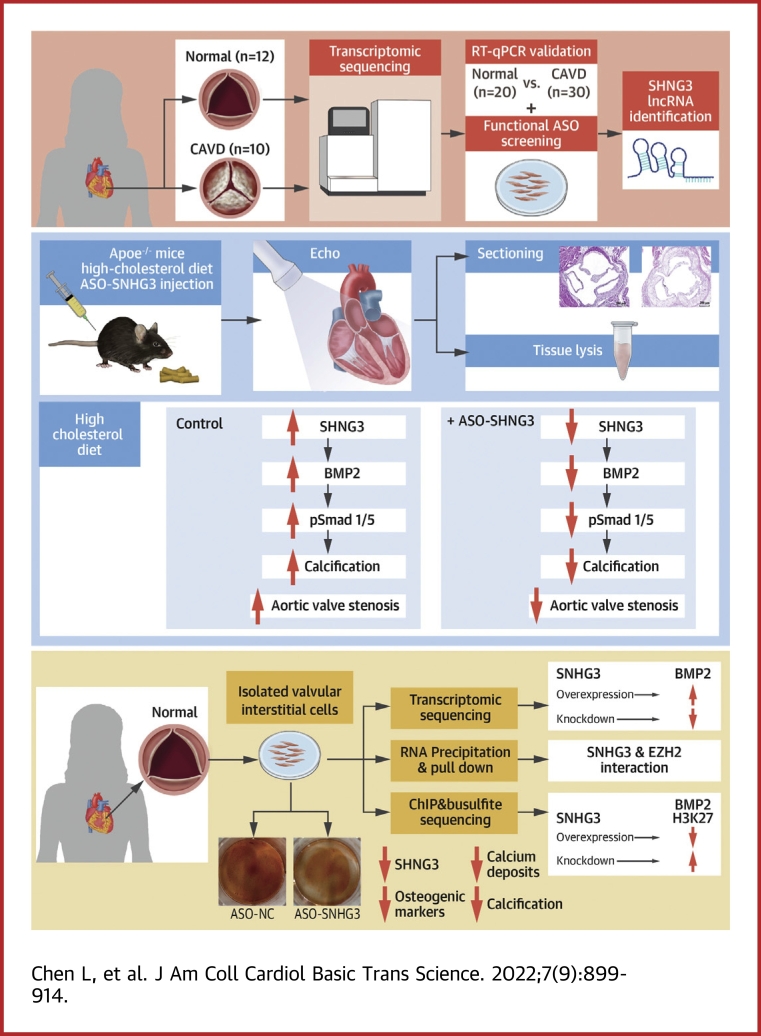
Visual Abstract
Key Words: BMP2 pathway, calcific aortic valve disease, long noncoding RNA, osteoblast differentiation, therapeutic target
Abbreviations and Acronyms: ADV, adenovirus; ALP, alkaline phosphatase; ApoE−/−, apolipoprotein E-deficient; ASO, antisense oligonucleotide; BMP, bone morphogenic proteins; CAVD, calcific aortic valve disease; EZH2, enhancer of zeste 2; FISH, fluorescence in situ hybridization; HCD, high-cholesterol diet; hVICs, human aortic valve interstitial cells; lncRNA, long noncoding RNA; ND, normal diet; PRC2, polycomb repressive complex 2; RT-qPCR, reverse-transcriptase-quantitative polymerase chain reaction; RUNX2, Runt-related transcription factor 2; SNHG3, small nucleolar RNA host gene 3; TGF, transforming growth factor
Highlights
-
•
The long noncoding RNA SNHG3 was upregulated in the leaflets of both patients and mice with calcific aortic valve disease.
-
•
SNHG3 can associate with EZH2 in the nucleus of hVICs to epigenetically upregulate BMP2, a key mediator of calcification.
-
•
SNHG3 promoted osteoblast differentiation of hVICs via upregulation of the BMP2 pathway.
-
•
SNHG3 silencing significantly ameliorated aortic valve calcification in experimental animals, providing a novel therapeutic target for CAVD.
Summary
Based on high-throughput transcriptomic sequencing, SNHG3 was among the most highly expressed long noncoding RNAs in calcific aortic valve disease. SNHG3 upregulation was verified in human and mouse calcified aortic valves. Moreover, in vivo and in vitro studies showed SNHG3 silencing markedly ameliorated aortic valve calcification. In-depth functional assays showed SNHG3 physically interacted with polycomb repressive complex 2 to suppress the H3K27 trimethylation BMP2 locus, which in turn activated BMP2 expression and signaling pathways. Taken together, SNHG3 promoted aortic valve calcification by upregulating BMP2, which might be a novel therapeutic target in human calcific aortic valve disease.
Calcific aortic valve disease (CAVD) is the most common heart valve disease and the most common cause of aortic valve replacement,1 and there are no effective treatments to halt or slow down the disease progression. However, the causes and pathogenesis of CAVD remain to be unexplored. CAVD is an active and progressive process associated with endothelial dysfunction, immune cell infiltration, calcium deposition, and extracellular matrix remodeling, which eventually result in human valve interstitial cells (hVICs) undergoing a phenotype transition to become osteoblast-like cells.2
Long noncoding RNAs (lncRNAs), a class of transcripts longer than 200 nucleotides, have recently emerged to play important roles in diverse cellular processes and the development of various diseases.3,4 LncRNAs can regulate gene expression at multiple levels by interacting with RNA, DNA, and proteins.5 The effects of lncRNAs in CAVD have been preliminarily explored. LncRNA H19 downregulates Notch1 to promote mineralization in vitro and has not been further verified in CAVD models.6 However, the role of lncRNAs in CAVD remains unelucidated.
We identified an lncRNA named small nucleolar RNA host gene 3 (SNHG3, NR_036473.1, NONHSAT001953) that was upregulated in calcific aortic valves. SNHG3 is located at chromosome 1p35 with approximately 2.3 kb nucleotides, which contributes to the development of Alzheimer disease7 and various malignant tumors.8, 9, 10 SNHG3 has not been previously reported to be associated with cardiovascular disease. Nevertheless, some studies have reported that SNHG3 can mediate molecular markers related to CAVD, such as Notch1 and Runt-related transcription factor 2 (RUNX2) in some tumors.9,11 Therefore, SNHG3 may be a new therapeutic target for CAVD.
Bone morphogenic proteins (BMPs), which belong to the transforming growth factor(TGF)-β superfamily, are recognized as potent pro-osteogenic factors.12,13 BMP2 was considered to be highly expressed and play a vital role in CAVD14,15 and phosphorylated Smad1 and 5 (pSmad1/5), as the main signal transducer of BMP2 signaling pathway also increased significantly in calcific aortic valves.16 Nevertheless, the mechanism of significant BMP2 upregulation during CAVD remains unclear.
In this study, we aimed to explain the role of lncRNAs in regulating the mechanism of CAVD. These findings provide new insights on SNHG3, which may promote CAVD progression and be considered a therapeutic target.
Methods
Detailed materials and methods are found in the Supplemental Appendix.
Clinical sample
This study was performed with the approval of the institutional ethics committees of the Fuwai Hospital, Chinese Academy of Medical Sciences (No. 2012-404), and complied with the Declaration of Helsinki. All the patients signed written informed consent before participating in the study. Human calcific aortic valve leaflets were obtained from patients with CAVD during aortic valve replacement. Control nonmineralized aortic valves were collected from the explanted hearts of patients who underwent heart transplantation procedures. The clinical characteristic patients for lncRNA Sequencing are shown in Supplemental Table 1 and for RT-qPCR analysis are shown in Supplemental Table 2. The clinical characteristics of patients for cell cultures are shown in Supplemental Table 3.
Animal experiments
All animal studies were carried out in accordance with protocols approved by the Ethics Committee of Fuwai Hospital, Chinese Academy of Medical Sciences for the Use and Care of Laboratory Animals (No. FW-2020-0022), and all the procedures complied with US National Institutes of Health (NIH Publication No.85-23, revised 1996) on the protection of animals used for scientific purposes. Adult apolipoprotein E-deficient (ApoE−/−) (C57BL/6 background) mice were purchased from Beijing Vital River Laboratory Animal Technology Co, Ltd, and housed in a pathogen-free, temperature-controlled environment under a 12:12 hour light-dark cycle.
