Abstract
Improvement of serologic assays for detection of antibodies against human herpesvirus 8 (HHV-8) is critical to better understand its epidemiology and biology. We produced the HHV-8 latent (ORF73) and lytic (ORF65, K8.1, and glycoprotein B) antigens in the Semliki Forest virus system and evaluated their performance in immunofluorescence assays (IFAs) and enzyme-linked immunosorbent assays (ELISAs). These assays were compared with other latent antigen-based assays, including an IFA based on primary effusion lymphoma (PEL) cells and an ELISA based on bacterially expressed ORF73 antigen, as well as with other lytic antigen-based assays, including an IFA based on induced PEL cells, a commercial ELISA based on purified virions, and ELISAs based on K8.1- and ORF65-derived oligopeptides. We used a panel of 180 serum specimens obtained from three groups expected to have high, intermediate, and low HHV-8 prevalences. Using three different evaluation methods, we found that (i) the performances of the lytic antigen-based ELISAs were almost equivalent, (ii) the lytic antigen-based assays were more sensitive than the latent antigen-based assays, and (iii) in general, IFAs were more sensitive than ELISAs based on the same open reading frame. We also found that serum specimens from healthy individuals contained antibodies cross-reactive with HHV-8 glycoprotein B that can potentially cause false-positive reactions in lytic PEL-based IFAs. Although this is not a substantial problem in most epidemiologic studies, it may confound the interpretation of data in studies that require high assay specificity. Because the K8.1-based IFA provides sensitivity similar to that of lytic PEL-based IFAs and improved specificity, it can be a useful alternative to the PEL-based IFAs.
Human herpesvirus 8 (HHV-8) was identified in AIDS-associated Kaposi's sarcoma (KS) by representational difference analysis (7). Subsequent studies demonstrated the association of this virus with all four forms of KS, primary effusion lymphoma (PEL), and multicentric Castleman's disease (reviewed in references 3, 37, and 38). Both sexual and nonsexual avenues of transmission of HHV-8 have been identified. Among men who have sex with men (MSM), HHV-8 is predominantly sexually transmitted (24, 26). Transmission through transplantation and intravenous drug use has been reported (6, 15, 22, 30, 35). It is likely that HHV-8 primary infection also occurs during childhood in areas of endemicity (5, 25, 33). Because HHV-8 is frequently secreted into saliva in MSM (2, 4,18, 32, 41), it may be transmitted from saliva as is Epstein-Barr virus (EBV).
Since HHV-8 is not readily isolated in cell culture, infection is usually determined by either PCR or serology. Although PCR detects HHV-8 DNA in almost all KS lesions, viral DNA loads in peripheral blood mononuclear cells are frequently not high enough to be detected (29, 39, 40, 42). To complement PCR and to better understand the natural history of HHV-8 infection, several serologic assays have been established. PEL cell lines harboring the HHV-8 genome were used as antigens in immunofluorescence assay (IFA) and immunoblotting (IB) (12, 20, 28, 39). HHV-8 ORF73 is the major component of latent nuclear antigens detected in these latent PEL-based assays. Seropositivity determined by these assays correlated well with KS development (17, 24). After treatment with 12-O-tetradecanoylphorbol-13-acetate (TPA), 10 to 30% of PEL cells produce cytoplasmic antigens that correspond to viral proteins associated with lytic infection. Although IFAs using TPA-treated PEL cells (lytic PEL-based IFAs) are the most sensitive assays (20) and have been used widely, they are possibly confounded by cross-reaction with antibodies against other herpesviruses. The lack of a matched HHV-8-negative PEL cell line hinders discrimination of nonspecific reactions against cellular components. We recently developed new IFAs based on HHV-8 antigens expressed with the recombinant Semliki Forest virus (rSFV) system (14). Expression of HHV-8 antigens at a high level and the availability of true negative controls in this system increased the specificity and sensitivity of the IFAs. IFA using rSFV K8.1-infected BHK21 cells (K8.1SFV IFA) was as sensitive as lytic PEL-based IFA, and the correlation coefficient of end-point titers and concordance between these two assays were >0.92 and 97% (κ = 0.93), respectively. Because K8.1 has no homolog in other herpesviruses, there is no potential cross-reaction with antibodies against other herpesviruses in this assay.
To obtain a high-throughput assay, an enzyme-linked immunosorbent assay (ELISA) based on purified HHV-8 virions was established (9), and it is commercially available. To increase sensitivity and specificity, several studies have examined the utility of defined HHV-8 antigens, including synthetic oligopeptides and recombinant proteins. The most useful antigens for serologic assays are derived from ORF65, ORF73, and K8.1 (13, 16, 19, 31, 39, 40, 43).
Assay performance greatly influences measured seroprevalence in low-risk populations. For example, seroprevalence in healthy populations in the United States has ranged from 0% in latent antigen-based assays to 20% in lytic PEL-based IFAs (reviewed in reference 8). A blinded comparison of three ELISAs and four PEL-based IFAs demonstrated their utility for epidemiologic investigations and limitations for diagnosing individuals (34). This made clear the need for both new assay development and standardization. Because the lack of a “gold standard” assay for HHV-8 has hampered comparison of assays newly developed in different laboratories, it is important to compare assays by using a single set of defined serum samples (23).
In this study, we developed rSFV-based ELISAs and compared them with previously reported assays. We also studied the specificity of the lytic PEL-based IFA.
MATERIALS AND METHODS
Plasmids.
pSCAx, pSCAβ, pSCA-K8.1A, pSCA-orf73Δ, and pSCA-Helper were described previously (14). pSCA-orf65 was constructed by PCR amplification of the orf65 gene with primers 5′-GATGAGGATCCTTCCAACTTTAAGGTGAGAGAC-3′ and 5′-CGCGGATCCAATCGTTGCCTATTT-3′ (orf65-derived sequences are underlined, and BamHI sequences are in boldface), followed by cloning into pSCAx. pSCA-gB, which carries the entire HHV-8 glycoprotein B (gB) open reading frame in pSCAx, was constructed from pcDNA3.1-gB1b-4 (1). For the expression of amino and carboxyl domains of ORF73, pBAD-orf73N and -orf73C were constructed by cloning fragments from the orf73 gene encoding amino acids 1 to 330 and 935 to 1162, respectively, into pBAD-Topo (Invitrogen, Carlsbad, Calif.). To obtain these fragments, the following primer sets were used: for orf73N, 5′-GACGACGACAAGATGGCGCCCCCGGGAATGCGCCTGAGG-3′ and 5′-CTGACTTTCCTTGCTAATCTC-3′; for orf73C, 5′-GACGACGACAAGGAGCCCATAATCTTGACAGGGTCGTCA-3′ and 5′-TTAATGATGATGATGATGATGTGTCATTTCCTGTGGAGAGTCC-3′ (orf73-derived sequences are underlined, and the His6 tag sequence is in italics).
rSFV production, IFAs, and IB.
