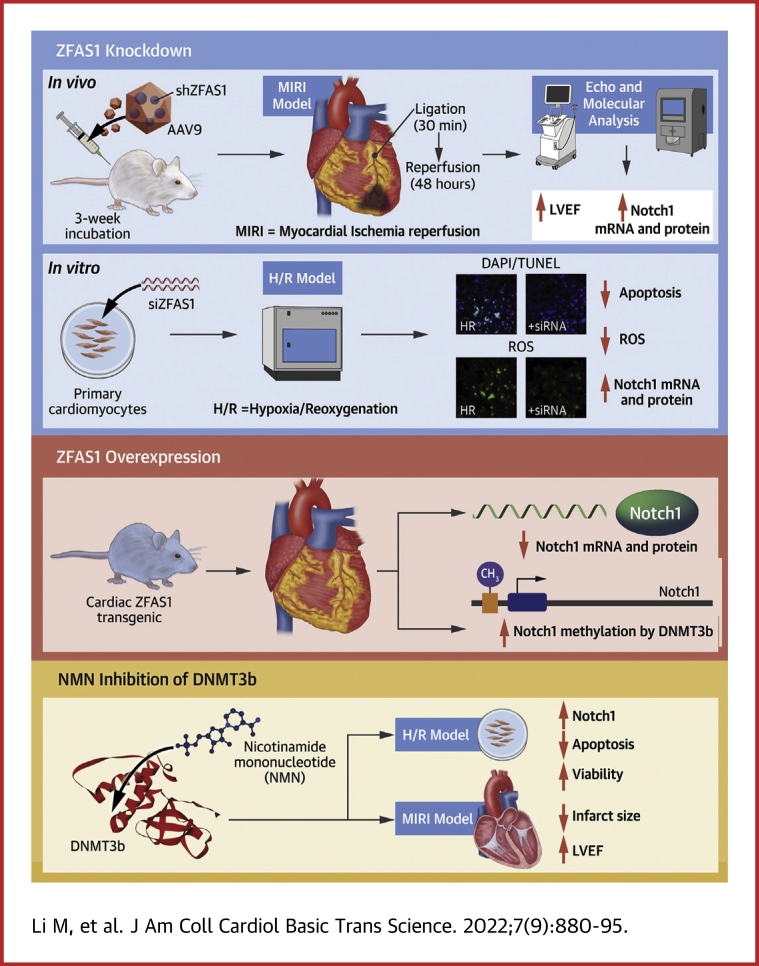
Visual Abstract
Key Words: DNA methylation, long noncoding RNA, myocardial ischemia-reperfusion injury, Notch1, ZFAS1
Abbreviations and Acronyms: AAV, adeno-associated virus; DNMT, DNA methyltransferase; HR, hypoxia/reoxygenation; lncRNA, long noncoding RNA; MI, myocardial infarction; MIRI, myocardial ischemia-reperfusion injury; NICD, Notch intracellular domain; NMCM, neonatal mouse cardiac myocytes; NMN, nicotinamide mononucleotide; ROS, reactive oxygen species; shZFAS1, short hairpin RNA ZFAS1; siZFAS1, small interfering RNA ZFAS1; TG, transgenic; WT, wild-type; ZFAS1, zinc finger antisense 1
Highlights
-
•
The increase of ZFAS1 expression in MIRI is an important cause of cardiomyocyte apoptosis and ROS production.
-
•
ZFAS1 can directly interact with the promoter region of Notch1, recruit DNMT3b to promote DNA methylation in the promoter region of Notch1, and trigger cardiomyocyte apoptosis and ROS production after MIRI.
-
•
Nicotinamide mononucleotide has the potential to attenuate the apoptosis of cardiomyocytes after MIRI by competitively binding to DNMT3b and inhibiting the DNA methylation of Notch1.
Summary
The most devastating and catastrophic deterioration of myocardial ischemia-reperfusion injury (MIRI) is cardiomyocyte death. Here we aimed to evaluate the role of lncRNA-ZFAS1 in MIRI and delineate its mechanism of action. The level of lncRNA-ZFAS1 was elevated in MIRI hearts, and artificial knockdown of lncRNA-ZFAS1 in mice improved cardiac function. Notch1 is a potential target of lncRNA-ZFAS1, and lncRNA-ZFAS1 could bind to the promoter region of Notch1 and recruit DNMT3b to induce Notch1 methylation. Nicotinamide mononucleotide could promote the expression of Notch1 by competitively inhibiting the expression of DNMT3b and improving the apoptosis of cardiomyocytes and cardiac function.
Coronary artery disease is one of the most common causes of mortality in the world.1 The standard treatment for repairing blood supply to the ischemic myocardium is recovery of reperfusion.2 However, reperfusion also induces major cardiac damage, which is commonly known as myocardial ischemia-reperfusion injury (MIRI).3 MIRI has been correlated with oxidative stress, autophagy, endoplasmic reticulum stress, apoptosis, calcium overload, and epigenetic alterations.4 Oxidative stress or reactive oxygen species (ROS) generation is the important initiating mechanism of MIRI, the occurrence of which is the key to distinguishing myocardial infarction (MI) from MIRI. MIRI is a complex pathologic condition involving various signaling pathways.
The Notch signaling pathway is involved in a variety of heart functions5, 6, 7 and is composed of 4 Notch proteins (Notch 1-4) and 5 ligands, namely, Jagged 1, Jagged 2, Delta-like 1, Delta-like 3, and Delta-like 4.8 The Notch protein is cleaved into the Notch intracellular domain (NICD) and released into the cytoplasm when the ligand on the neighboring cell surface interacts with the Notch receptor.9 After being cleaved, the NICD enters the nucleus and binds with the DNA-binding protein CSL (CBF1)/Su (H)/Lag-1, which triggers downstream gene transcription (such as Hes1 and Hey1).10 Inhibiting the Notch signaling has been shown to alter the energy supply of cardiomyocytes, leading to cardiac dysfunction.11 Notch1 gene inhibition can reduce the expression of Hey1 while enhancing Runx2 expression, resulting in aortic valve calcification.12 High glucose can increase the sensitivity to myocardial ischemia by inhibiting the Notch signaling pathway in mice.13 Importantly, Notch1, as a cardioprotective factor, can inhibit cardiomyocyte apoptosis and oxidative stress caused by MIRI,14 yet its upstream regulatory mechanism is still unclear.
DNA methylation is an epigenetic alteration correlated with changes in transcription.15 In cell proliferation, cell differentiation, apoptosis, and autoimmunity, DNA methylation is critical.16 The inhibition of Apaf1 expression is enhanced by DNA methylation of its promoter.17 Long noncoding (lnc) RNA, cardiomyocyte proliferation regulator, also participates in cardiomyocyte proliferation and cardiac repair by regulating the DNA methylation of MCM3.18 Transcriptome analysis in the human failing heart also changes the level of DNA methylation and is related to myocardial dysfunction.19,20 However, the function of DNA methylation in MIRI regulation is mostly unclear.
LncRNAs are differentially expressed in cardiac diseases,21,22 and our previous studies unraveled that zinc finger antisense 1 (ZFAS1) is deregulated in MI. Specifically, circulating ZFAS1 was proposed as a potential independent predictor of MI.23 ZFAS1 can act as an endogenous SERCA2a inhibitor to regulate contractile function24 and further induce mitochondrial-mediated apoptosis.25 However, the role of ZFAS1 in MIRI is currently unclear.
Nicotinamide mononucleotide (NMN) is an essential part in NAD+ synthesis,26 which has multiple pharmacologic effects on heart disease.27, 28, 29 NMN exerts acute myocardial protection by targeting SIRT1 to stimulate glycolysis.30 In aging mice, NMN and melatonin can protect against MIRI by activating SIRT3/FOXO1 and decreasing apoptosis.31 Importantly, NMN protects the MIRI by increasing the level of NAD+ in the heart.29 However, whether NMN is related to DNA methylation in MIRI remains unknown.
The general purpose of the present study was to determine the role and mechanism of ZFAS1 in MIRI regulation, as well as whether DNA methylation and Notch signaling pathways are key controllers in this process.
Methods
Detailed descriptions on the materials and methods used in this study are provided in the Supplemental Methods section.
Animals
As previously described, the MIRI model was used.32 Mice were intubated and anesthetized under direct vision, and their chests were opened to reveal the heart. The left anterior descending coronary artery was ligated with 7-0 line for 30 minutes to cause myocardial ischemia. The line was released for 48 hours to allow for reperfusion. NMN (500 mg/kg/d intraperitoneally) was administered to MIRI mice for 2 days. Use of the animals was approved by the Ethics Committee of Harbin Medical University and conformed to the Guide for the Care and Use of Laboratory Animals published by the U.S. National Institutes of Health.
