping
We used DGRP release 2 genotypes, and FlyBase R5 coordinates for gene models. As in Nelson et al., we used only homozygous positions and a minor allele frequency of R 25% to ensure that the minor allele was represented by many observations at a given polymorphic locus [43, 46]. The collected phenotype and genotype data were used as input into an association test via ordinary least-squares regression using the StatsModels module in Python [47]. The linear model was phenotype = β1 × genotype + β2 × diet + β3 × genotype × diet + intercept. Nominal p values denoted as “genotype” in Fig. 1a report the probability that β1 ≠ 0, and those denoted as “interaction” report the probability that β3 ≠ 0. Additional GWAS of purine metabolites that accounted for Wolbachia infection status was performed with the linear model being phenotype = β1 × genotype + β2 × diet + β3 × genotype × diet + Wolbachia + intercept. Raw data can be found in the online data repository.
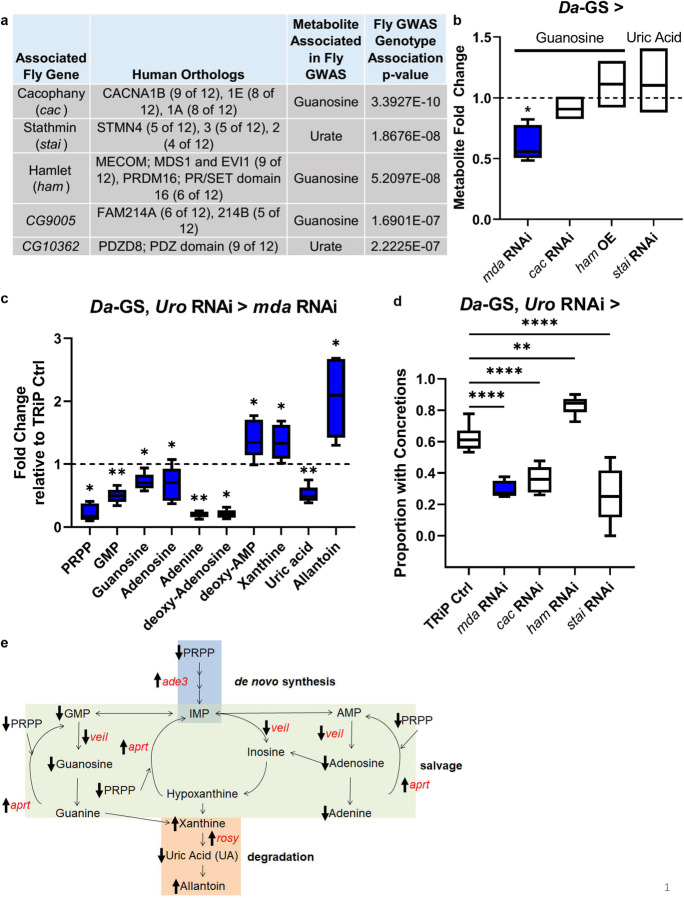
Fig. 1.
Whole-body mda RNAi reduces guanosine levels in wild-type and alters purine metabolism in model, preventing increased uric acid levels, and rescuing the concretion phenotype. a Purine GWAS candidate genes and associated metabolites. b Fold change of associated metabolite with inhibition (RNAi) or over expression (OE) of candidate gene in wild-type background compared to uninhibited isogenic control flies fed food with ethanol instead of RU486 (indicated by dashed line). Fly strains used, each crossed with Da-GS: mda RNAi—BL33362, cac RNAi—v104168/kk, ham OE—BL22209, and stai RNAi—BL53925. n = 5 biological replicates of 5 flies per condition. c Purine metabolites with significant fold changes in model with concurrent mda RNAi (BL33362) compared to background control, each crossed with Da-GS, uro RNAi. (TRiP empty vector control strain BL35785 represented by dashed black line.) d Proportion of flies with concretions in flies with Da-GS, uro RNAi crossed with TRiP BL35785 control or with concurrent candidate gene RNAi. Fly strains used same as in b, except ham RNAi—BL22209 (number of flies from left to right, n = 71, 38, 82, 73, and 43). Multiple comparisons made using Dunnett’s one-way ANOVA, with direct comparisons using two-tailed Student’s t test for first repetition, and one-tailed for subsequent repeats, in excel, with raw data and all repeats in Online Resource 23 and Table S4. e Model of purine metabolism, with metabolites in black and genes significantly altered with concurrent mda RNAi (BL33362) in the Da-GS, uro RNAi model in red, with arrows indicating direction of metabolite level or expression change. Mated females used for all experiments due to stronger phenotypic responses. Comparisons made using Student’s t test unless otherwise indicated. Significant differences for all tests are indicated by *: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Whiskers on boxplots drawn to largest or smallest values if within 1.5 times inter-quartile distance
To avoid the potential for false positives at a given nominal cutoff owing to p value inflation, we calculated false discovery rates (FDR) via permutation as follows: for given permutation i, we randomized all strain-metabolite value associations across DGRP lines, retaining the true diet assignment, and on this permuted dataset we carried out association tests for each marker in turn as above. To account for multiple testing across the 111 metabolites (14 purine metabolites), we kept the 111 metabolite values for a strain together by randomizing the strain assignments. The effect size was the coefficient for variables in each linear regression analysis. FDR values were calculated by taking the ratio of the number of permutation p values below a real p value, divided by the number of permutations run, over the total number of real p values below that same real p value and multiplying by 100, i.e., ((sum(permpvalues < = sortedrealpvalues[pvalue])/PERMs)/(sum(realpvalues < = sortedrealpvalues[pvalue])) × 100).
High-performance liquid chromatography (HPLC) mass spectrometry (MS)
High-performance liquid chromatography (HPLC) was performed using an Agilent 1260 UHPLC system and connected to a Phenomenex Luna NH2 column (2 × 100 mm, 3 μm, 100 Å) and a SecurityGuard NH2 guard column (4 × 2 mm ID). Mass spectrometry (MS) was performed using a 5500 Triple-Quadrupole LC–MS/MS mass spectrometer from Sciex fitted with a Turbo VTM ion source. Sciex’s Analyst v1.6.1 [68] was used for all forms of data acquisition, development of HPLC method, and optimization of analyte-specific MRM (multiple reaction monitoring) transitions. Skyline v4.1 was used for LC–MS/MS data analysis. For whole fly analysis, 5 flies (in sextuplicates) per condition were flash-frozen over liquid nitrogen and subsequently homogenized ultrasonically using a Fisher Scientific’s 550 Sonic Dismembrator with 50 μl of an 8:2 mixture of methanol/water (v/v), containing 2.5 μM of 2-chloroadenosine as internal standard. Three 20-s pulses at amplitude setting 3 of the instrument (on ice) were sufficient to completely homogenize fly specimens. The homogenates were then vortexed 5 times throughout ~ 30 min (each 1 min long). Subsequently, the samples were centrifuged at 10,000 rpm for 10 min, the supernatant was filtered, and 3 μl of each was injected for HPLC-MRM analysis (vide infra) without any additional processing. For the analysis of fly deposits, samples were collected as described above and prepared for HPLC-MRM analysis (vide infra) as described above for whole flies.
