Abstract
HRas, KRas, and NRas are GTPases with a common set of effectors that control many cell-signaling pathways, including proliferation through Raf kinase. Their G-domains are nearly identical in sequence, with a few isoform-specific residues that have an effect on dynamics and biochemical properties. Here, we use accelerated molecular dynamics (aMD) simulations consistent with solution x-ray scattering experiments to elucidate mechanisms through which isoform-specific residues associated with each Ras isoform affects functionally important regions connected to the active site. HRas-specific residues cluster in loop 8 to stabilize the nucleotide-binding pocket, while NRas-specific residues on helix 3 directly affect the conformations of switch I and switch II. KRas, the most globally flexible of the isoforms, shows greatest fluctuations in the switch regions enhanced by a KRas-specific residue in loop 7 and a highly dynamic loop 8 region. The analysis of isoform-specific residue effects on Ras proteins is supported by NMR experiments and is consistent with previously published biochemical data.
Significance
This work links specific residues unique to each Ras isoform to G-domain dynamics that impact functional roles such as hydrolysis of GTP and are in areas that affect effector binding, the Ras dimerization interface and interaction with the membrane. Given the importance of these regions in Ras function, the mechanistic work presented here contributes to the greater puzzle, currently being assembled, of how Ras signals through its various effectors and may give insight to isoform-specific selection of oncogenic mutations associated with cancers. For example, lower accessibility of NRas to the catalytic conformation necessary for GTP hydrolysis may synergize with oncogenic mutants of residue 61 to promote sustained signaling through Ras/Raf, providing understanding for this preferred locus in NRas-driven cancers.
Introduction
Ras GTPases are molecular switches that modulate signals through multiple effector proteins, including Raf and PI3K, and are involved in the control of cell proliferation, survival, and apoptosis (1). They are inactive when bound to GDP, active when bound to GTP, and switch between these states with the help of guanine nucleotide exchange factors and GTPase-activating proteins (GAPs) (2). The G-domain of Ras, residues 1–166, has two lobes: the effector lobe (residues 1–86) and the allosteric lobe (residues 87–166) (3). The effector lobe contains the P-loop (residues 10–17), switch I (residues 30–40), and switch II (residues 60–76), as well as sites of protein-protein interactions (4, 5, 6).
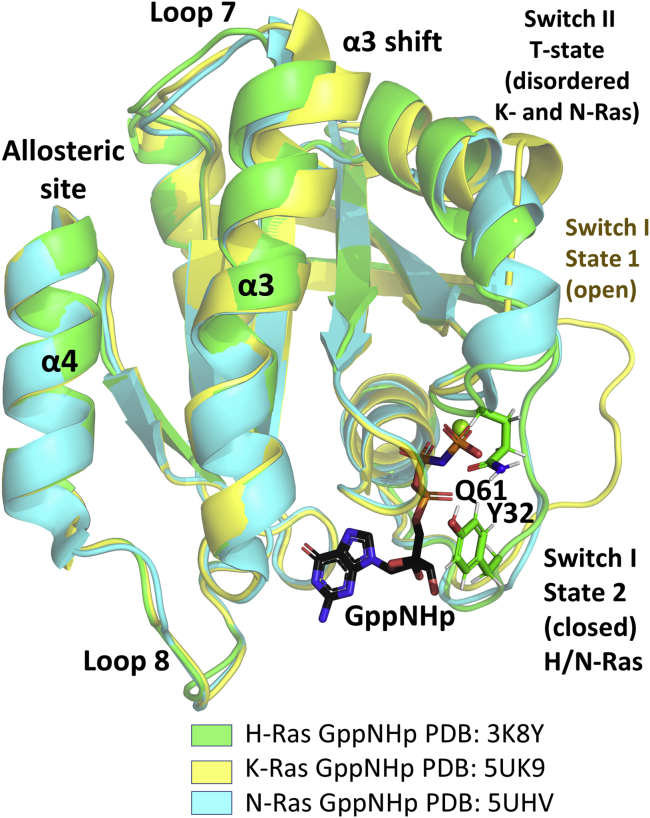
In its active GTP-bound form the switch regions are dynamic and can access different conformational states, several of which have been observed in crystal structures (Fig. 1). Switch I can access an open state 1, where T35 is away from the nucleotide (PDB: 4EFL, 5UK9) (7,8); and a closed state 2 where it interacts with the bound Mg2+ ion bridging to the γ-phosphate of GTP (PDB: 1CTQ, 3K8Y) (9,10). With switch I in the state 2 conformation, switch II can exist in a disordered T state (tardy) (PDB: 2RGE) (11), where GTP hydrolysis is expected to be very slow, or an R state (reactive), with a highly ordered active site (PDB: 3K8Y) (9). Ordering of the active site to the R state requires stabilization of switch I in state 2 (for instance, by binding of Raf-RBD (Ras-binding domain) or a GAP) and of switch II, including catalytic residue Q61. This can be promoted by GAP binding the free G-domain (12) or by binding of Ca2+ at a remote allosteric site in the presence of Raf-RBD (9,13). Thus, to hydrolyze GTP and stop the signaling events, Ras needs to have its switch I in state 2 and switch II in the R state. In uncomplexed Ras bound to GTP or its analogs, the various conformational states are sampled differently depending on isoform-specific amino acid residue differences in the allosteric lobe (14). These subtle differences in access to conformational space may be one of the ways in which H, K, and NRas are associated with distinct biological functions (15).
Figure 1.
Conformational states of the Ras catalytic G-domain observed in crystal structures. HRas (PDB: 3K8Y, green), KRas (PDB: 5UK9, yellow), and NRas (PDB: 5UHV, cyan) are superimposed. HRas is in state 2, with switch I positioned over the nucleotide, and switch II is in the R state, placing Q61 in the active site. KRas is in state 1 with switch I in the open conformation and switch II is in the T state. NRas is in state 2, with switch II in the T state. The GTP analog, GppNHp, is shown in black. To see this figure in color, go online.
There are four Ras isoforms, including HRas, NRas, KRas4B, and KRas4A, that are posttranslationally modified in different ways for attachment to the membrane through C-terminal hypervariable regions that are only 15% conserved (16). In contrast, their G-domains are very similar, with 95% sequence identity and all of the variation on the allosteric lobe (17). Here, we focus on the G-domains of the three most prominent isoforms, HRas, KRas4B (from here on referred to as KRas), and NRas. We have shown that these Ras proteins have different intrinsic hydrolysis rate constants and respond differently to binding Raf-RBD (18). HRas has the highest rate constant and it is unchanged by Raf-RBD, whereas KRas and NRas are slower enzymes. Interestingly, while the rate constant for KRas is increased to nearly that of HRas in the presence of Raf-RBD, for NRas it does not change upon binding the effector (18). Given identical active sites, these differences in the hydrolysis rate constants must be due to changes in accessibility of the catalytic conformation affected by isoform-specific residues in the allosteric lobe.
Here, we study the differences in dynamics and conformational states between the three Ras isoforms and propose mechanisms by which isoform-specific residues are responsible for these differences. For this, we first present wide-angle solution x-ray scattering (WAXS) data (19) and, as expected, find that conventional molecular dynamics (cMD) simulations do not represent the range of conformational states observed in solution for Ras proteins, at least not within the 600 ns timescale encompassed by our three 200 ns replicates. In contrast, 600 ns accelerated MD (aMD) simulations, where a boost potential is used to overcome energy barriers (20), show better agreement with the WAXS experiments. The aMD results show that each Ras isoform has a unique pattern of isoform-specific residue clusters that affect functionally important regions in distinct ways, with overall features consistent with the biochemical differences between the isoforms (18) and with conformational states assessed by NMR (8).
Methods
WAXS
Solution x-ray scattering data capture the behavior of a protein in solution and can quickly determine radius of gyration, molecular envelopes, and the pair distribution function (21). While solution x-ray scattering yields very low-resolution data, WAXS extends data out to higher resolution and is sensitive enough to detect structural changes in proteins (22). For the present experiments, HRas, KRas, and NRas (residues 1–166) were expressed in E. coli, purified, and the GDP exchanged for the GTP analog 5′-guanylyl imidodiphosphate (GppNHp) as published previously (23). Ras proteins were exchanged into stabilization buffer (20 mM HEPES [pH 7.5], 5 mM MgCl2, 50 mM NaCl, 1 mM DTT) and concentrated to 10 mg/mL for WAXS data collection. Two measurements were conducted for each experiment: protein in buffer solution and the buffer solution by itself, which was subtracted from the protein + buffer to obtain the protein scattering data. Data were collected at the G1 beamline at Cornell High Energy Synchrotron Source, Ithaca, NY, USA, with x rays at 9.846 keV, λ = 1.259 Å, and with a beam size of 250 ∗ 250 μm (24). Data were collected in the range of 0.13 < q < 0.68 1/Å and processed using the software RAW (25). The relative flatness of the observed intensities based on experimental WAXS data collected for each of the three Ras isoforms is calculated as follows,
where a and b are the 1/d indices in the range 1/d ∼0.03–0.09 and I is the observed intensity. The flattening of the overall intensity curve compared with the rigid protein structure is characteristic of an increase in protein flexibility. For comparing the relative flatness of the observed intensity curves between the three Ras isoforms the HRas curve was normalized to 1%.