Cardiac-specific ZFAS1 knock-in mice
As previously described, cardiac-specific ZFAS1 knock-in (transgenic [TG]) mice were created.32
Statistical analysis
Data are presented as mean ± SEM of at least 3 independent experiments. Unpaired Student's t-test was used for comparisons between 2 groups. One-way and 2-way analysis of variance tests were used to compare parameters among 3 or more independent groups, whereas the t-test was used to compare 2 groups. Pairwise comparisons between groups were made with the Bonferroni multiple comparisons test when analysis of variance yielded significant differences. A 2-tailed P value <0.05 was considered to indicate statistical significance. Pearson correlation test was used to analyze the correlation of the parameters in the 2 groups in Figure3B. The statistical analyses were performed with the use of Prism version 8.0 software (GraphPad Software).
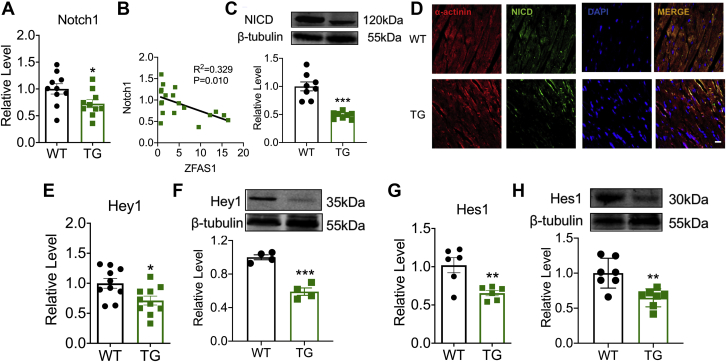
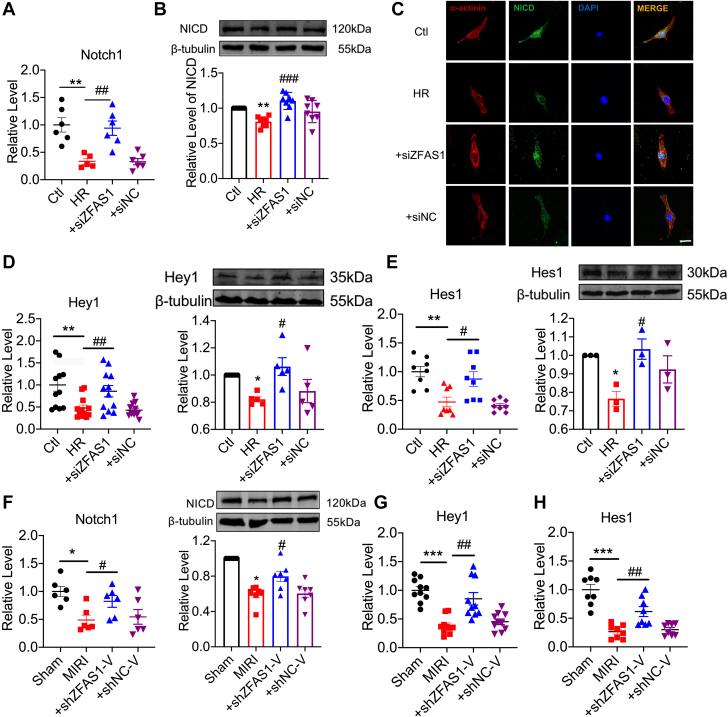
Figure 3.
ZFAS1 Is a Negative Regulator of Notch1
(A) The mRNA level of Notch1 in TG mice. n = 10. ∗P < 0.05 vs WT. (B) Correlation between ZFAS1 and Notch1 mRNA expression. n = 19. (C) Down-regulation of NICD expression in TG mice at protein levels. n = 8. ∗∗∗P < 0.001 vs WT. (D) Images of NICD (green) and α-actinin (red) immunofluorescence staining in NMCMs, Scale bar: 50 μm. n = 6. (E) The mRNA expression level of Hey1 in TG mice. n = 10. ∗P < 0.05 vs WT. (F) Down-regulation of Hey1 expression in TG mice at protein levels. n = 4. ∗∗∗P < 0.001 vs WT. (G) The level of Hes1 mRNA expression in TG mice. n = 6. ∗∗P < 0.01 vs WT. (H) Hes1 expression is dysregulated at the protein level in TG mice. n = 7. ∗∗P < 0.01 vs WT. NICD = Notch intracellular domain; NMCM = neonatal mouse cardiac myocytes; other abbreviations as in Figures 1 and 2.
Results
Pro–oxidative stress and proapoptosis effects of ZFAS1 in MIRI
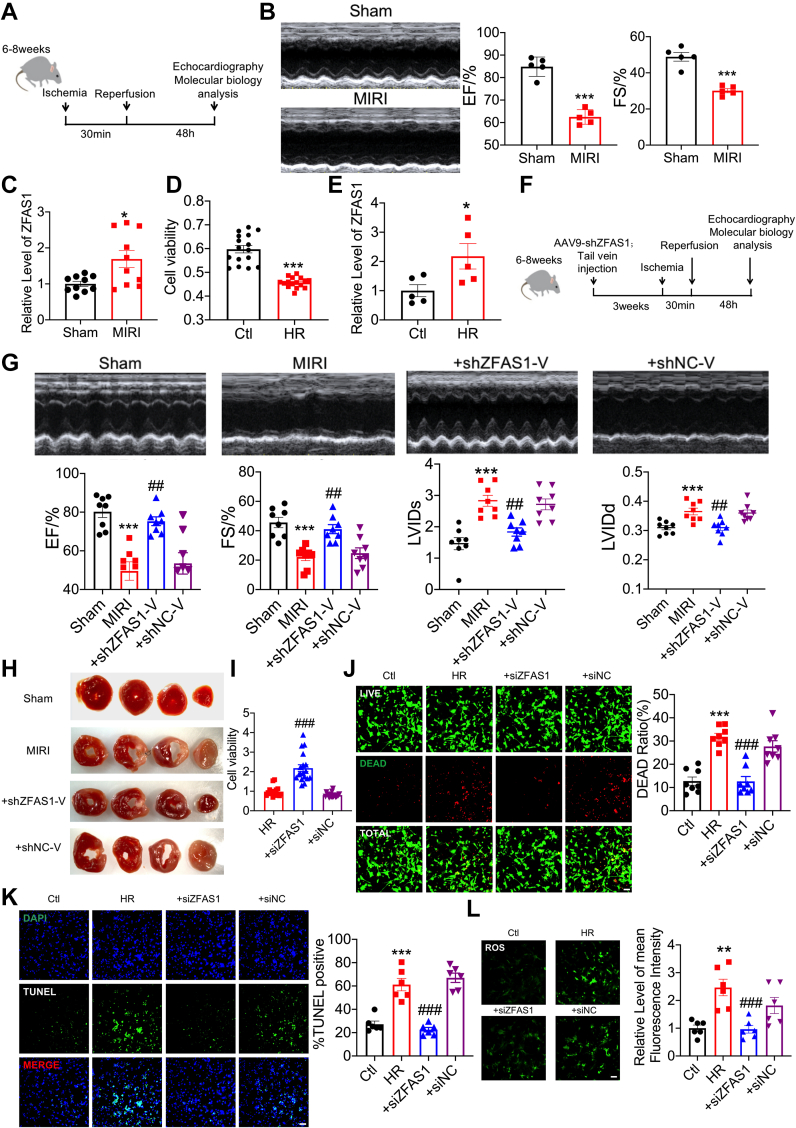
We constructed a mouse MIRI model to investigate the function of ZFAS1 in MIRI, and confirmed that cardiac function was clearly reduced in this condition (Figures 1A and 1B). MIRI mice had a larger proportion of ZFAS1 in their hearts (Figure 1C). In cardiomyocytes exposed to hypoxia/reoxygenation (HR) (Figure 1D), cell viability was severely reduced, and ZFAS1 expression was dramatically elevated (Figure 1E). Next, we inhibited with adeno-associated virus (AAV) vectors to examine ZFAS1 function (Figure 1F). Quantitative reverse-transcription polymerase chain reaction was used to confirm the efficiency of short hairpin RNA (sh) ZFAS1-V in knocking down endogenous ZFAS1 in MIRI mice (Supplemental Figure 1). ZFAS1 knockdown significantly improved MIRI-induced impairment of cardiac function (Figure 1G). Furthermore, shZFAS1-V reduced the infarct size of MIRI hearts (Figure 1H). Transfection of small interfering RNA (si) ZFAS1 mitigated the HR-induced reduction of cell viability (Figure 1I) and Live/Dead Viability/Cytotoxicity assay showed similar results (Figure 1J). Terminal deoxynucleotide transferase–mediated dUTP nick-end labeling (TUNEL) staining indicates that apoptosis was restored by siZFAS1 (Figure 1K). The enhanced ROS generation induced by HR was reduced by siZFAS1 (Figure 1L).
