Abstract
Background:
Bronchiolitis is the leading cause of hospitalization in U.S. infants and a major risk factor for childhood asthma. Growing evidence supports clinical heterogeneity within bronchiolitis. We aimed to identify endotypes of infant bronchiolitis by integrating clinical, virus, and serum proteome data, and examine their relationships with asthma development.
Methods:
This is a multicenter prospective cohort study of infants hospitalized for physician-diagnosis of bronchiolitis. We identified bronchiolitis endotypes by applying unsupervised machine learning (clustering) approaches to integrated clinical, virus (respiratory syncytial virus [RSV], rhinovirus [RV]), and serum proteome data measured at hospitalization. We then examined their longitudinal association with the risk for developing asthma by age 6 years.
Results:
In 140 infants hospitalized with bronchiolitis, we identified three endotypes: 1) clinicalatopicvirusRVproteomeNFκB-dysregulated, 2) clinicalnon-atopicvirusRSV/RVproteomeTNF-dysregulated, and 3) clinicalclassicvirusRSVproteomeNFκB/TNF-regulated endotypes. Endotype 1 infants were characterized by high proportion of IgE sensitization and RV infection. These endotype 1 infants also had dysregulated NFκB pathways (FDR<0.001) and significantly higher risks for developing asthma (53% vs. 22%; adjOR 4.04; 95%CI, 1.49–11.0; P=0.006), compared with endotype 3 (clinically resembling “classic” bronchiolitis). Likewise, endotype 2 infants were characterized by low proportion of IgE sensitization and high proportion of RSV or RV infection. These endotype 2 infants had dysregulated tumor necrosis factor (TNF)-mediated signaling pathway (FDR<0.001) and significantly higher risks for developing asthma (44% vs. 22%; adjOR 2.71; 95%CI, 1.03–7.11, P=0.04).
Conclusion:
In this multicenter cohort, integrated clustering of clinical, virus, and proteome data identified biologically distinct endotypes of bronchiolitis that have differential risks of asthma development.
Keywords: asthma, bronchiolitis, endotyping, infants, proteome, recurrent wheeze
BACKGROUND
Bronchiolitis is the most common virus-induced acute lower respiratory infection in infants. It is the leading cause of infant hospitalization in the U.S., accounting for 110,000 hospitalizations each year.1 In addition to the large acute morbidity burden, its chronic morbidity is also considerable. Among infants hospitalized with bronchiolitis (i.e., severe bronchiolitis), 30%−40% subsequently develop recurrent wheeze2–6 and 30% develop childhood asthma.5–11
While bronchiolitis is conventionally deemed as a single disease entity that has similar pathobiology,12 a growing body of evidence supports heterogeneity in its acute presentation and chronic morbidity risk.3,13–15 For example, recent epidemiology research has reported and validated clinically distinct subtypes (or phenotypes) of bronchiolitis13 with a different risk of developing recurrent wheeze3 and asthma14,15. Recent data have also suggested that these subtypes may correlate with treatment responses.16–18 Yet, these phenotypes were derived solely through major clinical features. Accordingly, little is known about the pathobiological processes underlying the heterogeneity and the mechanisms that link the two common conditions—infant bronchiolitis and childhood asthma. This insufficient understanding has hindered efforts to develop asthma prevention strategies. Proteomics is pertinent to addressing this knowledge gap by comprehensively characterizing proteins—the main regulator of cellular physiology, enabling to characterize the disease pathobiology.19 However, no study has yet determined biologically distinct subtypes (i.e., endotypes) of bronchiolitis based on proteome data or their integrated contribution to asthma development in later childhood.
To address this knowledge gap, we analyzed data from a multicenter prospective cohort study to identify endotypes of severe bronchiolitis by integrating clinical, virus, and serum proteome data, and investigate their longitudinal relationship with the development of recurrent wheeze and asthma.
METHODS
Study design, setting, and participants
We analyzed data from the 35th Multicenter Airway Research Collaboration (MARC-35) study—a multicenter prospective cohort study.20 Details of the study design, setting, participants, data collection, testing, and statistical analysis may be found in the Supplementary Methods. Briefly, investigators enrolled infants (age <1 year) hospitalized with attending physician-diagnosis of bronchiolitis at 17 sites across 14 U.S. states (Table S1) in 2011–2014. The diagnosis of bronchiolitis was made according to the American Academy of Pediatrics bronchiolitis guidelines,12 defined as an acute respiratory illness with a combination of rhinitis, cough, tachypnoea, wheezing, crackles, or chest retractions, regardless of previous breathing problem episodes. We excluded infants with a known heart-lung disease, immunodeficiency, immunosuppression, or gestational age of <32 weeks. All patients were treated at the discretion of the treating physicians.
Of 1,016 infants enrolled in the MARC-35 cohort, the current analysis investigated 140 infants who were selected for serum proteomic testing (Table S2). The institutional review board at each participating hospital approved the study with written informed consent obtained from the parent or guardian.
Data collection and measurement of virus and proteome
Clinical data (demographic characteristics; medical, environmental, and family history; and details of the acute illness and hospital course) were collected via structured interview and chart reviews using a standardized protocol.21 All data were reviewed at the EMNet Coordinating Center (Boston, Massachusetts, USA), and site investigators were queried about missing data and discrepancies identified by manual data checks.
In addition to the clinical data, investigators also collected nasopharyngeal airway and serum specimens by using standardized protocols.21,22 All sites used the same collection equipment and collected the samples within 24 hours of a child’s arrival on the medical ward or intensive care unit. Nasopharyngeal specimens were shipped in batches to Baylor College of Medicine (Houston, Texas, USA) where they were tested for respiratory viruses (e.g., respiratory syncytial virus [RSV] and rhinovirus [RV]) by using real-time polymerase chain reaction (RT-PCR).21–24 Serum specimens were shipped to Beth Israel Deaconess Medical Center (Boston, Massachusetts, USA) for proteomic profiling, as described in the Supplemental Methods. Briefly, serum proteins were measured with the use of the Olink multiplex platform (13 panels; Olink Bioscience, Uppsala, Sweden). The expression value of each protein was normalized to a unit on a log2 scale, proportional to its concentration. Serum specific IgE (sIgE) was measured at enrollment using two different assays (ImmunoCAP sIgE and ImmunoCAP ISAC; ThermoFisher Scientific; Waltham, MA, USA) at the Phadia Immunology Reference Laboratory.
Clinical outcome measures
The primary outcome was the development of asthma by age 6 years. Asthma was defined using a commonly used epidemiologic definition: physician-diagnosis of asthma, with either asthma medication use (e.g., albuterol inhaler, inhaled corticosteroids) or asthma-related symptoms (e.g., wheezing, nocturnal cough) in the preceding year.25 The secondary outcome was the development of recurrent wheeze by age 3 years. Recurrent wheeze was defined as having at least 2 corticosteroid-requiring exacerbations in 6 months or at least 4 wheezing episodes in 1 year that last at least 1 day and affect sleep.20
Statistical analysis
The objectives of the present study are: i) to identify biologically distinct endotypes among infants with severe bronchiolitis, and ii) to examine their relationships with the risk of developing asthma and recurrent wheeze. The analytic workflow is summarized in Figure 1. The details of the statistical analysis can be found in the Supplementary Methods.
Figure 1. Analytic workflow of endotyping.