Statistical analysis
Continuous data are presented as the mean ± SEM or mean ± SD for normally distributed data. The normality of the distribution of continuous data was confirmed by Shapiro-Wilk test and was visualized by a Q-Q plot. The Levene test was used to confirm the homogeneity of variance of continuous data. For normally distributed data, comparisons between the 2 groups were evaluated for significance using the unpaired Student’s t-test or Welch’s t-test, whereas comparisons among 3 or more groups were evaluated for significance using analysis of variance followed by least significance difference, Holm-Sidak, Dunnett’s, and Bonferroni multiple comparison post hoc test using the SPSS software. The data that are not normality distributed were compared using the Mann-Whitney U test (2 groups) or Kruskal-Wallis test (>2 groups). The counts of category data were compared using chi-square analysis between 2 independent groups. The association between the 2 continuous variables was evaluated using a 2-tailed Pearson’s correlation analysis. Statistical significance was set at P < 0.05. Differential expression analysis of lncRNAs in CAVD and the nonmineralized control group using the limma R package and the log2 fold change was computed as log2 (calcified aortic valve) minus log2 (nonmineralized aortic valve).
Results
SNHG3 is highly expressed in human CAVD
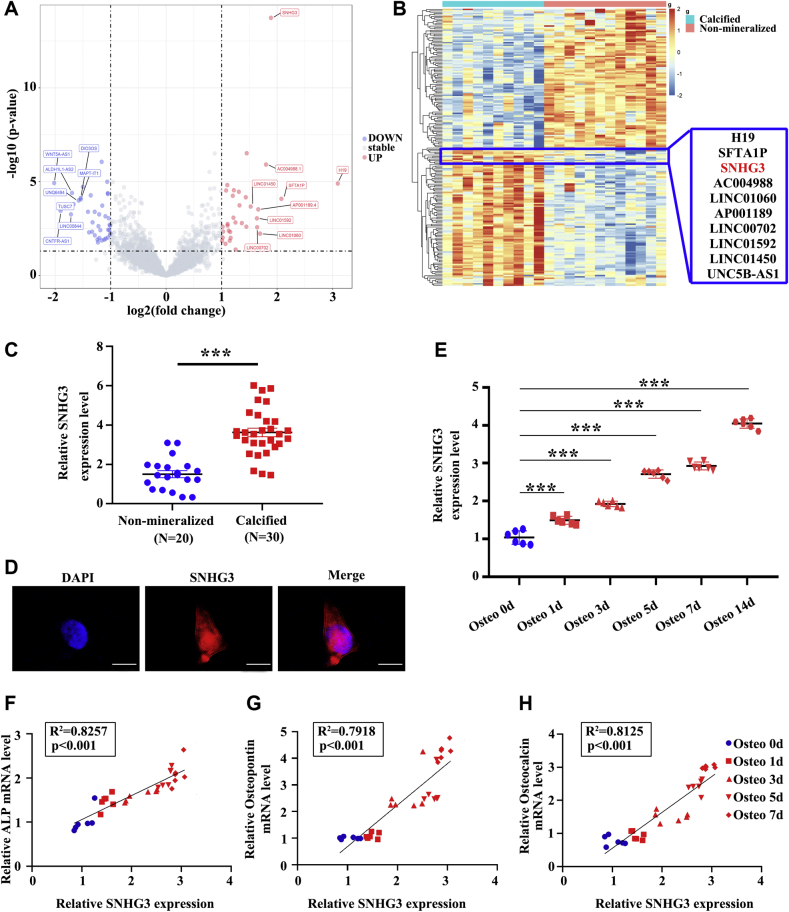
To identify important lncRNAs related to the development of CAVD, we performed transcriptomic sequencing on 10 patients with CAVD and 12 controls (nonmineralized aortic valves). The calcification staining for human aortic valve tissues are shown in Supplemental Figure 1. The volcano and plot of lncRNAs differentially expressed is presented in Figure 1A and the hierarchical cluster analysis and heat mapping is showed in Figure 1B. Compared with nonmineralized aortic valves, 33 lncRNAs were upregulated and 39 were significantly downregulated in CAVD (filtered by |fold change| >2 and P < 0.05) (Supplemental Table 4). We verified the top 10 highly expressed lncRNAs in 50 samples (30 cases of CAVD and 20 controls) by reverse-transcriptase quantitative polymerase chain reaction (RT-qPCR, the primers and probes for RT-qPCR are shown in Supplemental Table 5) and found that 7 of 10 of these lncRNAs were upregulated in patients with CAVD (Supplemental Figures 2A to 2J). Studies by Mathieu found H19 was upregulated in CAVD and promotes mineralization of hVICs6; we also found H19 was highly expressed in calcific aortic valve tissues through transcriptomic sequencing and validated in RT-qPCR assays in expanded samples of CAVD. Therefore, we used H19 as a positive control. Next, we isolated hVICs from nonmineralized aortic valves, which were confirmed by immunofluorescence staining with anti-alpha smooth muscle actin and anti-vimentin (Supplemental Figure 3). Functional antisense oligonucleotide (ASO) screening to identify calcification-related lncRNAs showed silencing of H19, SNHG3, and SFTA1P downregulated 3 osteogenic markers (alkaline phosphatase [ALP], RUNX2, and OSTEOPONTIN) in osteogenic medium-induced hVICs; the effect of SNHG3 knockdown was very prominent (Supplemental Figures 2K to 2M). Hence, in our subsequent experiments we focused on SNHG3.
Figure 1.
lncRNA SNHG3 Is Highly Expressed in CAVD and Associated With the Osteoblast Differentiation of Primary hVICs
(A and B) Volcano plot and hierarchical cluster heat map showing the differentially expressed lncRNAs from transcriptomic sequencing data. (C) Expression level of SNHG3 in nonmineralized aortic valves (n = 20) and calcified aortic valves (n = 30), unpaired 2-tailed Student’s t-test. (D) RNA-florescence in situ hybridization for SNHG3 in isolated hVICs (n = 6). Scale bar, 10 μm. (E) SNHG3 expression in hVICs 0, 1, 3, 5, 7, and 14 days after osteogenic induction (n = 6), repeated measures analysis of variance followed by Dunnett’s post hoc test. (F to H) Correlation analysis between SNHG3 and osteogenic differentiation markers (ALP, osteocalcin, and osteopontin) mRNA levels in hVICs 0, 1, 3, 5, and 7 days after osteogenic induction (n = 6), 2-tailed Pearson’s correlation analysis. Values are mean ± SEM, ∗∗∗P < 0.001 vs control. CAVD = calcific aortic valve disease; H&E = hematoxylin-eosin; HCD = high-cholesterol diet; hVIC = human aortic valve interstitial cell; lncRNA, long noncoding RNA; ND = normal diet; OPN = osteopontin; SNHG3 = small nucleolar RNA host gene 3.
SNHG3 (NR_036473.1, NONHSAT001953) is located on human chromosome 1 with 4 exons and low coding potential. RNA fluorescence in situ hybridization (RNA-FISH) showed that SNHG3 was mainly distributed in the cell nucleus (Figure 1D). We detected expression of SNHG3 and osteoblastic differentiation markers in osteogenic medium-induced hVICs. As shown in Figure 1E, SNHG3 expression was induced on stimulation of hVICs with the osteogenic medium. In addition, the level of SNHG3 was strongly positively correlated with 3 known osteoblastic differentiation markers (ALP, osteopontin, and osteocalcin) at 0, 1, 3, 5, and 7 days after osteogenic induction of hVICs (Figure 1F to 1H). These data indicated that SNHG3 may participate in CAVD progression through its association with osteoblast differentiation of hVICs.
