Summary
Molecular cloning of BRD4-L is a challenging technique, because the DNA insert is formed by a long, GC-rich sequence, which folds into secondary structures. The present protocol defines a specific strategy to amplify BRD4-L, followed by the successful cloning of the gene into an overexpression vector. Since there are no existing protocols nor commercially available plasmids, this work provides a useful tool for studies involving molecular cloning of BRD4-L and could potentially be applied to other challenging genes.
Subject areas: Cell biology, Molecular biology, Protein biochemistry, Protein expression and purification
Graphical abstract
Highlights
-
•
PCR amplification of a long, GC-rich sequence
-
•
Ligation of BRD4-L with the overexpression vector LentiV_Blast
-
•
Construction and sequencing of the plasmid LentiV_Blast-BRD4-L
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Molecular cloning of BRD4-L is a challenging technique, because the DNA insert is formed by a long, GC-rich sequence, which folds into secondary structures. The present protocol defines a specific strategy to amplify BRD4-L, followed by the successful cloning of the gene into an overexpression vector. Since there are no existing protocols nor commercially available plasmids, this work provides a useful tool for studies involving molecular cloning of BRD4-L and could potentially be applied to other challenging genes.
Before you begin
Protocol overview
The protocol bellow describes specific steps to introduce the long isoform of BRD4 (BRD4-L) into the lentiviral mammalian expression plasmid LentiV_Blast (Addgene, cat# 111887). However, we have also used the same protocol to introduce BRD4-L into the lentiviral mammalian expression plasmid LentiV_Puro (Addgene, cat# 111886) and the dox-inducible (Tet-On) lentiviral expression vector pLentiTRE/rtTA carrying both TRE and rtTA cassettes (kindly provided by Trudy Oliver, Huntsman Cancer Institute, Salt Lake City, UT).
Note: 2.5% Luria Broth Medium (LB Medium) and agarose/ampicillin plates were used in multiple steps throughout the experiment. Therefore, these components should be made prior to initiating the experiments, and stock availability should be ensured throughout the protocol, until the desired plasmid is confirmed by Sanger DNA sequencing.
Making 2.5% Luria Broth medium (LB medium)
Timing: 4–5 h
-
1.In a 1 L autoclave safe bottle, add:
-
a.25 g of Luria Broth Base (LB) powder (Invitrogen, cat# 12795-084).
-
b.Distilled water to 1,000 mL.
-
a.
-
2.
Mix LB powder and water.
Note: Powder will likely not dissolve completely until after the next step.
-
3.
Autoclave LB medium in a steam sterilizer at the temperature of 122°C, for 45 min.
Note: Leave the bottle cap slightly loose for pressure equalization to occur.
CRITICAL: Caution removing the hot liquid from the sterilizer, as it will be boiling hot.
-
4.
After LB medium cools down to approximately 25°C, tighten the cap and make sure to maintain this solution sterile while handling it.
CRITICAL: LB Medium has to be handled and kept in completely sterile environment throughout the experiments, otherwise, contamination might occur. Only open the LB medium bottle under a previously disinfected laminar flow hood and use sterilized instruments, such as sterile serological pipettes, to transfer LB medium into sterilized, clean flasks, prior to use.
-
5.
Store LB Medium at approximately 25°C, for up to a year.
Making agarose/ampicillin plates
Timing: 4–5 h
-
6.
Dilute ampicillin (VWR, cat# 71003-352) in UltraPure™ DNAse/RNase Free distilled water (Invitrogen™, cat# 10977015).
Note: In our lab, we chose to prepare ampicillin at a final concentration of 50 mg/mL. Separate ampicillin in aliquots and keep them at −20°C. For optimal performance, freeze-thaw cycles of the ampicillin aliquots is not recommended. Ampicillin should be stable in −20°C for up to 5 years.
-
7.In a 500 mL flask add:
-
a.6.25 g of LB powder.
-
b.2.5 g of Molecular Biology Grade Agarose LE (Thomas Scientific, cat# C9961-57).
-
c.Distilled water to 250 mL.
-
a.
-
8.
Heat up in the microwave until agarose is completely melted (approximately 2 min and 45 s).
Note: Make sure the solution does not boil by pausing the microwave and checking the solution continuously throughout the process.
-
9.
Let the agarose cool down to approximately 45°C–50°C, as higher temperatures can inactivate the ampicillin.
-
10.
Add ampicillin to the cooled agar to a final concentration of 100 μg/mL and mix.
CRITICAL: Beware that ampicillin is light and heat sensitive. Make sure to handle ampicillin with care, to avoid ampicillin degradation and posterior plate contamination.
-
11.
Pour approximately 20 mL of agarose/ampicillin mixture onto 10 cm petri dishes.
Note: The suggested volume of agarose/ampicillin mixture (250 mL) should be enough to pour 12 plates.
-
12.
Allow gel to solidify protected from light.
-
13.
After the agarose/ampicillin gel has solidified, wrap the plates with plastic wrap, to avoid gel dehydration.
-
14.
Store the plates in 4°C, for up to 2 months.
Note: Store the plates upside down (with the gel on the top plate) to avoid that condensation affects the proper consistency of the plates.