Optimization of analyte-specific MRM transitions, such as determination of suitable precursor and productions and optimal MS parameters for each transition (Q1, precursor ! Q3, product), was achieved by isocratic flow injection of the 1–10 μM solution (final) for each standard, diluted in 80% methanol. The most intense transition was used as a quantifier, whereas one or more additional transitions were used as a qualifier for each compound [25]. A final standard mixture of all compounds at 5 μM (containing the internal standard 2-chloroadenosine at 2.5 μM) was prepared before analysis and injected at the onset of each biological sample set.
Based on previous reports [48], the following HPLC program was developed: a solvent gradient of 20 mM ammonium acetate + 20 mM ammonium hydroxide (pH = ~ 9.5) + 5% acetonitrile (aqueous)–acetonitrile (organic) was used with 0.4 ml/min flow rate, starting with an acetonitrile content of 85% for 1 min, which was decreased to 30% over 3 min and then to 0% over 7 min and held at 0% for 2 min. The HPLC column was subsequently reconstituted to its initial condition (acetonitrile content of 85%) over the next 1 min and re-equilibrated for 7 min. Metabolome extracts from whole flies or deposits were analyzed by HPLC-MRM with positive/negative switching of source ion modes [48]. Source conditions were as follows: curtain gas (CUR) 20, collision gas (CAD) 7, ion source gas 1 (GS1) 30, ion source gas 2 (GS2) 30, ionspray voltage (IS) ± 4500 V, and source temperature (TEM) 450 °C. Quantification was based on the integration of analyte-specific peaks obtained from HPLC-MRM runs.
Gene expression analysis
To determine gene expression in a normal system, we sampled 3 biological replicates of 3–5 whole mated female flies after 2 weeks on both diets. We isolated RNA using Zymo Quick RNA MiniPrep kit (R1054) (Zymo Research, Irvine, CA). cDNA of isolated RNA was synthesized using the iScript™ cDNA Synthesis Kit from Bio-Rad (Bio-Rad 1,708,890) following the manufacturer’s protocol. For qRT-PCR, we quantified Cp values for 3 technical replicates per sample using the FastStart SYBR Green qPCR mix (Sigma-Aldrich 04,673,484,001) and followed the manufacturer’s instructions with a Roche LightCycler 480 II machine. Primer information can be found in Table S3. Bar plots show AL + values for all biological and technical replicates normalized to the respective AL-control group and the standard error. To validate the effects of RNAi or mutation on gene expression, we collected 3–5 whole females following 2 weeks on their diet. We then isolated RNA from these samples, synthesized cDNA, and performed qRT-PCR on the perturbed genes as described.
Western blot
Cells were lysed in RIPA buffer with 2 mM sodium vanadate and protease inhibitor cocktails (Roche, Mannheim, Germany). Cell lysates (10–15 μg) were separated by gradient 4–20% SDS-PAGE and transferred onto PVDF membranes (Sigma, St. Louis, MO). The blots were blocked with Odyssey Blocking buffer (LI-COR 927–50,000, Lincoln, NE) for 1 h at room temperature and then incubated with primary rabbit antibodies against FAM214A (1:1000 dilution, #HPA039369, Sigma-Aldrich, St. Louis, MO) overnight at 4 °C. Washes were performed with TBS 0.1% Tween-20 (TBST) before the addition of IRDye 680RD goat anti-rabbit IgG secondary antibody (1:10,000 dilution, LI-COR, Lincoln, NE) for 1 h at room temperature. For loading control, the blots were incubated with primary mouse monoclonal antibody against β-actin (1:5000 dilution, #A2228, Sigma-Aldrich, St. Louis, MO) followed by washing and incubating with IRDye 800CW anti-mouse IgG secondary antibody (1:10,000 dilution, LI-COR, Lincoln, NE). All blots were imaged on the Odyssey® CLx imaging system using 680 nm and 780 nm channels. Protein detection was performed using Image Studio Ver. 5.2. Expression of FAM214A was normalized to β-actin.
Cell transfection
HepG2 cells were cultured in the DMEM + 10% FBS + 1% pen-strep. Cells were transfected with (A) plasmids of pCMV3-hFAM214A and the empty vector using X-tremeGENE HP DNA transfection reagent (Sigma) or (B) siRNAs of human FAM214A siRNA and non-targeted (Thermo Fisher) control using X-tremeGENE siRNA transfection reagents (Sigma) for 48 h, and following manufacturer protocols.
Intracellular uric acid measurement
The intracellular uric acid levels were determined using Uric Acid Assay Kit (Sigma-Aldrich MAK077). Raw data are included as Online Resource 2.
Mouse bone marrow–derived macrophage (BMDM) differentiation
BMDMs were differentiated from bone marrows isolated from 10-week-old C57BL/6 mice in the presence of 20% L929 cell-conditioned media for 7 days, which represented M0 macrophages. These cells were subsequently treated with LPS (20 ng/ml) and INFγ (10 ng/ml) or IL-4 (20 ng/ml) for 24 h to polarize to M1 and M2 macrophages, respectively. Raw data are included as Online Resource 2.
Mouse liver explant IL-1β treatment
C57BL/6 J male mice were housed in static, polysulfone, microisolation cages and maintained on a 12:12-h light:dark cycle with caging, food (regular chow), and water bottles changed weekly. Mice at 10 weeks of age were sacrificed and livers were removed and cut into small pieces and placed in a DMEM culture medium containing 10% FBS for 2 days. The liver tissue explants were then treated with IL-1β (2 ng/ml) in fresh DMEM media containing 1% FBS for 6 h. The tissues were collected and washed with PBS 3 times and subjected to RNA extraction followed by quantitative RT-PCR analysis for FAM214A expression. Raw data are included as Online Resource 2.
Quantification and statistical analysis
Significance was determined between different gene expression (qRT-PCR) data and phenotypic data at specific time points using the Wilcoxon test of the pcr_test() function from the PCR package in R unless otherwise specified. The significance of differences between survival curves was assessed by the Cox proportional hazards test. p < 0.05 was considered statistically significant. Significant differences between experimental groups and controls were indicated by *: *p < 0.05, **p < 0.005, ***p < 0.0005, nc = no change, and ns = not significant. Correlation between traits was determined using the cor() function in R on all data points and calculating Pearson correlation coefficients. Data and scripts used for analysis are provided in Online Resources.