Figure 1.
Pro-Oxidative Stress and Proapoptotic Effects of ZFAS1 in MIRI
(A) Experimental timeline. (B) Echocardiographic detection of changes in cardiac function. n = 5, ∗∗∗P < 0.001 vs Sham. (C) qRT-PCR was used to detect ZFAS1 expression in MIRI mice. n = 10. ∗P < 0.05 vs Sham. (D) CCK8 assay to detect the effects of cell viability. n = 16. ∗∗∗P < 0.001 vs Ctl. (E) qRT-PCR was used to determine the level of ZFAS1 expression in HR-treated cells. n = 5. ∗P < 0.05 vs Ctl. (F) Experimental timeline. (G) Echocardiographic detection of changes in cardiac function. n = 8. ∗∗∗P < 0.001 vs Sham; ##P < 0.01 vs MIRI. (H) TTC detects the effect of shZFAS1-V on the infarct size of MIRI mice. n = 4. (I) CCK8 assay to detect the effects of cell viability. n = 18, ###P < 0.001 vs siZFAS1. (J) To determine the effects of siZFAS1 on cardiomyocytes cell viability. n = 8. ∗∗∗P < 0.001 vs Ctl; ###P < 0.001 vs HR. Magnification ×200. (K) TUNEL assay was used to investigate the effect on apoptosis. n = 6. ∗∗∗P < 0.001 vs Ctl; ###P < 0.001 vs HR. Magnification ×200. (L) Effects of siZFAS1 on the contents of ROS. n = 6. ∗∗P < 0.01 vs Ctl; ###P < 0.001 vs HR. Magnification ×200. P values were determined by means of t-tests and 1-way analyses of variance with Bonferroni multiple group comparisons. Data are presented as mean ± SEM. AAV = adeno-associated virus; Ctl = control cells; HR = hypoxia/reoxygenation-treated cells; MIRI = myocardial ischemia-reperfusion injury; qRT-PCR = quantitative reverse-transcription polymerase chain reaction; ROS = reactive oxygen species; shNC-V = the negative control shRNA engineered into the AAV9 vector; shZFAS1 = short hairpin RNA ZFAS1; siZFAS1, small interfering RNA ZFAS1; TTC = triphenyltetrazolium chloride; TUNEL = terminal deoxynucleotide transferase–mediated dUTP nick-end labeling; ZFAS1 = zinc finger antisense 1.
Intracellular Ca2+ overload occurs during both MI and MIRI, and our previous study verified that ZFAS1 could cause intracellular Ca2+ overload. Therefore, we conducted additional experiments to determine whether ZFAS1 promotes ROS levels after excluding the effect of ZFAS1 on calcium overload. BAPTA, a calcium chelator, was administrated to both the HR-treated cardiomyocytes and the ZFAS1-overexpressed cardiomyocytes. As shown in Supplemental Figure 2, the viability of cardiomyocytes was significantly increased after BAPTA administration, which was further increased after ROS was scavenged by N-acetyl-l-cysteine (NAC). As shown in Supplemental Figures 2E and 2F, the level of ROS did not change significantly after administration of BAPTA, whereas it was significantly decreased after administration of NAC. These data indicated that ZFAS1 still exerts an effect on ROS levels, excluding the regulation of calcium overload, and this further suggests that ZFAS1 has other underlying mechanisms in the regulation of MIRI.
The gene expression profile of ZFAS1-overexpression mice
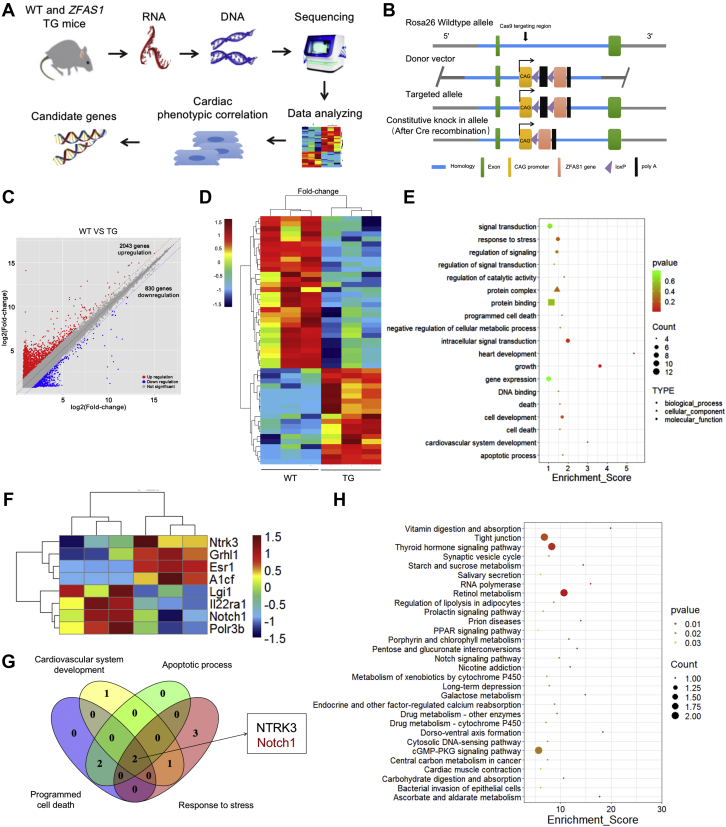
To understand the molecular processes supporting MIRI by ZFAS1 (Figure 2A), we constructed cardiac specific ZFAS1 knock-in TG mice (Figure 2B). ZFAS1 was upregulated in TG mice (Supplemental Figure 3). We compared the transcriptomes between TG and wild-type (WT) hearts by gene expression profiles (Figure 2C). The experiment revealed 30 down-regulated genes and 19 up-regulated genes in TG mice (Figure 2D). A Gene Ontology (GO) functional analysis of these 49 differentially expressed genes was then conducted with the cluster profile bioinformatics tool (Figure 2E). Next, we predicted and screened the direct target of ZFAS1 from 4 pathways, including the activation of cardiovascular phylogeny, stress response, programmed cell death cardiac development, and apoptosis process (Figure 2F), and identified that Notch1 and NTRK3 might be potential targets of ZFAS1 (Figure 2G). Notch signaling pathway is one of the top 30 rich Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, according to a KEGG pathway enrichment analysis of differentially expressed (Figure 2H). Therefore, our follow-up research mainly discusses the role of Notch1 in MIRI.
Figure 2.
The Gene Expression Profile of ZFAS1-Overexpression Mice
(A) Schematic diagram of RNA sequencing and target gene identification. (B) The production of cardiac-specific ZFAS1 knock-in mice is depicted throughout this diagram. (C) Volcano plot of the identified differentially expressed mRNAs. Red area:P < 0.05; fold change ≥2; blue area:P < 0.05, fold change ≤0.5. Benjamin-Hochberg was used to report the false discovery rate for the volcano plot and fold-change analyses. (D) Heat map of the differentially expressed mRNAs identified between TG and WT mice (red, up-regulated mRNAs; blue, down-regulated mRNAs). (E) The second level of differentially expressed genes identified with the use of Gene Ontology. (F) Heat map of mRNA involved in response to stress, apoptosis, protein binding, and other pathways. (G) Venn diagram for screening target genes. (H) Pathway enrichment analysis of differentially expressed genes (criterion: differentially expressed genes >2 and P ≤ 0.05) according to the Kyoto Encyclopedia of Genes and Genomes. TG = cardiac-specific ZFAS1 knock-in (transgenic); WT = wild-type; other abbreviations as in Figure 1.
ZFAS1 is a negative regulator of Notch1
Notch1 expression was identified in TG mice to examine the influence of ZFAS1 on Notch1. Notch1 mRNA levels were significantly lower in TG mice, which is negatively connected to ZFAS1 expression (Figures 3A and 3B). Similarly, NICD in TG mice was considerably lower than in WT mice (Figure 3C), which was verified by the immunofluorescence (Figure 3D). In TG mice, the mRNA and protein levels of Hes1 and Hey1, downstream regulators of the Notch signaling pathway, were both reduced compared with WT mice (Figures 3D to 3H).