CRITICAL: Write down the date of confection on the plates and make sure to use plates within 2 months of production. Old and dehydrated plates will affect the growth efficiency of E. coli bacteria. Troubleshooting 1.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-BRD4 long isoform (Dilution 1:50) | Cell Signaling Technology | Cat# 13440; RRID: AB_2687578 |
| Rabbit monoclonal anti-Vinculin (Dilution 1:50) | Cell Signaling Technology | Cat# 13901; RRID: AB_2728768 |
| Bacterial and virus strains | ||
| NEB 5-alpha Competent E.coli (High Efficiency) | New England Biolabs | Cat# C2987H |
| Chemicals, peptides, and recombinant proteins | ||
| Betaine solution (5 M) | Sigma-Aldrich | Cat# B0300-5VL |
| Ampicillin antibiotic, sodium salt 25 g | VWR | Cat# 71003-352 |
| Restriction enzyme - BamHI | New England Biolabs | Cat# R0136S |
| Restriction enzyme - EcoRI | New England Biolabs | Cat# R0101S |
| Restriction enzyme - MluI | New England BioLabs | Cat# R0198S |
| NEBuffer™ 3:1 (Provided with restriction enzyme) | New England Biolabs | Cat# B7203S |
| Luria Broth Base | Invitrogen | Cat# 12795-084 |
| SOC Outgrowth Medium | New England Biolabs | Cat# B9020 |
| Molecular Biology Grade Agarose LE | Thomas Scientific | Cat# C9961-57 |
| Phusion™ High-Fidelity DNA Polymerase (2 U/μL) | Thermo Scientific™ | Cat# F-530L |
| Phusion™ G-C Buffer (Provided with Polymerase) | Thermo Scientific™ | Cat# F-530L |
| 10 mM dNTP Mix | Invitrogen | Cat# 100004893 |
| Cold Fusion Cloning Kit with Competent Cells | System Biosciences | Cat# MC010A-1 |
| QIAquick Gel Extraction Kit | QIAGEN | Cat# 28704 |
| QIAprep Spin Miniprep Kit | QIAGEN | Cat# 27106 |
| CompactPrep Plasmid Maxi Kit | QIAGEN | Cat# 12863 |
| Blasticidin antibiotic, S HCl (10 mg/mL), liquid | Gibco, Fisher Scientific | Cat# A1113903 |
| Critical commercial assays | ||
| ProteinSimple Wes 66–440 kDa separation module | ProteinSimple (Bio-Techne) | Cat# SM-W008 |
| Deposited data | ||
| NCBI Reference Sequence for assembly NM_058243.3 | NCBI/NIH | https://www.ncbi.nlm.nih.gov/nuccore/NM_058243.3 |
| Oligonucleotides | ||
| Cold fusion cloning BRD4-L forward primer: 5′ GGTGTCGTGACG CGGCTATGTCTGCGGAGAGCGG 3′ |
This paper | CF-BRD4-L_pLV-F |
| Cold fusion cloning BRD4-L reverse primer: 5′ CCATGGTGGCGGATCCC TAGGTGCGCTCAGAAAAGA 3′ |
This paper | CF-BRD4-L_pLV-R |
| Recombinant DNA | ||
| LentiV_Blast mammalian stable overexpression plasmid | Addgene; (Tarumoto et al., 2020) | Cat# 111887; RRID: Addgene_111887 |
| pcDNA5-Flag-BRD4-WT transient expression plasmid | Addgene, a gift from Dr. Kornelia Polyak (Shu et al., 2016) | Cat# 90331; RRID: Addgene_90331 |
| LentiV_Puro mammalian stable overexpression plasmid | Addgene, a gift from Christopher Vakoc (Tarumoto et al., 2020) | Cat# 111886; RRID: Addgene_111886 |
| pLentiTRE/rtTA inducible stable overexpression plasmid | A gift from Trudy Oliver (Bieniasz et al., 2015) | N/A |
| LentiV_Blast-BRD4-L stable overexpression plasmid | This paper | LentiV_Blast-BRD4-L |
| LentiV_Puro-BRD4-L stable overexpression plasmid | This paper | LentiV_Puro-BRD4-L |
| pLentiTRE/rtTA-BRD4-L inducible overexpression plasmid | This paper | pLentiTRE/rtTA-BRD4-L |
| Software and algorithms | ||
| GenBank | NCBI/NIH | https://www.ncbi.nlm.nih.gov/genbank/ RRID: SCR_002760 |
| Oligo Calculator, version 3.27 | Northwestern University (Kibbe, 2007) | http://biotools.nubic.northwestern.edu/OligoCalc.html |
| Image Lab, version 6.0.1 | Bio-Rad | RRID: SCR_014210 |
| Compass software for SW, version 4.0.0 | ProteinSimple (Bio-Techne) | https://www.proteinsimple.com/compass/downloads/ |
| Other | ||
| UltraPure™ DNase/RNase Free distilled water | Invitrogen™ | Cat# 10977015 |
| GeneRuler 100 bp Plus DNA Ladder | Thermo Scientific™ | Cat# SM0323 |
| GeneRuler 1 kb Plus DNA Ladder | Thermo Scientific™ | Cat# SM1334 |
| DS-11+Spectrophotometer | DeNovix | N/A |
| Master Cycler Pro thermocycler | Eppendorf | N/A |
| Mini-sub Cell GT System gel running system | Bio-Rad | N/A |
| Chemidoc MP Imaging System | Bio-Rad | N/A |
| New Brunswick Innova 40 benchtop shaker | Innova | N/A |
| Eppendorf ThermoMixer C | Eppendorf | N/A |
| ProteinSimple Wes System | ProteinSimple (Bio-Techne) | N/A |
Materials and equipment
Recipes
Ampicillin solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Ampicillin powder | 50 mg/mL | 5 g |
| UltraPure™ water | N/A | To 100 mL |
| Total | N/A | 100 mL |
CRITICAL: To ensure complete efficiency, store protected from light.
2.5% Luria Broth Medium
| Reagent | Final concentration | Amount |
|---|---|---|
| Luria Broth Base Powder | 25 mg/mL | 25 g |
| ddH2O | N/A | To 1,000 mL |
| Total | N/A | 1,000 mL |
CRITICAL: Keep and handle in completely sterile environment to avoid contamination.
Agarose/ampicillin plates
| Reagent | Final concentration | Amount |
|---|---|---|
| Luria Broth Base Powder | 2.5 mg/mL | 6.25 g |
| Mol. Biology Grade Agarose | 1% | 2.5 g |
| Ampicillin | 100 μg/mL | 500 μL |
| ddH2O | N/A | To 250 mL |
| Total | N/A | 250 mL |
CRITICAL: Store plates wrapped in plastic, to avoid dehydration. Ensure the gel is on the top plate, to avoid that condensation drips on the gel.
Step-by-step method details
Cold fusion primers design
Timing: 4–6 h
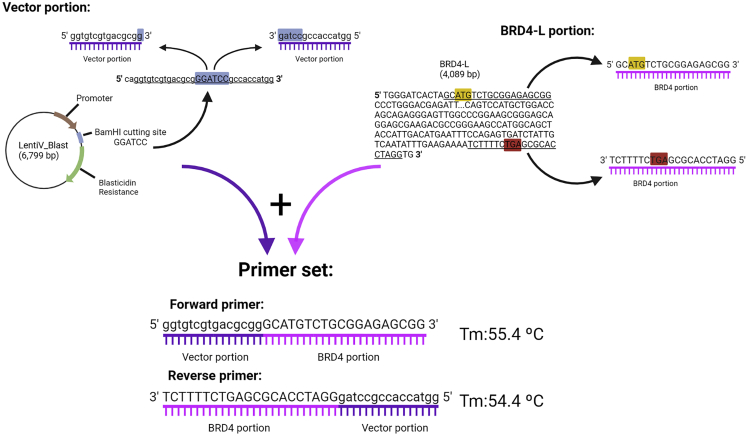
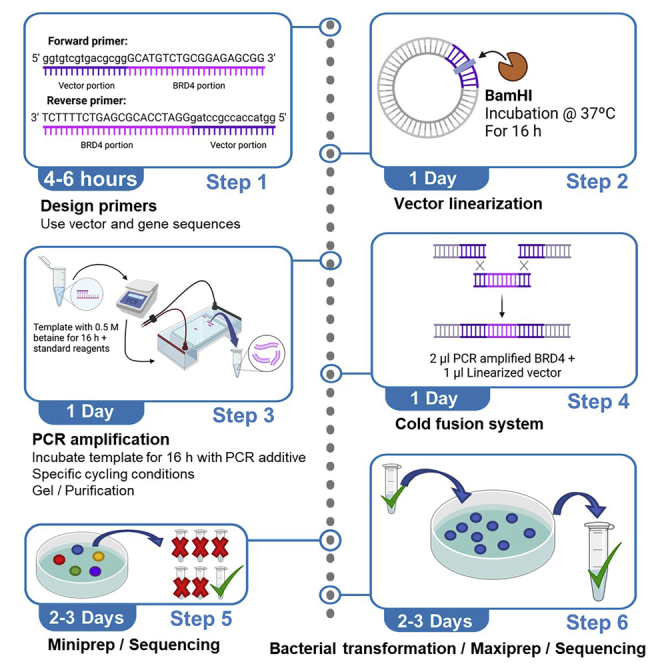
Following the instructions in the manufacturer’s protocol (Cold Fusion), we designed PCR (polymerase chain reaction) primers (Figure 1) that contained sequences of the cDNA that encoded BRD4-L (based on Reference Sequence NM_058243.3) in addition to sequences that encoded the overexpression vector (LentiV_Blast). We then used OligoCalc website (OligoCalc (Kibbe, 2007)) to analyze the full sequence of the prospective primers for potential hairpin formation and/or self-annealing. Targeting PCR efficiency due to maximum primer availability and specific template binding, we avoided using the primer pairs that were predicted to form hairpin or self-annealing. In addition, we only accepted forward and reverse primer pairs with differences in melting temperatures smaller than 5°C (please refer to the note below for detailed explanation). It is important to notice that, due to the limited options of primers, we accepted primers that allowed self-annealing, without hairpin formation.
