Abstract
The construction of human organs on demand remains a tantalizing vision to solve the organ donor shortage. Yet, engineering tissues that recapitulate the cellular and architectural complexity of native organs is a grand challenge. The use of organ building blocks (OBBs) composed of multicellular spheroids, organoids, and assembloids offers an important pathway for creating organ-specific tissues with the desired cellular to tissue-level organization. Here, we review the differentiation, maturation, and 3D assembly of OBBs into functional human tissues and, ultimately, organs for therapeutic repair and replacement. We also highlight future challenges and areas of opportunity for this nascent field.
1. Introduction
Every day over 150 people join the more than 100,000 patients on the U.S. transplant waitlist and nearly 20 people die as they wait for an available donor organ (Organdonor.gov). Post transplantation, many recipients will suffer acute rejection within their first year, while chronic rejection and side effects from immunosuppression remain lifelong threats. Moreover, the short viability window of life-saving donor organs limits their deployment to remote locations, resulting in geographical healthcare disparities. The creation of autologous, on-demand organs could overcome these challenges. This vision has been the holy grail of tissue engineering since its inception. However, despite decades of intense research, the biomanufacturing of functional organ-specific tissues remains elusive.
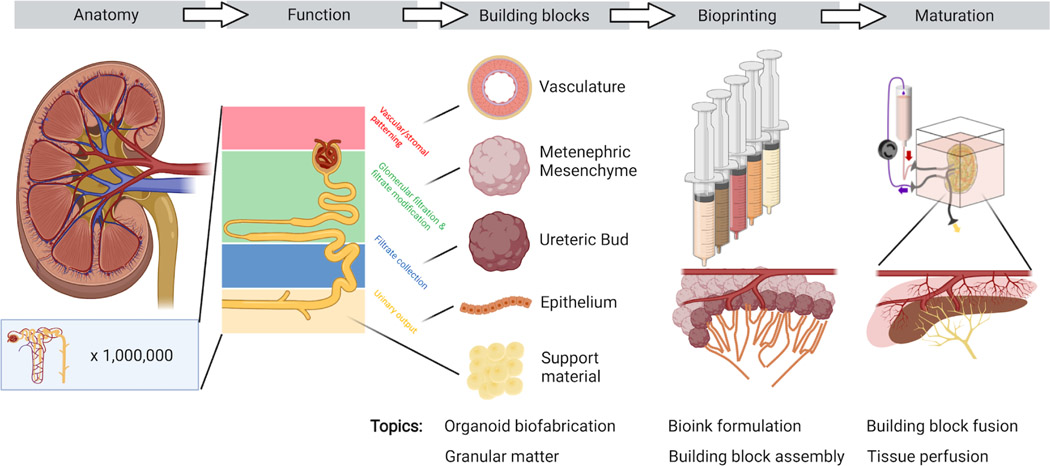
There are myriad difficulties in biomanufacturing human organs, including cell sourcing, vascularization, maturation, attaining physiologic levels of function, developing standards for quality control and safety, and translating to clinical practice. Despite these vast hurdles in writing human organs and organ-specific tissues, rapid advances in 3D imaging, genomics, and transcriptomics now allow us to read these tissues with unprecedented spatial and temporal detail. Emerging organ-specific and whole-embryo cell atlases offer precise instruction-sets for assembling de novo tissues (Srivatsan et al., 2021). Yet, paradoxically, the more detail obtained by reading organs, the more daunting the task of writing them seems. For example, single cell studies of adult kidneys reveal over 20 different cell types arranged into 1M+ nephrons per kidney (Park et al., 2018), which are responsible for blood filtration, reabsorption, secretion, and hemostasis (Fig. 1). Each nephron contains cells arranged into glomerular and tubular segments with invading and wrapping capillaries that carry out these critical functions. As another example, closer inspection of the adult human heart has revealed that the traditional delineation of left and right ventricular and atrial cardiomyocytes hides deeper levels of complexity. Using single cell RNA sequencing, researchers have uncovered the existence of five distinct and spatially localized classes of ventricular cardiomyocytes as well as five classes of atrial cardiomyocytes (Litviňuková et al., 2020). Indeed, writing de novo human organs that recapitulate their native counterparts would require the growth, differentiation, maturation, and precise patterning of not just a few, but tens of different cell types.
Figure 1:
Biomanufacturing human tissues from organ building blocks.
The grand challenge of organ engineering can therefore be framed as closing the gap between our ability to read organs and our ability to write them. Fortunately, advances in stem cell generation and differentiation coupled with advancements in biomanufacturing are opening new avenues to creating functional human tissues. To generate autologous human organs, one must produce 10–100 billion patient-specific cells, guide their differentiation into distinct and mature phenotypes, pattern these cells into precise 3D architectures, maintain and mature the construct, and surgically implant it. The advent of human induced pluripotent stem cells (hiPSCs), combined with their scalable production and differentiation in suspension culture bioreactors, offers a path to obtaining the necessary raw material (Kropp et al., 2017). Simultaneous innovations that reduce cell media costs are upending the economics of production of human tissues in bulk (Kuo et al., 2020). From a regulatory standpoint, the recent development of genomically engineered hypoimmunogenic hiPSCs can enable the creation of highly characterized and safe universal cell lines to fuel downstream differentiation and biofabrication pipelines (Deuse et al., 2019; Han et al., 2019; Xu et al., 2019). Deriving defined cell types from these pluripotent cells traditionally requires the sequential addition of growth factors or other small molecules; however, emerging efforts are harnessing genomic engineering to deterministically generate targeted cell types of interest. Traditionally, human cells have been seeded onto or into porous scaffolds to produce connective tissues or vascular grafts (Hibino et al., 2011; Kirkton et al., 2019; Niklason et al., 1999). However, this approach lacks the ability to precisely pattern dozens of distinct cell types into organ-specific micro- to macro-architectures that compose whole human organs.
3D bioprinting enables the programmatic patterning of cell-laden inks and multicellular organ building blocks (e.g., embryoid bodies, spheroids, and organoids). All complex solid organs can conceptually be broken down into spatially patterned, or ‘voxelated’, functional units each with a distinct cellular and extracellular makeup that bestows homeostatic or physiologic function to the organ. Hence, the roadmap for organ biomanufacturing can be described as a two-step process: (1) render discrete functional units with the requisite cellular components and self-assembled microarchitecture, and (2) assemble these functional units into a bulk organ-specific tissue replete with embedded vasculature required to achieve and sustain their function (Fig. 1). Within this framework, the key challenges are both maximizing the generation and self-assembly of functional human tissue blocks as well as spatially patterning and maturing these building blocks to produce therapeutic-scale tissues.
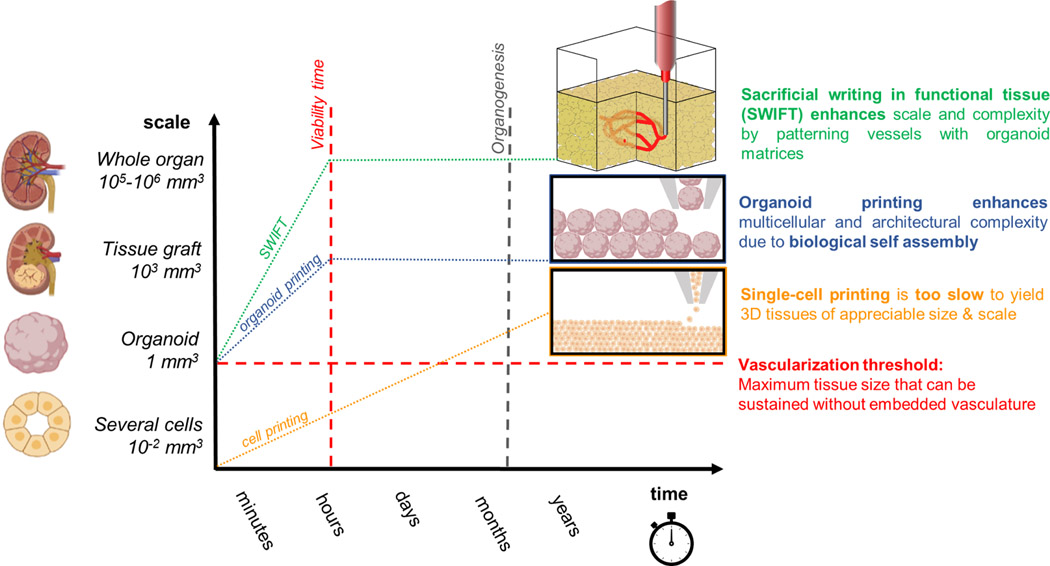
Constructing whole organs using single cells as building blocks is currently unfeasible due to inherent scaling limitations of 3D bioprinting (Fig. 2). The time required to “pick and place” each cell throughout tissue volumes over 1 mm3 vastly exceed the cell viability window (~1–2 hours) during bioprinting. By contrast, the use of multicellular organ building blocks (e.g., organoids) offers several advantages by both recapitulating the different cell types and microarchitectures within organ-specific tissues as well as vastly reducing the build times required to generate whole organs (~105-106 mm3 in volume) (Miller, 2014). Direct organoid printing is capable of generating human tissues at faster rates, since each building block is composed of more than 104 cells. However, when patterning tissues at the macroscale, one must simultaneously print the vascular network required to supply nutrients, clear waste products, and maintain the tissues as they undergo maturation (Kolesky et al., 2014; Miller et al., 2012; Skylar-Scott et al., 2019a). Indeed, embedding vascularization becomes necessary once the fabricated tissue volume exceeds roughly 1mm3, i.e., roughly the size of the individual organ building blocks. Tissue vascularization can be achieved rapidly using embedded printing methods, such as Sacrificial Writing Into Functional Tissue (SWIFT) (Skylar-Scott et al., 2019a), wherein a sacrificial ink is printed into a living tissue matrix composed of compacted organoids in the shape of a vascular tree. The subsequent removal of this sacrificial ink leaves behind an interconnected network of cylindrical vessels that can be lined with endothelium, through which nutrients and oxygen can flow.