Transfection, production, and infection of rSFV, IFAs using rSFV-infected cells, and IB were performed as described previously (14), except that rSFV infection of BHK21 cells for preparation of ELISA antigens was done at a multiplicity of infection of 0.5 to 1.0. Monoclonal antibody-enhanced IFAs (mIFAs) using untreated and TPA-induced BCBL-1 cells (latent and lytic mIFAs, respectively) were performed as described previously (20). Human serum specimens were coded, and two readers read the IFA slides independently. Antibodies against HHV-8 ORF65 were obtained from rabbits immunized with a bacterially expressed ORF65 protein (40). Antibodies against EBV gp110 (gB) and HHV-8 gB, ORF73, and K8.1 were described previously (1, 14, 36).
rSFV-based ELISAs.
BHK-21 cells infected with rSFV in one 10-cm-diameter dish were lysed in 2 ml of phosphate-buffered saline (PBS) containing 0.5% sodium deoxycholate (Dox) until they detached from the plate (≈10 min). The lysed cells were collected, sonicated twice for 1 min each (50% duty cycle, power 10; model W-375; Heat Systems-Ultrasonic, Inc., Farmingdale, N.Y.), and then centrifuged (10 min, 16,000 × g) to remove cell debris. Supernatants containing HHV-8 antigens and β-galactosidase were used as antigens for ELISA.
rSFV-based ELISAs were performed as follows. Antigens were diluted in carbonate buffer (0.1 M Na2HCO3-NaH2CO3, pH 9.4) to a final concentration of 3 μg/ml; 100 μl of the solution was incubated in each well of ELISA plates (Immulon 2HB; Dynex) at 4°C overnight. After coating, the plates were washed once with PBS and incubated with PBS containing 5% skim milk at 4°C for 2 h. Sera were diluted in PBS–5% skim milk with a 30-μg/ml concentration of a BHK-21 cell extract that was prepared in the same manner as the extracts from rSFV-infected cells, incubated with agitation at room temperature (RT) for 2 h to reduce cross-reaction with cellular components, and then reacted in the ELISA plate for 2 h at 37°C. After washing with PBS containing 0.05% Tween 20 (PBST), bound antibodies were incubated with peroxidase-conjugate goat anti-human immunoglobulin G (Kirkegaard & Perry Laboratories, Gaithersburg, Md.) for 1 h. The color reaction was developed for 1 h at RT with tetramethylbenzidine after PBST washing. Reactions were stopped with 50 μl of 1 M H2SO4. Plates were read at 450 nm against 630 nm with an ELISA plate reader. Optical density (OD) values specific to HHV-8 antigens were obtained by subtracting the OD values for the β-galactosidase extracts from those for the HHV-8 antigens.
Bacterial expression of ORF73.
Bacteria containing pBAD-orf73N or pBAD-orf73C were grown to mid-log phase. ORF73 expression was induced by addition of l-(+)-arabinose (final concentration of 0.05%) for 3 h. Cells from 1 liter of culture were harvested and sonicated. After centrifugation (27,000 × g for 1 h), the supernatant was passed through a 0.4-μm-pore-size filter and mixed with a 1.5-ml bed volume of Talon resin (Clontech, Palo Alto, Calif.). ORF73 antigens were eluted with a buffer containing 0.5 M imidazole and dialyzed against carbonate buffer. About 50 μg of partially purified antigens with ≈90% purity was obtained per liter of culture.
ELISA using bacterially expressed ORF73 as antigen (ORF73bac ELISA).
Microtiter plate wells were coated with 60 ng of partially purified antigen (4°C, overnight) and then blocked with 5% skim milk in PBST for 1 h at RT. Serum specimens were diluted 1:100 and applied to wells. Plates were incubated at RT for 2 h. Secondary antibody and color reactions were done as described for rSFV-based ELISAs.
Virion ELISA.
An ELISA kit using purified HHV-8 virions was purchased from a commercial source (Advanced Biotechnologies, Inc. Columbia, Md.) and used according to the manufacturer's instructions.
Human sera.
Serum specimens analyzed in two previous studies (14, 40) were used. The collection included sera from 56 human immunodeficiency virus (HIV)-positive, KS-positive (HIV+ KS+) patients; 61 HIV+ KS− patients; 3 HIV− KS+ patients, and 10 HIV− individuals with some risk factors, such as MSM (n = 2) and patients attending sexually transmitted disease clinics (n = 8). Fifty serum specimens obtained as out-of-date material from the Atlanta American Red Cross blood bank were used as healthy controls. Among the 61 HIV+ KS− patients, 13 developed KS within 5 years (Later KS), 25 did not develop KS in the 5 years after specimen collection, and 23 had no follow-up study. We refer to the HIV+ KS+, HIV− KS+, and HIV+ Later KS patients as the KS group (n = 72); the HIV+ KS− patients and individuals with risk factors as the intermediate group (n = 58); and blood donors as the control group (n = 50).
Statistical analyses.
Correlations between titers obtained by two different assays were evaluated by Pearson's correlation coefficient. The degree of concordance between two assays was assessed by the kappa statistic, which measures agreement beyond chance. Kappa (κ) equals 1.0 for perfect agreement; 0.4 to 0.6 and 0.6 to 0.8 were considered to represent fair and good agreements, respectively. Maximum-likelihood estimates of the parameters for receiver-operating characteristic (ROC) and drawing of ROC curves were obtained with ROCKIT (0.9B, IBM-compatible version) (27) and PlotROC software, respectively. The software was downloaded from a website of the Department of Radiology, University of Chicago (http://www-radiology.uchicago.edu/krl/top page11.htm). The area under an ROC curve equals the probability of correctly answering the two-alternative forced-choice problem. In other words, the area is proportional to the quality of the performance (accuracy) of a diagnostic test; an area of 0.5 represents a worthless test, and an area of 1.0 represents a perfect test.
RESULTS
Expression of HHV-8 antigens.
Expression of ORF65 and gB in the rSFV system was examined by IB and IFA (Fig. 1 and 2). Although ORF65 protein was barely detectable in TPA-induced BCBL-1 cells, it was easily detected as a band with an apparent molecular mass of 20 kDa both in 293T cells transfected with pSCA-orf65 and in BHK-21 cells infected with rSFV-orf65 (Fig. 1A). The molecular mass was consistent with that in a prior report (21). Although the rabbit antibody detected nonspecifically a band of ≈30 kDa (Fig. 1A, lanes 3 to 6), KS+ human sera did not detect the band (data not shown). Cytoplasmic localization of ORF65 was observed in the rSFV-infected cells (Fig. 2A and C). Two forms of gB, corresponding to the full-length and cleaved forms (1), were detected in 293T cells transfected with pSCA-gB and in BHK-21 cells infected with rSFV-gB as well as in TPA-induced BCBL-1 cells (Fig. 1B). HHV-8 gB was detected in the cytoplasm and on cell surfaces of BHK-21 cells infected with rSFV-gB (Fig. 2D and F) (data with paraformaldehyde fixation not shown), which was consistent with its expression in BCBL-1 cells (1). As shown here and previously (14), the expression levels of every HHV-8 antigen in rSFV-infected BHK-21 cells were significantly higher than in those in plasmid-transfected 293T cells or in TPA-induced BCBL-1 cells.
