Highlights
-
•
RNA-based therapeutics have the potential to reach previously “undruggable” pathways in cardiovascular disease
-
•
RNA-based therapeutics constitute a vast array of technologies, including unique forms, chemistries, and modalities of delivery
-
•
Rapid development of RNA-based vaccines was made possible by decades of foundational work
-
•
Specificity and efficacy of targeting and determination of mechanism(s) of action remain a distinct challenge
Key Words: cardiovascular disease, noncoding RNA, nucleic acid-based therapies, RNA, therapeutic modalities
Abbreviations and Acronyms: AAV, adeno-associated viral vector; ASO, antisense oligonucleotide; circRNA, circular RNA; CRISPR, clustered regularly interspaced short palindromic repeats; CVD, cardiovascular disease; DMD, Duchenne muscular dystrophy; HF, heart failure; lncRNA, long noncoding RNA; miRNA, microRNA; NA, nucleic acid; ncRNA, noncoding RNA
Summary
Cardiovascular disease (CVD) remains the largest cause of mortality worldwide. The development of new effective therapeutics is a major unmet need. The current review focuses broadly on the concept of nucleic acid (NA)–based therapies, considering the use of various forms of NAs, including mRNAs, miRNAs, siRNA, and guide RNAs, the latter specifically for the purpose of CRISPR-Cas directed gene editing. We describe the current state-of-the-art of RNA target discovery and development, the status of RNA therapeutics in the context of CVD, and some of the challenges and hurdles to be overcome.
Central Illustration
With cardiovascular disease (CVD) remaining the largest cause of mortality worldwide, responsible for close to 18 million deaths annually (World Health Organization statistics),1 the development of new assays for early diagnosis, more accurate prognosis, and effective preventative and curative therapies is a major unmet global need. Other than the relatively rare pathogenic sequence variants that lead to CVD, such as truncating mutations in the gene for Titin responsible for at least one-quarter of cases of familial idiopathic dilated cardiomyopathy, the molecular basis of CVD remains poorly understood. This is reflected in the increasing morbidity and mortality rates for CVD even in countries with highly developed health care and biomedical research.
In the context of CVD, the history of therapeutics for most disorders, including heart failure (HF), hypertension, lipids, and ischemia, has been limited primarily to small organic molecules, eg, cardiac glycosides, beta-blockers, angiotensin-converting enzyme inhibitors, angiotensin receptor blockers, calcium-channel blockers, nitrates, statins, and so on.2 And, more often than not, the most recently marketed “new” drugs for CVD are combinations or formulations of already approved therapeutics rather than novel compounds. Opening new doors to pathways previously considered “untargetable” are alternative therapeutic modalities, including peptides, antibodies, and nucleic acid (NA)-based therapies; one virtue of the latter approach is the high-fidelity nature of complementary base-pairing of therapeutic and target.3
The use of NA-based therapies is rapidly accelerating as evidenced by the number of clinical trials4, 5, 6, 7, 8, 9, 10 using this approach. Specifically, within the CVD space, NA-based therapies are currently undergoing testing in several areas including those of lipid management: apo(a), PCSK9; hypertension and HF: angiotensinogen, FOXO3, SERCA2A; angiogenesis: angiopoietin like proteins (ANGPTLS); and amyloidosis: transthyretin (TTR).9
For recent comprehensive reviews detailing noncoding RNA (ncRNA) therapeutics in animal models, see Braga et al,11 or De Majo and De Windt.12 For recent U.S. Food and Drug Administration (FDA) and/or European Medicines Agency approved NA-based therapies and/or clinical trials, see Winkle et al,13 Roberts et al14 and Dammes and Peer,4 Laina et al,15 Kaczmarek et al,7 Crooke et al,16 or Qadir et al17 (Table 1).
Table 1.
Currently Approved NA-Based Therapies and/or Clinical Trials in CVD and Related Diseases
| Drug/Stage | Disease | Therapeutic Modality | Target | Comments/Company |
|---|---|---|---|---|
| Preclinical | ||||
| CDR426D | Dilated cardiomyopathy | Interaction with ncRNA | undisclosed | Cardior |
| CDR348T | Hypertrophic cardiomyopathy | Modulation of ncRNA | undisclosed | Cardior |
| CDR641L | Hypertrophic cardiomyopathy | Interaction with ncRNA | undisclosed | Cardior |
| SRP-5051 | DMD | ASO | Dystrophin | Sarepta |
| SRP-5053 | DMD | ASO | Dystrophin | Sarepta |
| SRP-5045 | DMD | ASO | Dystrophin | Sarepta |
| SRP-5052 | DMD | ASO | Dystrophin | Sarepta |
| SRP-5044 | DMD | ASO | Dystrophin | Sarepta |
| xx | DMD | CRISPR-Cas9 | Dystrophin | Vertex (acquisition of Exonics Therapeutics) |
| Clinical trials | ||||
| CDR132L | Heart failure (HFpEF) | ASO | Interaction with miR-321 | Phase 1 (Cardior) |
| Pelacarsen (IONIS-APO(a)-LRx) | Lipid disorders | AS | Lp(a) | Phase 3 (Ionis) |
| Eplontersen (IONIS-TTR-LRx) | Amyloidosis | AS | transthyretin | Phase 2 (Ionis/AstraZeneca) |
| Olezarsen | Lipid disorders | AS | ApoC-III | Phase 3 (Ionis) |
| Vupanorsen (AKCEA-ANGPTL3-LRx) | Lipid disorders | LICA | ANGPTL3 | Phase 2 (Ionis/Pfizer) |
| IONIS-AGT-LRx | CHF/HFrER | LICA | Angiotensinogen | Phase 2 (Ionis) |
| Olpasiran (AMG-890) | Lipid disorders | siRNA | Lp(a) | Phase 2 (Amgen) |
| AZD8601 | Heart failure/ischemia | naked mRNA | VEGF-A | Phase 2 (AstraZeneca) |
| FDA/EMA approved | ||||
| Nusinersen (Spinraza) | SMA | AS0 | SMN2 (intron 7) | FDA approved (Biogen) |
| Waylivra (volanesorsen) | Triglycerides | ASO | APO-C3 | EMA approved (Ionis/Aksea) |
| Exondys 51 (eteplirsen) | DMD | ASO | Dystrophin exon 51 | FDA approved (Sarepta) |
| Vyondys 53 (golodirsen) | DMD | ASO | Dystrophin exon 53 | FDA approved (Sarepta) |
| Amondys 45 (casimersen) | DMD | ASO | Dystrophin exon 45 | FDA approved (Sarepta) |
| Leqvio (inclisiran) | Lipid disorders | siRNA | PCSK9 | FDA approved (Novartis) |
| Vitepso (Viltolarsen) | DMD | ASO | Dystrophin exon 53 | FDA approved (NS Pharma) |
AS = antisense; ASO = antisense oligonucleotide; CRISPR = clustered regularly interspaced short palindromic repeats; CVD = cardiovascular disease; DMD = Duchenne muscular dystrophy; EMA = European Medicines Agency; FDA = U.S. Food and Drug Administration; HFpEF = heart failure with preserved ejection fraction; HFrEF = heart failure with reduced ejection fraction; LICA = ligand conjugated antisense; ncRNA = noncoding RNA; VEGF = vascular endothelial growth factor.
The current review focuses broadly on the concept of NA-based therapies especially for CVD, considering the use of synthetic antisense oligonucleotides (ASOs), mRNAs, noncoding (nc)RNAs: siRNAs, micro (mi)RNAs, long noncoding (lnc) RNAs, and single guide (sg) RNAs, the latter specifically for the purpose of clustered regularly interspaced short palindromic repeats (CRISPR)-Cas directed gene editing. We describe the current state-of-the-art of RNA target discovery and development, the status of RNA therapeutics in the context of CVD, and some of the challenges and hurdles to be overcome (see Central Illustration).
