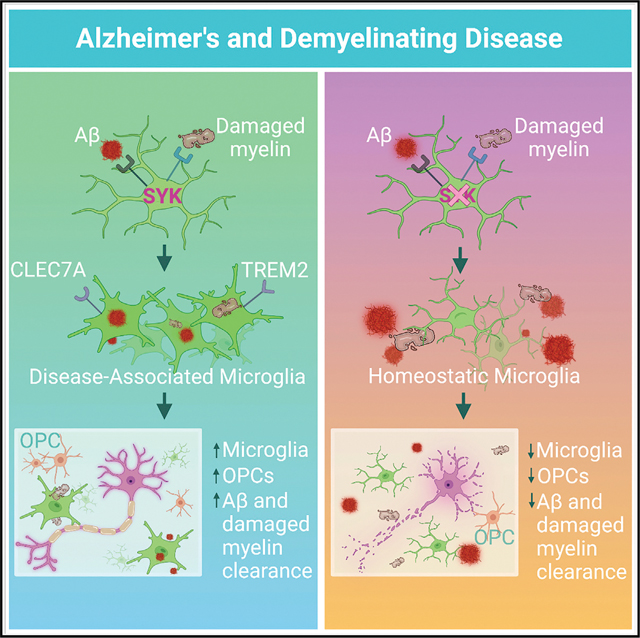
SUMMARY
Recent studies have begun to reveal critical roles for the brain’s professional phagocytes, microglia, and their receptors in the control of neurotoxic amyloid beta (Aβ) and myelin debris accumulation in neurodegenerative disease. However, the critical intracellular molecules that orchestrate neuroprotective functions of microglia remain poorly understood. In our studies, we find that targeted deletion of SYK in microglia leads to exacerbated Aβ deposition, aggravated neuropathology, and cognitive defects in the 5xFAD mouse model of Alzheimer’s disease (AD). Disruption of SYK signaling in this AD model was further shown to impede the development of disease-associated microglia (DAM), alter AKT/GSK3β-signaling, and restrict Aβ phagocytosis by microglia. Conversely, receptor-mediated activation of SYK limits Aβ load. We also found that SYK critically regulates microglial phagocytosis and DAM acquisition in demyelinating disease. Collectively, these results broaden our understanding of the key innate immune signaling molecules that instruct beneficial microglial functions in response to neurotoxic material.
In brief
SYK is a central intracellular regulator of microglial activation and phagocytosis that is deployed to limit Aβ pathology and demyelinating disease.
Graphical Abstract
INTRODUCTION
Neurodegenerative disorders, such as Alzheimer’s disease (AD), are major public health issues that are likely to increase in prevalence with the aging population. In general terms, neurodegenerative diseases are thought to be driven by the accumulation of neurotoxic material such as amyloid beta (Aβ) or myelin debris in the central nervous system (CNS) (Nussbaum and Ellis, 2003; Trapp and Nave, 2008).The buildup of neurotoxic agents is believed to cause neuronal damage and death, which can ultimately lead to various forms of neurological dysfunction that include cognitive decline, motor abnormalities, mental disorders, and loss of inhibition (Chung et al., 2018; Taylor et al., 2002; Vickers et al., 2009). Mounting evidence suggests that microglia, which are the professional phagocytes of the CNS, are critically involved in ensuring the proper containment and removal of neurotoxic material in neurodegenerative disease pathogenesis (Condello et al., 2015; Hickman et al., 2018; Lampron et al., 2015). Indeed, human genome-wide association studies (GWAS) have implicated mutations in microglial receptors in the development of several neurodegenerative diseases (Cooper-Knock et al., 2017; Efthymiou and Goate, 2017; IMSG, 2019).
Most notably, emerging evidence from both AD patients and neurodegenerative mouse models has identified key roles for TREM2, CD33, and CD22 in disease progression (Bemiller et al., 2017; Krasemann et al., 2017; Malik et al., 2013; Pluvinage et al., 2019; Ulland et al., 2017; Wang et al., 2015). Although there is growing interest in targeting these receptors to treat neurodegenerative disease, we currently lack knowledge of the major downstream signaling molecules and effector mechanisms employed by these receptors to influence disease pathogenesis. Identification of the intracellular mediators that coordinate neuroprotective microglial functions will help to uncover novel pathways that can be targeted to treat neurodegenerative disease, and will also offer new insights into the pathoetiology underlying neurodegeneration. Moreover, targeting major shared intracellular signaling pathways may prove more effective than targeting individual receptors in isolation.
In the studies presented here, we explored whether the intracellular signaling molecule, spleen tyrosine kinase (SYK), is involved in coordinating neuroprotective functions in microglia during neurodegenerative disease. SYK is perhaps best known for the critical role that it plays in mounting protective antifungal immune responses downstream of C-type lectin (CLEC) receptors expressed on innate immune cells (Lionakis et al., 2017; Mocsai et al., 2010). In addition, SYK has been identified as the central kinase that instructs signaling and effector functions downstream of TREM2, CD33, and CD22 receptors (Clark and Giltiay, 2018; Wissfeld et al., 2021; Yao et al., 2019). The activation of SYK in microglia surrounding Aβ and other forms of neurotoxic material (Schweig et al., 2017) spurred our interest in delineating whether SYK is a critical regulator of microglial responses in neurodegenerative disease. Though the ability of SYK to modulate Aβ and tau biology has been explored in previous in vitro studies using immortalized CNS lines and pharmacological agents with well-described off-target effects (Lawlor et al., 2018; Paris et al., 2014), the extent to which SYK influences in vivo microglial responses and neurodegenerative disease pathogenesis currently remains poorly understood.
Here, we show that microglia-specific deletion of SYK leads to elevated levels of Aβ deposition, exacerbated neuropathology, and cognitive impairment in the 5xFAD mouse model of AD. We further demonstrate that SYK is critically involved in both the compaction and phagocytosis of Aβ by microglia as well as the regulation of AKT/GSK3β-signaling. Interestingly, we identify SYK as a key intracellular regulator of disease-associated microglia (DAM) phenotype acquisition and further show that CLEC7A-induced activation of SYK in 5xFAD mice promotes improved clearance of Aβ. Moreover, we show that the neuroprotective effects of SYK on microglia are replicated in the context of demyelinating disease. Collectively, these findings define SYK as a central regulator of neuroprotective microglial responses in neurodegenerative disease.
RESULTS
SYK signaling in microglia limits Aβ accumulation
To investigate how SYK signaling in microglia impacts Aβ-mediated neurodegenerative disease, we first generated Sykfl/fl Cx3cr1ERT2Cre mice (hereafter referred to as SykΔMG mice) as a genetic tool to delete SYK from microglia. We then crossed SykΔMG mice with 5xFAD mice, an AD mouse model that develops aggressive Aβ pathology starting at 1.5 months of age (Richard et al., 2015). Importantly, SYK expression is unchanged between 5xFAD and non-5xFAD immune cells that have been described to modulate AD pathogenesis (Figures S1A and S1B). 5xFAD SykΔMG mice were given tamoxifen food for 2 weeks after weaning to induce deletion of SYK and then returned to normal chow to allow for peripheral Cx3cr1-expressing immune cells to turn over and regain Syk expression, while permitting long-lived microglia to remain SYK-deficient (Figures S1C–S1H). As controls, Cre-negative Sykfl/fl 5xFAD littermates (hereafter referred to as 5xFAD mice) were similarly fed tamoxifen for 2 weeks at weaning and then returned to normal chow for the remainder of the experiment. It is important to note that this genetic targeting strategy may also induce deletion of SYK in Cx3cr1-expressing CNS border-associated macrophages (BAMs), which do not undergo the frequent turnover characteristic of Cx3cr1-expressing peripheral immune cells (Wu et al., 2021).
Using these newly generated transgenic mouse lines, we found that SYK-deletion in 5xFAD SykΔMG mice leads to significantly elevated accumulation of Aβ in the cortex, hippocampus, and thalamus at 5 months of age (Figures 1A and 1B). Microglia help to limit the spread of pathological Aβ in the brain parenchyma by forming a barrier around Aβ deposits and promoting the physical compaction of Aβ into dense spherical plaques, which ultimately decreases Aβ interaction with susceptible neurons (Condello et al., 2015). Therefore, the lack of Aβ plaque sphericity is often used to identify potentially neurotoxic Aβ aggregates and to provide insights into the efficacy of microglial compaction (Condello et al., 2018; Wang et al., 2016). In addition to higher amounts of Aβ load, we observed that Aβ plaques in the cortex and hippocampus of 5xFAD SykΔMG mice exhibited lower sphericity than the plaques found in 5xFAD littermate controls (Figures 1C,1D, S2A, and S2B). Importantly, delayed deletion of SYK in 4-month-old 5xFAD mice yields similarly increased plaque load in the hippocampus and decreased plaque sphericity in the cortex when harvested at 8 months of age (Figures S2H–S2K). This suggests that microglial SYK remains influential in attenuating pathology after disease onset in 5xFAD mice. In further support of there being less-efficient compaction of Aβ into plaques by SYK-deficient microglia, we observed that Aβ aggregates in 5xFAD SykΔMG mice were more filamentous (increased 6E10 antibody labeling that detects filamentous Aβ; Lee et al., 2018) and less compact (decreased staining of Thioflavin S [ThioS] that detects inert plaques; Lee et al., 2018) than the Aβ deposits found in 5xFAD littermate controls (Figures 1E and 1F). Taken together, these findings suggest that SYK signaling in microglia plays a critical role in the control of Aβ accumulation and compaction.
Figure 1. Deletion of Syk in microglia leads to increased Aβ burden and altered plaque composition in 5xFAD mice.