-
1.Download the appropriate sequences.
-
a.Download the Reference Sequence for Homo sapiens bromodomain containing 4 (BRD4), transcript variant long, mRNA (GenBank: NM_058243.3) from the NCBI’s website, using the GenBank data base (BRD4-L RefSeq).
-
b.Download the sequence for LentiV_Blast (Addgene, cat# 111887) from Addgene’s website (LentiV_Blast).
-
a.
-
2.
Using Addgene’s sequence analyzer, identify in the sequence of overexpression vector (LentiV_Blast) the point of insertion of the gene to be overexpressed.
Note: In the sequence of LentiV_Blast, the point of insertion can be identified by the site including the sequence of the restriction enzyme of BamHI (GGATCC).
-
3.
Assemble prospective forward and reverse PCR primers, containing 18–20 bases complimentary to the gene of interest plus 15 bases corresponding to the vector.
CRITICAL: While selecting the 18–20 bases complimentary to the gene, make sure the forward primer includes the start codon (ATG) and the reverse primer includes one of the three stop codons (TAA, TAG and TGA).
-
4.
Use OligoCalc website (OligoCalc, (Kibbe, 2007)) to verify potential hairpin formation and self-annealing of the full sequence of the prospective primers.
-
5.
Make sure the difference in melting temperature between forward and reverse primers is smaller than 5°C.
Note: For melting temperature purposes, only the BRD4-L portion of the primer’s sequences should be considered.
-
6.
Order primer sets.
Note: Our lab uses custom DNA oligos from the company Integrated DNA Technologies (IDT). Primers are ordered with standard desalting purification and scale of 100 nmole.
Note: It is important to notice that the specific difference in primers melting temperature (Tm) of 5°C or less was determined in accordance to the instructions of the manufacturer of the DNA polymerase used (Phusion™, Thermo Scientific™, cat# F-530L - Phusion High-Fidelity DNA Polymerase). However, it is also relevant to understand that the Tm of a primer correlates with the temperature at which the primer anneals with the specific sequence of DNA we are intending to amplify during a PCR reaction, also known as annealing temperature (AT). AT is the temperature at which the primers bind, specifically, to the complimentary template region for which they were designed. To avoid unspecific annealing of the primers with the template, it is ideal that both the forward and the reverse primers anneal with the DNA at approximate temperatures. Therefore, a smaller difference in the Tm of the primers will increase the chances that the optimal temperature for annealing (AT) is similar between the forward and reverse primers pair.
Figure 1.
Designing cold fusion primers set
Tm: Melting temperature.
Vector linearization
Timing: 1 day
The overexpression plasmid LentiV_Blast is delivered to the lab as a circular molecule of DNA inserted into live bacteria. Addgene provides the bacteria as an agar stab and the tube with bacteria is maintained at 4°C for up to 2 weeks, during which the plasmid is amplified, and a glycerol stock is created. For detailed instructions of how to manipulate and maintain plasmids obtained by Addgene, please refer to the Addgene website here: Addgene - Bacteria stab. In order to prepare the plasmid to receive the insert of BRD4-L, we first had to grow the bacteria for 16 h in LB medium, containing 100 μg/mL of ampicillin. Then, we extracted the circular plasmids from the bacteria using QIAGEN’s CompactPrep Plasmid Maxi Kit (cat# 12863). Finally, we linearized the circular plasmid with the restriction enzyme BamHI (Figure 2).
-
7.
Using a disposable, sterile, pipette tip, collect the bacteria provided by Addgene from the agar stab.
Note: The bacteria should be visible to the naked eye, and it is enough to just dip the pipette tip into the stab, while touching the colony, without pipetting the content.
-
8.
Add the bacteria to a 500 mL flask containing 100 mL of 2.5% LB medium and 100 μg/mL of ampicillin, by dropping the pipette tip inside the flask.
-
9.
Cover the 500 mL flask with aluminum foil and grow the bacteria for 16 h, at 37°C, on a benchtop shaker incubator, using a speed of 1 g.
Note: Our lab uses New Brunswick Innova 40 benchtop shaker, in which the speed is converted to 240 rpm.
-
10.
Perform plasmid purification, using the CompactPrep Plasmid Maxi Kit (QIAGEN, cat# 12863).
Note: All of the sub-steps in this section were performed in accordance to manufacturer’s protocol (CompactPrep Plasmid Maxi Kit).
-
11.
Elute the plasmid in 70 μL of Buffer EB.
Note: Although QIAGEN’s CompactPrep Plasmid Maxi Kit protocol suggests plasmid elution with 100 μL or 200 μL of Buffer EB, our lab uses smaller volumes, to ensure the minimum concentration of 1 μg/μL.
-
12.
Measure the plasmid concentration with spectrophotometer.
Note: Our lab uses DS-11+ Spectrophotometer (DeNovix).
-
13.
Adjust the final concentration of the plasmid to 1 μg/μL.
-
14.
Using a thermocycler, digest 4 μg of LentiV_Blast with BamHI, at 37°C, for 16 h.
Note: Our lab uses Eppendorf Master Cycler Pro thermocycler.
CRITICAL: While adding the reagents for diagnostic digestion, make sure the restriction enzyme is the last component to be added to the digestion tube. Concerning incubation time, in previous experiments we attempted to digest LentiV_Blast with BamHI for 1 h and 3 h, however, we observed that this amount of time was not enough to digest the plasmid completely.
-
15.
Run the digestion product in 0.8% agarose gel (Thomas Scientific, cat# C9961-57), for 35–40 min, using a voltage of 110 V.
Note: Make sure to also run circular LentiV_Blast plasmid as control (Figure 2). Add 10 μL of DNA ladder (GeneRuler 1 kb Plus DNA Ladder, Thermo Scientific™, cat# SM1334) to each side of the gel.
-
16.
Remove the digested plasmid from the agarose gel, using the QIAquick Gel Extraction Kit (QIAGEN, cat# 28704), in accordance to manufacturer’s protocol (QIAquick Gel Extraction Kit).