ZFAS1 promotes cardiomyocyte apoptosis during MIRI by regulating the Notch signaling pathway
Next, we characterized the regulatory effects of ZFAS1 on the Notch signaling pathway in MIRI and found that the mRNA level of Notch1 was reduced during HR, which was reversed by siZFAS1 (Figure 4A). Figures 4B and 4C showed the results that inhibiting the expression of ZFAS1 remarkably increased NICD protein levels. In neonatal mouse cardiac myocytes (NMCM) exposed to HR damage, similar expression modifications of Hey1 and Hes1 were consistently found among constructs (Figures 4D and 4E). The cardiac expression of Notch1, Hes1, and Hey1 was prominently decreased in MIRI mice, and shZFAS1-V normalized these anomalies (Figures 4F to 4H). The Notch signaling pathway appears to be involved in the regulation of MI and MIRI. We conducted additional experiments to verify the regulation differences of the Notch signaling pathway between MI and MIRI mice, and the results showed that the expressions of Notch1, Hes1, and Hey1 were dramatically reduced in both the MI and the MIRI groups, and the decrease was more severe in the MIRI group. These data indicate that the Notch signaling pathway is a key target for MIRI regulation (Supplemental Figure 4).
Figure 4.
ZFAS1 Promotes Cardiomyocyte Apoptosis During MIRI by Regulating Notch1 Signaling Pathway
(A) Up-regulation of Notch1 expression at the mRNA level in HR-treated cardiomyocytes pretreated with siZFAS1. n = 6. ∗∗P < 0.01 vs Ctl; ##P < 0.01 vs HR. (B) Up-regulation of NICD expression at the protein level in HR-treated cardiomyocytes pretreated with siZFAS1. n = 7. ∗∗P < 0.01 vs Ctl; ###P < 0.001 vs HR. (C) Images of NICD (green) and α-actinin (red) immunofluorescence staining in NMCMs Scale bar: 50 μm. n = 6. (D) HR-treated cardiomyocytes pretreated with siZFAS1 showed up-regulation of Hey1 expression at both mRNA (n = 12) and protein (n = 5) levels. ∗P < 0.05; ∗∗P < 0.01 vs Ctl; #P < 0.05 vs HR; ##P < 0.01 vs HR. (E) In HR-treated cardiomyocytes pretreated with siZFAS1, Hes1 expression was elevated at both mRNA (n = 8) and protein (n = 3) levels. ∗P < 0.05; ∗∗P < 0.01 vs Ctl; #P < 0.05 vs HR; ##P < 0.01 vs HR. (F) In MIRI mice pretreated with shZFAS1-V, expressions of Notch1 mRNA (n = 6) and NICD protein (n = 7) were up-regulated. ∗P < 0.05 vs sham; #P < 0.05 vs MIRI. (G, H) In MIRI mice pretreated with shZFAS1-V, expressions of Hey1 (n = 10) and Hes1 (n = 8) were up-regulated at the mRNA level. ∗∗∗P < 0.001 vs Sham; ##P < 0.01 vs MIRI. P values were determined by means of t-tests and 1-way analyses of variance with Bonferroni multiple group comparisons. Data are presented as mean ± SEM. siNC = scrambled negative control siRNA.
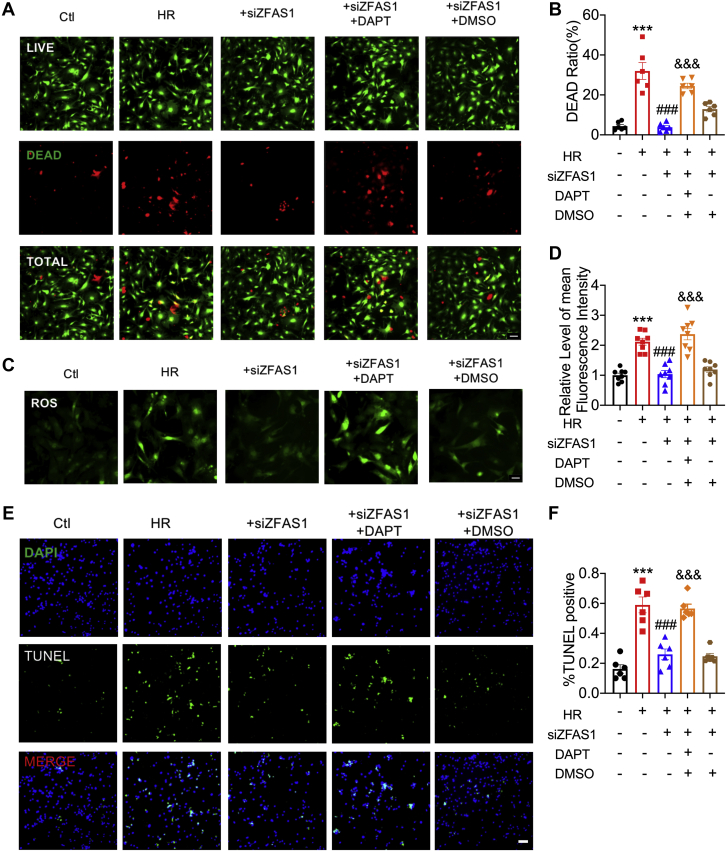
Inhibition of Notch signaling pathway promotes cardiomyocyte apoptosis
To elucidate whether ZFAS1 induces MIRI by targeting the Notch signaling pathway, we explored with the use of the γ-secretase inhibitor DAPT,33 an inhibitor of the Notch signaling pathway. DAPT could abolish the protective effect of siZFAS1 in HR-treated cardiomyocytes, as determined by Live/Dead Viability/Cytotoxicity assay and ROS staining (Figures 5A to 5D). TUNEL staining also showed that DAPT could reverse the protective effect of siZFAS1 on HR-treated cardiomyocytes (Figures 5E and 5F). It is further confirmed that the ZFAS1-Notch signal axis plays a key role in MIRI.
Figure 5.
Inhibition of Notch Signaling Pathway Promotes Cardiomyocyte Apoptosis
(A, B) The effects of the Notch signaling pathway inhibitor DAPT on cardiomyocyte cell viability. n = 6. Magnification ×200. (C, D) The effects of DAPT on the contents of ROS. n = 8. Magnification ×200. (E) (F) TUNEL assay was used to investigate the effect of DAPT on apoptosis. n = 6. Magnification ×200. Data are presented as mean ± SEM. ∗∗∗P < 0.01 vs Ctl; ###P < 0.001 vs HR; and &&&P < 0.001 vs siZFAS1; t-tests and 1-way analyses of variance with Bonferroni multiple group comparisons. DAPI = 4′,6′-diamidino-2-phenylindole; DMSO = dimethylsulfoxide; other abbreviations as in Figure 1.
Our previous study identified that SERCA2a was an important target of ZFAS1 in regulating MI, so does it have the same role in MIRI? The expression of SERCA2a was significantly decreased in HR-treated cardiomyocytes, and siZFAS1 normalized these anomalies (Supplemental Figures 5A and 5B). HR-treated cardiomyocytes experienced calcium overload, which was likewise alleviated by siZFAS1 (Supplemental Figure 5C). These data indicated that the ZFAS1-SERCA2a axis participated in the regulation of MIRI. To distinguish the main regulation mechanism of ZFAS1 in MIRI, cyclopiazonic acid (CPA), the SERCA2a inhibitor, and DAPT, the Notch signaling pathway inhibitor, were used. After ZFAS1 knockdown, CPA dramatically affected the viability of HR-treated cardiomyocytes. And administration of DAPT more dramatically reduced the viability of cardiomyocytes (Supplemental Figure 5D). Moreover, the SERCA2a inhibitor CPA significantly inhibited the protective effect of siZFAS1 on Hypoxia/reoxygenation-treated cardiomyocytes, whereas DAPT administration did not further inhibit this effect (Supplemental Figure 5E). These data indicated that the ZFAS1-Notch1 axis played a major role in MIRI rather than MI.