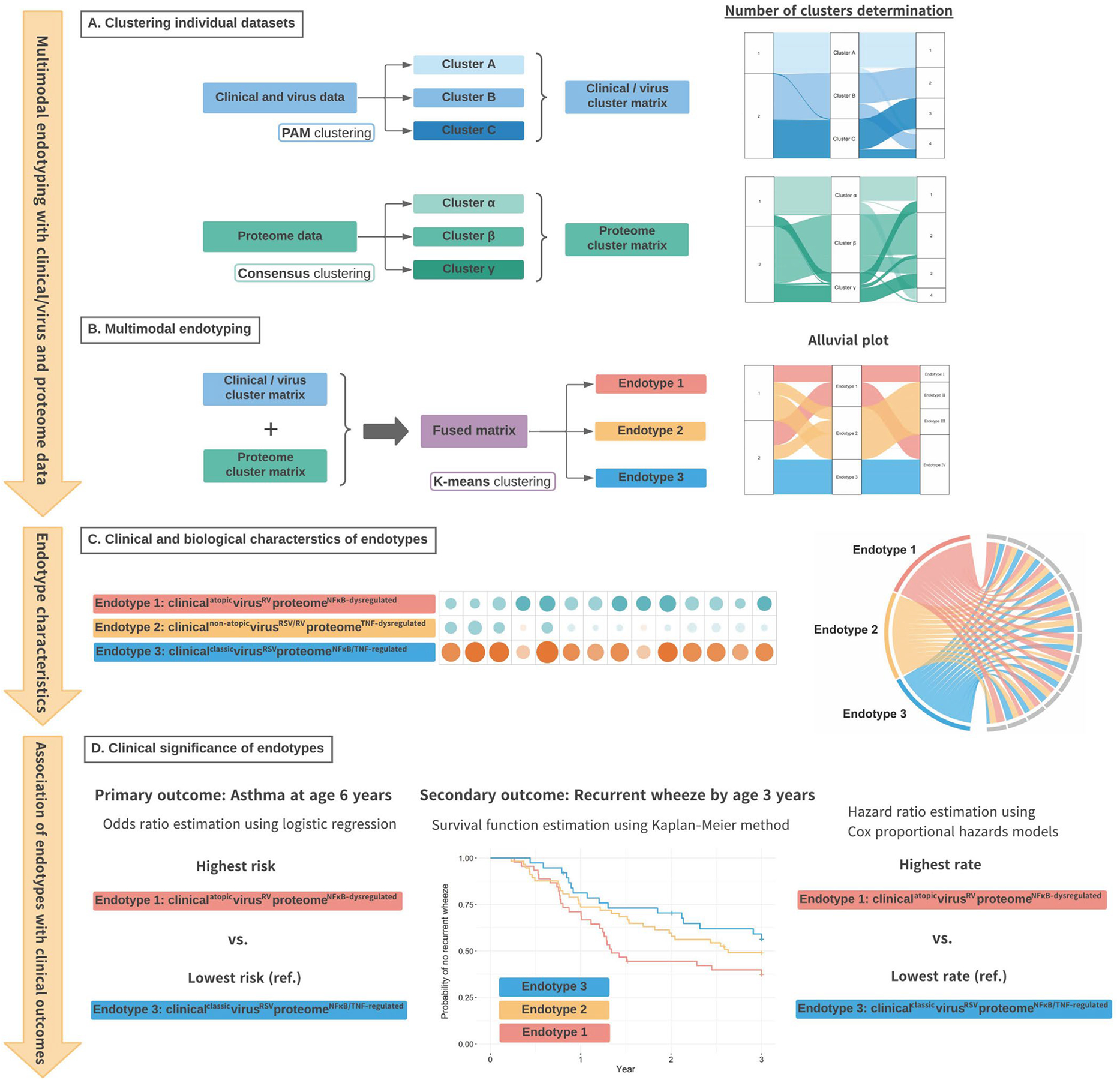
A. Of 1,016 infants (age <1 year) hospitalized with bronchiolitis, the current analysis investigated 140 infants who underwent serum proteomic testing. We first identified mutually exclusive clusters in the clinical and virus dataset (clusters A, B, and C) using the PAM clustering algorithm and separate clusters in the proteome dataset (clusters α, β, and γ) using the consensus clustering algorithm. We also generated alluvial plots to examine consistencies across different numbers (k=2–4) of the clinical/virus and proteomic clusters.
B. We computed a Gower distance from a fused matrix of the clinical/virus and proteome clusters, and identified three mutually exclusive endotypes by applying K-means clustering algorithm. We also generated an alluvial plot to examine consistencies across different numbers (k=2–4) of endotypes.
C. We conducted a functional pathway analysis to investigate whether proteins for specific biological pathways are enriched by comparing the reference endotype (endotype 3) with each of the other endotypes. We also generated chord diagrams to visualize the between-endotype differences in the major clinical and virus characteristics.
D. We constructed unadjusted and adjusted (random-effect) logistic regression models to determine the association of the endotypes with the risk of developing asthma by age 6 years (the primary outcome). We also used Kaplan-Meier estimator and Cox proportional hazards models to examine the longitudinal relationship of the endotypes with the rate of recurrent wheeze (the secondary outcome).
Abbreviation: PAM, partition around medoids
Briefly, we first computed a distance matrix for each of the datasets—1) clinical and virus data (including the genomic load of RSV and RV) and 2) proteome data—and derived mutually exclusive clusters for each dataset by using with partition around medoids (PAM)26 and consensus clustering27 methods, respectively. To choose an optimal number of clusters for each dataset, we used a combination of the silhouette widths (Figure S1A), relative change in the area under cumulative distribution function curve (Figure S1B), cluster size (Figure S2A–B), and clinical and biological plausibility based on a priori knowledge (Figure S2C–D).11,21 Second, we combined these clusters (i.e., the clinical/virus clusters and the proteome clusters) to derive a fused matrix, computed a Gower distance, and derived mutually exclusive multimodal endotypes by applying K-means clustering algorithm.28 To choose an optimal number of endotypes, we used a combination of the silhouette widths (Figure S1C), endotype size (Figure S2E), and clinical and biological plausibility (Figure S2F). We also visualized the three endotypes through t-distributed stochastic neighbor embedding (t-SNE)29 (Figure 2A) and chord diagrams (Figure 2B).30 Third, to examine the functional profile of each endotype, we conducted differential expression protein and functional pathway analyses31 by comparing the reference endotype with each of the other endotypes using pathfindR package.32 We also conducted joint pathway analyses with parallel serum metabolome data (n=112)33 using MetaboAnalyst 5.0.34 Fourth, to examine the longitudinal relationship of the endotypes with the asthma risk, we constructed random-effect logistic regression models accounting for patient clustering within sites. To examine the relationship with the rate of recurrent wheeze, we modeled the time to outcome by fitting Cox proportional hazards models. Patients who did not have an outcome were censored at their last follow-up interview or at the time of withdrawal during the 36-month follow-up period. We verified the proportionality of hazards assumption by examining Schoenfeld residuals.
Figure 2. Between-endotype differences in major clinical and virus variables, serum proteome profile, and enriched biological pathways.