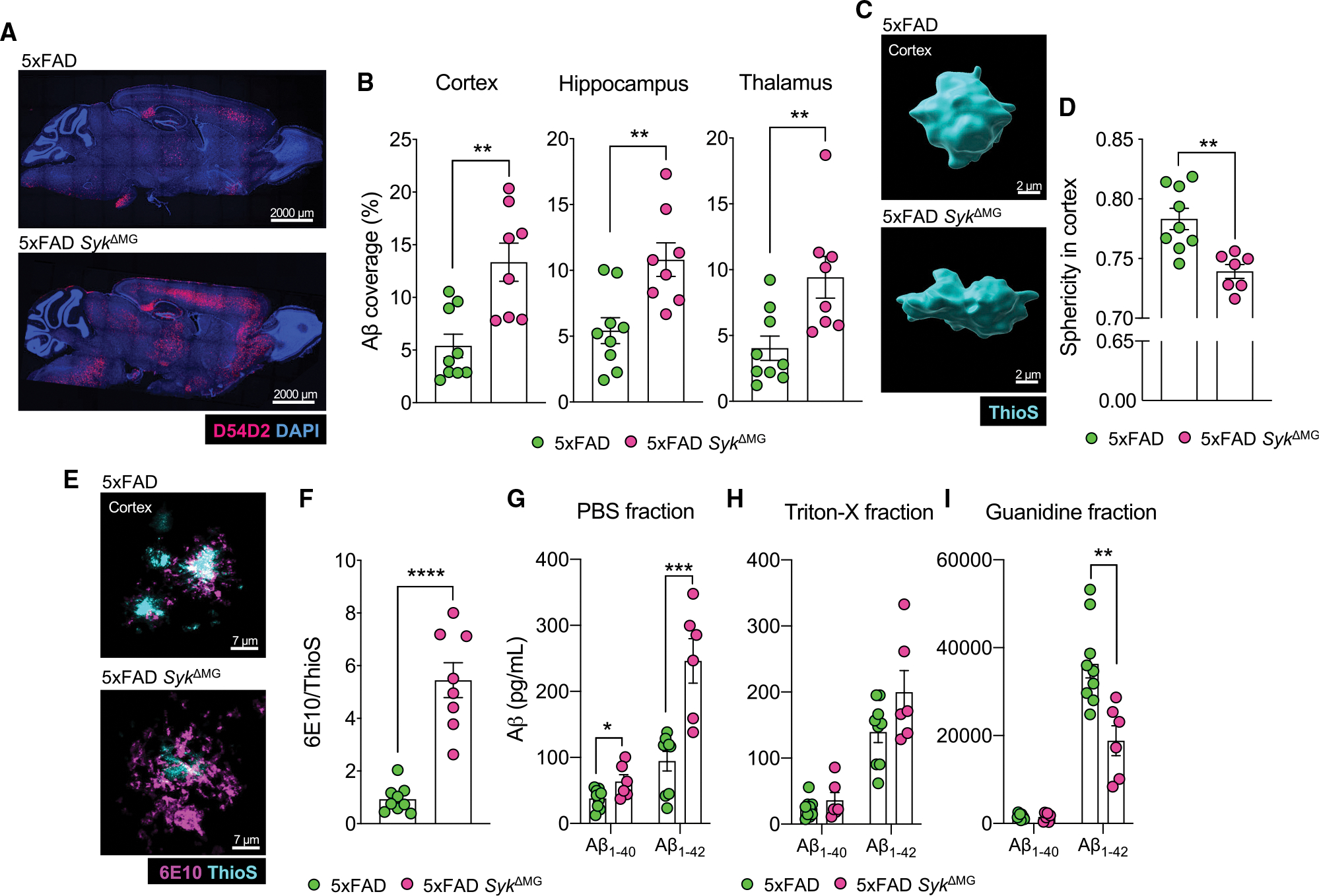
5xFAD Sykfl/flCx3cr1ERT2Cre (5xFAD SykΔMG mice) and Cre-negative 5xFAD Sykfl/fl littermate controls (5xFAD mice) received tamoxifen food for 2 weeks starting at 3 weeks of age and then mice were returned to regular food for the remainder of the experiment. Mice were later harvested at 5 months of age to evaluate amyloid beta (Aβ) load in the brain.
(A and B) Immunofluorescence staining of Aβ (D54D2, red; DAPI, blue) was performed on sagittal sections and the percent area covered by Aβ was quantified.
(C) Sphericity of ThioflavinS (ThioS)-labeled and Imaris-rendered Aβ plaques in the cortex.
(D) Quantification of sphericity with 1.00 being the most spherical, combined data from a total of 50–100 plaques from 3 matching brain sections per mouse.
(E) Representative images of Aβ plaque composition labeling 6E10 (purple) and ThioS (blue).
(F) Quantification represents the percent volume of the 6E10/ThioS ratio per field of view (FOV) from a total of 10–15 plaques from 3 brain sections per mouse.
(G–I) Soluble and insoluble fractions of Aβ1–40 and Aβ1–42 measured by ELISA.
Statistical significance between experimental groups was calculated by unpaired Student’s t test (B), (D), and (F)–(I). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Error bars represent mean ± SEM and each data point represents an individual mouse.
See also Figures S1 and S2.
Microglia are required for the consolidation of neurotoxic soluble Aβ oligomers into more inert insoluble fibrils and for construction into compact plaques (Brown et al., 2020; Huang et al., 2021; Martin et al., 1994). To evaluate whether the absence of SYK in microglia also affects the levels of both soluble and insoluble Aβ40 and Aβ42 in 5xFAD mice, we next evaluated Aβ load using an ELISA and various extraction techniques to isolate Aβ based on solubility. From these studies, we saw that the levels of soluble Aβ40 and Aβ42 were considerably higher in 5xFAD SykΔMG mice than in 5xFAD littermate controls (Figure 1G). Although we did not detect any appreciable differences in Aβ ELISA levels between experimental groups in the Triton X-100 extraction samples, we did observe reduced levels of insoluble Aβ42 in the guanidine fractions isolated from 5xFAD SykΔMG mice (Figures 1H and 1I), indicating that SYK centrally contributes to the ability of microglia to construct more inert Aβ structures. Altogether, these data indicate that SYK is critical for Aβ consolidation and plaque compaction by microglia in 5xFAD mice.
SYK deletion in microglia leads to worsened neuronal health and memory impairment in 5xFAD mice
Due to the influence of SYK signaling in microglia on Aβ accumulation and composition, and the propensity of Aβ to impair neuronal function (Colie et al., 2017; Jawhar et al., 2012), we next assessed neuronal health in 5xFAD and 5xFAD SykΔMG mice. We found that 5-month-old 5xFAD SykΔMG mice had a ~1.5-fold increase in plaque-associated dystrophic neurites in the cortex compared with 5xFAD controls (Figures 2A and 2B). Heightened accumulation of hyperphosphorylated tau, labeled with the AT8 antibody, was also observed around plaques in the cortex of 5xFAD SykΔMG mice (Figures 2C and 2D). This increase in hyperphosphorylated tau is likely indicative of neuronal debilitation (Gendron and Petrucelli, 2009; Kanno et al., 2014) that has culminated from the accumulation of neurotoxic Aβ. 5xFAD SykΔMG mouse neurons in the CA1 region of the hippocampus also displayed increased levels of cell death, as visualized by TUNEL staining (Figures 2E and 2F). These collective findings suggest that SYK activity in microglia helps to preserve neuronal health in 5xFAD mice.
Figure 2. Loss of Syk in microglia negatively affects neuronal health and exacerbates AD-related behaviors in 5xFAD mice.
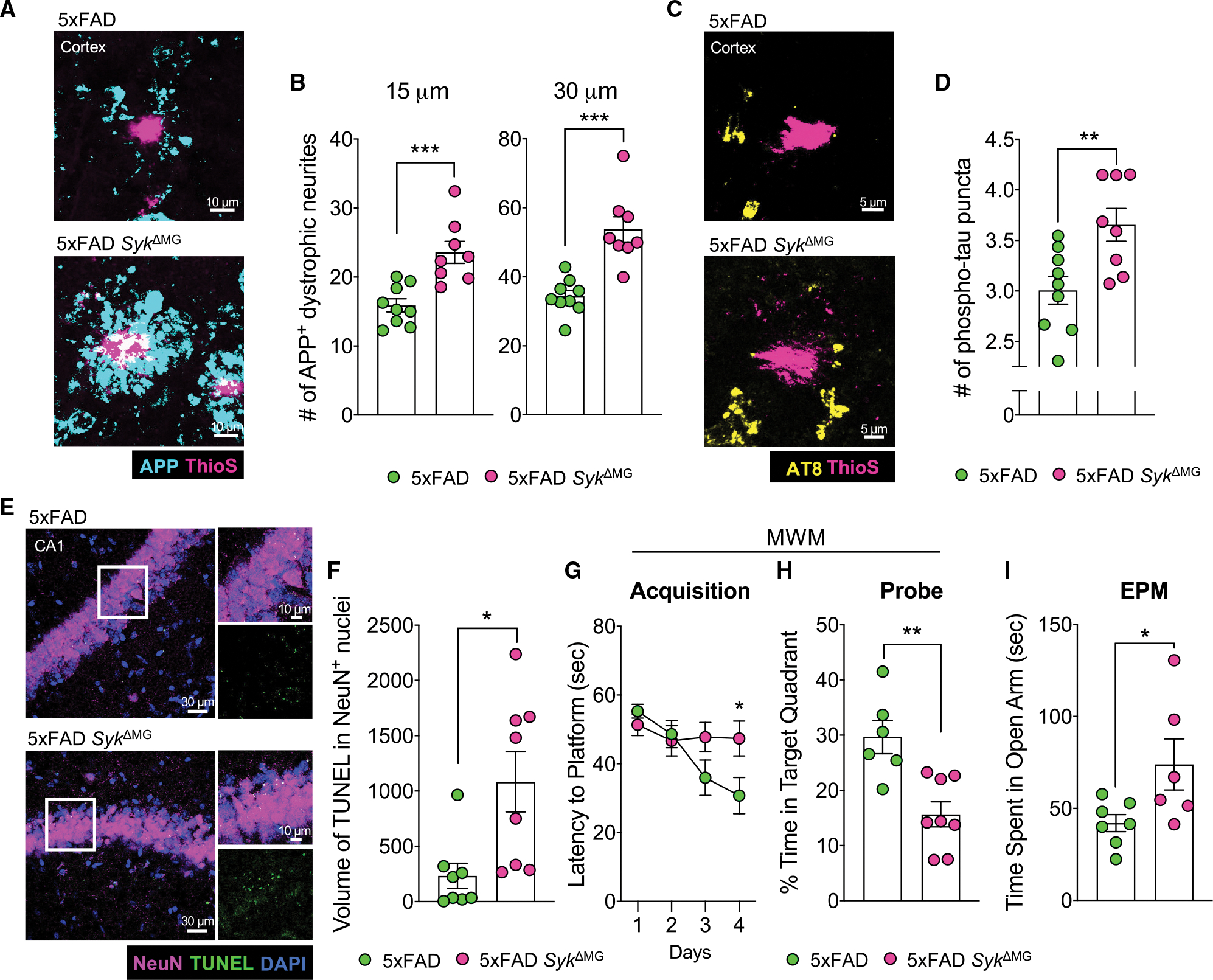
(A–F) Brains were harvested from 5xFAD SykΔMG mice and 5xFAD littermate controls at 5 months of age to evaluate neuronal health and cell death.