FIG. 1.
rSFV expression of HHV-8 ORF65 and gB. Cell lysates obtained from equal numbers (5 × 104 and 4 × 103 cells/lane in panels A and B, respectively) of the following cells were separated by sodium dodecyl sulfate–15% (A) or –8% (B) polyacrylamide gel electrophoresis: BCBL-1 cells untreated (lanes 1) or treated with TPA (lanes 2); BHK-21 cells infected with rSFV-βgal (lanes 3), rSFV-orf65 (lane 4 in panel A), or rSFV-gB (lane 4 in panel B); and 293T cells transfected with pSCA-βgal (lanes 5), pSCA-orf65 (lane 6 in panel A), or pSCA-gB (lane 6 in panel B). ORF65 and gB proteins were detected with rabbit antibodies against a bacterially expressed ORF65 protein (α-ORF65) and against an oligopeptide containing gB amino acids 828 to 845 (α-gB), respectively.
FIG. 2.
Localization of HHV-8 ORF65, and gB in rSFV-infected cells. BHK-21 cells infected with rSFV-ORF65 (A and C) and with rSFV-gB (D and F) were reacted with the same rabbit antibodies described for Fig. 1 (A, B, D, and E) and with KS+ human sera (C and F). BHK-21 cells infected with rSFV-βgal were used as a negative control (B and E).
The amino (ORF73N) and carboxyl (ORF73C) domains of ORF73 were expressed in Escherichia coli and affinity purified (data not shown). Because all KS+ human sera that reacted with ORF73C reacted with ORF73N in an ELISA, ORF73C was used for further assay comparisons.
Optimization of rSFV-based ELISAs.
To optimize antigen preparation from rSFV-infected cells, we tested various solubilization detergents. Of 17 detergents tested, Dox reproducibly resulted in the highest signal-to-noise ratio in an ELISA format (data not shown). Lysis of cells directly in the culture dish with Dox-containing PBS gave more consistent and efficient antigen preparation than harvesting the cells by trypsinization and subsequent Dox treatment (data not shown). The HHV-8 antigens were found mainly in the soluble fractions of the lysates (K8.1, >95%; ORF73, 85%). Therefore, the soluble fractions prepared from the lysates were used as the ELISA antigens.
Because an amount of coating antigen of greater than 300 ng per well led to signal saturation, this amount (equivalent to 2 × 103 rSFV-infected cells) was used for subsequent experiments. Twenty 96-well ELISA plates can be prepared from a single 10-cm-diameter culture dish of cells.
Comparison of the assays.
The following 11 assays were compared: IFAs using untreated and TPA-treated BCBL-1 cells (latent and lytic mIFAs), rSFV-based IFAs (K8.1SFV, ORF73SFV, and ORF65SFV IFAs), a commercial ELISA using HHV-8 virions (virion ELISA), ELISAs using oligopeptides with major immunogenic epitopes of ORF65 and K8.1 (ORF65pep and K8.1pep ELISAs), rSFV-based ELISAs (ORF73SFV and K8.1SFV ELISAs), and ORF73bac ELISA. Among these 11 assays, latent mIFA, ORF73SFV IFA and ELISA, and ORF73bac ELISA are latent antigen-based assays, and the others are lytic antigen-based assays. Human serum specimens from three groups, i.e., the KS, intermediate, and control groups, were used for the assay comparison. The prevalence of HHV-8 infection in these groups was expected to be very high, intermediate, and very low, respectively.
The results of assay comparisons were analyzed first in the context of interassay agreement and then in comparison to defined standards as described below.
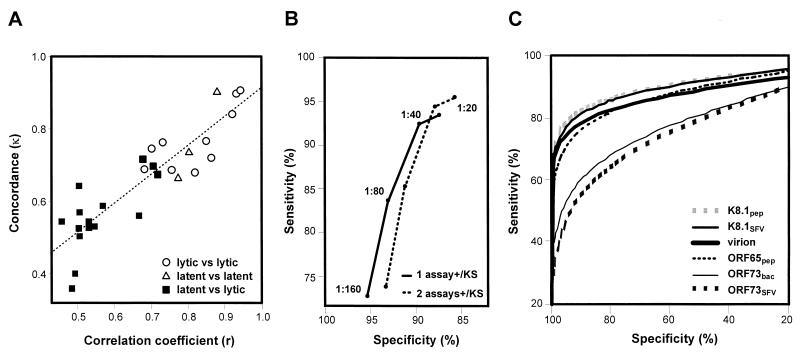
Pairwise interassay agreements were evaluated in terms of two statistical values: the correlation coefficient (r) of OD measurements (or IFA end-point titers) between two assays and the concordance kappa statistic (κ) of serologic status (seropositive or seronegative) determined by two assays (Table 1). An arbitrary cutoff for each ELISA was defined as the mean plus five standard deviations (SD) of OD values obtained from a panel of healthy blood donors whose HHV-8 serologic status was negative (end-point titer of <40) in K8.1SFV IFA (n = 49). As shown in Table 1, the ORF73-based ELISAs had good correlation and concordance. The K8.1- and virion-based ELISAs also showed very good correlations and concordances, which were better than those between these assays and ORF65pep ELISA. Lower concordances between K8.1SFV IFA and the lytic antigen-based ELISAs can be explained by the different serum dilutions in ELISAs (1:100) and IFAs (starting from 1:20), because their correlations were better for the sera with IFA end-point titers of >80 (Fig. 3). Some of the ELISAs were also evaluated with 1:40 serum dilutions; increased background obscured any possible sensitivity increase (data not shown).
TABLE 1.
Correlations and concordances between two assaysa
| Antigen type | Assay | Correlation coefficient (concordance kappa values) with:
|
|||||
|---|---|---|---|---|---|---|---|
| Latent antigen-based assays
|
Lytic antigen-based assays
|
||||||
| ORF73SFV ELISA | ORF73bac ELISA | Virion ELISA | K8.1SFV ELISA | K8.1pep ELISA | ORF65pep ELISA | ||
| Latent | ORF73SFV IFA | 0.77 (0.66) | 0.80 (0.73) | 0.68 (0.72) | 0.72 (0.67) | 0.71 (0.70) | 0.50 (0.64) |
| ORF73SFV ELISA | 0.88 (0.90) | 0.50 (0.53) | 0.53 (0.54) | 0.51 (0.50) | 0.46 (0.54) | ||
| Lytic | K8.1SFV IFA | 0.82 (0.68) | 0.86 (0.71) | 0.85 (0.77) | 0.68 (0.68) | ||
| Virion ELISA | 0.94 (0.91) | 0.92 (0.84) | 0.76 (0.69) | ||||
| K8.1SFV ELISA | 0.93 (0.90) | 0.70 (0.75) | |||||
| K8.1pep ELISA | 0.73 (0.76) | ||||||
The mean plus 5 SD of OD values obtained from seronegative blood donors was defined as the arbitrary cutoff for ELISAs. Seroreactivities at serum dilutions of 1:20 and 1:40 were used as cutoffs for ORF73SFV and K8.1SFV IFAs, respectively.