Results
Identifying variants associated with regulating purine metabolism
To identify regulators of purine metabolism, we used a previously published metabolomic dataset in female flies from the Drosophila Genetic Reference Panel (DGRP) on either high yeast extract food (AL, 5% yeast extract in food) or low yeast extract food (DR, same composition except 0.5% yeast extract in food) for GWAS. This form of DR, which restricts nutrients without causing malnutrition, has been shown to robustly extend lifespan and healthspan, as well as improve phenotypes associated with hyperuricemia [25, 49]. This allowed the discovery of genetic markers which associate with purine levels across multiple genotypes under standard or hyperuricemia-easing dietary conditions [34]. The screen identified 5 candidate genes that were associated with at least one purine metabolite (Fig. 1a). Three genes, cacophany (cac), the transcription factor hamlet (ham), and medusa (mda), associated with guanosine, while stathmin (stai) and CG10362 were both associated with urate, which can act as a toxic product of the purine degradation pathway (Fig. 1a). None of the genes was previously known to participate in purine metabolism, with cac, stai, and ham having neuron-related functions, and CG10362 being involved in intracellular signal transduction. Addition of Wolbachia infection status to the linear model did not remove the above candidates at the top of the candidate list, and added an additional candidate gene, sulfateless (sfl), to the top candidates with human orthologs (Table S5).
mda reduced guanosine in both a wild-type background and the hyperuricemia fly model
To validate the GWAS candidates, we measured the impact of whole-body RNAi using the driver line Da-GS on the candidate genes’ associated metabolite levels in mated female flies of a wild-type background using HPLC mass spectrometry (Fig. 1b). Inhibition of mda significantly decreased levels of its associated metabolite, guanosine (p = 1.59 × 10−02), validating the GWA result. We also used a fly model of elevated urate levels to determine if the candidate genes could impact their associated metabolite in a model of elevated uric acid (UA) levels [25]. In this hyperuricemia fly model, uricase (uro) is inhibited in the whole body using the driver line Da-GS to elevate uric acid and simulate human purine metabolism (Da-GS, uro RNAi recombinant line). This model recapitulates some hyperuricemia health outcomes, making it ideal for testing human genes associated with this condition [25]. The model exhibits a burden of concretions (stone-like deposits) in the kidney-like Malpighian tubule excretory system, triggered by a rich diet that associates with a significantly shortened lifespan. As in the wild-type background, when mda was inhibited in our hyperuricemia fly model, it also significantly decreased guanosine (p = 1.65 × 10−02; Fig. 1c). Additionally, mda inhibition nearly completely prevented the model’s increase in uric acid (p = 4.96 × 10−04; Fig. 1c). The modulation of other purine metabolites, in both the wild-type and our model, revealed mda to be a regulator of purine metabolism, particularly in a hyperuricemia context (Fig. 1c; Sup Fig. 1a, b). In addition to regulating purine metabolite levels, mda regulates expression of Xanthine dehydrogenase ortholog rosy (p = 6.37 × 10−05), GART ortholog ade3 (p = 1.0 × 10−02), aprt (p = 4.29 × 10−03), and the 5ʹ-nucleotide transferase ortholog veil (p = 2.9 × 10−04) (Sup Fig. 1c). Furthermore, inhibition of stai expression significantly altered its associated metabolites’ level in our hyperuricemia model (Sup Fig. 1a).
Inhibition of mda rescues the concretion and shortened lifespan phenotypes of the hyperuricemia fly model
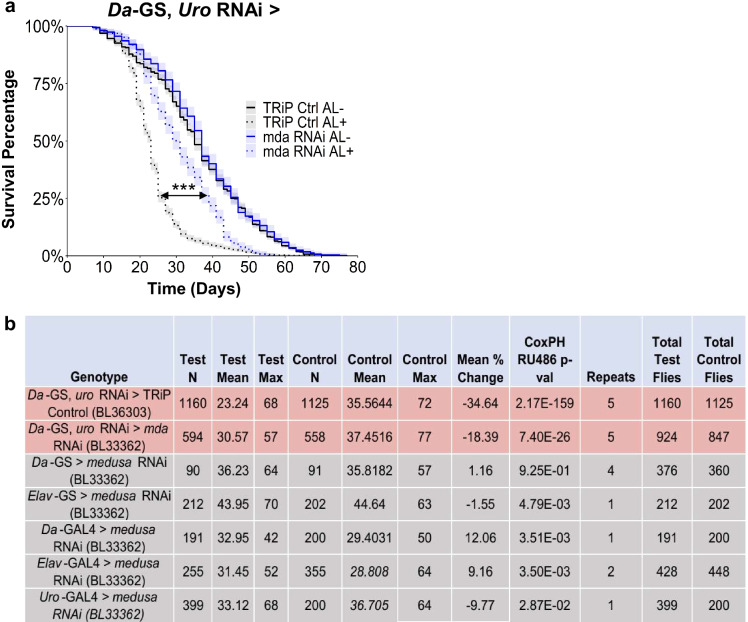
While mda inhibition altered purine metabolism, we determined whether it rescued the hyperuricemia model phenotypes of a shortened lifespan and concretion formation in the Malpighian tubules and hindguts [25] (Sup Fig. 1). Despite most candidates rescuing the concretion phenotype, with ham as the exception, only cac and mda also rescued the lifespan reduction (Figs. 1d and 2a; Sup Fig. 2). The transcription factor ham was the only candidate whose knockdown increased the formation of concretions, yet it still did not significantly impact the model’s lifespan phenotype (Fig. 1d; Sup Fig. 2). Since mda RNAi significantly altered its associated metabolite, as well as its impact on the model’s concretion and lifespan phenotypes, we focused on elucidating the mechanisms of mda. To identify the tissue where mda expression contributes to the shortened lifespan, we examined the role of mda expression in a wild-type background. Inhibiting mda in a wild-type background had minimal effects on lifespan, regardless of which tissue it was inhibited (Fig. 2b; Sup Fig. 3). Considering that mda significantly rescued the hyperuricemia model traits, we decided to further characterize the mechanisms by which it functions.
Fig. 2.