(A) The formation of dystrophic neurites surrounding plaques in the cortex was determined by staining for APP (blue) and Aβ using ThioS (pink).
(B) Quantification of APP+ puncta found within 15 and 30 μm of Aβ plaques from a total of ~40 plaques from 3 matching brain sections per mouse.
(C) Cortical sections were stained with AT8 (yellow) for phosphorylated tau (p-tau) puncta found within 15 μm of ThioS (pink)-stained Aβ plaques.
(D) Quantification of p-tau from a total of ~40 plaques from 3 matching sections per mouse.
(E) TUNEL assay (green) and NeuN staining (pink) in the hippocampal CA1 region.
(F) Quantification of volume of TUNEL+ stain found in NeuN+ nuclei from 2 corresponding brain sections per mouse.
(G and H) 4-month-old 5xFAD (n = 6) and 5xFAD SykΔMG (n = 8) mice were evaluated in the Morris water maze (MWM). Statistics for MWM acquisition were calculated on day 4. Combined data from 3 independent experiments.
(I) Performance in the elevated plus maze (EPM) was measured in 4-month-old 5xFAD and 5xFAD SykΔMG mice. Combined data from 2 independent experiments. Statistical significance between experimental groups was calculated by unpaired Student’s t test (B), (D), and (F)–(I). *p < 0.05, **p < 0.01, ***p < 0.001. Error bars represent mean ± SEM and each data point represents an individual mouse.
See also Figures S1 and S2.
To better understand how Syk deficiency in microglia impacts brain function, we evaluated the performance of 4-month-old 5xFAD and 5xFAD SykΔMG mice in the Morris water maze (MWM), which is commonly used to assess spatial learning and memory. In parallel with increased Aβ plaque load and neuronal death, we found that it took markedly longer for 5xFAD SykΔMG mice to find the hidden platform in comparison to 5xFAD littermate controls on day 4 of the MWM test (Figure 2G), which suggests that there is impaired spatial learning in 5xFAD SykΔMG mice. Importantly, these differences in performance were not due to altered bodyweight or defects in locomotor activity in 5xFAD SykΔMG mice, as we observed that body weight and travel velocities in the MWM were similar between the experimental mouse groups (Figures S2O and S2P). Moreover, when the platform was removed for a probe trial on day 5 of the MWM test, 5xFAD SykΔMG mice spent significantly less time than 5xFAD mice in the quadrant of the pool where the hidden platform had previously been located (Figure 2H), which is indicative of impaired spatial memory in 5xFAD SykΔMG mice.
Aβ deposition in 5xFAD mice has also been shown to spur the development of risk-taking and exploratory behaviors, as can be observed in some AD patients (Ha et al., 2012; Jawhar et al., 2012). Therefore, we evaluated the performance of 4-month-old 5xFAD SykΔMG mice and 5xFAD littermate controls in the elevated plus maze (EPM). In these studies, we observed that 5xFAD SykΔMG mice spent more time exploring the open arms of the maze compared to 5xFAD controls (Figure 2I and S2Q), which suggests that SYK deletion in microglia on the 5xFAD background also leads to greater levels of risk-taking and exploratory behaviors. Taken together, these data demonstrate a critical role for microglial SYK in preventing neuronal loss as well as limiting the development of memory impairment and risk-taking-related behaviors in Aβ-mediated neurodegenerative disease.
SYK regulates microglial proliferation and association with Aβ plaques
To gain insights into how SYK influences microglial biology in response to Aβ pathology, we next explored the impact of SYK deletion on microgliosis. Here, we found that 5xFAD littermate controls have significantly more cortical and hippocampal microglia than 5xFAD SykΔMG mice (Figures 3A, 3B, S2C, and S2D). We also observed impaired microglia clustering to Aβ plaques in the cortex and hippocampus of 5xFAD SykΔMG mice, with the number of plaque-associated microglia being 2-fold lower in 5xFAD SykΔMG mice than 5xFAD littermate controls (Figures 3A, 3C, 3D, S2C, and S2E). Interestingly, the reduction in microglia numbers observed in 5xFAD SykΔMG mice appears to be specific to Aβ-mediated pathology, as we did not observe any appreciable differences in microglia numbers between SykΔMG mice and Cre-negative Cx3cr1ERT2Cre Sykfl/fl mice (Sykcon) that do not express the 5xFAD transgenes (Figures S2R and S2S). In addition, 5xFAD mice that underwent delayed deletion of SYK at 4 months of age displayed a corresponding decrease in microglial number and association with Aβ plaques at 8 months of age compared with 5xFAD controls (Figures S2L–S2N). Thus, the criticality of SYK driving microglial responses exists during both disease onset and disease progression in 5xFAD mice. To distinguish what might contribute to the reduced numbers of microglia in 5xFAD SykΔMG mice, we evaluated the proliferative capacity of SYK-deficient microglia. We found that SYK deficiency in 5xFAD mice leads to reduced microglial proliferation, as illustrated by the ~3-fold decrease in Ki67+ microglia seen in the cortex and hippocampus of 5xFAD SykΔMG mice (Figures 3E and 3F). In contrast, in the absence of Aβ accumulation in mice that lack the 5xFAD transgenes, we did not detect any appreciable differences in microglial Ki67 staining between Sykcon and SykΔMG mice (Figures S2R and S2T).
Figure 3. Syk-deficiency limits microglial proliferation and association with Aβ plaques.
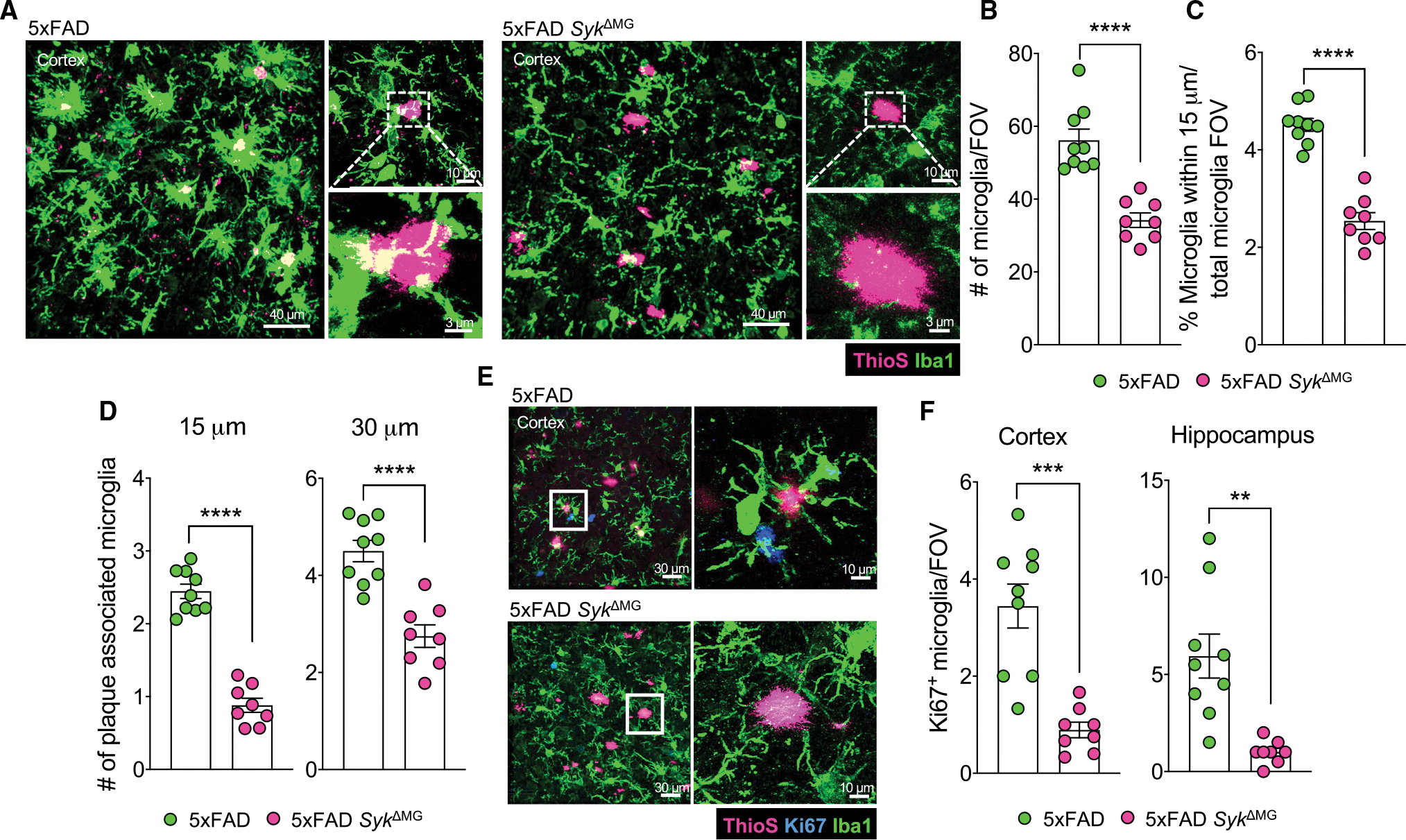
Brains were harvested from 5xFAD SykΔMG mice and 5xFAD littermate controls at 5 months of age to evaluate microgliosis.
(A–C) Microglia were imaged by labeling with Iba1 (green) surrounding Aβ plaques labeled with ThioS (pink) to assess microglial coverage and proximity to plaques. (A) Representative images of Iba1 and ThioS staining in the cortex. (B) Quantification of microglial numbers. (C) Quantification of microglial association with plaques as the percent of microglia within 15 μm of a plaque normalized to the total number of microglia.
(D) Quantification of the number of microglia within a 15 and 30 μm radius surrounding ThioS-labeled Aβ plaques. Each point represents an individual mouse with an average of 50–100 plaques from 3 matching brain sections per mouse.
(E) Representative images of microglial proliferation measured by evaluating Ki67 (blue) colocalization with Iba1+ (green) microglia in the cortex.
(F) Quantification of Ki67+ microglia.
Statistical significance between experimental groups was calculated by an unpaired Student’s t test (B)–(D), and (F). **p < 0.01, ***p < 0.001, ****p < 0.0001. Error bars represent mean ± SEM and each data point represents an individual mouse.