FIG. 3.
Correlations between lytic antigen-based assays. IFA end-point titers or OD measurements were compared among the indicated assays. (A) K8.1SFV IFA versus virion ELISA. (B) K8.1SFV IFA versus K8.1SFV ELISA. (C) K8.1SFV ELISA versus virion ELISA. (D) K8.1SFV ELISA versus K8.1pep ELISA. Closed squares, blood donors; gray triangles, intermediate group; open circles, KS group. Broken lines indicate the arbitrary cutoffs for ELISAs (the mean plus 5 SD of OD values from a panel of 49 healthy blood donors) and the cutoff for IFAs (serum dilution of 1:40, based on the results shown in Fig. 4B). Broken lines divide a set of serum specimens into four groups: specimens in the upper right quadrant of each panel were positive in both assays, the ones in the lower left quadrant were negative in both assays, the ones in the lower right quadrant were positive in one assay (x axis) but negative in the other assay (y axis), and the ones in upper left quadrant were negative in one assay (x axis) but positive in the other assay (y axis).
Correlation coefficients are independent of cutoffs, because they are based on comparison of signal strengths. In contrast, κ values are dependent on how the cutoffs are determined, because the value represents interassay agreement of serologic status that is classified based on the cutoffs. Interestingly, the κ and r values correlated well, suggesting that arbitrary cutoff determination does not change the overall interpretation (Fig. 4A). The lowest correlations were between latent antigen- and lytic antigen-based assays (Fig. 4A; Table 1).
FIG. 4.
Interassay comparisons. (A) Correlation coefficients (r) and concordance kappa statistics (κ) between two lytic antigen-based assays (○), between two latent antigen-based assays (▵), and between latent and lytic antigen-based assays (▪). Latent antigen-based assays include ORF73SFV IFA and ORF73SFV and ORF73bac ELISAs. Lytic antigen-based assays include K8.1SFV IFA and virion, K8.1SFV, K8.1pep, and ORF65pep ELISAs. A dashed line shows a linear regression between the r and κ values. (B) Specificity and sensitivity of K8.1SFV IFA using different serum dilutions as cutoffs to determine whether each specimen is seronegative or seropositive. To calculate specificity and sensitivity, the following conditions were used as a standard: (i) seropositive in one of the four lytic antigen-based assays (virion, K8.1SFV, K8.1pep, and ORF65pep ELISAs) or KS positive (including current KS and KS developed within 5 years) (solid line) or (ii) seropositive in two of the lytic antigen-based assays or KS positive (dashed line). (C) ROC analyses of ELISAs. The serologic status of each specimen determined by K8.1SFV IFA (cutoff serum dilution of 1:40) was used as a defined standard, based on the results shown in panel B.
Next, we evaluated each assay in comparison to a defined standard in two ways. In the first, on the assumption that all KS group sera contain antibodies against HHV-8, the sensitivity of each assay was calculated as a percentage of positives among the KS group serum specimens. HHV-8 antibodies were detected in 95.8, 94.4, and 83.3% of KS group specimens by K8.1SFV IFA using cutoff serum dilutions of 1:20, 1:40, and 1:80, respectively. The lytic antigen-based ELISAs detected antibodies in 78 to 88% of KS specimens, and latent antigen-based assays identified HHV-8 infection in less than two-thirds of KS specimens, using the mean plus 5 SD of control OD values as the cutoff (Table 2). Even when using the mean plus 2 SD in place of 5 SD as the cutoff, only 64 to 65% of KS specimens were positive in ORF73-based ELISAs, while 86 to 93% were positive in the lytic antigen-based ELISAs. ORF65SFV IFA detected antibodies in only 41% of KS specimens at a serum dilution of 1:20.
TABLE 2.
Comparison of HHV-8 serologic assay performances
| Antigen type | Assay | No. of sera analyzed
|
Cutoffd | % Positivity in KS groupf | % Sensitivityef | % Specificityef | ROCe
|
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| KSa | Intermediateb | Controlc | Total | Area | SE | ||||||
| Lytic | Lytic mIFA | 29 | 31 | 30 | 100 | 1:40 | 96.6 | 95.8 | 90.5 | ||
| K8.1SFV IFA | 72 | 56 | 50 | 178 | 1:40 | 94.4 | 100 | 100 | 100 | ||
| K8.1pep ELISA | 72 | 58 | 50 | 180 | 5 SD (2 SD) | 87.5 (93.1) | 80.9 (87.2) | 96.4 (85.7) | 92.1 | 2.3 | |
| K8.1SFV ELISA | 71 | 58 | 49 | 178 | 5 SD (2 SD) | 81.7 (87.5) | 75.5 (80.9) | 97.6 (93.9) | 92.0 | 2.3 | |
| ORF65pep ELISA | 72 | 58 | 50 | 180 | 5 SD (2 SD) | 80.6 (91.7) | 73.4 (86.2) | 95.2 (75.0) | 88.9 | 2.6 | |
| Virion ELISA | 72 | 58 | 50 | 180 | 5 SD (2 SD) | 77.8 (86.1) | 70.2 (80.9) | 98.8 (91.7) | 89.7 | 2.6 | |
| Latent | ORF73SFV IFA | 72 | 57 | 50 | 179 | 1:40 | 63.9 | 54.3 | 100 | ||
| Latent mIFA | 29 | 31 | 30 | 100 | 1:40 | 62.1 | 51.1 | 100 | |||
| ORF73bac ELISA | 72 | 58 | 50 | 180 | 5 SD (2 SD) | 52.8 (63.9) | 42.6 (53.2) | 98.8 (94.0) | 78.7 | 3.4 | |
| ORF73SFV ELISA | 66 | 58 | 49 | 173 | 5 SD (2 SD) | 50.0 (65.2) | 41.6 (53.9) | 96.1 (92.7) | 78.6 | 3.5 | |
| Lytic | ORF65SFV IFA | 29 | 31 | 29 | 99 | 1:20 | 41.1 | 35.4 | 100 | ||
KS group (HIV+ KS+, HIV− KS+, and HIV+ Later KS).
Intermediate group (HIV+ KS− and HIV− with risk factors.
Control group (healthy blood donors).
The mean plus 5 SD (or 2 SD) of OD values obtained from seronegative blood donors was defined as the arbitrary cutoff for ELISAs. A serum dilution of 1:40 was used as the cutoff for IFAs except ORF65SFV IFA. Because ORF65SFV IFA was performed only at a serum dilution of 1:20, this dilution was used as its cutoff.