cMD simulations
cMD simulations were performed for 200 ns production run for HRas, KRas, and NRas on the Discovery Cluster supercomputer at Northeastern University, Boston, MA, USA. The starting structures were obtained from x-ray crystal structures in their active conformations bound to the GTP analog GppNHp (HRas, PDB: 3K8Y; KRas, PDB: 6GOD; and NRas, PDB: 5UHV) or with GppCH2p (KRas, PDB: 5UK9). The simulations for HRas and NRas were started with structures in state 2. Two sets of simulations were performed for KRas: one starting from a structure in state 2 (PDB: 6GOD) and the other in state 1 (PDB: 5UK9). The GTP analogs were converted to GTP by replacing the β-γ bridging NH or CH2 groups with an oxygen atom. Crystallographic water molecules were retained for the simulation. The NAMD and VMD software packages were used to prepare, perform, and analyze all of the simulations with the CHARMM36 force field (26, 27, 28, 29). Each protein was placed in a TIP3P water box extending 10 Å from the protein molecule. Charges in the systems were neutralized by adding sodium and chloride ions. The simulations started with energy minimization for 5000 cycles, followed by gradual heating from 50 to 250 K before the production runs. Periodic boundary conditions were used at the x, y, and z directions. A time step of 1 fs was applied for the first 30 ns of the production run at isothermal-isobaric conditions of 300 K and 1 atm and then the time step was increased to 2 fs for the subsequent 170 ns. The switch function was used between 10 and 11 Å to calculate long-range nonbonded interactions. Electrostatic interactions were evaluated using particle mesh Ewald (30). Covalent bonds to hydrogen atoms were constrained using the SHAKE algorithm (31).
aMD simulations
In aMD a bias potential is added to increase conformational sampling by enhancing the escape rates from potential energy wells (20). The form of the modified potential is shown by Eq. 1, where a boost potential given by ΔV(r), shown in Eq. 2, is added to the true potential when the overall potential energy is below a user-calculated value. Dual boost potential was added to the total potential and dihedral energies (20,26). The form of the boost potential requires two parameters, E and α, as published by McCammon and colleagues and shown in Eqs. 3 and 4 (20). The first nanosecond of the simulation was done using cMD to determine E and α for the total potential energy and the dihedral energy of the protein. These values were added to the conformation file to continue the production run in aMD mode for a total of 200 ns simulation time. Boosting the potential for the total potential energy is done to increase the degree of diffusivity for the system, where the solvent molecules are also accelerated. The additional boost potential for the dihedral energy, shown in Eqs. 5 and 6, increases the conformational space sampled by the protein, as protein conformational changes occur mostly through torsional rotations.
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
The potential energy can be obtained by reweighting points in the phase space on the bias potential by the Boltzmann factor (32), with energy reweighting to account for the boost potential either using 1D or 2D data (33). To obtain an unbiased estimate of the population of conformational states, it is necessary to reweigh the conformations according to the potential energy difference between the biased and unbiased potential energy,
where A is any property of interest, is the unbiased potential energy, and is the aMD biased energy. However, this is only possible with sufficiently long simulations to provide equilibrium averages under the aMD potential energy function. Although three 200 ns aMD simulation replicates provided in this study for each isoform show transitions between the major conformational states observed in various crystal structures, there are very few transitions indicative of insufficient sampling (Figs. S1 and S2), such that Boltzmann reweighting the conformations would not provide more meaningful results. Thus, while we compare the aMD simulations directly between the three isoforms of Ras to get approximate information on atomic fluctuations of residues throughout the Ras structures, a qualitative sense of coverage of conformational space, or a feel for differences in interactions that we observed during the combined 600 ns aMD simulations for each isoform, we must acknowledge that there is an uncorrected bias that affects the absolute root mean-square fluctuation (RMSF) values, the accuracy of the fit to the WAXS data as reflected in the σ(r) values and the precise population of conformational states associated with each of the interactions we discuss in the results. Nevertheless, it is reassuring that the patterns of RMSFs that we observe for cMD are qualitatively retained in aMD. Even if at a qualitative level, our analysis provides useful insights that are consistent with the experimental observations provided by WAXS, NMR data, and previously published biochemical experiments as detailed below.
aMD validated by WAXS
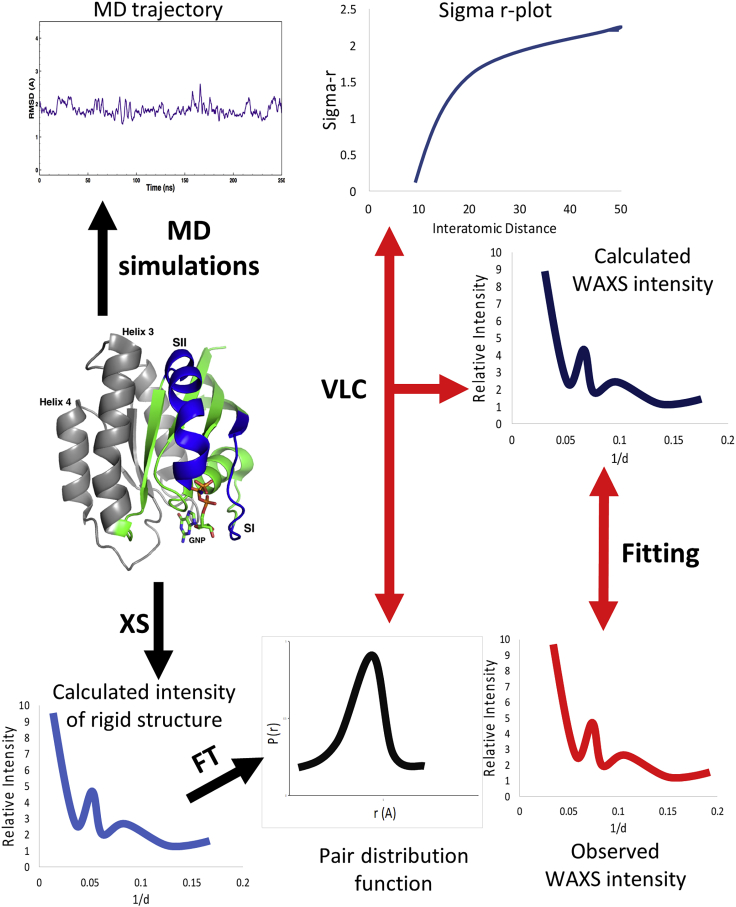
We previously published a method by which the extent of fluctuations sampled with MD simulations can be assessed experimentally by WAXS (19). Fig. 2 summarizes the process. Scattering from a protein is the Fourier transform of the pair distribution function p(r), obtained from a histogram of interatomic distances within the protein molecule (34). The p(r) for a flexible protein can be obtained by vector length convolution of p(r) for the rigid protein and the range of motions represented by σ(r) (calculated from an MD trajectory), shown as a function of the interatomic distances in a sigma-r plot (19,34). In this process, each interatomic vector in the pair correlation function of the rigid protein is replaced by a Gaussian distribution of vector lengths from the sigma-r plot (22,35). This results in a calculated scattering pattern that can be directly compared with the observed WAXS data, thereby assessing the extent to which the MD simulations represent the fluctuations observed in solution under the experimental conditions (19). For calculation of the sigma-r plot, a previously published Python script (34) was used to read the atomic coordinates from each frame of an MD trajectory (all frames included), calculate and accumulate all interatomic distances for each one, and then take the standard deviation of each interatomic distance. The values of the interatomic distances were listed in increasing order, and the standard deviations were averaged within intervals with a step size of 0.3 Å to determine the average r value for each interatomic distance interval. A scale factor is calculated to indicate the degree to which the fluctuations in the MD simulations are consistent with fluctuations underlying the experimental data. A scale factor greater than 1 indicates greater flexibility in solution than indicated by MD simulations, whereas a scale factor less than 1 implies that the protein is more flexible in the MD simulations. A scale factor close to 1 indicates excellent agreement between the MD simulations and experimental WAXS data. Keeping in mind that the solution scattering data are low resolution, this agreement reflects the range of conformations sampled as reflected by σ(r), even if the population of conformational states is not precisely correct due to the bias resulting from the boost potential.
Figure 2.
Combining crystal structure with MD simulations data to calculate a scattering intensity curve for comparison with experimentally observed WAXS data. Vector length convolution of the pair distribution function from a rigid structure and the pairwise interatomic variation with distance (sigma-r) taken from the MD simulations yield calculated intensities that can be compared directly with experimental data. To see this figure in color, go online.
H-bond analysis
Salt bridge and H-bond analysis was performed using the plugins in VMD (29). H-bonds are reported between interacting donor and accepted atoms that come within 3.0 Å of one another during the simulation. We recognize that the H-bonding angle is important in determining the strength of H-bonds. However, H-bonds in protein structures are known to exist within a wide range of angles (36) and here we use distance as a first approximation for interaction between donor and accepter atoms.