See also Figure S2.
The possibility also exists that increased microglial death could be contributing at some level to reduced numbers of microglia seen in the brains of 5xFAD SykΔMG mice (Figures 3A and 3B). Therefore, we performed TUNEL staining in the cortex of 5-month-old 5xFAD and 5xFAD SykΔMG mice. We did not, however, detect appreciable numbers of Iba1+ microglial cells that stained positive for TUNEL in either 5xFAD or 5xFAD SykΔMG mice (Figures S2W and S2X). This suggests that apoptosis is likely not a major driver of decreased microglial cell numbers in 5xFAD SykΔMG mice at this time point. In summary, these results suggest that SYK is critically involved in coordinating microgliosis in response to Aβ pathology.
Aβ-induced microglial activation is impaired in the absence of SYK
To ascertain whether SYK signaling also impacts microglia activation in response to Aβ, we first evaluated differences in microglia morphology via Sholl analysis. Homeostatic/resting microglia typically exhibit highly ramified processes, whereas activated microglia tend to retract these processes and acquire an ameboid morphology (Parakalan et al., 2012). We observed that non-plaque-associated microglia in the cortex and hippocampus of 5xFAD SykΔMG mice had significantly more ramified processes compared with 5xFAD controls (Figures 4A, 4B, S2F, and S2G). In contrast, resting morphological differences were not seen between Sykcon and SykΔMG mice in the absence of Aβ (Figures S2U and S2V), suggesting that SYK-dependent morphological differences in microglia are specific to Aβ-driven neurological disease. Taken together, these results suggest that microglia-intrinsic SYK signaling plays a central role in coordinating the ability of microglia to take on a morphologically activated state in response to Aβ pathology.
Figure 4. Defective activation of Syk-deficient microglia in 5xFAD mice.
5xFAD Sykfl/flCx3cr1ERT2Cre (5xFAD SykΔMG mice), Cre-negative 5xFAD Sykfl/fl littermate controls (5xFAD mice), Sykfl/fl Cx3cr1ERT2Cre (SykΔMG mice), and Cre-negative Sykfl/fl littermate controls (Sykcon mice) received tamoxifen food for 2 weeks starting at 3 weeks of age and then mice were returned to regular food for the remainder of the experiment. Brains were later harvested at 5 months of age to evaluate microglial activation.
(A) Imaris-rendered microglia morphology labeled with Iba1 (blue) in the cortex.
(B) Sholl analysis quantification from a total of 12 microglia from 3 matching brain sections per mouse (5xFAD n = 9, 5xFAD SykΔMG n = 8).
(C–F) Bulk RNA-seq performed on CD11b+-magnetic bead sorted microglia from 5-month-old mice. (C) Principal component (PC) analysis of sample clustering.
(D) Volcano plots depicting differentially expressed genes (FDR < 0.1). (E) KEGG term enrichment scatterplot highlighting major pathways that are repressed in 5xFAD SykΔMG microglia in comparison to 5xFAD microglia. (F) Heatmap representation of significantly downregulated (FDR < 0.1) stage 1 and 2 disease-associated microglia (DAM) genes between 5xFAD SykΔMG and 5xFAD groups.
(G and H) Mouse AKT pathway phosphorylation array conducted on microglia from 5-month-old 5xFAD SykΔMG and 5xFAD mice. (G) Representative membranes incubated with 5xFAD and 5xFAD SykΔMG microglia measuring AKT phosphorylation targets. (H) Quantification of dot pixel density normalized with respective positive and negative control sample dot pixel density. Data are plotted in membrane order of phosphorylated protein probes; n of 3 for each group.
Statistical significance between experimental groups was calculated by a two-way ANOVA with a Bonferroni post-hoc test (B) and an unpaired Student’s t test (H). *p < 0.05, **p < 0.01, ****p < 0.0001. Error bars represent mean ± SEM.
See also Figure S3.
Recent studies have also shown that microglia upregulate a unique transcriptional program in neurodegenerative disease. This activation-induced transition into DAM is thought to endow microglia with key neuroprotective machinery (Keren-Shaul et al., 2017). This progressive shift from resting-state microglia to DAM involves the coordinated downregulation of many homeostatic markers in stage 1, followed by an upregulation of genes related to microglial response to neurodegenerative pathology in stage 2. Therefore, we next aimed to elucidate whether SYK affects DAM acquisition in response to Aβ-driven neuropathology. To answer this question, we performed bulk RNA sequencing (RNA-seq) on magnetic bead-sorted CD11b+ cells isolated from the brains of 5-month-old Sykcon, SykΔMG, 5xFAD, and 5xFAD SykΔMG mice (Figure S3A). Principal component (PC) analysis revealed, as expected, that 5xFAD microglia form a distinct cluster from control microglia (Figure 4C), which is indicative of the altered transcriptional profile microglia take on in the presence of Aβ. In contrast, the loss of SYK in microglia blocked this transformation, as 5xFAD SykΔMG microglia clustered with unperturbed Sykcon and SykΔMG microglia, suggesting that 5xFAD SykΔMG are more similar to homeostatic microglia than those isolated from the 5xFAD mouse model of AD (Figure 4C). Upon further inspection, we observed 2,769 downregulated and 2,668 upregulated genes (false discovery rate [FDR] < 0.1) when comparing microglia isolated from 5xFAD SykΔMG and 5xFAD mice (Figures 4D and S3D). Moreover, KEGG analysis revealed that many of the repressed genes in 5xFAD SykΔMG microglia were related to neurodegeneration (Figure 4E). In contrast to the numerous transcriptional differences seen between 5xFAD and 5xFAD SykΔMG microglia, we only observed a marginal effect of SYK deletion when comparing SykΔMG and Sykcon microglia in the absence of Aβ pathology (Figures S3B and S3C). These findings suggest that SYK acts as a critical regulator of the transcriptional shift that microglia undergo in response to Aβ-associated neuropathology in 5xFAD mice.
Notably, we found that a large number of genes were markedly repressed in 5xFAD SykΔMG microglia (Figure 4F). More specifically, upon Syk deletion we observed a significant downregulation of stage 1 DAM genes between 5xFAD SykΔMG and 5xFAD microglia (Figure 4F). However, an even more striking downregulation of stage 2 DAM genes (i.e., Lpl, Cst7, Itgax, Axl, Clec7a, Csf1, and Ccl6) was observed in 5xFAD SykΔMG microglia relative to 5xFAD microglia (Figures 4F and S3D). Therefore, SYK appears to be especially critical for the ability of microglia to acquire the more activated stage 2 DAM transcriptional phenotype in 5xFAD mice. To validate this transcriptional block in DAM generation seen in SYK-deficient microglia at the protein level, we performed immunofluorescence staining to evaluate the expression levels of the signature microglial homeostatic marker, Tmem119. As DAM undergo transcriptional activation, homeostatic Tmem119 expression canonically decreases in stage 1 DAM (Krasemann et al., 2017). However, we observed that 5xFAD SykΔMG microglia retained higher Tmem119 expression compared to 5xFAD microglia (Figures S3E and S3F), suggesting their retention of a homeostatic state. In addition, we investigated the expression of stage 2 DAM marker, CLEC7A (Krasemann et al., 2017), on Iba1+ microglia surrounding Aβ plaques in 5xFAD and 5xFAD SykΔMG mice. These imaging studies revealed significantly reduced expression of CLEC7A on Iba1+ microglia surrounding plaques in 5xFAD SykΔMG mice (Figures S3G–S3I). Altogether, these findings reveal that SYK centrally contributes to the critical transformation of homeostatic microglia into DAM following exposure to Aβ.
The downstream signaling that coordinates DAM acquisition has remained poorly described, although it has been suggested that PI3K/AKT signaling can regulate many of the processes and pathways linked to microglial activation in AD (Chu et al., 2021). Therefore, we chose to investigate how the loss of SYK in microglia regulates PI3K/AKT signaling in response to Aβ pathology. Utilizing magnetic bead-sorted CD11b+ microglia isolated from the brains of 5-month-old 5xFAD and 5xFAD SykΔMG mice, we evaluated the phosphorylation status of 18 proteins that have been shown to be centrally involved in the PI3K/AKT signaling pathway. In these studies, we found that one particular arm of the AKT signaling pathway was differentially regulated between 5xFAD and 5xFAD SykΔMG microglia. More specifically, we observed that phosphorylation levels of both AKT (P-S473) and GSK3β (P-Ser9) were reduced in 5xFAD SykΔMG microglia when compared with 5xFAD control microglia (Figures 4G and 4H). These findings are notable as decreased phosphorylation of AKT (P-S473) and GSK3β (P-Ser9) has been observed in the brains of AD patients in comparison to age-matched controls (Mateo et al., 2006; Steen et al., 2005). Moreover, mutations in GSK3B have also been linked to both familial and sporadic forms of AD in humans (Schaffer et al., 2008). Our data indicate that 5xFAD SykΔMG microglia exhibit decreased AKT activation as well as diminished phosphorylation of GSK3β at Ser9 (Figures 4G and 4H). Phosphorylation of GSK3β at Ser9 leads to its potent inactivation (Doble and Woodgett, 2003; Steen et al., 2005), which ultimately indicates that there is increased activation of GSK3β in 5xFAD SykΔMG microglia. Given that GSK3β activation has been shown to contribute to Aβ accumulation, tau phosphorylation, and neuronal damage in models of AD (DaRocha-Souto et al., 2012; Hernandez et al., 2013; Hurtado et al., 2012; Reddy, 2013), SYK-related modulation of the GSK3β pathway may contribute to the exacerbation of disease seen in 5xFAD SykΔMG mice.
Extensive work has characterized microglial receptor TREM2 as influential in driving microglial acquisition of the DAM transcriptome (Keren-Shaul et al., 2017; Krasemann et al., 2017; Wang et al., 2015). Based on these findings, we wanted to determine if the impaired transcriptional shift we observed in our SYK-deficient 5xFAD microglia phenocopies the previously described deficiency in microglial transcriptional activation seen in TREM2-deficient 5xFAD microglia. To this end, we compared our RNA-seq dataset with a previously published dataset analyzing 5xFAD Trem2−/− microglia (Griciuc et al., 2019). We found that 25% of genes upregulated and ~60% of genes downregulated in 5xFAD Trem2−/− microglia were shared with 5xFAD SykΔMG microglia (FDR < 0.05) (Figure S3J). In addition, the genes downregulated by 5xFAD Trem2−/− and 5xFAD SykΔMG microglia share molecular function terms such as “signaling receptor binding” and “protein binding” (Figure S3K). These data suggest an important shared signaling axis between TREM2 and SYK. However, the transcriptional shift upon SYK deletion in 5xFAD SykΔMG microglia encompasses a substantial population of uniquely upregulated (>97%) and downregulated (>63%) differentially expressed genes not observed in 5xFAD mice that lack TREM2 (FDR < 0.05) (Figure S3J). Therefore, it is likely that TREM2 signaling through SYK is only partially regulating microglial DAM transition, and that SYK conceivably transmits signals from multiple receptors in addition to TREM2 in the 5xFAD mouse model.