Serologic status determined by K8.1SFV IFA at a serum dilution 1:40 was used as a standard for calculation, based on the results shown in Fig. 4B.
Values in parentheses were obtained using the 2-SD cutoff.
Alternatively, serologic status determined by K8.1SFV IFA was used as a standard to evaluate each assay, because K8.1SFV IFA is the most sensitive assay that uses a defined antigen. First, we employed ROC-type analysis to identify a cutoff that maximizes both sensitivity and specificity of K8.1SFV IFA. For this purpose, serologic results obtained by K8.1SFV IFA at different serum dilutions were compared with the combined results obtained from four different lytic antigen-based assays (virion, K8.1SFV, K8.1pep, and ORF65pep ELISAs), plus information on whether the specimen was obtained from the KS group (KS or Later KS) patients. As shown in Fig. 4B, the cutoff that gave the best combination of sensitivity and specificity (shortest distance to the top left corner) was a serum dilution of 1:40 when applying the standards of either (i) being positive in at least one of the four assays or in the KS group or (ii) being positive in at least two of the assays or in the KS group. Then, using the serologic status determined by K8.1SFV IFA at a serum dilution of 1:40 as the defined standard, the sensitivity and specificity of each assay were calculated. As shown in Table 2, the lytic antigen-based ELISAs, the latent antigen-based IFAs, the latent antigen-based ELISAs, and ORF65SFV IFA had sensitivities of 70 to 81%, 51 to 54%, 42 to 43%, and 35%, respectively, with >95% specificity, using the mean plus 5 SD of control OD values as the cutoff for the ELISAs. Even when using 2 SD in place of 5 SD for the cutoff, the ORF73-based ELISAs still had only 53 to 54% sensitivity. The order of sensitivities of the assays was similar to that determined by the positivity in the KS group based on each cutoff setting.
Finally, the ELISAs were analyzed by ROC analysis using the serologic status determined by K8.1SFV IFA as the standard (Fig. 4C). ROC analysis eliminates bias due to the arbitrarily determined cutoffs. Areas under the ROC curves of K8.1-based ELISAs, virion and ORF65pep ELISAs, and ORF73-based ELISAs were 92%, 89 to 90%, and 79%, respectively, with the values for ORF73-based ELISAs being significantly lower than the others (Table 2).
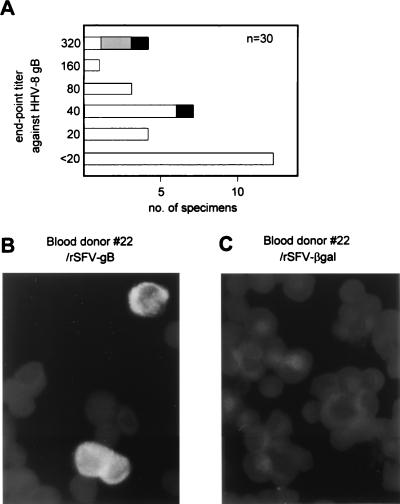
Antibodies recognizing HHV-8 gB in healthy blood donors.
The prevalence of antibodies against HHV-8 gB was determined by gBSFV IFA. While only 52% of the KS+ specimens were positive in this assay, other groups, including healthy controls, had similar levels of anti-HHV-8 gB seroprevalence. We determined the gBSFV IFA end-point titers of 30 blood donor specimens. Approximately 25% had titers of ≥80 (Fig. 5A; an example of IFA with a blood donor specimen is shown in Fig. 5B), although only four specimens were positive in at least one of the 11 other HHV-8 serologic assays. Interestingly, three out of four specimens that had end-point titers of 320 were positive in lytic mIFA, with only one of them being positive in K8.1SFV IFA. Anti-EBV VCA antibodies in most blood donor specimens (27 out of 30) were positive at a serum dilution of 1:250, with no significant correlation between EBV VCA and HHV-8 gB end-point titers (data not shown). The four serum specimens with HHV-8 gB end-point titers of 320 had intermediate EBV VCA titers (250 to 2,000).
FIG. 5.
Antibodies against HHV-8 gB in healthy blood donors. (A) gBSFV IFA end-point titers against HHV-8 gB of 30 blood donor serum specimens. Two serum specimens (black bars) were positive in both K8.1SFV IFA and lytic mIFA (titers of 20 and 40). Two other serum specimens (gray bar) were positive in lytic mIFA (titers of 40 and 80) but negative in all other HHV-8 serologic assays. All of the other 26 specimens (white bars) were negative in all of the other 11 HHV-8 serologic assays. (B and C) Example of IFA with a healthy blood donor serum specimen (RC22) that did not contain detectable antibodies against HHV-8 in any of the 11 other HHV-8 serologic assays. The specimen reacted with rSFV-gB-infected cells (B) but not with rSFV-βgal-infected cells (C).
DISCUSSION
Lytic antigen-based assays are more useful than latent antigen-based assays for detecting HHV-8 infections.
In this study, to improve HHV-8 serologic assays, we developed several IFAs and ELISAs, and compared the performance of a total of 11 assays, using a panel of 180 serum specimens obtained from groups with high, intermediate, and low HHV-8 prevalence.
Martin et al. (23) described a stepwise calibration using well-defined serum specimens for evaluation of a new ELISA. Although we did not take the same approach, we paid careful attention to the potential problems associated with the arbitrary setting of ELISA cutoffs. First, we compared the correlation coefficients with concordance statistics. The generally high correlation between these two parameters suggests that arbitrary determination of cutoffs may not be a major contributor to interassay discordance. Instead, we found that a match of antigen types, latent or lytic, was important to obtain concordant results. In addition, plots of IFA end-point titers against ELISA OD measurements suggest that IFAs are more sensitive than ELISAs, although it is formally possible that at low serum dilutions the IFAs gave nonspecific results. However, this is unlikely, because comparison of K8.1SFV IFA with the results from a combined data set (Fig. 4B) demonstrated that specificities of the IFA were 88 and 91% at serum dilutions of 1:20 and 1:40, respectively, and that the decrease in sensitivity exceeds the increase in specificity at higher serum dilution cutoffs.
We evaluated assay performance in three ways: by using (i) sensitivities in the KS group, (ii) conventional sensitivity and specificity calculations using a defined standard, and (iii) areas under ROC curves. To establish defined standards, we used the K8.1SFV IFA results instead of the PEL-based assay results because (i) our study indicates that latent mIFA has low sensitivity, (ii) lytic mIFA is potentially cross-reactive with antibodies against other herpesviruses, and (iii) the lack of negative control cell lines in the PEL-based assays introduces a degree of subjectivity into their interpretation.
Importantly, the three evaluation methods led to the same conclusions: (i) the performances of the lytic antigen-based ELISAs were almost equivalent, and (ii) the lytic antigen-based assays were more sensitive in identifying seropositives than the latent antigen-based assays. Although K8.1-based ELISAs were slightly better than virion and ORF65pep ELISAs in the ROC analysis, an analysis of more specimens will be needed to determine if the difference is statistically significant. In prior work, bacterially expressed K8.1 was found to be a less sensitive antigen than K8.1 expressed in eukaryotic cells (14, 16, 19). This may be attributed to our previous finding that KS+ sera contain antibodies that react more strongly with the glycosylated form of K8.1 (14).