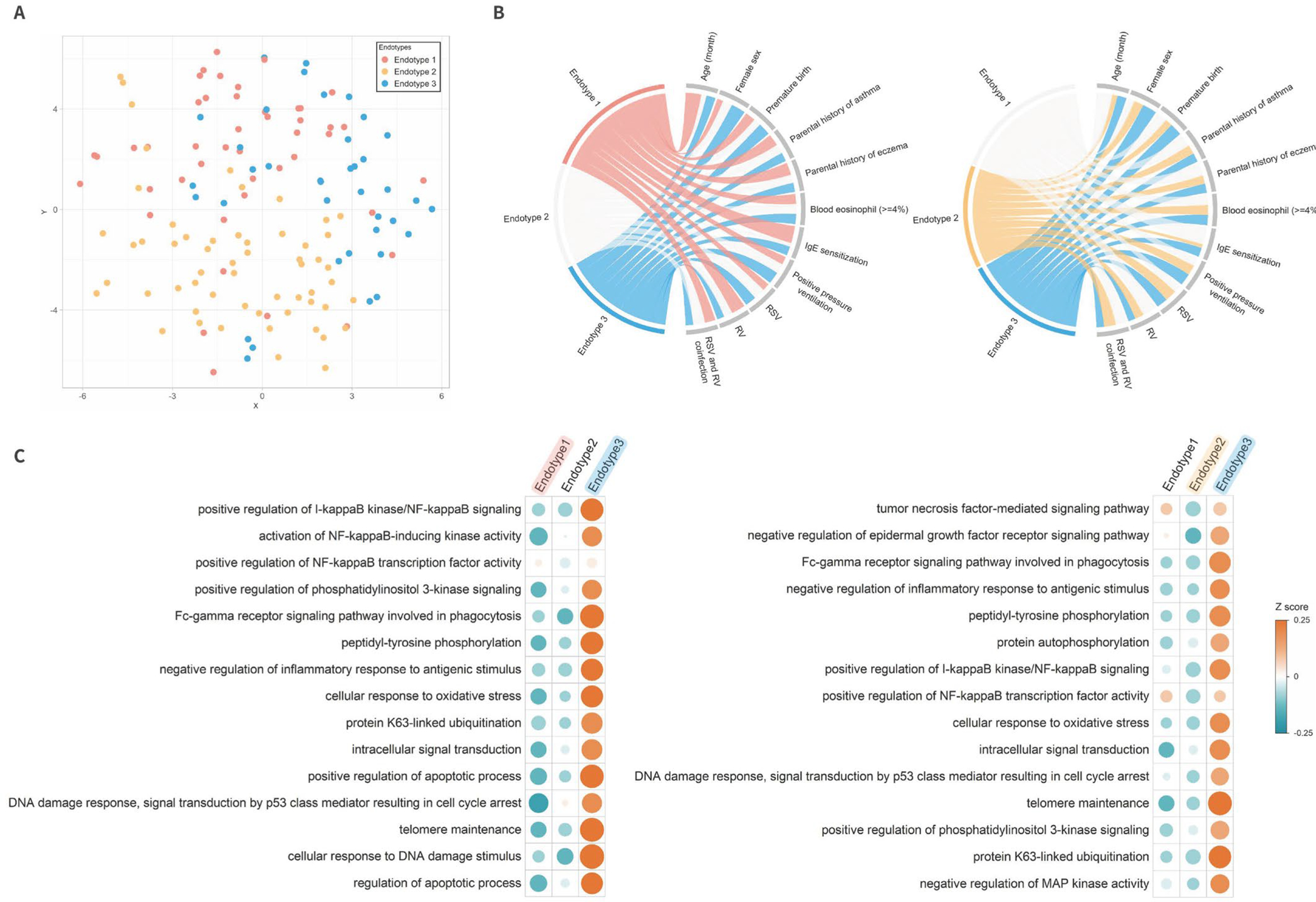
A. T-distributed stochastic neighbor embedding (t-SNE) of serum proteome according to endotypes
To visualize the overall serum proteome profile, the t-SNE method was applied to the three eigenvectors in the spectral clustering. Each dot represents the serum proteome profile of a single infant in a low-dimensional space. Colored dots indicate three endotypes: endotype 1 (red), endotype 2 (yellow), and endotype 3 (blue). The infants cluster together according to their endotypes.
B. Major clinical and virus characteristics according to endotypes
The ribbons connect each of the endotypes (endotypes 1–3) with the major clinical and virus characteristics. The width of the ribbon represents the proportion of infants within the endotypes who have the corresponding clinical or virus characteristic, which was scaled to a total of 100%. The left chord diagram represents the comparison between endotypes 1 and 3, while the right diagram represents the comparison between endotypes 2 and 3.
C. Functional pathway analysis for specific biological pathways of each endotype
To investigate whether proteins for specific biological pathways are enriched, we conducted a functional pathway analysis. The left half of the heatmap represents the pathways that distinguish endotype 1 from endotype 3 (the reference); the right half represents those pathways that distinguish endotype 2 from endotype 3 (all FDR<0.001; Table S3). The color bar indicates a Z score of each pathway. Upregulated pathways are displayed as orange, while downregulated pathways are displayed as green.
Abbreviations: DNA, deoxyribonucleic acid; IgE, immunoglobulin E; I-κB, inhibitor of κB; MAP, mitogen-activated protein; NFκB, nuclear factor-κB; RSV, respiratory syncytial virus; RV, rhinovirus.
In the sensitivity analysis, we first examined the endotype-outcome associations after excluding infants with a previous breathing problem. Second, we also examined the robustness of endotype-outcome associations by repeating the analysis using a different number of endotypes. We analyzed the data using R version 3.6.1 (R Foundation, Vienna, Austria). All P-values were two-tailed, with P<0.05 considered statistically significant. We corrected for multiple testing using the Benjamini-Hochberg false discovery rate (FDR) method.35
RESULTS
Of the infants enrolled in the MARC-35 cohort, the current study focused on 140 infants with severe bronchiolitis who underwent serum proteome testing. The analytic and non-analytic cohorts did not differ in patient characteristics (P≥0.05; Table S2), except for the proportion of RSV and RV. Among the analytic cohort, the median age was 3 (interquartile range [IQR], 1–6) months, 39% were female, and 37% were non-Hispanic white. Overall, 47% had solo-RSV infection, 18% had solo-RV infection, and 11% had RSV/RV coinfection (Table 1).
Table 1.
Baseline characteristics and clinical course of infants, according to endotypes
| Overall | Endotype 1 | Endotype 2 | Endotype 3 | ||
|---|---|---|---|---|---|
| (clinicalatopic virusRV proteomeNFκB-dysregulated) | (clinicalnon-atopic virusRSV/RV proteomeTNF-dysregulated) | (clinicalclassic virusRSV proteomeNFκB TNF-regulated) | |||
| Characteristics | (n=140; 100%) | (n=45; 32%) | (n=57; 41%) | (n=38; 27%) | P value |
| Demographics | |||||
| Age (month), median (IQR) | 3 (1–6) | 7 (5–9) | 2 (1–3) | 3 (1–6) | <0.001 |
| Female sex | 55 (39) | 11 (24) | 22 (39) | 22 (58) | 0.008 |
| Race/ethnicity | 0.08 | ||||
| Non-Hispanic white | 52 (37) | 15 (33) | 18 (32) | 19 (50) | |
| Non-Hispanic black | 35 (25) | 8 (18) | 21 (37) | 6 (16) | |
| Hispanic | 50 (36) | 20 (44) | 17 (30) | 13 (34) | |
| Other or unknown | 3 (2) | 2 (4) | 1 (2) | 0 (0) | |
| Prematurity (32.0–36.9 weeks) | 29 (21) | 10 (22) | 9 (16) | 10 (26) | 0.44 |
| C-section delivery | 51 (37) | 18 (41) | 19 (33) | 14 (37) | 0.74 |
| Previous breathing problems (count) | <0.001 | ||||
| 0 | 104 (74) | 17 (38) | 52 (91) | 35 (92) | |
| 1 | 28 (20) | 21 (47) | 5 (9) | 2 (5) | |
| 2 | 8 (6) | 7 (16) | 0 (0) | 1 (3) | |
| Previous ICU admission | 4 (3) | 3 (7) | 1 (2) | 0 (0) | 0.16 |
| History of eczema | 22 (16) | 11 (24) | 5 (9) | 6 (16) | 0.10 |
| Ever attended daycare | 23 (16) | 13 (29) | 5 (9) | 5 (13) | 0.02 |
| Cigarette smoke exposure at home | 15 (11) | 4 (9) | 7 (12) | 4 (11) | 0.86 |
| Parental history of eczema | 32 (23) | 13 (29) | 11 (20) | 8 (21) | 0.52 |
| Parental history of asthma | 50 (36) | 19 (42) | 19 (34) | 12 (32) | 0.55 |
| Clinical presentation | |||||
| Weight (kg), median (IQR) | 6.0 (4.6–7.8) | 7.8 (6.5–8.7) | 4.8 (4.0–5.7) | 6.0 (4.9–7.0) | <0.001 |
| Respiratory rate (per minute), median (IQR) | 48 (40–60) | 50 (42–60) | 48 (40–57) | 46 (39–60) | 0.43 |
| Oxygen saturation | 0.52 | ||||
| <90% | 14 (10) | 3 (7) | 5 (9) | 6 (16) | |
| 90–93% | 27 (20) | 7 (16) | 12 (22) | 8 (22) | |
| ≥94% | 95 (70) | 35 (78) | 37 (69) | 23 (62) | |
| Blood eosinophilia (≥4%) | 18 (15) | 6 (16) | 7 (13) | 5 (16) | 0.92 |
| IgE sensitization | 31 (22) | 18 (40) | 5 (9) | 8 (21) | 0.001 |
| Any food allergen | 28 (20) | 16 (36) | 5 (9) | 7 (18) | 0.003 |
| Aeroallergen | 3 (7) | 2 (4) | 0 (0) | 1 (3) | 0.30 |
| Clinical course | |||||
| Positive pressure ventilation use* | 27 (19) | 7 (16) | 11 (19) | 9 (24) | 0.65 |
| Intensive treatment use† | 57 (41) | 15 (33) | 25 (44) | 17 (45) | 0.47 |
| Length-of-day (day), median (IQR) | 2 (1–5) | 2 (1–4) | 2 (2–4) | 2 (1–5) | 0.90 |
| Corticosteroid use | 27 (19) | 13 (29) | 6 (11) | 8 (21) | 0.06 |
| Respiratory virus | |||||
| RSV only | 66 (47) | 9 (20) | 37 (65) | 20 (53) | 0.006 |
| RV only | 25 (18) | 11 (24) | 10 (18) | 4 (11) | 0.26 |
| RSV/RV coinfection | 15 (11) | 5 (11) | 7 (12) | 3 (8) | 0.79 |
| Other pathogen‡ only | 5 (4) | 3 (7) | 1 (2) | 1 (3) | 0.39 |
| Other pathogen‡ with RSV or RV coinfection | 26 (19) | 16 (36) | 1 (2) | 9 (24) | <0.001 |
Abbreviations: ICU, intensive care unit; IgE, immunoglobulin E; IQR, interquartile range; RSV, respiratory syncytial virus; RV, rhinovirus.
