Abstract
With the number of cancer cases projected to significantly increase over time, researchers are currently exploring “nontraditional” research fields in the pursuit of novel therapeutics. One emerging area that is steadily gathering interest revolves around cellular mechanical machinery. When looking broadly at the physical properties of cancer, it has been debated whether a cancer could be defined as either stiffer or softer across cancer types. With numerous articles supporting both sides, the evidence instead suggests that cancer is not particularly regimented. Instead, cancer is highly adaptable, allowing it to endure the constantly changing microenvironments cancer cells encounter, such as tumor compression and the shear forces in the vascular system and body. What allows cancer cells to achieve this adaptability are the particular proteins that make up the mechanical network, leading to a particular mechanical program of the cancer cell. Coincidentally, some of these proteins, such as myosin II, α-actinins, filamins, and actin, have either altered expression in cancer and/or some type of direct involvement in cancer progression. For this reason, targeting the mechanical system as a therapeutic strategy may lead to more efficacious treatments in the future. However, targeting the mechanical program is far from trivial. As involved as the mechanical program is in cancer development and metastasis, it also helps drive many other key cellular processes, such as cell division, cell adhesion, metabolism, and motility. Therefore, anti-cancer treatments targeting the mechanical program must take great care to avoid potential side effects. Here, we introduce the potential of targeting the mechanical program while also providing its challenges and shortcomings as a strategy for cancer treatment.
Keyword s: actin crosslinkers, cancer progression, cortical mechanics, mechanical adaptability, myosin II
Significance
The number of cancer cases are projected to significantly increase over time, and new strategies are needed to address this deadly disease. Cancer is inherently a disease of altered cell mechanics. Because cancer cells are subjected to numerous types of mechanical stresses during the course of cancer progression, they acquire a system of mechanoresponsive proteins, which allow them to adapt to this changing mechanical landscape. Here, we review the alterations in the cellular mechanical systems associated with cancer progression and how these systems are integrated with many other systems within the cell. We also review the current status of cancer therapeutics that target this machinery as well as the potential opportunity for new therapeutic development in this space.
Introduction
Cancer is a unique disease in which its pathology is not predominantly caused by environmental, pathogenic, or parasitic factors but rather inopportune mutations that render a normal cell malignant. As a result, normal cells begin to develop characteristics of unchecked growth and aberrant behavior, which then become detrimental as tumor formation commences. Being able to understand what gives rise to cancers, how they become metastatic, and the factors that promote or demote their cancerous behavior requires the unraveling of complicated biological and medical problems. When pathologists examine cancer tissue, they can decipher different cancer stages based on cell and tissue shape. Morphology and shape of cancer cells is regulated by many factors such as intracellular tension and altered nucleus function. It is, however, majorly governed by the cortical cytoskeleton, which helps define the stage of cancer progression. These elements are determined by mechanical factors. Specifically, a mechanical program grants these cancer cells the ability to adapt and navigate the constantly changing mechanical environments (i.e., TME). In fact, the mechanical surroundings and the mechanical stresses they impose on the cancer cells continuously evolve throughout the course of cancer development (Fig. 1). These stimuli, in turn, affect gene expression and tumor progression. The mechanical program renders the cancer cells the ability to constantly navigate and respond to mechanical stimuli and potentially acts as a node of communication to other cellular pathways that also drive cancer development.
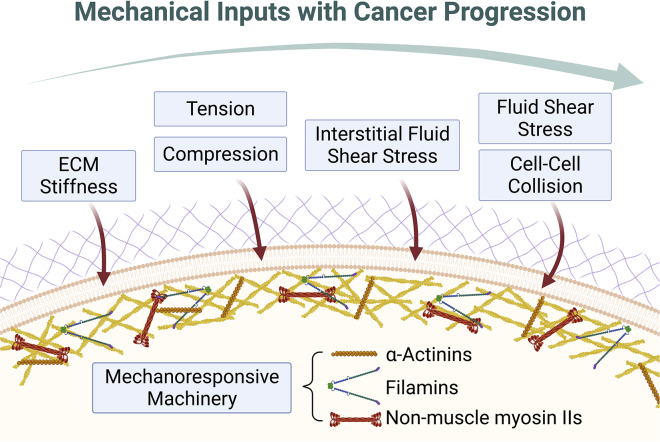
Figure 1.
The interactions between the external mechanical environment and the intrinsic mechanoresponsive machinery. During different stages of cancer progression, cancer cells experience various external mechanical inputs, starting from ECM stiffness and tension, but these mechanical inputs progressively expand to include fluid shear stress and cell-cell collisions and interactions as cancer progresses. While these forces have significant impact on gene expression and the development of cancer cells, these cells also harbor an intrinsic mechanoresponsive machinery that endows the cells with the ability to sense and respond to these mechanical stimuli. For a Figure360 author presentation of this figure, see https://doi.org/10.1016/j.bpj.2022.04.039.
For the purposes of this review, we point out that the mechanical program includes two sets of actin-associated mechanical proteins. One set can bear mechanical load without accumulating in response to mechanical stress (i.e., non-mechanoresponsive). In contrast, a separate set of proteins bears load and accumulates locally in response to the mechanical stresses (i.e., mechanoresponsive) (Fig. 1). As one example of these differential roles, in Dictyostelium, myosin II, α-actinin, cortexillin, and filamin can sense and respond to applied mechanical stress. In contrast, other actin-associated proteins, such as dynacortin, coronin, and fimbrin, can bear load but not accumulate in response to stress (1). Removal of these load-bearing cross-linkers shifts the applied stresses over to the mechanoresponsive proteins, such as myosin II, allowing it to accumulate to a greater extent in response to smaller stresses. In the human scenario, specific paralogs of α-actinins (ACTN4), filamins (filamin B [FLNB]), non-muscle myosin II (NMII) proteins are mechanoresponsive, while specific sister paralogs are not (NMIIB’s mechanoresponsiveness is unique in that it is cell-type specific). Interestingly, the expression levels of at least one these mechanoresponsive proteins are elevated in many different types of cancers (2,3). As a result, these changes reflect a reprogramming of cancer that favors increased mechanical adaptability required for growth and metastasis.
In this review, we discuss how we can consider cancer as a biophysical disease, where the mechanical properties of the surroundings and the cells themselves dictate different stages of cancer progression. We also focus on the mechanical program that confers the degree of adaptability necessary for cancer cells to navigate through complex mechanical environments, and we discuss the targetability of this program alongside potential challenges for future cancer therapeutic development.
The tumor mechanical microenvironment is constantly evolving
The tumor microenvironment (TME) is a dynamic landscape comprised of cancer cells surrounded by extracellular matrix (ECM) and adjacent stromal cells. The TME contains chemical signals that can promote growth and invasion of cancer cells through alternating their mechanical properties and behaviors. The presence of intratumoral hypoxia has been shown in several studies to associate with an increase in invasion and metastasis (4,5). Tumor cells respond to hypoxia through the upregulation of hypoxia-inducible factors (HIFs) 1 and 2. The upregulation of HIFs positively correlates with metastasis and poor prognosis in breast cancer (6, 7, 8, 9). Chronic hypoxia also causes HIF-dependent pulmonary endothelial cell death and increases microvascular permeability, predisposing lungs for metastases (10). Another study revealed that the enhanced motility and invasiveness of breast cancer cells in hypoxic conditions are facilitated by HIF-dependent RhoA-ROCK1 signaling. Under these conditions, phosphorylation of myosin phosphatase target subunit 1 (MYPT1) and myosin light chain (MLC) is increased, leading to changes in integral mechanical activities of these cells, including cell contraction, focal-adhesion formation, and matrix contraction (11). Other chemical signals, such as stress hormone signaling through β-adrenergic receptors, can also modulate the mechanics of cells involved in the tumor and TME. Soluble stress hormones act on Arp2/3, causing actin-filament rearrangements that subsequently increase the stiffness and reduce the deformability of macrophages. These changes can affect the migration and phagocytosis ability of these cells (12). Strikingly, the activation of β-adrenergic signaling also reduces deformability in various cancer cell lines, including ovarian, prostate, melanoma, and leukemia cells. This reduction is associated with actin-cytoskeleton remodeling and myosin II activity (13). These findings elucidate the consistent observation that β-adrenergic signaling is strongly associated with enhanced metastasis and cancer progression (14, 15, 16).
Apart from chemical stimuli, the TME also harbors constant physical and mechanical inputs, thereby forming the mechanical microenvironment. Remarkably, the ECM within the TME is typically thought to be a by-product of the tumor and is actively involved in promoting cancer growth and proliferation, enhancing invasion and dissemination, and altering the gene-expression profile of the tumor cells and, in turn, having profound influence on cancer progression (17).
As tumor formation occurs, certain signals stimulate increased matrix deposition, increasing the overall stiffness of the ECM surrounding the tumor. The expanding tumor gradually bears forces imposed by its surroundings, and, in return, the tumor experiences compression and tension, leading to activation of various growth factors that aid tumor progression (18,19). Simultaneously, the ECM can be remodeled as a consequence of tumor compression, which further exacerbates the tension placed on tumor cells (20). In the instance of invasion, invading cancer cells have to navigate against a progressively stiffened ECM toward the circulatory system (21,22). Throughout this process, these cells continue experiencing other various mechanical forces, ranging from interstitial compression to shear forces. As migrating cancer cells intravasate, they become exposed to shear forces from other adjacent cells within the circulatory system as well as from hydrodynamic flow (23, 24, 25).
Accumulating evidence shows that the tumor mechanical microenvironment can direct and reprogram cancer initiation, proliferation, and invasion. In fact, numerous studies confirm that the mechanical niche can significantly stimulate cancer development (26, 27, 28). Moreover, the mechanical microenvironment is a constantly evolving landscape throughout the course of cancer progression, which continuously challenges cancer cells to adapt to these mechanical cues.