SNHG3 is upregulated in experimental calcific aortic valve disease in mice
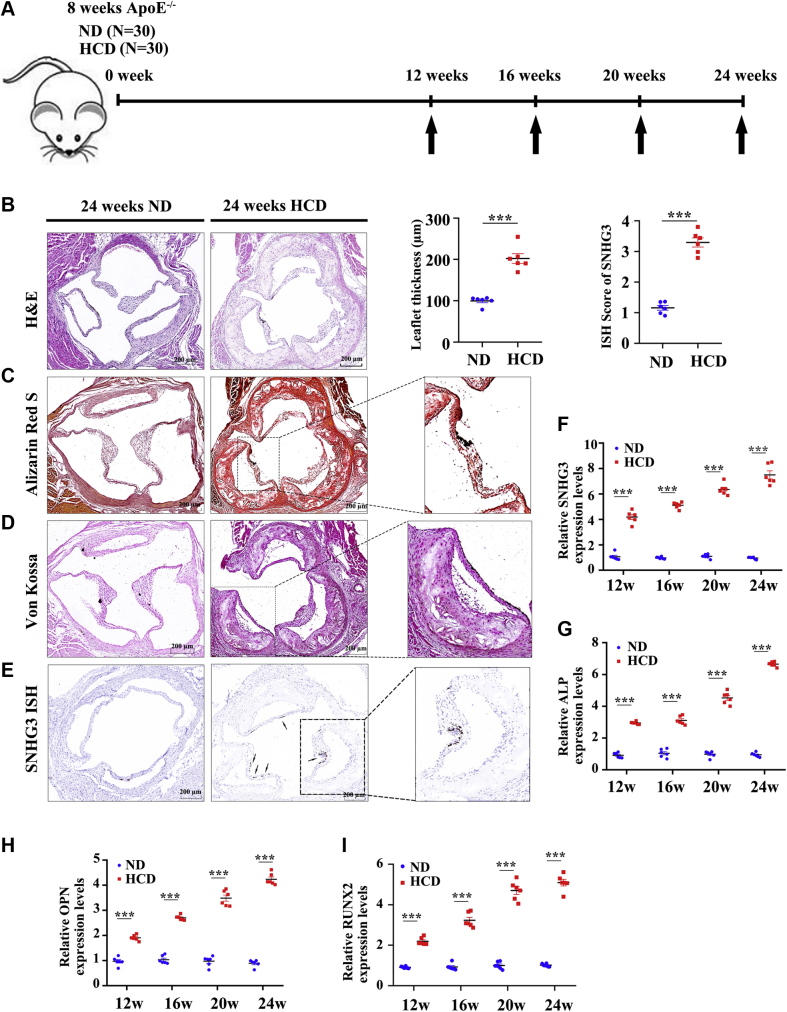
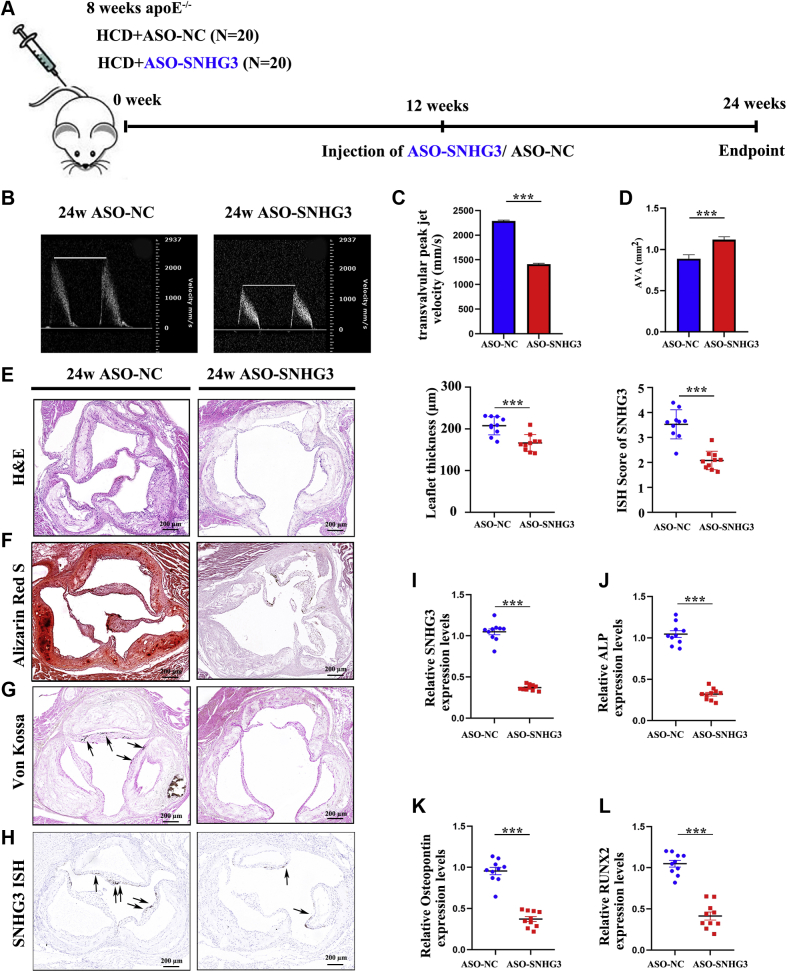
We built an animal model of CAVD by administering ApoE−/− mice with a high-cholesterol diet for 24 weeks (HCD group) and another group of ApoE−/− mice received a normal diet as a negative control group (ND group). From the 12th week of the high-cholesterol diet to the 24th week every 4 weeks, echocardiography was used to evaluate the degree of aortic valve stenosis in the mice of the 2 groups (Figure 2A), and the mice in the HCD group had a significant increase in transvalvular peak jet velocity and a decrease in aortic valve area, which were in a time-dependent manner (Supplemental Figure 4). We then collected the aortic valve and evaluated the morphology of the valve leaflet thickness and degree of calcification. Hematoxylin-eosin staining showed that aortic valve leaflet thickness was consistently increased in the HCD group compared with that in the ND group at the 24th week (Figure 2B). This is consistent with the increased calcium deposits in the aortic valve leaflets of the animals, as assessed by Alizarin Red S (Figure 2C) and Von Kossa staining (Figure 2D). Moreover, we also found that SNHG3 expression was significantly higher in the HCD group than in the ND group in situ hybridization with RNA scope (Figure 2E). In addition, we then collected aortic valves and extracted RNA from both groups to evaluate the level of SNHG3 and osteogenic differentiation markers at 12, 16, 20, and 24 weeks. RT-qPCR results showed that SNHG3 in the HCD group was significantly upregulated compared with that in the ND group and gradually increased from the 12th to 24th week of the high-cholesterol diet (Figure 2F). As expected, the levels of osteogenic differentiation markers showed an increasing trend similar to that of SNHG3 (Figures 2G-2I), which indicated that SNHG3 expression was strongly positively correlated with CAVD progression in vivo.
Figure 2.
SNHG3 Was Upregulated in Experimental CAVD in Mice
(A) Schematic diagram showing the experimental procedures for building CAVD model in ApoE−/− mice. (B-E) Representative H &E staining, Alizarin Red S staining, Von Kossa staining, and SNHG3 in situ hybridization with RNA scope in the HCD group and ND group at the 24th week (n = 6/group), and we analyzed mean aortic valve leaflet thickness and quantification of SNHG3 in aortic valve leaflet between 2 groups. Scale bar, 200 μm. (F to I) The transcript level of SNHG3 and osteogenic differentiation marker gene (ALP, OPN, and RUNX2) in aortic valve leaflets of HCD group and ND group treated mice from the 12th week to 24th week. Welch’s t-test for (B), unpaired 2-tailed Student’s t-test for (C), and 2-way ANOVA followed by Dunnett’s test for (F to I). Values are mean ± SEM. ∗∗∗P < 0.001 compared with ND group. Ad = adenovirus; ALP = alkaline phosphatase; ASO = antisense oligonucleotide; GFP = green fluorescent protein; RUNX2 = Runt-related transcription factor 2; other abbreviations as in Figure 1.