mda RNAi is sufficient to rescue the hyperuricemia model’s decreased lifespan phenotype. a Lifespan rescue of whole-body mda RNAi (BL33362) crossed with hyperuricemia model (Da-GS, uro RNAi, TRiP empty vector control strain BL36303; number of flies from top of legend to bottom, n = 1264, 1334, 561, and 585). b Breakdown of mda RNAi (BL33362) lifespan effects in hyperuricemia model background (shaded red) or tissue-specific inhibition in wild-type backgrounds (shaded gray). AL − / + indicates flies fed ethanol or RU486, respectively. All p values from coxph() function in survival package in R (survival curves and cox proportional hazards results can be found in Sup Figs. 2 and 3, and Table S2, respectively)
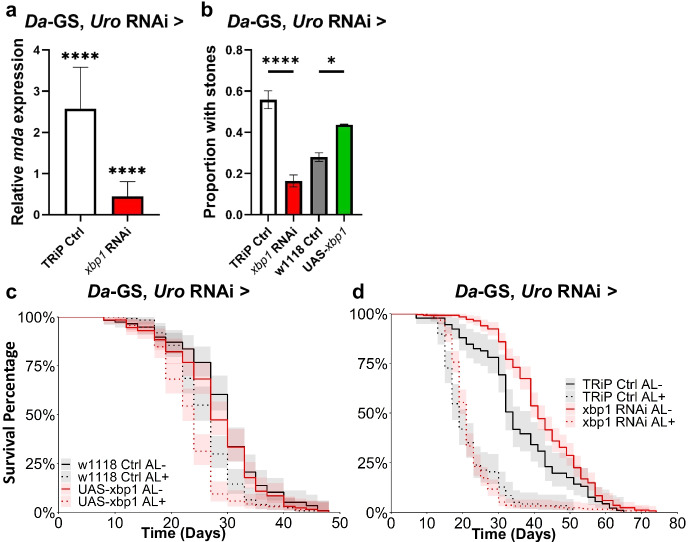
mda expression is regulated by xbp1
To identify transcriptional regulators of mda, we used a transcription factor (TF) analysis utilizing publicly available ChIP-Seq data from fly larvae [36–38]. This analysis showed mda to be a target for the endoplasmic reticulum (ER) stress response TF Xbp1 during development. We confirmed that xbp1RNAi inhibited adult mda expression in our hyperuricemia model; however, xbp1 overexpression (OE) did not have a significant effect (Fig. 3a; Sup Fig. 4a). Further, xbp1RNAi significantly rescued the concretion phenotype (Fig. 3b), while xbp1 OE did not tend to increase concretions. Despite these effects, neither inhibiting nor overexpressing xbp1 significantly impacted the model’s shortened lifespan (Fig. 3 c and d, respectively). To rule out pleiotropic effects of xbp1 inhibition, we tested whether xbp1 inhibition was sufficient to shorten lifespan in a wild-type background. Indeed, whole-body inhibition in a wild-type background significantly shortened lifespan (p < 2.00 × 10−16; Sup Fig. 4), and its inhibition in the model did not separately increase the hazard ratio (p = 4.61 × 10−01; Table S3). The lifespan effects of xbp1 inhibition are also confounded by prior results demonstrating that its inhibition can be detrimental to longevity [50]. Together these results show that xbp1 is necessary but not sufficient to induce mda expression and the knockdown of either of these genes rescues the concretion phenotype in the hyperuricemia model, but its pleiotropic effects confound any interpretation of its lifespan effects. Furthermore, mda is a more specific target for the downstream effects of hyperuricemia that is less likely to disrupt other key stress response pathways.
Fig. 3.
xbp1 regulates mda expression, but its modulation does not rescue lifespan. a mda expression levels with RNAi of xbp1 (BL36755) or mda RNAi (BL33362) crossed with hyperuricemia background (Da-GS, uro RNAi) relative to uninhibited isogenic control flies fed food with ethanol instead of RU486. 3 biological replicates of 3 flies, each, per condition. b Proportion of flies with concretions with xbp1 or mda RNAi in hyperuricemia background. (direct comparisons using two-tailed Student’s t test for first repetition, and one-tailed for subsequent repeats, in excel, with raw data and all repeats in File S1 and Table S4; TRiP empty vector control strain BL35785; number of flies from left to right, n = 61, 55, 75, and 48). c Lifespan with over expression (UAS) of xbp1 (BL60731) in hyperuricemia background. Number of flies from top of legend to bottom, n = 115, 124, 129, and 138. d Lifespan with inhibition of xbp1 in hyperuricemia background (Cox proportional hazards results can be found in Table S2; TRiP empty vector control strain BL35785; number of flies from top of legend to bottom, n = 90, 117, 248, and 246). Significant differences for all tests are indicated by *: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. AL − / + indicates flies fed ethanol or RU486, respectively. Error bars of a and b represent SEM between replicates
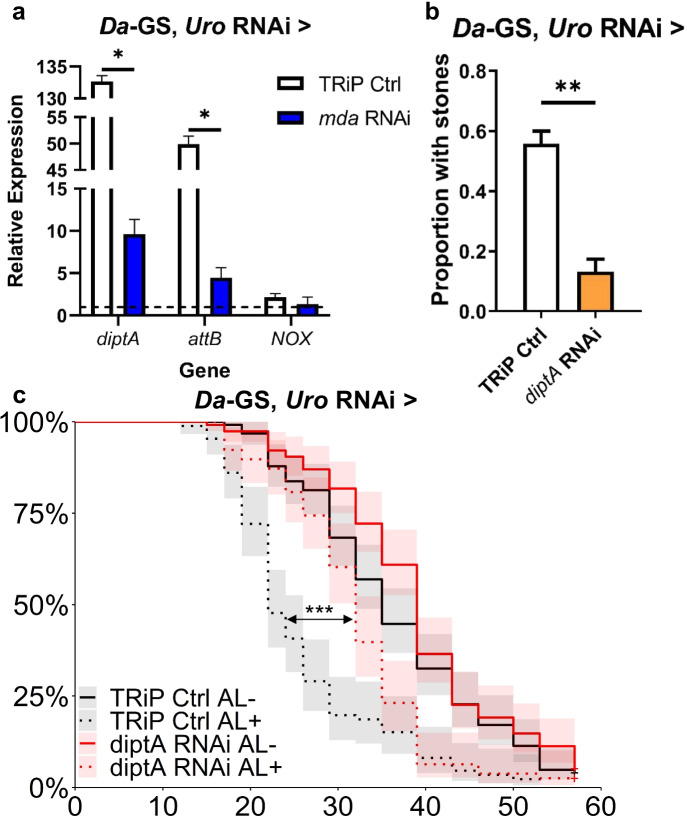
mda is necessary for innate immune response in the hyperuricemia model
Next, we sought to identify the pathways by which mda expression impacts lifespan and concretion phenotypes. To do so, we performed qPCR of genes shown to be upregulated in the hyperuricemia model [25]. We sought to determine if mda acts through reactive oxygen species gene NADPH-oxidase (NOX), inhibition of which has previously been shown to rescue the hyperuricemia phenotypes [25]. Since an inflammatory response was also seen in the fly model and connected with hyperuricemia, we also tested the role of innate immunity, using diptericin A (diptA) and attacin B (attB) as readouts of an activated inflammatory response. We found that inhibiting mda in our model almost completely negated the increased expression of diptA and attB in the hyperuricemia model but did not significantly prevent the increase in NOX (Fig. 4a). Furthermore, inhibition of diptA alone was sufficient to rescue the model’s concretion (p = 6.63 × 10−03) and lifespan (p = 3.10 × 10−07) phenotypes (Fig. 4b and c, respectively). This suggests that mda’s mechanism of action is through regulating the immune response in a hyperuricemia context, independently of the NOX pathway.