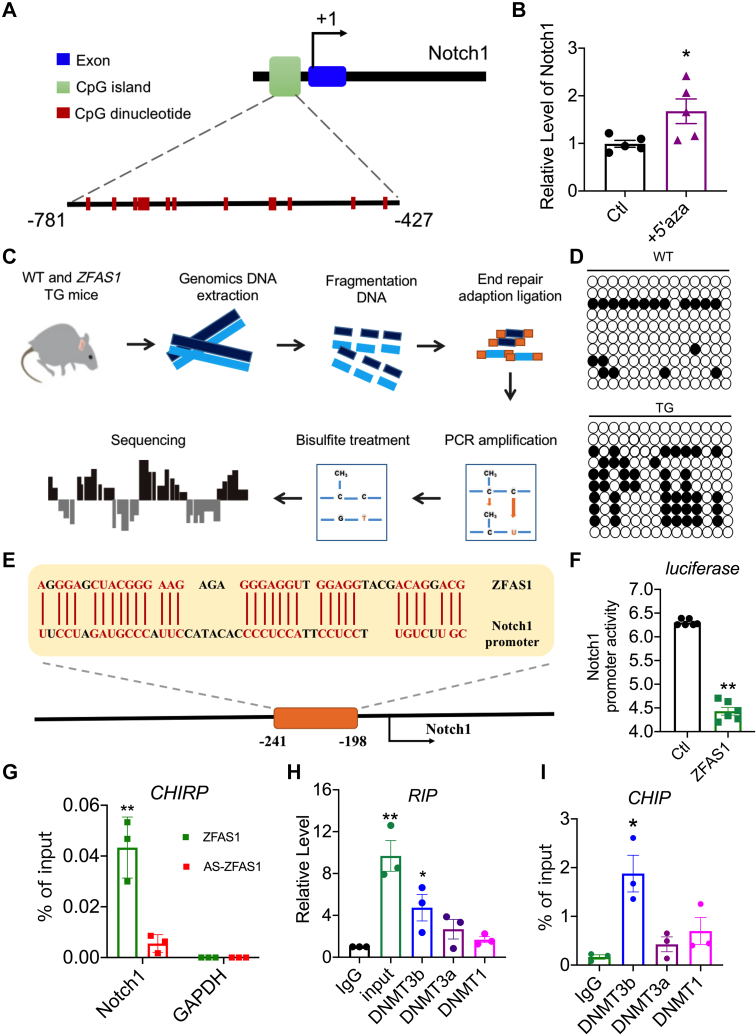
ZFAS1 promotes DNA methylation of Notch1
A question we asked was how ZFAS1 regulates the transcription of Notch1. ZFAS1 is localized within the nucleus of cardiomyocytes.24 These facts prompted us to conjecture that ZFAS1 might regulate Notch1 transcription through an epigenetic mechanism, such as DNA methylation. We found a substantial CpG island in the promoter region of Notch1, using the UCSC Genome Browser and Methprimer to predict the probable domain of Notch1 for DNA methylation (Figure 6A). The methylation inhibitor 5-aza-2′-deoxycytidine (5-aza) dramatically enhanced the transcription level of Notch1 in cardiomyocytes (Figure 6B). Next, In TG mice, bisulfite sequencing revealed a large increase in methylation at the Notch1 promoter region (Figures 6C and 6D). So how does ZFAS1 affect the methylation of Notch1 promoter region? GO analysis found that ZFAS1 has the potential to affect the DNA-binding and protein-binding pathways (Figure 2F). LncRNAs can limit gene transcription by binding to DNA and building RNA:DNA triplexes, which recruit DNA methyltransferases (DNMTs) and increase DNA methylation modification in target areas.34,35 Using Freiburg RNA Tools,36 we identified a binding site for ZFAS1 within the proximal promoter region of Notch1 (Figure 6E). The direct functional interaction between ZFAS1 and Notch1 promoter region was experimentally verified by means of luciferase reporter assay (Figure 6F). Subsequently, the direct physical interaction between ZFAS1 and the Notch1 promoter region was also verified by means of the ChIRP technique with ZFAS1 antisense as a negative control (Figure 6G). Furthermore, prediction with the use of RNA-Protein Interaction Prediction (RPISeq) database suggests that ZFAS1 has the potential to bind methylase DMNT3b (Supplemental Figure 6). The immunoprecipitation of DNMT3b carried a significant quantity of ZFAS1, and a CHIP experiment revealed that DNMT3b binding to Notch1 promoter CpG islands was considerable (Figure 6I). According to these findings, ZFAS1 binds to the Notch1 promoter area and recruits DNMT3b to enhance Notch1 DNA methylation.
Figure 6.
ZFAS1 Promotes DNA Methylation of Notch1
(A) The Notch1 gene’s genomic architecture is depicted in a diagram. (B) The expression level of Notch1 in NMCMs treated with 5-aza-2′-deoxycytidine (5′aza). n = 5. ∗P < 0.05 vs Ctl. (C) Schematic diagram of the bisulfite sequencing. (D) Bisulfite PCR analysis of the Notch1 promoter methylation level in WT and TG mice. (E) The binding region between ZFAS1 and Notch1 is depicted in this diagram. (F) The effect of ZFAS1 on Notch1 promoter activity in HEK293T cells with the use of a dual-luciferase reporter incorporating the Notch promoter. n = 3. ∗∗P < 0.01 vs Ctl. (G) The interaction of Notch1 with ZFAS1 in cardiomyocytes was examined with the use of the ChiRP assay. n = 3. ∗∗P < 0.01 vs AS-ZFAS1. (H) RIP analysis of specific associations of DNMT3b, DNMT3a, and DNMT1 with ZFAS1. n = 3. ∗P < 0.05 vs IgG; ∗∗P < 0.01 vs IgG. (I) ChIP analysis of associations of DNMT3b, DNMT3a, and DNMT1 with Notch1. n = 3. ∗P < 0.05 vs IgG. P values were determined by means of t-tests and 1-way analyses of variance with Bonferroni multiple group comparisons. Data are presented as mean ± SEM. DNMT = DNA methyltransferase; PCR = polymerase chain reaction; other abbreviations as in Figures 1 and 2.
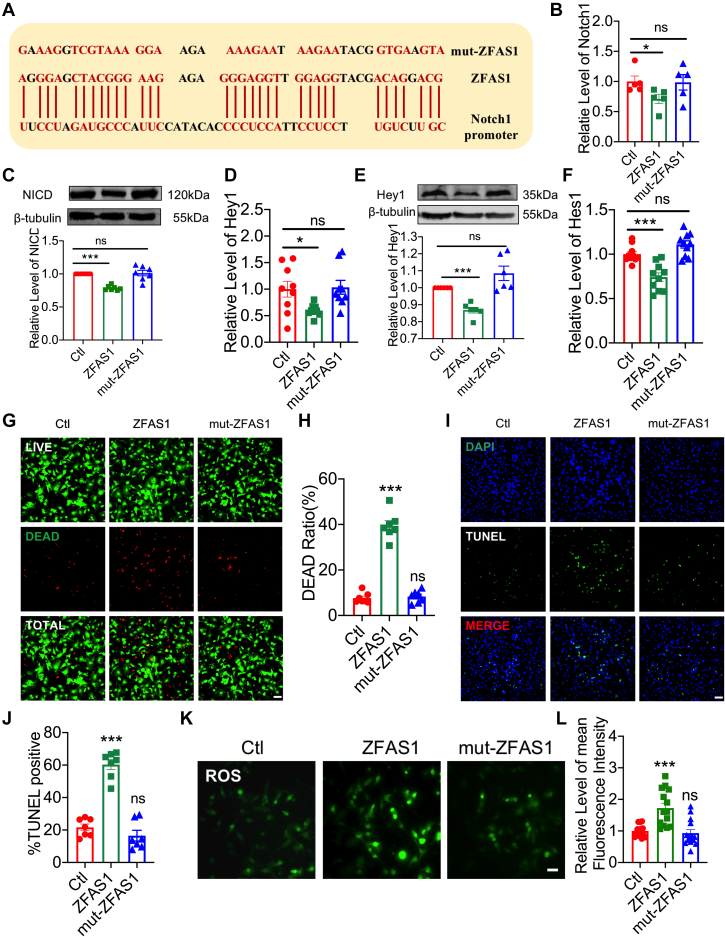
Loss of function of ZFAS1 mutation in the myocardium
Then, using nucleotide replacement, we engineered a mutation in the ZFAS1 sequence to disrupt its Notch1 binding site (mut-ZFAS1) (Figure 7A) and investigated the impact of mut-ZFAS1 on Notch1 function. As anticipated, ZFAS1 overexpression in NMCMs transfected with ZFAS1-carrying plasmid considerably decreased the transcript levels of Notch1, Hey1, and Hes1 (Figures 7B, 7D, and 7F), and mut-ZFAS1 failed to elicit any effects. Similar patterns of expression alterations were seen of NICD and Hey1 (Figures 7C and 7E) protein levels. However, NMCMs transfected with mut-ZFAS1–carrying plasmid lost this function. Furthermore, as shown in Figures 7G to 7L, the capacity of mut-ZFAS1 to modify cell viability, cell apoptosis, and ROS levels was likewise lost. The results indicate that the binding of ZFAS1 to the Notch1 promoter region is a critical step in its impact on MIRI.
Figure 7.