1H NMR of HRas, KRas, and NRas proteins
1H NMR on Ras spectra were obtained as described previously (8). Ras proteins used for NMR analyses were dialyzed into 40 mM HEPES (pH 7.4), 10 mM MgCl2, 150 mM NaCl, and 1 mM DTT, and concentrated to 1–2 mM. 1H NMR spectra were acquired at 700 MHz with a Bruker Avance II NMR spectrometer (Bruker Corperation, Billerica, MA, USA) equipped with a 5 mm triple resonance inverse probe at 37°C. For optimal detection of downfield exchangeable proton resonances, the 3-9-19 WATERGATE (37) pulse sequence (p3919fpgp) with gradients and additional flip-back pulse was used with the center of the maximal excitation region at 13.9 ppm. The calculated delay for binomial water suppression was 39 μs at 700 MHz. Routinely, 4000 scans were accumulated. Peak Z at 10.3 ppm and peak X at 10.9 ppm are associated with closed conformations of switch I and II, respectively (8).
Results
The extent of conformational variation observed in Ras crystal structures available in the PDB is not adequately represented by relatively short cMD simulations (38). To assess whether this can be better achieved with aMD for the three isoforms of Ras bound to GTP, we compared results from cMD and aMD with WAXS experimental data.
aMD results are more consistent with experimental data
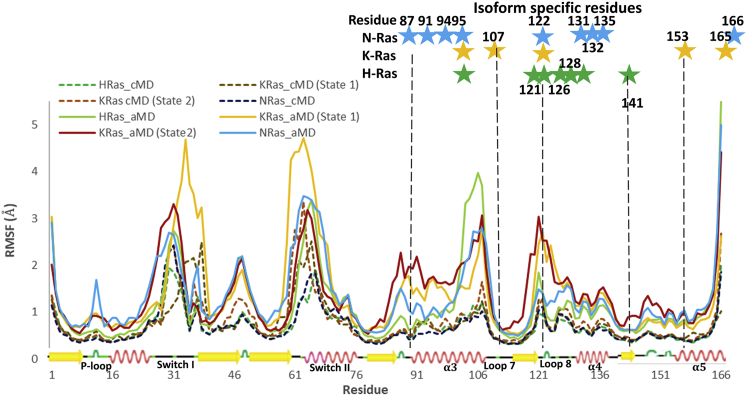
For each Ras isoform we ran three independent 200 ns simulations with cMD and aMD simulations, starting from their respective crystal structures. In aMD a boost potential is added to increase conformational sampling by enhancing the escape rates from potential energy wells (20). All 24 simulations were well behaved and converged on similar average RMSD values relative to the respective H, K, and NRas starting structures (Fig. S1). The RMSFs per residue were calculated after combining the three cMD (total 600 ns) and three aMD (total 600 ns) simulations for each isoform, with KRas simulations starting with state 2 as in HRas and NRas and with state 1 (Fig. 3). The locations of flexible residues vary little between the isoforms and are similar for cMD and aMD simulations, consistent with experiments and previous simulation analysis (7,9,11,38, 39, 40). Note, however, that the magnitudes of the fluctuations are significantly larger for aMD, suggesting a larger range of sampled conformations in areas known to be flexible, such as the switch regions and helix 3/loop 7. This is consistent with the expectation that the biased potential echoes the original shape of the conventional potential energy landscape (20). The cMD simulations show only slight differences in the residue fluctuations between the isoforms throughout most of the structure, although it does show greater flexibility in the switch regions of KRas relative to the other two isoforms as noted previously (38). Interestingly, the cMD and aMD simulations started from KRas in state 2 show very similar RMSFs for the switch II region, comparable with those obtained for HRas and NRas in aMD, but not cMD. Both sets of simulations, cMD and aMD, starting from KRas in state 1 show significantly increased fluctuations in both switch I and switch II relative to the comparable simulations in HRas and NRas, while increased fluctuations in switch I extend to include the initial part of β2 in the interswitch region. This again is consistent with aMD echoing features observed in cMD. Other than in the switch regions and loop 10, which is near the nucleotide-binding pocket, the aMD KRas simulations starting from state 1 and state 2 structures show very similar fluctuation patterns relative to the other isoforms in the functional regions analyzed below. Overall, by focusing on the aMD simulations more significant differences can be observed not only in the switch regions, but elsewhere in the structure. Furthermore, while enhancing the range of conformations for regions that are observed to have greater flexibility by cMD, aMD simulations maintain the integrity of the Ras core structure. Interestingly, while KRas, which is known to be more dynamic than HRas and NRas (8,40,41), consistently shows greater flexibility at the C-terminal end of switch I leading to β2, switch II, the N-terminal end of helix 3 and loop 8, HRas is the most flexible at the C-terminal end of helix 3 and loop 7 near the allosteric site (9). NRas, in turn, has the most flexible P-loop.
Figure 3.
RMSF per residue for combined 200 ns simulations done in triplicate with different initial velocities for HRas, KRas state 1 and state 2, and NRas using cMD (dotted lines) and aMD (solid lines). HRas is in green, KRas in yellow (state 1) and maroon (state 2), and NRas is in cyan. Isoform-specific residues in each Ras protein are indicated by stars at a location directly above its sequence number in the RMSF plot.
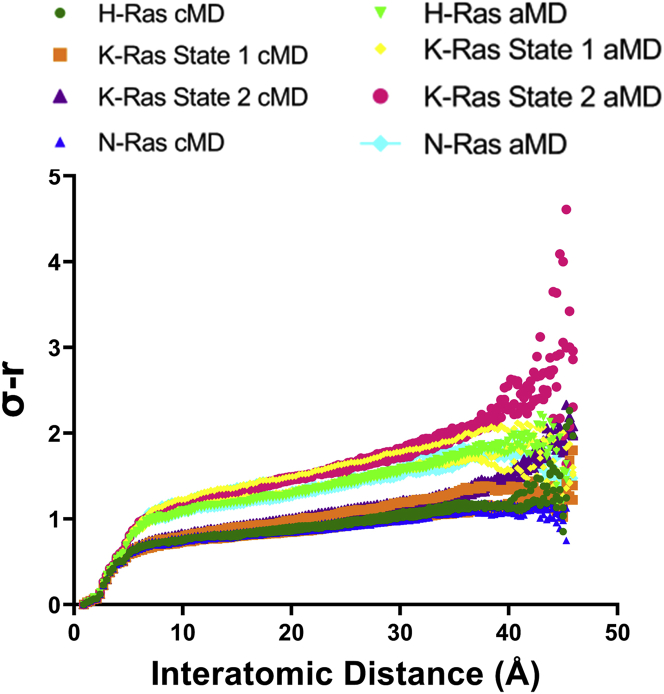
To justify the use of aMD over cMD in the present analysis, even with insufficient sampling of the landscape to appropriately correct for the boost potential bias (Fig. S2), we generated sigma-r plots (19) for each of the simulations (Fig. 4), representing the range of conformations within the MD trajectories. Note the consistently higher sigma-r values associated with aMD. This indicates greater flexibility in the aMD than in the cMD simulations. Furthermore, the triplicates for the cMD simulations have superimposable sigma-r plots. The small differences observed in the range of fluctuations in the individual aMD simulations are probably resulting from different access to certain conformational states associated with the randomly generated initial velocities at the start of each independent simulation run, reflecting the importance of independent replicates (42).
Figure 4.
Sigma-r plots for Ras proteins from each of the MD simulation replicates for HRas, KRas, and NRas. The darker colors are associated with cMD and the light colors with aMD, as indicated in the legend: HRas, green; KRas state 1, yellow; KRas state 2, maroon; NRas, blue. To see this figure in color, go online.