Data are no. (%) of infants unless otherwise indicated. Percentages may not equal 100, because of rounding and missingness.
Infants with bronchiolitis who underwent continuous positive airway ventilation and/or mechanical ventilation.
Infants with bronchiolitis who were admitted to ICU and/or who underwent positive pressure ventilation.
Adenovirus, bocavirus, Bordetella pertussis, enterovirus, human coronavirus NL63, OC43, 229E, or HKU1, human metapneumovirus, influenza A or B virus, Mycoplasma pneumoniae, and parainfluenza virus 1–3.
Integrated clustering of clinical, virus, and proteome data identified multimodal endotypes
First, by applying clustering approaches to the clinical/virus and proteome datasets, 3-class models led to an optimal fit for both the clinical/virus data (with the three clusters called A, B, and C; Figures 1, S1 and S2) and the proteome dataset (with the three clusters called α, β, and γ; Figure 1, S1 and S2). Second, by integrating these cluster data, the combination of average silhouette widths, endotype size, and clinical and biological plausibility demonstrated that a 3-class model was an optimal fit, with the three endotypes called 1, 2, and 3 (Figures 1, S1 and S2). The t-SNE plot shows that an infant’s serum proteome profile generally clustered together according to their endotypes, while there was a moderate overlap between endotypes 1 and 3 (Figure 2A).
The three distinct endotypes were chiefly characterized by their clinical characteristics, detected virus, and biological pathways: 1) clinicalatopicvirusRVproteomeNFκB-dysregulated (32%), 2) clinicalnon-atopicvirusRSV/RVproteomeTNF-dysregulated (41%), and 3) clinicalclassicvirusRSVproteomeNFκB/TNF-regulated (27%) (Table 1 and Figure 1). Descriptively, infants with an endotype 1 were characterized by older age at the index hospitalization and a high proportion of males, frequent previous breathing problems, daycare attendance, IgE sensitization, and RV infection (Table 1 and Figure 2B). Infants with an endotype 2 were characterized by a low proportion of IgE sensitization and a high proportion of RSV or RV infection. Infants with an endotype 3 were characterized by a high proportion of male sex, a low proportion of breathing problem history, and a high proportion of RSV infection. As the endotype 3 clinically resembled “classic” bronchiolitis,12 this group served as the reference group for the following analyses.
Endotypes had distinct biological function
To examine the biological significance of the endotypes, we conducted functional pathway analyses. Endotype 1 had 299 differentially enriched pathways (FDR<0.05) and endotype 2 had 260 differentially enriched pathways (FDR<0.05) when compared to the endotype 3 (Table S3). For example, endotype 1 (clinicalatopicvirusRVproteomeNFκB-dysregulated) infants had significantly dysregulated (more specifically, increased) NFκB and phosphatidylinositol 3-kinase (PI3K) signaling pathways, compared to those with endotype 3 (both FDR<0.001; Figure 2C left). In contrast, the endotype 2 (clinicalnon-atopicvirusRSV/RVproteomeTNF-dysregulated) infants had significantly dysregulated tumor necrosis factor (TNF)-mediated signaling and epidermal growth factor receptor (EGFR) signaling pathways, compared to those with endotype 3 (both FDR<0.001; Figure 2C right). The joint pathway analysis with parallel serum metabolome data showed consistent results with the functional pathway analyses (e.g., NFκB signaling pathway [endotype 1 vs. 3; FDR<0.001], EGFR signaling pathway [endotype 2 vs. 3; FDR=0.006]; Table S4).
Endotypes had differential risks for developing asthma and recurrent wheeze
These endotypes also had differential risks for subsequent airway comorbidities. Endotype 1 infants had a significantly higher risk of developing asthma when compared to endotype 3 infants (53% vs. 22%; adjOR, 4.04; 95%CI, 1.49–11.0; P=0.006; Figure 3). Likewise, endotype 2 infants also had a significantly higher risk (44% vs. 22%; adjOR, 2.71; 95%CI, 1.03–7.11; P=0.04). With regard to the recurrent wheeze, while the Kaplan-Meier curves did not show a statistically significant difference (PWilcoxon=0.09; Figure 4), endotype 1 infants did have a significantly higher rate than the endotype 3 infants (62% vs. 42%; adjHR, 1.87; 95% CI, 1.01–3.47; P=0.047; Figure 3).
Figure 3. Association of endotypes of infant bronchiolitis with risk for developing asthma and recurrent wheeze.