Figure 2:
Opportunity space for biomanufacturing human tissues at organ-scale.
2. Organ building blocks
Multicellular aggregates have been used for over a century to study spatial patterning in development (Wilson, 1907), with early studies of mammalian cell aggregates dating to the 1950s (Moscona, 1957). Until recently, multicellular spheroids have been largely generated from primary and immortalized human cells. For example, metabolically active hepatic spheroids have been produced from immortalized lines (Kelm et al., 2003) and primary hepatocytes (Landry et al., 1985). There is emerging interest in assembling these multicellular aggregates into larger tissues. In one of the earliest examples, spheroids were manually placed into rings to form a toroidal tissue architecture within a surrounding 3D matrix (Jakab et al., 2004). This simple yet non-trivial geometry demonstrated the feasibility of generating physiologically relevant forms using multicellular building blocks.
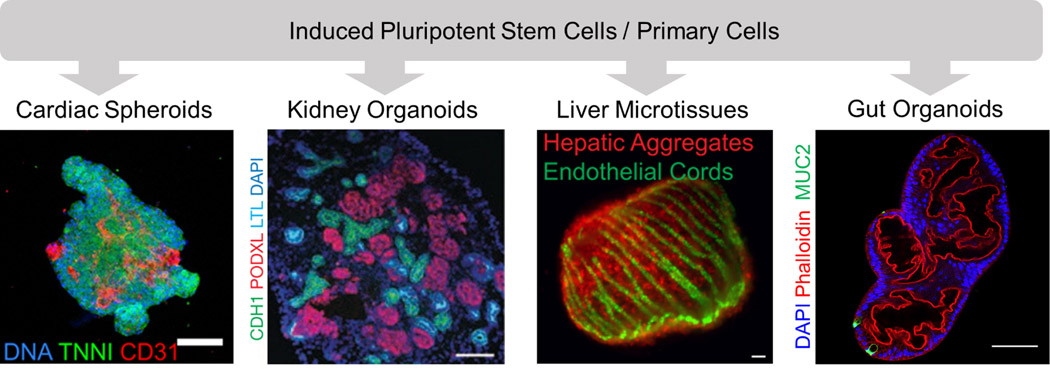
The advent of induced pluripotent stem cells (iPSCs) followed by the development of differentiation protocols that transform these cells into organoids, or “mini-organs”, has radically expanded the functional complexity and types of organ building blocks that can be generated (Giacomelli et al., 2017; Morizane et al., 2015; Sato et al., 2009; Taguchi et al., 2014; Takasato et al., 2015; Takebe et al., 2013). It is now possible to generate myriad tissue-specific organoids, including cardiac, kidney, liver, and gut spheroids/organoids with exquisite microarchitectures (Fig. 3). For example, hiPSC-derived cardiac cells co-cultured with hiPSC-derived endothelial cells yield cardiac spheroids (Giacomelli et al., 2017) that can be further assembled into contractile cardiac tissues that synchronously beat (Breckwoldt et al., 2017). Liver spheroids have also been generated from hiPSCs by combining hiPSC-derived hepatocytes with mesenchymal stem cells and endothelial cells (Takebe et al., 2013). When subjected to perfusion using a microfluidic platform, hepatic aggregates derived from iPSCs have been shown to retain albumin expression for over 28 days (Schepers et al., 2016). To optimize cell-cell interactions, hepatic aggregates have also been patterned from primary hepatocytes, endothelial, and stromal cells using a microfabrication method (Stevens et al., 2017). The resulting liver “seeds” can support microstructure formation, including self-organized bile ducts when implanted in animal models. While more advanced liver function will ultimately require an interconnected bile duct network, these seminal studies suggest that liver building blocks may retain structural plasticity which would facilitate functional adaptation in vivo. Akin to the liver, kidney organoids require interconnected tubule networks to achieve their ultimate function of filtration and filtrate modification. Several protocols for deriving nephron-rich organoids from metanephric mesenchyme have been recently developed; these organoids possess a cellular composition and structure that resembles a first-trimester kidney (Morizane et al., 2015; Taguchi et al., 2014; Takasato et al., 2014; Takasato et al., 2015). However, they generally lack perfusable glomeruli, collecting ducts, and a ureter, hence limiting their functional utility (Little and Combes, 2019; Takasato et al., 2015). Finally, gut organoids (Sato et al., 2009; Spence et al., 2011) have also been generated, and recent work by Brassard et al. has demonstrated that these progenitor cells, when printed into an extracellular matrix, will spontaneously assemble into an intestinal lumen (Brassard et al., 2020).
Figure 3:
Organ building blocks composed of multicellular spheroids and organoids. Scale bar, left to right: 100 μm, 50 μm, 400 μm, 100 μm. [(Left to right): Images reproduced with permission from Giacomelli et al., 2017; Morizane et al., 2015; Stevens et al., 2017. Far right image was provided courtesy of Joep Beumer, Hubrecht Institute.]
Despite the promise of organ building blocks for biomanufacturing, several hurdles remain. Bulk tissues and organs require a pervasive, hierarchical vascular network to maintain cell viability and function. While bioprinting methods are capable of patterning blood vessels roughly 100 μm and larger in size, vascularization of individual organ building blocks at the microscale is needed to fully drive bulk tissue maturation and function. Several strategies, including perfusion, growth factor addition, and matrix cues have been recently reported for inducing microvascularization within spheroids and organoids (Brassard and Lutolf, 2019). For example, Giacomelli et al. enriched iPSC-derived cardiac spheroids with iPSC-derived endothelial cells and demonstrated enhanced expression of cardiac maturation markers, such as genes encoding ion channels and Ca2+-handling proteins (Giacomelli et al., 2017). Beyond their role in perfusion, the co-culture of hiPSC-derived cardiomyocytes and cardiac fibroblasts with cardiac endothelial cells has also been shown to improve sarcomeric structure and contractility (Giacomelli et al., 2020). By contrast to direct addition of endothelial cells, physical cues can enhance endothelial cell differentiation and proliferation. Notably, kidney organoids that are subjected to flow during differentiation on an adherent matrix exhibit a three-fold enrichment in their endothelial progenitor cell population promoting both their vascularization at the microscale and maturation, as observed by significantly enhanced expression of glomerular, tubular, and endothelial markers (Homan et al., 2019).
The complex interplay between microvascular network formation and maturation raises two important questions: (1) what is the optimal differentiation state of organ building blocks prior to tissue assembly and (2) what is the appropriate balance between spontaneous self-assembly and deterministic biofabrication? At one extreme, a small number of undifferentiated cells could be aggregated and then guided through differentiation, growth, and morphogenesis to form the mature, functional tissue. As a key example of this approach, mouse ESC-derived cell aggregates cultured in gastruloid culture conditions and then exposed to cardiogenic factors form cardiovascular progenitors including first and second heart fields, which undergo morphogenesis resembling the formation of heart chambers (Rossi et al., 2021). Cardiac organoids, exhibiting chamber-like structures, have recently been produced using human iPSCs (Drakhlis et al., 2021; Silva et al., 2021). These strategies recapitulate key features of organogenesis (i.e., spontaneous cell assembly, tissue morphogenesis, and growth), but would require considerable time and resources to reach the scale of whole organs. To circumvent the need for tissue growth, Kupfer et al. deterministically 3D printed immature ESCs into an adult heart shape and then expanded and differentiated the ESCs into cardiovascular phenotypes (Kupfer et al., 2020). The resulting, perfusable heart chambers show continuous action potential propagation with a cardio-effective drug response. At the other extreme, hundreds of thousands of these organ building blocks can be fully differentiated and deterministically patterned into a large, cohesive cardiac tissue that synchronously beats after several days of maturation (Skylar-Scott et al., 2019a). Because normal tissue morphogenesis is bypassed in this approach, the primary challenge is to ensure adequate OBB fusion via cell-cell and cell-matrix interactions to generate the desired functional output akin to native organs as well as provide sufficient mechanical robustness for handling and suturing during implantation.
