Abstract
Background
Human papillomavirus–related biomarkers such as p16/Ki-67 “dual-stain” (DS) cytology have shown promising clinical performance for anal cancer screening. Here, we assessed the performance of automated evaluation of DS cytology (automated DS) to detect anal precancer in men who have sex with men (MSM) and are living with human immunodeficiency virus (HIV).
Methods
We conducted a cross-sectional analysis of 320 MSM with HIV undergoing anal cancer screening and high-resolution anoscopy (HRA) in 2009–2010. We evaluated the performance of automated DS based on a deep-learning classifier compared to manual evaluation of DS cytology (manual DS) to detect anal intraepithelial neoplasia grade 2 or 3 (AIN2+) and grade 3 (AIN3). We evaluated different DS-positive cell thresholds quantified by the automated approach and modeled performance compared with other screening strategies in a hypothetical population of MSM with HIV.
Results
Compared with manual DS, automated DS had significantly higher specificity (50.9% vs 42.2%; P < .001) and similar sensitivity (93.2% vs 92.1%) for detection of AIN2+. Human papillomavirus testing with automated DS triage was significantly more specific than automated DS alone (56.5% vs 50.9%; P < .001), with the same sensitivity (93.2%). In a modeled analysis assuming a 20% AIN2+ prevalence, automated DS detected more precancers than manual DS and anal cytology (186, 184, and 162, respectively) and had the lowest HRA referral rate per AIN2+ case detected (3.1, 3.5, and 3.3, respectively).
Conclusions
Compared with manual DS, automated DS detects the same number of precancers, with a lower HRA referral rate.
Keywords: anal cancer, p16/Ki-67 dual stain, human papillomavirus, screening, artificial intelligence
In this study of 320 men who have sex with men and are living with human immunodeficiency virus, we demonstrate that automated evaluation of p16/Ki-67 dual-stain cytology has high sensitivity for anal precancer and reduces unnecessary referral for high-resolution anoscopy, compared with manual evaluation.
Anal cancer incidence and mortality rates have been increasing in the United States [1], particularly among certain populations with elevated risk, most notably, human immunodeficiency virus (HIV)-positive men who have sex with men (MSM) [2, 3]. Most anal squamous cell carcinomas are caused by high-risk human papillomavirus (HPV) infection [4]. A majority of these infections will clear on their own without causing disease, but a subset will persist and progress to precancer, and an even smaller subset will progress to invasive cancer [5]. Individuals living with HIV are more likely to have persistent HPV infection, harbor a broader range of high-risk HPV types, and have higher rates of anal precancer and cancer than those without HIV [6].
The Anal Cancer/HSIL Outcomes Research (ANCHOR) study recently demonstrated that treating high-grade squamous intraepithelial lesions (HSILs) significantly reduced the risk of anal cancer in people with HIV, highlighting the importance of anal cancer screening [7]. While screening is conducted in some specialty clinics for MSM with HIV, there are no national screening guidelines; practices are not standardized and tend to vary across different clinical settings and providers [8]. Current practice may include anal cytology and high-resolution anoscopy (HRA) with targeted biopsies [8]; however, anal cytology has limited reproducibility and sensitivity and therefore needs to be repeated frequently [9]. Furthermore, performing HRA requires specialized training and expertise that is not widely available [9]. These challenges emphasize the need for objective biomarkers with high specificity to limit unnecessary HRA referral while maintaining high sensitivity for anal precancers.
Although cervical cancer screening is shifting toward primary HPV testing owing to its high sensitivity, the high prevalence of HPV in MSM living with HIV may limit the effectiveness of HPV testing for primary screening [10]. HPV triage biomarkers for cervical cancer screening may have applications as primary anal cancer screening tools, alone or in combination with HPV testing. p16/Ki-67 “dual-stain” (DS) cytology, a Food and Drug Administration–approved triage test for HPV-positive women in cervical cancer screening, has shown promising cross-sectional and long-term clinical performance in the detection of anal precancer [11, 12]. The DS technique is amenable to automated detection using scanned cytology slides. Previously, our group developed a deep learning approach for automated evaluation of DS cytology (hereafter, automated DS) for cervical and anal precancer detection [13]. In the current study, we performed an in-depth clinical analysis of this algorithm compared with manual evaluation of DS cytology (manual DS) for the detection of anal precancer in MSM living with HIV.
METHODS
Study Population
We enrolled MSM living with HIV who were aged ≥18 years and undergoing anal cancer screening and HRA at the Anal Cancer Screening Clinic at Kaiser Permanente Northern California (KPNC) in San Francisco. Participants with no history of anal cancer were identified through the Kaiser HIV registry. For this cross-sectional analysis, we used the baseline data from 363 men enrolled between August 2009 and June 2010. Only participants with manual and automated DS results of adequate slide quality were included in our analysis. This study was reviewed and approved by the National Cancer Institute and KPNC.