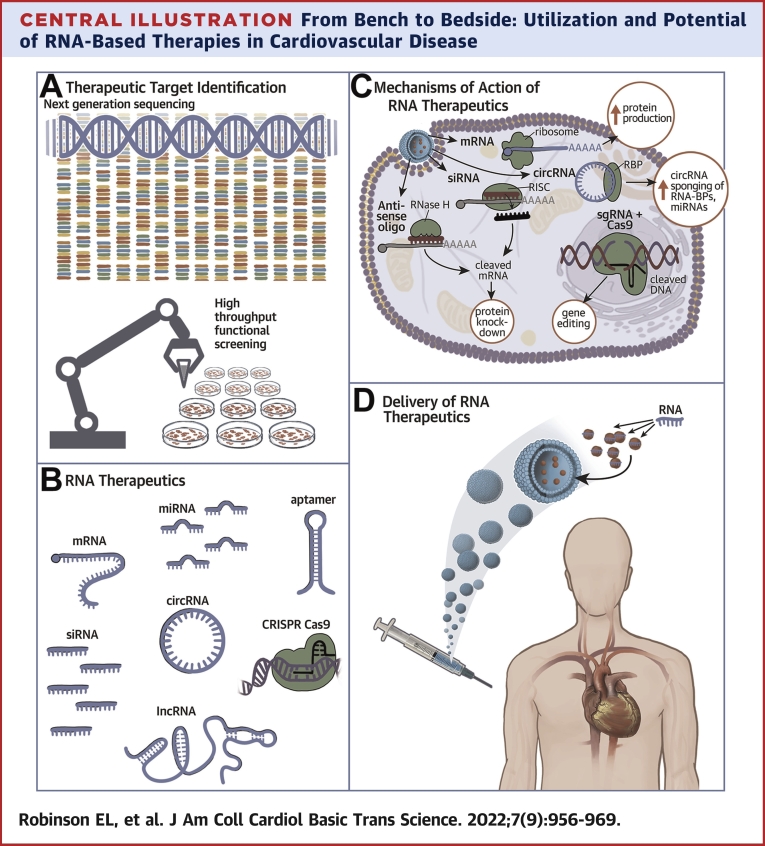
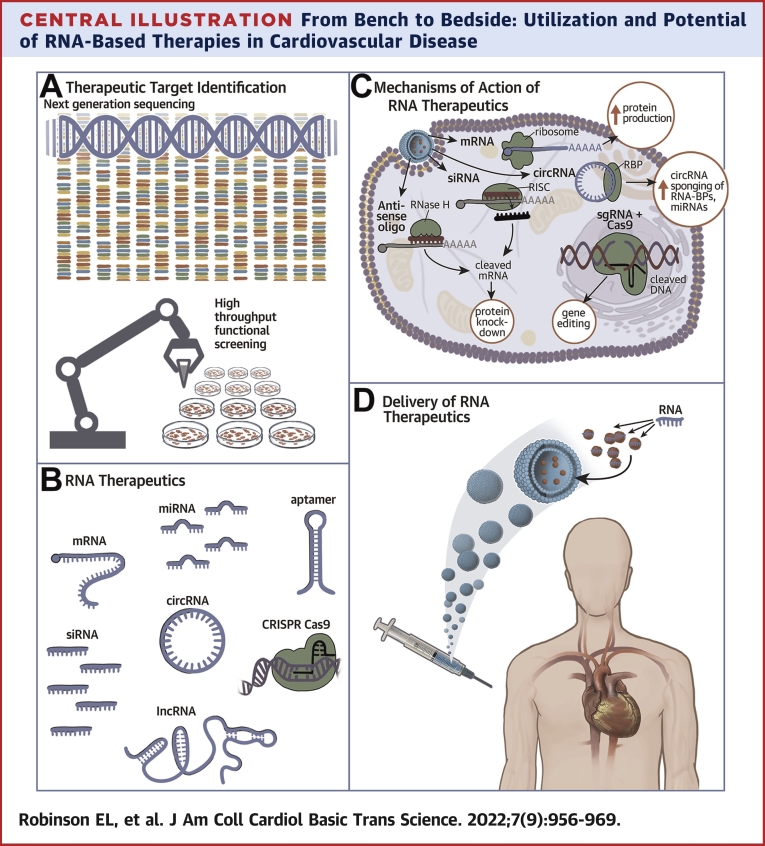
Central Illustration.
From Bench to Bedside: Utilization and Potential of RNA-Based Therapies in Cardiovascular Disease
(A) Therapeutic Target Identification: Molecular target drug discovery by next generation sequencing comparing diseased tissue with healthy tissue data. Candidates genes can be validated and evaluated for potential as therapeutic targets by high throughput cell based functional screens. (B) RNA Therapeutics. RNA molecules have different biological and therapeutic modalities. (C) Mechanisms of Action of RNA Therapeutics. Once inside the cell, nucleic acid therapeutics can exert their molecular modality through complementary base pairing to endogenous RNA molecules to inhibit their activity or target them for degradation by RNaseH. mRNA mimetics can increase protein production in the cell. circRNAs and other RNAs can act as sponges for RNA-BPs and miRNAs to sequester them and dampen their pathological activity in the cell. SgRNAs specifically binds to pathogenic genomic loci for editing, catalyzed by the dual RNA-guided DNA endonuclease enzyme, Cas9. (D) Delivery of RNA Therapeutics. An example of RNA therapeutic administration in humans, with the nuclei acid encapsulated in a lipid-based nanovesicle and administered via an injection. circRNA = circular RNA; CRISPR = Clustered Regularly Interspaced Short Palindromic Repeats; lncRNA = long non-coding RNA; miRNA = microRNA; mRNA = messenger RNA; RISC = RNA-induced silencing complex; RNA-BPs = RNA binding proteins; sgRNA = single guide RNA; siRNA = small interfering RNA.
RNA, the Basics
The original central dogma of molecular biology, as described by Francis Crick,18 dictates that DNA is transcribed into messenger RNA (mRNA), which is then translated into proteins. Molecular fidelity is maintained between DNA and RNA and between RNA and tRNA adaptors through Watson-Crick complementary base pairing. Until the completion of the Human Genome Project in 2003, it was firmly believed that this was the sole function of the genome—to be transcribed into messenger RNA and encoded for proteins. However, the final reporting of just ∼21,000 protein coding genes, accounting for just 1% of the human genome, brought to question why the overwhelming majority of the genome, then so-named “junk DNA,” had been conserved through evolution at a biologically energetic expensive cost.19
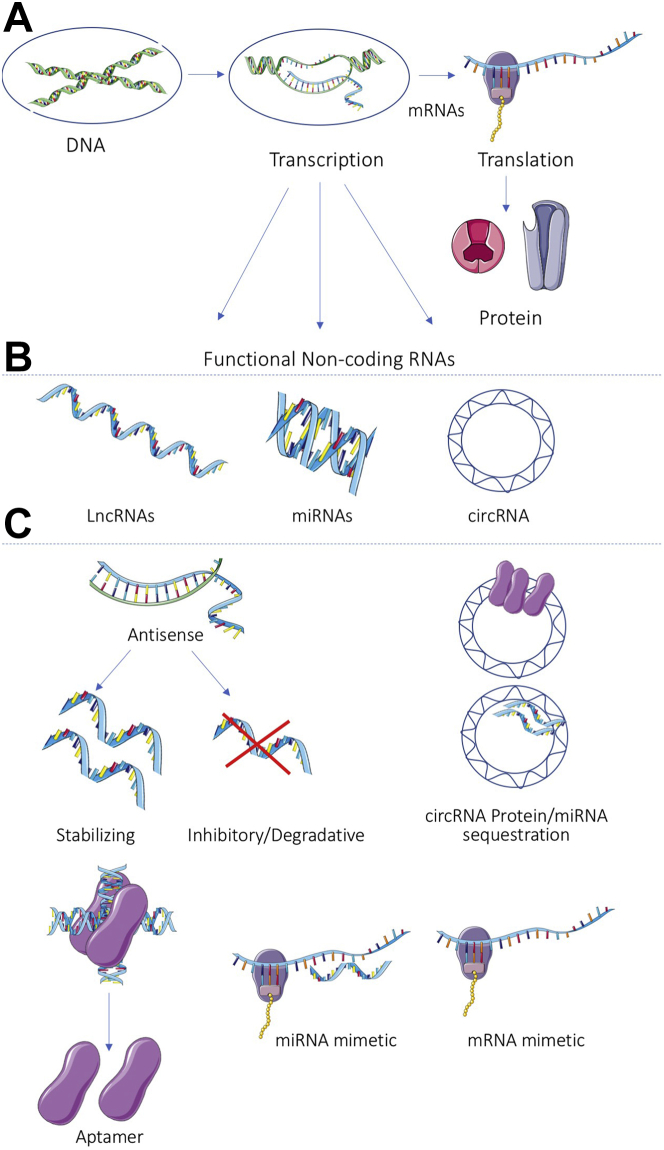
The revolution in next-generation deep genomic and RNA sequencing of the last decade has revealed that the portion of the human genome used to encode proteins is far outweighed by the proportion that is transcribed into ncRNAs. ncRNAs lack protein coding capability (see the next paragraph) but remain as functional RNA molecules of a number of subclasses. ncRNAs represent an extensive and heterogeneous family of biologically functional RNA molecules that have been shown to play essential roles in differentiation, development, homeostasis, and disease (Figures 1A and 1B).
Figure 1.
The Revised Central Dogma of Biology With Key Functional Modalities of NA Therapeutics Indicated
(A) Up to 76% of the human genome DNA, the genomic blueprint, is transcribed into RNA in the nucleus. 2%-3% is transcribed into mRNA, encoding for protein synthesis at the ribosome in the cytoplasm. (B) Other RNA encodes for functional non-coding RNAs including lncRNAs, miRNAs and circRNAs. (C) Functional RNAs, including lncRNAs and miRNAs, can influence cell biology by complementary base pairing to other RNA molecules and acting to stabilize, inhibit or degrade them. circRNAs can influence cell biology by binding and sequestering miRNAs and RNA binding proteins. Aptamer molecules, mRNA or miRNA mimetics are examples of nucleic acid therapeutic modalities that influence RNA or protein activity or abundance. circRNA = circular RNA; lncRNA = long noncoding RNA; miRNA = microRNA; ncRNA = noncoding RNA.
It should be noted that a number of lncRNAs have been found to encode cryptic, short, functionally relevant “micro” peptides. As a canonical example in the cardiovascular space, Olson’s group20, 21, 22, 23 have described a series of micropeptides that act similarly to or in opposition to phospholamban in their capacity to modulate SERCA2 medicated calcium flux. Thus, ncRNAs annotated as such are only truly “noncoding” until proven otherwise.24,25
The ncRNA revelation has led to a re-evaluation of the central dogma of molecular biology (Figure 1A).26 Categorization of ncRNA has been somewhat unstandardized, in part because of the explosive rate of discovery of ncRNAs, with different structures, functions, and genomic contexts by different laboratories, institutions, and fields. Although also ncRNAs, the core mediators of protein translational machinery, ribosomal (r)RNAs and transfer (t)RNAs, as well as RNA components of the spliceosome and nucleosome, snRNAs and snoRNAs, will not be discussed herein.