Substrate stiffness affects cancer development
Substrate stiffness is one of the major mechanical inputs experienced by cells and is largely defined by the composition of the ECM surrounding the tumor (27,29, 30, 31). Increasing evidence shows that substrate stiffness affects numerous signaling pathways and acts as a driver for cancer development. For example, increased ECM stiffness leads to enhanced invasion and metastasis in breast cancer cells in three-dimensional models (20,32,33). Other studies in colorectal cancer indicate that matrix stiffness leads to tumorigenesis and cancer progression (34, 35, 36). Specifically, increased activity of lysyl oxidase, an enzyme that catalyzes collagen cross-linking, leads to increased substrate stiffness and allows for progression of colorectal cancer (34). Similar observations were also made in other cancer types, such as glioblastoma and pancreatic and ovarian cancers (29,37,38). Interestingly, substrate stiffness can in turn influence the mechanical properties of the ECM by altering gene expression of cells. Expression of integrins mediated through YAP signaling, matrix metalloproteases, and ECM proteins are increased as substrate stiffness increases (39). It is important to note that YAP signaling may not mediate its effects on substrate stiffness in all cancer types, and YAP has a context-dependent role in mechanotransduction. For example, using tunable three-dimensional cultures, a study established that mechanotransduction occurs independently of YAP signaling in breast cancer (40). Regardless, this positive feedback between substrate stiffness and ECM mechanics highlights the cross communication between substrate stiffness, external mechanical forces, and gene expression.
Many cytoskeletal signaling and regulation pathways are directly altered by substrate stiffness during tumor progression. For example, ROCK1, a regulator of myosin II activity, and ROCK2, a regulator of cofilin, are implicated in modulating breast cancer motility in a substrate-stiffness-dependent manner (41). Using polyacrylamide gels mimicking different levels of substrate stiffness of breast tissue, the authors found that cancer cells cultured on stiff substrates migrated and invaded more than those cultured on soft substrates. These differences are accompanied by elevated expression of ROCK1 and ROCK2 along with enhanced activation of their downstream targets. Another study shows the critical role of myosin II, a central cytoskeletal protein, in promoting tumorigenesis on stiff substrates. In this scenario, the transforming growth factor-β1 pathway responds to elevated substrate stiffness and acts as a key driver for cancer proliferation in hepatocellular carcinoma and breast cancer cells. Furthermore, the addition of blebbistatin, a myosin II inhibitor, blocks the upregulation of the transforming growth factor-β1 pathway (42,43).
Other mechanical forces also influence cancer progression
In addition to substrate stiffness, cancer development is remarkably influenced by other mechanical inputs originating from its surroundings (26,27,44). A recent study demonstrated that cancer invasion is enhanced as its mechanical confinement increases. Specifically, the migration speed of colon and breast cancer cells is higher through narrow microchannels than through wide channels, both of which mimic the various dimensions of TME confinement (45). Furthermore, hemodynamic forces, immunological stress, collisions, and fluid forces greatly impact the gene-expression profile of cells during invasion. Shear stresses were found to act through ligand-dependent activation and phosphorylation of the MAPK/PI3K/Akt pathway, a prominent cancer proliferation pathway (46,47). While shear forces in the circulation are traditionally thought to impose cell death on circulating tumor cells, more intermediate shear forces can facilitate adhesion and extravasation of circulating tumor cells, enhancing their metastatic potential (48,49).
Beyond shear forces found in the circulatory system, fluidic forces in the tumor mechanical microenvironment can also modulate cancer progression, especially during invasion and metastasis. Using microfluidic platforms, a study found that hydrodynamic forces enhance epithelial-mesenchymal transition, a critical step of metastasis, in ovarian cancer cells (50). Fluid forces also increase glioblastoma cell invasion through CXCR4 signaling, a well-studied regulator of glioma invasion. Cells that experience high fluid pressure are correlated with cancer aggression in patient-derived glioma stem cells (51). Additionally, a separate study indicates that tissue fluidity endows glioblastoma cells with the ability to effectively infiltrate into normal brain matter, facilitating their aggressive behavior. The fluid property of the tumor mass, measured by magnetic resonance elastography, correlates significantly with the fingering growth capacity of these cells (52).
Altogether, there is an immense range of mechanical forces that cancer cells must endure during transformation and disease progression. These external forces have strong impacts on the development, progression, and aggressiveness of the tumor cells. However, reciprocity between the mechanical microenvironment and intracellular gene expression and signaling also exists. These cancer cells are not merely acted upon by their physical surrounding. Increasing evidence suggests that an intrinsic mechanical program lends these cells the ability to constantly sense and adapt to their surroundings.
An intrinsic program allows cancer to be mechanically adaptive
Previous studies found a positive correlation between increased tissue stiffness and cancer behavior, which suggests that cancer progression depends on tumor tissue stiffening (53,54). In addition, tissue stiffness can result from increased deposition of matrix proteins like type 1 collagen by cancer-associated fibroblasts (55, 56, 57, 58, 59). However, a recent study showed that inhibiting tumor tissue stiffness in liver of pancreatic ductal adenocarcinoma (PDAC) metastasis enhanced tumor growth and led to diminished survival in orthotopic PDAC mouse models (60). This study indicates that the tumor stroma has a protective role in PDAC, resulting in the opposite outcome from the prediction of a pro-tumorigenic influence on PDAC progression. These studies indicate that there has been a lack of consensus, correlating cancer tissue stiffness and progression.
This lack of consensus also follows the same trend in the correlation between cancer cell stiffness and invasiveness. Stiffer pancreatic cancer cells were shown to be more invasive (61). On the other hand, tumor cells and metastatic tumor cells have been reported to be soft (62,63). Also, recent studies revealed that soft cancer cells can evade T cell killing by impeding perforin-pore formation (64). Furthermore, cell softness can be used as a biomarker for cancer cells with tumor repopulating ability and stem cell markers (65). In the progression of cancer, tumor cell heterogeneity is important to note because some tumor cells are cancer stem cells or tumor stem cells that are intrinsically soft and more resistant to anti-cancer drugs. As a result, using cell softness as a biomarker for cancer cells with more aggressive characteristics remains unsettled. Studies have shown that more deformable cancer cells tend to be more invasive (29,66). On the other hand, there are studies where invasive cancer cells tend to be stiffer (13,61,67). Taken altogether, the relationship between cancer tissue and cell stiffness and invasive behavior depends on the context.
Given the context-dependent nature of cancer behavior, it is important to consider the external landscape experienced by cancer cells and how the internal mechanical machinery must respond to these inputs rather than focusing on individual mechanical phenotypes such as cell stiffness. As with healthy cells, cancer cells must be able to integrate chemical and physical signals from their external environments. Unlike their healthy counterparts, the survival of cancer cells is contingent upon their ability to adapt to mechanically distinct, yet ever-changing, microenvironments. This adaptation requires the mechanoresponsive machinery, which includes the proteins that are uniquely poised to sense and respond to mechanical stresses and that help establish the mechanical and force-producing activities of the cell. Collectively, the mechanoresponsive cytoskeletal machinery in humans encompasses NMII proteins (NMIIA, NMIIB, and NMIIC), specific paralogs of α-actinins (ACTN4, but not ACTN1), and FLN (FLNB, with FLNA to a much smaller extent) (2,68,69).
In cancer, NMII proteins play essential roles in tumor formation, growth, and metastasis, which is driven by NMII’s role in adhesion, mechanotransduction (70), motility, and contractility. A comprehensive review compiled a list of the different cancers with altered NMII expression and/or regulation (3). Interestingly, different cancers express specific subsets of NMII paralogs and exhibit different trends across cancer types and detection methods. For example, PDACs expresses NMIIA and NMIIC but not NMIIB (2). Additionally, driving myosin assembly reduces tumor cell dissemination and metastasis in in vitro and in vivo models (2,71). A recent study mimicked the mechanical properties of breast cancer tissue to investigate the roles of NMIIA and NMIIB in regulating cell’s response to substrate stiffness. In this study, the authors found that the signaling pathways (Rac1-NMIIA and PKCζ-NMIIB) triggered by substrate stiffness also regulate the distribution and activation of NMIIA and NMIIB (72). Note that each NMII isoform’s function does not scale with their cellular concentrations. The isoforms exist in different concentrations that can span 100-fold in range (e.g., NMIIC ranges from 5–10 nM when present, while NMIIA ranges from 500–750 nM in pancreatic cells). Yet, depletion of either has a similar magnitude in decrease of cortical tension (2).
ACTNs play a different role compared with NMII, but their role is just as pervasive across different cancer types. The ACTN family consists of four paralogs that act as actin cross-linkers: ACTN1–4. ACTN4 has been linked to metastasis through alterations in cell behaviors and differentiation (3). A group recently found that ACTN4 promotes proliferation, migration, and metastasis in osteosarcoma and augments invasive ability through the nuclear factor κB pathway (73). In addition, a recent study found that ACTN4 serum levels were significantly higher in cervical cancer patients than in the healthy control patients, suggesting that ACTN4 could be a prognostic biomarker for cervical cancer (74). In cancer stem cells, inhibition of ACTN4 led to increased sensitivity to anti-cancer drugs and reduced spheroid formation, proliferation ability, and tumor formation in vivo (75). Moreover, ACTN4 in breast cancer promotes invasive ability by directing NMIIA localization and distribution and NMIIB mRNA and protein expression (76). In PDAC cells, ACTN4 has been shown to interact with the GTPase dynamin 2 to alter invasive ability and ECM remodeling. The disruption of the dynamin 2 and ACTN4 interaction prevents migration and invasion due to a compromised actin-rich structure present at the basal surface of cells (77).
The FLN proteins are a family of actin cross-linkers that associate with actin filaments (F-actins) to make a dense gel meshwork (78). Human cells encode for three isoforms: FLNA, FLNB, and FLNC (79). Collectively, FLNs are found in the cell cortex, the F-actin-rich region underlying the plasma membrane (79, 80, 81). Recently, work discovered that FLNB knockout in HeLa cells altered the expression of genes involved in apoptosis, tumorigenesis, and metastases (82). Additionally, FLNB expression is upregulated specifically in pancreatic cancerous glands, whereas FLNA is elevated across the entire pancreatic tissue and stroma (2). A recent clinical study found that FLNA expression increased after chemotherapy in patients with colorectal cancer (83). Furthermore, increased FLNA expression induced resistance to a tyrosine kinase receptor inhibitor that is used to treat non-small cell lung cancer (84). Finally, the correlated expression of FLNA and clusterin, a secreted glycoprotein, has the potential to be a hepatocellular carcinoma biomarker (85).