3. Bioprinting 3D tissues from organ building blocks
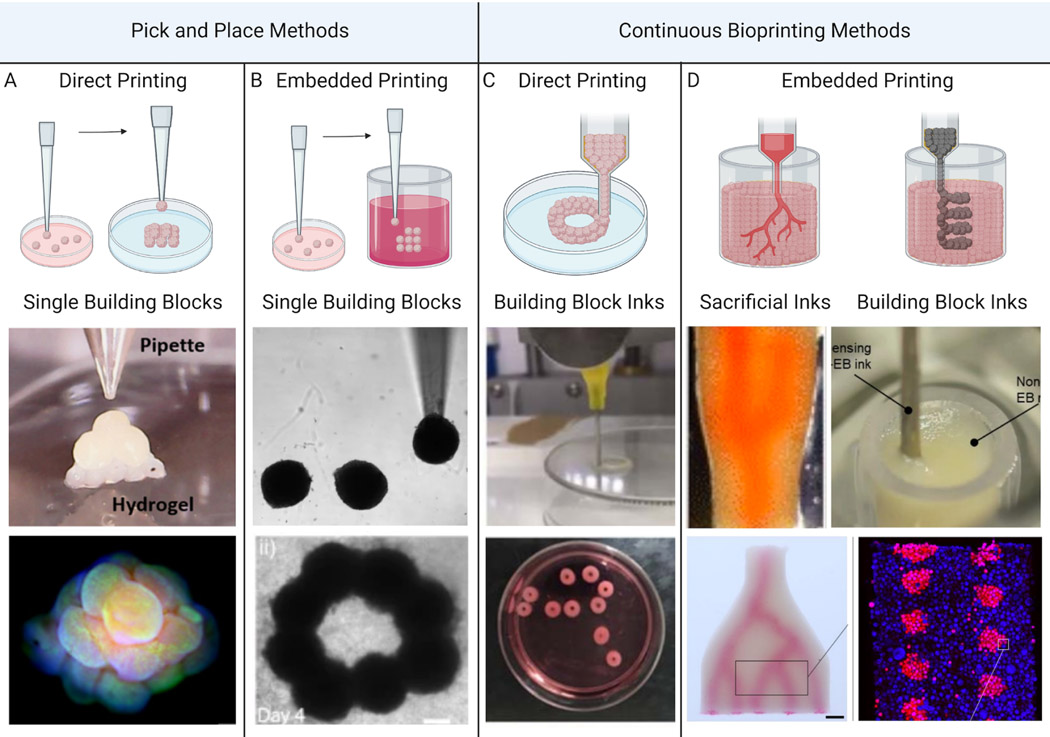
Bioprinting organ-specific tissues from multicellular building blocks derived from hiPSCs enables the construction of patient-specific tissues, thereby reducing the risk of immune rejection and enhancing potential for their clinical translation. Several methods have recently emerged for printing tissues from organ building blocks (Fig. 4). Unlike individual cells, spheroid- and organoids-based building blocks are granular in nature, i.e., their characteristic size exceeds roughly 100 μm in diameter, and, hence, their interactions are dominated by gravitational forces. Direct printing of individual spheroids or organoids relies on their granular nature to enable their sequential pick up and placement (Fig. 4a–b). For example, Ayan et al. showed that different spheroids, including those composed of human umbilical vein endothelial cells (HUVECs), mouse fibroblast cells (3T3), mouse mammary carcinoma cells (4T1), human dermal fibroblasts (HDFs), and human mesenchymal stem cells could be retrieved from a media bath by aspiration using a glass pipette and then placed onto a matrix-coated substrate (Fig. 4a) (Ayan et al., 2020a). Using this direct “pick and place” approach, they stacked individual spheroids (~300 μm in diameter) with ~35 μm precision to form tissues in small pyramids and rings. In the presence of HDFs and fibrin, HUVEC-laden spheroids exhibited angiogenic sprouting behavior, which intensified as the distance between adjacent spheroids decreased likely due to increased cellular signaling.
Figure 4.
Biomanufacturing of human tissues using organ building blocks: (a) direct pick and placement of individual spheroids or organoids, (b) embedded pick and placement of individual spheroids or organoids within a support matrix, (c) direct printing of spheroid- or organoid-based inks, and (d) embedded printing of sacrificial inks within a tissue matrix composed of densely packed spheroids or organoids (left) or embedded printing of spheroid- or organoid-based inks in either support (not shown) or tissue (right) matrices [Images (from left to right) adapted with permission from Ayan et al. (2020a), Daly et al. (2021), Goulart et al. (2020), Skylar-Scott et al. (2019b)].
Expanding upon this approach, Daly et al. and others demonstrated an embedded “pick and place” printing method for creating more complex 3D tissue geometries (Fig. 4b) (Ayan et al., 2020b; Daly et al., 2021). This technique relies on similar aspiration pipettes to pick individual spheroids from a media bath and place them within a shear-thinning, self-healing hydrogel support matrix whereupon they remain suspended in 3D space. Using this approach, individual spheroids were used to construct tissue pyramids and layered rings within the hydrogel matrix, from which the tissue could ultimately be harvested (Daly et al., 2021). As one example, spheroids composed of varying mixture ratios of cardiac fibroblasts and iPSC-derived cardiomyocytes were used to create a tissue construct to model post myocardial infarction-scarring. Localized regions rich in cardiac fibroblast spheroids serve as scars that reduce conduction velocity and mechanical contractility compared to control tissues composed solely of cardiomyocyte spheroids. Upon exposing the scarred tissue constructs to Hippo pathway-inhibiting miRNAs for several days, cardiomyocyte proliferation and improved tissue functionality were observed. These examples highlight the ability of pick and place methods to produce precisely constructed microtissues for investigating heterogeneous interactions between individual organ building blocks and for therapeutic drug screening. However, building tissues by patterning OBBs one at a time is slow, and thus not well suited for organ-scale biomanufacturing.
Direct bioprinting of OBB-laden inks offers a higher throughput method that unlocks therapeutic-scale biofabrication (Fig. 4c). Goulart et al. demonstrated the advantage of this method for constructing liver tissues by formulating inks composed of hiPSC-derived hepatic spheroids compared to hepatic cells (Goulart et al., 2020). These inks were printed into toroidal tissue constructs (5 mm high and 10 mm in diameter). After 18 days of culture post-printing, the viability and hepatic metabolic function of 3D liver tissue printed from spheroid-laden inks were significantly higher than those produced from cell-laden inks. Hence, the pre-organization of hepatocytes into organ building blocks with defined microenvironments promoted physiological function at the tissue-level. While this method is promising for the scalable generation of organ-scale tissues, two major challenges of direct OBB printing arise once the tissue thickness exceeds a few millimeters. First, as printed tissues become taller, they often lose their shape due to gravity-induced slumping. Second, when the printed tissues exceed ~1 mm in thickness, they will undergo necrosis due to a lack of perfusion. While this scale limitation can be addressed by implanting organoids or bioprinted tissues into a host to stimulate neovascularization (Mansour et al., 2018; Takebe et al., 2013), vascular ingrowth is too slow (~ 1 mm/day) to rescue therapeutic-scale tissues.