Messenger RNAs
mRNAs are the classic protein coding single-stranded RNAs that are transcribed from protein coding genes in a complementary base pair manner by RNA polymerase II. mRNA goes through a number of post-transcriptional modifications before translation, including 5′ capping, 3′ polyadenylation, and splicing of noncoding introns to form a mature mRNA. Different isoforms of a protein can be formed from the same gene and by the highly regulated process of alternative splicing. Stability, localization, and function of mRNAs can be altered further by post-transcriptional modifications, such as N6-methylation of adenosine or by RNA editing (A-to-I) with adenosine being replaced by inosine.27
MicroRNAs
MicroRNAs (miRNAs) are single-stranded 21-23 nt molecules that regulate gene expression post-transcriptionally by complementary base pairing to mRNAs or other ncRNA targets, preventing translation, activity, and/or priming them for degradation. Mature miRNAs are processed from pri-miRNAs and pre-miRNAs through a Drosha/DICER-dependent mechanisms.28,29
Further, miRNAs are known to be key regulators of cardiovascular development, homeostasis, and disease with functional studies in vitro, ex vivo, and in vivo demonstrating that modulation of miRNA activity affects cellular biochemistry and cardiovascular function, thereby presenting themselves as therapeutic targets to prevent or treat CVD decline.30,31 That said, compared with ASO and antisense RNAs, miR-based therapeutics remain a substantial challenge due, at least in part, to a lack of understanding of their mechanism(s) of action.32 Vagaries related to therapeutic efficacy include binding specificity/affinity, off-target effects, the number of miR binding sites per target mRNA, and ability to bind to AGO/GW182 complex.
Long ncRNAs
ncRNAs >200 base pairs in length are described as lncRNAs. However, lncRNAs continue to be further divided into categories according to their genomic context or function. For example, intergenic or intervening lncRNAs are located between annotated genes; intragenic lncRNAs can be found within described genes, such as within introns; antisense lncRNAs are encoded on the opposite genomic DNA strand to protein coding genes (antisense RNAs); and enhancer or promoter encoded lncRNAs are known as enhancer lncRNAs.33 Not unlike proteins, lncRNAs can form higher order structures and defined structural motifs that regulate and determine their function. X-ray crystallography can be used to determine lncRNA structure, either in isolation or in complex with bound protein partners. In silico programs are also being developed to predict complex lncRNA structures and potential interaction partners.
Predictions of the number of lncRNAs encoded by the human genome are in the tens of thousands, yet only hundreds have been functionally validated and characterized, possibly representing <1% of all lncRNAs. Further, there is a lack of documentation of annotated and functionally validated in a centralized database. A number of large-scale projects center around annotating lncRNAs including FANTOM, NONCODE, GENCODE, and Refseq; the pros and cons of each have been summarized previously.34 Other smaller databases exist that document, eg, lncRNA function or lncRNAs with roles in disease.35,36
In genomic structure and context, lncRNAs largely resemble mRNAs. They are transcribed by RNA polymerase II, have promoter and enhancer regions, have sequences resembling 5′ and 3′ UTRs, have introns and exons, and many are polyadenylated. Many have microRNA binding sites and can undergo RNA modifications, such as N6-methyladenosine. With these properties in mind, lncRNAs are amenable to modulation by different RNA technologies and modalities, just as are mRNAs.
Circular RNAs
Advances in next-generation sequencing technologies have enabled detection of 5′ back-spliced circularized RNAs from exonic and intronic regions from up to 20% of both protein coding genes and lncRNAs alike, the so-called circular RNAs (circRNA).37 circRNAs have been described as at least 2.5× more stable than their linear ncRNA counterparts and are also found in the circulation.38 Current known functionality of circRNAs includes acting as sponges of microRNAs and proteins, scaffolds for RNA binding proteins (RBPs), and regulation of translation. Their stability and functional properties make circRNAs not only a target but also as potential therapeutic agents.39
The first full landscape of expressed circRNAs in the mammalian heart was described by Tan et al40 in 2017, where more than 15,000 circRNAs were detected in the human heart. A number have since been described as functionally important or as promising circulating biomarkers of CVD.41 The therapeutic potential of RNA as drugs and disease targets is now in the spotlight in both basic and translational research, with basic discovery research identifying new potentially pathology-driving or protective genes and RNAs and translational research assessing and optimizing their delivery and disease-curbing capacity.
Modalities in RNA Therapy Technology
A number of molecular tools have been developed to manipulate RNA activity or abundance both in vitro and in vivo. The mechanism(s)—or modalities—by which they exert their action is based on their chemistry and some approaches are described in the following text, as well as depicted in Figure 1C.
The RNA Interference Approach: RNA Antisense Technology
The concept of RNA interference is a naturally occurring molecular phenomenon, with miRNAs and some lncRNAs exerting their function with high specificity by complementary base-pairing to their RNA targets. Basic and translational scientists have taken advantage of this phenomenon for functional experimentation and assessment of the therapeutic potential of RNA molecules. RNA interference can, in theory, reduce activity and levels of all species of RNA—miRNA, mRNA, lncRNA, and circRNA—with obstacles dependent on the form of functional RNA target. For example, targeting circRNAs remains a challenge.42 Current approaches to circumvent this issue includes targeting the unique 3′-5′ back spliced junction.
Small interfering (si)RNA was first described in 1998, and its discoverers were awarded the Nobel Prize in Physiology or Medicine in 2006. Fast forward, the FDA has recently approved the first therapeutic siRNA, Onpattro (patisiran), to treat nerve damage caused by a rare disease, hereditary transthyretin-mediated amyloidosis (hATTR), affecting ∼50,000 people worldwide. The major cause of hATTR is the buildup of amyloid protein in peripheral nerves, heart, and other organs, and the antisense drug attenuates that issue.
Antisense RNAs Affecting Splicing or Stability
Not all antisense RNAs are interfering; instead, they may alter the stability and/or splicing properties of their target counterpart. Due to their exceptional chemical specificity, antisense RNAs can distinguish between “healthy” and disease-causing RNA molecules, so as to target just the pathological entity. One example is nusinersen (Spinraza), which was approved in 2016 for spinal muscular atrophy, a disease caused by deletion or deleterious mutations in the gene Survival Motor Neuron 1 (SMN1), many of which result in the exclusion of exon 7 from SMN1.43 Nusinersen is an 18-mer antisense NA that does not bind SMN1 but instead binds to the pre-mRNA of SMN2, a nearly identical relative, with functional redundancy. Nusinersen binding to SMN2 pre-mRNA promotes inclusion of exon 7 and increase production of full-length SNM2, rescuing some of the biological effects of a lack of functional SMN1.44
The previous example demonstrates the use of NAs as effective in combating faulty disease-causing genes. In the case of complex diseases, such as CVD, monogenic hereditary complex diseases are rare in comparison to polygenic, epigenetic, risk factor-driven etiologies. Formulation of effective therapies for such multifactoral conditions is far more challenging, and RNA therapies for such as these are only just emerging. Other state-of-the-art, translational examples are described in the following text.
RNA Mimetics
RNA mimetics are exogenously sourced RNA or RNA-like molecules. Although many examples have been used in preclinical functional investigations, their pharmacokinetic properties have hindered their progress through to clinical trials compared with antisense RNAs. Chemical modifications to the backbone and NAs (see the following text) can improve their stability and half-life in vivo and reduce the risk of RNA toxicity of RNA. As an example, a miR-30c mimetic reduced systemic and multiorgan hyperlipidemia and atherosclerosis in mice fed a high-fat diet in part through inhibition of microsomal triglyceride transfer protein.45
RNA and RBP Sequestration/Sponging
A further modality of action of RNA molecules is that of RBP “sponging,” in essence, functional sequestration. Once again, this is a naturally occurring phenomenon and an intrinsic property of some virally-encoded RNA molecules.46 For example, a number of viral RNA 3′UTRs have high affinity for the mammalian HuR family of proteins, which are mRNA binding proteins regulating gene expression in a number of different contexts.47,48 Other endogenous RNAs bind and regulate proteins important in the heart.49 A number of bioinformatic approaches and in silico analyses have been to predict and identify RNA-protein interactions. Both linear RNA and circRNA are being harnessed for their RBP sequestration properties in RNA technology and therapeutics. As an example, Lavenniah and team,50 recently developed adeno-associated viral vector (AAV)–encoded artificial circRNA miRNA sponges (termed circmiRs) for pro-hypertrophic miRs, eg, miR-132 and miR-212, which proved antihypertrophic in mice subject to transverse aortic constriction.
Aptamers
Aptamers are single-stranded oligonucleotides of ∼25-80 bases that form complex structures, bind proteins, and have the capacity to disrupt multiprotein complexes and/or inhibit protein function. Aptamers can be composed of ribonucleotides or deoxyribonucleotides. For example, Pegaptanib (Macugen, Eyetech Pharmaceuticals/Pfizer) is an FDA-approved drug for macular degeneration and is an RNA aptamer that binds and interrupts the function of the pathogenic vascular endothelial growth factor isoform 165.51 Aptamers provide a further form of pharmacological modality to specifically target proteins or RNAs otherwise undruggable by conventional small molecule compounds.