Aβ pathology promotes increased lipid droplet formation and ROS production in SYK-deficient microglia
Our RNA-seq findings revealed that 5xFAD SykΔMG microglia exhibit a prominent reduction in lipoprotein lipase (Lpl) expression (Figure 4F), a DAM marker known to be critical for regulating cellular lipid homeostasis and lipid droplet accumulation (Loving et al., 2021). Recent work in microglial biology studying aging and neurodegeneration has identified a population of lipid-droplet-accumulating microglia (LDAM), which display impaired phagocytosis and increased reactive oxygen species (ROS) production (Marschallinger et al., 2020). Therefore, we chose to evaluate lipid homeostasis in SYK-deficient microglia using BODIPY, a fluorescent dye that detects lipid droplets (Marschallinger et al., 2020). In these studies, we observed a significant increase in BODIPY fluorescence in 5xFAD SykΔMG CD11b+CD45int microglia, indicating an increase in lipid droplet accumulation in SYK-deficient microglia (Figures S4A and S4B). Previous work has defined several dysfunctions in LDAM, including their increased production of ROS (Marschallinger et al., 2020). Consistently, 5xFAD SykΔMG microglia also displayed an increase in CellROX fluorescence, a cell-permeant dye that fluoresces when oxidized by ROS, when compared with control 5xFAD microglia (Figures S4C and S4D). These collective findings suggest that SYK may act to partially limit microglial transition to an LDAM-like state.
Phagocytosis of Aβ is coordinated by SYK
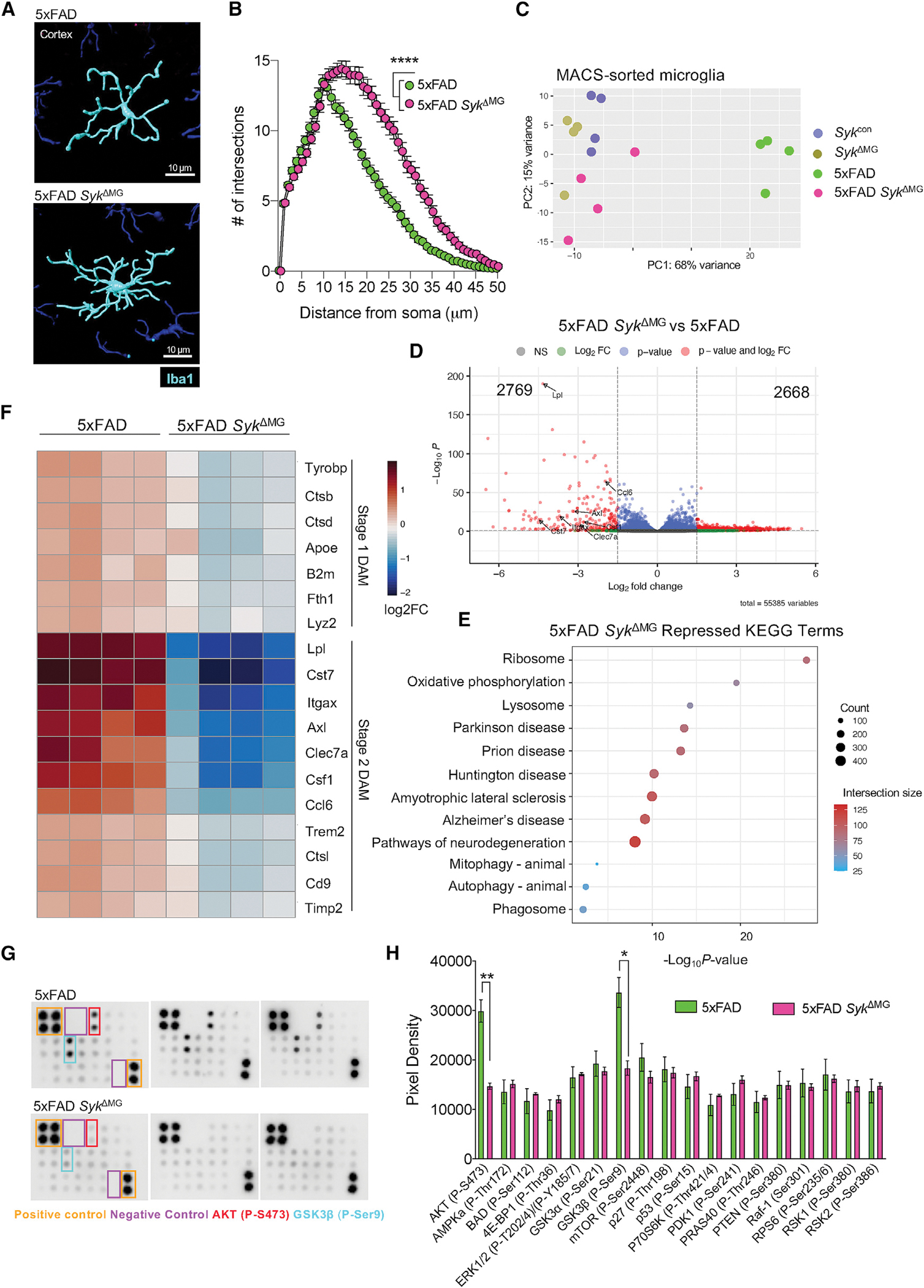
Given that the loss of Syk limits microglia DAM marker expression and augments Aβ accumulation, we hypothesized that SYK may also play critical roles in microglial phagocytosis of Aβ in 5xFAD mice, which could help to explain the elevated deposition of Aβ seen in 5xFAD SykΔMG mice (Figure 1). To investigate this possibility, we measured Aβ engulfment by microglia using immunofluorescence staining and saw that 5xFAD microglia engulfed more than twice the amount of Aβ than 5xFAD SykΔMG microglia (Figures 5A and 5B). Similarly, approximately twice the volume of Aβ was engulfed within CD68, a well-described marker of microglial phagolysosomes (Walker and Lue, 2015), in 5xFAD microglia compared with 5xFAD SykΔMG microglia (Figures 5C and 5D). Notably, we noticed that 5xFAD SykΔMG and 5xFAD microglia upregulated CD68 to comparable total levels across the cortex (Figures S4E and S4F). However, though much of the engulfed Aβ detected in 5xFAD microglia colocalized with CD68, there was far less Aβ engulfed within CD68 in SYK-deficient 5xFAD microglia (Figures 5C and 5D).
Figure 5. SYK is critical for microglial uptake and phagocytosis of Aβ.
(A–F) Brains were harvested from 5xFAD SykΔMG mice and 5xFAD littermate controls at 5 months of age to evaluate microglial phagocytosis.
(A) Imaris-rendered Aβ plaques (ThioS, pink) and Iba1+ cells (green) with the completely localized Aβ-microglia (engulfed) channel in blue.
(B) Percent area of engulfment quantification from a total of ~20 plaques from 3 matching brain sections per mouse.
(C) Imaris-rendered Aβ plaques (ThioS, pink) and CD68 (yellow) with the completely localized Aβ-CD68 (engulfed) channel in blue.
(D) Percent area of engulfment quantification from a total of ~20 plaques from 3 matching brain sections per mouse.
(E and F) Mice received intraperitoneal injections of Methoxy-X04 and then brains were harvested 3 h later to evaluate microglial phagocytosis of Methoxy-X04+ labeled Aβ. (E) and (F) Representative flow cytometry plots and quantification of the percentage of CD11bhiCD45int cells that had taken up Methoxy-X04+ labeled Aβ.
(G and H) WT and Sykfl/fl LysMCre bone marrow-derived macrophages (BMDMs) pre-treated with vehicle or 10 μM Tideglusib, a GSK3β inhibitor, for 1 h prior to treatment with 10 μM CypHer5E-tagged Aβ oligomers. Aβ phagocytosis by BMDMs was determined 24 h later by measuring CypHer5E fluorescence by flow cytometry. (G) and (H) Representative flow cytometry plots and quantification of percent CypHer5E CD11bhiF4/80hi cells.
(I–L) 10-week-old 5xFAD and 5xFAD SykΔMG mice received bilateral intrahippocampal injections of vehicle and CLEC7A agonist pustulan. Seven days post injection (dpi) brains were harvested to measure Aβ load between matched vehicle and pustulan injected hippocampal hemispheres. (J) Representative immunofluorescence staining of D54D2-labeled Aβ (pink) in hippocampal sections. (K and L) Mouse-matched quantification of Aβ in vehicle- and pustulan-injected hippocampal hemispheres.
Statistical significance between experimental groups was calculated by unpaired Student’s t test (B), (D), and (F), one-way ANOVA with multiple comparisons (H), and paired Student’s t test (K) and (L). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Error bars represent mean ± SEM and each data point represents an individual mouse.
See also Figure S4.
To further substantiate a role for microglial SYK in Aβ phagocytosis, we next explored this in a secondary experimental system. In these studies, 5-month-old 5xFAD SykΔMG mice and 5xFAD littermate controls received an intraperitoneal (i.p.) injection of Methoxy-X04, a brain penetrant dye that labels fibrillar Aβ. After 3 h, we harvested brains from these mice and used flow cytometry to quantify the percentage of microglia that had taken up Methoxy-X04-labeled Aβ. Approximately 20% of 5xFAD microglia had ingested Aβ (Methoxy-X04+) while almost none of the 5xFAD SykΔMG microglia contained Methoxy-X04-stained Aβ (Figures 5E and 5F). In total, these combined results provide evidence that SYK promotes the phagocytic capacity of microglia in response to Aβ.