Embedded 3D printing is an emerging technique that is uniquely capable of printing large-scale tissues replete with vascular networks using soft and biological inks. This method involves printing one ink into a bath, or matrix, comprising a second viscoelastic ink (Wu et al., 2011). The viscoelastic matrix is tailored to be liquid-like enough to enable a printer nozzle to translate through the matrix without tearing it while simultaneously being solid-like enough to adequately support the printed tissue to prevent it from sagging, sinking, or floating. The printed ink can be a sacrificial material that, upon removal, forms a vascular network within a tissue matrix composed of organ building blocks. Alternatively, the ink can be a cell- or OBB-laden bio-ink that builds a tissue within a sacrificial or living matrix (Fig. 4d). First demonstrated by patterning a sacrificial ink in the form of a hierarchical branching vascular tree within an acellular hydrogel matrix (Wu et al., 2011), this technique has undergone several advances that have enabled the construction of vascularized human tissues at scale. For example, SWIFT allows complex and branched vascular networks to be embedded within a living tissue matrix composed of OBBs composed of embryoid bodies, organoids, or spheroids derived from hiPSCs (Skylar-Scott et al., 2019a). To attain physiologically relevant scales, these organ building blocks are cultured in batches of 100,000’s, which contain up to half a billion cells. Next, they are mixed with an ECM solution of collagen and Matrigel and compacted into a granular tissue matrix with a total volume of ~2.5mL that contains ~200 million cells/mL. A sacrificial gelatin ink is then patterned within the tissue matrix via embedded 3D printing in the form of a vascular network. Upon warming the tissue to 37°C, the sacrificial ink liquifies and can be readily removed from the tissue, leaving behind a perfusable vascular network that sustains the tissue during fusion of the organ building blocks and subsequent tissue maturation. Using SWIFT, vascularized cardiac tissues were generated and underwent fusion to form a synchronously beating tissue after one week of perfusion. With further development, the SWIFT technique could enable biofabrication of vascularized human tissues at organ scale.
Freeform reversible embedding of suspension hydrogels (FRESH) is another important embedded 3D printing technique that enables patterning soft inks at bulk scale with high resolution (Hinton et al., 2015; Lee et al., 2019). This method has been used to create 3D collagen scaffolds in the form of a miniature human ventricle model that can be filled with hiPSC-derived cardiomyocytes. Extending this approach, Kupfer et al. demonstrated the feasibility of in situ differentiation of printed stem cells after allowing a period of growth and densification (Kupfer et al., 2020). In this case, a bio-ink containing hiPSCs and an ECM composed of methacrylated gelatin (10% or 15%), fibronectin (0–190 ug/mL), laminin (0–190 ug/mL), and ColMA (0–5 mg/mL) are printed in a gelatin microparticle support bath via FRESH. The printed structure is based on an MRI scan of a human heart scaled down to the size of a murine heart modified with a single input and output vessel connecting two internal chambers with a wall thickness <0.5mm, so vascularization was not required. After 2 weeks in culture to promote hiPSC proliferation, the estimated density of the tissue was ~100 million cells/mL, similar to that of native tissue. Cardiac differentiation was then induced post-printing whereupon contiguous electrical function and synchronous beating was observed up to 6 weeks. Looking ahead, co-writing organ building blocks and vascular networks via embedded 3D printing offers a promising approach for constructing whole organs.
4. Enhancing tissue function via building block fusion and maturation
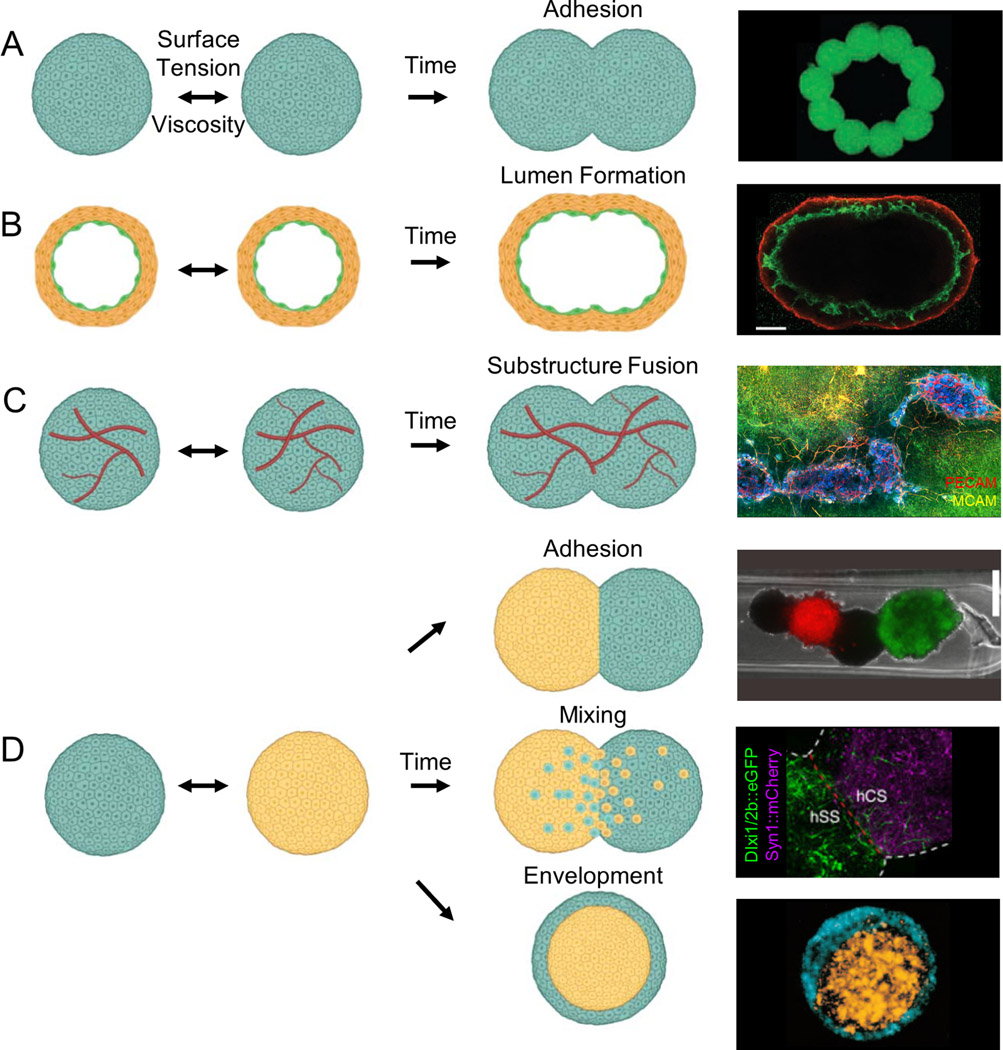
Once assembled into a 3D tissue, organ building blocks must fuse and form a cohesive network. To date, the fusion of these building blocks has been primarily explored in simple two-body systems composed of like or unlike spheroids (or organoids) (Fig. 5). The viscosity and surface tension of spheroids have been experimentally measured, which suggests that fusion of two spheroids of the same composition is akin to the coalescence of two viscous droplets (Jakab et al., 2010) (Fig. 5a). The differential adhesion hypothesis posits that cells will adhere to other like cells provided all cells remain in a motile state and express similar adhesion molecules. Remarkably, the dynamics of spheroid fusion can, in many cases, be predicted by measuring the physical properties of multicellular aggregates and applying this model (Jakab et al., 2008; Mironov et al., 2009). For example, spontaneous rounding of tissue fragments, driven by their surface tension, has been observed experimentally (Foty et al., 1996; Jakab et al., 2008), and the time required for fusion has been shown to correlate with cadherin expression (Foty and Steinberg, 2005). However, organ building blocks are more complex than liquid droplets, so this simple model is not fully predictive. For example, Kosheleva et al. compared the fusion of two mesenchymal spheroids to the fusion of two epithelial spheroids and found that even though the epithelial spheroids had lower surface tension, they underwent faster fusion than their mesenchymal counterparts (Kosheleva et al., 2020).
Figure 5.
Fusion of spheroid, organoid, and assembloid-based organ building blocks. (a) Homotypic spheroid fusion leading to simple adhesion [image adapted with permission from Jakab et al., 2004]. (b) Homotypic spheroid fusion of two luminal spheroids generates a spheroid with a single lumen [image adapted with permission from Fleming et al., 2010]. (c) Homotypic spheroid fusion with substructure interconnection of vascular networks [image adapted with permission from Homan et al., 2019]. (d) Heterotypic spheroid fusion leading to simple adhesion, mixing, or envelopment [images (top to bottom) adapted with permission from Kim et al., 2018; Birey et al., 2017; Foty et al., 1996].