RNA Chemistries
There are a number of chemical modifications relevant to stabilization including, most importantly, to the nucleotide backbone. Primary among these is the use of phosphorothioate linkages, which renders nucleotides more stable but can also affect target binding affinity. It is possible to use modified nucleobases, including 5-methylcytidine and 5-menthyluridine, or modified ribose substitutions including 2′-O-methoxyethyl (2′-MOE), 2′-O-methyl (2′-OMe), and 2′-Fluoro (2′-F). Importantly, 2′OMe modification also acts to decrease immune response. Other important modifications include “bridged NAs,” such as locked nucleic acids and 2′-O, 4′C-ethylene-bridged nucleic acids. Worth mentioning are phosphorodiamidate morpholino oligonucleotides, which have been used clinically by Sarepta Therapeutics for treatment of Duchenne muscular dystrophy (DMD).
RNA Therapeutic Delivery
Effective delivery of therapeutic NAs, in virtually any form, requires chemical modification. There are 3 essential reasons chemical modification is necessary: 1) stabilization, ie, resistance to nucleases such as RNase H; 2) enhanced PK/PD and distribution; and 3) reduction of immunogenic potential. The latter aspect has been addressed by numerous investigators, including Drs. Karikó and Weissman,52, 53, 54, 55 who have recently regained notoriety for their contributions to RNA chemistry underlying the effectiveness of COVID vaccines.
In terms of delivery of NAs and ASOs, this is most often facilitated by the use of lipid conjugates, including cholesterol and its variants, as well as a number of versions of N-acetylgalactosamine. There are also permutations of antibody and peptide conjugates, nanocarriers, and lipoplexes. Of note is the use of pRNA nanoparticles, derivative of the phi29 bacteriophage, a modality that has shown dramatic efficacy in the delivery of RNA strands and aptamers in preclinical models of cancer.56, 57, 58
For recent reviews of these and other chemical modifications and delivery modalities, see, eg, Roberts et al,14 Kowalski et al,59 Yu et al,60 and Anderson et al.61
It should also be mentioned that NA therapeutics can be delivered by routes other than direct injection into target tissues, ie, the heart, by intravenous or systemic delivery, or by intramuscular delivery, as would be the case for RNA-based vaccines. For example, NA therapeutics have also been delivered by inhalation,62 orally,63,64 and intradermally.65
Despite their advantageous, highly selective target specificity, one major technological gap retarding the realization of anti-CVD RNA therapeutics in the clinic is that of cell-, tissue-, or organ-specific targeting. Significant strides have been taken in experimental cardiology to address this, including use of vectors presenting with certain tropism, specific promoter-driven vector expression, and nanocarrier technology. In this context, AAV-based technology is currently the only vector for gene therapy approved in the United States and Europe. In particular, AAV gene therapy has proven successful in the treatment of certain genetic disorders including spinal muscular atrophy and lipoprotein lipase expression to treat lipoprotein lipase deficiency, see Table 2. Unfortunately, the recent death of 3 children enrolled in the NCT03199469 trial using AAV8 vector to overexpress MTM1 to treat X-linked myotubular myopathy (XLMTM) has raised red flags as to the pharmaco-safety of AAV-based gene therapy. Major hurdles remain including acute toxicity and immediate and long-term immune resistance to AAVs and other expression vectors. This may be due in part to pre-existing immunity and in particular, cytotoxic T-cell activation and neutralizing antibody production by the human adaptive immune system. Efforts to overcome immune resistance to AAVs will be necessary before this therapeutic modality to be of greater use for NA-based therapies.
Table 2.
Functional Gene Candidates in Cardiovascular Disease Identified by RNA Sequencing
| Specimen | Form of Sequencing | Identified RNA Target | First Author |
|---|---|---|---|
| Mouse heart tissue from control and infarcted region of I/R mice | Single-cell SORT sequencing | mRNA: cytoskeleton-associated protein 4 (CKAP4) | Gladka et al94 |
| WT (PKP2-corrected) and c.2013delC hiPSC-derived epicardial cells | Single-cell SORT sequencing | mRNA: Transcription Factor AP-2-Alpha (TFAP2A) | Kohela et al95 |
| Human primary cardiac fibroblasts and single-cell RNA-seq on hearts from PlnR9C/+ and WT mice (cardiac fibrosis phenotype) | Total RNA-seq (human primary cardiac fibroblasts) and single-cell RNA-seq of noncardiomyocytes | mRNA: Interleukin-11 | Schafer et al96 |
| Human LV tissue from HF and donor hearts | Total RNA-seq and small RNA-seq overlaid with histology for fibrosis (Masson Trichrome) | mRNA: COL1A1 | Hua et al97 |
| LV cardiomyocyte nuclei from TAC or sham-operated mice and human end-stage failing (DCM) and donor hearts | PCM1+ single nuclear RNA seq | L(i)ncRNAs: Gas5 and Sghrt | See et al67 |
| Primary HCASMC | Total RNA-seq | LncRNA: smooth muscle–induced lncRNA enhances replication (SMILR) | Ballantyne et al98 |
| HCASMCs | Total RNA-seq | LncRNA: smooth muscle and endothelial cell-enriched migration/differentiation-associated long NonCoding RNA (SENCR) | Bell et al99 |
| Angiotensin-II treated rat vascular smooth muscle cells (VSMC) | Small RNA-seq | MiRNA: miR-132/212 | Jin et al100 |
| Engineered heart tissue from rat hearts under a mechanical stress model of cardiac hypertrophy | Small RNA-sequencing | MiRNA: miR-21-5p | Hirt et al101 |
| GFP-labeled fibroblasts from different regions in mouse myocardial infarction model (LAD) | Single-cell and bulk RNA-sequencing | mRNA: CTHRC1 (Collagen Triple Helix Repeat Containing 1) | Ruiz-Villalba et al102 |
| Mouse hearts +/- TAC +/- JQ1 (BET protein inhibitor) | Single-cell RNA-seq and single-cell ATAC-seq | MRNA: Meox1 | Alexanian et al103 |
| Carotid artery tissue from rat models of type I (streptozotocin) and type II (Zucker diabetic fatty rat) diabetes | mRNA and miRNA-sequencing | MiRNA: miR-29c and miR-204 | Torella et al104 |
| Human LV samples from control, hypertrophic and dilated cardiomyopathy and ischemic heart disease patients. Sham and TAC mouse LV samples. Human embryonic stem cell-derived cardiomyocytes through 28 days of differentiation. | CircRNA sequencing | CircRNA: circSLC8A1-1 | Tan et al,40 Lim et al105 |
| Peripheral blood cells from healthy and heart failure patients | CircRNA sequencing | CircRNA: hsa_circ_0097435 | Han et al106 |
DCM = dilated cardiomyopathy; HCASMC = human coronary artery smooth muscle cell; lncRNA = long noncoding RNA; LAD = left anterior descending; LV = left ventricular; TAC = transverse aortic constriction; WT = wild type; other abbreviations as in Table 1.
State of the Art: From Cells to Humans/Bench to Bedside
Although the history of drug discovery is filled with eclectic stories of serendipitous findings of molecules with a therapeutic benefit, current socioeconomic health crises, such as CVD, require fast, effective, well-validated drug targets and therapies with a known mode of biological action.66 Both commercial and academic laboratories alike are turning to high-throughput approaches to drug discovery in general. In the case of RNA therapeutics, this predominantly takes the form of the following: 1) deep RNA sequencing; and/or 2) cellular screening.
Deep sequencing for RNA target discovery
Bulk and single-cell RNA technology has rapidly become a standard and affordable experimental read out in basic and translational research, allowing us to determine the alteration in the RNA landscape in CVD and models. Cellular and preclinical models in experimental cardiology are then employed to validate the functional relevance of the candidates to disease initiation and progression. Once new potential RNA targets have been identified, the ability to modulate them in vivo effectively and safely must be established. Although next-generation sequencing technology has largely surpassed gene array approaches for transcriptome analyses, commercial services and platforms continue to be developed, such as circRNA, lncRNA, and mRNA arrays provided by companies such as Arraystar Inc, which target annotated noncoding and protein coding RNAs in the mouse, rat, or human genome.
Only within the last 5 years have the first single-cell data sets from human heart tissue emerged.67,68 Caveats and limitations for single-cell sequencing include the size of healthy cardiomyocytes, requiring adaptation of many standard flow cytometry nozzles, and microfluidics-based systems and the availability of fresh heart tissue. One way to circumvent the aforementioned limitations includes the employment of single nuclear isolation and sequencing (snRNA-seq), which can be performed from archived frozen tissue. A caveat, snRNA-seq gives a different readout than that of cellular RNA sequencing; thus, further bioinformatic considerations need to be made.
Table 2 summarizes some important functional candidate genes that have been identified and experimentally validated from RNA-sequencing approaches in the field of CVD, with representation of different forms of RNA target.
Cellular screening for RNA target discovery
Although next-generation sequencing has the potential to reveal the relative abundance of all RNA species in a biological sample, not all differentially expressed genes may be disease-driving or protective against disease stimuli. Many differentially expressed transcripts are likely to be an artifact or consequence of the main pathological driver causing remodeling of the organ or tissue. Ergo, RNA-seq data set–derived candidate genes should be validated for their functional relevance and potential as a drug target.