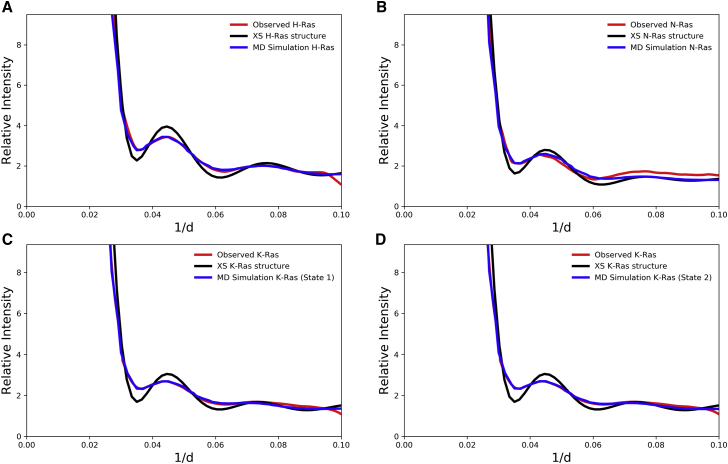
X-ray scattering from a protein in solution is the Fourier transform of the pair distribution function p(r), obtained from a histogram of interatomic distances within the protein molecule (34). It is possible to obtain a calculated scattering curve for a flexible protein by vector length convolution of p(r) for the rigid protein and the range of motions represented by sigma-r values calculated from an MD trajectory, shown as a function of the interatomic distances in a sigma-r plot (19,34) (Figs. 2 and 4). The average of the three sigma-r plots for each Ras isoform was used for vector length convolution with the respective crystal structure to obtain a calculated scattering curve for direct comparison with WAXS data (Fig. 5). In all cases, the calculated intensity curves from aMD simulations show better fits to the experimentally observed intensity curves out to 1/d = 0.1, corresponding to a resolution of 10 Å. In particular, HRas and KRas show excellent fit between the calculated intensity using aMD simulations (blue curve) and the observed intensity (red curve) (Fig. 5, A and B). The rigid Ras proteins (black curve) show sharper features than the flexible Ras proteins obtained from the aMD trajectories and the experimental data. Normalizing HRas to 1%, the overall flatness of the intensity curve calculated for KRas is 0.79% and for NRas it is 0.88%, with lower numbers indicating greater flatness associated with more flexible structures. The flattening of the curve is more prominent in KRas than HRas, indicating that KRas is more flexible in solution relative to HRas. The experimental scattering curve for NRas indicates that NRas is intermediate between HRas and KRas, which is in rough agreement with the RMSFs shown in Fig. 3, particularly in the helix 3 and loop 8 regions.
Figure 5.
Observed and calculated scattering curves for (A) HRas, (B) NRas (C) KRas state 1, and (D) KRas state 2. The observed experimental curves are in red, those calculated from the aMD trajectories are in blue, and those from the rigid crystal structures are in black. To see this figure in color, go online.
We have previously shown that 90 ns aMD simulations are more representative of active HRas fluctuations in solution than 90 ns cMD based on comparison with WAXS data (19). Here, we show that this is also the case for HRas, KRas, and NRas, with 200 ns simulations in triplicates, totaling 600 ns for each isoform. This is quantified by the scale factors that we obtain by scaling the sigma-r plots obtained from the MD simulations to optimize the match between calculated and observed intensity curves (Table 1). This scale factor corresponds to the degree to which the range of fluctuations in the MD simulations agree with those reflected by the WAXS data. The scale factors are all close to 1 in aMD, indicating an excellent representation of the range of conformational states found in solution, while the sigma-r values for cMD need to be significantly scaled to match the experimental data. The difference between scale factors obtained with cMD and aMD is largest for KRas, the most flexible of the three isoforms. Furthermore, since the scale factor for the KRas state 1 simulation series shows better agreement with WAXS data, we used it for the analysis that follows, although we checked that the conclusions regarding influence of isoform-specific residues are essentially the same for the two simulation sets.
Table 1.
Scale factors for HRas, KRas, and NRas comparing cMD and aMD simulations
| Protein | Method | Scale factor | Error |
|---|---|---|---|
| HRas | cMD | 1.61 | 0.022 |
| KRas state 1 | 1.91 | 0.023 | |
| KRas state 2 | 1.60 | 0.020 | |
| NRas | 1.63 | 0.072 | |
| HRas | aMD | 1.05 | 0.020 |
| KRas state 1 | 1.08 | 0.025 | |
| KRas state 2 | 1.15 | 0.019 | |
| NRas | 1.03 | 0.074 |
The error is the mean absolute error between the observed and calculated intensity curves.
Keeping in mind that the solution scattering data are low resolution, the agreements with the WAXS data do not mean that the aMD simulations perfectly reflect the balance of conformational states in solution. Rather, the results indicate that aMD better represents the range of accessed conformations given by σ(r), recognizing that, due to insufficient sampling for meaningful reweighting of the relative energies, we do not have accurate quantitative information on the population of conformational states. It has previously been noted that distinct MD simulation packages generate distinct conformational ensembles while agreeing equally well with experimental data (43). The authors conclude that agreement with experiments is necessary but not always sufficient to validate atomistic simulations. Here, we show that triplicate 200 ns aMD simulations show significantly better agreement with WAXS data than do the triplicate 200 ns cMD simulations. Thus, we go forward with analysis based on the aMD simulations while recognizing the limitations imparted by the biased potential in terms of obtaining quantitative information on the population of conformational states.
HRas samples R state more frequently than KRas and NRas in our simulations
The HRas, NRas, and one of the two KRas crystal structures used for the simulations have closed switch I in state 2, whereas one KRas simulation set started with switch I in the open state 1 conformation. All proteins remain with switch I near their starting positions during cMD, with no transitions observed between states 1 and state 2. There are also no transitions observed between R and T state conformations of switch II/helix 3/loop 7 in our cMD simulations. In contrast, aMD captures a larger range of conformational states, particularly in switch I, where transitions between state 2 and state 1 are observed, albeit very few of those transitions take place (Figs. S1 and S2). The three isoforms undergo transitions between R and T states in all four sets of aMD simulations, with prominent T state conformation regardless of starting structure. HRas, which has a higher intrinsic hydrolysis rate constant than KRas and NRas (18), has its average structure from the simulations, with helix 3 shifted toward the allosteric site and away from switch II. KRas and NRas on the other hand sample the R state less frequently, with average simulation structures showing helix 3 leaning toward switch II in a manner characteristic of the T state.
Isoform-specific residues centered on helix 3 affect functionally important regions
Analysis of the experimentally validated aMD simulations presented here expands on previous insights regarding the role of isoform-specific residues on structure and dynamics (18), adding further mechanistic information to allosteric effects on the active site and beyond. Since the largest differences in dynamics between the isoforms exist within the active site and the allosteric site, the residues that flank the water-mediated H-bond network associated with these sites were explored. Many of the isoform-specific residues, such as those on helices 4 and 5, are in areas that interact with the plasma membrane (14,44) or are near the Ras dimerization interface in the presence of Raf (45,46). Interestingly, several helix 3 residues are involved in the allosteric pathways linking the active site to helices 4 and 5 and to the allosteric site (9,47). Analysis of the aMD trajectories suggests that helix 3 is at the center of communication between the effector and allosteric lobes, with nuances driven by isoform-specific interactions.
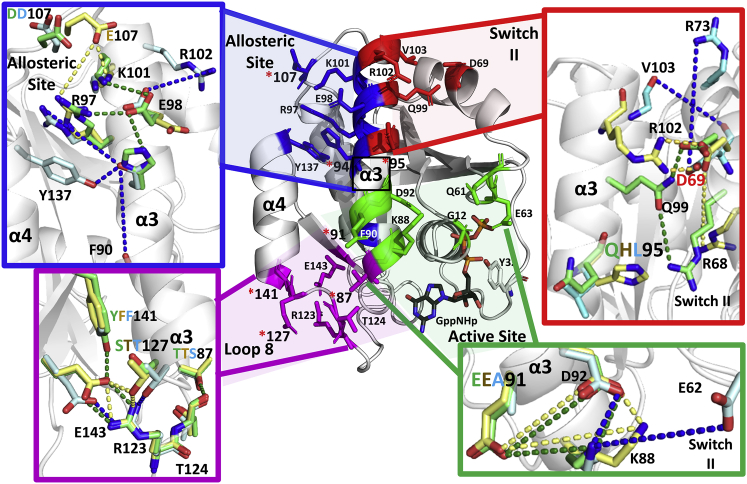
Helix 3, composed of residues 87–103, is adjacent to switch I and switch II and can be divided into 4 regions that contribute to functional areas containing isoform-specific residues (Fig. 6). When observed from an orientation with helix 3 at the center, Ras can be viewed as forming four quadrants, each impacting allosteric networks linking the effector and allosteric lobes: loop 8 (magenta), switch I (green), allosteric site (blue), and switch II (red) (Fig. 6). In all, 14 of the 17 G-domain residues (1–166) that are not identical between the three isoforms of Ras are in these 4 areas, with significant impact on isoform-specific modulation of active site dynamics. The remaining 3 are on helices 4 or 5, at the surface that becomes the dimer interface in the presence of Raf (46). The loop 8 region (Fig. 6) is adjacent to the nucleotide-binding pocket NKxD and ExSAK conserved active site motifs and contains residues involved in a water-mediated allosteric network linking switch I to helix 5 (47). This region includes isoform-specific residues A/P/P121, A/S/T122, E/D/D126, S/T/T127, R/K/K128, and Y/F/F141 (HRas/KRas/NRas). All of these residues are distinct in HRas, and identical in KRas and NRas, with residue 122 being the one exception. Residue 122, which is specific to each of the isoforms, is chemically unique in HRas (A) and contains a hydroxyl group in the other two isoforms: KRas (S) and NRas (T). Overall, the loop 8 region is unique in HRas compared with KRas and NRas and it is poised to impact the nucleotide-binding pocket and switch I.
Figure 6.