Clinical Evaluation
Two anal swab specimens for liquid cytology were collected during clinical examination and transferred to PreservCyt medium (Hologic). Results were reported, according to the Bethesda system [14], as negative for intraepithelial lesion or malignancy (NILM); atypical squamous cells of undetermined significance (ASC-US); atypical squamous cells, cannot rule out HSIL (ASC-H); low-grade squamous intraepithelial lesion (LSIL); or HSIL [14]. All patients underwent a digital anorectal examination followed by HRA. Biopsies of suspicious lesions identified by HRA were performed, and biopsy specimens were reviewed by KPNC pathologists, according to clinical practice. Results were reported as negative, condyloma, or anal intraepithelial neoplasia (AIN) grades 1–3.
Clinical End Points
We created composite cytologic and histologic end points to maximize disease ascertainment and minimize misclassification by HRA, as described elsewhere [12]. We classified precancers as having either HSIL with cytology and/or AIN grade 2 or 3 (AIN2+) or AIN grade 3 (AIN3) with histology. Cytologic diagnoses of NILM, ASC-US, ASC-H, or LSIL and histologic results of negative, condyloma, or AIN1 were categorized as AIN less than grade 2 (<AIN2).
p16/Ki-67 DS Biomarker Testing and Automation
p16/Ki-67 dual immunostaining was performed on residual cytologic specimens by Roche MTM Laboratories, using the CINtec PLUS Kit according to the manufacturer’s guidelines. Trained cytotechnologists reviewed all cases and considered a slide positive if ≥1 anal squamous epithelial cell stained positive for both p16 and Ki-67. The number of positive cells was also reported in semiquantitative categories as 0, 1, 2–5, 6–50, or >50 cells.
Development and validation of the automated DS technology is described in detail elsewhere [13]. Briefly, the deep learning approach (convolutional neural network with 4 layers) for automation of DS technology uses an algorithm to quantify the number of DS-positive cells on a ThinPrep slide by detecting the number of tiles above a specific likelihood threshold (0.5). A slide is considered positive if the number of DS-positive tiles exceeds a predetermined threshold of ≥3 tiles per cell, as described elsewhere (hereafter referred to as a “3-cell threshold”) [13].
HPV DNA Testing
HPV DNA testing was performed on the second liquid cytologic specimen, using the cobas 4800 HPV assay (Roche Molecular Systems), which provides results for HPV-16, HPV-18, and 12 other pooled high-risk HPV genotypes (type 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, and 68; “other high-risk 12”) [15].
Statistical Analyses
We used descriptive statistics and χ2 tests to summarize demographics and risk factors by disease status. We calculated the percentage of patients with a positive test result (ie, positivity), sensitivity, specificity, and predictive values for detection of AIN2+ and AIN3 for manual and automated DS alone and in combination with HPV results, including triage of HPV-positive results (ie, positive if both HPV and DS results are positive), partial HPV-16/18 genotyping with DS triage for the other 12 high-risk HPV types (ie, positive for HPV-16/18 and DS positive for the remainder of those positive for another high-risk HPV type), and cotesting with HPV-16/18 partial genotyping (ie, positive if either HPV-16/18 or DS positive).
We used the McNemar χ2 test to compare the paired positivity, sensitivity, and specificity of automated and manual DS for detection of AIN2+ and AIN3; differences in predictive values were evaluated using the R package DTComPair and the generalized score statistic. We performed a receiver operating characteristic (ROC) curve analysis to evaluate the performance of automated DS at different thresholds for detecting AIN2+ and AIN3, and we calculated the respective area under the ROC curve (AUC). For comparison, we plotted the coordinates for manual DS sensitivity and specificity at the 1-cell and >50-cell thresholds, which our group previously demonstrated approximates the performance of HSIL cytology in cervical cancer screening [16], a result that may be used for clinical decision making in anal cancer screening [17].
We modeled the performance of automated DS in a hypothetical population of 1000 MSM living with HIV assuming a lower (20%) and higher (40%) prevalence of AIN2+. Estimates were selected based on a systematic review and meta-analysis of the prevalence of AIN2+ in studies restricted to MSM with HIV, which ranged from 7.5% to 54.5% [18]. We compared different automated and manual DS thresholds alone and with HPV testing and applied external summary performance estimates of cytology (ASC-US or worse [ASC-US+] and HSIL or worse [HSIL+] thresholds) from a meta-analysis of studies of MSM with HIV [9]. We calculated hypothetical HRA referral rates based on test positivity, and the number of AIN2+ cases detected based on sensitivity and specificity estimates for each test. All analyses were performed using Stata (version 17.0) and R (version 4.1.1) software. Differences were considered statistically significant at P < .05 for all tests of significance.