Fig. 4.
mda inhibition dampens hyperuricemia model’s immune response, which is sufficient to rescue fly model’s concretion accumulation and decreased lifespan. a Diptericin A (diptA), Attacin B (attB), or NADPH-oxidase (NOX) expression levels in hyperuricemia model (Da-GS, uro RNAi crossed with TRiP empty vector control BL35785) alone or with concurrent mda RNAi (BL33362) normalized to uninhibited isogenic control flies fed food with ethanol instead of RU486 (indicated by dashed line). 3 biological replicates of 3 flies, each, per condition. b Proportion of flies with concretions in flies with Da-GS, uro RNAi crossed with TRiP empty vector control BL35785 or with concurrent inhibition of diptA (BL67221) (direct comparisons using two-tailed Student’s t test for first repetition, and one-tailed for subsequent repeats, in excel, with raw data and all repeats in File S1 and Table S4; number of flies from left to right, n = 61 and 45). c Lifespan with inhibition of diptA in hyperuricemia background (TRiP empty vector control strain BL35785; Cox proportional hazards results can be found in Table S2; number of flies from top of legend to bottom, n = 123, 86, 115, and 78). AL − / + indicates flies fed ethanol or RU486, respectively. Significant differences for all tests are indicated by *: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Error bars represent SEM between replicates
mda homolog FAM214A regulates intracellular urate in HepG2 cells
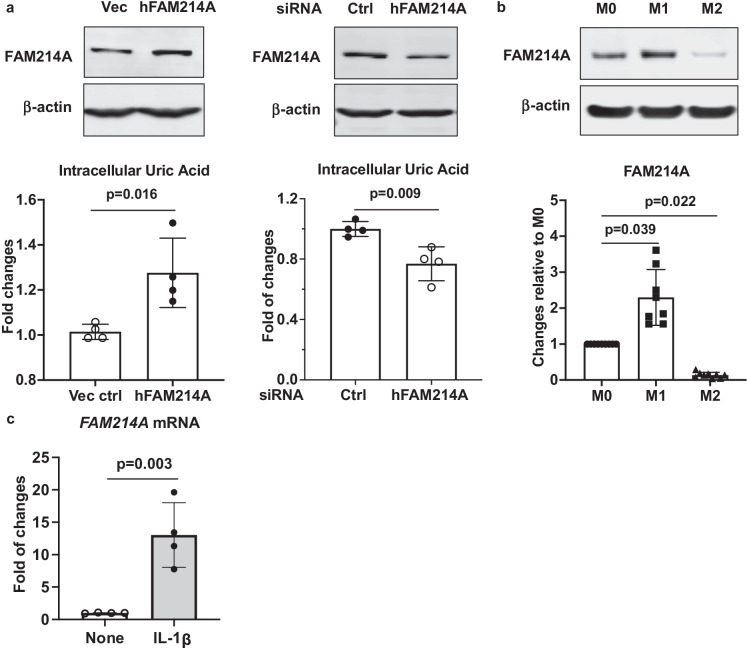
To determine whether the effects of mda are conserved, we probed the ability of mda to regulate intracellular urate (UA) in human cells known to produce uric acid. We first looked for changes in cell urate level when FAM214A was overexpressed or knocked down in human hepatocyte cancer cell line HepG2. Cells with overexpression of FAM214A had a significant increase in urate (p = 1.60 × 10−02), while FAM214A knockdown led to a significant decrease (p = 9.00 × 10−03) (Fig. 5a). To determine whether the immune function of mda might potentially be conserved in mammals, we then assayed FAM214A levels in a murine immune cell model, bone marrow–derived monocytes (BMDM), employing SDS-PAGE/Western Blot. FAM214A was low in resting macrophages (M0), increased in cells differentiated to be pro-inflammatory (M1 macrophages), and FAM214A protein was sparse in macrophages differentiated to be anti-inflammatory (M2 macrophages) (p = 3.9 × 10−02 and 2.2 × 10−02, respectively, when compared to M0) (Fig. 5b). To determine if FAM214A expression is responsive to inflammatory signals, we treated mouse liver tissue explants with IL-1β ex vivo. Treated livers had significantly higher levels of FAM214A expression (p = 3.00 × 10−03; Fig. 5c). These results suggest that mda/FAM214A regulates uric acid metabolism and the innate immune response can be conserved in flies and mammals.
Fig. 5.
mda homolog FAM214A expression influences intracellular uric acid (UA) levels, is differentially expressed in bone marrow–derived macrophages (BMDM), and is upregulated in response to IL-1β. a Both “gain-of-function” and “loss-of-function” approaches were used to overexpress and knockdown FAM214A expression in human HepG2 cells via transient transfection. The data were presented as fold changes relative to the vector control or control siRNA (bottom panels). FAM214A protein expression was examined by Western blot analysis (top panels). Unpaired Student’s t test was used for the statistical analysis. b Top panel shows FAM214A protein expression in mouse BMDMs that were undifferentiated (M0) or differentiated to pro-inflammatory (M1) or anti-inflammatory (M2) states, and here examined by Western blot analysis. Bottom panel shows fold of changes of FAM214A expression (normalized to β-actin by semi-quantitative densitometry analyses of Western blot images from 9 different mouse donors) in M1 and M2 relative to M0 BMDMs. Kruskal Wallis with Dunn’s multiple comparisons test was used for the statistical analysis. c Upregulation of FAM214A mRNA expression by IL-1β ex vivo in mouse liver tissue explants (n = 4). Unpaired Student’s t test was used for the statistical analysis. Error bars represent SD between replicates
Discussion
The end product of purine metabolism is uric acid in humans and closely related primates, as opposed to allantoin in the majority of organisms. This is due to the lack of human expression of a functional uricolytic uricase gene [51, 52]. The evolutionary benefits of humans losing the uricase gene remain speculative [52–54]. However, the detrimental health outcomes arising from a pathological elevation of circulating urate burden given the current epidemic of obesity, in over 20% of Americans, render this an important area for drug discovery [5, 55]. Beyond the more acute impacts of related pathologies due to crystallization of sodium urate and uric acid, the shortened maximum fly and human lifespan caused by elevated urate burden incentivize pursuing new drug targets [25]. Many recently identified genes that impact hyperuricemia in the fly, such as IGF-1, FOXO, and NOX, have pleiotropic effects [56–58]. As such, identifying downstream “effector” genes offers the potential to treat not only the pathological outcomes but also the effects of human purine metabolism on aging processes and lifespan.
Our findings raise new questions on the connections between lifespan, hyperuricemia, and pathologies due to the deposition of crystalline monosodium urate in the synovial joint or as uric acid in the renal urine collecting system. In this study, all the GWAS candidate genes examined significantly altered the concretion phenotype. However, all but two failed to rescue shortened lifespan in the fly model of hyperuricemia. Interestingly, despite only mda significantly changing the levels of its associated metabolite, guanosine, all candidate genes altered other purine metabolites in the fly model. These results support new approaches to studying and potentially treating hyperuricemia. Treatments that target transporter gene candidates from GWAS of human gout patients, such as SLC2A9 and its gene product GLUT9, may be impacting only limited health aspects of the disease [7]. Furthermore, since uric acid is the end product of human purine metabolism, these observations point to the existence of alternative lifespan-specific pathways of purine metabolites, potentially through treatments that abrogate uric acid–induced inflammatory responses.
The gene that we focused on, mda, has been uncharacterized except for its linkage to tunicamycin-induced ER stress [35]. medusa was also identified in a ChIP-Seq of 3rd-instar fly larvae as a target of the known ER stress response transcription factor Xbp1, providing further support for its role in ER stress [36–38]. We discovered that mda regulates purine metabolism in a hyperuricemia context in vivo in the fly, modulating the levels of guanosine and certain other purine metabolites, at least partly by effects on expression of the Xanthine dehydrogenase ortholog rosy, GART ortholog ade3, aprt, and the 5ʹ-nucleotide transferase ortholog veil. We demonstrated that mda is an “effector” gene in the xbp1 ER stress response pathway which regulates purine metabolism changes and the inflammatory response in a hyperuricemia context (Fig. 6). Despite inhibition of xbp1 being sufficient to rescue the concretion phenotype of our model, we did not see a similar rescue of the model’s shortened lifespan. This could be due to the lifespan shortening effects of inhibiting xbp1 in a wild-type background, which would confound any rescue of the model lifespan. Studies of the human mda ortholog FAM214A, which had an unknown function, suggest that mda also regulates intracellular urate levels, that mda contributes to the fly pro-inflammatory response to hyperuricemia, and that FAM214A could impact pathophysiology in gout. Our results further suggested that the mda human ortholog FAM214A could function in regulating purine metabolism.