Interactions around helix 3 influenced by isoform-specific residues. The main panel shows a view of helix 3 at the center flanked by four major regions that contain isoform-specific residues (PDB: 3K8Y): loop 8 (magenta) switch I (green), allosteric site (blue), and switch II (red). Residues that influence the differences in dynamics are shown in sticks. Isoform-specific residues are designated by red stars. Each quadrant is magnified in the peripheral panels, with residue side chains and their favored interactions shown color-coded for each of the isoforms: HRas, green; KRas, yellow; NRas, cyan.
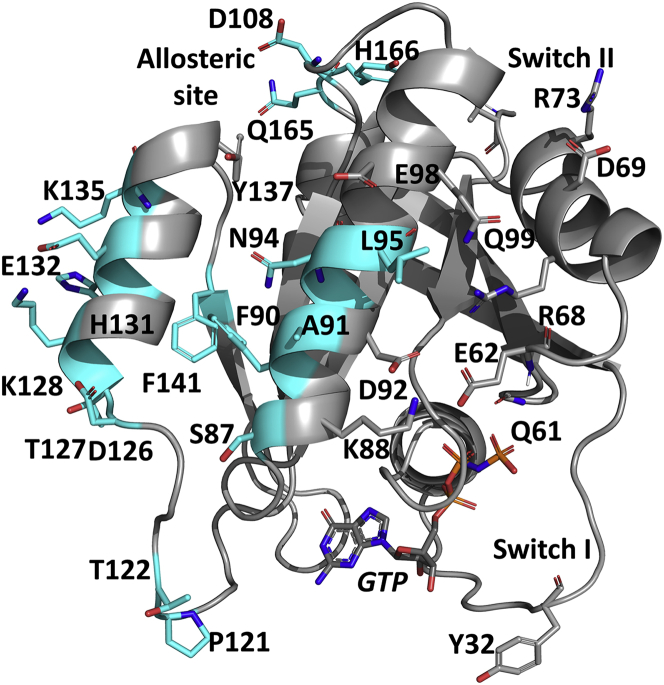
Residue T/T/S87 on helix 3 faces loop 8 and is S in NRas and T in the other two isoforms, with a hydroxyl group present in all cases. Residue 87 is at the N-terminal end of helix 3, at the beginning of a continuous surface of isoform-specific residues unique to NRas that also includes E/E/A91, H/H/N94, and Q/H/L95 on helix 3, with significant impact on switch I and switch II. Other residues specific to NRas include conservative mutations Q/Q/H131, D/D/E132, and R/R/K135 on helix 4 and H/H/Y166, which increases bulk at the C-terminus of helix 5 near the allosteric site (Fig. 7). All of these NRas-specific residues are identical in HRas and KRas, except for residue 95, which is hydrophilic in HRas and KRas and with a unique hydrophobic character in NRas. Here, we focus on the effects of the helix 3 NRas-specific residues. Helix 4 residue H131 is adjacent to residues that affect the loop 8 region and may impact the distinct switch I behavior discussed below for NRas. Residues E132, K135, and Y166 are near the dimer interface and may also contribute through allosteric connections to the unique switch conformations for NRas. Overall, NRas-specific residues are situated to impact switch I and switch II and their connection to the allosteric site. Unlike HRas and NRas, which have several isoform-specific residues that cluster to affect loop 8 and the connection between helix 3 and the switch regions, respectively, the three residues unique to KRas, other than S122 and H95, are distant from each other on the G-domain structure. Their common feature is that they are located near the Ras dimer interface consisting of helices 4 and 5. Residue E107 (D in HRas and NRas) is on the allosteric site loop 7, whereas D153 (E in HRas and NRas) and K165 (Q in HRas and NRas) are near the N- and C-terminal ends of helix 5, respectively.
Figure 7.
The structure of NRas (PDB: 5UHV) with residues that are different in at least one isoform shown in sticks colored cyan. Conserved residues that behave differently in H, K, and NRas due to the isoform-specific residues are in gray sticks. To see this figure in color, go online.
While useful to look at the four quadrants of Ras through the lens focused on helix 3, the dynamics of each quadrant are linked to the others. Here, we explore the connections between loop 8 and switch I and between the allosteric site and switch II, as well as factors that differently affect the link between switch I and switch II.
Loop 8 and switch I
Loop 8 is adjacent to the nucleotide-binding pocket and contributes to a conserved salt bridge between R123 and E143 of the ExSAK nucleotide-binding motif. In HRas the R123:E143 salt bridge is highly prominent and is at the center of an H-bonding network supported by HRas-specific residues. Alanine at residues 121 and 122 allows loop 8 flexibility for the backbone carbonyl oxygen of A120 to bridge to the Nε group of R123 through a water molecule on one side, while the longer side chain of E126 (D in KRas and NRas) bridges through a different water molecule to the terminal NH2 of R123 on the other side from the A120 interaction. This network is extended through the R123:E143 salt bridge with direct H-bonds to the hydroxyl groups of Y141 and S127 side chains on opposite sides of the E143 side chain. In KRas and NRas, the more restricted P121 backbone keeps loop 8 slightly more open and D126 is too short, such that their water-mediated interactions with R123 are weakened. Concomitantly, E143 is shifted due to steric hindrance with F141, which now is within van der Waal’s contact of the side-chain methyl group of T127, further weakening the tight H-bonding network prevalent in HRas (Fig. 6). The prominence of the R123:E143 salt bridge and surrounding interactions in HRas results in reduced dynamics in the P-loop and helix 1, the N-terminal end of helix 3, loop 8 itself, and the N-terminal end of helix 4 compared with KRas and NRas (Fig. 3). The loop 8 region is poised in HRas to form an anchor to the nucleotide-binding pocket, including stabilization of the GTP γ-phosphate, which interacts with the P-loop, promoting interaction with Y32 in switch I in the canonical state 2/R state that we associate with hydrolysis of GTP (9). Weakening of the loop 8 H-bonding network in KRas and NRas results in increased dynamics adjacent to the guanine base of the nucleotide and the P-loop regions (Fig. 3), with significant impact on switch I and associated allosteric networks (47). The increased flexibility in the P-loop is consistent with the fact that we were not able to use 31P NMR to study state 1 and state 2 in KRas (8) as has been done by others for HRas (39) and successfully reproduced in in our laboratory (see SI of (8) for KRas and HRas 31P NMR spectra).
Residue A91 in NRas is among several that are chemically different from those found in the other two isoforms, with significant consequences to interactions near the active site. In HRas and KRas, K88 comes within interaction distance of three negatively charged residues: E91 and D92 on helix 3 and E62 on switch II (Fig. 6). The K88-E91 salt bridge is prominent in these two isoforms, followed by the K88-D92. In NRas, with no negatively charged side chain at residue 91, the major interaction of K88 is with D92, followed by interaction with E62. While in HRas the K88-E62 interaction is of little consequence to the stability of switch II due to a robust H-bond network between helix 3 and switch II residues, which includes interaction between D92 and R68, in NRas the K88-E62 interaction contributes to the disruption between helix 3 and switch II driven by isoform-specific residues N94 and L95 (described below). These unique interactions in NRas, promote a conformation of the Y32 side chain turned toward helix 1, not observed for HRas and KRas. This would be expected to decrease access of Y32 to the active site and increase exposure of the nucleotide and the P-loop residues to solvent. In the GTP-bound state this conformation is unique to NRas as a result of isoform-specific residues (Fig. 7). The combination of the loop 8 residues resulting in increased dynamics in the nucleotide-binding pocket and the shifts in NRas due to the absence of E91, leads to a conformation of switch I that increases the active site solvent exposure, while its backbone remains in state 2 with the characteristic interaction between T35 and Mg2+ ion in the active site.
Allosteric site and switch II
The allosteric site is a charged pocket at the interface of helix 3, loop 7, and helix 4, previously observed to bind calcium and acetate in HRas (9). This pocket involves residues R97, K101, and D/E/D107, and is flanked by Y137 in helix 4, and E98 and R102 in helix 3 (Fig. 6). This region is relatively dynamic (Fig. 3) and interactions within it directly impact the access to the R and T states associated with shifts of helix 3/loop 7 toward helix 4 (R state) or switch II (T state) (5,9). This is modulated by isoform-specific residue 94 (H94 in HRas and KRas, N94 in NRas) and 107 (D107 in H-Ras and N-Ras, E107 in K-Ras) affecting K101 and R102 salt bridges with negatively charged residues E98 on helix 3 and D69 on switch II (Fig. 6, top two quadrants). In KRas, E107 reaches into the allosteric site making significant interactions with R97 and K101. While both interactions are diminished in our simulations in HRas, NRas retains good sampling of the D107-K101 salt bridge, possibly due to the presence of Y166, which H-bonds directly with S106 and may stabilize loop 7. The diminished D107-R97 interaction increases the prominence of the R97-E98 salt bridge in both HRas and NRas relative to KRas. With E98 shifted toward the allosteric site in HRas, it rarely interacts with R102, located at the very top of helix 3. Thus, R102 faces the solvent and is highly dynamic in HRas, interacting only rarely with the side chain of D69 in switch II. As observed in Fig. 3, residues at the top of helix 3 and in loop 7 have high RMSFs in HRas compared with the same region in KRas and NRas, primarily due to the relative lack of interactions between R102 and either E98 in the allosteric site or D69 in switch II. While D69 is exposed to solvent in HRas, other nearby residues such as Q99 and Q95 form favorable H-bond interactions with switch II (9), helping to maintain the integrity of the switch. In KRas, the more prominent interaction between E107 and R97 releases E98 which shits its position toward the top of helix 3, where it interacts with R102 more frequently in our simulations. However, R102 can also swing toward switch II to interact with D69, promoting the T state and distancing Q61 from the active site. In NRas, isoform-specific residue N94 interacts with the backbone carbonyl group of F90, with R97, and, to a lesser extent, with E98. This series of interactions, not present in HRas, allow E98 to interact with R102 as in KRas, and D69 participates in a salt bridge interaction with R73 (Fig. 6), which in concert with the NRas isoform-specific residues in helix 3 facing switch II impair the H-bonding networks integral to the R state.