RESULTS
Study Population Characteristics
Of the 363 individuals enrolled in the study, 43 (11.8%) were excluded, leaving 320 individuals included in our analysis (Figure 1). Based on a combined cytologic and histologic end point, 232 men (72.5%) had <AIN2 and 88 (27.5%) had AIN2+. Of those with AIN2+, 50 (56.8%) had AIN3. Most were white (83.4%) and non-Hispanic (70.9%), with a mean age of 52.8 years. The overall high-risk HPV positivity rate was 80.9%, with 45.9% testing positive for HPV-16 and/or HPV-18. All cases tested HPV positive, with 65.9% positive for HPV-16/18. HPV positivity was significantly lower among those with <AIN2 (73.7% for all high-risk HPV types and 35.7% for HPV-16/18; P < .001). Those with <AIN2 had higher CD4 cell counts than those with AIN2+ (≥350/µL in 81.9% vs 70.5%, respectively; P = .03) and were less likely to have ever smoked (43.1% vs 55.7%; P = .04) (Table 1).
Figure 1.
CONSORT diagram illustrating the study population. Abbreviations: <AIN2, anal intraepithelial neoplasia (AIN) less than grade 2 by cytology and histology; AIN2+, AIN grade 2 or 3 by cytology and/or histology; DS, dual-stain cytology; HIV+, human immunodeficiency virus positive; MSM, men who have sex with men.
Table 1.
Characteristics of the Study Populationa
| Characteristic | Study Participants, No. (Column %)b | P Value | ||
|---|---|---|---|---|
| Total (n = 320) | <AIN2 (n = 232) |
AIN2+ (n = 88) |
||
| Age, mean (SD), y | 52.8 (9.1) | 53.1 (9.4) | 51.8 (8.3) | .30 |
| Race | ||||
| White | 267 (83.4) | 190 (81.9) | 77 (87.5) | .23 |
| Nonwhite | 41 (12.8) | 32 (13.8) | 9 (10.2) | .39 |
| Data missing | 12 (3.8) | 10 (4.3) | 2 (2.3) | … |
| Ethnicity | ||||
| Non-Hispanic | 227 (70.9) | 165 (71.1) | 62 (70.5) | .91 |
| Hispanic | 29 (9.1) | 24 (10.3) | 5 (5.7) | .19 |
| Data missing | 64 (20.0) | 43 (18.5) | 21 (23.9) | … |
| HPV statusc | ||||
| Positive | 259 (80.9) | 171 (73.7) | 88 (100.0) | <.001d |
| Negative | 59 (18.4) | 59 (25.4) | 0 (0.0) | |
| Data missing | 2 (0.6) | 2 (0.9) | 0 (0.0) | … |
| HPV genotypee | ||||
| HPV-16/18f | 119 (45.9) | 61 (35.7) | 58 (65.9) | <.001d |
| HPV-16 positiveg | 97 (37.5) | 45 (26.3) | 52 (59.1) | <.001d |
| HPV-18 positive, HPV-16 negativeh | 22 (8.5) | 16 (9.4) | 6 (6.8) | .49 |
| Other HR-HPVi | 140 (54.1) | 110 (64.3) | 30 (34.1) | <.001d |
| HIV viral load | ||||
| <75 Copies/mL | 277 (86.6) | 201 (86.6) | 76 (86.4) | .95 |
| ≥75 Copies/mL | 31 (9.7) | 25 (10.8) | 6 (6.8) | .29 |
| Data missing | 12 (3.8) | 6 (2.6) | 6 (6.8) | … |
| CD4 cell count | ||||
| ≥350/µL | 252 (78.8) | 190 (81.9) | 62 (70.5) | .03d |
| <350/µL | 56 (17.5) | 36 (15.5) | 20 (22.7) | .13 |
| Data missing | 12 (3.8) | 6 (2.6) | 6 (6.8) | … |
| Any prior anal cancer screening | ||||
| Yes | 160 (50.0) | 114 (49.1) | 46 (52.3) | .62 |
| No | 15 (4.7) | 13 (5.6) | 2 (2.3) | |
| Data missing | 145 (45.3) | 105 (45.3) | 40 (45.5) | … |
| Ever smoker | ||||
| Yes | 149 (46.6) | 100 (43.1) | 49 (55.7) | .04d |
| No | 119 (37.2) | 98 (42.2) | 21 (23.9) | |
| Data missing | 52 (16.3) | 34 (14.7) | 18 (20.5) | … |
| Lifetime no. of male partners | ||||
| 0–4 | 46 (14.4) | 38 (16.4) | 8 (9.1) | .10 |
| 5–39 | 112 (35.0) | 88 (37.9) | 24 (27.3) | .07 |
| ≥40 | 101 (31.6) | 66 (28.4) | 35 (39.8) | .05 |
| Data missing | 61 (19.1) | 40 (17.2) | 21 (23.9) | … |
Abbreviations: AIN, anal intraepithelial neoplasia; <AIN2, AIN less than grade 2 by cytology and histology; AIN2+, AIN grade 2 or 3 by cytology and/or histology; HIV, human immunodeficiency virus; HPV, human papillomavirus; HR-HPV, high-risk human papillomavirus; SD, standard deviation.