An emerging relevant family of proteins that might play an important role in the mechanical program are the dynamins. The dynamin superfamily consists of dynamins 1, 2, and 3. The superfamily encompasses multi-domain GTPases, which have a modular structure, are greater than 70 kDa, and have a low affinity for guanine nucleotides (86). The dynamin-superfamily proteins mediate membrane fission and fusion during endocytosis and organelle biogenesis and organize the cytoskeleton, among others (87). Dynamins 1, 2, and 3 have different expression levels in tissue. A recent study used qRT-PCR and immunohistochemistry to determine the expression of dynamins 1, 2, and 3 in tissue sections of human hepatocellular carcinoma HCC. Specifically, the group found that the expressions of dynamins 1, 2, and 3 were upregulated in patients with human hepatocellular carcinoma (88). Another group inhibited dynamin in pediatric acute leukemia cell lines and found suppressed cell proliferation and induced caspase-dependent apoptotic cell death (89). One study demonstrated how dynamin GTPase bundles F-actins by forming a helical structure (90). Furthermore, they found that the assembly and disassembly cycles of dynamin generate mechanically stiff actin bundles (90,91).
Overall, NMII, ACTN, and FLN proteins are heavily implicated in cancer progression and serve as biomarkers of cancer formation and progression. Altogether, these observations further highlight the role these mechanoresponsive proteins have in allowing cancer cells to endure and survive in a broad range of changing mechanical environments. Unsurprisingly, upregulation of this mechanical program is associated with poor clinical outcomes (88,92, 93, 94, 95, 96, 97). Pancreatic cancer seems to be particularly vulnerable, as multiple mechanoresponsive proteins are upregulated in PDAC progression.
Targeting the mechanical-adaptability program and its challenges
The mechanical-adaptability program embedded in the mechanical network is an alluring area for cancer therapeutics. Its direct role in tumor metastasis and proliferation makes it an appealing target (98). Unsurprisingly, proteins within this mechanical program have altered expression in many forms of cancer (98). By manipulating this program, one can imagine modifying these proteins to diminish or even reverse cancer progression. Here, we present various studies targeting mechanical program components in the hopes of remedying cancer. These targets include F-actins, α-actinin, NMII, and 14-3-3 (3,71,99,100). However, from a cell-biology standpoint, a system-wide inhibition of individual mechanical proteins should be handled with caution, as perturbing a single component of the mechanical system can lead to unforeseen consequences. Due to this, extensive consideration must be addressed before attempting to modify the mechanical network. The mechanical network is anything but an isolated system, as it is integrated with many other processes and functions of the cell that can also be affected. Such diverse processes include signaling, gene expression, and cell metabolism (101, 102, 103, 104, 105, 106, 107, 108, 109). As a result, solving one process’s defects can introduce a new problem in another cellular process. It is therefore imperative to understand the downstream effects of manipulating mechanical-network components in treating cancer.
Targeting the mechanical program through actin
Actin is a globular protein that assembles into filaments to create a polymeric network in the cell’s cortex and cytoplasm. Actin serves as one of the main structural components of the cell, contributing to numerous cellular capabilities such as cell motility, division, adhesion, and signaling (71,98). In the context of cancer, actin reorganization is especially involved in tumor metastasis and proliferation (98). The Yu et al. study explored targeting actin as a potential therapeutic. Tumor cells were exposed to magnetic compounds that bind to the cell’s actin cytoskeleton. Upon applying an alternating magnetic current, the compounds aggregate and disrupt the cell’s actin, leading to downstream cell-cycle arrest and death (110). The main obstacle of this approach lies with the challenges of targeted drug delivery to tumor cells. For another potential therapeutic angle, Yamaguchi and Condeelis summarize the roles of several actin remodeling proteins in the cancer-progression process. These proteins include N-WASP, cofilin, and cortactin, all of which have altered expression levels in different cancers, leading to enhanced metastatic capabilities (98). Collectively, these changes in expression of this network of proteins suggests the target potential of these proteins for cancer intervention.
The following studies reveal the challenges of targeting actin and the regulators of actin assembly within a tumor environment. Absi et al. found that knocking down N-WASP or Rho-subfamily GTPase CDC42 in breast cancer cells diminished the rapid F-actin remodeling and accumulation at the immune synapse region in response to natural killer cells (111). As a result of impairing the actin response, natural killer cells were then able to recognize cancerous cells and mark them for removal (111). Consequently, researchers exploring therapeutics must recognize actin’s role in other processes; otherwise, unaccounted effects such as increased metastasis and immune evasion may arise. Outside of conventional cancer-related consequences, Park et al. introduced a cytoskeletal and metabolic link while investigating actin’s role in glycolytic activity (101). Actin assembly sequesters TRIM21, an E3 ubiquitin ligase that mediates phosphofructokinase-protein degradation and renders it inactive. Actin polymer sequestration of TRIM21 increases phosphofructokinase-protein levels, thereby enhancing the glycolytic activity in non-small cell lung cancer (101,112). This study complements the exhibition of aerobic glycolysis, i.e., the Warburg effect, of cancer cells where glycolysis is favored despite having access to the more ATP efficient metabolic process of oxidative phosphorylation (113). In short, actin association with metabolism emphasizes the overlap that must be considered when targeting the mechanical program’s components that associate with actin assembly and organization.
Targeting the mechanical program through myosin II
As introduced previously, myosin II is another key component of the mechanical-adaptability program. In cancer, myosin II plays an array of roles in cancer formation and disease progression (70). Myosin II has three paralogs (NMIIA, NMIIB, and NMIIC) with distinct roles in cell motility and spatial-force generation with differing impacts, depending on cell type (114). NMII depletion reduced cell migration and cell-to-cell adhesion in mammalian cells (115). Despite not being a defined “genetic” cancer driver, NMII proteins lay at the intersection of many different pathways that allow cancer cells to exploit the myosin II machinery (70,115). For example, the Rho-ROCK pathway in cancer has been shown to induce tumor cell migration and invasion via altered ROCK function, which increases actin-myosin assembly and contraction (116). Additionally, NMIIA has been implicated as a tumor suppressor in squamous cell carcinomas, in which its expression improved p53 stability and nuclear accumulation (114,117,118). These observations emphasize the potential of altering NMII directly instead of going after the cancer-altered pathways. However, unsurprisingly, NMII modulation is anything but simple.
Myosin II proteins are integrated with several cellular processes that include membrane-associated proteins, RNA-interacting proteins, nuclear proteins, and metabolic enzymes (119). This concept of an integrated contractility network has been explored in Dictyostelium discoideum and lends insight into the intricacy of the mammalian system. For example, Ren et al. found several metabolism-related proteins, including adenine nucleotide translocase and methylmalonate semialdehyde dehydrogenase, that, upon overexpression, serve as genetic suppressors of myosin II mutants (120). A follow-up proteomic study identified interactions between metabolic components and the cytoskeleton, and methylmalonate semialdehyde dehydrogenase turned up again as a biochemical interactor of the contractility network (121).
To further complicate the therapeutic potential of NMII, studies show the ambiguous nature of NMII in different cell types. In glioblastoma, in vivo modeling revealed individual NMIIA or NMIIB knockout failed to suppress tumor proliferation and invasion individually (108). When NMIIA was deleted alone, tumor invasion was reduced, but tumor lethality increased because proliferation was enhanced. However, when both NMIIA and NMIIB were deleted together, tumor invasiveness and proliferation were reduced. Similarly, as described by Wang et al., NMIIA’s role as an oncogene or tumor suppressor still remains unresolved. In their review, they discuss that high expressions of NMIIA in gastric and esophageal cancer are correlated with poor prognosis and metastasis. In contrast, they also outline that in skin, head, and neck squamous carcinomas, NMIIA levels were reduced. They conclude that NMIIA is important in post-transcriptional activity and nuclear retention of p53 (122). Thus, NMIIs can be either cancer promoting or inhibitory depending on the context. This differential impact undoubtedly reflects the cell type and its particular state, as well as the tumor cell’s environment, which is constantly evolving. Thus, while it is possible to target components of the mechanical network, we must consider unintended consequences due to interactions with other cellular processes.
Variable results in different experimental systems
Given the complexity of different experimental systems, the results of studies on the mechanical-adaptability program with cultured cells and those with in vivo systems may not always align. For example, when Ivkovik et al. used blebbistatin to inhibit NMIIA in an ex vivo glioma model, they were able to block cancer invasion (123). As a follow up on this study, another group found that blebbistatin did indeed block rat glioma cell invasion in the brain cortex (124). Despite these results, however, Picariello et al. found that in their glioma rat models, deleting NMIIA shifts the glioma cells’ stiffness toward optimal proliferation—killing the glioma rat models faster despite blocking glioma invasion (108). This underlines the difficulty in targeting a component of the mechanical-adaptability program due to their role in numerous cellular functions. Additionally, cancer progression adds another layer of complexity into interpreting results across different studies. On this topic, Singh and Settleman discussed the topic of cancers and their K-ras “addiction” and how K-ras, despite being the most frequent mutant oncogenic event promoting tumorigenesis, is perhaps not required for tumor maintenance during malignant cancer progression (125). This implies that the biological context of primary tumor cells may be substantially different than that of malignant cancer cells. While the aforementioned appear to be tall obstacles, it is overall worth targeting the mechanical-adaptability program as it results in desirable therapeutic outcomes, and current ongoing research will only strengthen the anti-cancer therapeutic pipeline.
Updates on therapeutic development targeting the mechanical program
Given the significance of the mechanical-adaptability machinery in cancer development and its potential targetability, developing therapies against this machinery has been a focus for researchers in cancer mechanobiology. Recent studies show promise on this front (Fig. 2). Metformin, a classical anti-diabetic drug, demonstrates anti-cancer activity through acting on tumor-associated fibroblasts (126, 127, 128, 129, 130, 131). Investigators performed proteomic analysis of tumor-associated fibroblasts treated with metformin and found that proteins involved in F-actin depolymerization and cortical cytoskeletal regulation constituted a high percentage of the dysregulated proteins (131). Cisplatin and paclitaxel, both well-known chemotherapeutic drugs, have also been reported to affect cell mechanics. These drugs have long been thought to retard cancer progression through DNA damage. However, recent studies reveal they also influence actin stress-fiber formation and cytoskeleton organization in several cancers, leading to increased stiffness in these cells (132, 133, 134, 135).