SNHG3 promotes osteoblast differentiation of hVICs
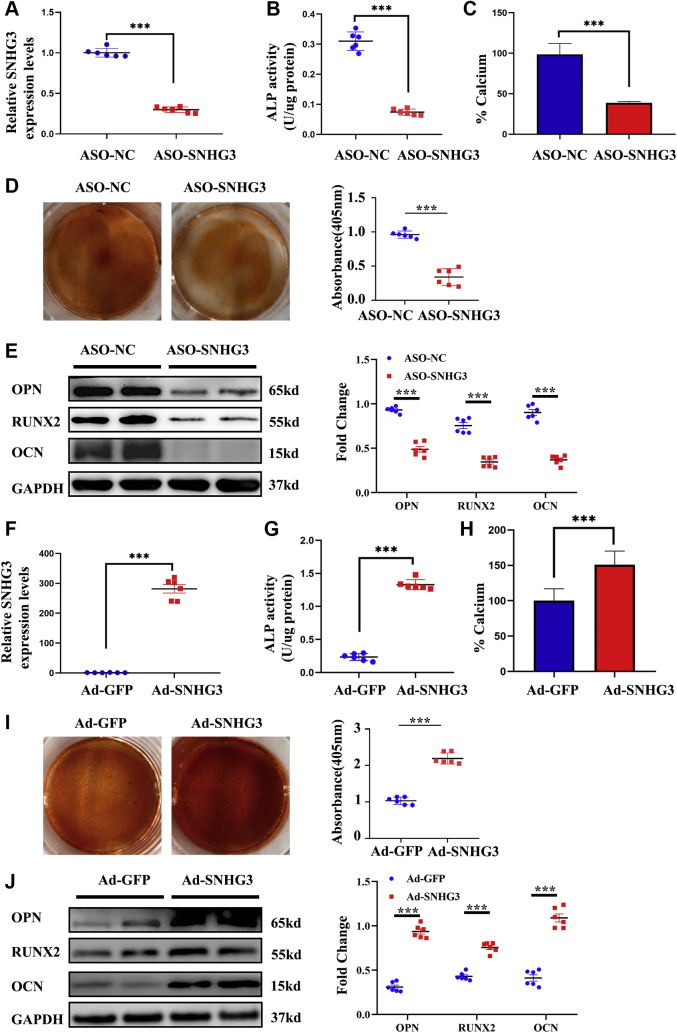
Considering that hVICs express SNHG3 and that its level is increased and correlated with osteoblastic differentiation markers during CAVD, we hypothesized that SNHG3 might contribute to reprogramming hVICs toward an osteogenic phenotype. Human VIC cultures were treated with a mineralizing medium for 7 days, and osteogenic genes were measured after silencing SNHG3 with an ASO. SNHG3 was downregulated approximately 75% by ASO (Figure 3A). HVICs were collected to detect ALP activity, calcified nodule formation, calcium ion concentration in cultures, and protein levels of osteoblastic differentiation markers. The results showed that ASO-mediated SNHG3 silencing in hVICs negated the osteoblastic differentiation medium-induced increase in ALP activity (Figure 3B), calcium ion concentration (Figure 3C), calcified nodule formation (Figure 3D), and protein levels of osteoblastic differentiation markers (Figure 3E) compared with the control cells. In contrast, the overexpression of SNHG3 through an adenovirus (ADV) vector (Figure 3F) resulted in further increases in ALP activity (Figure 3G), calcium ion concentration (Figure 3H), calcified nodule formation (Figure 3I), and protein levels of osteoblastic differentiation markers in hVICs (Figure 3J). These results indicate that SNHG3 plays a positive role in the osteoblast differentiation of hVICs.
Figure 3.
SNHG3 Promotes Osteogenic Medium-Induced Osteoblast Differentiation in Vitro
(A) Silencing of SNHG3 with ASO (n = 6/group). (B to E) The ALP activity evaluated by spectrophotometry, calcium content in cell cultures measured by Arsenazo III, mineralized bone matrix formation evaluated by Alizarin Red staining, and osteogenic differentiation markers (RUNX2, OCN, and OPN) protein levels measured by western blot were significantly decreased in the ASO-SNHG3 group compared with the ASO-NC group. (F) SNHG3 increased in hVICs transfected with through Ad (n = 6/group). (G to J) Overexpression of SNHG3 in hVICs resulted in further in increases in ALP activity, calcium ion concentration, calcified nodule formation, and protein levels of osteogenic differentiation markers. GAPDH was used as internal control. Unpaired 2-tailed Student’s t-test for A, D, and G to J, and Welch’s t-test for B, C, and F. Values are means ± SEM. ∗∗∗P < 0.001 vs ASO-NC or Ad-GFP. OCN = osteocalcin; other abbreviations as in Figures 1 and 2.
Lack of SNHG3 alleviates aortic valve calcification in vivo
We tested the therapeutic potential of SNHG3 inhibition in an animal model of aortic valve calcification. ApoE−/− mice were fed a high-cholesterol diet for 24 weeks to develop aortic valve calcification and were randomly treated for 12 weeks with twice-a-week injection of ASO against a scrambled sequence (ASO-NC group) or mus-SNHG3 (ASO-SNHG3 group) at the 12th week of a high-cholesterol diet (Figure 4A). Echocardiographic assessment of heart aortic valve stenosis in mice treated with ASO-SNHG3 showed a significant decrease in transvalvular peak jet velocity and a significant increase in aortic valve area at the time of euthanization (Figures 4B to 4D, Supplemental Table 6), which means that SNHG3 silencing can ameliorate the aortic valve calcification in a mice model. In vivo repression of SNHG3 consistently decreased aortic valve leaflet thickness compared with that in mice treated with ASO-NC, as assessed by hematoxylin-eosin staining (Figure 4E). This is in line with the decreased calcium deposits in the aortic valve leaflets of mice treated with ASO-SNHG3 when compared with ASO-NC, as assessed by Alizarin Red S (Figure 4F) and von Kossa staining (Figure 4G). As expected, ASO-mediated silencing of SNHG3 assessed by in situ hybridization with RNA scope (Figure 4H) and RT-qPCR (Figure 4I) led to a significant decrease in osteogenic differentiation markers (Figures 4J to 4L) in aortic valve leaflets of mice. However, SNHG3 silencing failed to modulate the levels of glucose, total cholesterol, low-density lipoprotein, and triglycerides (Supplemental Table 7), which suggests that SNHG3 silencing ameliorates aortic valve calcification in ApoE−/− mice independently of metabolic regulation. These results further verify the role of SNHG3 in osteogenic differentiation and provide potent evidence for a therapeutic strategy targeting SNHG3 in CAVD treatment.
Figure 4.
In Vivo Targeting SNHG3 Reduces HCD-Induced Aortic Valve Calcification in Apoe−/− Mice
(A) Schematic representation of the experimental design that HCD-induced ApoE−/− mice were assigned randomly into injection of ASO against a scrambled sequence (ASO-NC group) or against SNHG3 (ASO-SNHG3 group) for 12 weeks with twice a week at the 12th week of HCD. (B) Representative echocardiographic images of transvalvular peak jet velocity in ApoE−/− mice from both groups. (C and D) Echocardiographic data in ASO-NC group and ASO-SNHG3 group at 24th week of HCD: transvalvular peak jet velocity, aortic valve area. Representative H and E staining (E), Alizarin Red S staining (F), Von Kossa staining (G), and SNNG3 in situ hybridization with RNA scope (H) in the ASO-NC group and ASO-SNHG3 group at the 24th week of HCD. The mean aortic valve leaflet thickness and quantification of SNHG3 in aortic valve leaflets between 2 groups were analyzed. Scale bar, 200 μm. (I to L) The transcript level of SNHG3 and osteogenic differentiation markers (ALP, OPN, RUNX2) in aortic valve leaflets of ASO-NC group and ASO-SNHG3 group (n = 10/group). Unpaired 2-tailed Student’s t-test for (C to H) and (J to L), and Welch’s t-test for (I). Values are mean ± SEM. ∗∗∗P < 0.001 compared with ASO-NC group. ApoE−/−, apolipoprotein E-deficient; other abbreviations as in Figures 1 and 2.