We next explored what contributes to defective Aβ phagocytosis in 5xFAD SykΔMG mice. We turned our attention to GSK3β signaling as this was found to be profoundly affected in 5xFAD SykΔMG microglia (Figures 4G and 4H). To this end, we pre-treated wild-type (WT) and SYK-deficient bone marrow-derived macrophages (BMDMs) with the GSK3β inhibitor Tideglusib and then evaluated phagocytosis of CypHer5E-tagged Aβ oligomers. CypHer5E fluoresces in a low pH environment such as the phagolysosome; therefore, we analyzed CypHer5E fluorescence by flow cytometry as a readout for Aβ phagocytosis. In these studies, we found that GSK3β inhibition with Tideglusib treatment significantly increased Aβ phagocytosis in SYK-deficient macrophages (Figures 5G and 5H). This suggests that the dysregulated GSK3β signaling that unfolds in the absence of SYK can contribute to defective phagocytosis of Aβ by macrophages.
Exogenous activation of the CLEC7A-SYK signaling pathway promotes improved clearance of Aβ
Thus far, we have demonstrated that SYK-deficiency impairs microglial responses to Aβ pathology in 5xFAD mice. However, to reinforce the integral role for SYK in driving microglial responses in this environment, we investigated if the reciprocal activation of SYK signaling would enhance protective microglial activities in the AD brain. To achieve this, we chose to induce SYK activation through CLEC7A, a receptor shown to be important for microglial activation in response to AD pathology. CLEC7A is a canonical fungal pathogen receptor that signals through SYK in the periphery and has recently been identified to be upregulated in stage 2 DAM (Drummond et al., 2011; Keren-Shaul et al., 2017). In our studies presented here, we have identified SYK as critical for Aβ phagocytosis (Figures 5A–5H); therefore, we aimed to determine if CLEC7A-stimulated SYK signaling could enhance microglia-mediated phagocytosis of Aβ. Thus, we injected pustulan, a β-D-glucan and ligand for CLEC7A, into the hippocampus of 2-month-old 5xFAD and 5xFAD SykΔMG mice. We chose the hippocampus due to its reliable accumulation of Aβ in 5xFAD mice. As an internal control, one hemisphere of the hippocampus received a vehicle injection, while the other hippocampal hemisphere received a pustulan injection. After 7 days, we harvested the brains from the injected mice and investigated levels of Aβ between the vehicle and pustulan-injected hippocampal hemispheres using immunofluorescence (Figure 5I). Strikingly, 5xFAD mice displayed decreased Aβ load in the pustulan-injected hippocampal hemisphere compared to the vehicle-injected hippocampal hemisphere (Figures 5J and 5K), indicating that pustulan-induced microglial CLEC7A activation can boost Aβ clearance in the 5xFAD brain. In contrast, pustulan treatment in 5xFAD SykΔMG mice did not promote Aβ clearance in the hippocampus (Figure 5J and 5L), suggesting that SYK is necessary for the protective CLEC7A-driven phagocytic response by microglia. Altogether, our data suggest that CLEC7A signals through SYK to enhance the phagocytosis of Aβ.
DAM generation is regulated by SYK in demyelinating neuroinflammatory disease
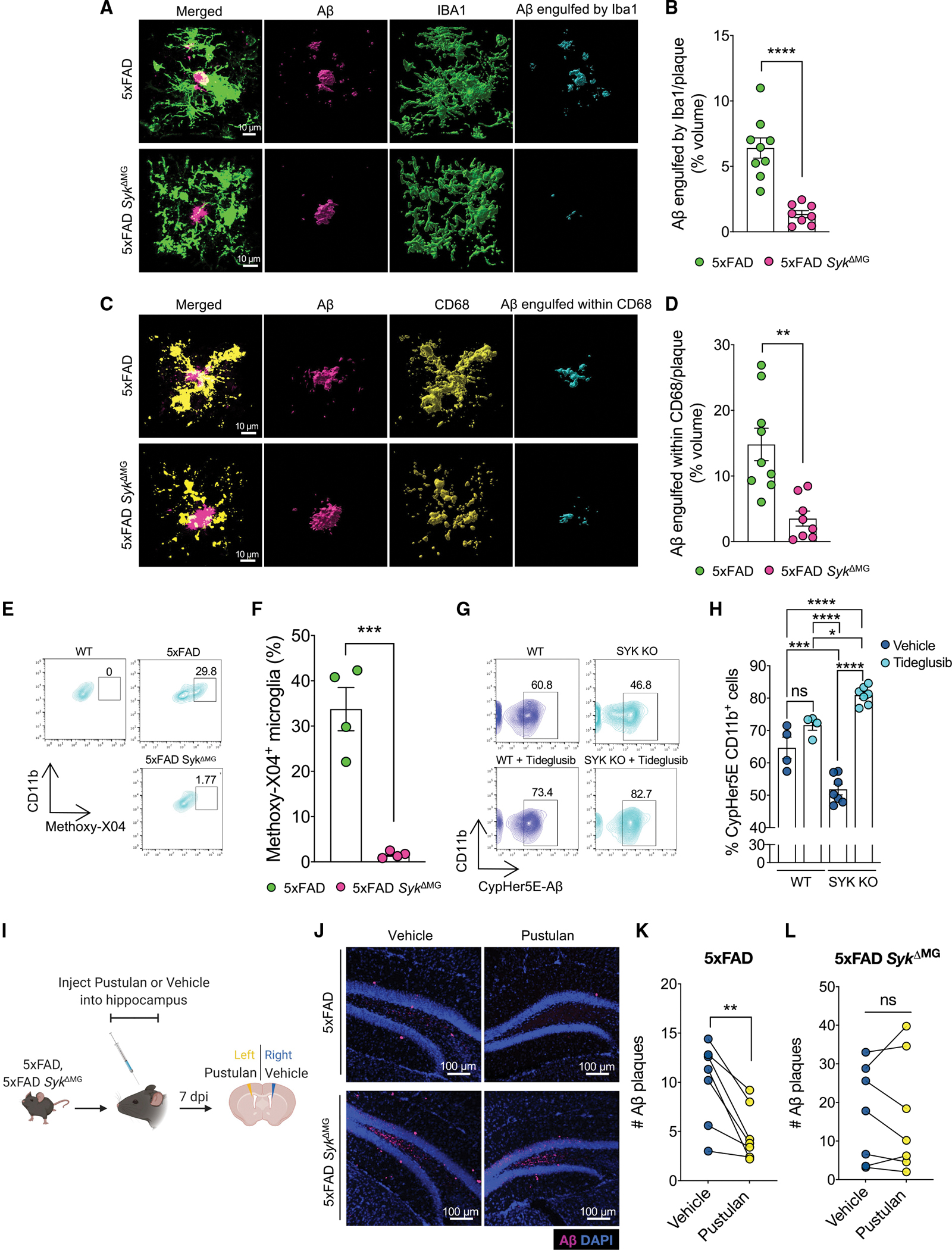
Next, we investigated whether SYK also influences DAM generation and microglial biology in other models of neurological disease. As a first approach, we explored the impact of SYK deletion in microglia on demyelinating neuroinflammatory disease progression in the experimental autoimmune encephalomyelitis (EAE) mouse model of multiple sclerosis (MS). Importantly, microglia have previously been described to play pivotal roles in EAE disease progression (Plastini et al., 2020). In particular, the clearance of myelin debris by microglia is believed to be critically required to limit neuronal damage in EAE (Cignarella et al., 2020; Takahashi et al., 2007; Weinger et al., 2011). In our studies, we found that SykΔMG mice develop exacerbated paralyzing disease and more severe demyelination in comparison to Sykcon littermate controls (Figures 6A–6C). Ablation of SYK in SykΔMG mice was also found to have an effect on T cell responses in the EAE disease model. More specifically, we observed a modest, albeit statistically significant, increase in total T cell numbers, and more T cells producing GM-CSF, IFN-γ, and IL-17 in the spinal cords of SykΔMG EAE mice relative to Sykcon EAE controls during the effector phase of EAE disease (>30 days post-immunization) (Figures S5A–S5E). Moreover, we detected less splenic CD4+ T cells making GM-CSF, IFNγ, and IL-17 in SykΔMG EAE mice compared to Sykcon EAE controls when mice were harvested during the EAE effector phase (Figures S5F–S5H). These collective findings point toward key neuroprotective roles for SYK in microglia during demyelinating neuroinflammatory disease.
Figure 6. Syk-deletion in microglia impedes the formation of DAM in EAE.
(A–C) SykΔMG mice and Sykcon littermate controls received tamoxifen food for 2 weeks starting at 3 weeks of age and then mice were returned to regular food for the remainder of the experiment. Mice were then immunized with MOG + CFA and pertussis toxin at 8–14 weeks of age to induce experimental autoimmune encephalomyelitis (EAE). Control mice did not receive EAE induction. (A) Severity of hindlimb paralysis was assessed using a 5-point clinical scoring system. (B) and (C) Representative images and quantification of spinal cords stained with Luxol fast blue (LFB).
(D–J) Syk+/+ Cx3cr1ERT2Cre and Sykfl/fl Cx3cr1ERT2Cre were crossed onto the Ai6-ZsGreen reporter background (denotated as Sykcon–Ai6 and SykΔMG–Ai6 mice) to isolate microglia in the EAE disease model. Sykcon–Ai6 and SykΔMG–Ai6 mice were pre-treated with tamoxifen and EAE was induced as described in (A)–(C). Spinal cords were harvested from mice on day 35 post-immunization and single-cell RNA-sequencing was performed on FACS-sorted ZsGreen+ microglia.
(D) Uniform Manifold Approximation and Projection (UMAP) representation of combined Sykcon–Ai6 and SykΔMG–Ai6 microglia cell populations.
(E) Dot plot representation of cluster defining genes for each cell population.
(F) UMAP representation of pseudotime cellular trajectory profiles showing microglia maturation trajectories.
(G) UMAP representation of the cell populations present in each of the clusters.
(H) Breakdown of cluster proportions.
(I) Feature plots depicting several DAM genes.
(J) Plotted KEGG and GO terms related to phagocytosis using defining genes of the DAM cluster.
Statistical significance between experimental groups was calculated by non-parametric Mann-Whitney U-test (A) and unpaired Student’s t test (C). **p < 0.01. Error bars represent mean ± SEM and each data point represents an individual mouse (C).
See also Figure S5.