Univariate analyses of study population demographics and risk factors dichotomized by case status. All information was collected from a self-administered questionnaire completed by study participants or extracted from electronic medical records, and χ2 tests were used to identify statistically significant differences (P < .05) between participants with <AIN2 and those with AIN2+.
Data represent no. (column %) except where identified as mean (SD).
Defined as positive for HPV type 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, or 68; otherwise considered negative.
Significant at P < .05 for comparison between <AIN2 and AIN2+ groups.
HPV genotypes for participants with positive HPV results; percentages represent proportion of those with positive results.
Positive for HPV-16 or HPV-18, regardless of other positive HPV results.
Positive for HPV-16, regardless of other positive HPV results.
Positive for HPV-18 but negative for HPV-16.
Negative for HPV-16 or HPV-18 but positive for any of the other 12 high-risk HPV types: 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, or 68.
Performance of Manual and Automated DS
The rate of positivity was significantly higher for manual DS (67.2%) than for automated DS (61.3%) (P = .001; Table 2). The sensitivity for detecting AIN2+ was similar for automated and manual DS (93.2% and 92.1%, respectively), whereas the specificity of automated DS was significantly higher (50.9% vs 42.2%; P < .001). Likewise, the positive predictive value of automated DS was significantly higher than that of manual DS (41.8% vs 37.7%; P < .001), whereas the negative predictive values were similar. For detecting AIN3, automated DS, compared with manual DS, had comparable sensitivity (96.0% vs 94.0%, respectively), significantly higher specificity (45.2% vs 37.8%; P < .001), and significantly higher positive predictive value (24.5% vs 21.9%; P < .001).
Table 2.
Performance of Manual and Automated Dual-Stain Cytology for Detection of Anal Precancer Among 320 Human Immunodeficiency Virus–Positive Men Who Have Sex With Mena
| Variable | DS Result, No./Total No. (%) | |
|---|---|---|
| Manual DS | Automated DS | |
| Threshold | 1 Cell positive | 3 Cells positive |
| Positivity | 215/320 (67.2)a | 196/320 (61.3)a |
| Detection of AIN2+ | ||
| Sensitivity | 81/88 (92.1) | 82/88 (93.2) |
| Specificity | 98/232 (42.2)a | 118/232 (50.9)a |
| PPV | 81/215 (37.7)b | 82/196 (41.8)b |
| NPV | 98/105 (93.3) | 118/124 (95.2) |
| Detection of AIN3 | ||
| Sensitivity | 47/50 (94.0) | 48/50 (96.0) |
| Specificity | 102/270 (37.8)a | 122/270 (45.2)a |
| PPV | 47/215 (21.9)b | 48/196 (24.5)b |
| NPV | 102/105 (97.1) | 122/124 (98.4) |
Abbreviations: AIN2+, anal intraepithelial neoplasia (AIN) grade 2 or 3 by cytology and/or histology; AIN3, AIN grade 3 by cytology and/or histology; DS, dual-stain cytology; NPV, negative predictive value; PPV, positive predictive value.
Significant at P < .05 for comparison between automated and manual DS (McNemar χ2 test).
Significant at P < .05; predictive values were evaluated with the R package DTComPair, using the generalized score statistic.
Evaluating Different Thresholds for Automated DS
We evaluated the absolute number of DS-positive cells quantified by the automated algorithm compared with the semiquantitative categories derived from manual DS. In general, there was a shift toward higher positive DS cell counts for the automated approach within each manual DS category, and at each threshold, automated DS had higher positivity (Supplementary Table 1).
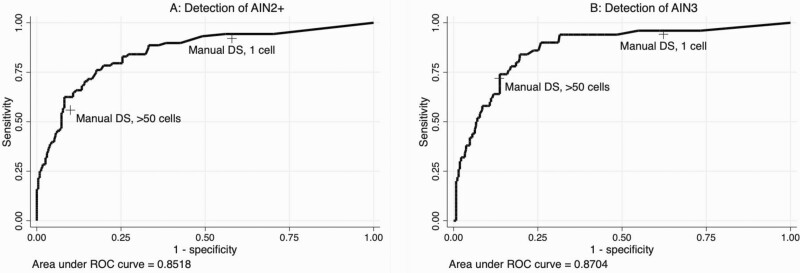
We generated an ROC curve evaluating the number of cells positive with automated DS for detection of AIN2+ (AUC, 0.85) and AIN3 (AUC, 0.87) (Figure 2). For comparison, we plotted the sensitivity and specificity coordinates for manual DS at the 1-cell and >50-cell thresholds, demonstrating that automated DS improves detection of AIN2+ and AIN3 compared with manual DS.
Figure 2.