-
17.
Elute the linearized vector in 30 μL of UltraPure™ water.
Note: Although QIAGEN’s QIAquick Gel Extraction Kit protocol suggests plasmid elution with 50 μL of water, our lab uses smaller volumes, to ensure the optimal concentration.
-
18.
Measure the linearized vector’s concentration with spectrophotometer.
-
19.
If possible, adjust the final concentration of the linearized vector to 25 ng/μL.
Plasmid linearization by restriction enzyme digestion
| Reagent | Amount |
|---|---|
| Plasmid (Concentration 1 μg/μL) | 4 μL |
| 3.1 NEBuffer™ | 5 μL |
| ddH2O (To 50 μL) | 37 μL |
| BamHI enzyme | 4 μL |
Enzymatic digestion cycling conditions
| Steps | Temperature | Time | Cycles |
|---|---|---|---|
| Enzymatic digestion | 37°C | 16 h | 1 |
| Hold | 4°C | forever | |
Figure 2.
Agarose gel electrophoresis of digested plasmid (linearized vector) and control uncut plasmid (circular plasmid)
The reference band for estimating the plasmid size (6,799 bp) has been identified on the ladder as 7,000 base pairs (bp). Reference ladder used is GeneRuler 1 kb Plus DNA Ladder (Thermo Scientific™, cat# SM1334), 10 μL were added to each side of the gel.
PCR amplification of BRD4-L
Timing: 1 day
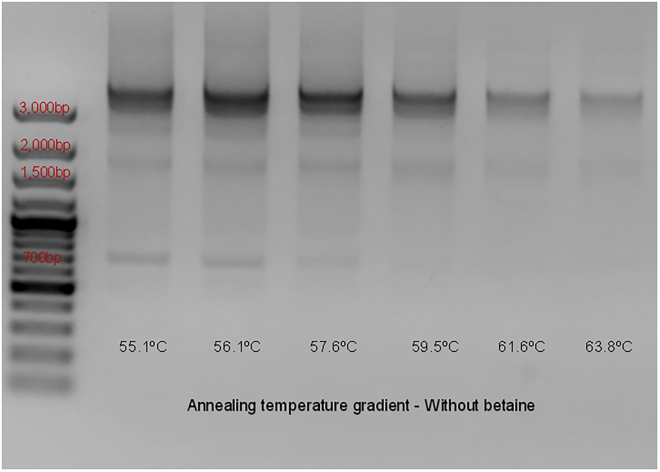
The PCR amplification of BRD4-L can be very challenging, because this is a large gene (4,089 bp) which contains several G-C rich regions. Some of the BRD4-L fragments present a G-C content as high as 66.1%. This becomes especially complicated when using cold-fusion primers, which can be prone to self-annealing. After receiving the primers, we performed an annealing temperature (AT) gradient experiment and verified that these primers produced unspecific amplifications bands of sizes ∼700 bp and between 1,500 bp and 2,000 bp (Figure 3). In addition, after sequencing bands that appeared consistent with BRD4-L amplification (∼4,000 bp) we verified that there were multiple sites with severe changes of the expected sequences, with over 300 bp mismatch.
Figure 3.
Agarose gel electrophoresis of the first attempt to amplify BRD4-L via PCR, by performing an annealing temperature (AT) gradient, without betaine
For illustration purposes, only the lower range gradient (10°C lower than the mid-point) is observed above. No amplification was observed with AT higher than 63.8°C. Non-specific amplification can be identified around sizes 700 bp and 1,700 bp. The reference band for estimating the product size (4,089 bp) has been identified on the ladder above 3,000 base pairs (bp). Reference ladder used is GeneRuler 100 bp Plus DNA Ladder (Thermo Scientific™, cat# SM0323), 10 μL were added to the left side of the gel.
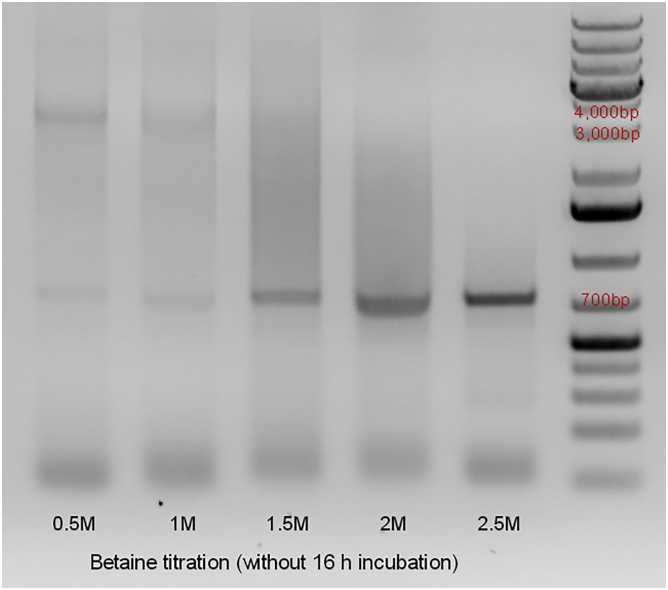
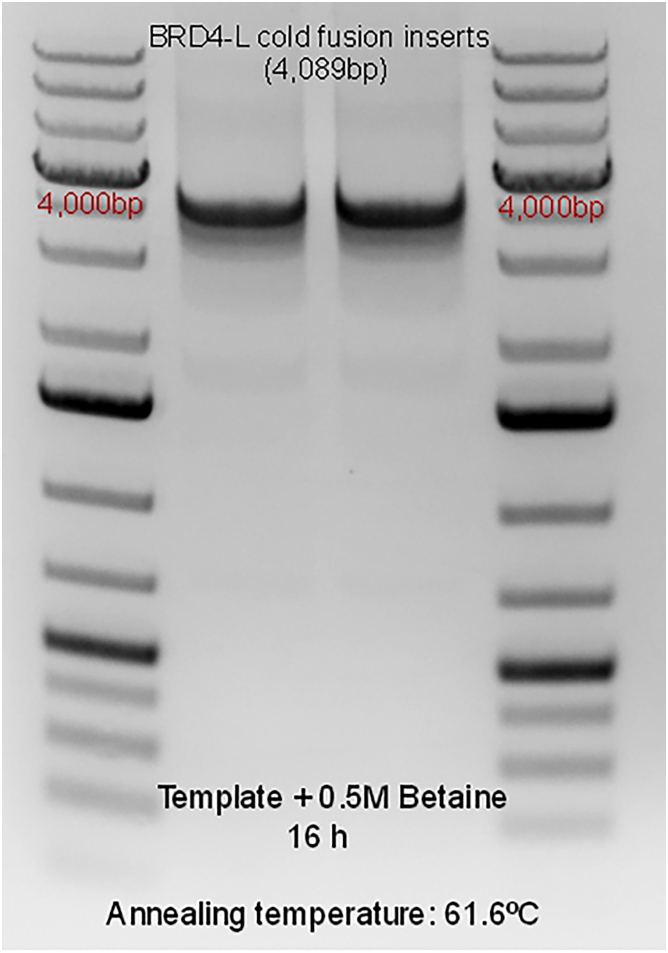
After researching different strategies involved in the PCR amplification of G-C rich templates (Mamedov et al., 2008; Green and Sambrook, 2019), we decided to investigate the use of betaine (Sigma-Aldrich, cat# B0300-5VL), as a PCR additive in the amplification of BRD4. Betaine has been widely used in amplification of G-C rich sequences and it acts by reducing the melting temperature of the DNA, decreasing the formation of secondary structures formed by the regions of DNA rich in G-C (Green and Sambrook, 2019) and stabilizing the DNA by equalizing the contribution of GC- and AT-base pairs (5M Betaine - Sigma-Aldrich). Our approach with betaine included an attempt to titrate the ideal molar concentration of betaine that would allow for amplification of BRD4-L (Figure 4). However, the use of betaine as a mere additive to the PCR reactive failed to amplify BRD4-L in multiple efforts, producing inefficient BRD4-L amplification and non-specific amplification. Taking into consideration the fact that the template might have multiple secondary structures prior to the PCR reactions that might not be undone in the process of denaturation, and considering the mode of action of betaine, we attempted to incubate the template in betaine for 16 h (Figure 5). After 16 h incubation of the template, BRD4-L was successfully amplified via PCR reaction in every attempt (Figure 5), including with the use of cold fusion primers designed for different vectors (data not shown).