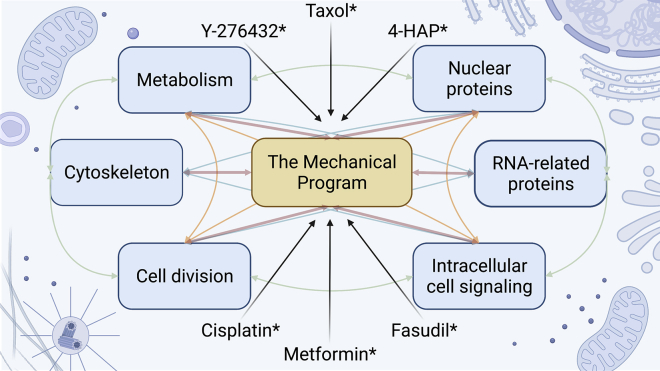
Figure 2.
The challenges of targeting the mechanical program due to its intricate relationship with other cellular processes. With more interest in looking at the components of the mechanical program for cancer therapeutics, a holistic view of the system is required. The mechanical program, like other cellular processes, affect different processes and vice versa. As a consequence, modulating one process that results in tumor-suppressive effects may result in pro-tumor behavior in another. Those marked with asterisks (∗) are example drugs that modulate the mechanical program.
Another prominent group of anti-cancer drugs targeting the adaptability program works within the Rho/ROCK pathway, which regulates many cellular processes, including actin remodeling and cell migration, morphology, and proliferation. F-actin and NMII proteins are downstream targets of the Rho/ROCK signaling cascade. Recent findings show that ROCK1 and ROCK2 are upregulated in numerous cancers (136, 137, 138) and are associated with tumor size, cancer aggression, and metastasis (137,139). Many ROCK inhibitors show significant effects in cancer treatment. For example, the ROCK inhibitor Fasudil reduces proliferation and dissemination of fibrosarcoma, melanoma, breast, and bladder cancer cells (140, 141, 142, 143). This compound also reduces extravasation of PDAC cells and their attachment to secondary sites (144). Another example ROCK inhibitor is Y-27632. This compound is reported to attenuate the growth and invasion phenotypes of bladder, ovarian, and skin cancers and works in part by blocking ROCK-mediated phosphorylation of myosin light-chain kinase, a direct activator of myosin II (145, 146, 147, 148). Interestingly, one study shows that Y-27632 increases breast cancer cells’ rigidity (145). This observation is somewhat non-intuitive, as one initially assumes that ROCK inhibition would lead to inactive myosin II, which in turn would lead to softer cells. While the effects in this study are fairly small, they highlight the necessity of confirming assumptions when manipulating a cell’s mechanical system.
Since several components of the mechanical program exert tumor-suppressor functions, inhibiting this system may lead to cancer enhancement instead. A recent study identified 4-hydroxyacetophenone (4-HAP) as an activator of the system, reducing metastatic potential of PDAC. 4-HAP activates NMIIB and NMIIC by promoting their assembly and thereby increasing cell stiffness and cortical tension. The increased NMII assembly, in turn, reduces invasive phenotypes and blocks dissemination in PDAC cancer cells. 4-HAP also reduced metastasis in mouse models of pancreatic and colorectal cancers (2,71). Importantly, 4-HAP does not appear to impact NMIIA, which may allow this molecule to bypass some of the consequences of targeting all NMII paralogs simultaneously. Moreover, by driving NMIIB and/or NMIIC assembly, 4-HAP is not expected to disrupt NMII’s tumor-suppressive roles that have been suggested in a number of scenarios, including in squamous cell carcinomas and glioblastoma, as discussed above. Taken together, these studies suggest that targeting the mechanical program has the potential to develop future therapeutics in cancer treatment.
Conclusion
Cancer is a complex disease with devastating consequences. For decades, scientists and researchers have relentlessly investigated new approaches in therapeutic development to tackle this group of diseases. Thus far, current therapeutic approaches focus mainly on a handful of systems controlling cell cycle, replication, immune evasion, etc. To more effectively treat cancer, simultaneously targeting multiple systems facilitating the progression of cancer is needed. In this review, we have discussed the impact of the surrounding mechanical environment on cancer cells as well as how the external mechanical stimuli can lead to changes in gene expression, drive tumor development, and facilitate invasion and metastasis (Fig. 1). These stimuli pose mechanical challenges but also force the cancer cells to develop systems that allow them to adapt and survive. The reciprocity between cancer cells and their mechanical surroundings acts similar to a feedback system that endows cancer with the immense ability to adapt to different environments through various stages of progression. At the heart of this feedback system is the mechanical program comprised of the F-actins and the mechanoresponsive proteins, including NMII, ACTN, FLN, and no doubt others. This program renders cancer cells their ability to sense, respond, and adapt to the numerous mechanical stimuli coming from their constantly evolving surroundings.
Due to its significance in facilitating cancer adaptability and progression, this mechanical program poses as an ideal target for treatment development. As mentioned previously, promising compounds that target different central components of this system have produced encouraging results for anti-cancer activity. However, challenges remain to be overcome (Fig. 2). For one, some components in this system display both pro- and anti-cancer activity. Moreover, the mechanical system is inextricably connected to numerous other cellular pathways, such as signal transduction, metabolism, RNA regulation, etc. Therefore, more studies are necessary to determine whether prospective therapeutics targeting this system can do so efficiently without disturbing overall cellular balances. Nevertheless, the mechanical program is an attractive avenue for cancer therapeutic development, especially for combination therapies in the future. Now is the time for scientists to consider cancer a biophysical disease, as both external and intrinsic mechanical factors play significant roles in shaping and advancing the disease.
Author contributions
L.T.S.N., M.A.C.J., E.P., and D.N.R. conceived of the vision for the review. L.T.S.N., M.A.C.J., and E.P. reviewed the literature and drafted the review. D.N.R. provided a sounding board and helped edit the manuscript.
Declaration of interests
D.N.R. is exploring a start-up company. All other authors have no competing interests.
Acknowledgments
We thank the members of the D.N.R. lab for helpful feedback on this review article. Our research in this area is supported by NIH R01 GM66817 and a Johns Hopkins Discovery grant. All figures in this paper were made on BioRender.com.
Editor: Meyer B. Jackson.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2022.04.039.
Supporting material
References
- 1.Luo T., Mohan K., et al. Robinson D.N. Molecular mechanisms of cellular mechanosensing. Nat. Mater. 2013;12:1064–1071. doi: 10.1038/nmat3772. https://www.ncbi.nlm.nih.gov/pubmed/24141449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Surcel A., Schiffhauer E.S., et al. Robinson D.N. Targeting mechanoresponsive proteins in pancreatic cancer: 4-hydroxyacetophenone blocks dissemination and invasion by activating MYH14. Cancer Res. 2019;79:4665–4678. doi: 10.1158/0008-5472.CAN-18-3131. https://www.ncbi.nlm.nih.gov/pubmed/31358530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parajon E., Surcel A., Robinson D.N. The mechanobiome: a goldmine for cancer therapeutics. Am. J. Physiol. Cell Physiol. 2021;320:C306–C323. doi: 10.1152/ajpcell.00409.2020. https://www.ncbi.nlm.nih.gov/pubmed/33175572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brahimi-Horn M.C., Chiche J., Pouyssegur J. Hypoxia and cancer. J. Mol. Med. (Berl) 2007;85:1301–1307. doi: 10.1007/s00109-007-0281-3. https://www.ncbi.nlm.nih.gov/pubmed/18026916 [DOI] [PubMed] [Google Scholar]
- 5.Vaupel P., Hockel M., Mayer A. Detection and characterization of tumor hypoxia using pO2 histography. Antioxid. Redox Signal. 2007;9:1221–1236. doi: 10.1089/ars.2007.1628. https://www.ncbi.nlm.nih.gov/pubmed/17536958 [DOI] [PubMed] [Google Scholar]
- 6.Semenza G.L. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene. 2010;29:625–634. doi: 10.1038/onc.2009.441. https://www.ncbi.nlm.nih.gov/pubmed/19946328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bos R., van der Groep P., et al. van der Wall E. Levels of hypoxia-inducible factor-1alpha independently predict prognosis in patients with lymph node negative breast carcinoma. Cancer. 2003;97:1573–1581. doi: 10.1002/cncr.11246. https://www.ncbi.nlm.nih.gov/pubmed/12627523 [DOI] [PubMed] [Google Scholar]