Although switch II is conserved among the three isoforms, it behaves differently in the three Ras proteins due to key isoform-specific residues on helix 3. The first residue at this location is residue 95, unique in H, K, and NRas (Fig. 6). In HRas this residue is Q95, which sustains interactions with nearby Q99 and R68, both of which, along with Y96, are involved in the allosteric network linking the allosteric and active sites (9). KRas features H95, which interacts with Q99 but not with R68. The absence of this interaction, in concert with KRas isoform-specific residue E107, which enhances the R102-D69 interaction as described above, appears to favor the T state in KRas relative to HRas, and switch II shifts outward allowing an interaction between Y71 and D38 in switch I not observed in the other two isoforms during our simulations. This, could be associated with a sharp kink in helix 3 that is unique to KRas and may contribute to the high RMSF values seen for switch II, not only in the aMD simulations but also in the cMD simulations (Fig. 3). As switch II is a driver of switch I conformation in KRas (48), it is not surprising that switch I exhibits high RMSF values in KRas as well. NRas, with L95 unable to participate in any H-bond interactions with either Q99 or R68 in switch II, shows reduced connectivity between switch II and the allosteric site and, without these key interactions to promote the R state, NRas is almost always in the T state during our simulations. In short, the interactions at the allosteric site provide a molecular basis for how the E98-R102 and D69-R102 pairs influenced by isoform-specific residues affect access to R state and T state associated with the placement of catalytic Q61 in the active site, which we propose is essential for hydrolysis of GTP (9,49).
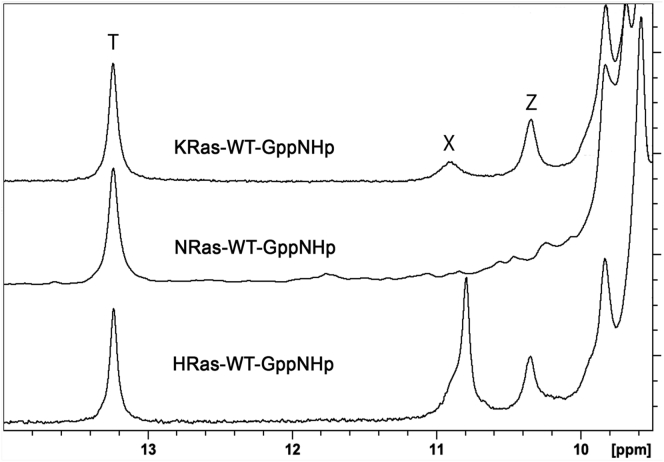
1H NMR spectrum in line with aMD simulations
To test whether the aMD simulation analysis described above is consistent with an experimental probe of active site dynamics, we turned to proton NMR data in the downfield region of the spectrum as we previously described for HRas and KRas (8), with added NMR data for NRas (Fig. 8). Note that here we add a new spectrum for KRas with no contribution from the GDP-bound form, and summarize the peaks associated with the Ras isoforms bound to the GTP analog GppNHp. In brief, we take a resonance peak at 13.2 ppm (peak T) as a sensor of Ras bound to GppNHp; a resonance at 10.9 ppm (peak X) as a sensor of the switch II shielding of the protonated γ-phosphate of GppNHp (49); and a resonance at 10.3 ppm (peak Z) as a sensor of switch I shielding of the P-loop, as we described previously in detail (8). The aMD simulation analysis described above reveals a relatively stable active site for HRas supported by HRas-specific residues in the loop 8 region, such that its state 2 is characterized not only by the T35 H-bond to the Mg2+ ion but also by a Y32 side chain turned toward the nucleotide interacting with the γ-phosphate of GTP. This protected active site results in strong downfield NMR resonance peaks, particularly the X peak, due to a relatively stable switch II, presumably associated with the R state. For KRas, the peaks are present, although with a less prominent X peak, consistent with a more open switch II and more prominent T state, while a Z peak similar to that of HRas is indicative of similar sampling of state 1 in the two isoforms. Interestingly, the NRas NMR spectrum is flat in the region where the X and Z peaks are expected. As discussed above, diminished connection between switch I and the N-terminal end of helix 3 due to NRas-specific residue A91 and between helix 3 and switch II due to NRas-specific residue L95, promotes a more open state 2, with Y32 interacting with helix 1 residues and a prominent T state. This leads to an active site highly exposed to solvent, including the P-loop, which is highly dynamic in NRas (Fig. 3), and the γ-phosphate of GTP, consistent with the absence of the X and Z peaks in the NMR spectrum. In all, the NMR spectra shown in Fig. 8 support our MD simulation analysis of active site dynamics in the three Ras isoforms.
Figure 8.
Downfield proton NMR spectra for HRas, NRas, and KRas. The T peak is associated with binding of the GTP analog GppNHp, shifted further downfield when Ras is bound to GDP. Peak Z is a sensor for switch I, present when Ras is in state 2 but not state 1. The X peak is an indicator of switch II closed over the active site. It is weakened or absent when switch II is shifted away from the active site.
Discussion
It is now accepted that while HRas, KRas, and NRas have essentially the same 3D structure, with highly similar sequences, their dynamics and access to functionally relevant conformational states vary substantially. Experimentally, this is reflected in distinct GTP hydrolysis rate constants (18), different levels of hydrogen-deuterium exchange observed by mass spectrometry (50), and distinct proton NMR signatures in the downfield region associated with switch I and II conformational states (Fig. 8) (8). Computationally, cMD has been used to study differences between the Ras isoforms, establishing KRas as the most flexible of the isoforms and delineating important regions of correlated motions in the active, GTP-bound state (38,40,48). Importantly, the conformational transition between the GDP- and GTP-bound states of HRas has been studied using aMD, shown to sample a wide range of conformational states exemplified in various crystal structures in the PDB (51). This supported our choice of aMD for observing transitions between state 1 and state 2 and R and T states, not accessible by cMD for Ras bound to GTP in relatively short simulations as presented in this study. Here, we contribute to the existing body of work by adding WAXS as experimental evidence that the flexibility or range of conformational states observed in solution for the three Ras isoforms is better captured by aMD than cMD, and analyze the aMD simulations of the G-domains with focus on how isoform-specific residues lead to distinct sets of interactions that affect their structure and dynamics. Our comparative analysis is based on simulations that started with our crystal structures of each wild-type Ras, HRas (PDB: 3K8Y) (9), KRas (PDB: 5UK9) (8), and NRas (PDB: 5UHV) (18), and an additional structure of wild-type KRas present in the PDB (PDB: 6GOD) (52), without having to make any in silico reversions other than the substitution of the β- and γ-phosphate bridging group with an oxygen atom to obtain GTP. This is important because short simulations that start with mutant structures reverted back to wild-type may retain some features of the original structure that could bias the simulation results.
We have previously described the use of WAXS data to show that HRas-GDP is globally more rigid in solution than HRas-GppNHp and that, while cMD appropriately captures the flexibility of HRas-GDP, aMD is needed to match the WAXS data in the case of HRas-GppNHp (19). Here, we use aMD to capture conformations not accessed in comparably short cMD simulations, with scale factors shown in Table 1 that are near 1. The greater global flexibility of the GTP-bound state is supported experimentally, with HDX-MS results showing that HRas, KRas, and NRas bound to GppNHp undergo increased hydrogen-deuterium exchange relative to their respective GDP-bound forms (50). The global features of our simulations are consistent with experimental WAXS data and the results of our analysis are supported by the NMR spectra presented here.
Although aMD has limitations due to the boost potential bias already discussed, it succeeds in traveling across the conformational landscape more easily than cMD within a short simulation time, revealing possible conformations that do not come up within an equivalent time frame in cMD. Therefore, there is added value for a biological analysis of these isoforms of Ras. Although several of the conformations observed in the aMD simulations are similar to those seen in crystal structures, some are not represented in the PDB. Thus, the aMD simulations provided more information about the overall accessible conformations than cMD as presented here. Possibly, running long multi-microsecond simulations would yield sufficient sampling of conformational states for a quantitative assessment of the population distribution. However, being able to attain dynamics information with relatively short aMD simulations is an important factor for the wider use of simulation tools in the analysis of structural data.