Figure 4.
Agarose gel electrophoresis of the attempt to amplify BRD4-L via PCR, by performing betaine titration, without incubating the template in betaine for 16 h (overnight)
Non-specific amplification can be identified around sizes 700 bp. The reference band for estimating the product size (4,089 bp) has been identified on the ladder above 4,000 base pairs (bp). Reference ladder used is GeneRuler 1 kb Plus DNA Ladder (Thermo Scientific™, cat# SM1334), 10 μL were added to the right side of the gel.
Figure 5.
Agarose gel electrophoresis of the specific amplification of BRD4-L via PCR, after incubation of the DNA template in 0.5 M betaine for 16 h (overnight)
The reference band for estimating the product size (4,089 bp) has been identified on the ladder as 4,000 base pairs (bp). Reference ladder used is GeneRuler 1 kb Plus DNA Ladder (Thermo Scientific™, cat# SM1334), 10 μL were added to each side of the gel.
Therefore, to overcome issues encountered around the amplification of BRD4-L, we have determined that the following measures are necessary for a successful BRD4-L PCR:
-
a.
Incubate the template with 0.5 M betaine (Sigma-Aldrich, cat# B0300-5VL) for 16 h, prior to PCR reaction. This procedure was established as critical based on our lab experience.
-
b.
Use Phusion™ G-C Buffer instead of Phusion™ HF Buffer (both are provided with the enzyme). Troubleshooting 2.
-
c.
Use initial denaturation time of 3 min, which is longer, to ensure that the entire template gets denatured. This modification is suggested in the Phusion™ polymerase protocol (Phusion High-Fidelity DNA Polymerase).
-
d.
Be aware that the annealing temperature (AT) might be different than what is recommended to be used with Phusion™ polymerase, because of the presence of betaine in the PCR mix. For our specific primers, the advised AT was 65.1°C, however, we found after gradient experiments that AT of 61.6°C produced better results. In order to find the ideal AT for your primers, we suggest performing a gradient experiment (Figure 3) using the temperature calculated via Thermo Fisher Scientific’s temperature calculator (Tm calculator) as mid-point. ThermoFisher Scientific’s Tm calculator will estimate the ideal AT for your primer pair, based on the Tm for each primer. It is ideal to test temperatures that range from approximately 10°C higher to 10°C lower than the mid-point. Troubleshooting 3.
-
e.
To avoid amplification of unspecific products, we found that shorter annealing times (Mamedov et al., 2008) and a reduced number of cycles (Phusion High-Fidelity DNA Polymerase) decreased the chances of our primers biding to unspecific targets. We found these steps to be critical to eliminate non-specific products and to increase efficiency in our reactions. Troubleshooting 4.
PCR steps are as follows:
-
20.
In a 0.2 mL PCR tube, combine the DNA template (pcDNA5-Flag-BRD4-WT, transient expression plasmid, Addgene, cat# 90331) with 5 M betaine and UltraPure™ water.
-
21.
Vortex and spin.
-
22.
Place the tube in 4°C for 16 h.
-
23.
Add the PCR reagents to the same tube.
Note: Add all the reagents to the tube in which the template was incubated with 0.5 M betaine. Additional 5 M betaine and ddH2O will be necessary.
CRITICAL: Make sure the polymerase is the last reagent to be added to the tube.
-
24.
Vortex and spin.
-
25.
Using a thermocycler, conduct PCR in accordance to the PCR cycling conditions indicated below.
-
26.
Run the PCR product in 0.8% agarose gel, for 35–40 min, using a voltage of 110 V (Figure 5).
-
27.
Remove the PCR product from the agarose gel, using the QIAquick Gel Extraction Kit, in accordance to manufacturer’s protocol (QIAquick Gel Extraction Kit).
-
28.
Elute the PCR product in 30 μL of UltraPure™ water.
Note: Although QIAGEN’s QIAquick Gel Extraction Kit protocol suggests plasmid elution with 50 μL of water, our lab uses smaller volumes, to ensure the optimal concentration.
-
29.
Measure the PCR product’s concentration with spectrophotometer.
Template incubation with betaine
| Reagent | Amount |
|---|---|
| pcDNA5-Flag-BRD4-WT (Template) (Concentration 10 ng/μL) | 3 μL |
| 5 M Betaine | 1.5 μL |
| ddH2O (To 15 μL) | 10.5 μL |
PCR reaction master mix
| Reagent | Amount |
|---|---|
| Template incubated with betaine (DNA + betaine + water) | 15 μL |
| 5 M Betaine | Add 3.5 μL (Final 5 μL) |
| ddH2O (To 50 μL) | Add 15 μL (Final 25.5 μL) |
| Phusion™ G-C Buffer | 10 μL |
| 100 μM Reverse primer | 2.5 μL |
| 100 μM Forward primer | 2.5 μL |
| 10 mM dNTP | 1 μL |
| Phusion™ High-Fidelity DNA Polymerase (2 U/μL) | 0.5 μL |
PCR cycling conditions
| Steps | Temperature | Time | Cycles |
|---|---|---|---|
| Initial Denaturation | 98°C | 3 min | 1 |
| Denaturation | 98°C | 10 s | 20 cycles |
| Annealing | 61.6°C | 5 s | |
| Extension | 72°C | 2 min 40 s | |
| Final extension | 72°C | 5 min | 1 |
| Hold | 4°C | forever | |
CRITICAL: Cycling times and the number of cycles are critical for proper BRD4-L amplification. Troubleshooting 4.
Ligating BRD4-L and linearized LentiV_blast with Cold Fusion™
Timing: 1 day
After succeeding in the challenging task of amplifying BRD4-L via PCR, we decided to use Cold Fusion™ cloning kit (System Biosciences, cat# MC010A-1) to ligate our BRD4-L PCR inserts onto LentiV_Blast linearized vectors. We have found Cold Fusion™ to be a simple and quick technique that provided us with a ligation strategy that diminished the chances of promoting alterations to the BRD4-L insert that might impair its expression.