- 8.Zhang H., Wong C.C.L., et al. Semenza G.L. HIF-1-dependent expression of angiopoietin-like 4 and L1CAM mediates vascular metastasis of hypoxic breast cancer cells to the lungs. Oncogene. 2012;31:1757–1770. doi: 10.1038/onc.2011.365. https://www.ncbi.nlm.nih.gov/pubmed/21860410 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Gilkes D.M., Chaturvedi P., et al. Semenza G.L. Collagen prolyl hydroxylases are essential for breast cancer metastasis. Cancer Res. 2013;73:3285–3296. doi: 10.1158/0008-5472.CAN-12-3963. https://www.ncbi.nlm.nih.gov/pubmed/23539444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reiterer M., Colaco R., et al. Branco C. Acute and chronic hypoxia differentially predispose lungs for metastases. Sci. Rep. 2019;9:10246. doi: 10.1038/s41598-019-46763-y. https://www.ncbi.nlm.nih.gov/pubmed/31308473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilkes D.M., Xiang L., et al. Semenza G.L. Hypoxia-inducible factors mediate coordinated RhoA-ROCK1 expression and signaling in breast cancer cells. Proc. Natl. Acad. Sci. U S A. 2014;111:E384–E393. doi: 10.1073/pnas.1321510111. https://www.ncbi.nlm.nih.gov/pubmed/24324133 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Kim T.H., Ly C., et al. Rowat A.A.C. Stress hormone signaling through beta-adrenergic receptors regulates macrophage mechanotype and function. FASEB J. 2019;33:3997–4006. doi: 10.1096/fj.201801429RR. https://www.ncbi.nlm.nih.gov/pubmed/30509116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim T.H., Gill N.K., et al. Rowat A.C. Cancer cells become less deformable and more invasive with activation of beta-adrenergic signaling. J. Cell Sci. 2016;129:4563–4575. doi: 10.1242/jcs.194803. https://www.ncbi.nlm.nih.gov/pubmed/27875276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sloan E.K., Priceman S.J., et al. Cole S.W. The sympathetic nervous system induces a metastatic switch in primary breast cancer. Cancer Res. 2010;70:7042–7052. doi: 10.1158/0008-5472.CAN-10-0522. https://www.ncbi.nlm.nih.gov/pubmed/20823155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Creed S.J., Le C.P., et al. Sloan E.K. β2-adrenoceptor signaling regulates invadopodia formation to enhance tumor cell invasion. Breast Cancer Res. 2015;17:145. doi: 10.1186/s13058-015-0655-3. https://www.ncbi.nlm.nih.gov/pubmed/26607426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le C.P., Nowell C.J., et al. Sloan E.K. Chronic stress in mice remodels lymph vasculature to promote tumour cell dissemination. Nat. Commun. 2016;7:10634. doi: 10.1038/ncomms10634. https://www.ncbi.nlm.nih.gov/pubmed/26925549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Northcott J.M., Dean I.S., et al. Weaver V.M. Feeling stress: the mechanics of cancer progression and aggression. Front. Cell Dev. Biol. 2018;6:17. doi: 10.3389/fcell.2018.00017. https://www.ncbi.nlm.nih.gov/pubmed/29541636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J.H., Sakamoto H., et al. Lee R.T. Biomechanical regulation of human monocyte/macrophage molecular function. Am. J. Pathol. 2000;156:1797–1804. doi: 10.1016/S0002-9440(10)65051-1. https://www.ncbi.nlm.nih.gov/pubmed/10793091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalli M., Li R., Mills G.B., Stylianopoulos T., Zervantonakis I.K. Mechanical stress signaling in pancreatic cancer cells triggers p38 MAPK- and JNK-dependent cytoskeleton remodeling and promotes cell migration via Rac1/cdc42/myosin II. Mol. Cancer Res. 2021;20:485–497. doi: 10.1158/1541-7786.mcr-21-0266.MCR-21-0266. https://www.ncbi.nlm.nih.gov/pubmed/34782370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levental K.R., Yu H., Kass L., Lakins J.N., Egeblad M., Erler J.T., Fong S.F., Csiszar K., Giaccia A., Weninger W., Yamauchi M., Gasser D.L., Weaver V.M. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. https://www.ncbi.nlm.nih.gov/pubmed/19931152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wyckoff J.B., Jones J.G., Condeelis J.S., Segall J.E. A critical step in metastasis: in vivo analysis of intravasation at the primary tumor. Cancer Res. 2000;60:2504–2511. https://www.ncbi.nlm.nih.gov/pubmed/10811132 [PubMed] [Google Scholar]
- 22.Condeelis J., Pollard J.W. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–266. doi: 10.1016/j.cell.2006.01.007. https://www.ncbi.nlm.nih.gov/pubmed/16439202 [DOI] [PubMed] [Google Scholar]
- 23.Chen M.B., Whisler J.A., Jeon J.S., Kamm R.D. Mechanisms of tumor cell extravasation in an in vitro microvascular network platform. Integr. Biol. (Camb). 2013;5:1262–1271. doi: 10.1039/c3ib40149a. https://www.ncbi.nlm.nih.gov/pubmed/23995847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar S., Weaver V.M. Mechanics, malignancy, and metastasis: the force journey of a tumor cell. Cancer Metastasis Rev. 2009;28:113–127. doi: 10.1007/s10555-008-9173-4. https://www.ncbi.nlm.nih.gov/pubmed/19153673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu H., Mouw J.K., Weaver V.M. Forcing form and function: biomechanical regulation of tumor evolution. Trends Cell Biol. 2011;21:47–56. doi: 10.1016/j.tcb.2010.08.015. https://www.ncbi.nlm.nih.gov/pubmed/20870407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amos S.E., Choi Y.S. The cancer microenvironment: mechanical challenges of the metastatic cascade. Front. Bioeng. Biotechnol. 2021;9:625859. doi: 10.3389/fbioe.2021.625859. https://www.ncbi.nlm.nih.gov/pubmed/33644019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riehl B.D., Kim E., et al. Lim J.Y. The role of microenvironmental cues and mechanical loading milieus in breast cancer cell progression and metastasis. Front. Bioeng. Biotechnol. 2021;8:608526. doi: 10.3389/fbioe.2020.608526. https://www.ncbi.nlm.nih.gov/pubmed/33585411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das J., Chakraborty S., Maiti T.K. Mechanical stress-induced autophagic response: a cancer-enabling characteristic? Semin. Cancer Biol. 2020;66:101–109. doi: 10.1016/j.semcancer.2019.05.017. https://www.ncbi.nlm.nih.gov/pubmed/31150765 [DOI] [PubMed] [Google Scholar]
- 29.Swaminathan V., Mythreye K., et al. Superfine R. Mechanical stiffness grades metastatic potential in patient tumor cells and in cancer cell lines. Cancer Res. 2011;71:5075–5080. doi: 10.1158/0008-5472.can-11-0247. http://www.ncbi.nlm.nih.gov/pubmed/21642375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu C., Pei H., Tan F. Matrix stiffness and colorectal cancer. Onco Targets Ther. 2020;13:2747–2755. doi: 10.2147/OTT.S231010. https://www.ncbi.nlm.nih.gov/pubmed/32280247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jain R.K., Martin J.D., Stylianopoulos T. The role of mechanical forces in tumor growth and therapy. Annu. Rev. Biomed. Eng. 2014;16:321–346. doi: 10.1146/annurev-bioeng-071813-105259. https://www.ncbi.nlm.nih.gov/pubmed/25014786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaudhuri O., Koshy S.T., et al. Mooney D.J. Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium. Nat. Mater. 2014;13:970–978. doi: 10.1038/nmat4009. https://www.ncbi.nlm.nih.gov/pubmed/24930031 [DOI] [PubMed] [Google Scholar]
- 33.Peela N., Sam F.S., et al. Nikkhah M. A three dimensional micropatterned tumor model for breast cancer cell migration studies. Biomaterials. 2016;81:72–83. doi: 10.1016/j.biomaterials.2015.11.039. https://www.ncbi.nlm.nih.gov/pubmed/26724455 [DOI] [PubMed] [Google Scholar]
- 34.Baker A.M., Bird D., et al. Erler J.T. Lysyl oxidase enzymatic function increases stiffness to drive colorectal cancer progression through FAK. Oncogene. 2013;32:1863–1868. doi: 10.1038/onc.2012.202. https://www.ncbi.nlm.nih.gov/pubmed/22641216 [DOI] [PubMed] [Google Scholar]
- 35.Kim Y., Roh S., et al. Kim J.C. Differential expression of the LOX family genes in human colorectal adenocarcinomas. Oncol. Rep. 2009;22:799–804. doi: 10.3892/or_00000502. https://www.ncbi.nlm.nih.gov/pubmed/19724858 [DOI] [PubMed] [Google Scholar]
- 36.Krndija D., Schmid H., et al. von Wichert G. Substrate stiffness and the receptor-type tyrosine-protein phosphatase alpha regulate spreading of colon cancer cells through cytoskeletal contractility. Oncogene. 2010;29:2724–2738. doi: 10.1038/onc.2010.25. [DOI] [PubMed] [Google Scholar]
- 37.Ulrich T.A., de Juan Pardo E.M., Kumar S. The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res. 2009;69:4167–4174. doi: 10.1158/0008-5472.CAN-08-4859. https://www.ncbi.nlm.nih.gov/pubmed/19435897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rice A.J., Cortes E., et al. Del Rio Hernandez A. Matrix stiffness induces epithelial-mesenchymal transition and promotes chemoresistance in pancreatic cancer cells. Oncogenesis. 2017;6:e352. doi: 10.1038/oncsis.2017.54. https://www.ncbi.nlm.nih.gov/pubmed/28671675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nukuda A., Sasaki C., et al. Haga H. Stiff substrates increase YAP-signaling-mediated matrix metalloproteinase-7 expression. Oncogenesis. 2015;4:e165. doi: 10.1038/oncsis.2015.24. https://www.ncbi.nlm.nih.gov/pubmed/26344692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee J.Y., Chang J.K., et al. Chaudhuri O. YAP-independent mechanotransduction drives breast cancer progression. Nat. Commun. 2019;10:1848. doi: 10.1038/s41467-019-09755-0. https://www.ncbi.nlm.nih.gov/pubmed/31015465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng Y., Chen Z., et al. Liu Y. ROCK isoforms differentially modulate cancer cell motility by mechanosensing the substrate stiffness. Acta Biomater. 2019;88:86–101. doi: 10.1016/j.actbio.2019.02.015. https://www.ncbi.nlm.nih.gov/pubmed/30771534 [DOI] [PubMed] [Google Scholar]
- 42.Pang M., Teng Y., et al. Xiong C. Substrate stiffness promotes latent TGF-β1 activation in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2017;483:553–558. doi: 10.1016/j.bbrc.2016.12.107. https://www.ncbi.nlm.nih.gov/pubmed/28025149 [DOI] [PubMed] [Google Scholar]
- 43.Lin F., Zhang H., et al. Xiong C. Substrate stiffness coupling TGF-β1 modulates migration and traction force of MDA-MB-231 human breast cancer cells in vitro. Acs Biomater. Sci. Eng. 2018;4:1337–1345. doi: 10.1021/acsbiomaterials.7b00835. https://www.ncbi.nlm.nih.gov/pubmed/33418664 [DOI] [PubMed] [Google Scholar]
- 44.Tang K., Li S., et al. Liu Y. Shear stress stimulates integrin β1 trafficking and increases directional migration of cancer cells via promoting deacetylation of microtubules. Biochim. Biophys. Acta Mol. Cell Res. 2020;1867:118676. doi: 10.1016/j.bbamcr.2020.118676. https://www.ncbi.nlm.nih.gov/pubmed/32044386 [DOI] [PubMed] [Google Scholar]