Focus on the contribution of the isoform-specific residues in the aMD simulation analysis revealed that these residues cluster in functionally important regions that connect the two lobes of the Ras G-domain centered around helix 3. HRas, with its isoform-specific residues located in the loop 8 region, which includes both the NKxD and the ExSAK nucleotide-binding motifs, has the most stable active site where the nucleotide is anchored and serves as a docking area for switch I residues Y32 and T35. This is reflected in lower RMSF for the P-loop, helix 1, and N-terminal portions of helix 3 adjacent to loop 8, as well as the N-terminal portion of switch II in HRas, relative to the other two isoforms (Fig. 3). Consistently, our NMR results indicate protected P-loop and γ-phosphate of GTP in HRas (Fig. 8). Interestingly, HRas has the highest fluctuations in the C-terminal end of helix 3/loop 7 due to the disordered nature of R102 and D69 in this isoform stemming from the E98-R97 interaction in the allosteric site. Fluctuations in this region of KRas and NRas are attenuated by the presence of isoform-specific residues E107 in KRas and N94, L95, and possibly Y166 in NRas. In KRas the presence of E107 positions this residue to interact more optimally with K101, shifting allosteric site interactions such that E98 interacts with R102. In NRas, the E98-R102 interaction is enhanced by the fact that N94 interacts with F90 and by the diminished connection between helix 3 and switch II, strongly promoting the T state. KRas has highly dynamic switch I and switch II, loop 8, and the N-terminal end of helix 3, all areas surrounding the nucleotide-binding pocket. Its P-loop also exhibits significantly higher RMSF values, albeit not as high as NRas (Fig. 3). This reflects the arrangement of residues in loop 8, similar to NRas but without the switch I and II stabilizing features promoted by helix 3 NRas-specific residues that favor a more open state 2 in switch I and stabilizing interactions within switch II, such as that between D69 and R73 (Fig. 7). NRas shows significantly increased P-loop dynamics and this is explained by the helix 3 NRas-specific residues that lead to a relative lack of connection between the switch regions and helix 3, with an enhanced K88 salt bridge with E62 in switch II promoted by A91 in an open state 2 conformation that exposes the nucleotide and the P-loop. Previous simulations have established R102 as a driver of motions for the switch regions in K-Ras (48). This residue is modulated by isoform-specific residues in Ras proteins, with significant impact on the behavior of switch II, which in turn affects switch I. The features we describe for the three isoforms are in line with the NMR spectra presented in Fig. 8, and consistent with previously published Ras intrinsic GTP hydrolysis behavior in the absence and presence of Raf-RBD (18). In the context of the results presented here, the greater hydrolysis rate constant for HRas (0.016 min−1) relative to KRas (0.006 min−1) and NRas (0.006 min−1) (18) can be ascribed to loop 8 residues specific for HRas, which anchors GTP and with it Y32 and Q61 in the active site, favoring the R state conformation associated with GTP hydrolysis (9,49). Binding of Raf-RBD to both KRas and NRas stabilizes switch I in a catalytically competent conformation as it does in HRas (9,13,53). In KRas, this leads to a rate constant for GTP hydrolysis similar to HRas, providing an anchor for switch II to adopt the R state. In NRas the disconnection between helix 3 and switch II due to L95, with an enhanced T state, prevents the binding of Raf-RBD from having a similar effect on the GTP hydrolysis rate constant, which remains slow. Thus, here we add mechanistic understanding of our previously published observation that the stabilization of switch I in the active site in the presence of Raf-RBD is rate-limiting in KRas, whereas in NRas attaining the R state associated with switch II is the rate-limiting step in the intrinsic hydrolysis of GTP on Ras (18). The variation in hydrolysis rate constants in the absence of GAPs (54) or of membrane components and ligand binding in the allosteric site (9,13) are useful to validate the analysis presented here, although other factors are needed to achieve the 3–5 orders of magnitude necessary for a biologically relevant increase in GTP hydrolysis. Furthermore, the fact that there seems to be greater difficulty in attaining the R state in NRas due to isoform-specific residues in helix 3, may have consequences for signaling through Ras/Raf/MEK/ERK where switch II is not part of the Ras/effector interface and where an allosteric switch mechanism may be of key importance in turning the signal off (9,13,45,46). This could be related to the prominence of Q61 mutations in NRas-mutant cancers, such as melanomas, in association with signaling through Raf (55). One last point to note is that NRas was observed to have slightly lower affinity for Raf-RBD than the other isoforms (200 vs. 100 nM for HRas and KRas) (18). This could be due to a more open state 2 conformation of switch I where Y32 interacts with helix 1, as the Y32 side chain must change conformation to close over the nucleotide when the two proteins interact (13).
To conclude, we present a series of aMD simulations starting from crystal structures of the active states of each Ras isoform, providing mechanistic understanding of the influence of isoform-specific residues on Ras dynamics, consistent with experimental results. It is clear that isoform-specific residues coevolved in clusters to fine-tune dynamics in a way that affects functionally important regions connected to the active site. These differences affect biochemical properties of the G-domain and it will be exciting to learn going forward of mechanistic connections to signaling outputs.
Author contributions
A.Y.V. and H.G. performed the research, analyzed the data, and wrote the manuscript. H.Z., D.R., and S.P. performed the research and analyzed the data. L.M. designed the research and analyzed the data. C.M. designed the research, analyzed the data, and wrote the manuscript. All authors read and commented on the final version of the manuscript.
Acknowledgments
This research was funded by a grant from the National Science Foundation (MCB-2121426, to C.M.). The MD simulations were performed on the Discovery Cluster supercomputer at Northeastern University. We thank Kendra Marcus for help with NAMD and various analysis tools as well as for general discussions on Ras proteins. Thanks to Benoit Roux and Chris Chipot for critical discussion of the Boltzman reweighting of the aMD simulations.
Declaration of interests
The authors declare no competing interests.
Editor: Alemayehu Gorfe.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2022.07.005.
Supporting material
Data availability
The MD trajectories will be made available upon request. Please contact Carla Mattos (c.mattos@northeastern.edu).
References
- 1.Bar-Sagi D., Hall A. Ras and Rho GTPases: a family reunion. Cell. 2000;103:227–238. doi: 10.1016/s0092-8674(00)00115-x. [DOI] [PubMed] [Google Scholar]
- 2.Bourne H.R., Sanders D.A., McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 3.Buhrman G., O'Connor C., et al. Mattos C. Analysis of binding site hot spots on the surface of Ras GTPase. J. Mol. Biol. 2011;413:773–789. doi: 10.1016/j.jmb.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vetter I.R., Arndt A., et al. Wittinghofer A. Structural view of the Ran–importin β interaction at 2.3 Å resolution. Cell. 1999;97:635–646. doi: 10.1016/s0092-8674(00)80774-6. [DOI] [PubMed] [Google Scholar]
- 5.Johnson C.W., Mattos C. The allosteric switch and conformational states in Ras GTPase affected by small molecules. Enzymes. 2013;33 Pt A:41–67. doi: 10.1016/B978-0-12-416749-0.00003-8. [DOI] [PubMed] [Google Scholar]
- 6.Wittinghofer A., Vetter I.R. Structure-function relationships of the G domain, a canonical switch motif. Annu. Rev. Biochem. 2011;80:943–971. doi: 10.1146/annurev-biochem-062708-134043. [DOI] [PubMed] [Google Scholar]
- 7.Muraoka S., Shima F., et al. Kataoka T. Crystal structures of the state 1 conformations of the GTP-bound H-Ras protein and its oncogenic G12V and Q61L mutants. FEBS Lett. 2012;586:1715–1718. doi: 10.1016/j.febslet.2012.04.058. [DOI] [PubMed] [Google Scholar]
- 8.Parker J.A., Volmar A.Y., et al. Mattos C. K-ras populates conformational states differently from its isoform H-ras and oncogenic mutant K-RasG12D. Structure. 2018;26:810–820.e4. doi: 10.1016/j.str.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 9.Buhrman G., Holzapfel G., et al. Mattos C. Allosteric modulation of Ras positions Q61 for a direct role in catalysis. Proc. Natl. Acad. Sci. USA. 2010;107:4931–4936. doi: 10.1073/pnas.0912226107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheidig A.J., Burmester C., Goody R.S. The pre-hydrolysis state of p21(ras) in complex with GTP: new insights into the role of water molecules in the GTP hydrolysis reaction of ras-like proteins. Structure. 1999;7:1311–1324. doi: 10.1016/s0969-2126(00)80021-0. [DOI] [PubMed] [Google Scholar]