The Function of mut-ZFAS1 in the Myocardium
(A) Nucleotide sequence of mut-ZFAS1 after mutating ZFAS1 and Notch1 binding site. (B, C) Notch1 mRNA (n = 5) and NICD protein (n = 7) levels in ZFAS1- and mut-ZFAS1–transfected cardiomyocytes. (D, E) Hey1 mRNA (n = 9) and protein (n = 6) levels in ZFAS1- and mut-ZFAS1–transfected cardiomyocytes. (F) Hes1 mRNA expression in cardiomyocytes transfected with ZFAS1 and mut-ZFAS1. n = 10. (G, H) The effects of ZFAS1 and mut-ZFAS1 on cardiomyocyte cell viability. n = 7. (I, J) The effects of ZFAS1 and mut-ZFAS1 on cardiomyocytes were detected by means of TUNEL staining. n = 7. Magnification ×200. (K, L) The contents of ROS of cardiomyocytes transfected with ZFAS1 and mut-ZFAS1. n = 13. Magnification ×200. Data are presented as mean ± SEM. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; t-tests and one-way analyses of variance with Bonferroni multiple group comparisons. Abbreviations as in Figures 1 and 3.
NMN is a potential therapeutic strategy to ameliorate MIRI
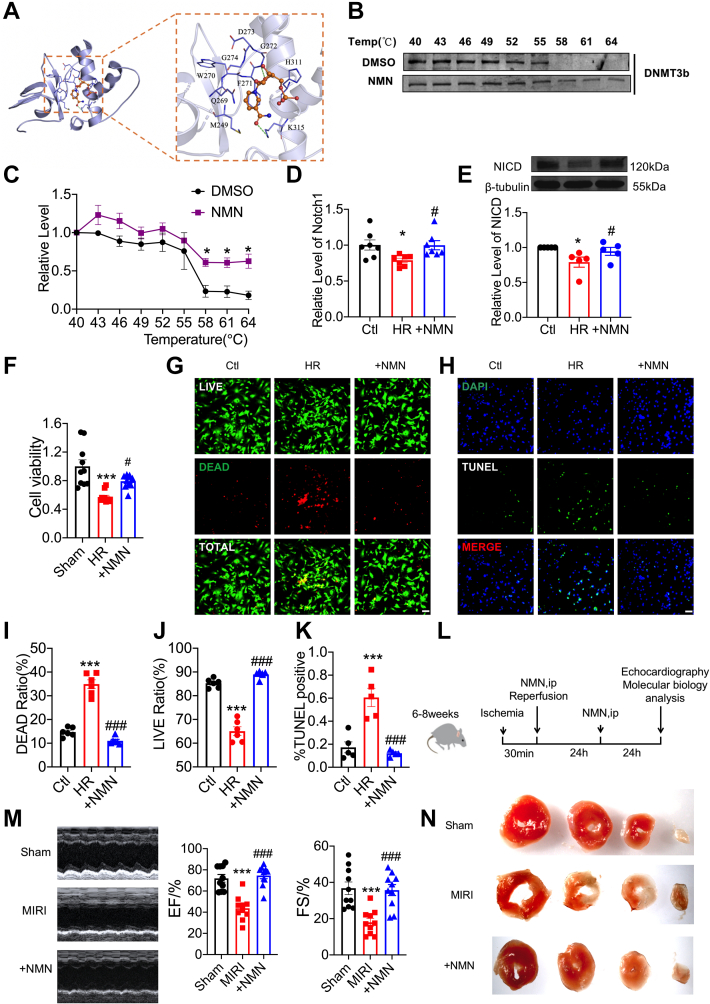
With the mechanism of DNA methylation in MIRI clarified, is there a drug that can interfere with the DNA methylation process of Notch1 to improve MIRI? We know that DNMT3b plays an important role in bridging ZFAS1 to DNA methylation of Notch1. Therefore, we searched for a known drug that can target DNMT3b. Molecular docking revealed that the amino acid residues G272 and K315 of DNMT3b can form hydrogen bond interactions with NMN, and amino acid residues H311, D273, G274, W270, F271, Q269, and M249 can form hydrophobic interactions with NMN (Figure 8A). Meanwhile, we discovered that NMN treatment strongly shifted the DNMT3b melting curve compared with dimethylsulfoxide in cells (Figures 8B and 8C). Surprisingly, NMN promotes the expression of Notch1 under HR conditions (Figures 8D and 8E). In addition, NMN improved the viability of cardiomyocytes under HR conditions (Figures 8F and 8G) and mitigated the apoptosis of cardiomyocytes induced by HR (Figure 8H). Furthermore, NMN reduces the infarct size of MIRI hearts and improves impaired heart function (Figures 8L to 8N). The above results indicated that NMN can relieve MIRI, likely by inhibiting the DNA methylation of the Notch1 promoter.
Figure 8.
NMN Is a Potential Therapeutic Strategy to Ameliorate MIRI
(A) Schematic diagram of the binding site of NMN and DNMT3b by molecular docking. (B) CETSA was used to detect the binding of NMN to DNMT3b. (C) Statistical results of CETSA: the protein level of DNMT3b at different temperatures after treatment with NMN and DMSO. n = 3. ∗P < 0.05 vs DMSO. (D) NMN promotes the mRNA expression of Notch1 in HR-treated cardiomyocytes. n = 8. ∗P < 0.05 vs Ctl; #P < 0.05 vs HR. (E) NMN promotes the protein expression of NICD in HR-treated cardiomyocytes. n = 5. ∗P < 0.05 vs Ctl; #P < 0.05 vs HR. (F) NMN increased the cell viability of HR-treated cells according to CCK8 assay. n = 10. ∗∗∗P < 0.001 vs Ctl; #P < 0.05 vs HR. (G, I, J) The effects of NMN on HR-treated cardiomyocytes cell viability. n = 6, ∗∗∗P < 0.001 vs Ctl, ###P < 0.001 vs MIRI, Magnification × 200. (H, K) The effects of NMN on HR-treated cardiomyocytes were detected with the use of TUNEL staining. n = 5. ∗∗∗P < 0.001 vs Ctl; ###P < 0.001 vs MIRI., Magnification ×200. (L) Experimental timeline. (M) Repair of cardiac function by NMN in MIRI mice. n = 10. ∗∗∗P < 0.001 vs Sham; ###P < 0.001 vs MIRI. (N) The effect of NMN on the infarct size of MIRI mice according to TTC. Data are presented as mean ± SEM. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; t-tests and one-way analyses of variance with Bonferroni multiple group comparisons. ip = intraperitoneal; NMN = nicotinamide mononucleotide; other abbreviations as in Figures 1, 5, and 6.
As a downstream effector of NMN, Sirt1 has been identified as a key regulator for MIRI.29 So additional experiments were done to determine whether Sirt1 is involved in the NMN-Notch1 regulation pathway in MIRI (Supplemental Figure 7). The Sirt1 inhibitor selisistat had no effect on Notch1 expression in HR-treated cardiomyocytes after NMN injection. Moreover, Sirt1 agonist SRT-2104 showed no effects on Notch1 in the normal cardiomyocytes. Therefore, we think that Sirt1 is not involved in the NMN-Notch1 regulation pathway in MIRI.
Discussion
Involvement of ZFAS1 in MIRI
ZFAS1 is closely associated with cardiovascular diseases. ZFAS1 was shown to be a possible biomarker for MI based on current evidence.23 Our research group discovered that as a SERCA2a inhibitor, ZFAS1 can cause intracellular Ca2+ excess and contractile failure.24 Furthermore, ZFAS1 produced intracellular Ca2+ excess, resulting in cardiomyocyte death, according to our findings.25 Wu et al37 also demonstrated that ZFAS1 promotes the functional availability of miR-150 by acting as a competing endogenous RNA to promote apoptosis. In the present investigation, we discovered that ZFAS1 was elevated during MIRI and that it played a role in myocardial injury by increasing oxidative stress and apoptosis.
Mechanisms underlying Notch1 regulation of MIRI
In cardiovascular diseases, Notch signaling is crucial. Notch gene re-expression after the myocardial injury is an adaptive response secondary to myocardial injury.38 Olmesartan can improve ventricular remodeling in chronic pressure overload mice by activating the Dll4-Notch1 signaling pathway. Also, Notch signaling protects against MIRI in part caused by antioxidative and antinitrative actions mediated by PTEN/Akt.14 Our present work conducted gene expression profiling experiments on ZFAS1 knock-in mice and determined that ZFAS1 can target Notch1 and regulate the Notch signaling pathway. Further verification also confirmed that ZFAS1 can inhibit the Notch signaling pathway by regulating Notch1. The inhibitory effect of MIRI on the Notch signaling pathway can be restored by knocking down ZFAS1; however DAPT, a Notch signaling pathway inhibitor, can reverse this effect even more. According to our findings, ZFAS1 functions as an upstream factor in the Notch signaling pathway and is essential in MIRI.