-
30.
Adjust the concentration of the BRD4-L insert, ensuring it is within the desired range (20–200 ng/μL).
-
31.
Adjust the concentration of the LentiV_Blast linearized vector, ensuring it is within the desired range (10–100 ng/μL).
-
32.Perform Cold Fusion™ cloning, in accordance to the manufacturer’s protocol (Cold Fusion), making the following changes:
-
a.Ensure the proportion of BRD4-L insert:LentiV_Blast linearized vector is 3:2 molar ratio. Troubleshooting 5.
-
b.Add 250 μL pre-warmed SOC Medium (NEB, cat# B9020) and incubate the tube at 37°C, shaking at a speed of 0.24 g, for 1 h.Note: Our lab uses Eppendorf ThermoMixer C in which the referred speed converts to 1,200 rpm.
-
c.Pre-warm a culture plate containing ampicillin in a 37°C incubator for 1 h.Note: For instructions on how to make agarose/ampicillin plates, please refer to the “recipes” section.
-
d.Spread the entire volume of the reaction (260 μL) onto the pre-warmed culture plate containing ampicillin. Troubleshooting 6.
-
a.
-
33.
Incubate the plate with Cold Fusion™ competent cells spread in 37°C for 16–20 h.
Note: Make sure the gel is on the top plate, i.e., the bacteria will growth “upside-down”.
-
34.
After bacterial growth, wrap the plates in plastic film and place them in 4°C until ready to perform miniprep.
Note: Alternatively, set up culture growth for 16 h, for miniprep execution on the next day. To ensure cloning efficiency, it is not recommended that the miniprep be performed further than a week after bacterial growth.
Note: After multiple attempts, our lab defined that the ideal proportion of DNA (BRD4-L) to Vector (LentiV_Blast) is in fact 3:2 molar ratio and not 1:1 or 1:2, as suggested by the cold fusion cloning kit’s protocol.
Pause point: After bacterial growth, you can wrap the plates with plastic film and store them in 4°C until ready to perform miniprep, however, it is advised that miniprep be performed within 1 week, to ensure successful cloning.
CRITICAL: Make sure to observe the changes to the manufacture’s protocol, as our lab has found them to be critical for the success of Cold Fusion™ cloning.
Cold fusion reaction mix
| Reagent | Amount |
|---|---|
| BRD4-L PCR amplified insert (25 ng/μL) | 2 μL |
| LentiV_Blast linearized vector (25 ng/μL) | 1 μL |
| 5× cloning master mix | 2 μL |
| ddH2O (To 10 μL) | 5 μL |
CRITICAL: Make sure the concentrations of the linearized vector (10–100 ng/μL) and the DNA insert (20–200 ng/μL) are within appropriate range. Use 3:2 molar ratio of DNA insert to linearized vector.
Miniprep, clone screening, and sequencing
Timing: 2–3 days
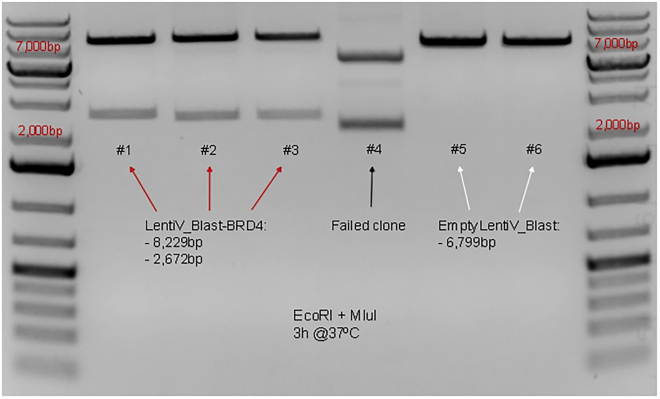
If the processes of vector linearization, PCR amplification and Cold Fusion™ cloning are completed successfully, the steps to be followed for miniprep and clone screening should occur smoothly. The most critical point for clone screening is to define the appropriate combination of restriction enzymes to use with your new plasmid (LentiV_Blast-BRD4-L) that will allow for identification of specific band sizes. Specific band sizes for diagnostic digestion of LentiV_Blast-BRD4-L should be different from empty LentiV_Blast vectors and from plasmids with improperly amplified BRD4-L, i.e., BRD4-L inserts missing fragments. While performing diagnostic digestion of LentiV_Blast-BRD4-L, we determined that the combination of the enzymes EcoRI and MluI digest the plasmid creating bands of sizes 8,229 bp and 2,672 bp. The same combination of restriction enzymes should not cut empty LentiV_Blast vectors, producing a single band (Figure 6).
-
35.
Add 2 mL of 2.5% LB Media with 100 μg/mL of ampicillin to 15 mL polypropylene tubes.
-
36.
Cover tubes with aluminum foil.
-
37.
Incubate 15 mL tubes for 16 h in a benchtop shaker at 1 g, set for temperature of 36°C.
Note: Our lab uses New Brunswick Innova 40 benchtop shaker, in which the speed is converted to 240 rpm. Troubleshooting 7.
CRITICAL: Bacterial growth, prior to plasmid isolation via miniprep, should be performed at 36°C. We have found that temperatures higher than 36°C (37°C, for example) promote occurrence of multiple mutations and poor colony yield.
-
38.
Perform miniprep using the QIAprep Spin Miniprep Kit (QIAGEN, cat# 27106), in accordance to manufacturer’s protocol (QIAprep Spin Miniprep Kit).
-
39.
Elute the plasmid in 30 μL of EB buffer.
Note: Although QIAGEN’s QIAquick Gel Extraction Kit protocol suggests plasmid elution with 50 μL of water, our lab uses smaller volumes, to ensure the optimal concentration.
-
40.
Measure the plasmid concentration with spectrophotometer.
-
41.Screen clones performing enzymatic digestion with the restriction enzymes EcoRI (NEB, cat# R0101S) and MluI (NEB, cat# R0198S):
-
a.Using a thermocycler, digest 500 ng of LentiV_Blast-BRD4-L with EcoRI and MluI, at 37°C, for 3 h.
-
b.Run the digestion product in 0.8% agarose gel (Thomas Scientific, cat# C9961-57), for 35–40 min, using voltage of 100 V.
-
c.Detect which clones yielded plasmids that appear to have the full length LentiV_Blast-BRD4-L plasmid (Figure 6).
-
a.
Note: We observed that plasmids with yield concentration of 500 ng/μL or higher were more likely to have the full sequence of BRD4-L and, therefore, were more likely to be identified as successful clones after diagnostic digestion.
-
42.
Send the plasmid candidates for sequencing, to ensure that the correct sequence for BRD4-L has been cloned into LentiV_Blast.
Note: All of our sequencing experiments were performed by The OMRF Sanger DNA Sequencing facility.
CRITICAL: To avoid formation of secondary structures and ensure that the sequencing process occurs smoothly, betaine should be added to the LentiV_Blast-BRD4-L plasmid to a concentration of 0.1 M, prior to Sanger DNA sequencing.