- 11.Buhrman G., Wink G., Mattos C. Transformation efficiency of RasQ61 mutants linked to structural features of the switch regions in the presence of Raf. Structure. 2007;15:1618–1629. doi: 10.1016/j.str.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheffzek K., Ahmadian M.R., et al. Wittinghofer A. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 13.Fetics S.K., Guterres H., et al. Mattos C. Allosteric effects of the oncogenic RasQ61L mutant on Raf-RBD. Structure. 2015;23:505–516. doi: 10.1016/j.str.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parker J.A., Mattos C. The ras-membrane interface: isoform-specific differences in the catalytic domain. Mol. Cancer Res. 2015;13:595–603. doi: 10.1158/1541-7786.MCR-14-0535. [DOI] [PubMed] [Google Scholar]
- 15.Castellano E., Santos E. Functional specificity of ras isoforms: so similar but so different. Genes Cancer. 2011;2:216–231. doi: 10.1177/1947601911408081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hancock J.F., Parton R.G. Ras plasma membrane signalling platforms. Biochem. J. 2005;389(Pt 1):1–11. doi: 10.1042/BJ20050231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prior I.A., Lewis P.D., Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson C.W., Reid D., et al. Mattos C. The small GTPases K-Ras, N-Ras, and H-Ras have distinct biochemical properties determined by allosteric effects. J. Biol. Chem. 2017;292:12981–12993. doi: 10.1074/jbc.M117.778886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou H., Guterres H., et al. Makowski L. Predicting X-ray solution scattering from flexible macromolecules. Protein Sci. 2018;27:2023–2036. doi: 10.1002/pro.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wereszczynski J., McCammon J.A. Accelerated molecular dynamics in computational drug design. Methods Mol. Biol. 2012;819:515–524. doi: 10.1007/978-1-61779-465-0_30. [DOI] [PubMed] [Google Scholar]
- 21.Förster F., Webb B., et al. Sali A. Integration of small-angle X-ray scattering data into structural modeling of proteins and their assemblies. J. Mol. Biol. 2008;382:1089–1106. doi: 10.1016/j.jmb.2008.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makowski L. Characterization of proteins with wide-angle X-ray solution scattering (WAXS) J. Struct. Funct. Genomics. 2010;11:9–19. doi: 10.1007/s10969-009-9075-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson C.W., Buhrman G., et al. Mattos C. Expression, purification, crystallization and X-ray data collection for RAS and its mutants. Data Brief. 2016;6:423–427. doi: 10.1016/j.dib.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Acerbo A.S., Cook M.J., Gillilan R.E. Upgrade of MacCHESS facility for X-ray scattering of biological macromolecules in solution. J. Synchrotron Radiat. 2015;22:180–186. doi: 10.1107/S1600577514020360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nielsen S.S., Toft K.N., et al. Arleth L. BioXTAS RAW, a software program for high-throughput automated small-angle X-ray scattering data reduction and preliminary analysis. J. Appl. Crystallogr. 2009;42:959–964. [Google Scholar]
- 26.MacKerell A.D., Bashford D., et al. Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 27.Brooks B.R., Brooks C.L., 3rd, et al. Karplus M. CHARMM: the biomolecular simulation program. J. Comput. Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phillips J.C., Braun R., et al. Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 30.Steinbach P.J., Brooks B.R. New spherical-cutoff methods for long-range forces in macromolecular simulation. J. Comput. Chem. 1994;15:667–683. [Google Scholar]
- 31.Ryckaert J.-P., Ciccotti G., Berendsen H.J. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 32.Hamelberg D., Mongan J., McCammon J.A. Accelerated molecular dynamics: a promising and efficient simulation method for biomolecules. J. Chem. Phys. 2004;120:11919–11929. doi: 10.1063/1.1755656. [DOI] [PubMed] [Google Scholar]
- 33.Miao Y., Sinko W., et al. McCammon J.A. Improved reweighting of accelerated molecular dynamics simulations for free energy calculation. J. Chem. Theory Comput. 2014;10:2677–2689. doi: 10.1021/ct500090q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou H., Li S., Makowski L. Visualizing global properties of a molecular dynamics trajectory. Proteins. 2016;84:82–91. doi: 10.1002/prot.24957. [DOI] [PubMed] [Google Scholar]
- 35.Makowski L., Gore D., et al. Fischetti R.F. X-ray solution scattering studies of the structural diversity intrinsic to protein ensembles. Biopolymers. 2011;95:531–542. doi: 10.1002/bip.21631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baker E.N., Hubbard R.E. Hydrogen bonding in globular proteins. Prog. Biophys. Mol. Biol. 1984;44:97–179. doi: 10.1016/0079-6107(84)90007-5. [DOI] [PubMed] [Google Scholar]
- 37.Sklenar V., Piotto M., et al. Saudek V. Gradient-tailored water suppression for H-1-N-15 Hsqc experiments optimized to retain full sensitivity. J. Magn. Reson. 1993;102:241–245. [Google Scholar]
- 38.Gorfe A.A., Grant B.J., McCammon J.A. Mapping the nucleotide and isoform-dependent structural and dynamical features of Ras proteins. Structure. 2008;16:885–896. doi: 10.1016/j.str.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spoerner M., Hozsa C., et al. Kalbitzer H.R. Conformational states of human rat sarcoma (Ras) protein complexed with its natural ligand GTP and their role for effector interaction and GTP hydrolysis. J. Biol. Chem. 2010;285:39768–39778. doi: 10.1074/jbc.M110.145235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kapoor A., Travesset A. Differential dynamics of RAS isoforms in GDP- and GTP-bound states. Proteins. 2015;83:1091–1106. doi: 10.1002/prot.24805. [DOI] [PubMed] [Google Scholar]
- 41.Lukman S., Grant B.J., et al. McCammon J.A. The distinct conformational dynamics of K-Ras and H-Ras A59G. PLoS Comput. Biol. 2010;6:e1000922. doi: 10.1371/journal.pcbi.1000922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knapp B., Ospina L., Deane C.M. Avoiding false positive conclusions in molecular simulation: the importance of replicas. J. Chem. Theory Comput. 2018;14:6127–6138. doi: 10.1021/acs.jctc.8b00391. [DOI] [PubMed] [Google Scholar]
- 43.Childers M.C., Daggett V. Validating molecular dynamics simulations against experimental observables in light of underlying conformational ensembles. J. Phys. Chem. B. 2018;122:6673–6689. doi: 10.1021/acs.jpcb.8b02144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gorfe A.A., Hanzal-Bayer M., et al. McCammon J.A. Structure and dynamics of the full-length lipid-modified H-Ras protein in a 1, 2-dimyristoylglycero-3-phosphocholine bilayer. J. Med. Chem. 2007;50:674–684. doi: 10.1021/jm061053f. [DOI] [PubMed] [Google Scholar]
- 45.Cookis T., Mattos C. Crystal structure reveals the full ras-raf interface and advances mechanistic understanding of Raf activation. Biomolecules. 2021;11:996. doi: 10.3390/biom11070996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Packer M.R., Parker J.A., et al. Mattos C. Raf promotes dimerization of the Ras G-domain with increased allosteric connections. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2015648118. e2015648118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kearney B.M., Johnson C.W., et al. Mattos C. DRoP: a water analysis program identifies Ras-GTP-specific pathway of communication between membrane-interacting regions and the active site. J. Mol. Biol. 2014;426:611–629. doi: 10.1016/j.jmb.2013.10.036. [DOI] [PubMed] [Google Scholar]
- 48.Vatansever S., Gümüş Z.H., Erman B. Intrinsic K-Ras dynamics: a novel molecular dynamics data analysis method shows causality between residue pair motions. Sci. Rep. 2016;6:37012. doi: 10.1038/srep37012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Knihtila R., Holzapfel G., et al. Mattos C. Neutron crystal structure of RAS GTPase puts in question the protonation state of the GTP gamma-phosphate. J. Biol. Chem. 2015;290:31025–31036. doi: 10.1074/jbc.M115.679860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harrison R.A., Lu J., et al. Engen J.R. Structural dynamics in ras and related proteins upon nucleotide switching. J. Mol. Biol. 2016;428:4723–4735. doi: 10.1016/j.jmb.2016.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grant B.J., Gorfe A.A., McCammon J.A. Ras conformational switching: simulating nucleotide-dependent conformational transitions with accelerated molecular dynamics. PLoS Comput. Biol. 2009;5:e1000325. doi: 10.1371/journal.pcbi.1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cruz-Migoni A., Canning P., et al. Rabbitts T.H. Structure-based development of new RAS-effector inhibitors from a combination of active and inactive RAS-binding compounds. Proc. Natl. Acad. Sci. USA. 2019;116:2545–2550. doi: 10.1073/pnas.1811360116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tran T.H., Chan A.H., et al. Simanshu D.K. KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation. Nat. Commun. 2021;12:1176. doi: 10.1038/s41467-021-21422-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scheffzek K., Shivalingaiah G. Ras-specific GTPase-activating proteins-structures, mechanisms, and interactions. Cold Spring Harb. Perspect. Med. 2019;9:a031500. doi: 10.1101/cshperspect.a031500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang A.X., Qi X.Y. Targeting RAS/RAF/MEK/ERK signaling in metastatic melanoma. IUBMB Life. 2013;65:748–758. doi: 10.1002/iub.1193. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MD trajectories will be made available upon request. Please contact Carla Mattos (c.mattos@northeastern.edu).