Receiver operating characteristic (ROC) curve analysis of “positive” tiles identified by automated dual-stain (DS) technology for the detection of anal intraepithelial neoplasia (AIN) grade 2 or 3 (AIN2+) (A) and AIN grade 3 (AIN3) (B). Manual DS sensitivity and specificity estimates at the 1-cell and 50-cell thresholds are plotted for reference.
We used the ROC curves to determine a threshold at which the performance of automated DS was equivalent to or better than that of manual DS at the 50-cell threshold, to approximate the high specificity of cytologic HSIL, which may be used for clinical decision making [16, 17]. For AIN2+ detection, a threshold of 58 positive cells for automated DS achieved higher sensitivity and specificity (62.5% and 91.8%, respectively) compared with manual DS at the 50-cell threshold (55.7% and 90.1%, respectively) (Figure 2A). For AIN3, at the 50-cell threshold, automated DS achieved slightly higher sensitivity than manual DS (74.0% vs 72.0%, respectively), with equivalent specificity (86.3% vs 86.7%) (Figure 2B).
Automated and Manual DS With Different HPV Testing Strategies
We evaluated the performance of automated and manual DS in combination with 3 different HPV testing strategies (Table 3). For both automated and manual DS approaches, primary HPV testing with DS triage (ie, with results considered positive if both HPV and DS results are positive), compared with DS approaches alone, had significantly lower positivity and significantly higher specificity at equal sensitivity for AIN2+ and AIN3. Partial HPV-16/18 genotyping with DS triage of other high-risk HPV types had higher positivity and sensitivity but equal or worse specificity, respectively, than automated and manual DS alone. Similarly, cotesting with HPV-16/18 partial genotyping and DS had significantly higher positivity and high sensitivity but the lowest specificity for any testing strategy (Table 3).
Table 3.
Performance of Automated and Manual Dual-Stain Cytology in Various Combinations With Human Papillomavirus Testing and Partial Genotypinga
| DS Testing Strategy | Positivity, % | AIN2+ | AIN3 | ||
|---|---|---|---|---|---|
| Sensitivity, % | Specificity, % | Sensitivity, % | Specificity, % | ||
| Automated evaluation (3-cell threshold) | |||||
| HPV and DSb | 57.2c | 93.2 | 56.5c | 96.0 | 50.0c |
| HPV-16/18 with DS triage of other HR- HPVd | 64.1 | 96.6 | 47.8 | 98.0 | 41.8 |
| HPV-16/18 or DSe | 68.1c | 96.6 | 42.7c | 98.0 | 37.4c |
| Manual evaluation (1-cell threshold) | |||||
| HPV and DSb | 61.9f | 92.1 | 49.6f | 94.0 | 44.1f |
| HPV-16/18 with DS triage of other HR-HPVd | 67.2 | 95.5 | 43.0 | 98.0 | 38.1 |
| HPV-16/18 or DSe | 72.5f | 95.5 | 36.2f | 98.0 | 32.2f |
Abbreviations: AIN2+, anal intraepithelial neoplasia (AIN) grade 2 or 3 by cytology and/or histology; AIN3, AIN grade 3 by cytology and/or histology; DS, dual-stain cytology; HPV, human papillomavirus; HR-HPV, high-risk HPV.
Automated and manual evaluation of DS combined with 3 different strategies incorporating HPV testing with or without partial HPV-16/18 genotyping, in the detection AIN2+ and AIN3 among 320 men who have sex with men and are living with human immunodeficiency virus.
Considered positive if both HPV (types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, or 68) and DS results are positive; otherwise considered negative.
Significant at P < .05 (McNemar χ2 test) for difference between HPV testing with automated DS and automated DS alone.
Considered positive if HPV-16 or HPV-18 results are positive, with DS triage of the other 12 high-risk HPV types.
Considered positive if either HPV-16 or HPV-18 results are positive or if DS results are positive; otherwise considered negative.
Significant at P < .05 (McNemar χ2 test) for difference between HPV testing with manual DS and manual DS alone.
Modeled Performance of Different Testing Strategies for Anal Cancer Screening
To put our findings in clinical context and to compare our results with the performance of anal cytologic testing, we modeled the performance of different screening approaches in a hypothetical population of 1000 MSM with HIV, assuming 2 different population prevalence estimates for AIN2+ (20% and 40%; Table 4) and using external estimates of sensitivity and specificity for cytology based on a meta-analysis in MSM living with HIV [9]. In these strategies, participants positive with individual tests or test strategies would be referred to HRA.
Table 4.