Diagnostic digestion using restriction enzymes
| Reagent | Amount |
|---|---|
| LentiV_Blast-BRD4-L | 500 ng (volume may vary) |
| 3.1 NEBuffer™ | 5 μL |
| ddH2O | To 50 μL |
| EcoRI enzyme | 1 μL |
| MluI enzyme | 1 μL |
Enzymatic digestion cycling conditions
| Steps | Temperature | Time | Cycles |
|---|---|---|---|
| Enzymatic digestion | 37°C | 3 h | 1 |
| Enzyme inactivation | 80°C | 20 min | 1 |
| Hold | 4°C | forever | |
Figure 6.
Agarose gel electrophoresis of the diagnostic digestion of LentiV_Blast-BRD4-L plasmids
Red arrows indicate miniprep of successful clones (#1–#3). Black arrow indicates a failed clone (#4). White arrows indicate that the isolated clones presented empty LentiV_Blast vectors, with no BRD4-L inserts (#5-#6). The reference bands for estimating the expected band sizes after diagnostic digestion (8,229 bp and 2,672 bp) have been identified on the ladder as 2,000 and 7,000 base pairs (bp). Reference ladder used is GeneRuler 1 kb Plus DNA Ladder (Thermo Scientific™, cat# SM1334), 10 μL were added to each side of the gel.
Bacterial transformation, maxiprep, and sequencing
Timing: 2–3 days
Once we obtained a LentiV_Blast-BRD4-L plasmid that was identified as successful clone by diagnostic digestion and then confirmed by Sanger DNA sequencing, we moved on to the last step of cloning. At the last step, we introduced the LentiV_Blast-BRD4-L into DH5-alpha competent E. coli (NEB, cat# C2987H) and grew these cells onto agarose/ampicillin plates. We finally expanded the bacterial colony for 16 h, extracted the final copies of plasmid using the CompactPrep Plasmid Maxi Kit (QIAGEN, cat# 12863) and confirmed its sequence again.
-
43.
Once confirmed by Sanger DNA sequencing, use UltraPure™ water to dilute the LentiV_Blast-BRD4-L plasmid to 5 ng/μL.
-
44.
While keeping the entire system in ice, add 1 μL of LentiV_Blast-BRD4-L plasmid (5 ng/μL) to a 1.5 mL tube containing 50 μL of DH5-alpha competent E. coli (NEB, cat# C2987H).
CRITICAL: DH5-alpha competent E. coli are very sensitive to temperature changes, store them in −80°C until ready to use. The thaw process should be conducted in ice, when you are ready to introduce the plasmid.
-
45.
Incubate in ice for 30 min.
-
46.
Transfer the 1.5 mL tube containing DH5-alpha cells and plasmid to a water bath set for 42°C for exact 30 s.
CRITICAL: Do not exceed the timing for incubation at 42°C (30 s).
-
47.
Immediately cool down the solution by immersing it in ice for 2 min.
-
48.
Add 250 μL of pre-warmed (37°C for 1 h) SOC Medium (NEB, cat# B9020).
-
49.
Incubate the tube at 37°C, shaking at a speed of 0.24 g, for 1 h.
Note: Our lab uses Eppendorf ThermoMixer C in which the referred speed is 1,200 rpm.
-
50.
Spread 50 μL of the solution containing competent E. coli onto a pre-warmed (37°C for 1 h) agarose/ampicillin plate.
-
51.
Incubate the agarose/ampicillin plate with the DH5-alpha cell spread for 16 h, in 37°C.
Note: Make sure the gel is on the top plate, i.e., the bacteria will growth “upside-down”.
-
52.
After bacterial growth, wrap the plates in plastic film and place them in 4°C until ready to perform maxiprep.
Note: Alternatively, set up culture growth for 16 h, for maxiprep execution on the next day.
Pause point: After bacterial growth, you can wrap the plates with plastic film and store them in 4°C until ready to perform maxiprep, however, it is advised that maxiprep be performed within 1 week, to ensure successful cloning.
-
53.
Using a disposable, sterile, pipette tip, pick one of the bacterial colonies on the agarose/ampicillin plate and drop the pipette tip into a 500 mL flask containing 100 mL of 2.5% LB medium, containing 100 μg/mL of ampicillin.
Note: To add the bacteria, drop the pipette tip inside the flask, without pipetting up and down the LB media.
-
54.
Cover the flask with aluminum foil.
-
55.
Incubate the 500 mL flask for 16 h on a benchtop shaker at 1 g, set for temperature 36°C.
Note: Our lab uses New Brunswick Innova 40 benchtop shaker, in which the speed is converted to 240 rpm. Troubleshooting 8.
CRITICAL: Bacterial growth, prior to plasmid isolation via maxiprep, should be performed at 36°C. We have found that temperatures higher than 36°C (37°C, for example) promote occurrence of multiple mutations and poor colony yield.
-
56.
Perform plasmid purification using the CompactPrep Plasmid Maxi Kit, in accordance to manufacturer’s protocol (CompactPrep Plasmid Maxi Kit).
-
57.
Elute the plasmid in 70 μL of EB buffer.
Note: Although QIAGEN’s QIAquick Gel Extraction Kit protocol suggests plasmid elution with 100 μL–200 μL of EB buffer, our lab uses smaller volumes, to ensure the optimal concentration.
-
58.
Measure the plasmid concentration with spectrophotometer.
-
59.
Send the plasmid for sequencing, to ensure that the correct sequence for BRD4-L has been cloned into LentiV_Blast.
Note: All of our sequencing experiments were performed by The OMRF Sanger DNA Sequencing facility.
CRITICAL: To avoid formation of secondary structures and ensure that the sequencing process occurs smoothly, 0.1 M Betaine should be added to the LentiV_Blast-BRD4-L plasmid prior to Sanger DNA sequencing
Expected outcomes
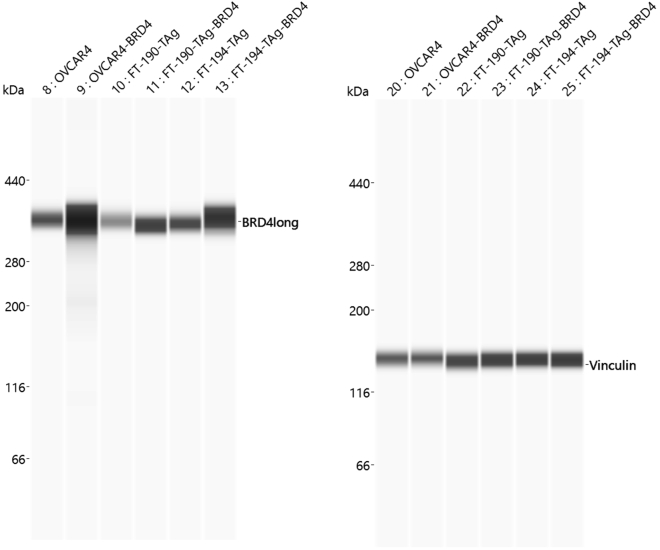
If the gene BRD4-L is successfully amplified and cloned into the overexpression plasmid LentiV_Blast, you should be able to overexpress BRD4-L in all mammalian cell lines of your choice. After lentiviral infection of mammalian cells with LentiV_Blast-BRD4-L, the transformed cells should survive selection with Blasticidin antibiotic (Fisher Scientific, cat# A1113903), and the resulting selected cells should overexpress BRD4-L, which can be confirmed via protein expression assay (Figure 7).