Fig. 6.
mda regulates hyperuricemia model phenotypes through altering purine metabolism and upregulating immune response
Precisely how FAM214A regulates urate levels has not yet been determined, but we speculate cytoskeletal functions play a role. Human FAM214A Q32MH5 has binary interactions with 6 proteins, including keratin, type I cytoskeletal 40, and ADIP and afadin- and alpha-actinin-binding protein, which localizes at and is required for the formation of the leading edge of moving cells and promotes cell movement, potentially through activation of Rac signaling [59–63]. Importantly, one mechanistic theory of uric acid generation in some cells is that it occurs in a “purinosome” structure of multiple purine metabolism enzymes macroaggregated at the leading edge of moving cells [64, 65]. Whether FAM214A is centrally involved as a scaffolding protein or chaperone in purinosome assembly and function remains to be tested.
Rescue of the fly hyperuricemia model concretion phenotype by mda knockdown, changes to purine metabolite levels in the fly model, and partial rescue of shortened fly lifespan compared to little impact on wild-type lifespan mark mda as an intriguing potential drug target. Indeed, mda may be a novel example of a gene involved in hyperuricemia that facilitates purinosome formation and promotes tissue damage. Since closely related primates and humans have relatively high levels of circulating urate compared to most other species, our findings may shed new light on approaches to lengthen and improve the health quality of human lifespan.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
T.A.H. is supported by NIH and NIA award F31AG062112. K.A.W. was supported by NIH and NIA award F31AG052299 and is currently supported by NIH/NIA training grant T32AG000266-23. C.S.N. was supported by NIH/NIA F32 award AG047024. R.T and R.L.B were supported by NIH awards AR060772 and AR075990, and also by the VA Research Service. We thank the Bloomington Drosophila Stock Center, the Vienna Drosophila Stock Center for providing the flies used.
Author contribution
T.A.H., S.L., D.P., R.B.B., R.T., and P.K. designed research; T.A.H., R.L.B., T.G., K.A.W., N.B., J.N.B., K.J., J.N.B., M.S., and D.R. performed research; T.A.H., R.L.B., R.B.B., R.T., and P.K. analyzed data; T.A.H., C.S.N., and T.O. provided R and Python code; T.A.H., R.T., and P.K. wrote the paper.
Funding
This work was funded by grants from the American Federation of Aging Research (R.B.B. and P.K.); NIH grants R56AG038688, R21AG054121, AG045835 (P.K.), and R01AG049494 (D.P.); and the Larry L. Hillblom Foundation.
Declarations
Conflict of interest
R.T. declares a research grant from Astra-Zeneca, and consulting honoraria from Astra-Zeneca, Fortress, Dyve, Selecta, Allena, and SOBI. The other authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dalbeth N, Merriman TR, Stamp LK. Gout Lancet. 2016;388(10055):2039–2052. doi: 10.1016/S0140-6736(16)00346-9. [DOI] [PubMed] [Google Scholar]
- 2.Chen-Xu M, Yokose C, Rai SK, Pillinger MH, Choi HK. Contemporary prevalence of gout and hyperuricemia in the United States and decadal trends: the National Health and Nutrition Examination Survey, 2007–2016. Arthritis Rheumatol. 2019;71(6):991–999. doi: 10.1002/art.40807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Safiri S, Kolahi AA, Cross M, Carson-Chahhoud K, Hoy D, Almasi-Hashiani A, et al. Prevalence, incidence, and years lived with disability due to gout and its attributable risk factors for 195 countries and territories 1990–2017: a systematic analysis of the Global Burden of Disease Study 2017. Arthritis Rheumatol. 2020;72(11):1916–1927. doi: 10.1002/art.41404. [DOI] [PubMed] [Google Scholar]
- 4.Dehlin M, Jacobsson L, Roddy E. Global epidemiology of gout: prevalence, incidence, treatment patterns and risk factors. Nat Rev Rheumatol. 2020;16(7):380–390. doi: 10.1038/s41584-020-0441-1. [DOI] [PubMed] [Google Scholar]
- 5.Zhu Y, Pandya BJ, Choi HK. Comorbidities of gout and hyperuricemia in the US general population: NHANES 2007–2008. Am J Med. 2012;125(7). [DOI] [PubMed]
- 6.Broughton S, Partridge L. Insulin/IGF-like signalling, the central nervous system and aging. Biochem J. 2009;418(1):1–12. doi: 10.1042/BJ20082102. [DOI] [PubMed] [Google Scholar]
- 7.Sulem P, Gudbjartsson DF, Walters GB, Helgadottir HT, Helgason A, Gudjonsson SA, et al. Identification of low-frequency variants associated with gout and serum uric acid levels. Nat Genet. 2011;43(11):1127–1130. doi: 10.1038/ng.972. [DOI] [PubMed] [Google Scholar]
- 8.Perez-Ruiz F, Aniel-Quiroga MA, Herrero-Beites AM, Chinchilla SP, Erauskin GG, Merriman T. Renal clearance of uric acid is linked to insulin resistance and lower excretion of sodium in gout patients. Rheumatol Int. 2015;35(9):1519–1524. doi: 10.1007/s00296-015-3242-0. [DOI] [PubMed] [Google Scholar]
- 9.Clarson LE, Chandratre P, Hider SL, Belcher J, Heneghan C, Roddy E, et al. Increased cardiovascular mortality associated with gout: a systematic review and meta-analysis. Eur J Prev Cardiol. 2015;22(3):335–343. doi: 10.1177/2047487313514895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubchak N, Falasca GF. New and improved strategies for the treatment of gout. Int J Nephrol Renovasc Dis. 2010;3:145–166. doi: 10.2147/IJNRD.S6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu J, Hou X, Yuan X, Cui L, Liu Z, Li X, et al. Knockout of the urate oxidase gene provides a stable mouse model of hyperuricemia associated with metabolic disorders. Kidney Int. 2018;93(1):69–80. doi: 10.1016/j.kint.2017.04.031. [DOI] [PubMed] [Google Scholar]
- 12.Adachi S-I, Yoshizawa F, Yagasaki K. Hyperuricemia in type 2 diabetic model KK-Ay/Ta mice: a potent animal model with positive correlation between insulin resistance and plasma high uric acid levels. BMC Res Notes. 2017;10(1):1–5. doi: 10.1186/s13104-017-2897-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pendse J, Ramachandran P V., Na J, Narisu N, Fink JL, Cagan RL, et al. A Drosophila functional evaluation of candidates from human genome-wide association studies of type 2 diabetes and related metabolic traits identifies tissue-specific roles for dHHEX. BMC Genomics. 2013;14(1). [DOI] [PMC free article] [PubMed]
- 14.Palanker Musselman L, Fink JL, Narzinski K, Ramachandran PV, Sukumar Hathiramani S, Cagan RL, et al. A high-sugar diet produces obesity and insulin resistance in wild-type Drosophila. Dis Model Mech. 2011;4(6):842–849. doi: 10.1242/dmm.007948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pineda C, Fuentes-Gómez AJ, Hernández-Díaz C, Zamudio-Cuevas Y, Fernández-Torres J, López-Macay A, et al. Animal model of acute gout reproduces the inflammatory and ultrasonographic joint changes of human gout. Arthritis Res Ther. 2015;17:37. doi: 10.1186/s13075-015-0550-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448(7150):151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 17.Kanbay M, Jensen T, Solak Y, Le M, Roncal- C, Rivard C, et al. Uric acid in metabolic syndrome: from an innocent bystander to a central player. Eur J Intern Med. 2016;29:3–8. doi: 10.1016/j.ejim.2015.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Battelli MG, Bortolotti M, Polito L, Bolognesi A. The role of xanthine oxidoreductase and uric acid in metabolic syndrome. Biochim Biophys Acta - Mol Basis Dis. 2018;1864(8):2557–2565. doi: 10.1016/j.bbadis.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 19.Shen C, Guo Y, Luo W, Lin C, Ding M. Serum urate and the risk of Parkinson’s disease: results from a meta-analysis. Can J Neurol Sci. 2013. [DOI] [PubMed]