Simulated Clinical Performance of Screening Strategies to Detect Anal Precancer in a Hypothetical Population of 1000 Men Who Have Sex With Men and Are Living with Human Immunodeficiency Virusa
| Cytologic Screening Strategy |
Sensitivity, % | Specificity, % | Value for 20% AIN2+ Prevalence/Value for 40% AIN2+ Prevalence | |||
|---|---|---|---|---|---|---|
| Test Positivity, % |
No. Referred to HRA | No. of Precancer Cases Detected | No. Referred to HRA per Precancer Case Detected | |||
| ASC-US+ | 80.8 | 54.0 | 53.0; 59.9 | 530; 599 | 162; 323 | 3.3; 1.9 |
| HSIL+ | 40.0 | 92.5 | 14.0; 20.5 | 140; 205 | 80; 160 | 1.8; 1.3 |
| Manual DS (1-cell threshold) | 92.1 | 42.2 | 64.6; 71.5 | 646; 715 | 184; 368 | 3.5; 1.9 |
| Manual DS (>50-cell threshold) | 55.7 | 90.1 | 19.0; 28.2 | 190; 282 | 111; 223 | 1.7; 1.3 |
| Automated DS (3-cell threshold) | 93.2 | 50.9 | 57.9; 66.8 | 579; 668 | 186; 373 | 3.1; 1.8 |
| Automated DS (>58-cell threshold) | 62.5 | 91.8 | 19.1; 29.9 | 191; 299 | 125; 250 | 1.5; 1.2 |
| HPV and automated DSb | 93.2 | 56.5 | 53.4; 63.4 | 534; 634 | 186; 373 | 2.9; 1.7 |
| HPV-16/18 with automated DS triage of other HR-HPVc | 96.6 | 47.8 | 61.1; 69.9 | 611; 699 | 193; 386 | 3.2; 1.8 |
Abbreviations: AIN2+, anal intraepithelial neoplasia grade 2 or 3 by cytology and/or histology; ASC-US+, cytology result worse than atypical squamous cells of undetermined significance; DS, dual-stain technique; HPV, human papillomavirus; HR-HPV, high-risk HPV; HRA, high-resolution anoscopy; HSIL+, cytology result worse than high-grade squamous intraepithelial lesion.
Population prevalence estimates of AIN2+ for men who have sex with men and are living with human immunodeficiency virus, selected from a systematic review and meta-analysis [17].
Considered positive if both HPV and DS results are positive.
Considered positive if either HPV-16/18 or DS results are positive.
For the 20% prevalence scenario, automated DS at a 3-cell threshold detected more cases of AIN2+ than cytology (ASC-US+) or manual DS. with a sensitivity of 93.2% (vs 80.8% and 92.1%, respectively) and had lower HRA referral rate (ie, lower test positivity) than manual DS (57.9% vs 64.6%, respectively). Although the proportion referred to HRA based on a positive cytologic (ASC-US+) result was slightly lower than that with automated DS (53.0%), the ratio of HRA referrals per cases detected was lower for automated DS owing to its superior sensitivity. Primary HPV testing with automated DS triage resulted in a lower HRA referral rate than automated DS alone (53.4% vs 57.9%, respectively) for the same number of cases detected (n = 186). Patterns were similar in the 40% AIN2+ prevalence setting, with greater efficiency observed for all strategies than with a setting with 20% prevalence, demonstrated by lower ratios of HRA referrals per case of AIN2+. For strategies designed to maximize specificity (ie, prioritizing HRA referral for only those individuals with the highest risk of precancer), including cytology (HSIL+), >50 cells positive with manual DS, and >58 cells positive with automated DS, automated DS referred a higher or nearly equivalent number of individuals to HRA than cytology (HSIL+) or manual DS but detected the most AIN2+ cases in both the higher- and lower-prevalence settings.
DISCUSSION
The high burden of HPV-associated anal disease among MSM living with HIV and the limitations of cytology and high-quality HRA practice underscore the need for molecular biomarkers for anal cancer screening. To our knowledge, we are the first to develop and clinically validate an algorithm for automated evaluation of DS cytology slides. Our data demonstrate that p16/Ki-67 DS cytology can detect precancer with high sensitivity and specificity and that using an automated approach further improved specificity. Primary HPV testing with automated DS triage had similar sensitivity and improved specificity compared with automated DS alone, while algorithms involving HPV-16/18 partial genotyping in conjunction with automated DS had much lower specificity.
Our findings are in line with findings of a prior study by our group that showed similar sensitivity and improved specificity of automated compared with manual DS evaluation in HPV-positive women undergoing cervical cancer screening [13]. Of note, we applied an automated DS algorithm that was primarily trained for cervical cytology. This algorithm showed good performance in anal cytology, demonstrating the robustness of the approach. While manual DS has well-established superior performance compared with cytology for HPV triage in cervical cancer screening, few studies have evaluated manual DS for anal cancer screening, with estimates varying from 41% to 84% for sensitivity and from 65% to 72% for specificity [19–21]. Differences in performance may be attributed to the composition of study participants (ie, differences in HIV prevalence or recruitment approach), disease prevalence and outcome definitions, and/or differences in slide preparation (ie, using the same slide for cytology and DS testing) and/or adequacy (eg, low cellularity) [19–21].