Many organ building blocks contain substructures, such as microvascular, tubular, and biliary features that must ultimately form interconnected networks throughout the biofabricated tissues. Since cells typically adhere most strongly to other like cells, organ building blocks that contain a mixture of endothelial and supporting cells can spontaneously form lumens. Fleming et al. demonstrated that two hollow spheroids, each containing a vascular lumen surrounded by smooth muscle cells, can fuse to form a single continuous lumen (Fleming et al., 2010). Using this principle, the authors fused multiple spheroids into a single, elongated vessel (Fig. 5b). The process of lumen fusion is biologically relevant – for example, the aorta is formed by the fusion of two dorsal aortae (Fleming et al., 2010). While the model implies that substructure fusion is a secondary phenomenon arising from random movement of differentially adhesive cells, some biological fusion processes are cell-directed through chemical gradients, such as vascular anastomosis. Fusion of microvessels between organ building blocks is necessary to develop a cohesive, perfusable capillary network. While such studies are limited, early examples suggest that vascular fusion is possible. When both microvascularized kidney organoids (Homan et al., 2019) and cardiac spheroids (Polonchuk et al., 2021) fuse together, they appear to form a continuous vasculature network (Fig. 5c). Kim et al. also observed fusion of vascular networks between more complex organ building blocks, i.e., core-shell spheroids (Kim et al., 2019). Interestingly, core-shell spheroids with endothelial cells surrounding stem cells rapidly induced vessel network formation relative to homogenous spheroids, highlighting the potential for directing tissue fusion through building block design.
The fusion of organ building blocks may be further complicated or enhanced when they are printed within an extracellular matrix, e.g., via FRESH. Interactions between the matrix and embedded spheroids (or organoids) might dictate fusion dynamics. For example, spheroids have been shown to undergo fusion in a collagen matrix with subsequent contraction that depends on the collagen concentration (Jakab et al., 2004). Cell-matrix adhesions may stabilize shape of spheroids by balancing cell-cell adhesions. For example, vascular spheroids undergoing fusion within a matrix remained ovoid, while they formed spheres absent this matrix (Fleming et al., 2010). Clearly, additional studies that elucidate the competing cell-cell versus cell-matrix interactions that guide the fusion of organ building blocks within biofabricated human tissues are needed.
The ability to pattern and guide the fusion of different OBBs is important for generating complex tissues. The differential adhesion hypothesis predicts that mismatches in their respective surface tensions would lead to envelopment of the high surface tension spheroid (or organoid) by the lower surface tension counterpart, which has been experimentally observed (Fig. 5d) (Foty et al., 1996). However, more complex geometries, known as assembloids, have also been formed by fusing two or more different spheroids (or organoids) together (Jakab et al., 2004; Birey et al, 2017]). These assembloids, sometimes in contrast predictions from the differential adhesion hypothesis, can exist in metastable states and give rise to complex biological interactions. As a simple functional demonstration, Kim et al. fused cardiac myocyte spheroids alternating with cardiac fibroblast spheroids in a line and demonstrated the cardiac fibroblasts support action potential propagation but with a significant conduction delay (Fig. 5d) (Kim et al., 2018). Interestingly, heterotypic fusion of hiPSC-derived dorsal forebrain organoids and ventral forebrain organoids recapitulated in vivo-like migration and functional integration of interneurons from the ventral to dorsal region (Fig. 5d) (Birey et al., 2017). The differentiation or maturation state of these organ building blocks can also affect cell behavior and, hence, fusion. Lindberg et al. found that fusion of mesenchymal stromal cell spheroids and articular chondrocyte spheroids as well as matrix deposition depends on the timing of contact relative to their maturation state (Lindberg et al., 2021). Hajdu et al. found that their maturation state determines whether spheroids simply adhere or undergo envelopment, and that fusion kinetics are related to extracellular matrix production (Hajdu et al., 2010). Collectively, these studies highlight the need for cell type-dependent fusion studies especially in heterotypic assemblies.
After assembly, exogenous cues can be provided through embedded vasculature to further promote tissue fusion and maturation. For example, the addition of VEGF to fusing vascularized cardiac spheroids increased fusion efficiency by more than two-fold as well as increased vascular fusion between spheroids (Polonchuk et al., 2021). The construction of human-scale heterogeneous tissues may be seen as a natural extension of assembloids, but would require fusion of millions of homotypic and heterotypic spheroids to form patterned tissue. Further work is needed to determine how chemo-mechanical cues, including growth factors, matrix composition, luminal flow, and stretch, support the maturation of spheroids, organoids, assembloids, and, ultimately, biofabricated human organs.
Ultimately, fabricated organ-specific tissues must be functionally integrated with the host tissue; interconnection with the cardiovascular system is critical to perfuse implanted tissues and facilitate tissue function. Small tissue (millimeter scale) constructs might be sufficiently sustained via host-derived microvascular invasion and perfusion, eliminating the need for surgical anastomosis. For example, human iPSC-derived kidney organoids implanted under the renal capsule in mouse were microvascularized by the host after 10–20 days (Sharmin et al., 2016). Interestingly, iPSC-derived glomeruli were preferentially vascularized by the angiogenic host endothelial cells rather than HUVECs co-implanted with the kidney organoids, possibly indicating the importance of vascular flow or HUVEC incompatibility. Human liver seed grafts were also microvascularized by the host after 80 days; grafts contained red blood cells and microvasculature was formed by both human and mouse endothelial cells, suggesting functional anastomosis (Stevens et al., 2017). However, large tissue grafts will ultimately require hierarchical vasculature with a large vascular inlet and outlet to be surgically anastomosed to the host to facilitate immediate perfusion. In addition, some organs will require additional anastomoses, such as the kidney to the ureter or the liver to the bile duct. Many potential challenges with anastomosis of engineered constructs have been identified or solved through the development of tissue engineered vascular grafts (Kirkton et al., 2019; Patterson et al., 2012). For example, risks include thrombosis, intimal hyperplasia, atherosclerosis, or infection (Pashneh-Tala et al., 2016). Design considerations, such as graft compliance and geometry, at the site of anastomosis could alleviate some potential modes of failure when implanting engineered tissues (Pashneh-Tala et al., 2016).
5. Future Challenges and Opportunities
Despite the rapid progress, there are several remaining challenges and opportunities for biomanufacturing human tissues and organs at scale. One important hurdle is that the generation of clinical grade hiPSCs is costly, with estimates of $1M per iPSC line (Bravery, 2015). Depending on the differentiation protocol and the size of tissue required, the cost of growth factors and culture supplements are often prohibitive for commercialization (Bravery, 2015). The discovery of low-cost small molecules, such as CHIR-99021, which acts as a Wnt pathway agonist (Sato et al., 2004), can replace costly growth factors to enable affordable large-scale cultures. Importantly, the fabrication of autologous tissues requires the scalable generation of patient-specific iPSCs, in which each distinct cell line could require modified differentiation protocols to generate the requisite OBBs. This issue could be solved in part by genetic editing of HLAs in a standard bank of iPSCs to provide hypo-immunogenic cell lines for broader patient use (Deuse et al., 2019; Han et al., 2019; Xu et al., 2019).
Beyond cost, adapting culture protocols and platforms to be compliant with current good manufacturing practices (cGMP) at scale is another major challenge. Suspension bioreactors are currently the optimal solution for mass cGMP production due to volume efficiency, scalability, and amenability to automated culture, all of which can improve reproducibility (Kropp et al., 2017). However, many hiPSC protocols begin with 2D adherent culture, necessitating either protocol adaptation or the use of less efficient, more costly scaling methods. In addition, many differentiation protocols rely on subjective measures of cell health and differentiation efficiency, such as evaluating confluency and phenotype to determine timing and composition of medium changes (Morizane et al., 2015). Combined with patient-to-patient variability, sex differences, heterogeneity with a single hiPSC lines, and genetic drift in subcultures, batch-to-batch reproducibility is a major hurdle (Colter et al., 2021). Biosensors could aid in real-time monitoring of batch quality and control, and a variety of genetic controls such as microRNA switches could be applied to purify differentiated cell populations (Miki et al., 2015). In addition to cells, the scalability and manufacturability of any exogenous extracellular matrix (ECM) or scaffolding materials must be considered. Multiple biomaterials have been developed as scaffolds and applied clinically, as thoroughly reviewed elsewhere (Sadtler et al., 2016). In general, synthetic polymers (e.g. polyethylene glycol) offer more control over material properties at a lower cost while biologically-derived materials (e.g. collagen I or fibrin) offer more complex instructive cues critical for some cell culture protocols (Sadtler et al., 2016). Synthetic materials have been modified to incorporate biologically active groups to create scaffolds with tailored mechanical and biological properties. These materials could replace Matrigel in iPSC and organoid culture by mimicking properties such as heparin-like growth factor binding (Aisenbrey and Murphy, 2020). Alternately, patient-specific, biologically-derived ECM materials could be obtained directly from the patient, for example, by using omentum tissue derived from a biopsy (Shevach et al., 2015).