Having seen that SYK ablation in SykΔMG mice leads to more severe demyelinating neuroinflammatory disease, we next wanted to better understand how SYK influences microglial responses in EAE. Given our results in the 5xFAD model, we were particularly interested in investigating whether SYK also instructs DAM generation and modulates microglial transcriptional expression of phagocytic machinery in this separate model of neurodegenerative disease. To accomplish this in a comprehensive and unbiased fashion, we conducted single-cell RNA-sequencing (scRNA-seq) on sorted spinal cord macrophages. Due to the known infiltration of peripheral myeloid cells in the EAE model (Constantinescu et al., 2011), we crossed Syk+/+ Cx3cr1ERT2Cre and Sykfl/fl Cx3cr1ERT2Cre mice with the Ai6-ZsGreen reporter mouse line to target Ai6-ZsGreen expression to Cx3cr1-expressing cells. This Ai6-ZsGreen model system has been adopted by others in the field to purify microglia in settings where peripheral-derived myeloid cells are expected to infiltrate the CNS (Batista et al., 2020; Whittaker Hawkins et al., 2017).
In our studies, we purified ZsGreen+ cells from the spinal cords of Ai6-ZsGreen Syk+/+ Cx3cr1ERT2Cre (Sykcon–Ai6) and Ai6-ZsGreen Sykfl/fl Cx3cr1ERT2Cre (SykΔMG–Ai6) mice at day 35 post-EAE induction using flow cytometry-based cell sorting (Figure S5I). Utilizing scRNA-seq, we uncovered 6 unique microglia populations, including homeostatic microglia, highly metabolic microglia, M1-like microglia, and M2-like microglia (Figure 6D). We also identified a unique population of microglia that we denoted as CD36hi microglia, as Cd36 was one of the top-defining genes of this cluster and it failed to conform with other known microglia types (Figures 6D and 6E). The final microglia cluster highly expressed canonical DAM genes, including Cst7, Lpl, Spp1, Tyrobp, and Itgax (Figures 6D and 6E).
To understand if microglia followed a trajectory in their maturation state during EAE and if this was influenced by SYK, we performed a pseudotime analysis of the microglia transcriptional data. After establishing homeostatic microglia as the earliest point in pseudotime, three potential pathways were revealed: a homeostatic to highly metabolic and M1-like microglia pathway, a homeostatic to CD36hi pathway, and a homeostatic to DAM and M2-like pathway (Figure 6F). We observed that Sykcon–Ai6 microglia tend to follow the homeostatic to DAM and M2-like pathways more than SykΔMG–Ai6 microglia (Figures 6F and 6G). In contrast, SykΔMG–Ai6 microglia, when compared to Sykcon–Ai6 microglia, more commonly followed the homeostatic to CD36hi trajectory (Figures 6F and 6G). These pathway biases are confirmed by the proportion of cells in each cluster by sample, where the Sykcon–Ai6 samples have a higher proportion of DAM microglia and the SykΔMG–Ai6 samples have a higher proportion of homeostatic and CD36hi microglia (Figure 6H).
To further examine the failure of SykΔMG–Ai6 microglia to take on a DAM transcriptional signature, we generated feature plots to visualize gene expression of DAM markers Cst7, Lpl, Itgax, and Spp1 by cluster (Figure 6I). We observed that SykΔMG microglia in the DAM cluster had much lower average expression of these DAM genes when compared with microglia obtained from Sykcon–Ai6 mice (Figure 6I). Finally, to better understand the biological processes potentially being driven by the DAM cluster, we used DAM-defining genes to plot KEGG and GO terms related to phagocytosis. These shared terms included “phagosome,” “abnormal phagocyte morphology,” and “microglia pathogen phagocytosis pathway” (Figure 6J). In summary, our collective EAE findings corroborate our 5xFAD data characterizing SYK as a pivotal intracellular regulator of DAM generation and promoter of neuroprotective functions in microglia during neurodegenerative disease.
Defective SYK signaling in microglia during demyelinating disease causes damaged myelin debris accumulation and impaired oligodendrocyte proliferation
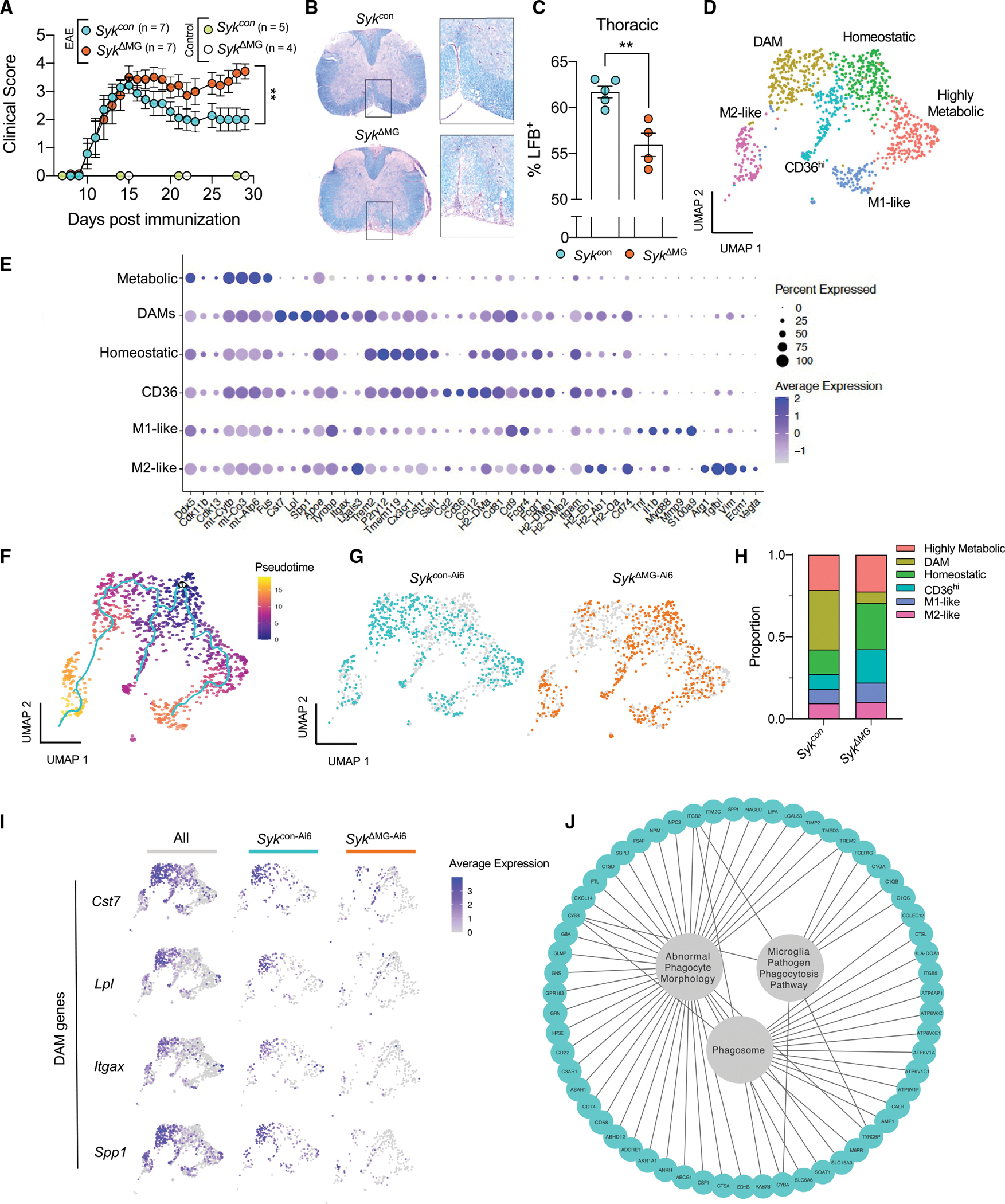
To further validate the ability of SYK to modulate microglial responses in a separate model of demyelinating disease that does not involve autoimmune attack, we explored the effects of cuprizone intoxication on neurological disease in SykΔMG mice and Sykcon littermate controls. Cuprizone is toxic to myelinating oligodendrocytes, including those found in the corpus callosum, thus 5 continuous weeks of feeding cuprizone to mice leads to localized areas of demyelination (Zhan et al., 2020). Importantly, in the absence of cuprizone, we did not observe any appreciable differences in myelin basic protein (MBP) levels between SykΔMG and Sykcon mice, indicating that baseline corpus callosum myelination at steady-state is not affected in SYK-deficient mice (Figures S6A and S6B). However, we noticed that the corpus callosum of SykΔMG mice had significantly fewer microglia than Sykcon controls during both cuprizone-induced demyelination and remyelination (Figures S6C and S6D). We determined that the decreased number of microglia in SykΔMG mice is likely not due to differential apoptosis, as the levels of TUNEL staining in Iba1+ cells was found to be similar between the experimental groups(Figures S6E and S6F). Consistent with previous studies, we found that feeding Sykcon mice a cuprizone diet leads to increased staining of the phagocytic marker CD68 in Iba1+ cells (Figures S6G and S6H) (Cignarella et al., 2020). In contrast, CD68 positivity was substantially decreased in Iba1+ cells found in the corpus callosum of cuprizone-fed SykΔMG mice during the demyelinating phase (Figures S6G and S6H), suggesting impaired phagocytic microglial response compared to Sykcon mice. Indeed, increased damaged myelin basic protein (dMBP) accumulation was evident during both demyelination and remyelination in the SykΔMG corpus callosum compared with Sykcon mice (Figures 7A and 7B). Microglia are established to phagocytose damaged myelin in the cuprizone model of demyelination; therefore, this accumulation is likely due to a microglial phagocytic deficit (Gudi et al., 2014). These results provide evidence that SYK signaling in microglia is critically involved in the clearance of myelin debris independent of the robust autoimmune response associated with EAE.
Figure 7. Disruption of SYK signaling in microglia during demyelinating disease leads to accumulation of damaged myelin debris and impaired oligodendrogenesis.
SykΔMG mice and Sykcon littermate controls received tamoxifen food for 2 weeks starting at 3 weeks of age and then mice were returned to regular food. Adult (8–12 month old) mice were later fed a diet consisting of 0.3% cuprizone for 5 weeks to induce demyelination. Mice were then either harvested after 5 weeks of cuprizone treatment (demyelination group) or returned to normal chow for one additional week before being harvested (remyelination group). Control mice were not introduced to the cuprizone diet.
(A) Representative images of microglia labeled with Iba1 (green) and damaged myelin basic protein (dMBP; pink) staining in the corpus callosum (CC).
(B) Quantification of dMBP volume in the CC.
(C) Representative images of oligodendrocyte lineage markers in the CC.
(D and E) Quantification of the number of Pdgfra+ Olig2+ oligodendrocyte precursor cells (D) and number of CC1+ Olig2+ mature oligodendrocytes (E) in the CC.