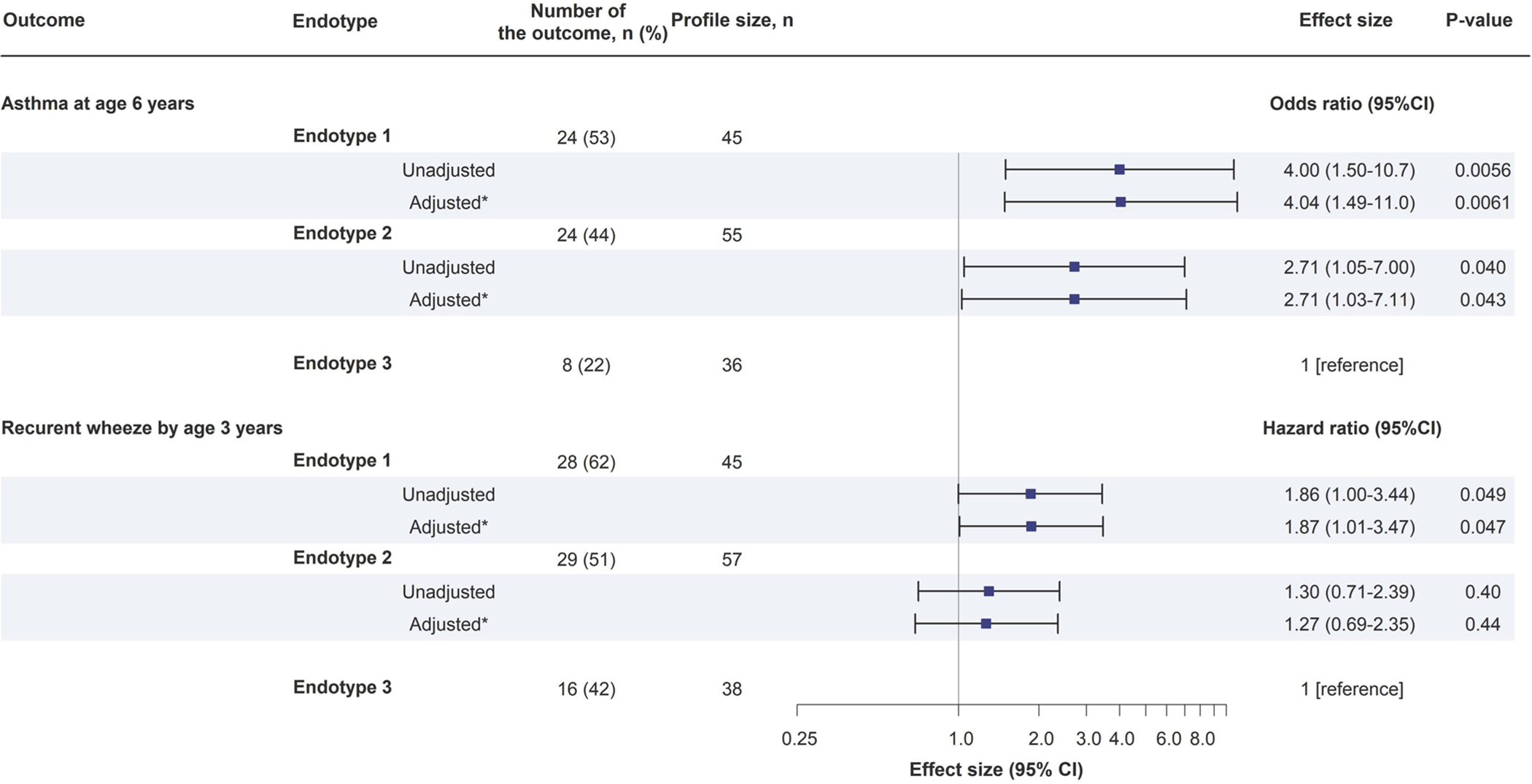
To examine the association of endotypes (endotype 3 as the reference) with the risk of developing childhood asthma and the rate of recurrent wheeze, logistic regression models and Cox proportional hazards models were fit.
* Random-effect logistic regression model and Cox proportional hazards model accounting for patient clustering within hospitals
Abbreviation: CI, confidence interval
Figure 4. Kaplan-Meier curves for development of recurrent wheeze by age 3 years, according to endotypes.
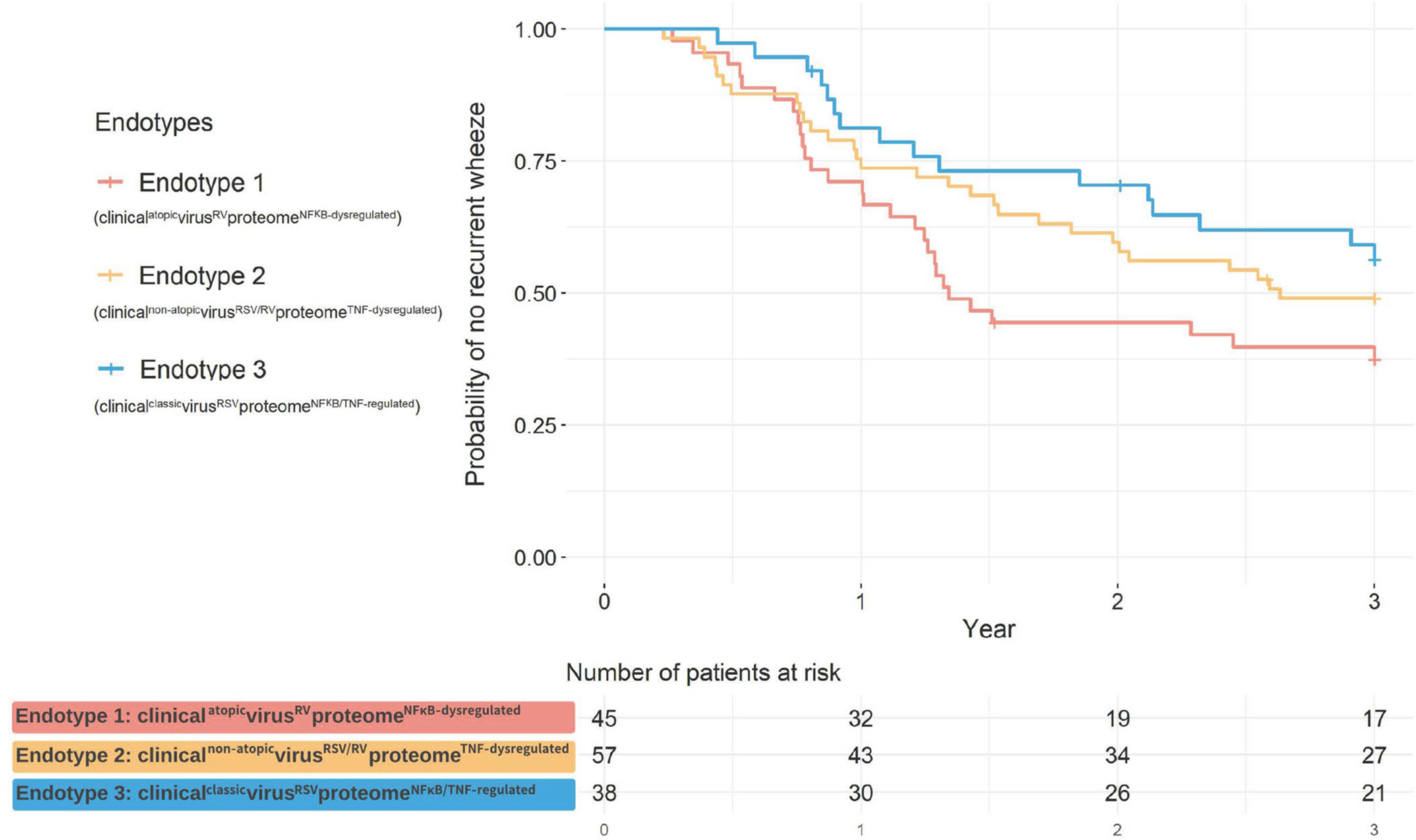
The Kaplan-Meier curves for recurrent wheeze outcome did not significantly differ between the endotypes (PWilcoxon=0.09). However, compared to the endotype 3 infants, the rate of recurrent wheeze was significantly higher among the endotype 1 infants. (adjHR, 1.87; 95% CI, 1.01–3.47; P=0.047). The corresponding HRs are presented in Figure 3.
Sensitivity analysis
In the analysis limiting to infants without a previous breathing problem, the endotype-outcome associations remained consistent (Figure S3). For example, the asthma risk was significantly higher in endotype 1 (adjOR, 3.18; 95%CI, 1.02–9.92; P=0.046) and endotypes 2 (adjOR, 3.18; 95%CI, 1.17–8.63; P=0.022), compared to endotype 3. Next, different numbers of endotypes were examined. Alluvial plot (Figure S2C) demonstrates a consistency of the original endotypes (endotypes 1–3) across the different numbers chosen. For example, with the use of 4-class model, the endotypes I-III had 100% concordance with the original endotype 1 or 2 (Table S5). Additionally, endotype I (concordant with endotype 1) had a significantly higher risk for developing asthma when compared to endotype IV (67% vs. 32%; adjOR, 4.25; 95%CI, 1.33–13.6; P=0.02; Figure S4).
DISCUSSION
By integrating the clinical, virus, and proteome data from a multicenter prospective cohort study of 140 infants with severe bronchiolitis, we identified three clinically and biologically distinct endotypes. Endotype 1 was characterized by a high proportion of IgE sensitization and RV infection with unique proteome signatures, such as dysregulated (increased) NFκB and PI3K signaling pathways. In contrast, endotype 2 was characterized by a low proportion of IgE sensitization with dysregulated TNF-mediated signaling pathway. Infants with either endotype 1 or 2 had a significantly higher risk for subsequently developing asthma compared to the reference endotype 3, which resembled “classic” bronchiolitis. The sensitivity analysis showed the robustness of the findings. To the best of our knowledge, this is the first investigation that has identified biologically distinct proteomics endotypes in infants with severe bronchiolitis and demonstrated their longitudinal relationship with the risk of chronic respiratory outcomes.