Even with the generation of an efficacious therapeutic tissue, safety remains a critical concern. Aside from potential tissue failure, hiPSC tumorigenicity poses a significant risk, which has been demonstrated in animal studies (Lee et al., 2013). Reprogramming of hiPSCs from somatic cells and subsequent subculture compromises genomic integrity, often introducing chromosomal abnormalities in a subpopulation of cells (Hussein et al., 2011). In addition, undifferentiated cells in the engineered tissue could form teratomas (Ben-David and Benvenisty, 2011). There is a growing interest in genetically engineered drug sensitivities or “kill-switches” into hiPSC populations as safeguards to eliminate undifferentiated cells on demand (Ando et al., 2015; Martin et al., 2020). In addition, banked hiPSCs that have undergone extensive whole-genomic analysis could reduce variability and provide a foundation for comparing clinical outcomes (Merkle et al., 2022).
High-throughput, scalable biofabrication methods are needed to fabricate human tissues and, ultimately, whole organs in a practical time scale necessary to maintain cell viability and minimize cost. To date, most extrusion-based bioprinting methods have used printheads composed of one or a few nozzles (Kang et al., 2016; Kolesky et al., 2016). Notably, the build time required to print a full-size human heart composed of 100 μm layers is nearly 100 h using a single 250 μm nozzle (Mirdamadi et al., 2020). Since single nozzles only print one voxel of material at a time, the fabrication time scales roughly with the cube of the tissue volume. Volumetric printing methods can decouple the relationship between printing speed and tissue volume by parallelizing voxel formation. For example, multimaterial, multinozzle printheads are capable of simultaneously printing arrays of voxels in a 2D plane and rapidly switching between bio-inks would facilitate rapid fabrication of intricately patterned tissues (Skylar-Scott et al., 2019b). Light-based bioprinting approaches, such as stereolithography (SLA) and digital light processing (DLP), print layer-by-layer by applying light to the surface of a photo-reactive resin bed. To date, these methods have been used to rapidly pattern complex, biologically relevant shapes, such as vascular networks at the centimeter scale within an hour (Grigoryan et al., 2019). In computed axial lithography (CAL), light is delivered to a liquid resin bed as a series of 2D images at different angles; only regions in which images are superimposed receive enough energy to solidify (Kelly et al., 2019). This approach has been applied to pattern spheroids derived from human liver epithelial organoids into a gel containing a perfusable vascular network (Bernal et al., 2022). However, each of these light-based printing methods require resins that are compatible with photo-crosslinking, which limits the use of biological matrices and light-sensitive cells. The incorporation of multicellular spheroid, organoid, or assembloid-based OBBs within these photopolymerizable resins will enhance light scattering, limiting the feature resolution and cellular density within the printed tissues. New advances in scalable, high resolution bioprinting technologies coupled with industrial standardization and cGMP protocols are needed for biomanufacturing clinical-grade human tissues and organs.
A final challenge involves delineating the appropriate balance of organ building block maturation before and after tissue biofabrication, which may vary by tissue type. To date, methods for controlling maturation of large-scale human tissues after their assembly have not yet been widely explored. Techniques developed for single organoids or microtissues could be applied to larger tissues as well as bioreactors developed to sustain whole organs prior to transplantation. However, it is unclear whether full maturation in vitro is necessary or desirable before implantation. Looking ahead, scientific and technological advances are anticipated across multiple areas – stem cell scale up and differentiation, building block design, fusion, and maturation, and biofabrication – and their convergence may soon provide a viable pathway for de novo biomanufacturing of organ-specific tissues for therapeutic use.
Lewis and colleagues review the differentiation, maturation, and 3D assembly of organ building blocks into functional human tissues and, ultimately, organs for therapeutic repair and replacement. They also highlight future challenges and areas of opportunity for this nascent field.
Acknowledgements:
The authors are grateful for funding provided to the Lewis Lab from ONR Vannevar Bush Faculty Fellowship Program (N00014-21-1-2958), Wellcome Leap’s HOPE program (Contract # N00014-16-1-2823), NIDDK (Re)Building a Kidney Consortia (Grant# 1UC2DK126023-01), NSF CELL-MET (#EEC-1647837), and the Wyss Institute 3D Organ Engineering Initiative and to the Skylar-Scott Lab from BASE Initiative at Stanford. The authors thank Joep Beumer, Hubrecht Institute, for providing an image of a gut organoid. Figures created with BioRender.com.
Footnotes
Declaration of Interests: J.A.L. is a cofounder of AcousticaBio and serves as a scientific advisor for Autodesk, Azul3D, Desktop Health (a subsidiary of Desktop Metal), Mooji Meats, and Trestle Biotherapeutics. M.A.S.S. owns stock in Formlabs, consults for 3D Systems, and is a scientific advisor for AcousticaBio and Mooji Meats. M.A.S.S., S.U., and J.A.L. are inventors of the SWIFT biomanufacturing method and have filed a patent on this work. M.A.S.S. and J.A.L. are inventors on a broader set of patents focused on vascularized human organoids and tissues. Trestle Biotherapeutics has licensed these patents from Harvard University.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aisenbrey EA, and Murphy WL (2020). Synthetic alternatives to Matrigel. Nat. Rev. Mater. 5, 539–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando M, Nishimura T, Yamazaki S, Yamaguchi T, Kawana-Tachikawa A, Hayama T, Nakauchi Y, Ando J, Ota Y, Takahashi S, et al. (2015). A safeguard system for induced pluripotent stem cell-derived rejuvenated T cell therapy. Stem Cell Reports 5, 597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayan B, Heo DN, Zhang Z, Dey M, Povilianskas A, Drapaca C, and Ozbolat IT (2020a). Aspiration-assisted bioprinting for precise positioning of biologics. Sci. Adv. 6, eaaw5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayan B, Celik N, Zhang Z, Zhou K, Kim MH, Banerjee D, Wu Y, Costanzo F, and Ozbolat IT (2020b). Aspiration-assisted freeform bioprinting of pre-fabricated tissue spheroids in a yield-stress gel. Commun. Phys. 3, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David U, and Benvenisty N. (2011). The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat. Rev. Cancer 11, 268–277. [DOI] [PubMed] [Google Scholar]
- Bernal PN, Bouwmeester M, Madrid‐Wolff J, Falandt M, Florczak S, Rodriguez NG, Li Y, Größbacher G, Samsom R, Wolferen M, et al. (2022). Volumetric Bioprinting of Organoids and Optically Tuned Hydrogels to Build Liver‐Like Metabolic Biofactories. Adv. Mater. 2110054. [DOI] [PubMed] [Google Scholar]
- Birey F, Andersen J, Makinson CD, Islam S, Wei W, Huber N, Fan HC, Metzler KRC, Panagiotakos G, Thom N, et al. (2017). Assembly of functionally integrated human forebrain spheroids. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brassard JA, and Lutolf MP (2019). Engineering Stem Cell Self-organization to Build Better Organoids. Cell Stem Cell 24, 860–876. [DOI] [PubMed] [Google Scholar]
- Brassard JA, Nikolaev M, Hübscher T, Hofer M, and Lutolf MP (2020). Recapitulating macro-scale tissue self-organization through organoid bioprinting. Nat. Mater. 1–8. [DOI] [PubMed] [Google Scholar]
- Bravery CA (2015). Do human leukocyte antigen-typed cellular therapeutics based on induced pluripotent stem cells make commercial sense? Stem Cells Dev. 24, 1–10. [DOI] [PubMed] [Google Scholar]
- Breckwoldt K, Letuffe-Brenière D, Mannhardt I, Schulze T, Ulmer B, Werner T, Benzin A, Klampe B, Reinsch MC, Laufer S, et al. (2017). Differentiation of cardiomyocytes and generation of human engineered heart tissue. Nat. Protoc. 12, 1177–1197. [DOI] [PubMed] [Google Scholar]