- 45.Holle A.W., Govindan Kutty Devi N., et al. Spatz J.P. Cancer cells invade confined microchannels via a self-directed mesenchymal-to-amoeboid transition. Nano Lett. 2019;19:2280–2290. doi: 10.1021/acs.nanolett.8b04720. https://www.ncbi.nlm.nih.gov/pubmed/30775927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laakkonen P., Waltari M., et al. Alitalo K. Vascular endothelial growth factor receptor 3 is involved in tumor angiogenesis and growth. Cancer Res. 2007;67:593–599. doi: 10.1158/0008-5472.can-06-3567. [DOI] [PubMed] [Google Scholar]
- 47.Maimari N., Pedrigi R.M., et al. Krams R. Integration of flow studies for robust selection of mechanoresponsive genes. Thromb. Haemost. 2016;115:474–483. doi: 10.1160/TH15-09-0704. https://www.ncbi.nlm.nih.gov/pubmed/26842798 [DOI] [PubMed] [Google Scholar]
- 48.Follain G., Osmani N., et al. Goetz J.G. Hemodynamic forces tune the arrest, adhesion, and extravasation of circulating tumor cells. Dev. Cell. 2018;45:33–52.e12. doi: 10.1016/j.devcel.2018.02.015. https://www.ncbi.nlm.nih.gov/pubmed/29634935 [DOI] [PubMed] [Google Scholar]
- 49.Headley M.B., Bins A., et al. Krummel M.F. Visualization of immediate immune responses to pioneer metastatic cells in the lung. Nature. 2016;531:513–517. doi: 10.1038/nature16985. https://www.ncbi.nlm.nih.gov/pubmed/26982733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rizvi I., Gurkan U.A., et al. Hasan T. Flow induces epithelial-mesenchymal transition, cellular heterogeneity and biomarker modulation in 3D ovarian cancer nodules. Proc. Natl. Acad. Sci. U S A. 2013;110:E1974–E1983. doi: 10.1073/pnas.1216989110. https://www.ncbi.nlm.nih.gov/pubmed/23645635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cornelison R.C., Brennan C.E., et al. Munson J.M. Convective forces increase CXCR4-dependent glioblastoma cell invasion in GL261 murine model. Sci. Rep. 2018;8:17057. doi: 10.1038/s41598-018-35141-9. https://www.ncbi.nlm.nih.gov/pubmed/30451884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Streitberger K.J., Lilaj L., et al. Sack I. How tissue fluidity influences brain tumor progression. P Natl. Acad. Sci. U S A. 2020;117:128–134. doi: 10.1073/pnas.1913511116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paszek M.J., Zahir N., et al. Weaver V.M. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16169468 [DOI] [PubMed] [Google Scholar]
- 54.Ondeck M.G., Kumar A., et al. Engler A.J. Dynamically stiffened matrix promotes malignant transformation of mammary epithelial cells via collective mechanical signaling. Proc. Natl. Acad. Sci. U S A. 2019;116:3502–3507. doi: 10.1073/pnas.1814204116. https://www.ncbi.nlm.nih.gov/pubmed/30755531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wullkopf L., West A.K.V., et al. Erler J.T. Cancer cells' ability to mechanically adjust to extracellular matrix stiffness correlates with their invasive potential. Mol. Biol. Cell. 2018;29:2378–2385. doi: 10.1091/mbc.E18-05-0319. https://www.ncbi.nlm.nih.gov/pubmed/30091653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cox T.R., Erler J.T. Remodeling and homeostasis of the extracellular matrix: implications for fibrotic diseases and cancer. Dis. Model. Mech. 2011;4:165–178. doi: 10.1242/dmm.004077. https://www.ncbi.nlm.nih.gov/pubmed/21324931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Provenzano P.P., Inman D.R., et al. Keely P.J. Mammary epithelial-specific disruption of focal adhesion kinase retards tumor formation and metastasis in a transgenic mouse model of human breast cancer. Am. J. Pathol. 2008;173:1551–1565. doi: 10.2353/ajpath.2008.080308. https://www.ncbi.nlm.nih.gov/pubmed/18845837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu P., Weaver V.M., Werb Z. The extracellular matrix: a dynamic niche in cancer progression. J. Cell Biol. 2012;196:395–406. doi: 10.1083/jcb.201102147. https://www.ncbi.nlm.nih.gov/pubmed/22351925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nissen N.I., Karsdal M., Willumsen N. Collagens and Cancer associated fibroblasts in the reactive stroma and its relation to Cancer biology. J. Exp. Clin. Cancer Res. 2019;38:115. doi: 10.1186/s13046-019-1110-6. https://www.ncbi.nlm.nih.gov/pubmed/30841909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang H., Torphy R.J., et al. Collisson E.A. Pancreatic ductal adenocarcinoma progression is restrained by stromal matrix. J. Clin. Invest. 2020;130:4704–4709. doi: 10.1172/JCI136760. https://www.ncbi.nlm.nih.gov/pubmed/32749238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nguyen A.V., Nyberg K.D., et al. Rowat A.C. Stiffness of pancreatic cancer cells is associated with increased invasive potential. Integr. Biol. 2016;8:1232–1245. doi: 10.1039/c6ib00135a. https://www.ncbi.nlm.nih.gov/pubmed/27761545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guck J., Schinkinger S., et al. Bilby C. Optical deformability as an inherent cell marker for testing malignant transformation and metastatic competence. Biophys. J. 2005;88:3689–3698. doi: 10.1529/biophysj.104.045476. https://www.ncbi.nlm.nih.gov/pubmed/15722433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cross S.E., Jin Y.S., et al. Gimzewski J.K. Nanomechanical analysis of cells from cancer patients. Nat. Nanotechnol. 2007;2:780–783. doi: 10.1038/nnano.2007.388. https://www.ncbi.nlm.nih.gov/pubmed/18654431 [DOI] [PubMed] [Google Scholar]
- 64.Liu Y., Zhang T., et al. Huang B. Cell softness prevents cytolytic T-cell killing of tumor-repopulating cells. Cancer Res. 2021;81:476–488. doi: 10.1158/0008-5472.CAN-20-2569. https://www.ncbi.nlm.nih.gov/pubmed/33168645 [DOI] [PubMed] [Google Scholar]
- 65.Lv J., Liu Y., et al. Huang B. Cell softness regulates tumorigenicity and stemness of cancer cells. EMBO J. 2021;40:e106123. doi: 10.15252/embj.2020106123. https://www.ncbi.nlm.nih.gov/pubmed/33274785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu W., Mezencev R., et al. Sulchek T. Cell stiffness is a biomarker of the metastatic potential of ovarian cancer cells. PLoS One. 2012;7:e46609. doi: 10.1371/journal.pone.0046609. https://www.ncbi.nlm.nih.gov/pubmed/23056368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu H.W., Chen Y., et al. Kuo J.C. β- PIX controls intracellular viscoelasticity to regulate lung cancer cell migration. J. Cell Mol. Med. 2015;19:934–947. doi: 10.1111/jcmm.12441. https://www.ncbi.nlm.nih.gov/pubmed/25683605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schiffhauer E.S., Luo T., et al. Robinson D.N. Mechanoaccumulative elements of the mammalian actin cytoskeleton. Curr. Biol. 2016;26:1473–1479. doi: 10.1016/j.cub.2016.04.007. https://www.ncbi.nlm.nih.gov/pubmed/27185555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kothari P., Johnson C., et al. Robinson D.N. How the mechanobiome drives cell behavior, viewed through the lens of control theory. J. Cell Sci. 2019;132:jcs234476. doi: 10.1242/jcs.234476. https://www.ncbi.nlm.nih.gov/pubmed/31477578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Halder D., Mallick D., et al. Jana S.S. Nonmuscle Myosin II in cancer cell migration and mechanotransduction. Int. J. Biochem. Cell Biol. 2021;139:106058. doi: 10.1016/j.biocel.2021.106058. https://www.ncbi.nlm.nih.gov/pubmed/34400319 [DOI] [PubMed] [Google Scholar]
- 71.Bryan D.S., Stack M., et al. Weichselbaum R.R. 4-Hydroxyacetophenone modulates the actomyosin cytoskeleton to reduce metastasis. Proc. Natl. Acad. Sci. U S A. 2020;117:22423–22429. doi: 10.1073/pnas.2014639117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peng Y., Chen Z., et al. Liu Y. Non-muscle myosin II isoforms orchestrate substrate stiffness sensing to promote cancer cell contractility and migration. Cancer Lett. 2022;524:245–258. doi: 10.1016/j.canlet.2021.10.030. https://www.ncbi.nlm.nih.gov/pubmed/34715250 [DOI] [PubMed] [Google Scholar]
- 73.Huang Q., Li X., et al. Lin J. ACTN4 promotes the proliferation, migration, metastasis of osteosarcoma and enhances its invasive ability through the NF-κB pathway. Pathol. Oncol. Res. 2020;26:893–904. doi: 10.1007/s12253-019-00637-w. https://www.ncbi.nlm.nih.gov/pubmed/30879239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ma X., Xue H., et al. Zuo Y. Serum actinin-4 levels as a potential diagnostic and prognostic marker in cervical cancer. Dis. Markers. 2020;2020:5327378. doi: 10.1155/2020/5327378. https://www.ncbi.nlm.nih.gov/pubmed/32855746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jung J., Kim S., et al. Ko J. α-Actinin-4 regulates cancer stem cell properties and chemoresistance in cervical cancer. Carcinogenesis. 2020;41:940–949. doi: 10.1093/carcin/bgz168. https://www.ncbi.nlm.nih.gov/pubmed/31584624 [DOI] [PubMed] [Google Scholar]
- 76.Barai A., Mukherjee A., et al. Sen S. α-Actinin-4 drives invasiveness by regulating myosin IIB expression and myosin IIA localization. J. Cell Sci. 2021;134:jcs258581. doi: 10.1242/jcs.258581. https://www.ncbi.nlm.nih.gov/pubmed/34730180 [DOI] [PubMed] [Google Scholar]