SNHG3 activates BMP2 signaling pathway to promote osteoblast differentiation of hVICs
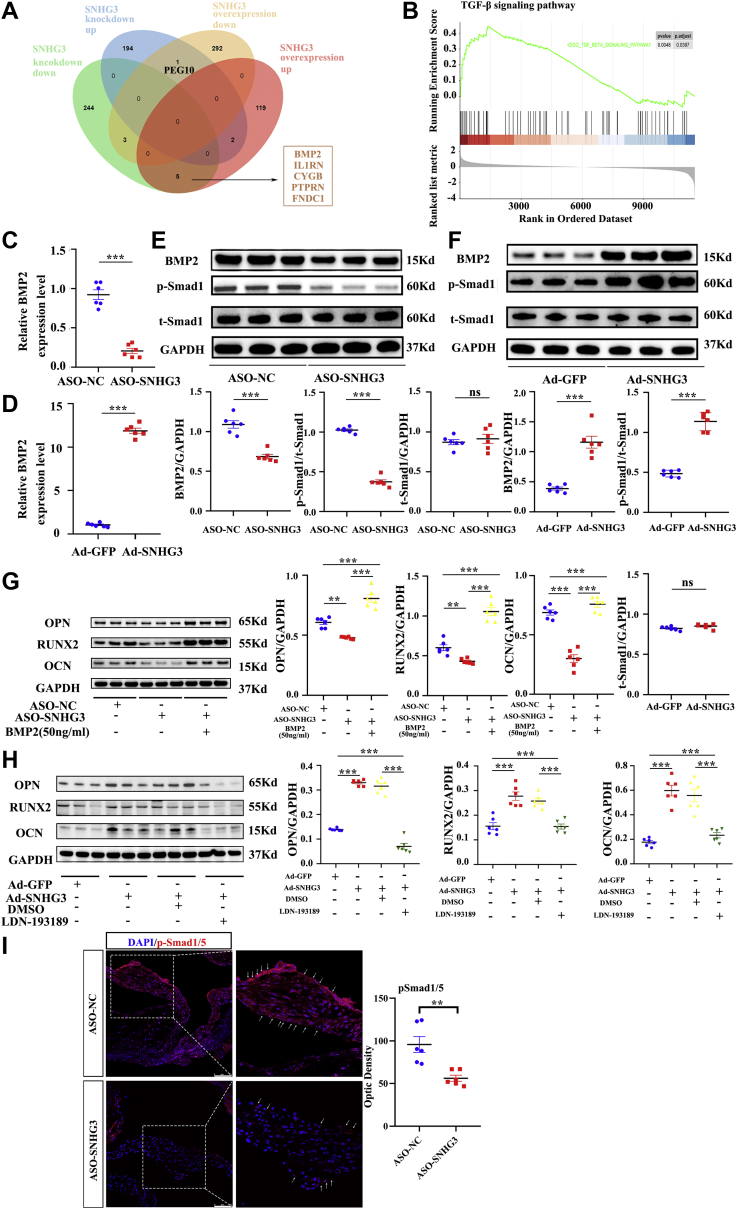
To identify the molecular mechanism by which lncRNA SNHG3 promotes aortic valve calcification, we examined the mRNA expression profiles of hVICs after knockdown of SNHG3 with ASO and overexpression of SNHG3 with ADV. A total of 126 upregulated and 296 downregulated genes were identified in the SNHG3-silenced hVICs compared with the control cells, and 197 upregulated genes and 252 downregulated genes were identified in the SNHG3-overexpressed hVICs (filtered by |fold change| >2 and P < 0.05) (Figure 5A). To identify coordinated changes in the expression of functionally related genes, we performed Kyoto Encyclopedia of Genes and Genomes enrichment analysis and gene set enrichment analysis.17,18 We found several pathways associated with aortic valve calcification, such as the calcium, MAPK, and TGF-β signaling pathways (Figure 5B, Supplemental Figures 5A and 5B). In addition, we found that the gene set of aortic valve stenosis and increased bone mineral density were upregulated when SNHG3 was overexpressed in hVICs (Supplemental Figures 5C and 5D). Among them, the TGF-β signaling pathway was significantly downregulated in the case of SNHG3 knockdown (Supplemental Figure 6A) and was significantly upregulated under conditions of SNHG3 overexpression (Supplemental Figure 6B). We also screened the differentially expressed genes between 2 RNA sequences, and BMP2 was also significantly downregulated as a result of SNHG3 knockdown and was significantly upregulated in the SNHG3-overexpression group (Figure 5A).
Figure 5.
SNHG3 Activates BMP2 Signaling Pathway to Promote Osteogenic Differentiation of hVICs
(A) Differentially expressed genes (filtered by |fold change| >2-fold; P < 0.05) in the case of SNHG3 knockdown and SNHG3 overexpression through 2 RNA sequencing. (B) Gene set enrichment analysis revealed the TGF-β signaling pathway was significantly upregulated in condition of SNHG3 overexpression with adenovirus compared with Ad-GFP. (C and D) The mRNA level of BMP2 in hVICs transfected with ASO-SNHG3 or Ad-SNHG3 (n = 6/group). (E and F) The protein level of BMP2 and pSmad1 in hVICs transfected with ASO-SNHG3 or Ad-SNHG3 (n = 6/group). (G) The down-regulation of protein levels of osteoblastic differentiation markers (OPN, RUNX2, OCN) induced by ASO-SNHG3 in hVICs were efficiently reversed by adding BMP2 stimulation (50 ng/mL) (n = 6/group). (H) The upregulation of protein levels of osteoblastic differentiation markers caused by overexpression of SNHG3 was partially reversed by the BMP2 pathway inhibitor LDN-193189 (100 ng/mL) (n = 6/group). (I) Representative phosphorylated smad1/5 immunofluorescence staining in red and quantification in ApoE−/− mice treated with ASO-SNHG3 and ASO-NC aortic valve leaflets. Nuclei are stained blue with DAPI. Scale bar, 75 μm. Values are mean ± SEM. Unpaired 2-tailed Student’s t-test for C to F and I. One-way ANOVA followed by Holm-Sidak post hoc test for G and H, ∗∗P < 0.01, ∗∗∗P < 0.001. DAPI = 4′,6-diamidino-2-phenylindole; DMSO = dimethyl sulfoxide; TGF = transforming growth factor; other abbreviations as in Figures 1, 2, and 3.
BMP2 is a member of the TGF-β superfamily that plays essential roles in aortic valve calcification, and the BMP2 signaling pathway is also a part of the TGF-β signaling pathway involved in osteogenic differentiation of hVICs.15,16 We hypothesized that SNHG3 might activate the BMP2 signaling pathway to promote osteoblast differentiation of hVICs. Next, we confirmed the link between SNHG3 and BMP2 by RT-qPCR (Figures 5C and 5D) and western blot and found the level of BMP2 expression and its downstream effector smad1 phosphorylation (Figures 5E and 5F) dependent on SNHG3. Furthermore, we explored the relationships among SNHG3, the BMP2 signaling pathway, and osteoblastic differentiation using 2 rescue experiments. The down-regulation of protein levels of osteoblastic differentiation markers induced by ASO-SNHG3 in hVICs was efficiently reversed by BMP2 stimulation (Figure 5G). In addition, the upregulation of protein levels of osteoblastic differentiation markers caused by overexpression of SNHG3 with ADV was partially reversed by the BMP2 signaling pathway inhibitor LDN-193189 (Figure 5H). To further confirm the in vitro results, we examined the levels of p-Smad1/5 in an HCD-induced aortic valve calcification model treated with ASO-SNHG3. We found p-Smad1/5 was decreased more in the ASO-SNHG3 group than that in the ASO-NC group (Figure 5I). These analyses show that SNHG3 activates the BMP2 signaling pathway to promote osteoblast differentiation of hVICs.