The heterogeneity in the literature surrounding the performance of manual DS underscores the importance of an automated approach, which would reduce subjective interpretation and could be used for quality control of a manual DS program. Automation would also allow for many slides to be evaluated in a shorter time frame, with a subset identified for manual review, particularly in settings with few skilled cytotechnologists. Furthermore, automated DS provides fully quantitative results, so thresholds can potentially be shifted to meet the needs of the clinical setting. For example, if HRA capacity is limited, a higher threshold could be used to optimize specificity and refer only the highest-risk patients. However, in settings where it is desirable to maximize sensitivity, the positivity threshold could be lowered.
In the current study, we primarily focused on automated thresholds with higher specificity, because lowering the threshold to 1 or 2 positive cells would not have changed the sensitivity but would have lowered specificity. In our modeled analysis, we demonstrated that an automated threshold of 58 DS-positive cells had higher sensitivity than HSIL cytology for AIN2+, with approximately equivalent specificity. This has potential clinical implications, because HSIL is often used as a cytologic indicator of underlying anal precancer risk to guide clinical management. More research is needed to evaluate a head-to-head comparison of automated DS and anal cytology within the same study population, as we could only model this scenario based on published estimates for cytology.
The strengths of our study include a large, clinical population of MSM living with HIV, with cytology and HRA performed in all participants. We created combined cytologic and histologic end points to account for potential disease misclassification resulting from HRA; however, misclassification is still possible. Our study population was overwhelmingly white and non-Hispanic with well-controlled HIV infection, and our results may not be generalizable to all MSM living with HIV. More research is needed in other populations at risk of anal cancer, such as MSM without HIV and women living with HIV, who may also benefit from anal cancer screening. Future studies are also required to assess issues related to implementation of this technology in a clinical setting.
In conclusion, automated evaluation of p16/Ki-67 DS cytology is a promising biomarker for anal cancer screening in MSM living with HIV. In particular, it shows great clinical utility for optimizing sensitivity and specificity, either alone or in combination with primary HPV screening. Future research should evaluate the performance of automated DS for long-term risk stratification in prospective studies, as well as in other populations at risk for development of anal cancer.
Supplementary Material
Notes
Financial support. This work was supported by the Intramural Research Program of the National Institute of Health and the National Cancer Institute (grant Z01 CP010124-21).
Contributor Information
Camryn M Cohen, Division of Cancer Epidemiology & Genetics, National Cancer Institute, Rockville, Maryland, USA.
Nicolas Wentzensen, Division of Cancer Epidemiology & Genetics, National Cancer Institute, Rockville, Maryland, USA.
Bernd Lahrmann, Steinbeis Transfer Center for Medical Systems Biology, Heidelberg, Germany.
Diane Tokugawa, Kaiser Permanente, Permanente Medical Group Regional Laboratory, Berkeley, California, USA.
Nancy Poitras, Kaiser Permanente, Permanente Medical Group Regional Laboratory, Berkeley, California, USA.
Liam Bartels, Steinbeis Transfer Center for Medical Systems Biology, Heidelberg, Germany; Hamamatsu Tissue Imaging and Analysis Center, BIOQUANT, University Heidelberg, Heidelberg, Germany; National Center of Tumor Diseases, Medical Oncology, University Hospital Heidelberg, Heidelberg, Germany.
Alexandra Krauthoff, Steinbeis Transfer Center for Medical Systems Biology, Heidelberg, Germany; Hamamatsu Tissue Imaging and Analysis Center, BIOQUANT, University Heidelberg, Heidelberg, Germany; National Center of Tumor Diseases, Medical Oncology, University Hospital Heidelberg, Heidelberg, Germany.
Andreas Keil, Steinbeis Transfer Center for Medical Systems Biology, Heidelberg, Germany.
Felipe Miranda, Steinbeis Transfer Center for Medical Systems Biology, Heidelberg, Germany.
Philip E Castle, Division of Cancer Epidemiology & Genetics, National Cancer Institute, Rockville, Maryland, USA; Division of Cancer Prevention, National Cancer Institute, Rockville, Maryland, USA.
Thomas Lorey, Kaiser Permanente, Permanente Medical Group Regional Laboratory, Berkeley, California, USA.
Brad Hare, Permanente Medical Group, San Francisco, California, USA.
Teresa M Darragh, University of California at San Francisco, San Francisco, California, USA.
Niels Grabe, Steinbeis Transfer Center for Medical Systems Biology, Heidelberg, Germany; Hamamatsu Tissue Imaging and Analysis Center, BIOQUANT, University Heidelberg, Heidelberg, Germany; National Center of Tumor Diseases, Medical Oncology, University Hospital Heidelberg, Heidelberg, Germany.
Megan A Clarke, Division of Cancer Epidemiology & Genetics, National Cancer Institute, Rockville, Maryland, USA.