The regulatory mechanisms of ZFAS1 on Notch1
Based on our previous work, we know that ZFAS1 is distributed in the nucleus and cytoplasm,24 and previous research mainly explored its role in the cytoplasm. The methylation of genomic loci controlled by nuclear lncRNA is a newly recognized defining attribute of lncRNAs, according to a vast amount of experimental evidence.39 Our present study confirmed that Notch1 is hypermethylated in the heart tissue of ZFAS1 TG mice. By binding to DNA and generating RNA:DNA triplexes that can recruit DNMTs and promote DNA methylation of particular areas, lncRNAs can suppress gene transcription.40,41 Intriguingly, our observations found by means of ChiRP assay that ZFAS1 can bind to the Notch1 promoter, and by means of ChIP and RIP assays, we determined that ZFAS1 recruited DNMT3b in contrast to DNMT1 and DNMT3a. We specifically mutated the region where ZFAS1 binds to Notch1 and found that its recruitment of DNMT3b mainly relies on its binding to the Notch1 promoter region. Despite this, our findings suggest that the interaction between lncRNA and DNA promoters can recruit DNMTs and contribute to DNA methylation and subsequent molecular processes.
The regulatory mechanism of NMN in MIRI
It has been reported that NMN can cooperate with melatonin to protect MIRI,31 and it can protect MIRI by regulating mitochondrial ROS and redox.29 We found that NMN can significantly attenuate the apoptosis of HR-induced cardiomyocytes and improve the cardiac function of MIRI mice. Unlike in previous studies, we think that this effect is related to the molecular conformation of NMN and DNMT3b and the regulation of the methylation process.
Comparison with published studies on ZFAS1
Researchers have gradually recognized the regulatory role of ZFAS1 in heart disease. For MI, because of the absence of oxygen, cellular metabolism shifts to anaerobic respiration, producing lactate and causing a drop in intracellular pH, which activates the Na+-Ca2+ exchanger and causes intracellular Ca2+ overload. Thus, impairment of intracellular Ca2+ homeostasis is a key process in causing the dysfunction of MI. Our previous research confirmed that unusually high expression of ZFAS1 can lead to intracellular Ca2+ overload and, as a result, myocardial dysfunction in MI. MIRI, a completely different disease from MI, occurred caused by the production of a burst of detrimental oxidative stress and the accumulation of ROS, which mediates myocardial injury and cardiomyocyte death contributing to intracellular Ca2+ overload and damaged cell membrane. Thus, oxidative stress or ROS generation is the important initiating mechanism of MIRI, the occurrence of which is the key to distinguishing MI from MIRI. And our data demonstrated that ZFAS1 has a regulatory effect on MIRI and indicated that ZFAS1 exerts an effect on ROS levels, excluding the regulation of calcium overload. Moreover, although our data demonstrated that the ZFAS1-SERCA2a axis participated in the regulation of MIRI, we clarified that targeting the Notch signaling pathway is the main regulation mechanism of ZFAS1 in MIRI. Furthermore, ZFAS1 is reported to inhibit in lipopolysaccharide (LPS)–treated cardiomyocytes, and overexpression of ZFAS1 prevents LPS-induced apoptosis.42 However, both our previous studies and the findings by Huang et al43 and Wu et al37 have verified that knockdown of ZFAS1 could protect the cardiomyocytes from apoptosis. Here, we propose that LPS-induced cellular damage differs mechanistically from that of MIRI. The beneficial effects of inhibition of ZFAS1 and activation of Notch signaling on cardiomyocyte apoptosis are well established.
Study limitations
We are aware that the preapplication of AAV9-shZFAS1 may be a limitation of this study. This 3-week advance delivery of AAV9-shZFAS1 has limitations for clinical treatment of MIRI, and the protective impact of knockdown on ZFAS1 on MIRI was established only at the mechanistic level in our investigation, but it opens up the possibility of rectifying the functional impairment of the heart induced by ischemia-reoxygenation. In a follow-up study, we would consider establishing nanoparticles or exosome-encapsulated shZFAS1 or mut-ZFAS1 to provide the possibility for clinical treatment of heart diseases.
Conclusions
As a basis of these observations, we suggest the following paradigm for ZFAS1 to regulate apoptosis in cardiac ischemia-reperfusion injury: MIRI → ZFAS1↑ → Notch1 DNA methylation ↑ → Notch signaling pathway ↓ → cell apoptosis ↑ → heart function ↓. As shown in the Visual Abstract, up-regulated ZFAS1 facilitates DNMT-Notch1 interaction and promotes DNA methylation–mediated Notch1 down-regulation, according to our findings, which explains how up-regulated ZFAS1 underpins the genesis of MIRI. By interfering with this epigenetic process, NMN could be used to treat MIRI as a potential therapeutic therapy.
Perspectives.
COMPETENCY IN MEDICAL KNOWLEDGE: The present study identified the ZFAS1-Notch1 axis as a molecular integrator and therapeutic target of oxidative stress–induced cardiac dysfunction in MIRI. Indeed, genetic inhibition of ZFAS1 or activation of Notch1 significantly attenuated the typical hallmarks of maladaptive remodeling and directly improved cardiac function in a relevant preclinical animal model. NMN can activate Notch1 through epigenetic regulation and improve cardiac function after MIRI.
TRANSLATIONAL OUTLOOK: ZFAS1 could be considered as a novel therapeutic target for maintaining cardiac function in MIRI. NMN or other forms of ZFAS1 inhibitor could be developed into a novel therapeutic agent for ameliorating cardiac dysfunction after MIRI.
Funding Support and Author Disclosures
The National Key R & D Program of China (2017YFC1702003) and the National Natural Science Foundation of China (81970320, 81773735, 81961138018, 91949130, and 82003749) both contributed to this research. The authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Acknowledgments
The authors thank all of their colleagues in the Department of Pharmacology at Harbin Medical University.
Footnotes
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
Appendix
For an expanded Methods section as well as supplemental figures, please see the online version of this paper.
Contributor Information
Ying Zhang, Email: jennying223@126.com.
Yong Zhang, Email: hmuzhangyong@hotmail.com.
Appendix
References
- 1.Benjamin E.J., Blaha M.J., Chiuve S.E., et al. Heart disease and stroke statistics—2017 update: a report from the American Heart Association. Circulation. 2017;135:e146–e603. doi: 10.1161/CIR.0000000000000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeWood M.A., Spores J., Notske R., et al. Prevalence of total coronary occlusion during the early hours of transmural myocardial infarction. N Engl J Med. 1980;303:897–902. doi: 10.1056/NEJM198010163031601. [DOI] [PubMed] [Google Scholar]
- 3.Perrelli M.G., Pagliaro P., Penna C. Ischemia/reperfusion injury and cardioprotective mechanisms: role of mitochondria and reactive oxygen species. World J Cardiol. 2011;3:186–200. doi: 10.4330/wjc.v3.i6.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okura T., Gong L., Kamitani T., et al. Protection against Fas/APO-1– and tumor necrosis factor–mediated cell death by a novel protein, sentrin. J Immunol. 1996;157:4277–4281. [PubMed] [Google Scholar]
- 5.MacGrogan D., Munch J., de la Pompa J.L. Notch and interacting signalling pathways in cardiac development, disease, and regeneration. Nat Rev Cardiol. 2018;15:685–704. doi: 10.1038/s41569-018-0100-2. [DOI] [PubMed] [Google Scholar]
- 6.Daudet N., Zak M. Notch signalling: the multitask manager of inner ear development and regeneration. Adv Exp Med Biol. 2020;1218:129–157. doi: 10.1007/978-3-030-34436-8_8. [DOI] [PubMed] [Google Scholar]
- 7.Gordon W.R., Vardar-Ulu D., Histen G., Sanchez-Irizarry C., Aster J.C., Blacklow S.C. Structural basis for autoinhibition of Notch. Nat Struct Mol Biol. 2007;14:295–300. doi: 10.1038/nsmb1227. [DOI] [PubMed] [Google Scholar]
- 8.d’Souza B., Miyamoto A., Weinmaster G. The many facets of Notch ligands. Oncogene. 2008;27:5148–5167. doi: 10.1038/onc.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luxan G., d’Amato G., MacGrogan D., de la Pompa J.L. Endocardial Notch signaling in cardiac development and disease. Circ Res. 2016;118:e1–e18. doi: 10.1161/CIRCRESAHA.115.305350. [DOI] [PubMed] [Google Scholar]