- 20.Pu Z, Xu W, Lin Y, He J, Huang M. Oxidative stress markers and metal ions are correlated with cognitive function in Alzheimer’s disease. Am J Alzheimers Dis Other Demen. 2017;32(6):353–359. doi: 10.1177/1533317517709549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Y, Zhao M, Pu Z, Xu G, Li X. Relationship between oxidative stress and inflammation in hyperuricemia analysis based on asymptomatic young patients with primary hyperuricemia. Medicine (United States) 2018;97(49):1–8. doi: 10.1097/MD.0000000000013108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joosten LAB, Johnson RJ. Leo A. B. Joosten *, Tania O. Cri. 2020;16(February).
- 23.Labat-Robert J, Robert L. Longevity and aging. Role of free radicals and xanthine oxidase. A review. Pathol Biol. 2014;62(2):61–6. 10.1016/j.patbio.2014.02.009. [DOI] [PubMed]
- 24.Battelli MG, Polito L, Bortolotti M, Bolognesi A. Xanthine oxidoreductase-derived reactive species: physiological and pathological effects. Oxid Med Cell Longev. 2016;2016. [DOI] [PMC free article] [PubMed]
- 25.Lang S, Hilsabeck TA, Wilson KA, Sharma A, Bose N, Brackman DJ, et al. A conserved role of the insulin-like signaling pathway in diet-dependent uric acid pathologies in Drosophila melanogaster. PLoS Genet. 2019. [DOI] [PMC free article] [PubMed]
- 26.Dow JAT, Romero MF. Drosophila provides rapid modeling of renal development, function, and disease. Am J Physiol - Ren Physiol. 2010;299(6). [DOI] [PMC free article] [PubMed]
- 27.Chung VY, Konietzny R, Charles P, Kessler B, Fischer R, Turney BW. Proteomic changes in response to crystal formation in Drosophila Malpighian tubules. Fly (Austin) 2016;10(2):91–100. doi: 10.1080/19336934.2016.1171947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sayer JA. Progress in understanding the genetics of calcium-containing nephrolithiasis. J Am Soc Nephrol. 2017;28(3):748–759. doi: 10.1681/ASN.2016050576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knauf F, Preisig PA. Drosophila: a fruitful model for calcium oxalate nephrolithiasis. Kidney Int. 2011;80(4):327–329. doi: 10.1038/ki.2011.166. [DOI] [PubMed] [Google Scholar]
- 30.Hirata T, Cabrero P, Berkholz DS, Bondeson DP, Ritman EL, Thompson JR, et al. In vivo Drosophilia genetic model for calcium oxalate nephrolithiasis. Am J Physiol - Ren Physiol. 2012;303(11):1555–1562. doi: 10.1152/ajprenal.00074.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chi T, Kim MS, Lang S, Bose N, Kahn A, Flechner L, et al. A Drosophila model identifies a critical role for zinc in mineralization for kidney stone disease. PLoS One. 2015;10(5). [DOI] [PMC free article] [PubMed]
- 32.Chen YH, Liu HP, Chen HY, Tsai FJ, Chang CH, Lee YJ, et al. Ethylene glycol induces calcium oxalate crystal deposition in Malpighian tubules: a Drosophila model for nephrolithiasis/urolithiasis. Kidney Int. 2011;80(4):369–377. doi: 10.1038/ki.2011.80. [DOI] [PubMed] [Google Scholar]
- 33.MacKay TFC, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, et al. The Drosophila melanogaster Genetic Reference Panel. Nature. 2012. [DOI] [PMC free article] [PubMed]
- 34.Jin K, Wilson KA, Beck JN, Nelson CS, Brownridge GWGW, Harrison BR, et al. Genetic and metabolomic architecture of variation in diet restriction-mediated lifespan extension in Drosophila. PLoS Genet. 2020;16(7):1–22. doi: 10.1371/journal.pgen.1008835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chow CY, Wolfner MF, Clark AG. Using natural variation in Drosophila to discover previously unknown endoplasmic reticulum stress genes. Proc Natl Acad Sci [Internet]. 2013;110(22):9013–8. Available from: http://www.pnas.org/cgi/doi/10.1073/pnas.1307125110 [DOI] [PMC free article] [PubMed]
- 36.Nègre N, Brown CD, Ma L, Bristow CA, Miller W, Wagner U, et al. NIH Public Access. 2011;471(7339):527–531. doi: 10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slattery M, Ma L, Spokony RF, Arthur RK, Kheradpour P, Kundaje A, et al. Diverse patterns of genomic targeting by transcriptional regulators in Drosophila melanogaster. Genome Res. 2014;24(7):1224–1235. doi: 10.1101/gr.168807.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ni X, Zhang YE, Nègre N, Chen S, Long M, White KP. Adaptive evolution and the birth of CTCF binding sites in the Drosophila genome. PLoS Biol. 2012. [DOI] [PMC free article] [PubMed]
- 39.Gregan J, Rabitsch PK, Sakem B, Csutak O, Latypov V, Lehmann E, et al. Novel genes required for meiotic chromosome segregation are identified by a high-throughput knockout screen in fission yeast. Curr Biol. 2005;15(18):1663–1669. doi: 10.1016/j.cub.2005.07.059. [DOI] [PubMed] [Google Scholar]
- 40.Nagase T, Nakayama M, Nakajima D, Kikuno R, Ohara O. Prediction of the coding sequences of unidentified human genes. XX. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 2001;8(2):85–95. [DOI] [PubMed]
- 41.Bandyopadhyay S, Chiang C-Y, Srivastava J, Gersten M, White S, Bell R, et al. A human MAP kinase interactome HHS public access. Nat Methods [Internet]. 2010;7(10):801–5. Available from: http://www.nature.com/authors/editorial_policies/license.html#terms. [DOI] [PMC free article] [PubMed]
- 42.Akagi K, Wilson KA, Katewa SD, Ortega M, Simons J, Hilsabeck TA, et al. Dietary restriction improves intestinal cellular fitness to enhance gut barrier function and lifespan in D. melanogaster. PLoS Genet. 2018. [DOI] [PMC free article] [PubMed]
- 43.Wilson KA, Beck JN, Nelson CS, Hilsabeck TA, Promislow D, Brem RB, et al. GWAS for lifespan and decline in climbing ability in flies upon dietary restriction reveal decima as a mediator of insulin-like peptide production. Curr Biol. 2020;30(14):2749–2760.e3. doi: 10.1016/j.cub.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma A, Akagi K, Pattavina B, Wilson KA, Nelson C, Watson M, et al. Musashi expression in intestinal stem cells attenuates radiation-induced decline in intestinal permeability and survival in Drosophila. Sci Rep. 2020;10(1):1–16. doi: 10.1038/s41598-020-75867-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katewa SD, Akagi K, Bose N, Rakshit K, Camarella T, Zheng X, et al. Peripheral circadian clocks mediate dietary restriction-dependent changes in lifespan and fat metabolism in Drosophila. Cell Metab. 2016;23(1):143–154. doi: 10.1016/j.cmet.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelson CS, Beck JN, Wilson KA, Pilcher ER, Kapahi P, Brem RB. Cross-phenotype association tests uncover genes mediating nutrient response in Drosophila. BMC Genomics. 2016;17(1). [DOI] [PMC free article] [PubMed]
- 47.Seabold, S., Perktold J. statsmodels: econometric and statistical modeling with python. 9th Python in Science Conference; 2010.