- 77.Burton K.M., Cao H., et al. Razidlo G.L. Dynamin 2 interacts with alpha-actinin 4 to drive tumor cell invasion. Mol. Biol. Cell. 2020;31:439–451. doi: 10.1091/mbc.E19-07-0395. https://www.ncbi.nlm.nih.gov/pubmed/31967944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang K., Singer S.J. Interaction of filamin with f-actin in solution. Proc. Natl. Acad. Sci. U S A. 1977;74:2021–2025. doi: 10.1073/pnas.74.5.2021. https://www.ncbi.nlm.nih.gov/pubmed/325564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stossel T.P., Condeelis J., et al. Shapiro S.S. Filamins as integrators of cell mechanics and signalling. Nat. Rev. Mol. Cell Biol. 2001;2:138–145. doi: 10.1038/35052082. http://www.ncbi.nlm.nih.gov/pubmed/11252955 [DOI] [PubMed] [Google Scholar]
- 80.Feng Y., Walsh C.A. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat. Cell Biol. 2004;6:1034–1038. doi: 10.1038/ncb1104-1034. https://www.ncbi.nlm.nih.gov/pubmed/15516996 [DOI] [PubMed] [Google Scholar]
- 81.Glogauer M., Arora P., et al. McCulloch C.A. The role of actin-binding protein 280 in integrin-dependent mechanoprotection. J. Biol. Chem. 1998;273:1689–1698. doi: 10.1074/jbc.273.3.1689. https://www.ncbi.nlm.nih.gov/pubmed/9430714 [DOI] [PubMed] [Google Scholar]
- 82.Ma H.R., Cao L., et al. Qian Z. Filamin B extensively regulates transcription and alternative splicing, and is associated with apoptosis in HeLa cells. Oncol. Rep. 2020;43:1536–1546. doi: 10.3892/or.2020.7532. https://www.ncbi.nlm.nih.gov/pubmed/32323860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yesilkaya F., Tastekin D., et al. Pence S. Examination of the expression levels of MACC1, Filamin A and FBXW7 genes in colorectal cancer patients. North. Clin. Istanb. 2019;7:1–5. doi: 10.14744/nci.2019.26780. https://www.ncbi.nlm.nih.gov/pubmed/32232196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cheng L., Tong Q. Interaction of FLNA and ANXA2 promotes gefitinib resistance by activating the Wnt pathway in non-small-cell lung cancer. Mol. Cell Biochem. 2021;476:3563–3575. doi: 10.1007/s11010-021-04179-1. https://www.ncbi.nlm.nih.gov/pubmed/34018148 [DOI] [PubMed] [Google Scholar]
- 85.Patarat R., Riku S., et al. Puttipanyalears C. The expression of FLNA and CLU in PBMCs as a novel screening marker for hepatocellular carcinoma. Sci. Rep. 2021;11:14838. doi: 10.1038/s41598-021-94330-1. https://www.ncbi.nlm.nih.gov/pubmed/34290294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ramachandran R., Schmid S.L. The dynamin superfamily. Curr. Biol. 2018;28:R411–R416. doi: 10.1016/j.cub.2017.12.013. https://www.ncbi.nlm.nih.gov/pubmed/29689225 [DOI] [PubMed] [Google Scholar]
- 87.Jimah J.R., Hinshaw J.E. Structural insights into the mechanism of dynamin superfamily proteins. Trends Cell Biol. 2019;29:257–273. doi: 10.1016/j.tcb.2018.11.003. https://www.ncbi.nlm.nih.gov/pubmed/30527453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tian M., Yang X., et al. Guo S. The expression of dynamin 1, 2, and 3 in human hepatocellular carcinoma and patient prognosis. Med. Sci. Monit. 2020;26:e923359. doi: 10.12659/MSM.923359. https://www.ncbi.nlm.nih.gov/pubmed/32573516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.von Beek C., Alriksson L., et al. Pejler G. Dynamin inhibition causes context-dependent cell death of leukemia and lymphoma cells. PLoS One. 2021;16:e0256708. doi: 10.1371/journal.pone.0256708. https://www.ncbi.nlm.nih.gov/pubmed/34492077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang R., Lee D.M., et al. Chen E.H. Dynamin regulates the dynamics and mechanical strength of the actin cytoskeleton as a multifilament actin-bundling protein. Nat. Cell Biol. 2020;22:674–688. doi: 10.1038/s41556-020-0519-7. https://www.ncbi.nlm.nih.gov/pubmed/32451441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gu C., Yaddanapudi S., et al. Sever S. Direct dynamin-actin interactions regulate the actin cytoskeleton. EMBO J. 2010;29:3593–3606. doi: 10.1038/emboj.2010.249. https://www.ncbi.nlm.nih.gov/pubmed/20935625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang Z., Zhu Z., et al. Sun S. NMIIA promotes tumorigenesis and prevents chemosensitivity in colorectal cancer by activating AMPK/mTOR pathway. Exp. Cell Res. 2021;398:112387. doi: 10.1016/j.yexcr.2020.112387. https://www.ncbi.nlm.nih.gov/pubmed/33220257 [DOI] [PubMed] [Google Scholar]
- 93.Pecci A., Ma X., et al. Adelstein R.S. MYH9: structure, functions and role of non-muscle myosin IIA in human disease. Gene. 2018;664:152–167. doi: 10.1016/j.gene.2018.04.048. https://www.ncbi.nlm.nih.gov/pubmed/29679756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Honda K. The biological role of actinin-4 (ACTN4) in malignant phenotypes of cancer. Cell Biosci. 2015;5:41. doi: 10.1186/s13578-015-0031-0. https://www.ncbi.nlm.nih.gov/pubmed/26288717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Park S., Kang M., et al. Ko J. α-Actinin-4 promotes the progression of prostate cancer through the Akt/GSK-3β/β-catenin signaling pathway. Front. Cell Dev Biol. 2020;8:588544. doi: 10.3389/fcell.2020.588544. https://www.ncbi.nlm.nih.gov/pubmed/33363146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu X., Chu K.M. α-Actinin-4 promotes metastasis in gastric cancer. Lab. Invest. 2017;97:1084–1094. doi: 10.1038/labinvest.2017.28. https://www.ncbi.nlm.nih.gov/pubmed/28581489 [DOI] [PubMed] [Google Scholar]
- 97.Kamil M., Shinsato Y., et al. Arita K. High filamin-C expression predicts enhanced invasiveness and poor outcome in glioblastoma multiforme. Br. J. Cancer. 2019;120:819–826. doi: 10.1038/s41416-019-0413-x. https://www.ncbi.nlm.nih.gov/pubmed/30867563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yamaguchi H., Condeelis J. Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim. Biophys. Acta. 2007;1773:642–652. doi: 10.1016/j.bbamcr.2006.07.001. https://www.ncbi.nlm.nih.gov/pubmed/16926057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Foerster F., Braig S., et al. Vollmar A.M. Targeting the actin cytoskeleton: selective antitumor action via trapping PKCɛ. Cell Death Dis. 2014;5:e1398. doi: 10.1038/cddis.2014.363. https://www.ncbi.nlm.nih.gov/pubmed/25165884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hsu K.S., Kao H.Y. Alpha-actinin 4 and tumorigenesis of breast cancer. Vitam Horm. 2013;93:323–351. doi: 10.1016/B978-0-12-416673-8.00005-8. https://www.ncbi.nlm.nih.gov/pubmed/23810014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Park J.S., Burckhardt C.J., et al. Danuser G. Mechanical regulation of glycolysis via cytoskeleton architecture. Nature. 2020;578:621–626. doi: 10.1038/s41586-020-1998-1. https://www.ncbi.nlm.nih.gov/pubmed/32051585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ong M.S., Deng S., et al. Yap C.T. Cytoskeletal proteins in cancer and intracellular stress: a therapeutic perspective. Cancers. 2020;12 doi: 10.3390/cancers12010238. https://www.ncbi.nlm.nih.gov/pubmed/31963677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.DeWane G., Salvi A.M., DeMali K.A. Fueling the cytoskeleton - links between cell metabolism and actin remodeling. J. Cell Sci. 2021;134:jcs248385. doi: 10.1242/jcs.248385. https://www.ncbi.nlm.nih.gov/pubmed/33558441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Angstadt S., Zhu Q., et al. Anders R.A. Pancreatic ductal adenocarcinoma cortical mechanics and clinical implications. Front. Oncol. 2022;12:809179. doi: 10.3389/fonc.2022.809179. https://www.ncbi.nlm.nih.gov/pubmed/35174086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fife C.M., McCarroll J.A., Kavallaris M. Movers and shakers: cell cytoskeleton in cancer metastasis. Br. J. Pharmacol. 2014;171:5507–5523. doi: 10.1111/bph.12704. https://www.ncbi.nlm.nih.gov/pubmed/24665826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Etienne-Manneville S. Actin and microtubules in cell motility: which one is in control? Traffic. 2004;5:470–477. doi: 10.1111/j.1600-0854.2004.00196.x. https://www.ncbi.nlm.nih.gov/pubmed/15180824 [DOI] [PubMed] [Google Scholar]
- 107.Blanchoin L., Boujemaa-Paterski R., et al. Plastino J. Actin dynamics, architecture, and mechanics in cell motility. Physiol. Rev. 2014;94:235–263. doi: 10.1152/physrev.00018.2013. https://www.ncbi.nlm.nih.gov/pubmed/24382887 [DOI] [PubMed] [Google Scholar]
- 108.Picariello H.S., Kenchappa R.S., et al. Rosenfeld S.S. Myosin IIA suppresses glioblastoma development in a mechanically sensitive manner. Proc. Natl. Acad. Sci. U S A. 2019;116:15550–15559. doi: 10.1073/pnas.1902847116. https://www.ncbi.nlm.nih.gov/pubmed/31235578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bai H., Zhu Q., et al. Anders R.A. Yes-associated protein impacts adherens junction assembly through regulating actin cytoskeleton organization. Am. J. Physiol. Gastrointest. Liver Physiol. 2016;311:G396–G411. doi: 10.1152/ajpgi.00027.2016. https://www.ncbi.nlm.nih.gov/pubmed/27229120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yu Q., Zhang B., et al. Liu Y. Actin cytoskeleton-disrupting and magnetic field-responsive multivalent supramolecular assemblies for efficient cancer therapy. ACS Appl. Mater. Inter. 2020;12:13709–13717. doi: 10.1021/acsami.0c01762. https://www.ncbi.nlm.nih.gov/pubmed/32118400 [DOI] [PubMed] [Google Scholar]
- 111.Al Absi A., Wurzer H., et al. Thomas C. Actin cytoskeleton remodeling drives breast cancer cell escape from natural killer-mediated cytotoxicity. Cancer Res. 2018;78:5631–5643. doi: 10.1158/0008-5472.CAN-18-0441. https://www.ncbi.nlm.nih.gov/pubmed/30104240 [DOI] [PubMed] [Google Scholar]
- 112.Ayad N.M.E., Weaver V.M. Tension in tumour cells keeps metabolism high. Nature. 2020;578:517–518. doi: 10.1038/d41586-020-00314-y. https://www.ncbi.nlm.nih.gov/pubmed/32094916 [DOI] [PubMed] [Google Scholar]