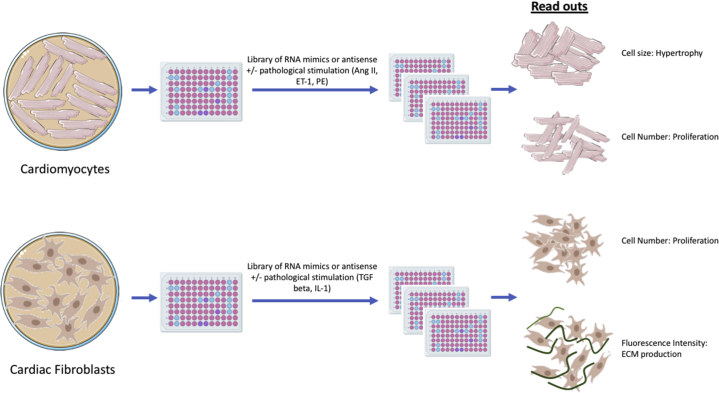
To bypass the expensive, time-consuming, and analytically heavy task of RNA-seq, direct functional screening of a library of genes known to have a role in a particular disease-related read-out is a more efficient alternative. The approach to gene manipulation in the screen could be by knocking down a set of RNA targets using antisense technology or a genetic approach, such as CRISPR-Cas9, or alternatively, overexpression through expression vectors or transfection of RNA mimics.69 In the case of CVD, the functional read out may be proliferation or migration of fibroblast cells, regeneration, hypertrophy, apoptosis, or stiffening of cardiomyocytes or activation of macrophages. The intervention may be a transfection of antisense RNAs, RNA mimetics, or overexpression vectors with or without additional pathology-relevant treatment of cells such as transforming growth factor-beta (fibroblasts), phenylephrine (cardiomyocyte hypertrophy), or lipopolysaccharide (mimicking infection on white blood cells). Readouts should be amenable to high-throughput measurement and analysis, such as high content imaging and automated analysis. Figure 2 shows an example of a high-throughput screening pipeline in the context of searching for new targets in cardiovascular biology, the process of which is further explained with examples by Eulalio et al.70
Figure 2.
Schematic of an Example High-Throughput Functional Screen for New RNA Targets in the Field of Cardiovascular Disease, With Readouts Measured by Cellular Imaging and (Semi) Automated Analysis
IL = interleukin; ECM = extracellular matrix; ET-1 = endothelin 1; PE = phenylephrine; TGF = transforming growth factor.
In one study, Eulalio et al70 screened a library of 875 mature human miRNAs in neonatal rat ventricular myocytes, measuring immunostaining of the proliferation-linked antigen Ki-67 and for 5-ethynyl-2′-deoxyuridine (incorporation, the latter of which reflects DNA synthesis. A subsequent series of screens with readouts including phospho-Histone 3 Serine 10 (H3S10) and Aurora B kinase immunostaining, resulted in focusing on miR-590-3p and miR-199a-3p as inducers of cell cycle re-entry and proliferation in adult rodent cardiomyocytes.70 In vivo, intramyocardial injection of miR-590 or miR-199a miRNA mimetics or AAV9s expressing miR-590 or -199a increased DNA synthesis and H3S10 phosphorylation and increased heart size in the absence of cardiomyocyte hypertrophy.71 Importantly, in the context of CVD, overexpression of miR-590 or -199a reduced infarct size in a mouse model of myocardial infarction. A follow-up study optimized miRNA mimetic administration in swine and similarly found that intracardiac injection of either miR-590 or -199a was able to improve cardiac function and survival, induce cardiomyocyte proliferation, and reduce infarct size in this large animal model.72
In another study using a similar high-throughput high imaging content screen with neonatal rat ventricular myocytes, fibroblasts, or bone marrow–derived macrophages, Verjans et al73 screened 194 microRNAs selected based on previous data showing differential expression in human samples and animal models of inflammatory cardiac disease. A handful of microRNAs involved in regulation of cardiomyocyte size, collagen production by fibroblasts, and roundness of macrophages were identified as new functional target miRNAs for further investigation.
Translational success stories
One of the more contemporary and transformative forms of NA-based therapy is the use of CRISPR-Cas endonuclease gene editing technology. Although CRISPR-Cas technology has significant potential across many diseases, one that has attracted significant attention is editing of the dystrophin gene as it relates to the treatment of DMD, a process that has been referred to as “myoediting.” For an excellent overview of the subject, see Zhang et al.74 Thus, NA-based therapies of DMD will be considered as an exemplar of CRISPR-Cas–mediated gene editing in the context of CVD.
Muscular dystrophies constitute a broad range of genetic-based diseases with varying degrees of phenotypic penetrance. DMD is an X-linked recessive monogenic disorder that can, depending on the specific mutation, have both severe skeletal and cardiac muscle phenotypes.75 Among the 7,000+ recognized mutations in dystrophin, certain mutational “hot spots” account for a substantial proportion of the mutations. By using a complex array of small guide (sg)RNAs and viral vectors (AAV9) to express Cas9 and taking advantage of the proximity of protospacer adjacent motifs to naturally occurring exon/intron splice junctions, CRISPR gene editing can be used to cause exon deletion or skipping, resulting in the removal of premature stop codons within the target gene, in this case, dystrophin. Fortunately, deletion or editing of specific exons, such as exons 44, 45, or 51,76, 77, 78 is permissive of virtually full-length, but more importantly, functional dystrophin protein expression resulting in reversal of the cardiac and skeletal muscle DMD phenotypes.
Unfortunately, CRISPR-Cas gene editing is not always as precise or as directed (off-target effects) as would be desirable for human clinical usage. Cells such as cardiomyocytes, which are virtually nonproliferative, present a limitation as to the type of gene editing that can be used to, most commonly, nonhomologous end joining. Recently, the Olson group has used an adenine base editor (adenine base editors convert G:C base pairs to A:T base pairs) in conjunction with engineered versions of CRISPR (nCas9 or dCas9), to perform “precise” correction of exon deletion mutations via what is called a prime editing system.78
Olson et al have provided a robust proof of principle that gene editing demonstrates significant promise as a therapeutic modality.76 However, the technique, although robust, is not applicable to all variants of diminished dystrophin expression. Further, it should be noted that the previously cited work is still quite preliminary and is in the preclinical stage, with questions remaining before clinical implementation including issues of permissible level of AAV9 dose, durability of response, and the potential for immune reaction.
In contrast, several other DMD therapies are already FDA approved, including eteplirsen (EXONDYS 51), golodirsen (VYONDYS 53), and casimersen (AMONDYS 45), all ASOs developed by Sarepta Therapeutics, and another dozen or so exon-specific DMD therapeutics undergoing development. However, in contrast to myoediting approaches, treatments such as EXONDYS 51 only rescue dystrophin expression to ∼0.44% of normal. Similarly, VYONDYS 53 and AMODYS 45 on average replace ∼1%-2% of normal dystrophin, which is sufficient to have a detectable phenotypic effect. In each case, all of the approved therapeutics require a weekly infusion regimen. Other companies in the preclinical DMD NA therapy space include PepGen, Entrada Therapeutics, and Vertex Pharmaceuticals.
An exciting development at the intersection of CVD, HF, and NA-based therapeutics is work performed by Dr Thomas Thum and colleagues, work that has been actively pursued in both the academic and biotechnology sectors. In particular, Thum’s group has demonstrated in preclinical models, rodents and pigs,79, 80, 81 and now in a first-in human Phase 1b, randomized, double-blinded, placebo-controlled clinical trial82 that antisense targeting of miR-132 has salutary properties in patients with HF.
For quite some time, it has been known that the miR-212/132 family is important for biological processes relevant to a broad spectrum of diseases, including cancer, fibrosis, and neurological and cardiovascular diseases. In particular, there are established links between the gene expression regulatory effects of miR-132 and a number of signaling pathways, to name a few, the SIRT1, NF-kB, FoxA1, FoxO3, PTEN, HDACs, and calcineurin/NFAT. In turn, many of these pathways can affect cardiac biology in terms of modulating cellular hypertrophy and autophagy.80
Working initially in cell culture models, Ucar et al80 demonstrated that miR-212/132 null mice were substantially protected from pressure overload–induced HF, reducing end-systolic and -diastolic dimensions, preserving fractional shortening (%), and reducing myocardial fibrosis. Progressing to large animal HF and post-MI models, the group demonstrated favorable pharmacokinetics (PK/PD) for an miR-132 anti-miR with salutary physiological effects similar to those seen in rodents.79,81 Ultimately, these studies led to the previously noted first-in-human Phase 1B dose-ranging clinical trial with the ASO antimiR (designated CDR132L, Cardior Pharmaceuticals). In particular, 116 patients were screened for inclusion criteria with 28 patients ultimately randomized to 1 of 4 dose levels (0.32 to 10 mg/kg) vs placebo. Inclusion criteria included New York Heart Association functional class 1-3 chronic HF with a left ventricular ejection fraction between >30% and <50% or an N-terminal pro–B-type natriuretic peptide >125 ng/L with major exclusion criteria of patients with nonischemic HF, alcoholic cardiomyopathy, or dysfunction secondary to AF. Preliminary results demonstrated that treatment was safe and well-tolerated, with patients receiving the anti-miR having fewer treatment emergent adverse events (TEAE), with a general improvement in biomarkers and a signal (not statistically significant) toward an increase in LVEF. Overall, the results are encouraging and suggest that reduction in miR-132 expression may be of benefit in chronic HF patients when combined with standard of care.