- 10.Kopan R., Ilagan M.X. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jabs M., Rose A.J., Lehmann L.H., et al. Inhibition of endothelial notch signaling impairs fatty acid transport and leads to metabolic and vascular remodeling of the adult heart. Circulation. 2018;137:2592–2608. doi: 10.1161/CIRCULATIONAHA.117.029733. [DOI] [PubMed] [Google Scholar]
- 12.Garg V. In: Etiology and Morphogenesis of Congenital Heart Disease: From Gene Function and Cellular Interaction to Morphology. Nakanishi T., Markwald R.R., Baldwin H.S., Keller B.B., Srivastava D., Yamagishi H., editors. Springer Open; Tokyo, Japan: 2016. Notch signaling in aortic valve development and disease; pp. 371–376. [PubMed] [Google Scholar]
- 13.Wu L., Xu H., Zhang W., Chen Z., Li W., Ke W. Circular RNA circCCDC9 alleviates ischaemic stroke ischaemia/reperfusion injury via the Notch pathway. J Cell Mol Med. 2020;24:14152–14159. doi: 10.1111/jcmm.16025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pei H., Yu Q., Xue Q., et al. Notch1 cardioprotection in myocardial ischemia/reperfusion involves reduction of oxidative/nitrative stress. Basic Res Cardiol. 2013;108:373. doi: 10.1007/s00395-013-0373-x. [DOI] [PubMed] [Google Scholar]
- 15.Beerman I., Bock C., Garrison B.S., et al. Proliferation-dependent alterations of the DNA methylation landscape underlie hematopoietic stem cell aging. Cell Stem Cell. 2013;12:413–425. doi: 10.1016/j.stem.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 16.van Noesel M.M., van Bezouw S., Salomons G.S., et al. Tumor-specific down-regulation of the tumor necrosis factor–related apoptosis-inducing ligand decoy receptors DcR1 and DcR2 is associated with dense promoter hypermethylation. Cancer Res. 2002;62:2157–2161. [PubMed] [Google Scholar]
- 17.Fu W.N., Bertoni F., Kelsey S.M., et al. Role of DNA methylation in the suppression of Apaf-1 protein in human leukaemia. Oncogene. 2003;22:451–455. doi: 10.1038/sj.onc.1206147. [DOI] [PubMed] [Google Scholar]
- 18.Ponnusamy M., Liu F., Zhang Y.H., et al. Long noncoding RNA CPR (cardiomyocyte proliferation regulator) regulates cardiomyocyte proliferation and cardiac repair. Circulation. 2019;139:2668–2684. doi: 10.1161/CIRCULATIONAHA.118.035832. [DOI] [PubMed] [Google Scholar]
- 19.Movassagh M., Choy M.K., Knowles D.A., et al. Distinct epigenomic features in end-stage failing human hearts. Circulation. 2011;124:2411–2422. doi: 10.1161/CIRCULATIONAHA.111.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meder B., Haas J., Sedaghat-Hamedani F., et al. Epigenome-wide association study identifies cardiac gene patterning and a novel class of biomarkers for heart failure. Circulation. 2017;136:1528–1544. doi: 10.1161/CIRCULATIONAHA.117.027355. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y., Sun L., Xuan L., et al. Long noncoding RNA CCRR controls cardiac conduction via regulating intercellular coupling. Nat Commun. 2018;9:4176. doi: 10.1038/s41467-018-06637-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bai X., Yang C., Jiao L., et al. LncRNA MIAT impairs cardiac contractile function by acting on mitochondrial translocator protein TSPO in a mouse model of myocardial infarction. Signal Transduct Target Ther. 2021;6:172. doi: 10.1038/s41392-021-00538-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y., Sun L., Xuan L., et al. Reciprocal changes of circulating long noncoding RNAs Zfas1 and CDR1as predict acute myocardial infarction. Sci Rep. 2016;6 doi: 10.1038/srep22384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y., Jiao L., Sun L., et al. LncRNA ZFAS1 as a SERCA2a inhibitor to cause intracellular Ca2+ overload and contractile dysfunction in a mouse model of myocardial infarction. Circ Res. 2018;122:1354–1368. doi: 10.1161/CIRCRESAHA.117.312117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiao L., Li M., Shao Y., et al. LncRNA-ZFAS1 induces mitochondria-mediated apoptosis by causing cytosolic Ca2+ overload in myocardial infarction mice model. Cell Death Dis. 2019;10:942. doi: 10.1038/s41419-019-2136-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berger F., Lau C., Dahlmann M., Ziegler M. Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J Biol Chem. 2005;280:36334–36341. doi: 10.1074/jbc.M508660200. [DOI] [PubMed] [Google Scholar]
- 27.Hong W., Mo F., Zhang Z., Huang M., Wei X. Nicotinamide mononucleotide: a promising molecule for therapy of diverse diseases by targeting NAD+ metabolism. Front Cell Dev Biol. 2020;8:246. doi: 10.3389/fcell.2020.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang R., Shen Y., Zhou L., et al. Short-term administration of nicotinamide mononucleotide preserves cardiac mitochondrial homeostasis and prevents heart failure. J Mol Cell Cardiol. 2017;112:64–73. doi: 10.1016/j.yjmcc.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto T., Byun J., Zhai P., Ikeda Y., Oka S., Sadoshima J. Nicotinamide mononucleotide, an intermediate of NAD+ synthesis, protects the heart from ischemia and reperfusion. PLoS One. 2014;9 doi: 10.1371/journal.pone.0098972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiss T., Nyul-Toth A., Balasubramanian P., et al. Nicotinamide mononucleotide (NMN) supplementation promotes neurovascular rejuvenation in aged mice: transcriptional footprint of SIRT1 activation, mitochondrial protection, antiinflammatory, and antiapoptotic effects. Geroscience. 2020;42:527–546. doi: 10.1007/s11357-020-00165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hosseini L., Vafaee M.S., Badalzadeh R. Melatonin and nicotinamide mononucleotide attenuate myocardial ischemia/reperfusion injury via modulation of mitochondrial function and hemodynamic parameters in aged Rats. J Cardiovasc Pharmacol Ther. 2020;25:240–250. doi: 10.1177/1074248419882002. [DOI] [PubMed] [Google Scholar]
- 32.Takahara S., Inoue S.I., Miyagawa-Tomita S., et al. New Noonan syndrome model mice with RIT1 mutation exhibit cardiac hypertrophy and susceptibility to beta-adrenergic stimulation–induced cardiac fibrosis. EBioMedicine. 2019;42:43–53. doi: 10.1016/j.ebiom.2019.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J., Zheng Y., Cheng X., et al. Inhibition of microRNA-34a suppresses epileptiform discharges through regulating notch signaling and apoptosis in cultured hippocampal neurons. Neurochem Res. 2019;44:1252–1261. doi: 10.1007/s11064-019-02772-x. [DOI] [PubMed] [Google Scholar]
- 34.Liu S., Wang Z., Chen D., et al. Annotation and cluster analysis of spatiotemporal- and sex-related lncRNA expression in rhesus macaque brain. Genome Res. 2017;27:1608–1620. doi: 10.1101/gr.217463.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmitz K.M., Mayer C., Postepska A., Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24:2264–2269. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith C., Heyne S., Richter A.S., Will S., Backofen R. Freiburg RNA Tools: a web server integrating INTARNA, EXPARNA and LOCARNA. Nucleic Acids Res. 2010;38:W373–W377. doi: 10.1093/nar/gkq316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu T., Wu D., Wu Q., et al. Knockdown of long noncoding RNA–ZFAS1 protects cardiomyocytes against acute myocardial infarction via antiapoptosis by regulating miR-150/CRP. J Cell Biochem. 2017;118:3281–3289. doi: 10.1002/jcb.25979. [DOI] [PubMed] [Google Scholar]
- 38.Gude N.A., Emmanuel G., Wu W., et al. Activation of Notch-mediated protective signaling in the myocardium. Circ Res. 2008;102:1025–1035. doi: 10.1161/CIRCRESAHA.107.164749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L., Zhao Y., Bao X., et al. LncRNA DUM interacts with DNMTs to regulate DPPA2 expression during myogenic differentiation and muscle regeneration. Cell Res. 2015;25:335–350. doi: 10.1038/cr.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de la Pompa J.L., Epstein J.A. Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev Cell. 2012;22:244–254. doi: 10.1016/j.devcel.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luxan G., Casanova J.C., Martinez-Poveda B., et al. Mutations in the Notch pathway regulator MIB1 cause left ventricular noncompaction cardiomyopathy. Nat Med. 2013;19:193–201. doi: 10.1038/nm.3046. [DOI] [PubMed] [Google Scholar]
- 42.Chen D.D., Wang H.W., Cai X.J. Transcription factor Sp1 ameliorates sepsis-induced myocardial injury via ZFAS1/Notch signaling in H9C2 cells. Cytokine. 2021;140 doi: 10.1016/j.cyto.2021.155426. [DOI] [PubMed] [Google Scholar]
- 43.Huang P., Yang D., Yu L., Shi Y. Downregulation of lncRNA ZFAS1 protects H9C2 cardiomyocytes from ischemia/reperfusion–induced apoptosis via the miR5903p/NFκB signaling pathway. Mol Med Rep. 2020;22:2300–2306. doi: 10.3892/mmr.2020.11340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.