- 113.Liberti M.V., Locasale J.W. The Warburg effect: how does it benefit cancer cells? Trends Biochem. Sci. 2016;41:211–218. doi: 10.1016/j.tibs.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ouderkirk J.L., Krendel M. Non-muscle myosins in tumor progression, cancer cell invasion, and metastasis. Cytoskeleton. 2014;71:447–463. doi: 10.1002/cm.21187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vicente-Manzanares M., Ma X., et al. Horwitz A.R. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chin V.T., Nagrial A.M., et al. Pajic M. Rho-associated kinase signalling and the cancer microenvironment: novel biological implications and therapeutic opportunities. Expert Rev. Mol. Med. 2015;17:e17. doi: 10.1017/erm.2015.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schramek D., Sendoel A., et al. Fuchs E. Direct in vivo RNAi screen unveils myosin IIa as a tumor suppressor of squamous cell carcinomas. Science. 2014;343:309–313. doi: 10.1126/science.1248627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Anne Conti M., Saleh A.D., et al. Adelstein R.S. Conditional deletion of nonmuscle myosin II-A in mouse tongue epithelium results in squamous cell carcinoma. Sci. Rep. 2015;5:14068. doi: 10.1038/srep14068. https://www.ncbi.nlm.nih.gov/pubmed/26369831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nguyen L.T.S., Robinson D.N. The unusual suspects in cytokinesis: fitting the pieces together. Front. Cell Dev Biol. 2020;8:441. doi: 10.3389/fcell.2020.00441. https://www.ncbi.nlm.nih.gov/pubmed/32626704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ren Y., West-Foyle H., et al. Robinson D.N. Genetic suppression of a phosphomimic myosin II identifies system-level factors that promote myosin II cleavage furrow accumulation. Mol. Biol. Cell. 2014;25:4150–4165. doi: 10.1091/mbc.e14-08-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kothari P., Srivastava V., et al. Robinson D.N. Contractility kits promote assembly of the mechanoresponsive cytoskeletal network. J. Cell Sci. 2019;132:jcs226704. doi: 10.1242/jcs.226704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang Y., Liu S., et al. Yang J. Myosin heavy chain 9: oncogene or tumor suppressor gene? Med. Sci. Monit. 2019;25:888–892. doi: 10.12659/MSM.912320. https://www.ncbi.nlm.nih.gov/pubmed/30739906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ivkovic S., Beadle C., et al. Rosenfeld S.S. Direct inhibition of myosin II effectively blocks glioma invasion in the presence of multiple motogens. Mol. Biol. Cell. 2012;23:533–542. doi: 10.1091/mbc.E11-01-0039. https://www.ncbi.nlm.nih.gov/pubmed/22219380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Beadle C., Assanah M.C., et al. Canoll P. The role of myosin II in glioma invasion of the brain. Mol. Biol. Cell. 2008;19:3357–3368. doi: 10.1091/mbc.E08-03-0319. https://www.ncbi.nlm.nih.gov/pubmed/18495866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Singh A., Settleman J. Oncogenic K-ras "addiction" and synthetic lethality. Cell Cycle. 2009;8:2676–2678. doi: 10.4161/cc.8.17.9336. https://www.ncbi.nlm.nih.gov/pubmed/19690457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Suissa S., Azoulay L. Metformin and cancer: mounting evidence against an association. Diabetes Care. 2014;37:1786–1788. doi: 10.2337/dc14-0500. [DOI] [PubMed] [Google Scholar]
- 127.Wheaton W.W., Weinberg S.E., et al. Chandel N.S. Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis. Elife. 2014;3:e02242. doi: 10.7554/eLife.02242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Podhorecka M., Ibanez B., Dmoszynska A. Metformin - its potential anti-cancer and anti-aging effects. Postep Hig Med. Dosw. 2017;71:170–175. doi: 10.5604/01.3001.0010.3801. [DOI] [PubMed] [Google Scholar]
- 129.Song Z., Wei B., et al. Chen L. Metformin suppresses the expression of Sonic hedgehog in gastric cancer cells. Mol. Med. Rep. 2017;15:1909–1915. doi: 10.3892/mmr.2017.6205. [DOI] [PubMed] [Google Scholar]
- 130.Whitburn J., Edwards C.M., Sooriakumaran P. Metformin and prostate cancer: a new role for an old drug. Curr. Urol. Rep. 2017;18:46. doi: 10.1007/s11934-017-0693-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chen G.X., Yu C., et al. Zhang S.Y. Metformin suppresses gastric cancer progression through calmodulin-like protein 3 secreted from tumor-associated fibroblasts. Oncol. Rep. 2019;41:405–414. doi: 10.3892/or.2018.6783. [DOI] [PubMed] [Google Scholar]
- 132.Kopf-Maier P., Muhlhausen S.K. Changes in the cytoskeleton pattern of tumor cells by cisplatin in vitro. Chem. Biol. Interact. 1992;82:295–316. doi: 10.1016/0009-2797(92)90002-3. https://www.ncbi.nlm.nih.gov/pubmed/1606625 [DOI] [PubMed] [Google Scholar]
- 133.Kung M.L., Hsieh C.W., et al. Hsieh S. Nanoscale characterization illustrates the cisplatin-mediated biomechanical changes of B16-F10 melanoma cells. Phys. Chem. Chem. Phys. 2016;18:7124–7131. doi: 10.1039/c5cp07971c. https://www.ncbi.nlm.nih.gov/pubmed/26886764 [DOI] [PubMed] [Google Scholar]
- 134.Raudenska M., Kratochvilova M., et al. Masarik M. Cisplatin enhances cell stiffness and decreases invasiveness rate in prostate cancer cells by actin accumulation. Sci. Rep. 2019;9:1660. doi: 10.1038/s41598-018-38199-7. https://www.ncbi.nlm.nih.gov/pubmed/30733487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Vassilopoulos A., Xiao C., et al. Deng C.X. Synergistic therapeutic effect of cisplatin and phosphatidylinositol 3-kinase (PI3K) inhibitors in cancer growth and metastasis of Brca1 mutant tumors. J. Biol. Chem. 2014;289:24202–24214. doi: 10.1074/jbc.M114.567552. https://www.ncbi.nlm.nih.gov/pubmed/25006250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kalender M.E., Demiryurek S., et al. Camci C. Association between the Thr431Asn polymorphism of the ROCK2 gene and risk of developing metastases of breast cancer. Oncol. Res. 2009;18:583–591. doi: 10.3727/096504010x12767359113767. https://www.ncbi.nlm.nih.gov/pubmed/20939434 [DOI] [PubMed] [Google Scholar]
- 137.Kamai T., Tsujii T., et al. Oshima H. Significant association of Rho/ROCK pathway with invasion and metastasis of bladder cancer. Clin. Cancer Res. 2003;9:2632–2641. https://www.ncbi.nlm.nih.gov/pubmed/12855641 [PubMed] [Google Scholar]
- 138.Lane J., Martin T.A., et al. Jiang W.G. The expression and prognostic value of ROCK I and ROCK II and their role in human breast cancer. Int. J. Oncol. 2008;33:585–593. https://www.ncbi.nlm.nih.gov/pubmed/18695890 [PubMed] [Google Scholar]
- 139.Liu S., Goldstein R.H., et al. Rosenblatt M. Inhibition of rho-associated kinase signaling prevents breast cancer metastasis to human bone. Cancer Res. 2009;69:8742–8751. doi: 10.1158/0008-5472.CAN-09-1541. https://www.ncbi.nlm.nih.gov/pubmed/19887617 [DOI] [PubMed] [Google Scholar]
- 140.Miyamoto C., Maehata Y., et al. Lee M.C.i. Fasudil suppresses fibrosarcoma growth by stimulating secretion of the chemokine CXCL14/BRAK. J. Pharmacol. Sci. 2012;120:241–249. doi: 10.1254/jphs.12177fp. https://www.ncbi.nlm.nih.gov/pubmed/23099322 [DOI] [PubMed] [Google Scholar]
- 141.Guerra F.S., Oliveira R.G.d., et al. Fernandes P.D. ROCK inhibition with Fasudil induces beta-catenin nuclear translocation and inhibits cell migration of MDA-MB 231 human breast cancer cells. Sci. Rep. 2017;7:13723. doi: 10.1038/s41598-017-14216-z. https://www.ncbi.nlm.nih.gov/pubmed/29057980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Xia Y., Cai X.Y., et al. Wu G. Rho kinase inhibitor Fasudil suppresses the vasculogenic mimicry of B16 mouse melanoma cells both in vitro and in vivo. Mol. Cancer Ther. 2015;14:1582–1590. doi: 10.1158/1535-7163.MCT-14-0523. https://www.ncbi.nlm.nih.gov/pubmed/25934709 [DOI] [PubMed] [Google Scholar]
- 143.Lee M.H., Kundu J.K., et al. Shim J.H. Targeting ROCK/LIMK/cofilin signaling pathway in cancer. Arch. Pharm. Res. 2019;42:481–491. doi: 10.1007/s12272-019-01153-w. https://www.ncbi.nlm.nih.gov/pubmed/31030376 [DOI] [PubMed] [Google Scholar]
- 144.Vennin C., Chin V.T., et al. Timpson P. Transient tissue priming via ROCK inhibition uncouples pancreatic cancer progression, sensitivity to chemotherapy, and metastasis. Sci. Transl Med. 2017;9:eaai8504. doi: 10.1126/scitranslmed.aai8504. https://www.ncbi.nlm.nih.gov/pubmed/28381539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cascione M., De Matteis V., et al. Rinaldi R. Morphomechanical and structural changes induced by ROCK inhibitor in breast cancer cells. Exp. Cell Res. 2017;360:303–309. doi: 10.1016/j.yexcr.2017.09.020. https://www.ncbi.nlm.nih.gov/pubmed/28935466 [DOI] [PubMed] [Google Scholar]
- 146.Jiang L., Wen J., Luo W. Rhoassociated kinase inhibitor, Y27632, inhibits the invasion and proliferation of T24 and 5367 bladder cancer cells. Mol. Med. Rep. 2015;12:7526–7530. doi: 10.3892/mmr.2015.4404. https://www.ncbi.nlm.nih.gov/pubmed/26459851 [DOI] [PubMed] [Google Scholar]
- 147.Li Y., Li X., et al. Zhu Y. Visfatin derived from ascites promotes ovarian cancer cell migration through Rho/ROCK signaling-mediated actin polymerization. Eur. J. Cancer Prev. 2015;24:231–239. doi: 10.1097/CEJ.0000000000000064. https://www.ncbi.nlm.nih.gov/pubmed/25055182 [DOI] [PubMed] [Google Scholar]
- 148.Routhier A., Astuccio M., et al. Bryan B. Pharmacological inhibition of Rho-kinase signaling with Y-27632 blocks melanoma tumor growth. Oncol. Rep. 2010;23:861–867. https://www.ncbi.nlm.nih.gov/pubmed/20127030 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.