Bronchiolitis has been conventionally viewed as a single disease entity with similar pathobiological mechanisms. Indeed, current national guidelines for the diagnosis and management of bronchiolitis is based on this major assumption.12 Nevertheless, concordant with the results of the present study, recent research has suggested the complexity of infant bronchiolitis, as reflected by the heterogeneity in clinical characteristics,3,13,36,37 transcriptome,38 microRNAome,39 metabolome,40–44 and microbiome21,45–50. The proteomic approach adds to these previous reports through comprehensively profiling proteins that are the main effectors of cellular physiology.51 Recent studies have suggested the role of the proteome in the pathobiology of respiratory diseases, including severe bronchiolitis52 and asthma53–55. For example, a previous analysis has shown that virus-specific (RSV vs. RV) circulating proteome signatures were associated with acute severity in infants with bronchiolitis.52 Additionally, in an analysis of the U-BIOPRED study, sputum proteome testing in adults with moderate-to-severe asthma has demonstrated different proteomics endotypes with unique airway inflammatory profiles.53 Furthermore, another study investigating the serum proteome in adults has reported that patients with non-atopic asthma had unique proteome signatures (e.g., down-regulated Igκ chain C-region) compared to healthy controls.54 The integrated proteomic approach in the current study corroborates these earlier findings and extends them by demonstrating distinct bronchiolitis endotypes that have differential risks of developing recurrent wheeze and asthma.
The exact mechanisms underlying the observed endotypes—particularly endotype 1 characterized by a high proportion of IgE sensitization, RV infection, unique proteomic profiles (e.g., dysregulated NFκB and PI3K signaling pathways), and the highest asthma risk—warrant further clarification. Consistent with this endotype, previous research has also shown the interaction between parental atopy, allergic sensitization, and early-life RV infection (particularly RV-C species) on an increased risk of asthma development.20,56–59 Additionally, our previous study of nasal microRNA and mRNA in 32 infants with bronchiolitis has reported that RV infection was related to upregulated NFκB family and downregulated IκB family with elevated levels of NFκB-induced type-2 cytokines.39 Furthermore, the literature has also demonstrated that the NFκB pathways regulate immune responses and airway inflammation in asthma by regulating the gene expression of inflammatory factors.60–62 For example, the NFκB pathway upregulates the production of type-2 cytokines—e.g., interleukin 4 (IL-4), IL-5, and IL-13.60 Likewise, the inhibition of IκB kinase—a key regulator of all inducible NFκB signaling pathways—induces inflammatory mediators in allergic asthma (e.g., eotaxin, IgE, IL-4).62,63 In addition the NFκB pathways, endotype 1 also had dysregulated PI3K signaling pathway. In agreement with this finding, our previous analysis of nasopharyngeal (i.e., not serum) transcriptome and metabolome data of infants with bronchiolitis has shown that an endotype with atopy and RV coinfection had enriched PI3K-Akt-mTOR signaling pathway and highest asthma risk.64 The dysregulation of PI3K increases allergen-induced inflammation via TH2 cytokines activation and leads to induction of interferon γ-induced protein 10—a mediator of RV-induced inflammation in allergic asthma.65
In addition to endotype 1, we also observed an increased risk of asthma among infants with endotype 2—characterized by a low proportion of IgE sensitization and dysregulated TNF and EGFR pathways. Consistently, dysregulation of TNF-mediated pathway disturbs the balance and composition of TNF receptors-associated signaling complexes (e.g., linear ubiquitin chain assembly complex) through ubiquitination, contributing to inflammatory airway diseases (e.g., asthma, acute respiratory distress syndrome).66,67 Previous research has also suggested the role of TNF in neutrophilic inflammation in asthma68,69 and identified the TNF-mediated pathway as a potential therapeutic target against T2-low asthma.70 In addition, the current study has also revealed that endotype 2 had dysregulated EGFR signaling pathway. Recent research has shown that the EGFR signaling pathway is dysregulated in patients with IL-17 high asthma71 and that those with neutrophilic asthma had differentially expressed genes regulating epidermal growth factor, compared to eosinophilic asthma.72 Notwithstanding the complexity of these mechanisms, the identification of bronchiolitis endotypes and their longitudinal relationship with chronic respiratory outcomes is an important finding. Our data should advance research into the development of endotype-specific strategies for asthma prevention.
The current study has several potential limitations. First, the study did not have “healthy controls”. Yet, the objective of the study was not to derive endotypes related to incident bronchiolitis (i.e., bronchiolitis yes vs. no) but to determine the relationship of each bronchiolitis endotype with the risk for developing recurrent wheeze and asthma. Second, the current study had overrepresentation of RV infection, which might have led to selection bias. Third, the serum proteome was measured at a single time point. Although longitudinal proteome measurements are important, the study objective was to identify bronchiolitis endotypes at the time of hospitalization. Regardless, even with single time point data, the study successfully identified biologically distinct endotypes that are associated with the risk of recurrent wheeze and asthma development. Fourth, the serum proteome may not directly reflect the intracellular signaling pathways of molecules in the respiratory system. Fifth, it is possible that asthma diagnosis is misclassified and that some children are going to develop asthma at a later age. To address these potential limitations, the cohort is currently being followed up to age 9 years. Sixth, the sample size of the current analysis is relatively small; therefore, it is possible that the study did not identify more-granular endotypes with distinct mechanisms. In addition, the study lacks validation data. Both internal and external validation to confirm the inference is our future focus of research. Seventh, the study design that focused on the winter bronchiolitis seasons precluded us from enrolling patients in the other seasons, during which RV infection is more common. Additionally, the impact of the COVID-19 pandemic on the asthma incidence is unclear. Lastly, our inferences may not be generalizable to infants beyond severe bronchiolitis (i.e., infants with mild-to-moderate bronchiolitis or other types of acute respiratory infection). Nonetheless, our observations remain directly relevant for the 110,000 infants hospitalized annually in the U.S.1—a large population with a substantial morbidity burden.
CONCLUSIONS
By integrating the clinical, virus, and serum proteome data from a multicenter prospective cohort study of infants hospitalized for bronchiolitis, we identified three biologically distinct and clinically meaningful endotypes. These endotypes—characterized by distinct clinical and virus characteristics and host immune response signatures—had differential risks for developing recurrent wheeze and childhood asthma. While external validation is warranted, our data indicate a complex interplay between the respiratory virus and systemic immune response and their integrated contributions to chronic airway morbidities. For clinicians, our findings provide an evidence base for the early identification of high-risk children during a critical period of airway development—early infancy. For researchers, our data should advance research into accurately defining bronchiolitis endotypes, which will, in turn, accelerate precision medicine and the development of endotype-specific strategies for asthma prevention.
Supplementary Material
Funding/Support:
This study was supported by grants (UG3/UH3 OD-023253, R01 AI-127507, R01 AI-134940, and R01 AI-137091) from the National Institutes of Health (Bethesda, MD). The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funding organization was not involved in the collection, management, or analysis of the data; preparation or approval of the manuscript; or decision to submit the manuscript for publication.
Abbreviations:
- CI
confidence interval
- EGFR
epidermal growth factor receptor
- FDR
false discovery rate
- HR
hazard ratio
- ICU
intensive care unit
- IgE
immunoglobulin E
- IκB
inhibitor of nuclear factor-κB
- IL
interleukin
- IQR
interquartile range
- MAP
mitogen-activated protein
- MARC
multicenter airway research collaboration
- NFκB
nuclear factor-κB
- OR
odds ratio
- PAM
partition around medoids
- PI3K
phosphatidylinositol 3-kinase
- RSV
respiratory syncytial virus
- RT-PCR
real-time polymerase chain reaction
- RV
rhinovirus
- Th2
T helper cell type 2
- TNF
tumor necrosis factor
- t-SNE
t-distributed stochastic neighbor embedding
Footnotes
Conflict of Interest Disclosures: The authors have no conflicts of interest relevant to this article to disclose.