Another anti-miR–based therapy relevant to CVD is miR-92a-3p (MRG-110, miRagen Therapeutics), which recently underwent a first-in-human clinical trial.83 An extensive literature indicates that miR-92 and the miR-17-92a cluster is involved in numerous biological processes, including cardiovascular effects, angiogenesis, and wound healing. Several studies by Dimmeler’s group have detailed the effects of miR-92a over and under expression,31,84, 85, 86, 87, 88 including in a cell-type specific manner.88 In particular, inhibition of miR-92a promotes angiogenesis while preventing neointimal formation as well as affecting expression of genes associated with autophagy, including ATG4a, ATG9b and LC3B-1/II, and others associated with metabolism, including cd36, Fabp4, Atp1b2, and cytochrome c subunits.
In large animal models,85,89 inhibition of miR-92a had protective effects against ischemia/reperfusion injury, promoted angiogenesis, and reduced adverse ventricular remodeling postinfarct. Unfortunately, these studies were too small to draw conclusions about preservation of cardiac function or to conclude that there may be a survival advantage.
A further success story demonstrating advancement from basic experimental investigations into functional RNAs to commercialization of RNA therapeutics is that of the Swiss biotech company, HAYA Therapeutics, spearheaded by Drs. Samir Ounzain and Daniel Blessing. The company was founded on sound preclinical data showing that ASO-mediated inhibition of a super enhancer-associated fibroblast-enriched lncRNA, Wispr, reduced fibrosis and cardiac dysfunction caused by MI in mice.90 Further, HAYA is employing high-throughput screening to look for novel antifibrosis targets in multiple tissues and cells, focusing on noncoding RNA targets and therapies.
Towards Tailored Medicine
Importantly, not all forms of CVD are created equal, and a “one-size-fits all” approach to current treatment options is unlikely to be the most effective way to reduce symptoms across the clinical spectrum of CVD. For example, sex differences—in clinical presentation, etiology, and response to treatment—have been noted in hypertension, HF, atherosclerosis, coronary heart disease, and myocardial infarction.91 A deeper understanding disease mechanisms and development of therapies for distinct types of HF, such as HF with preserved ejection fraction, which is an increasing health burden especially in obese older women, is needed. Empagliflozin, a sodium glucose co-transporter 2 small molecule inhibitor, was recently approved as the first anti-HF with preserved ejection fraction medication following a series of EMPEROR-Preserved (Empagliflozin Outcome Trial in Patients With Chronic Heart Failure With Preserved Ejection Fraction) clinical trials.92,93 As the molecular underpinnings of this poorly understood heterogeneous syndrome are being unveiled, including transcriptional disease drivers, the horizon may be filled with novel RNA targets with therapeutic potential for patients with diastolic dysfunction.
With falling costs and increasing sensitivity and speed of next-generation RNA sequencing, rapid identification of the RNA profile of individual patients is becoming a reality. Although obtaining a full cardiac transcriptome sourced from an endomyocardial biopsy has its challenges and risks, the use of surrogate biopsies, such as peripheral white blood cells, to obtain RNA and protein that reflects that of the heart is a current area of translational research.
Basic and translational scientists, synthetic chemists, and drug development teams need to communicate and collaborate to identify novel candidate RNAs and proteins for RNA targeting in an attempt to generate and implement more effective, specific, and safe molecular medicines. In many ways, the future of medicine is likely to revolve around genomics and its ability to facilitate NA based therapeutics (Central Illustration).
Funding Support and Author Disclosures
Dr Robinson is funded by the American Heart Association grant #829504. Dr Port is partially funded by a supplement to the National Institutes of Health Training T32 Training Grant 3T32GM136444-02S1; and has a financial interest in Veridian Therapeutics, Inc.
Acknowledgments
SMART Servier Medical Art (smart.servier.com) and bioRENDER (app.biorender.com) are the sources of the schematic biomolecule images used in Figures 1 and 2.
Footnotes
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
Contributor Information
Emma Louise Robinson, Email: emma.2.robinson@cuanschutz.edu.
J. David Port, Email: david.port@cuanschutz.edu.
References
- 1.Cardiovascular diseases (CVDs) https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds)
- 2.Yancy Clyde W., Jessup M., Bozkurt B., et al. 2017 ACC/AHA/HFSA focused update of the 2013 ACCF/AHA Guideline for the Management of Heart Failure. J Am Coll Cardiol. 2017;70:776–803. doi: 10.1016/j.jacc.2017.04.025. [DOI] [PubMed] [Google Scholar]
- 3.Gupta S.K., Foinquinos A., Thum S., et al. Preclinical development of a microRNA-based therapy for elderly patients with myocardial infarction. J Am Coll Cardiol. 2016;68:1557–1571. doi: 10.1016/j.jacc.2016.07.739. [DOI] [PubMed] [Google Scholar]
- 4.Dammes N., Peer D. Paving the road for RNA therapeutics. Trends Pharmacol Sci. 2020;41:755–775. doi: 10.1016/j.tips.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kulkarni J.A., Witzigmann D., Thomson S.B., et al. The current landscape of nucleic acid therapeutics. Nat Nanotechnol. 2021;16:630–643. doi: 10.1038/s41565-021-00898-0. [DOI] [PubMed] [Google Scholar]
- 6.Huang C.K., Kafert-Kasting S., Thum T. Preclinical and clinical development of noncoding rna therapeutics for cardiovascular disease. Circ Res. 2020;126:663–678. doi: 10.1161/CIRCRESAHA.119.315856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaczmarek J.C., Kowalski P.S., Anderson D.G. Advances in the delivery of RNA therapeutics: from concept to clinical reality. Genome Med. 2017;9:60. doi: 10.1186/s13073-017-0450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Damase T.R., Sukhovershin R., Boada C., Taraballi F., Pettigrew R.I., Cooke J.P. The limitless future of RNA therapeutics. Front Bioeng Biotechnol. 2021;9:628137. doi: 10.3389/fbioe.2021.628137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landmesser U., Poller W., Tsimikas S., Most P., Paneni F., Luscher T.F. From traditional pharmacological towards nucleic acid-based therapies for cardiovascular diseases. Eur Heart J. 2020;41:3884–3899. doi: 10.1093/eurheartj/ehaa229. [DOI] [PubMed] [Google Scholar]
- 10.Ren L., Colafella K.M.M., Bovee D.M., Uijl E., Danser A.H.J. Targeting angiotensinogen with RNA-based therapeutics. Curr Opin Nephrol Hypertens. 2020;29:180–189. doi: 10.1097/MNH.0000000000000586. [DOI] [PubMed] [Google Scholar]
- 11.Braga L., Ali H., Secco I., Giacca M. Non-coding RNA therapeutics for cardiac regeneration. Cardiovasc Res. 2021;117:674–693. doi: 10.1093/cvr/cvaa071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Majo F., De Windt L.J. RNA therapeutics for heart disease. Biochem Pharmacol. 2018;155:468–478. doi: 10.1016/j.bcp.2018.07.037. [DOI] [PubMed] [Google Scholar]
- 13.Winkle M., El-Daly S.M., Fabbri M., Calin G.A. Noncoding RNA therapeutics - challenges and potential solutions. Nat Rev Drug Discov. 2021;20:629–651. doi: 10.1038/s41573-021-00219-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts T.C., Langer R., Wood M.J.A. Advances in oligonucleotide drug delivery. Nat Rev Drug Discov. 2020;19:673–694. doi: 10.1038/s41573-020-0075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laina A., Gatsiou A., Georgiopoulos G., Stamatelopoulos K., Stellos K. RNA therapeutics in cardiovascular precision medicine. Front Physiol. 2018;9:953. doi: 10.3389/fphys.2018.00953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crooke S.T., Witztum J.L., Bennett C.F., Baker B.F. RNA-targeted therapeutics. Cell Metab. 2018;27:714–739. doi: 10.1016/j.cmet.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 17.Qadir M.I., Bukhat S., Rasul S., Manzoor H., Manzoor M. RNA therapeutics: Identification of novel targets leading to drug discovery. J Cell Biochem. 2020;121:898–929. doi: 10.1002/jcb.29364. [DOI] [PubMed] [Google Scholar]
- 18.Crick F.H. On protein synthesis. Symp Soc Exp Biol. 1958;12:138–163. [PubMed] [Google Scholar]
- 19.Ohno S. So much “junk” DNA in our genome. Brookhaven Symp Biol. 1972;23:366–370. [PubMed] [Google Scholar]
- 20.Anderson D.M., Anderson K.M., Chang C.L., et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson D.M., Makarewich C.A., Anderson K.M., et al. Widespread control of calcium signaling by a family of SERCA-inhibiting micropeptides. Sci Signal. 2016;9:ra119. doi: 10.1126/scisignal.aaj1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson B.R., Makarewich C.A., Anderson D.M., et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science. 2016;351:271–275. doi: 10.1126/science.aad4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Small E.M., Thatcher J.E., Sutherland L.B., et al. Myocardin-related transcription factor-a controls myofibroblast activation and fibrosis in response to myocardial infarction. Circ Res. 2010;107:294–304. doi: 10.1161/CIRCRESAHA.110.223172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu P., Mo Y., Peng M., et al. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol Cancer. 2020;19:22. doi: 10.1186/s12943-020-1147-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartford C.C.R., Lal A. When long noncoding becomes protein coding. Mol Cell Biol. 2020;40(6):e00528–e00619. doi: 10.1128/MCB.00528-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Azodi M., Kamps R., Heymans S., Robinson E.L. The missing “lnc” between genetics and cardiac disease. Noncoding RNA. 2020;6(1):3. doi: 10.3390/ncrna6010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christofi T., Zaravinos A. RNA editing in the forefront of epitranscriptomics and human health. J Transl Med. 2019;17:319. doi: 10.1186/s12967-019-2071-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Da Costa Martins P.A., De Windt L.J. MicroRNAs in control of cardiac hypertrophy. Cardiovasc Res. 2012;93:563–572. doi: 10.1093/cvr/cvs013. [DOI] [PubMed] [Google Scholar]