SNHG3 associates with enhancer of zeste 2 to regulate BMP2 expression epigenetically
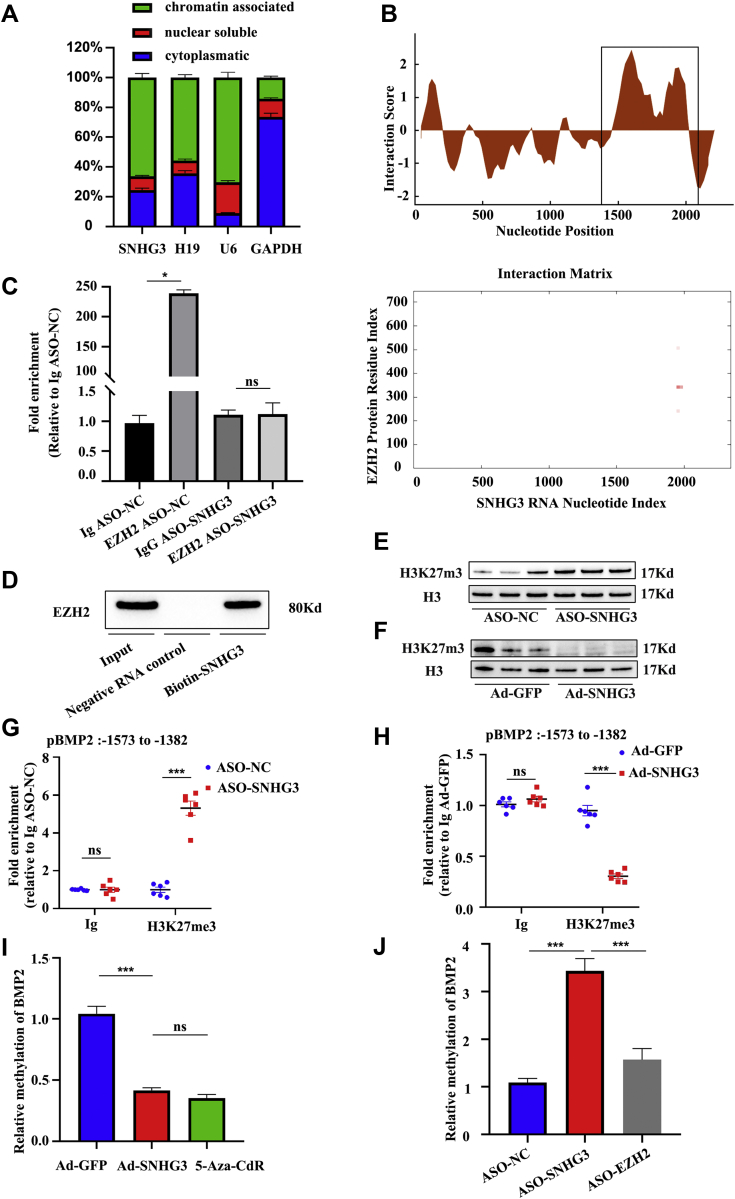
We performed subcellular fractionation of hVICs to understand the molecular role of SNHG3 in the upregulation of BMP2. SNHG3 was localized in the cytoplasm and the nuclear fraction, with most nuclear SNHG3 being bound to chromatin (Figure 6A), in accordance with SNHG3 FISH (Supplemental Figure 7), which suggests a potential involvement of SNHG3 in the epigenetic regulation of BMP2 expression. The cytoplasmic/nuclear distribution was not affected by the pro-calcification stimulation (Supplemental Figure 8). A common function of lncRNAs is their association with regulatory proteins (transcription factors and chromatin remodelers) to tether them as ribonucleoprotein complexes to their target sites. In this context, SNHG3 was previously shown to interact with enhancer of zeste 2 (EZH2) in cancer cells,19 an enzymatic catalytic subunit of the histone methyltransferase polycomb repressive complex 2 (PRC2) that primarily tri-methylates lysine residue 27 on histone H3 (H3K27me3)19,20 establishing suppressive histone marks. Our in silico analysis revealed a high propensity for interaction between the central region of SNHG3 and EZH2 (Figure 6B). Sequence-based prediction of SNHG3-EZH2 interaction probabilities was 0.85 with random forest and 0.96 with support vehicle machine (Supplemental Figure 9A). The CatRAPID fragment tool revealed that the EZH2 region (amino-acid residues 326-377) and SNHG3 region (1932-2025 base pairs [bp]) had the highest interaction propensity, discriminative power, and normalized score, which means that these 2 parts have the greatest possibility of interaction with each other (Supplemental Figure 9B). We identified a common motif with 2 paired 4-nt loop secondary structures in the SNHG3 region (1932-2025 bp) that is a typical feature responsible for the interaction with EZH2 (Supplemental Figure 9C).21,22 The EZH2 region (amino-acid residues 326-377) included a Thr-345, a conservative cyclin-dependent kinase phosphorylation site, which can be phosphorylated by AKT1 and promote maintenance of H3K27m3 levels at EZH2-target loci, thus leading to epigenetic gene silencing (Supplemental Figure 9D).23 RNA immunoprecipitation with specific antibodies against EZH2 confirmed this interaction in vitro, as indicated by ∼240-fold enrichment of SNHG3 over immunoglobulin G control (Figure 6C). SNHG3 silencing abolished the enrichment, suggesting that the interaction between SNHG3 and EZH2 is specific. In addition, biotinylated SNHG3 probes were used to pull down EZH2 (Figure 6D). SNHG3 silencing specifically increased trimethylation at H3K27 sites (Figure 6E), whereas SNHG3 overexpression reduced H3K27 trimethylation globally (Figure 6F). To clarify whether the BMP2 H3K27 trimethylation or mRNA expression is dependent on SNHG3, chromatin immunoprecipitation-qPCR results confirmed that ASO-SNHG3 treatment increased H3K27 trimethylation at the BMP2 locus and decreased BMP2 mRNA levels, which were partially rescued by the knockdown of EZH2 (Figures 6G and 5C). Overexpression of SNHG3 decreased BMP2 H3K27 trimethylation and strengthened its expression (Figures 6H and 5D). In addition, the methylation status of the BMP2 promoter was remarkably decreased by SNHG3-overexpression or methyltransferase inhibitor, 5-Aza-CdR in hVICs (Figure 6I). In contrast, the ectopic SNHG3 silencing increased the methylation level of the BMP2 promoter region, which was also alleviated by EZH2 knockdown (Figure 6J). Our data suggest that SNHG3 functions as a key activator of BMP signaling by preventing PRC2-mediated epigenetic repression of the BMP2 locus.
Figure 6.
Nuclear SNHG3 Interacts With EZH2 to Regulate BMP2 Signaling Pathway
(A) Cytoplasmatic, nuclear soluble, and chromatin-associated abundance of SNHG3 and controls in subcellular fractions of hVICs (n = 6). (B) Interaction score and binding propensities for EZH2 interaction with SNHG3 predicted by CatRAPID. (C) Validation of direct binding between SNHG3 and EZH2 by RNA immunoprecipitation in nuclear lysates of hVICs in the presence and absence (ASO-SNHG3) of SNHG3 compared with controls (n = 3/group). Kruskal-Wallis test followed by Bonferroni post hoc test. (D) Pull down assay using biotin-SNHG3 followed by western blotting for EZH2 from the nuclear lysates of hVICs. (E and F) Methyl transferase activity, as assessed by immunoblotting using anti-H3K27me3 antibody, for knockdown or overexpression of SNHG3 (n = 6/group). (G) ChIP-qPCR validation for H3K27me3 in the BMP2 promoter and SNHG3 and EZH2 knockdown. (H) ChIP-qPCR validation for H3K27me3 in the BMP2 promoter after overexpression SNHG3 (n = 6/group). (I) The methylation level of BMP 2 promoter assessed by BSP in SNHG3-upregulated or methyltransferase inhibitor (5-Aza-CdR)-treated hVICs (n = 6/group). (J) The methylation level of BMP 2 promoter assessed by BSP in SNHG3-downregulated and 5-Aza-CdR-treated hVICs (n = 6/group). Values are mean ± SEM. Two-way ANOVA with Holm-Sidak post hoc test for G and H. One-way ANOVA followed by Holm-Sidak post hoc test for I and J, ∗P < 0.05, ∗∗∗P < 0.001. ANOVA = analysis of variance; BSP = bisulfite sequencing PCR analysis; ChIP = chromatin immunoprecipitation; EZH2 = enhancer of zeste 2; Ig = immunoglobulin; other abbreviations as in Figures 1 and 2.
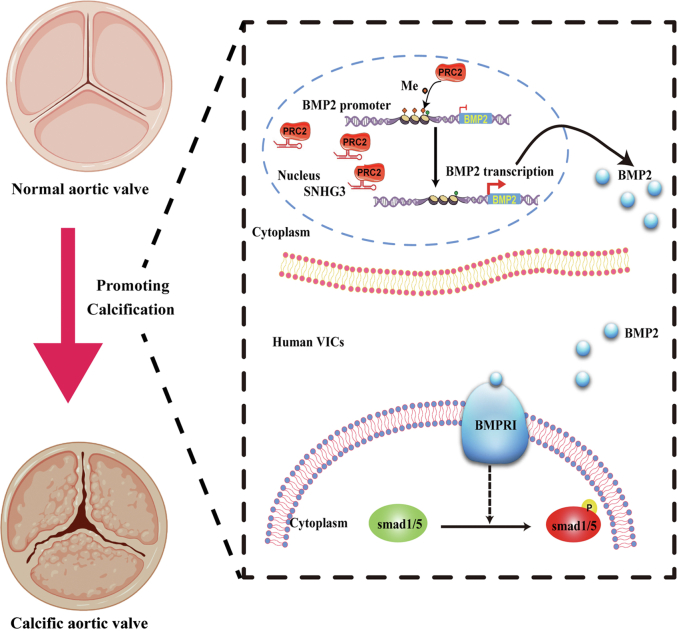
In summary, we demonstrate that lncRNA SNHG3 acts as a decoy lncRNA and physically interacts with EZH2, a core component of PRC2, to suppress the trimethylation of the BMP2 promoter, resulting in the upregulation of the BMP2 signaling pathway, thereby promoting osteoblast differentiation of hVICs in CAVD progression (Figure 7).
Figure 7.
Schematic Model Showing the Positive Effects of LNCRNA SNHG3 Promoting Aortic Valve Calcification Through Associating With PRC2 To Upregulate BMP2 Pathway
SNHG3 interacts with PRC2 to decrease the trimethylation levels of BMP2 promoter region, resulting in upregulating the BMP2 expression at the transcriptional level, and thereby promotes osteogenic differentiation of hVICs in progression of aortic valve calcification. PRC2 = polycomb repressive complex 2; other abbreviations as in Figures 1 and 2.