References
- 1. Siegel RL, Miller KD, Jemal A.. Cancer statistics, 2018. CA Cancer J Clin 2018; 68:7–30. [DOI] [PubMed] [Google Scholar]
- 2. Silverberg MJ, Lau B, Justice AC, et al. Risk of anal cancer in HIV-infected and HIV-uninfected individuals in North America. Clin Infect Dis 2012; 54:1026–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clifford GM, Georges D, Shiels MS, et al. A meta-analysis of anal cancer incidence by risk group: toward a unified anal cancer risk scale. Int J Cancer 2021; 148:38–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Saraiya M, Unger ER, Thompson TD, et al. US assessment of HPV types in cancers: implications for current and 9-valent HPV vaccines. J Natl Cancer Inst 2015; 107:djv086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Palefsky JM, Rubin M.. The epidemiology of anal human papillomavirus and related neoplasia. Obstet Gynecol Clin North Am 2009; 36:187–200. [DOI] [PubMed] [Google Scholar]
- 6. Looker KJ, Rönn MM, Brock PM, et al. Evidence of synergistic relationships between HIV and human papillomavirus (HPV): systematic reviews and meta-analyses of longitudinal studies of HPV acquisition and clearance by HIV status, and of HIV acquisition by HPV status. J Int AIDS Soc 2018; 21:e25110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. University of California, San Francisco. Treating anal cancer precursor lesions reduces cancer risk for people with HIV. Press release. 8 October 2021. Available at https://www.ucsf.edu/news/2021/10/421591/treating-anal-cancer-precursor-lesions-reduces-cancer-risk-people-hiv. Accessed 22 October 2021.
- 8. Palefsky JM. Screening to prevent anal cancer: current thinking and future directions. Cancer Cytopathol 2015; 123:509–10. [DOI] [PubMed] [Google Scholar]
- 9. Clarke MA, Wentzensen N.. Strategies for screening and early detection of anal cancers: a narrative and systematic review and meta-analysis of cytology, HPV testing, and other biomarkers. Cancer Cytopathol 2018; 126:447–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Berry JM, Palefsky JM, Jay N, Cheng SC, Darragh TM, Chin-Hong PV.. Performance characteristics of anal cytology and human papillomavirus testing in patients with high-resolution anoscopy-guided biopsy of high-grade anal intraepithelial neoplasia. Dis Colon Rectum 2009; 52:239–47. [DOI] [PubMed] [Google Scholar]
- 11. Clarke MA, Cheung LC, Lorey T, et al. 5-Year prospective evaluation of cytology, human papillomavirus testing, and biomarkers for detection of anal precancer in human immunodeficiency virus-positive men who have sex with men. Clin Infect Dis 2019; 69:631–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wentzensen N, Follansbee S, Borgonovo S, et al. Human papillomavirus genotyping, human papillomavirus mRNA expression, and p16/Ki-67 cytology to detect anal precursors in HIV-infected MSM. AIDS 2012; 26:2185–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wentzensen N, Lahrmann B, Clarke MA, et al. Accuracy and efficiency of deep-learning-based automation of dual stain cytology in cervical cancer screening. J Natl Cancer Inst 2021; 113:72–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Solomon D, Davey D, Kurman R, et al. The 2001 Bethesda system: terminology for reporting results of cervical cytology. JAMA 2002; 287:2114–9. [DOI] [PubMed] [Google Scholar]
- 15. Wentzensen N, Follansbee S, Borgonovo S, et al. Analytic and clinical performance of cobas HPV testing in anal specimens from HIV-positive men who have sex with men. J Clin Microbiol 2014; 52:2892–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Clarke MA, Cheung, LC, Castle PE, et al. Five-year risk of cervical precancer following p16/Ki-67 dual-stain triage of HPV-positive women. JAMA Oncol 2019; 5:181–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hillman, RJ, Cuming T, Darragh T, et al. 2016 IANS international guidelines for practice standards in the detection of anal cancer precursors. J Low Genit Tract Dis 2016; 20:283–91. [DOI] [PubMed] [Google Scholar]
- 18. Wei F, Gaisa MM, D’Souza, G, et al. Epidemiology of anal human papillomavirus infection and high-grade squamous intraepithelial lesions in 29 900 men according to HIV status, sexuality, and age: a collaborative pooled analysis of 64 studies. Lancet HIV 2021; 8:e531–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Clifford, GM, Siproudhis L, Piroth L, et al. Determinants of high-grade anal intraepithelial lesions in HIV-positive MSM. AIDS 2018; 32:2363–71. [DOI] [PubMed] [Google Scholar]
- 20. Fengyi J, Roberts JM, Grulich AE, et al. The performance of human papillomavirus biomarkers in predicting anal high-grade squamous intraepithelial lesions in gay and bisexual men. AIDS 2017; 31:1303–11. [DOI] [PubMed] [Google Scholar]
- 21. Serrano-Villar S, Hernández-Novoa B, de Benito A, et al. Screening for precancerous lesions with P16/Ki67 immunostaining in HIV-infected MSM. PLoS One 2017; 12:e0188851. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.