In most cases, IFAs performed better than ELISAs when antigens derived from the same open reading frame were used. The one exception was ORF65SFV IFA, which performed poorly in comparison with the corresponding ELISA, ORF65pep ELISA. Assays using bacterially and recombinant baculovirus-expressed ORF65 have performed relatively well (13, 39, 43). Since the ORF65 expression level in rSFV-infected cells was equivalent to that of K8.1 (data not shown), the amount of protein may not be the cause of the low sensitivity.
Comparison of several HHV-8 antigens expressed as glutathione S-transferase fusion proteins in E. coli suggested that the latent ORF73 antigen performed better than the tested lytic antigens in ELISAs and IB assays (16). In other work, latent PEL-based IFAs showed high concordances with virion ELISA and with an ELISA using bacterially expressed ORF65 antigens (34, 39). In contrast, IB assays with baculovirus-expressed ORF65 and K8.1 proteins were more sensitive than an IB assay with ORF73 (43). Comparison of ELISAs based on recombinant ORF73 and ORF65 proteins by ROC analysis using KS+ MSM and HIV− female intravenous drug users as defined true positive and negative standards showed better performance of the ORF65-based ELISA (13). ROC analysis using KS patients and hemophilia study subjects as defined positive and negative standards showed that K8.1- and virion-based ELISAs were significantly better than an ORF73-based ELISA (11). Because of the high-level ORF73 expression in the rSFV-based system and a strong correlation between ORF73SFV and ORF73bac ELISAs, it is unlikely that insufficient antigen or posttranslational modification of ORF73 can explain the poor performance of latent antigen-based assays in this study. Higher sensitivity of IFAs than ELISAs, methods used to define assay cutoffs, and posttranslation modifications of lytic antigens in mammalian cells may explain the discrepancies among the prior studies. It is likely that latent antigen-based assays markedly underestimate HHV-8 prevalence.
Our results suggest that there is still room to further optimize the performance of the ELISAs to bring them up to the sensitivity levels of IFAs. Sensitivity may be increased by refinement of ELISA conditions (10), use of combinations of multiple antigens in a single ELISA or use of combined results from multiple assays (11, 16, 40), or an additional step of signal amplification. Although the obvious candidates have been evaluated, it is also possible that additional antigenic proteins will be identified for serologic assays.
PEL-based lytic IFAs have a potential specificity problem.
Sixty percent of blood donor specimens reacted in gBSFV IFA. However, it is unlikely that these reactivities were specific to HHV-8 infection for the following reasons: (i) most blood donor specimens (26 of 30) were negative in all 11 other HHV-8 serologic assays; (ii) because gBSFV IFA detected antibodies against HHV-8 gB in only half of the KS+ specimens, it is unlikely that gBSFV IFA is more sensitive than the other assays; and (iii) rabbit antibodies against EBV gB reacted with TPA-induced BCBL-1 cells at a low serum dilution (data not shown). Because gB is one of the most highly conserved herpesvirus glycoproteins, the simplest explanation would be that antibodies against gBs of other herpesviruses cross-react in gBSFV IFA.
PEL-based lytic IFAs have been found to give consistently higher seroprevalence results than other assays. In this study, we found that three out of four serum specimens with the highest titer in gBSFV IFA were positive in lytic mIFA, and only one of them was positive in K8.1SFV IFA. In other words, 2 out of 28 blood donors were positive by lytic mIFA in the absence of corroborating evidence from any other assay. Because gB is expressed at a high level in TPA-induced BCBL-1 cells (1), it is possible that antibodies cross-reactive with HHV-8 gB produced false-positive results in lytic PEL-based IFAs. This potential false positivity is not a substantial problem in most epidemiologic studies, because lytic PEL-based IFAs generally agree with lytic antigen-based assays (14, 40, 43). However, it is essential to further define the specificity of lytic PEL-based IFAs for studies in which the effect of low specificity can become the main source of seropositivity and confound data interpretations. This would include: (i) determination of more precise seroprevalences in low-prevalence populations; (ii) evaluation of risks associated with HHV-8 transmission, for example, via transfusions or organ transplantation, because donors are frequently from low-prevalence populations; and (iii) diagnosis of individuals. If lytic PEL-based IFA is used for such purposes, it would be better to confirm positives with a second assay. Because K8.1SFV IFA provides sensitivity similar to that of lytic PEL-based IFAs and is more specific, it represents a useful alternative to PEL-based assays.
ACKNOWLEDGMENTS
We thank M. K. Offermann, C. Gunthel, S. Dollard, and the Atlanta HHV-8 Working Group for serum specimens; S. Dollard and Q. Zheng for use of peptide ELISA data; D. Burns, J. B. Black, C.-P. Pau, and D. Sanchez-Martinez for rabbit antibodies; Y.-X. Meng for pcDNA3.1-gB1b-4; and M. Cannon and other members of the Herpesvirus Section at the Centers for Disease Control and Prevention for useful discussion.
J.L.C. was supported by a fellowship through a Collaborative Research and Development Agreement between the Centers for Disease Control and Prevention and Biokit, S.A.