Figure 7.
Overexpression of BRD4-L in mammalian cells
ProteinSimple Wes blots show that the mammalian cell lines Ovcar4, FT-190-TAg and FT-194-TAg when infected with the lentiviral plasmid LentiV_Blast-BRD4-L successfully overexpressed BRD4-L, after selection with Blasticidin (Left panel). A consistent amount of protein was assayed by Wes for each cell line pair (parental and overexpression), as can be confirmed by the loading control Vinculin (Right panel). Antibodies used: Rabbit monoclonal anti-BRD4 long isoform, Cell Signaling, cat# 13440 – dilution 1:50; Rabbit monoclonal anti-Vinculin, Cell Signaling, cat# 13901 – dilution 1:50. ProteinSimple Wes 66–440 kDa separation module (cat# SM-W008).
Limitations
Limitations to the protocol have been described throughout the protocol, associated to specific crucial steps to avoid certain limitations. Although we believe that the strategies implemented in this protocol can be applied to other long genes, with G-C rich sequences, we have not attempted to amplify genes other than BRD4-L with the methods described here.
Troubleshooting
While amplifying BRD4-L and ligating BRD4-L insert with linearized LentiV_Blast vector, several steps were found to be critical. See below potential issues that may arise while attempting to clone long, G-C rich genes into overexpression plasmids, and essential steps that must be taken to resolve the problems.
Problem 1
No bacterial colonies were observed on agarose/ampicillin plates after bacterial growth of Cold Fusion™ competent cells, or later after introducing the plasmids into DH5-alpha competent cells.
Potential solution
Check when your plates were made, old plates will affect the yield of your cloning experiments.
Problem 2
No BRD4-L (or your gene of interest) amplification observed on the gel.
Potential solution
Make sure you use Phusion™ G-C buffer and not the standard Phusion™ HF buffer.
Problem 3
No BRD4-L (or your gene of interest) amplification observed on the gel.
Potential solution
Make sure to run a temperature gradient for annealing temperatures, every time you order a new primer set. Annealing temperature gradient experiments will help you determine the appropriate annealing temperature for each primer set and save time troubleshooting experiments.
Problem 4
No BRD4-L (or your gene of interest) amplification observed on the gel or multiple unspecific bands detected.
Potential solution
Adjust the cycling times and shorten the cycle repetition. We found that shorter annealing times and less cycle repetitions (20 cycles) were two great strategies that, combined, eliminated unspecific products nearly completely.
Problem 5
No colony formation after bacterial growth of Cold Fusion™ competent cells.
Potential solution
Make sure you used 3:2 molar ratio of DNA insert to linearized vector.
Problem 6
No colony formation after bacterial growth of Cold Fusion™ competent cells.
Potential solution
Make sure to add and spread the entire volume of Cold Fusion™ reaction, i.e., 260 μL.
Problem 7
Colonies appear to have yielded the full length LentiV_Blast-BRD4-L during diagnostic digestion, but showed altered fragments after DNA sequencing.
Potential solution
Make sure the bacterial clones are set up to grow in 36°C. Bacteria will grow slower at this temperature and will be less likely to alter your plasmid’s sequence due to replication mistakes.
Problem 8
The plasmid was successfully confirmed after miniprep, but the sequencing results after maxiprep suggest that the sequence for BRD4-L (or your gene of interest) has changed.
Potential solution
Make sure the bacterial clones are set up to grow for 16 h, in 36°C. Bacteria will grow slower at this temperature and will be less likely to alter your plasmid’s sequence due to replication mistakes.
Alternatives: At the time of this publication, no alternatives to the commercial kits used in this protocol had been tested.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Magdalena Bieniasz (Magdalena-Bieniasz@omrf.org).
Materials availability
At the time of submission of this publication, plasmids generated in this study have not yet been deposited into repositories, but are available upon request.
Acknowledgments
The authors would like to acknowledge the following funding support: Dr. Bieniasz’s National Institute of Health (NIH) grant R21 CA264573-01A. The authors would also like to acknowledge The OMRF Sanger DNA Sequencing facility and Sheryl Christofferson for all the sequencing services provided, which were crucial to build the plasmids properly. Finally, the authors would like to thank Trudy Oliver, from the Huntsman Cancer Institute, Salt Lake City, UT for kindly providing the pLentiTRE/rtTA inducible overexpression plasmid.
Author contributions
A.L.D.-B. is responsible for executing the experiments and processing all of the results performed in this study, and for writing the manuscript. M.C., L.W., and L.W. assisted with some of the technical provisions, in particular, confection of LB Medium, and agarose and ampicillin plates, and assisted in scientific discussions involving the protocols and the majority of experiments involved in this study. M.B. supervised the project and edited the manuscript. All authors have reviewed, edited, and approved the final version of this manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Ana Luiza Drumond-Bock, Email: analuiza-bock@omrf.org.
Magdalena Bieniasz, Email: magdalena-bieniasz@omrf.org.
Data and code availability
All of the data reported in this study are available upon request.
No codes were generated in this study.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Bieniasz M., Radhakrishnan P., Faham N., De La O J.-P., Welm A.L. Preclinical efficacy of ron kinase inhibitors alone and in combination with PI3K inhibitors for treatment of sfRon-expressing breast cancer patient-derived xenografts. Clin. Cancer Res. 2015;21:5588–5600. doi: 10.1158/1078-0432.ccr-14-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M.R., Sambrook J. Polymerase chain reaction (PCR) amplification of GC-rich templates. Cold Spring Harb. Protoc. 2019;2019 doi: 10.1101/pdb.prot095141. pdb.prot095141. [DOI] [PubMed] [Google Scholar]
- Kibbe W.A. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res. 2007;35:W43–W46. doi: 10.1093/nar/gkm234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamedov T.G., PIenaar E., Whitney S.E., Termaat J.R., Carvill G., Goliath R., Subramanian A., Viljoen H.J. A fundamental study of the PCR amplification of GC-rich DNA templates. Comput. Biol. Chem. 2008;32:452–457. doi: 10.1016/j.compbiolchem.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu S., Lin C.Y., He H.H., Witwicki R.M., Tabassum D.P., Roberts J.M., Janiszewska M., Huh S.J., Liang Y., Ryan J., et al. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature. 2016;529:413–417. doi: 10.1038/nature16508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarumoto Y., Lin S., Wang J., Milazzo J.P., Xu Y., Lu B., Yang Z., Wei Y., Polyanskaya S., Wunderlich M., et al. Salt-inducible kinase inhibition suppresses acute myeloid leukemia progression in vivo. Blood. 2020;135:56–70. doi: 10.1182/blood.2019001576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All of the data reported in this study are available upon request.
No codes were generated in this study.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.