- 29.Xun Y., Tang Y., Hu L., et al. Purification and identification of miRNA target sites in genome using DNA affinity precipitation. Front Genet. 2019;10:778. doi: 10.3389/fgene.2019.00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu N., Bezprozvannaya S., Williams A.H., et al. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonauer A., Carmona G., Iwasaki M., et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–1713. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 32.Kilikevicius A., Meister G., Corey D.R. Reexamining assumptions about miRNA-guided gene silencing. Nucleic Acids Res. 2022;50(2):617–634. doi: 10.1093/nar/gkab1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim T.-K., Hemberg M., Gray J.M., et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uszczynska-Ratajczak B., Lagarde J., Frankish A., Guigo R., Johnson R. Towards a complete map of the human long non-coding RNA transcriptome. Nat Rev Genet. 2018;19:535–548. doi: 10.1038/s41576-018-0017-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen G., Wang Z., Wang D., et al. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res. 2013;41:D983–D986. doi: 10.1093/nar/gks1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quek X.C., Thomson D.W., Maag J.L., et al. lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015;43:D168–D173. doi: 10.1093/nar/gku988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carrara M., Fuschi P., Ivan C., Martelli F. Circular RNAs: methodological challenges and perspectives in cardiovascular diseases. J Cell Mol Med. 2018;22:5176–5187. doi: 10.1111/jcmm.13789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bai Y., Zhang Y., Han B., et al. Circular RNA DLGAP4 ameliorates ischemic stroke outcomes by targeting miR-143 to regulate endothelial-mesenchymal transition associated with blood-brain barrier integrity. J Neurosci. 2018;38:32–50. doi: 10.1523/JNEUROSCI.1348-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan W.L., Lim B.T., Anene-Nzelu C.G., et al. A landscape of circular RNA expression in the human heart. Cardiovasc Res. 2017;113:298–309. doi: 10.1093/cvr/cvw250. [DOI] [PubMed] [Google Scholar]
- 41.Vausort M., Salgado-Somoza A., Zhang L., et al. Myocardial infarction-associated circular RNA predicting left ventricular dysfunction. J Am Coll Cardiol. 2016;68:1247–1248. doi: 10.1016/j.jacc.2016.06.040. [DOI] [PubMed] [Google Scholar]
- 42.Tsitsipatis D., Gorospe M. Practical guide for circular RNA analysis: Steps, tips, and resources. Wiley Interdiscip Rev RNA. 2021;12:e1633. doi: 10.1002/wrna.1633. [DOI] [PubMed] [Google Scholar]
- 43.Ottesen E.W. SS-N1 makes the first FDA-approved drug for spinal muscular atrophy. Transl Neurosci. 2017;8:1–6. doi: 10.1515/tnsci-2017-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hua Y., Sahashi K., Hung G., et al. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010;24:1634–1644. doi: 10.1101/gad.1941310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soh J., Iqbal J., Queiroz J., Fernandez-Hernando C., Hussain M.M. MicroRNA-30c reduces hyperlipidemia and atherosclerosis in mice by decreasing lipid synthesis and lipoprotein secretion. Nat Med. 2013;19:892–900. doi: 10.1038/nm.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Z., Nagy P.D. Diverse roles of host RNA binding proteins in RNA virus replication. RNA Biol. 2011;8:305–315. doi: 10.4161/rna.8.2.15391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Antic D., Keene J.D. Insights from model systems. Embryonic lethal abnormal visual RNA-binding protiens involved in growth, differentiation, and posttranscriptional gene expression. Am J Hum Genet. 1997;61:273–278. doi: 10.1086/514866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mukherjee N., Corcoran D.L., Nusbaum J.D., et al. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples Pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liao Y., Castello A., Fischer B., et al. The cardiomyocyte RNA-binding proteome: links to intermediary metabolism and heart disease. Cell Rep. 2016;16:1456–1469. doi: 10.1016/j.celrep.2016.06.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lavenniah A., Luu T.D.A., Li Y.P., et al. Engineered circular RNA sponges act as miRNA inhibitors to attenuate pressure overload-induced cardiac hypertrophy. Mol Ther. 2020;28:1506–1517. doi: 10.1016/j.ymthe.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ng E.W.M., Shima D.T., Calias P., Cunningham E.T., Guyer D.R., Adamis A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discov. 2006;5:123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- 52.Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 53.Karikó K., Weissman D. Naturally occurring nucleoside modifications suppress the immunostimulatory activity of RNA: implication for therapeutic RNA development. Curr Opin Drug Discov Devel. 2007;10:523–532. [PubMed] [Google Scholar]
- 54.Karikó K. In vitro-Transcribed mRNA therapeutics: out of the shadows and into the spotlight. Mol Ther. 2019;27:691–692. doi: 10.1016/j.ymthe.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.deLorimier E., Coonrod L.A., Copperman J., et al. Modifications to toxic CUG RNAs induce structural stability, rescue mis-splicing in a myotonic dystrophy cell model and reduce toxicity in a myotonic dystrophy zebrafish model. Nucleic Acids Res. 2014;42:12768–12778. doi: 10.1093/nar/gku941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shu Y., Cinier M., Shu D., Guo P. Assembly of multifunctional phi29 pRNA nanoparticles for specific delivery of siRNA and other therapeutics to targeted cells. Methods. 2011;54:204–214. doi: 10.1016/j.ymeth.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoo J.Y., Yeh M., Kaur B., Lee T.J. Targeted delivery of small noncoding RNA for glioblastoma. Cancer Lett. 2021;500:274–280. doi: 10.1016/j.canlet.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y., Leonard M., Shu Y., et al. Overcoming tamoxifen resistance of human breast cancer by targeted gene silencing using multifunctional pRNA nanoparticles. ACS Nano. 2017;11:335–346. doi: 10.1021/acsnano.6b05910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kowalski P.S., Rudra A., Miao L., Anderson D.G. Delivering the messenger: advances in technologies for therapeutic mRNA delivery. Mol Ther. 2019;27:710–728. doi: 10.1016/j.ymthe.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu A.M., Choi Y.H., Tu M.J. RNA Drugs and RNA targets for small molecules: principles, progress, and challenges. Pharmacol Rev. 2020;72:862–898. doi: 10.1124/pr.120.019554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anderson Brooke A., Freestone Graeme C., Low A., et al. Towards next generation antisense oligonucleotides: mesylphosphoramidate modification improves therapeutic index and duration of effect of gapmer antisense oligonucleotides. Nucleic Acids Res. 2021;49:9026–9041. doi: 10.1093/nar/gkab718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chow M.Y.T., Qiu Y., Lam J.K.W. Inhaled RNA therapy: from promise to reality. Trends Pharmacol Sci. 2020;41:715–729. doi: 10.1016/j.tips.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ball R.L., Bajaj P., Whitehead K.A. Oral delivery of siRNA lipid nanoparticles: fate in the GI tract. Sci Rep. 2018;8:2178. doi: 10.1038/s41598-018-20632-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O'Driscoll C.M., Bernkop-Schnürch A., Friedl J.D., Préat V., Jannin V. Oral delivery of non-viral nucleic acid-based therapeutics - do we have the guts for this? Eur J Pharm Sci. 2019;133:190–204. doi: 10.1016/j.ejps.2019.03.027. [DOI] [PubMed] [Google Scholar]
- 65.Gallant-Behm C.L., Piper J., Lynch J.M., et al. A MicroRNA-29 Mimic (Remlarsen) represses extracellular matrix expression and fibroplasia in the skin. J Invest Dermatol. 2019;139:1073–1081. doi: 10.1016/j.jid.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 66.Schlueter P.J., Peterson R.T. Systematizing serendipity for cardiovascular drug discovery. Circulation. 2009;120:255–263. doi: 10.1161/CIRCULATIONAHA.108.824177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.See K., Tan W.L.W., Lim E.H., et al. Single cardiomyocyte nuclear transcriptomes reveal a lincRNA-regulated de-differentiation and cell cycle stress-response in vivo. Nat Commun. 2017;8:225. doi: 10.1038/s41467-017-00319-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nomura S., Satoh M., Fujita T., et al. Cardiomyocyte gene programs encoding morphological and functional signatures in cardiac hypertrophy and failure. Nat Commun. 2018;9 doi: 10.1038/s41467-018-06639-7. 4435-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu S., Li W., Liu J., et al. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9 library. Nat Biotechnol. 2016;34:1279–1286. doi: 10.1038/nbt.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eulalio A., Mano M., Dal Ferro M., et al. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492:376–381. doi: 10.1038/nature11739. [DOI] [PubMed] [Google Scholar]