REFERENCES
- 1.Baghian A, Luftig M, Black J B, Meng Y-X, Pau C-P, Voss T, Pellett P E, Kousoulas K G. Glycoprotein B of human herpesvirus 8 is a component of the virion in a cleaved form composed of amino- and carboxyl-terminal fragments. Virology. 2000;269:18–25. doi: 10.1006/viro.2000.0198. [DOI] [PubMed] [Google Scholar]
- 2.Blackbourn D J, Lennette E T, Ambroziak J, Mourich D V, Levy J A. Human herpesvirus 8 detection in nasal secretions and saliva. J Infect Dis. 1998;177:213–216. doi: 10.1086/517356. [DOI] [PubMed] [Google Scholar]
- 3.Blauvelt A. The role of human herpesvirus 8 in the pathogenesis of Kaposi's sarcoma. Adv Dermatol. 1999;14:167–206. [PubMed] [Google Scholar]
- 4.Boldogh I, Szaniszlo P, Bresnahan W A, Flaitz C M, Nichols M C, Albrecht T. Kaposi's sarcoma herpesvirus-like DNA sequences in the saliva of individuals infected with human immunodeficiency virus. Clin Infect Dis. 1996;23:406–407. doi: 10.1093/clinids/23.2.406. [DOI] [PubMed] [Google Scholar]
- 5.Bourboulia D, Whitby D, Boshoff C, Newton R, Beral V, Carrara H, Lane A, Sitas F. Serologic evidence for mother-to-child transmission of Kaposi sarcoma-associated herpesvirus infection. JAMA. 1998;280:31–32. doi: 10.1001/jama.280.1.31-a. [DOI] [PubMed] [Google Scholar]
- 6.Cannon M J, Dollard S M, Smith D K, Kelin R S, Schuman P, Rich J D, Vlahov D, Pellett P E for the HIV Epidemiology Research Study Group. Bloodborne and sexual transmission of human herpesvirus 8 in US women with or at risk for human immunodeficiency virus infection. N Engl J Med. 2001;344:637–643. doi: 10.1056/NEJM200103013440904. [DOI] [PubMed] [Google Scholar]
- 7.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 8.Chatlynne L G, Ablashi D V. Seroepidemiology of Kaposi's sarcoma-associated herpesvirus (KSHV) Semin Cancer Biol. 1999;9:175–185. doi: 10.1006/scbi.1998.0089. [DOI] [PubMed] [Google Scholar]
- 9.Chatlynne L G, Lapps W, Handy M, Huang Y Q, Masood R, Hamilton A S, Said J W, Koeffler H P, Kaplan M H, Friedman-Kien A, Gill P S, Whitman J E, Ablashi D V. Detection and titration of human herpesvirus-8-specific antibodies in sera from blood donors, acquired immunodeficiency syndrome patients, and Kaposi's sarcoma patients using a whole virus enzyme-linked immunosorbent assay. Blood. 1998;92:53–58. [PubMed] [Google Scholar]
- 10.Edelman D C, Ketema F, Saville R D, Herman J, Sill A M, Gill P S, Blattner W A, Constantine N T. Specifics on the refinement and application of two serological assays for the detection of antibodies to HHV-8. J Clin Virol. 2000;16:225–237. doi: 10.1016/s1386-6532(99)00085-2. [DOI] [PubMed] [Google Scholar]
- 11.Engels E A, Whitby D, Goebel P B, Stossel A, Waters D, Pintus A, Contu L, Biggar R J, Goedert J J. Identifying human herpesvirus 8 infection: performance characteristics of serologic assays. J Acquir Immune Defic Syndr. 2000;23:346–354. doi: 10.1097/00126334-200004010-00011. [DOI] [PubMed] [Google Scholar]
- 12.Gao S-J, Kingsley L, Li M, Zheng W, Parravicini C, Ziegler J, Newton R, Rinaldo C R, Saah A, Phair J, Detels R, Chang Y, Moore P S. KSHV antibodies among Americans, Italians and Ugandans with and without Kaposi's sarcoma. Nat Med. 1996;2:925–928. doi: 10.1038/nm0896-925. [DOI] [PubMed] [Google Scholar]
- 13.Goudsmit J, Renwick N, Dukers N H, Coutinho R A, Heisterkamp S, Bakker M, Schulz T F, Cornelissen M, Weverling G J. Human herpesvirus 8 infections in the Amsterdam Cohort Studies (1984–1997): analysis of seroconversions to ORF65 and ORF73. Proc Natl Acad Sci USA. 2000;97:4838–4843. doi: 10.1073/pnas.97.9.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inoue N, Mar E-C, Dollard S C, Pau C-P, Zheng Q, Pellett P E. New immunofluorescence assays for detection of human herpesvirus 8-specific antibodies. Clin Diagn Lab Immunol. 2000;7:427–435. doi: 10.1128/cdli.7.3.427-435.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones D, Ballestas M E, Kaye K M, Gulizia J M, Winters G L, Fletcher J, Scadden D T, Aster J C. Primary-effusion lymphoma and Kaposi's sarcoma in a cardiac-transplant recipient. N Engl J Med. 1998;339:444–449. doi: 10.1056/NEJM199808133390705. [DOI] [PubMed] [Google Scholar]
- 16.Katano H, Iwasaki T, Baba N, Terai M, Mori S, Iwamoto A, Kurata T, Sata T. Identification of antigenic proteins encoded by human herpesvirus 8 and seroprevalence in the general population and among patients with and without Kaposi's sarcoma. J Virol. 2000;74:3478–3485. doi: 10.1128/jvi.74.8.3478-3485.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kedes D H, Operskalski E, Busch M, Kohn R, Flood J, Ganem D. The seroepidemiology of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat Med. 1996;2:918–924. doi: 10.1038/nm0896-918. [DOI] [PubMed] [Google Scholar]
- 18.Koelle D M, Huang M-L, Chandran B, Vieira J, Piepkorn M, Corey L. Frequent detection of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in saliva of human immunodeficiency virus-infected men: clinical and immunologic correlates. J Infect Dis. 1997;176:94–102. doi: 10.1086/514045. [DOI] [PubMed] [Google Scholar]
- 19.Lang D, Hinderer W, Rothe M, Sonneborn H-H, Neipel F, Raab M, Rabenau H, Masquelier B, Fleury H. Comparison of the immunoglobulin-G-specific seroreactivity of different recombinant antigens of the human herpesvirus 8. Virology. 1999;260:47–54. doi: 10.1006/viro.1999.9804. [DOI] [PubMed] [Google Scholar]
- 20.Lennette E T, Blackbourn D J, Levy J A. Antibodies to human herpesvirus type 8 in the general population and in Kaposi's sarcoma patients. Lancet. 1996;348:858–861. doi: 10.1016/S0140-6736(96)03240-0. [DOI] [PubMed] [Google Scholar]
- 21.Lin S-F, Sun R, Heston L, Gradoville L, Shedd D, Haglund K, Rigsby M, Miller G. Identification, expression, and immunogenicity of Kaposi's sarcoma-associated herpesvirus-encoded small viral capsid antigen. J Virol. 1997;71:3069–3076. doi: 10.1128/jvi.71.4.3069-3076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luppi M, Barozzi P, Schulz T F, Setti G, Staskus K, Trovato R, Narni F, Donelli A, Maiorana A, Marasca R, Sandrini S, Torelli G. Bone marrow failure associated with human herpesvirus 8 infection after transplantation. N Engl J Med. 2000;343:1378–1385. doi: 10.1056/NEJM200011093431905. [DOI] [PubMed] [Google Scholar]
