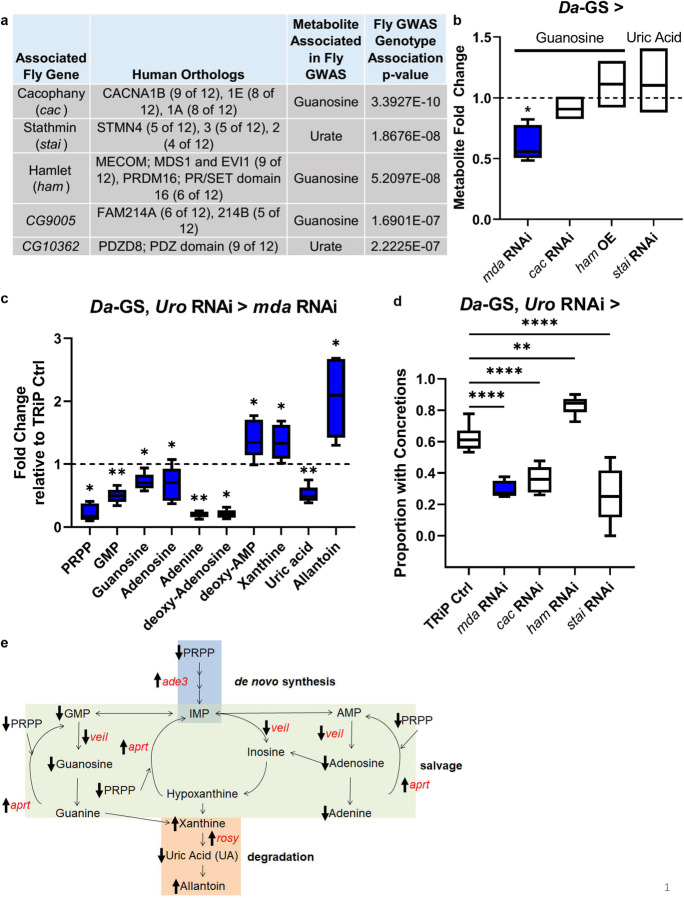
Fig. 1.
Whole-body mda RNAi reduces guanosine levels in wild-type and alters purine metabolism in model, preventing increased uric acid levels, and rescuing the concretion phenotype. a Purine GWAS candidate genes and associated metabolites. b Fold change of associated metabolite with inhibition (RNAi) or over expression (OE) of candidate gene in wild-type background compared to uninhibited isogenic control flies fed food with ethanol instead of RU486 (indicated by dashed line). Fly strains used, each crossed with Da-GS: mda RNAi—BL33362, cac RNAi—v104168/kk, ham OE—BL22209, and stai RNAi—BL53925. n = 5 biological replicates of 5 flies per condition. c Purine metabolites with significant fold changes in model with concurrent mda RNAi (BL33362) compared to background control, each crossed with Da-GS, uro RNAi. (TRiP empty vector control strain BL35785 represented by dashed black line.) d Proportion of flies with concretions in flies with Da-GS, uro RNAi crossed with TRiP BL35785 control or with concurrent candidate gene RNAi. Fly strains used same as in b, except ham RNAi—BL22209 (number of flies from left to right, n = 71, 38, 82, 73, and 43). Multiple comparisons made using Dunnett’s one-way ANOVA, with direct comparisons using two-tailed Student’s t test for first repetition, and one-tailed for subsequent repeats, in excel, with raw data and all repeats in Online Resource 23 and Table S4. e Model of purine metabolism, with metabolites in black and genes significantly altered with concurrent mda RNAi (BL33362) in the Da-GS, uro RNAi model in red, with arrows indicating direction of metabolite level or expression change. Mated females used for all experiments due to stronger phenotypic responses. Comparisons made using Student’s t test unless otherwise indicated. Significant differences for all tests are indicated by *: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Whiskers on boxplots drawn to largest or smallest values if within 1.5 times inter-quartile distance