- 71.Lesizza P., Prosdocimo G., Martinelli V., Sinagra G., Zacchigna S., Giacca M. Single-dose intracardiac injection of pro-regenerative MicroRNAs improves cardiac function after myocardial infarction. Circ Res. 2017;120:1298–1304. doi: 10.1161/CIRCRESAHA.116.309589. [DOI] [PubMed] [Google Scholar]
- 72.Gabisonia K., Prosdocimo G., Aquaro G.D., et al. MicroRNA therapy stimulates uncontrolled cardiac repair after myocardial infarction in pigs. Nature. 2019;569:418–422. doi: 10.1038/s41586-019-1191-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Verjans R., Derks W.J.A., Korn K., et al. Functional screening identifies microRNAs as multi-cellular regulators of heart failure. Sci Rep. 2019;9:6055. doi: 10.1038/s41598-019-41491-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Y., Long C., Bassel-Duby R., Olson E.N. Myoediting: toward prevention of muscular dystrophy by therapeutic genome editing. Physiol Rev. 2018;98:1205–1240. doi: 10.1152/physrev.00046.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Long C., McAnally J.R., Shelton J.M., Mireault A.A., Bassel-Duby R., Olson E.N. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science. 2014;345:1184–1188. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Olson E.N. Toward the correction of muscular dystrophy by gene editing. Proc Natl Acad Sci U S A. 2021;118(22) doi: 10.1073/pnas.2004840117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Min Y.L., Chemello F., Li H., et al. Correction of three prominent mutations in mouse and human models of duchenne muscular dystrophy by single-cut genome editing. Mol Ther. 2020;28:2044–2055. doi: 10.1016/j.ymthe.2020.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chemello F., Chai A.C., Li H., et al. Precise correction of Duchenne muscular dystrophy exon deletion mutations by base and prime editing. Sci Adv. 2021;7(18) doi: 10.1126/sciadv.abg4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Batkai S., Genschel C., Viereck J., et al. CDR132L improves systolic and diastolic function in a large animal model of chronic heart failure. Eur Heart J. 2021;42:192–201. doi: 10.1093/eurheartj/ehaa791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ucar A., Gupta S.K., Fiedler J., et al. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat Commun. 2012;3:1078. doi: 10.1038/ncomms2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Foinquinos A., Batkai S., Genschel C., et al. Preclinical development of a miR-132 inhibitor for heart failure treatment. Nat Commun. 2020;11:633. doi: 10.1038/s41467-020-14349-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Taubel J., Hauke W., Rump S., et al. Novel antisense therapy targeting microRNA-132 in patients with heart failure: results of a first-in-human Phase 1b randomized, double-blind, placebo-controlled study. Eur Heart J. 2021;42:178–188. doi: 10.1093/eurheartj/ehaa898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Abplanalp W.T., Fischer A., John D., et al. Efficiency and target derepression of anti-miR-92a: results of a first in human study. Nucleic Acid Ther. 2020;30:335–345. doi: 10.1089/nat.2020.0871. [DOI] [PubMed] [Google Scholar]
- 84.Daniel J.M., Penzkofer D., Teske R., et al. Inhibition of miR-92a improves re-endothelialization and prevents neointima formation following vascular injury. Cardiovasc Res. 2014;103:564–572. doi: 10.1093/cvr/cvu162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hinkel R., Penzkofer D., Zuhlke S., et al. Inhibition of microRNA-92a protects against ischemia/reperfusion injury in a large-animal model. Circulation. 2013;128:1066–1075. doi: 10.1161/CIRCULATIONAHA.113.001904. [DOI] [PubMed] [Google Scholar]
- 86.Lucas T., Bonauer A., Dimmeler S. RNA therapeutics in cardiovascular disease. Circ Res. 2018;123:205–220. doi: 10.1161/CIRCRESAHA.117.311311. [DOI] [PubMed] [Google Scholar]
- 87.Lucas T., Schafer F., Muller P., Eming S.A., Heckel A., Dimmeler S. Light-inducible antimiR-92a as a therapeutic strategy to promote skin repair in healing-impaired diabetic mice. Nat Commun. 2017;8:15162. doi: 10.1038/ncomms15162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rogg E.M., Abplanalp W.T., Bischof C., et al. Analysis of cell type-specific effects of microRNA-92a provides novel insights into target regulation and mechanism of action. Circulation. 2018;138:2545–2558. doi: 10.1161/CIRCULATIONAHA.118.034598. [DOI] [PubMed] [Google Scholar]
- 89.Bellera N., Barba I., Rodriguez-Sinovas A., et al. Single intracoronary injection of encapsulated antagomir-92a promotes angiogenesis and prevents adverse infarct remodeling. J Am Heart Assoc. 2014;3 doi: 10.1161/JAHA.114.000946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Micheletti R., Plaisance I., Abraham B.J., et al. The long noncoding RNA Wisper controls cardiac fibrosis and remodeling. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aai9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mosca L., Barrett-Connor E., Wenger N.K. Sex/gender differences in cardiovascular disease prevention: what a difference a decade makes. Circulation. 2011;124:2145–2154. doi: 10.1161/CIRCULATIONAHA.110.968792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Khumbhani D.J. Empagliflozin outcome trial in patients with chronic heart failure with preserved ejection fraction - EMPEROR-Preserved. https://www.acc.org/Latest-in-cardiology/clinical-trials/2021/08/25/23/07/emperor-preserved Available at:
- 93.Butler J., Filippatos G., Siddiqi T.J., et al. Empagliflozin, health status, and quality of life in patients with heart failure and preserved ejection fraction: the EMPEROR-Preserved trial. Circulation. 2022;145:184–193. doi: 10.1161/CIRCULATIONAHA.121.057812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gladka M.M., Molenaar B., de Ruiter H., et al. Single-cell sequencing of the healthy and diseased heart reveals cytoskeleton-associated protein 4 as a new modulator of fibroblasts activation. Circulation. 2018;138(2):166–180. doi: 10.1161/CIRCULATIONAHA.117.030742. [DOI] [PubMed] [Google Scholar]
- 95.Kohela A., van Kampen S.J., Moens T., et al. Epicardial differentiation drives fibro-fatty remodeling in arrhythmogenic cardiomyopathy. Sci Transl Med. 2021;13(612) doi: 10.1126/scitranslmed.abf2750. [DOI] [PubMed] [Google Scholar]
- 96.Schafer S., Viswanathan S., Widjaja A.A., et al. IL-11 is a crucial determinant of cardiovascular fibrosis. Nature. 2017;552(7683):110–115. doi: 10.1038/nature24676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hua X., Wang Y.Y., Jia P., et al. Multi-level transcriptome sequencing identifies COL1A1 as a candidate marker in human heart failure progression. BMC Med. 2020;18(1):2. doi: 10.1186/s12916-019-1469-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ballantyne M.D., Pinel K., Dakin R., et al. Smooth muscle enriched long noncoding RNA (SMILR) regulates cell proliferation. Circulation. 2016;133(21):2050–2065. doi: 10.1161/CIRCULATIONAHA.115.021019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bell R.D., Long X., Lin M., et al. Identification and initial functional characterization of a human vascular cell-enriched long noncoding RNA. Arterioscler Thromb Vasc Biol. 2014;34(6):1249–1259. doi: 10.1161/ATVBAHA.114.303240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jin W., Reddy M.A., Chen Z., Putta S., Lanting L., Kato M., Park J.T., Chandra M., Wang C., Tangirala R.K., Natarajan R. Small RNA sequencing reveals microRNAs that modulate angiotensin II effects in vascular smooth muscle cells. J Biol Chem. 2012;287(19):15672–15683. doi: 10.1074/jbc.M111.322669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hirt M.N., Werner T., Indenbirken D., et al. Deciphering the microRNA signature of pathological cardiac hypertrophy by engineered heart tissue- and sequencing-technology. J Mol Cell Cardiol. 2015;81:1–9. doi: 10.1016/j.yjmcc.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 102.Ruiz-Villalba A., Romero J.P., Hernández S.C., et al. Single-cell RNA sequencing analysis reveals a crucial role for CTHRC1 (collagen triple helix repeat containing 1) cardiac fibroblasts after myocardial infarction. Circulation. 2020;142(19):1831–1847. doi: 10.1161/CIRCULATIONAHA.119.044557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Alexanian M., Przytycki P.F., Micheletti R., et al. A transcriptional switch governs fibroblast activation in heart disease. Nature. 2021;595(7867):438–443. doi: 10.1038/s41586-021-03674-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Torella D., Iaconetti C., Tarallo R., et al. miRNA regulation of the hyperproliferative phenotype of vascular smooth muscle cells in diabetes. Diabetes. 2018;67(12):2554–2568. doi: 10.2337/db17-1434. [DOI] [PubMed] [Google Scholar]
- 105.Lim T.B., Aliwarga E., Luu T.D.A., et al. Targeting the highly abundant circular RNA circSlc8a1 in cardiomyocytes attenuates pressure overload induced hypertrophy. Cardiovasc Res. 2019;115(14):1998–2007. doi: 10.1093/cvr/cvz130. [DOI] [PubMed] [Google Scholar]
- 106.Han J., Zhang L., Hu L., et al. Circular RNA-expression profiling reveals a potential role of hsa_circ_0097435 in heart failure via sponging multiple microRNAs. Front Genet. 2020;11:212. doi: 10.3389/fgene.2020.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]