- 48.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, et al. Skyline-an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26(7):966–8. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson KA, Chamoli M, Hilsabeck TA, Pandey M, Bansal S, Chawla G, et al. Evaluating the beneficial effects of dietary restrictions: a framework for precision nutrigeroscience. Cell Metab. 2021;33(11):2142–2173. doi: 10.1016/j.cmet.2021.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luis NM, Wang L, Ortega M, Deng H, Katewa SD, Li PWL, et al. Intestinal IRE1 is required for increased triglyceride metabolism and longer lifespan under dietary restriction. Cell Rep. 2016;17(5):1207–1216. doi: 10.1016/j.celrep.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Friedman TB, Polanco GE, Appold JC, Mayle JE. On the loss of uricolytic activity during primate evolution-I. Silencing of urate oxidase in a hominoid ancestor. Comp Biochem Physiol -- Part B Biochem. 1985. [DOI] [PubMed]
- 52.Oda M, Satta Y, Takenaka O, Takahata N. Loss of urate oxidase activity in hominoids and its evolutionary implications. Mol Biol Evol. 2002;19(5):640–653. doi: 10.1093/oxfordjournals.molbev.a004123. [DOI] [PubMed] [Google Scholar]
- 53.Johnson RJ, Titte S, Cade JR, Rideout BA, Oliver WJ. Uric acid, evolution and primitive cultures. Semin Nephrol. 2005;25(1):3–8. doi: 10.1016/j.semnephrol.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 54.Johnson RJ, Andrews P, Benner S a, Oliver W. Theodore E. Woodward award. The evolution of obesity: insights from the mid-Miocene. Trans Am Clin Climatol Assoc [Internet]. 2010;121:295–305; discussion 305–8. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2917125&tool=pmcentrez&rendertype=abstracthttp://www.ncbi.nlm.nih.gov/pmc/articles/PMC2917125/pdf/tacca121000295.pdf. [PMC free article] [PubMed]
- 55.Zhu Y, Pandya BJ, Choi HK. Prevalence of gout and hyperuricemia in the US general population: The National Health and Nutrition Examination Survey 2007–2008. Arthritis Rheum [Internet]. 2011;63(10):3136–41. Available from: http://onlinelibrary.wiley.com/doi/10.1002/art.30520/abstracthttp://onlinelibrary.wiley.com/doi/10.1002/art.30520/abstract;jsessionid=B514A57E9629D361D47B62BBBDFB0A7F.f04t03http://onlinelibrary.wiley.com/store/10.1002/art.30520/asset/30520_ftp.pdf [DOI] [PubMed]
- 56.Ashpole NM, Logan S, Yabluchanskiy A, Mitschelen MC, Yan H, Farley JA, et al. IGF-1 has sexually dimorphic, pleiotropic, and time-dependent effects on healthspan, pathology, and lifespan. GeroScience. 2017;39(2):129–145. doi: 10.1007/s11357-017-9971-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24(50):7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- 58.Tsuchiya K, Tanaka J, Shuiqing Y, Welch CL, Depinho RA, Tabas I, et al. FoxOs integrate pleiotropic actions of insulin in vascular endothelium to protect mice from atherosclerosis. Cell Metab. 2012;15(3):372–381. doi: 10.1016/j.cmet.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bateman A, Martin MJ, Orchard S, Magrane M, Agivetova R, Ahmad S, et al. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49(D1):D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Honda T, Shimizu K, Kawakatsu T, Fukuhara A, Irie K, Nakamura T, et al. Cdc42 and Rac small G proteins activated by trans- interactions of nectins are involved in activation of c-Jun N-terminal kinase, but not in association of nectins and cadherin to form adherens junctions, in fibroblasts. Genes Cells. 2003;8(5):481–491. doi: 10.1046/j.1365-2443.2003.00649.x. [DOI] [PubMed] [Google Scholar]
- 61.Ooshio T, Irie K, Morimoto K, Fukuhara A, Imai T, Takai Y. Involvement of LMO7 in the association of two cell-cell adhesion molecules, nectin and E-cadherin, through afadin and α-actinin in epithelial cells. J Biol Chem [Internet]. 2004;279(30):31365–73. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0021925818361945. [DOI] [PubMed]
- 62.Asada M, Irie K, Morimoto K, Yamada A, Ikeda W, Takeuchi M, et al. ADIP, a novel afadin- and α-actinin-binding protein localized at cell-cell adherens junctions. J Biol Chem. 2003;278(6):4103–4111. doi: 10.1074/jbc.M209832200. [DOI] [PubMed] [Google Scholar]
- 63.Tsurumi H, Kurihara H, Miura K, Tanego A, Ohta Y, Igarashi T, et al. Afadin is localized at cell-cell contact sites in mesangial cells and regulates migratory polarity. Lab Investig. 2016;96(1):49–59. doi: 10.1038/labinvest.2015.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.An S, Kumar R, Sheets ED, Benkovic SJ. Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science. 2008;320(5872):103–6. doi: 10.1126/science.1152241. [DOI] [PubMed] [Google Scholar]
- 65.Hong Z, Frencha JB, Fangc Y, Benkovic SJ. The purinosome, a multi-protein complex involved in the de novo biosynthesis of purines in humans. Chem Commun. 2013;49(40). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.