REFERENCES
- 1.Fujiogi M, Goto T, Yasunaga H, et al. Trends in bronchiolitis hospitalizations in the United States: 2000–2016. Pediatrics. 2019;144(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hasegawa K, Piedra PA, Bauer CS, et al. Nasopharyngeal CCL5 in infants with severe bronchiolitis and risk of recurrent wheezing: A multi-center prospective cohort study. Clin Exp Allergy. 2018;48(8):1063–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dumas O, Hasegawa K, Mansbach JM, Sullivan AF, Piedra PA, Camargo CA Jr. Severe bronchiolitis profiles and risk of recurrent wheeze by age 3 years. J Allergy Clin Immunol. 2019;143(4):1371–1379. e1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Midulla F, Nicolai A, Ferrara M, et al. Recurrent wheezing 36 months after bronchiolitis is associated with rhinovirus infections and blood eosinophilia. Acta Paediatrica. 2014;103(10):1094–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Régnier SA, Huels J. Association between respiratory syncytial virus hospitalizations in infants and respiratory sequelae: systematic review and meta-analysis. Pediatr Infect Dis J. 2013;32(8):820–826. [DOI] [PubMed] [Google Scholar]
- 6.Henderson J, Hilliard TN, Sherriff A, Stalker D, Al Shammari N, Thomas HM. Hospitalization for RSV bronchiolitis before 12 months of age and subsequent asthma, atopy and wheeze: a longitudinal birth cohort study. Pediatr Allergy Immunol. 2005;16(5):386–392. [DOI] [PubMed] [Google Scholar]
- 7.Liu L, Pan Y, Zhu Y, et al. Association between rhinovirus wheezing illness and the development of childhood asthma: a meta-analysis. BMJ Open. 2017;7(4):e013034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koponen P, Helminen M, Paassilta M, Luukkaala T, Korppi M. Preschool asthma after bronchiolitis in infancy. Eur Respir J. 2012;39(1):76–80. [DOI] [PubMed] [Google Scholar]
- 9.Hasegawa K, Jartti T, Bochkov YA, et al. Rhinovirus species in children with severe bronchiolitis: multicenter cohort studies in the United States and Finland. Pediatr Infect Dis J. 2019;38(3):e59–e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Törmänen S, Lauhkonen E, Riikonen R, et al. Risk factors for asthma after infant bronchiolitis. Allergy. 2018;73(4):916–922. [DOI] [PubMed] [Google Scholar]
- 11.Hasegawa K, Dumas O, Hartert TV, Camargo CA. Advancing our understanding of infant bronchiolitis through phenotyping and endotyping: clinical and molecular approaches. Expert Rev Respir Med. 2016;10(8):891–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ralston SL, Lieberthal AS, Meissner HC, et al. Clinical practice guideline: the diagnosis, management, and prevention of bronchiolitis. Pediatrics. 2014;134(5):e1474–e1502. [DOI] [PubMed] [Google Scholar]
- 13.Dumas O, Mansbach JM, Jartti T, et al. A clustering approach to identify severe bronchiolitis profiles in children. Thorax. 2016;71(8):712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujiogi M, Dumas O, Hasegawa K, Jartti T, Camargo CA. Identifying and predicting severe bronchiolitis profiles at high risk for developing asthma: Analysis of three prospective cohorts. EClinicalMedicine. 2022;43:101257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dumas O, Erkkola R, Bergroth E, et al. Severe bronchiolitis profiles and risk of asthma development in Finnish children [published online ahead of print, 2021 Oct 5]. J Allergy Clin Immunol. 2021;S0091–6749(21)01513-X. [DOI] [PubMed] [Google Scholar]
- 16.Erkkola RA, Virta LJ, Vahlberg T, Jartti T. Prednisolone for the first rhinovirus induced wheezing reduces use of respiratory medication. Pediatr Allergy Immunol. 2022;33(1):e13668. [DOI] [PubMed] [Google Scholar]
- 17.Hurme P, Homil K, Lehtinen P, et al. Efficacy of inhaled salbutamol with and without prednisolone for first acute rhinovirus-induced wheezing episode. Clin Exp Allergy. 2021;51(9):1121–1132. [DOI] [PubMed] [Google Scholar]
- 18.Jartti T, Nieminen R, Vuorinen T, et al. Short- and long-term efficacy of prednisolone for first acute rhinovirus-induced wheezing episode. J Allergy Clin Immunol. 2015;135(3):691–8. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aslam B, Basit M, Nisar MA, Khurshid M, Rasool MH. Proteomics: technologies and their applications. J Chromatogr Sci. 2017;55(2):182–196. [DOI] [PubMed] [Google Scholar]
- 20.Hasegawa K, Mansbach JM, Bochkov YA, et al. Association of rhinovirus C bronchiolitis and immunoglobulin E sensitization during infancy with development of recurrent wheeze. JAMA Pediatr. 2019;173(6):544–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hasegawa K, Mansbach JM, Ajami NJ, et al. Association of nasopharyngeal microbiota profiles with bronchiolitis severity in infants hospitalised for bronchiolitis. European Respiratory Journal. 2016;48(5):1329–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hasegawa K, Jartti T, Mansbach JM, et al. Respiratory syncytial virus genomic load and disease severity among children hospitalized with bronchiolitis: multicenter cohort studies in the United States and Finland. J Infect Dis. 2015;211(10):1550–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mansbach JM, Piedra PA, Teach SJ, et al. Prospective multicenter study of viral etiology and hospital length of stay in children with severe bronchiolitis. Arch Pediatr Adolesc Med. 2012;166(8):700–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mansbach JM, Piedra PA, Stevenson MD, et al. Prospective multicenter study of children with bronchiolitis requiring mechanical ventilation. Pediatrics. 2012;130(3):e492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Camargo CA Jr., Ingham T, Wickens K, et al. Cord-blood 25-hydroxyvitamin D levels and risk of respiratory infection, wheezing, and asthma. Pediatrics. 2011;127(1):e180–187. [DOI] [PubMed] [Google Scholar]
- 26.Reynolds AP, Richards G, de la Iglesia B, Rayward-Smith VJ. Clustering rules: a comparison of partitioning and hierarchical clustering algorithms. J. Math. Model. Algorithms 2006;5(4):475–504. [Google Scholar]
- 27.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hartigan JA, Wong MA. Algorithm AS 136: A k-means clustering algorithm. J R Stat Soc Series C Stat Methodol. 1979;28(1):100–108. [Google Scholar]
- 29.Van der Maaten L, Hinton G. Visualizing data using t-SNE. J. Mach. Learn. Res 2008;9(11):2579–2605. [Google Scholar]
- 30.Gu Z, Gu L, Eils R, Schlesner M, Brors B. circlize implements and enhances circular visualization in R. Bioinformatics. 2014;30(19):2811–2812. [DOI] [PubMed] [Google Scholar]
- 31.Wu X, Hasan MA, Chen JY. Pathway and network analysis in proteomics. J Theor Biol. 2014;362:44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ulgen E, Ozisik O, Sezerman OU. pathfindR: an R package for comprehensive identification of enriched pathways in omics data through active subnetworks. Front Genet. 2019;10:858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujiogi M, Camargo CA Jr., Raita Y, et al. Integrated associations of nasopharyngeal and serum metabolome with bronchiolitis severity and asthma: A multicenter prospective cohort study. Pediatr Allergy Immunol. 2021;32(5):905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pang Z, Chong J, Zhou G, et al. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Research. 2021;49(W1):W388–W396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289–300. [Google Scholar]
- 36.Jartti T, Smits HH, Bønnelykke K, et al. Bronchiolitis needs a revisit: Distinguishing between virus entities and their treatments. Allergy. 2019;74(1):40–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Midulla F, Nenna R, Scagnolari C, et al. How respiratory syncytial virus genotypes influence the clinical course in infants hospitalized for bronchiolitis. J Infect Dis. 2018;219(4):526–534. [DOI] [PubMed] [Google Scholar]