- Colter J, Murari K, Biernaskie J, and Kallos MS (2021). Induced pluripotency in the context of stem cell expansion bioprocess development, optimization, and manufacturing: a roadmap to the clinic. Npj Regen. Med. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly AC, Davidson MD, and Burdick JA (2021). 3D bioprinting of high cell-density heterogeneous tissue models through spheroid fusion within self-healing hydrogels. Nat. Commun. 12, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuse T, Hu X, Gravina A, Wang D, Tediashvili G, De C, Thayer WO, Wahl A, Garcia JV, Reichenspurner H, et al. (2019). Hypoimmunogenic derivatives of induced pluripotent stem cells evade immune rejection in fully immunocompetent allogeneic recipients. Nat. Biotechnol. 37, 252–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakhlis L, Biswanath S, Farr CM, Lupanow V, Teske J, Ritzenhoff K, Franke A, Manstein F, Bolesani E, Kempf H, et al. (2021). Human heart-forming organoids recapitulate early heart and foregut development. Nat. Biotechnol. 39, 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming PA, Argraves WS, Gentile C, Neagu A, Forgacs G, and Drake CJ (2010). Fusion of uniluminal vascular spheroids: A model for assembly of blood vessels. Dev. Dyn. 239, 398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foty RA, and Steinberg MS (2005). The differential adhesion hypothesis: A direct evaluation. Dev. Biol. 278, 255–263. [DOI] [PubMed] [Google Scholar]
- Foty RA, Pfleger CM, Forgacs G, and Steinberg MS (1996). Surface tensions of embryonic tissues predict their mutual envelopment behavior. Development 122, 1611–1620. [DOI] [PubMed] [Google Scholar]
- Giacomelli E, Bellin M, Sala L, van Meer BJ, Tertoolen LGJ, Orlova VV, and Mummery CL (2017). Three-dimensional cardiac microtissues composed of cardiomyocytes and endothelial cells co-differentiated from human pluripotent stem cells. Dev. 144, 1008–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomelli E, Meraviglia V, Campostrini G, Cochrane A, Cao X, van Helden RWJ, Krotenberg Garcia A, Mircea M, Kostidis S, Davis RP, et al. (2020). Human-iPSC-Derived Cardiac Stromal Cells Enhance Maturation in 3D Cardiac Microtissues and Reveal Non-cardiomyocyte Contributions to Heart Disease. Cell Stem Cell 26, 862–879.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulart E, de Caires-Junoir LC, Telles-Silva KA, Araujo BHS, Rocco SA, Sforca M, de Sousa IL, Kobayashi GS, Musso CM, Assoni AF, et al. (2020). 3D bioprinting of liver spheroids derived from human induced pluripotent stem cells sustain liver function and viability in vitro. Biofabrication 12, 015010. [DOI] [PubMed] [Google Scholar]
- Grigoryan B, Paulsen SJ, Corbett DC, Sazer DW, Fortin CL, Zaita AJ, Greenfield PT, Calafat NJ, Gounley JP, Ta AH, et al. (2019). Multivascular networks and functional intravascular topologies within biocompatible hydrogels. Science 464, 458–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdu Z, Mironov V, Mehesz AN, Norris RA, Markwald RR, and Visconti RP (2010). Tissue spheroid fusion-based in vitro screening assays for analysis of tissue maturation. J. Tissue Eng. Regnerative Med. 4, 689–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Wang M, Duan S, Franco PJ, Kenty JHR, Hedrick P, Xia Y, Allen A, Ferreira LMR, Strominger JL, et al. (2019). Generation of hypoimmunogenic human pluripotent stem cells. Proc. Natl. Acad. Sci. U. S. A. 116, 10441–10446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibino N, Villalona G, Pietris N, Duncan DR, Schoffner A, Roh JD, Yi T, Dobrucki LW, Mejias D, Sawh‐Martinez R, et al. (2011). Tissue‐engineered vascular grafts form neovessels that arise from regeneration of the adjacent blood vessel. FASEB J. 25, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton TJ, Jallerat Q, Palchesko RN, Park JH, Grodzicki MS, Shue H, Ramadan MH, Hudson AR, and Feinberg AW (2015). Three-dimensional printing of complex biological structures by freeform reversible embedding of suspended hydrogels. Sci. Adv. 1, e15700758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homan KA, Gupta N, Kroll KT, Kolesky DB, Skylar-Scott M, Miyoshi T, Mau D, Valerius MT, Ferrante T, Bonventre JV, et al. (2019). Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat. Methods 16, 255–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein SM, Batada NN, Vuoristo S, Ching RW, Autio R, Narvä E, Ng S, Sourour M, Hämälä R, Olsson C, et al. (2011). Copy number variation and selection during reprogramming to pluripotency. Nature 471, 58–62. [DOI] [PubMed] [Google Scholar]
- Jakab K, Neagu A, Mironov V, Markwald RR, and Forgacs G. (2004). Engineering biological structures of prescribed shaped using self-assembling multicellular systems. Proc. Natl. Acad. Sci. U. S. A. 101, 2864–2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakab K, Damon B, Marga F, Doaga O, Mironov V, Kosztin I, Markwald R, and Forgacs G. (2008). Relating cell and tissue mechanics: Implications and applications. Dev. Dyn. 237, 2438–2449. [DOI] [PubMed] [Google Scholar]
- Jakab K, Norotte C, Marga F, Murphy K, Vunjak-Novakovic G, and Forgacs G. (2010). Tissue engineering by self-assembly and bio-printing of living cells. Biofabrication 2, 0022001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HW, Lee SJ, Ko IK, Kengla C, Yoo JJ, and Atala A. (2016). A 3D bioprinting system to produce human-scale tissue constructs with structural integrity. Nat. Biotechnol. 34, 312–319. [DOI] [PubMed] [Google Scholar]
- Kelly BE, Bhattacharya I, Heidari H, Shusteff M, Spadaccini CM, and Taylor HK (2019). Volumetric additive manufacturing via tomographic reconstruction. Science 363, 1075–1079. [DOI] [PubMed] [Google Scholar]
- Kelm JM, Timmins NE, Brown CJ, Fussenegger M, and Nielsen LK (2003). Method for generation of homogeneous multicellular tumor spheroids applicable to a wide variety of cell types. Biotechnol. Bioeng. 83, 173–180. [DOI] [PubMed] [Google Scholar]
- Kim EM, Lee Y. Bin, Kim S. jeong, Park J, Lee J, Kim SW, Park H, and Shin H. (2019). Fabrication of core-shell spheroids as building blocks for engineering 3D complex vascularized tissue. Acta Biomater. 100, 158–172. [DOI] [PubMed] [Google Scholar]
- Kim TY, Kofron CM, King ME, Markes AR, Okundaye AO, Qu Z, Mende U, and Choi BR (2018). Directed fusion of cardiac spheroids into larger heterocellular microtissues enables investigation of cardiac action potential propagation via cardiac fibroblasts. PLoS One 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkton RD, Santiago-Maysonet M, Lawson JH, Tente WE, Dahl SLM, Niklason LE, and Prichard HL (2019). Bioengineered human acellular vessels recellularize and evolve into living blood vessels after human implantation. Sci. Transl. Med. 11, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesky DB, Truby RL, Gladman AS, Busbee TA, Homan KA, and Lewis JA (2014). 3D bioprinting of vascularized, heterogeneous cell-laden tissue constructs. Adv. Mater. 26, 3124–3130. [DOI] [PubMed] [Google Scholar]
- Kolesky DB, Homan KA, Skylar-Scott MA, and Lewis JA (2016). Three-dimensional bioprinting of thick vascularized tissues. Proc. Natl. Acad. Sci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosheleva NV, Efremov YM, Shavkuta BS, Zurina IM, Zhang D, Zhang Y, Minaev NV, Gorkun AA, Wei S, Shpichka AA, et al. (2020). Cell spheroid fusion: beyond liquid drops model. Sci. Rep. 10, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropp C, Massai D, and Zweigerdt R. (2017). Progress and challenges in large-scale expansion of human pluripotent stem cells. Process Biochem. 59, 244–254. [Google Scholar]
- Kuo HH, Gao X, DeKeyser JM, Fetterman KA, Pinheiro EA, Weddle CJ, Fonoudi H, Orman MV, Romero-Tejeda M, Jouni M, et al. (2020). Negligible-Cost and Weekend-Free Chemically Defined Human iPSC Culture. Stem Cell Reports 14, 256–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupfer ME, Lin WH, Ravikumar V, Qiu K, Wang L, Gao L, Bhuiyan DB, Lenz M, Ai J., Mahutga RR., et al. (2020). In Situ Expansion, Differentiation, and Electromechanical Coupling of Human Cardiac Muscle in a 3D Bioprinted, Chambered Organoid. Circ. Res. 127, 207–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J, Bernier D, Ouellet C, Goyette R, and Marceau N. (1985). Spheroidal aggregate culture of rat liver cells: Histotypic reorganization, biomatrix deposition, and maintenance of functional activities. J. Cell Biol. 101, 914–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A, Hudson AR, Shiwarski DJ, Tashman JW, Hinton TJ, Yerneni S, Bliley JM, Campbell PG, and Feinberg AW (2019). 3D bioprinting of collagen to rebuild components of the human heart. Science 365, 482–487. [DOI] [PubMed] [Google Scholar]