Discussion
At present, there is an unmet medical requirement to discover novel potential therapeutic targets for treating CAVD. In this study, our data generated the following novel findings: 1) SNHG3 expression is significantly higher in human calcific aortic valves than that in nonmineralized aortic valves; 2) SNHG3 can promote CAVD with evidence from in vitro and in vivo models; and 3) the first evidence that SNHG3 promotes aortic valve calcification through its epigenetic reprogramming of BMP2 gene. SNHG3 interacts with EZH2 to suppress the trimethylation of the BMP2 promoter, resulting in the upregulation of the BMP2 pathway during aortic valve calcification progression. Taken together, our results confirm that SNHG3 might be a novel therapeutic target for the treatment of aortic valve calcification.
LncRNAs are located in the nucleus mainly and subcellular localization patterns of lncRNAs reveal fundamental insights into their biology and foster hypotheses for potential molecular roles.24 SNHG3 is mainly localized in the nucleus rather than the cytoplasm in gastric cells,25 and we found a similar result when we performed an RNA-FISH assay and cytoplasmic and nuclear RNA qPCR assays in hVICs, indicating that SNHG3 might regulate gene expression at the transcriptional level. Recently, a number of lncRNAs, termed epi-lncRNAs, have been involved in epigenetic regulation by directly interacting with epigenetic modifiers in the nucleus.26,27 Mechanistically, different lncRNAs interact with the epigenetic modifier PRC2 to repress or promote the binding and methylation of chromatin at specific subsets of genes.28,29 The SNHG3 interaction with epigenetic modifiers to regulate gene expression is supported by the evidence generated in our study. SNHG3 is associated with PRC2 enzymatic catalytic subunit EZH2 in cancer cells.25 It was unknown whether this link exists in the hVICs, and we performed in silico and in vitro binding assays that demonstrated that SNHG3 physically interacts with the PRC2 core subunit EZH2. The predicted secondary SNHG3 structure (region: 1932-2025 bp) with 2 paired 4-nt motifs is responsible for its interaction with EZH2. The binding region of EZH2 with SNH3 contains Thr-345, which is essential for maintaining the H3K27m3 level at EZH2-target loci. Therefore, SNHG3 binds with EZH2 in the nucleus and occupies the Thr-345 phosphorylation site of CDK1 to decrease H3K27m3 levels at EZH2-target loci, ultimately leading to epigenetic gene upregulation. Moreover, SNHG3 binds EZH2 to suppress promoter-specific H3K27me3 levels. This negative regulation of PRC2 function is consistent with previous studies showing that other lncRNAs interact with EZH2.21,28 Here, we performed chromatin immunoprecipitation-qPCR and bisulfite sequencing PCR analyses, which demonstrated that SNHG3 can specifically alter the BMP2 promoter region trimethylation level. To the best of our knowledge, the present study revealed that SNHG3 interacts with the epigenetic modifier PRC2 to alter the methylation level of the target gene.
The master osteoblast protein BMP2, a secreted ligand of the TGF-β superfamily, closely binds to BMPRI/II to promote osteoblast differentiation of hVICs in CAVD.30,31 Previous studies reported that lipid accumulation,32 oxidative stress,33 and inflammation34 can mediate valvular calcification via BMP2.35 BMP2 can also upregulate a chondro-osteogenic pathway involving the transcription factor RUNX2,15 which promotes CAVD progression. Here, we performed gain and loss of function assays, transcriptional sequencing bioinformatics analysis, and rescue experiments to verify that SNHG3 can regulate the BMP2 pathway to induce an osteogenic program in the aortic valve.
CAVD is the most prevalent form of aortic valve disease worldwide because of the lack of effective pharmacotherapy. It is necessary to understand the mechanism of its progression to develop new pharmacotherapies. The complex and multifaceted pathobiology process involving fibro-calcific remodeling, lipid accumulation and oxidation,32 chronic inflammation,36 and osteogenic differentiation of hVICs have been found to contribute to CAVD, suggesting that targeting these biological processes may result in the development of novel therapeutic interventions to halt disease progression or treat CAVD.37 lncRNAs play a prominent role in the regulation of osteoblast differentiation in many cell lines. For instance, lncRNA TUG1 can promote osteoblast differentiation by sponging miR-204-5p to upregulate RUNX2 in human hVICs,38 whereas silencing of lncRNA NONHSAT009968 reduces staphylococcal protein A-inhibited osteogenic differentiation in human bone mesenchymal stem cells.39 To the best of our knowledge, this is the first study to report that the silencing of SNHG3 significantly inhibited osteogenic differentiation by epigenetic regulation of the BMP2 pathway in vitro and in vivo. Therefore, ASO-delivery of SNHG3 may represent a novel RNA-based therapy for CAVD.
Study limitations
First, we only explored the epigenetic regulatory function of SNHG3 in the nucleus. It should be noted that apart from the cell nucleus, SNHG3 is also expressed in the cytoplasm of hVICs. Further studies are necessary to investigate the other roles of SNHG3 in hVICs. Second, we only identified that SNHG3 could interact with EZH2 to suppress trimethylation of the BMP2 promoter, resulting in the upregulation of BMP2. Whether SNHG3 could function as an alternative way to regulate the other key regulators in CAVD progression requires further investigation. In addition, to expand the scope of screening candidate genes, we analyzed the differentially expressed genes without adjusting for type I error using a false discovery rate, which did not affect SNHG3 was screened out as the key lncRNA in our research.
Conclusions
In the present study, we identified SNHG3 as a novel positive regulator of osteogenic differentiation in CAVD pathogenesis. Moreover, SNHG3 physically interacts with the PRC2 core subunit EZH2 in the nucleus of hVICs to suppress the trimethylation of the BMP2 promoter, resulting in the upregulation of the BMP2 signaling pathway during CAVD. SNHG3 silencing significantly alleviates aortic valve calcification in experimental animals, providing a novel therapeutic target for CAVD.
Perspectives.
COMPETENCY IN MEDICAL KNOWLEDGE: Long noncoding RNA SNHG3 is significantly upregulated in patients with calcific aortic valve. SNHG3 interacts with PRC2 to decrease the trimethylation levels of BMP2 promoter region, resulting in upregulating the BMP2 expression at the transcriptional level, and thereby promotes osteogenic differentiation of human aortic valve interstitial cells in progression of aortic valve calcification.
TRANSLATIONAL OUTLOOK: SNHG3 silencing significantly alleviates aortic valve calcification in experimental animals, providing a novel therapeutic target for CAVD.
Funding Support and Author Disclosures
This work was supported by research grants from the National Nature Science Foundation of China (81900352 to Dr Liu, 81770233 to Dr Zheng, and 81830072 to Dr Zheng). All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Footnotes
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
Appendix
For supplemental material, tables, and figures, please see the online version of this paper.