- 38.Fujiogi M, Camargo CA, Bernot JP, et al. In infants with severe bronchiolitis: dual-transcriptomic profiling of nasopharyngeal microbiome and host response. Pediatr Res. 2020;88(2):144–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hasegawa K, Pérez-Losada M, Hoptay CE, et al. RSV vs. rhinovirus bronchiolitis: difference in nasal airway microRNA profiles and NFκB signaling. Pediatr Res. 2018;83(3):606–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turi KN, Romick-Rosendale L, Gebretsadik T, et al. Using urine metabolomics to understand the pathogenesis of infant respiratory syncytial virus (RSV) infection and its role in childhood wheezing. Metabolomics. 2018;14(10):135–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fujiogi M, Camargo CA Jr., Raita Y, et al. Association of rhinovirus species with nasopharyngeal metabolome in bronchiolitis infants: A multicenter study. Allergy. 2020;75(9):2379–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujiogi M, Camargo CA Jr., Raita Y, et al. Respiratory viruses are associated with serum metabolome among infants hospitalized for bronchiolitis: A multicenter study. Pediatr Allergy Immunol. 2020;31(7):755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujiogi M, Camargo CA Jr., Raita Y, et al. Association of endemic coronaviruses with nasopharyngeal metabolome and microbiota among infants with severe bronchiolitis: a prospective multicenter study. Pediatr Res. 2021;89(7):1594–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu Z, Camargo CA Jr., Raita Y, et al. Metabolome subtyping of severe bronchiolitis in infancy and risk of childhood asthma. J Allergy Clin Immunol. 2022;149(1):102–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosas-Salazar C, Tang ZZ, Shilts MH, et al. Upper respiratory tract bacterial-immune interactions during respiratory syncytial virus infection in infancy [published online ahead of print, 2021 Sep 14]. J Allergy Clin Immunol. 2021;S0091–6749(21)01391–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hasegawa K, Mansbach JM, Ajami NJ, et al. Serum cathelicidin, nasopharyngeal microbiota, and disease severity among infants hospitalized with bronchiolitis. J Allergy Clin Immunol. 2017;139(4):1383–1386. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hasegawa K, Mansbach JM, Ajami NJ, et al. The relationship between nasopharyngeal CCL5 and microbiota on disease severity among infants with bronchiolitis. Allergy. 2017;72(11):1796–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toivonen L, Camargo CA Jr., Gern JE, et al. Association between rhinovirus species and nasopharyngeal microbiota in infants with severe bronchiolitis. J Allergy Clin Immunol. 2019;143(5):1925–1928. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Steenhuijsen Piters WAA, Heinonen S, Hasrat R, et al. Nasopharyngeal microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am J Respir Crit Care Med. 2016;194(9):1104–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mansbach JM, Hasegawa K, Henke DM, et al. Respiratory syncytial virus and rhinovirus severe bronchiolitis are associated with distinct nasopharyngeal microbiota. J Allergy Clin Immunol. 2016;137(6):1909–1913. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bowler RP, Wendt CH, Fessler MB, et al. New strategies and challenges in lung proteomics and metabolomics. An official American Thoracic Society workshop report. Ann Am Thorac Soc. 2017;14(12):1721–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ooka T, Raita Y, Ngo D, et al. Proteome signature difference between respiratory viruses is associated with severity of bronchiolitis. Pediatr Allergy Immunol. 2021;32(8):1869–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schofield JPR, Burg D, Nicholas B, et al. Stratification of asthma phenotypes by airway proteomic signatures. J Allergy Clin Immunol. 2019;144(1):70–82. [DOI] [PubMed] [Google Scholar]
- 54.Ejaz S, Nasim F-u-H, Ashraf M, Ahmad S. Serum proteome profiling to identify proteins promoting pathogenesis of non-atopic asthma. Protein Pept Lett. 2018;25(10):933–942. [DOI] [PubMed] [Google Scholar]
- 55.Xu P, Wang L, Chen D, et al. The application of proteomics in the diagnosis and treatment of bronchial asthma. Ann Transl Med. 2020;8(4):132–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rubner FJ, Jackson DJ, Evans MD, et al. Early life rhinovirus wheezing, allergic sensitization, and asthma risk at adolescence. J Allergy Clin Immunol. 2017;139(2):501–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jackson DJ, Gangnon RE, Evans MD, et al. Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. Am J Respir Crit Care Med. 2008;178(7):667–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kusel MM, Kebadze T, Johnston SL, Holt PG, Sly PD. Febrile respiratory illnesses in infancy and atopy are risk factors for persistent asthma and wheeze. Eur Respir J. 2012;39(4):876–882. [DOI] [PubMed] [Google Scholar]
- 59.Kusel MM, de Klerk NH, Kebadze T, et al. Early-life respiratory viral infections, atopic sensitization, and risk of subsequent development of persistent asthma. J Allergy Clin Immunol. 2007;119(5):1105–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Das J, Chen CH, Yang L, Cohn L, Ray P, Ray A. A critical role for NF-kappa B in GATA3 expression and TH2 differentiation in allergic airway inflammation. Nat Immunol. 2001;2(1):45–50. [DOI] [PubMed] [Google Scholar]
- 61.Park SJ, Lee KS, Lee SJ, et al. l-2-oxothiazolidine-4-carboxylic acid or α-lipoic acid attenuates airway remodeling: involvement of nuclear factor-κB (NF-κB), nuclear factor rrythroid 2p45-related factor-2 (Nrf2), and hypoxia-inducible factor (HIF). Int J Mol Sci. 2012;13(7):7915–7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yuan F, Liu R, Hu M, et al. JAX2, an ethanol extract of Hyssopus cuspidatus Boriss, can prevent bronchial asthma by inhibiting MAPK/NF-κB inflammatory signaling. Phytomedicine. 2019;57:305–314. [DOI] [PubMed] [Google Scholar]
- 63.Athari SS. Targeting cell signaling in allergic asthma. Signal Transduct Target Ther. 2019;4(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Raita Y, Pérez-Losada M, Freishtat RJ, et al. Integrated omics endotyping of infants with respiratory syncytial virus bronchiolitis and risk of childhood asthma. Nat Commun. 2021;12(1):3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cakebread JA, Haitchi HM, Xu Y, Holgate ST, Roberts G, Davies DE. Rhinovirus-16 induced release of IP-10 and IL-8 is augmented by Th2 cytokines in a pediatric bronchial epithelial cell model. PloS one. 2014;9(4):e94010–e94010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Webster JD, Vucic D. The balance of TNF mediated pathways regulates inflammatory cell death signaling in healthy and diseased tissues. Front Cell Dev Biol. 2020;8:365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bradley J TNF-mediated inflammatory disease. J Pathol. 2008;214(2):149–160. [DOI] [PubMed] [Google Scholar]
- 68.Kikuchi S, Kikuchi I, Hagiwara K, Kanazawa M, Nagata M. Association of tumor necrosis factor-α and neutrophilic inflammation in severe asthma. Allergol Int. 2005;54(4):621–625. [Google Scholar]
- 69.Silvestri M, Bontempelli M, Giacomelli M, et al. High serum levels of tumour necrosis factor-α and interleukin-8 in severe asthma: markers of systemic inflammation? Clin Exp Allergy. 2006;36(11):1373–1381. [DOI] [PubMed] [Google Scholar]
- 70.Kyriakopoulos C, Gogali A, Bartziokas K, Kostikas K. Identification and treatment of T2-low asthma in the era of biologics. ERJ Open Res. 2021;7(2):00309–02020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Davies ER, Perotin J-M, Kelly JFC, et al. Involvement of the epidermal growth factor receptor in IL-13-mediated corticosteroid-resistant airway inflammation. Clin Exp Allergy. 2020;50(6):672–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Östling J, van Geest M, Schofield JPR, et al. IL-17-high asthma with features of a psoriasis immunophenotype. J Allergy Clin Immunol. 2019;144(5):1198–1213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