- Lee AS, Tang C, Rao MS, Weissman IL, and Wu JC (2013). Tumorigenicity as a clinical hurdle for pluripotent stem cell therapies. Nat. Med. 19, 998–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg GCJ, Cui X, Durham M, Veenendaal L, Schon BS, Hooper GJ, Lim KS, and Woodfield TBF (2021). Probing Multicellular Tissue Fusion of Cocultured Spheroids—A 3D-Bioassembly Model. Adv. Sci. 8, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little MH, and Combes AN (2019). Kidney organoids: accurate models or fortunate accidents. Genes Dev. 33, 1319–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litviňuková M, Talavera-López C, Maatz H, Reichart D, Worth CL, Lindberg EL, Kanda M, Polanski K, Heinig M, Lee M, et al. (2020). Cells of the adult human heart. Nature 588, 466–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour AA, Gonçalves JT, Bloyd CW, Li H, Fernandes S, Quang D, Johnston S, Parylak SL, Jin X, and Gage FH (2018). An in vivo model of functional and vascularized human brain organoids. Nat. Biotechnol. 36, 432–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RM, Fowler JL, Cromer MK, Lesch BJ, Ponce E, Uchida N, Nishimura T, Porteus MH, and Loh KM (2020). Improving the safety of human pluripotent stem cell therapies using genome-edited orthogonal safeguards. Nat. Commun. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle FT, Ghosh S, Genovese G, Handsaker RE, Kashin S, Meyer D, Karczewski KJ, O’Dushlaine C, Pato C, Pato M, et al. (2022). Whole-genome analysis of human embryonic stem cells enables rational line selection based on genetic variation. Cell Stem Cell 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki K, Endo K, Takahashi S, Funakoshi S, Takei I, Katayama S, Toyoda T, Kotaka M, Takaki T, Umeda M, et al. (2015). Efficient Detection and Purification of Cell Populations Using Synthetic MicroRNA Switches. Cell Stem Cell 16, 699–711. [DOI] [PubMed] [Google Scholar]
- Miller JS (2014). The Billion Cell Construct: Will Three-Dimensional Printing Get Us There? PLoS Biol. 12, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen D-HT, Cohen DM, Toro E, Chen AA, Galie PA, Yu X, et al. (2012). Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues. Nat. Mater. 11, 768–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirdamadi E, Tashman JW, Shiwarski DJ, Palchesko RN, and Feinberg AW (2020). FRESH 3D Bioprinting a Full-Size Model of the Human Heart. ACS Biomater. Sci. Eng. 6, 6453–6459. [DOI] [PubMed] [Google Scholar]
- Mironov V, Visconti RP, Kasyanov V, Forgacs G, Drake CJ, and Markwald RR (2009). Organ printing: Tissue spheroids as building blocks. Biomaterials 30, 2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morizane R., Lam AQ., Freedman BS., Kishi S., Valerius MT., and Bonventre JV. (2015). Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat. Biotechnol. 33, 1193–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscona A. (1957). The development in vitro of chimeric aggregates of dissociated embryonic chick and mouse cells. Proc. Natl. Acad. Sci. 43, 184–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niklason LE, Gao J, Abbott WM, Hirschi KK, Houser S, Marini R, and Langer R. (1999). Functional arteries grown in vitro. Science 284, 489–493. [DOI] [PubMed] [Google Scholar]
- Organdonor.gov Organ Donation and Transplantation.
- Park J, Shrestha R, Qiu C, Kondo A, Huang S, Werth M, Li M, Barasch J, and Suszták K. (2018). Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science 360, 758–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashneh-Tala S, MacNeil S, and Claeyssens F. (2016). The tissue-engineered vascular graft - Past, present, and future. Tissue Eng. - Part B Rev. 22, 68–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson JT, Gilliland T, Maxfield MW, Church S, Naito Y, Shinoka T, and Breuer CK (2012). Tissue-engineered vascular grafts for use in the treatment of congenital heart disease: From the bench to the clinic and back again. Regen. Med. 7, 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polonchuk L, Surija L, Lee MH, Sharma P, Ming CLC, Richter F, Ben-Sefer E, Rad MA, Sarmast HMS, Al Shamery W, et al. (2021). Towards engineering heart tissues from bioprinted cardiac spheroids. Biofabrication 13, 045009. [DOI] [PubMed] [Google Scholar]
- Rossi G, Broguiere N, Miyamoto M, Boni A, Guiet R, Girgin M, Kelly RG, Kwon C, and Lutolf MP (2021). Capturing Cardiogenesis in Gastruloids. Cell Stem Cell 28, 230–240.e1–e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadtler K, Singh A, Wolf MT, Wang X, Pardoll DM, and Elisseeff JH (2016). Design, clinical translation and immunological response of biomaterials in regenerative medicine. Nat. Rev. Mater. 1. [Google Scholar]
- Sato N, Meijer L, Skaltsounis L, Greengard P, and Brivanlou AH (2004). Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat. Med. 10, 55–63. [DOI] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, Van De Wetering M, Barker N, Stange DE, Van Es JH, Abo A, Kujala P, Peters PJ, et al. (2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265. [DOI] [PubMed] [Google Scholar]
- Schepers A, Li C, Chhabra A, Seney BT, and Bhatia S. (2016). Engineering a perfusable 3D human liver platform from iPS cells. Physiol. Behav. 176, 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharmin S, Taguchi A, Kaku Y, Yoshimura Y, Ohmori T, Sakuma T, Mukoyama M, Yamamoto T, Kurihara H, and Nishinakamura R. (2016). Human induced pluripotent stem cell-derived podocytes mature into vascularized glomeruli upon experimental transplantation. J. Am. Soc. Nephrol. 27, 1778–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevach M, Zax R, Abrahamov A, Fleischer S, Shapira A, and Dvir T. (2015). Omentum ECM-based hydrogel as a platform for cardiac cell delivery. Biomed. Mater. 10, 034106. [DOI] [PubMed] [Google Scholar]
- Silva AC, Matthys OB, Joy DA, Kauss MA, Natarajan V, Lai MH, Turaga D, Blair AP, Alexanian M, Bruneau BG, et al. (2021). Co-emergence of cardiac and gut tissues promotes cardiomyocyte maturation within human iPSC-derived organoids. Cell Stem Cell 28, 2137–2152.e6. [DOI] [PubMed] [Google Scholar]
- Skylar-Scott MA, Uzel SGM, Nam LL, Ahrens JH, Truby RL, Damaraju S, and Lewis JA (2019a). Biomanufacturing of organ-specific tissues with high cellular density and embedded vascular channels. Sci. Adv. 5, eaaw2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skylar-Scott MA, Mueller J, Visser CW, and Lewis JA (2019b). Voxelated soft matter via multimaterial multinozzle 3D printing. Nature 575, 330–335. [DOI] [PubMed] [Google Scholar]
- Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, Wells SI, Zorn AM, et al. (2011). Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470, 105–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivatsan SR, Regier MC, Barkan E, Franks JM, Packer JS, Grosjean P, Duran M, Saxton S, Ladd JJ, Spielmann M, et al. (2021). Embryo-scale, single-cell spatial transcriptomics. 117, 111–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens KR, Scull MA, Ramanan V, Fortin CL, Chaturvedi RR, Knouse KA, Xiao JW, Fung C, Mirabella T, Chen AX, et al. (2017). In situ expansion of engineered human liver tissue in a mouse model of chronic liver disease. Sci. Transl. Med. 9, 5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi A, Kaku Y, Ohmori T, Sharmin S, Ogawa M, Sasaki H, and Nishinakamura R. (2014). Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell 14, 53–67. [DOI] [PubMed] [Google Scholar]
- Takasato M, Er PX, Becroft M, Vanslambrouck JM, Stanley EG, Elefanty AG, and Little MH (2014). Directing human embryonic stem cell diffentiation towards a renal lineage generates a self-organizing kidney. Nature Cell Biology 16 118–126.. [DOI] [PubMed] [Google Scholar]
- Takasato M, Er PX, Chiu HS, Maier B, Baillie GJ, Ferguson C, Parton RG, Wolvetang EJ, Roost MS, Chuva de Sousa Lopes SM, et al. (2015). Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564–568. [DOI] [PubMed] [Google Scholar]
- Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, Zhang R-R, Ueno Y, Zheng Y-W, Koike N, et al. (2013). Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 499, 481–484. [DOI] [PubMed] [Google Scholar]
- Wilson HV (1907). On some phenomena of coalescence and regeneration in sponges. J. Elisha Mitchell Sci. Soc. 23, 161–174. [Google Scholar]
- Wu W, Deconinck A, and Lewis JA (2011). Omnidirectional printing of 3D microvascular networks. Adv. Mater. 23, 3–5. [DOI] [PubMed] [Google Scholar]
- Xu H, Wang B, Ono M, Kagita A, Fujii K, Sasakawa N, Ueda T, Gee P, Nishikawa M, Nomura M, et al. (2019). Targeted Disruption of HLA Genes via CRISPR-Cas9 Generates iPSCs with Enhanced Immune Compatibility. Cell Stem Cell 24, 566–578.e7. [DOI] [PubMed] [Google Scholar]