Appendix
References
- 1.Yadgir S., Johnson C.O., Aboyans V., et al. Global, regional, and national burden of calcific aortic valve and degenerative mitral valve diseases, 1990-2017. Circulation. 2020;141:1670–1680. doi: 10.1161/CIRCULATIONAHA.119.043391. [DOI] [PubMed] [Google Scholar]
- 2.Goody P.R., Hosen M.R., Christmann D., et al. Aortic valve stenosis: from basic mechanisms to novel therapeutic targets. Arterioscler Thromb Vasc Biol. 2020;40:885–900. doi: 10.1161/ATVBAHA.119.313067. [DOI] [PubMed] [Google Scholar]
- 3.Kumar S., Boon R.A., Maegdefessel L., Dimmeler S., Jo H. Role of noncoding RNAs in the pathogenesis of abdominal aortic aneurysm. Circ Res. 2019;124:619–630. doi: 10.1161/CIRCRESAHA.118.312438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poller W., Dimmeler S., Heymans S., et al. Non-coding RNAs in cardiovascular diseases: diagnostic and therapeutic perspectives. Eur Heart J. 2018;39:2704–2716. doi: 10.1093/eurheartj/ehx165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bar C., Chatterjee S., Thum T. Long noncoding RNAs in cardiovascular pathology, diagnosis, and therapy. Circulation. 2016;134:1484–1499. doi: 10.1161/CIRCULATIONAHA.116.023686. [DOI] [PubMed] [Google Scholar]
- 6.Hadji F., Boulanger M.C., Guay S.P., et al. Altered DNA methylation of long noncoding RNA H19 in calcific aortic valve disease promotes mineralization by silencing NOTCH1. Circulation. 2016;134:1848–1862. doi: 10.1161/CIRCULATIONAHA.116.023116. [DOI] [PubMed] [Google Scholar]
- 7.Arisi I., D'Onofrio M., Brandi R., et al. Gene expression biomarkers in the brain of a mouse model for Alzheimer's disease: mining of microarray data by logic classification and feature selection. J Alzheimers Dis. 2011;24:721–738. doi: 10.3233/JAD-2011-101881. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J., Dong B., Hao J., Yi S., Cai W., Luo Z. LncRNA Snhg3 contributes to dysfunction of cerebral microvascular cells in intracerebral hemorrhage rats by activating the TWEAK/Fn14/STAT3 pathway. Life Sci. 2019;237 doi: 10.1016/j.lfs.2019.116929. [DOI] [PubMed] [Google Scholar]
- 9.Dacheng W., Songhe L., Weidong J., Shutao Z., Jingjing L., Jiaming Z. LncRNA SNHG3 promotes the growth and metastasis of colorectal cancer by regulating miR-539/RUNX2 axis. Biomed Pharmacother. 2020;125 doi: 10.1016/j.biopha.2020.110039. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y., Li L., Lu K.X., Yu L.B., Meng J., Liu C.Y. LncRNA SNHG3 is responsible for the deterioration of colorectal carcinoma through regulating the miR-370-5p/EZH1 axis. Eur Rev Med Pharmacol Sci. 2021;25:6131–6137. doi: 10.26355/eurrev_202110_26891. [DOI] [PubMed] [Google Scholar]
- 11.Zhang L., Li G., Wang X., Zhang Y., Huang X., Wu H. lncRNA SNHG3 acts as oncogene in ovarian cancer through miR-139-5p and Notch1. Oncol Lett. 2021;21:122. doi: 10.3892/ol.2020.12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yetkin E., Waltenberger J. Molecular and cellular mechanisms of aortic stenosis. Int J Cardiol. 2009;135:4–13. doi: 10.1016/j.ijcard.2009.03.108. [DOI] [PubMed] [Google Scholar]
- 13.Watson K.E., Boström K., Ravindranath R., Lam T., Norton B., Demer L.L. TGF-beta 1 and 25-hydroxycholesterol stimulate osteoblast-like vascular cells to calcify. J Clin Invest. 1994;93:2106–2113. doi: 10.1172/JCI117205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song R., Fullerton D.A., Ao L., Zheng D., Zhao K.S., Meng X. BMP-2 and TGF-beta1 mediate biglycan-induced pro-osteogenic reprogramming in aortic valve interstitial cells. J Mol Med (Berl) 2015;93:403–412. doi: 10.1007/s00109-014-1229-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang X., Meng X., Su X., et al. Bone morphogenic protein 2 induces Runx2 and osteopontin expression in human aortic valve interstitial cells: role of Smad1 and extracellular signal-regulated kinase 1/2. J Thorac Cardiovasc Surg. 2009;138:1008–1015.e1. doi: 10.1016/j.jtcvs.2009.06.024. [DOI] [PubMed] [Google Scholar]
- 16.Wirrig E.E., Hinton R.B., Yutzey K.E. Differential expression of cartilage and bone-related proteins in pediatric and adult diseased aortic valves. J Mol Cell Cardiol. 2011;50:561–569. doi: 10.1016/j.yjmcc.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mootha V.K., Lindgren C.M., Eriksson K.F., et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 18.Subramanian A., Tamayo P., Mootha V.K., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu C., Qu K., Zhong F.L., Artandi S.E., Chang H.Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Z., Zhang X.J., Ji Y.X., et al. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat Med. 2016;22:1131–1139. doi: 10.1038/nm.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu L., Murat P., Matak-Vinkovic D., Murrell A., Balasubramanian S. Binding interactions between long noncoding RNA HOTAIR and PRC2 proteins. Biochemistry. 2013;52:9519–9527. doi: 10.1021/bi401085h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen S., Bohrer L.R., Rai A.N., et al. Cyclin-dependent kinases regulate epigenetic gene silencing through phosphorylation of EZH2. Nat Cell Biol. 2010;12:1108–1114. doi: 10.1038/ncb2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fort A., Hashimoto K., Yamada D., et al. Deep transcriptome profiling of mammalian stem cells supports a regulatory role for retrotransposons in pluripotency maintenance. Nat Genet. 2014;46:558–566. doi: 10.1038/ng.2965. [DOI] [PubMed] [Google Scholar]
- 25.Xuan Y., Wang Y. Long non-coding RNA SNHG3 promotes progression of gastric cancer by regulating neighboring MED18 gene methylation. Cell Death Dis. 2019;10:694. doi: 10.1038/s41419-019-1940-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grote P., Wittler L., Hendrix D., et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li L., Liu B., Wapinski O.L., et al. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013;5:3–12. doi: 10.1016/j.celrep.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Viereck J., Buhrke A., Foinquinos A., et al. Targeting muscle-enriched long non-coding RNA H19 reverses pathological cardiac hypertrophy. Eur Heart J. 2020;41:3462–3474. doi: 10.1093/eurheartj/ehaa519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salerno D., Chiodo L., Alfano V., et al. Hepatitis B protein HBx binds the DLEU2 lncRNA to sustain cccDNA and host cancer-related gene transcription. Gut. 2020;69:2016–2024. doi: 10.1136/gutjnl-2019-319637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomez-Stallons M.V., Wirrig-Schwendeman E.E., Hassel K.R., Conway S.J., Yutzey K.E. Bone morphogenetic protein signaling is required for aortic valve calcification. Arterioscler Thromb Vasc Biol. 2016;36:1398–1405. doi: 10.1161/ATVBAHA.116.307526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caira F.C., Stock S.R., Gleason T.G., et al. Human degenerative valve disease is associated with up-regulation of low-density lipoprotein receptor-related protein 5 receptor-mediated bone formation. J Am Coll Cardiol. 2006;47:1707–1712. doi: 10.1016/j.jacc.2006.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nsaibia M.J., Boulanger M.C., Bouchareb R., et al. OxLDL-derived lysophosphatidic acid promotes the progression of aortic valve stenosis through a LPAR1-RhoA-NF-kappaB pathway. Cardiovasc Res. 2017;113:1351–1363. doi: 10.1093/cvr/cvx089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y., Gu J., Du A., et al. SPARC-related modular calcium binding 1 regulates aortic valve calcification by disrupting BMPR-II/p-p38 signalling. Cardiovasc Res. 2022;118:913–928. doi: 10.1093/cvr/cvab107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang X., Fullerton D.A., Su X., Ao L., Cleveland J.C., Jr., Meng X. Pro-osteogenic phenotype of human aortic valve interstitial cells is associated with higher levels of Toll-like receptors 2 and 4 and enhanced expression of bone morphogenetic protein 2. J Am Coll Cardiol. 2009;53:491–500. doi: 10.1016/j.jacc.2008.09.052. [DOI] [PubMed] [Google Scholar]
- 35.Wozney J.M., Rosen V., Celeste A.J., et al. Novel regulators of bone formation: molecular clones and activities. Science. 1988;242:1528–1534. doi: 10.1126/science.3201241. [DOI] [PubMed] [Google Scholar]
- 36.Towler D.A. Oxidation, inflammation, and aortic valve calcification peroxide paves an osteogenic path. J Am Coll Cardiol. 2008;52:851–854. doi: 10.1016/j.jacc.2008.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freeman R.V., Otto C.M. Spectrum of calcific aortic valve disease: pathogenesis, disease progression, and treatment strategies. Circulation. 2005;111:3316–3326. doi: 10.1161/CIRCULATIONAHA.104.486738. [DOI] [PubMed] [Google Scholar]
- 38.Yu C., Li L., Xie F., et al. LncRNA TUG1 sponges miR-204-5p to promote osteoblast differentiation through upregulating Runx2 in aortic valve calcification. Cardiovasc Res. 2018;114:168–179. doi: 10.1093/cvr/cvx180. [DOI] [PubMed] [Google Scholar]
- 39.Cui Y., Lu S., Tan H., Li J., Zhu M., Xu Y. Silencing of long non-coding RNA NONHSAT009968 ameliorates the staphylococcal protein A-inhibited osteogenic differentiation in human bone mesenchymal stem cells. Cell Physiol Biochem. 2016;39:1347–1359. doi: 10.1159/000447839. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.