- 23.Martin J N, Amad Z, Cossen C, Lam P K, Kedes D H, Page-Shafer K A, Osmond D H, Forghani B. Use of epidemiologically well-defined subjects and existing immunofluorescence assays to calibrate a new enzyme immunoassay for human herpesvirus 8 antibodies. J Clin Microbiol. 2000;38:696–701. doi: 10.1128/jcm.38.2.696-701.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin J N, Ganem D E, Osmond D H, Page-Shafer K A, Macrae D, Kedes D H. Sexual transmission and the natural history of human herpesvirus 8 infection. N Engl J Med. 1998;338:948–954. doi: 10.1056/NEJM199804023381403. [DOI] [PubMed] [Google Scholar]
- 25.Mayama S, Cuevas L E, Sheldon J, Omar O H, Smith D H, Okong P, Silvel B, Hart C A, Schulz T F. Prevalence and transmission of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in Ugandan children and adolescents. Int J Cancer. 1998;77:817–820. doi: 10.1002/(sici)1097-0215(19980911)77:6<817::aid-ijc2>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 26.Melbye M, Cook P M, Hjalgrim H, Begtrup K, Simpson G R, Biggar R J, Ebbesen P, Schulz T F. Risk factors for Kaposi's-sarcoma-associated herpesvirus (KSHV/HHV-8) seropositivity in a cohort of homosexual men, 1981–1996. Int J Cancer. 1998;77:543–548. doi: 10.1002/(sici)1097-0215(19980812)77:4<543::aid-ijc12>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 27.Metz C E, Herman B A, Roe C A. Statistical comparison of two ROC-curve estimates obtained from partially-paired datasets. Med Decision Making. 1998;18:110–121. doi: 10.1177/0272989X9801800118. [DOI] [PubMed] [Google Scholar]
- 28.Moore P S, Gao S-J, Dominguez G, Cesarman E, Lungu O, Knowles D M, Garber R, Pellett P E, McGeoch D J, Chang Y. Primary characterization of a herpesvirus agent associated with Kaposi's sarcoma. J Virol. 1996;70:549–558. doi: 10.1128/jvi.70.1.549-558.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore P S, Kingsley L A, Holmberg S D, Spira T, Gupta P, Hoover D R, Parry J P, Conley L J, Jaffe H W, Chang Y. Kaposi's sarcoma-associated herpesvirus infection prior to onset of Kaposi's sarcoma. AIDS. 1996;10:175–180. doi: 10.1097/00002030-199602000-00007. [DOI] [PubMed] [Google Scholar]
- 30.Parravicini C, Olsen S J, Capra M, Poli F, Sirchia G, Gao S-J, Berti E, Nocera A, Rossi E, Bestetti G, Pizzuto M, Galli M, Moroni M, Moore P S, Corbellino M. Risk of Kaposi's sarcoma-associated herpes virus transmission from donor allografts among Italian posttransplant Kaposi's sarcoma patients. Blood. 1997;90:2826–2829. [PubMed] [Google Scholar]
- 31.Pau C-P, Lam L L, Spira T J, Black J B, Stewart J A, Pellett P E, Respess R A. Mapping and serodiagnostic application of a dominant epitope within the human herpesvirus 8 ORF 65-encoded protein. J Clin Microbiol. 1998;36:1574–1577. doi: 10.1128/jcm.36.6.1574-1577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pauk J, Huang M-L, Brodie S J, Wald A, Koelle D M, Schacker T, Celum C, Selke S, Corey L. Mucosal shedding of human herpesvirus 8 in men. N Engl J Med. 2000;343:1369–1377. doi: 10.1056/NEJM200011093431904. [DOI] [PubMed] [Google Scholar]
- 33.Plancoulaine S, Abel L, van Beveren M, Tregouet D A, Joubert M, Tortevoye P, de The G, Gessain A. Human herpesvirus 8 transmission from mother to child and between siblings in an endemic population. Lancet. 2000;356:1062–1065. doi: 10.1016/S0140-6736(00)02729-X. [DOI] [PubMed] [Google Scholar]
- 34.Rabkin C S, Schulz T F, Whitby D, Lennette E T, Magpantay L I, Chatlynne L, Biggar R J. Interassay correlation of human herpesvirus 8 serologic tests. HHV-8 Interlaboratory Collaborative Group. J Infect Dis. 1998;178:304–309. doi: 10.1086/515649. [DOI] [PubMed] [Google Scholar]
- 35.Regamey N, Tamm M, Wernli M, Witschi A, Thiel G, Cathomas G, Erb P. Transmission of human herpesvirus 8 infection from renal-transplant donors to recipients. N Engl J Med. 1998;339:1358–1363. doi: 10.1056/NEJM199811053391903. [DOI] [PubMed] [Google Scholar]
- 36.Sanchez-Martinez D, Patton J L, Stewart J A, Pellett P E. Detection of Epstein-Barr virus-specific antibodies by means of baculovirus-expressed EBV gp125. J Virol Methods. 1995;52:145–153. doi: 10.1016/0166-0934(94)00157-c. [DOI] [PubMed] [Google Scholar]
- 37.Sarid R, Olsen S J, Moore P S. Kaposi's sarcoma-associated herpesvirus: epidemiology, virology, and molecular biology. Adv Virus Res. 1999;52:139–232. doi: 10.1016/s0065-3527(08)60299-7. [DOI] [PubMed] [Google Scholar]
- 38.Schulz T F. Epidemiology of Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. Adv Cancer Res. 1999;76:121–160. doi: 10.1016/s0065-230x(08)60775-7. [DOI] [PubMed] [Google Scholar]
- 39.Simpson G R, Schulz T F, Whitby D, Cook P M, Boshoff C, Rainbow L, Howard M R, Gao S-J, Bohenzky R A, Simmonds P, Lee C, de Ruiter A, Hatzakis A, Tedder R S, Weller I V D, Weiss R A, Moore P S. Prevalence of Kaposi's sarcoma associated herpesvirus infection measured by antibodies to recombinant capsid protein and latent immunofluorescence antigen. Lancet. 1996;348:1133–1138. doi: 10.1016/S0140-6736(96)07560-5. [DOI] [PubMed] [Google Scholar]
- 40.Spira T J, Lam L, Dollard S C, Meng Y-X, Pau C P, Black J B, Burns D, Cooper B, Hamid M, Huong J, Kite-Powell K, Pellett P E. Comparison of serologic assays and PCR for diagnosis of human herpesvirus 8 infection. J Clin Microbiol. 2000;38:2174–2180. doi: 10.1128/jcm.38.6.2174-2180.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vieira J, Huang M-L, Koelle D M, Corey L. Transmissible Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in saliva of men with a history of Kaposi's sarcoma. J Virol. 1997;71:7083–7087. doi: 10.1128/jvi.71.9.7083-7087.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whitby D, Howard M R, Tenant-Flowers M, Brink N S, Copas A, Bishoff C, Hatzioannou T, Suggett F E A, Aldam D M, Denton A S, Miller R F, Weller I V D, Weiss R A, Tedder R S, Schultz T F. Detection of Kaposi's sarcoma associated herpesvirus in peripheral blood of HIV-infected individuals and progression to Kaposi's sarcoma. Lancet. 1995;346:799–802. doi: 10.1016/s0140-6736(95)91619-9. [DOI] [PubMed] [Google Scholar]
- 43.Zhu L, Wang R, Sweat A, Goldstein E, Horvat R, Chandran B. Comparison of human sera reactivities in immunoblots with recombinant human herpesvirus (HHV)-8 proteins associated with the latent (ORF73) and lytic (ORFs 65, K8.1A, and K8.1B) replicative cycles and in immunofluorescence assays with HHV-8-infected BCBL-1 cells. Virology. 1999;256:381–392. doi: 10.1006/viro.1999.9674. [DOI] [PubMed] [Google Scholar]