Statistical significance between experimental groups was calculated by one-way ANOVA with multiple comparisons (B), (D), and (E). ns = not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Data are mean ± SEM and combined from two independent experiments and each data point represents an individual mouse.
See also Figure S6.
The inability to phagocytose myelin debris in the cuprizone model is known to obstruct aspects of oligodendrocyte biology, including the differentiation of oligodendrocyte precursor cells (OPCs) into myelin-producing oligodendrocytes (Back et al., 2005). Therefore, we hypothesized that any phagocytic deficits seen in SykΔMG mice in the cuprizone model would subsequently manifest as impaired OPC proliferation and/or differentiation into mature oligodendrocytes during the remyelination phase that follows cuprizone cessation. Consistent with this hypothesis, we found that SYK deficiency in cuprizone-treated SykΔMG mice leads to greatly reduced numbers of OPCs (Olig2+Pdgfrα+ cells) and oligodendrocytes (Olig2+CC1+cells) during the remyelination phase of the cuprizone model (Figures 7C–7E). We also noted that OPCs in cuprizone-treated SykΔMG mice had severely impaired proliferation during demyelination (Figures S6I and S6J), which likely accounts for the decreased numbers of OPCs and oligodendrocytes seen in cuprizone-treated SykΔMG mice. In comparison, we observed comparable numbers of OPCs and oligodendrocytes in SykΔMG mice and Sykcon littermate controls that were fed normal chow (Figures 7C–7E), suggesting that SYK deficiency in microglia does not appreciably affect oligodendrocyte-lineage cell numbers under homeostatic conditions. Our findings indicate that disruption of SYK signaling in microglia causes prominent defects in the clearance of damaged myelin in the cuprizone model of demyelinating disease. Moreover, they suggest that the lack of neuroprotective functions in SYK-deficient microglia can ultimately lead to impaired oligodendrocyte generation during remyelination. Altogether, these cuprizone data support our 5xFAD and EAE findings that define a critical role for SYK in promoting protective microglial responses that limit neurodegenerative disease progression.
DISCUSSION
In the studies presented here, we have identified SYK as a pivotal intracellular regulator of neuroprotective microglial responses in mouse models of both AD and MS. SYK is perhaps best known for its essential roles in the generation of protective immunity against many fungal infections as well as in the regulation of T cell and B cell receptor signaling (Cornall et al., 2000; Latour et al., 1997; Malik et al., 2018). However, in recent years there has been growing appreciation for the critical involvement of SYK in models of sterile inflammation (Chung et al., 2019). For instance, the SYK homolog in Drosophila, known as Shark, was shown to be critical for glial phagocytosis of axonal debris (Ziegenfuss et al., 2008). In addition, there have been a handful of recent studies that have used pharmacological inhibitors and in vitro cell culture systems to explore how SYK affects CNS biology (Paris et al., 2014). Beyond these initial studies, we lack more integrated knowledge concerning how SYK signaling affects neurological health and disease.
The role of SYK in demyelinating neuroinflammatory disease also currently remains poorly described. This is surprising given that mutations in various SYK-related molecules have been identified as prominent MS genetic risk factors (Ramagopalan et al., 2010). For instance, mutations in multiple upstream activators of SYK, including CD37, TREM2, numerous CLEC receptors (e.g., CLEC16A, CLECL1, and CLEC2D), and Fc receptor-like proteins (i.e., FCRL2 and FCRL3) have been strongly linked to MS in GWAS studies (IMSG, 2019; IMSG, 2011). Likewise, mutations in downstream molecules involved in SYK signaling, including BCL10 and MALT1, have also been associated with MS risk (Mc Guire et al., 2013; Molinero et al., 2012).
In summary, although pivotal roles for microglia have recently been uncovered in AD, MS, and many other neurodegenerative disorders (Krasemann et al., 2017), the key signaling pathways that microglia leverage to instruct neuroprotective functions remain poorly defined. Through the studies presented here, we have identified SYK as an instrumental regulator of neuroprotective microglial responses in mouse models of AD and MS. Moreover, our studies suggest that targeting SYK may offer novel strategies to boost microglial protective responses, including phagocytosis of neurotoxic material, to treat neurodegenerative disease.
Limitations of the study
Future studies are needed to fully define all of the key microglial receptors that rely on SYK to influence neurodegenerative disease progression. Based on receptor structure alone, there are a wide range of microglial receptors that could potentially leverage SYK to coordinate their effects on neuropathogenesis. Most notably, and of relevance to neurodegenerative disease research, this list includes TREM2, CLEC7A, CD33, CD22, FcgR, and complement receptor 3 (CR3) (Clark and Giltiay, 2018; Hadas et al., 2012; Pluvinage et al., 2019; Wissfeld et al., 2021; Yao et al., 2019; Ye et al., 2020). However, various other CLEC receptors, SIGLEC receptors, and integrin receptors could also potentially leverage SYK signaling to influence neurological disease (Mocsai et al., 2010).
Though previous in vitro studies have shown that TREM2 activation can provoke SYK signaling (Yao et al., 2019), it still remains to be seen whether TREM2 relies exclusively on SYK to coordinate its effects on in vivo Alzheimer’s-related disease progression. Therefore, additional in vivo studies in relevant disease models are needed to address this. Likewise, even though our studies provide promising early evidence that promoting SYK activation via CLEC7A engagement can boost the clearance of Aβ, future studies are needed to better characterize the role of CLEC7A in AD-related disease. In particular, it still remains to be seen why CLEC7A is so highly expressed by microglia in response to neurodegenerative disease pathology in mice (Deczkowska et al., 2018; Keren-Shaul et al., 2017; Krasemann et al., 2017). Therefore, although our early studies suggest the SYK is a major regulator of neuroprotective microglial responses in models of AD and MS, future studies are needed to better characterize both the upstream and downstream players that coordinate the effects of SYK on neurodegenerative disease progression.
It is also important to note that replacing one allele of Cx3cr1 with Cre-recombinase in Cx3cr1ERT2cre mice may potentially affect aspects of microglial biology. However, in a recent study it was shown that targeting microglia with this strategy actually leads to improved clearance of Aβ and ameliorated disease progression in the APP/PS1 mouse model of AD (Hickman et al., 2019). Therefore, our observation of worsened disease status in 5xFAD SykΔMG is likely not explained by this variable. In addition, future studies are also needed to ascertain whether SYK deletion in long-lived BAMs contributes at any level to the phenotypes seen in our SykΔMG studies.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, John R. Lukens (Jrl7n@virginia.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Bulk and Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication (accession number: GEO: GSE212310) and can be found in the key resources table. All original code has been deposited at Zenodo and is publicly available as of the date of publication (Zenodo: https://doi.org/10.5281/zenodo.7026051) and is listed in the key resources table. Any additional information required to reanalyze the data reported in this paper will be shared by the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| SYK (D3Z1E) XP Rabbit Antibody | Cell Signaling Technology | Cat# 13198 RRID: AB_2687924 |
| StarBright Blue 700 Goat Anti-Rabbit IgG | Bio-Rad | Cat# 12004161 RRID: AB_2721073 |
| β-actin (D6A8) Rabbit mAb (HRP Conjugate) | Cell Signaling Technology | Cat# 12620 RRID: AB_2797972 |
| β-Amyloid (D54D2) XP Rabbit mAb | Cell Signaling Technology | Cat# 8243 RRID: AB_2797642 |
| Purified (azide-free) anti-beta-Amyloid, 1-16 | BioLegend | Cat# 803004 RRID: AB_2715854 |
| Iba1 antibody | Abcam | Cat# ab5076 RRID: AB_2224402 |
| Ki-67 Monoclonal Antibody (SolA15), Alexa Fluor 600, eBioscience | Thermo Fisher Scientific | Cat# 606-5698-80 RRID: AB_2896285 |
| Anti-mDectin-1-IgG | InvivoGen | Cat# mabg-mdect RRID: AB_2753143 |
| Anti-TMEM119 antibody [29–3] – Microglial marker | Abcam | Cat# ab209064 RRID: AB_2800343 |
| Rat Anti-Mouse CD68 Monoclonal antibody, Unconjugated, Clone FA-11 | Bio-Rad | Cat# MCA1957 RRID: AB_322219 |
| Amyloid beta precursor protein antibody [Y188] | Abcam | Cat# ab32136 RRID: AB_2289606 |
| Phospho-Tau (Ser202, Thr205) Monoclonal Antibody (AT8) | Thermo Fisher Scientific | Cat# MN1020 RRID: AB_223647 |
| Anti-NeuN | Millipore | Cat# MAB377 RRID: AB_2298772 |
| Mouse PDGF R alpha Antibody | R and D Systems | Cat# AF1062 RRID: AB_2236897 |
| Anti-APC (Ab-7) Mouse mAb (CC-1) | Millipore | Cat# OP80 RRID: AB_2057371 |
| Anti-Olig-2 Antibody | Millipore | Cat# AB9610 RRID: AB_570666 |
| Anti-Myelin Basic Protein | Millipore | Cat# AB5864 RRID: AB_2140351 |
| Rat Anti-Myelin Basic Protein Monoclonal Antibody, Unconjugated, Clone 12 | Abcam | Cat# ab7349 RRID: AB_305869 |
| Donkey anti-Rabbit IgG (H + L) Antibody, Alexa Fluor 488 Conjugated | Thermo Fisher Scientific | Cat# A-21206 RRID: AB_2535792 |
| Donkey anti-Rabbit IgG (H + L) Antibody, Alexa Fluor 594 Conjugated | Thermo Fisher Scientific | Cat# A-21207 RRID: AB_141637 |
| Donkey anti-Rabbit IgG (H + L) Antibody, Alexa Fluor 647 Conjugated | Thermo Fisher Scientific | Cat# A-31573 RRID: AB_2536183 |
| Donkey anti-mouse IgG (H + L) Antibody, Alexa Fluor 488 Conjugated | Thermo Fisher Scientific | Cat# A21202 RRID: AB_141607 |
| Donkey anti-mouse IgG (H + L) Antibody, Alexa Fluor 647 Conjugated | Thermo Fisher Scientific | Cat# A-31571 RRID: AB_162542 |
| Donkey anti-goat IgG (H + L) Antibody, Alexa Fluor 488 Conjugated | Thermo Fisher Scientific | Cat# A-11055 RRID: AB_2534102 |
| Donkey anti-goat IgG (H + L) Antibody, Alexa Fluor 546 Conjugated | Thermo Fisher Scientific | Cat# A-11056 RRID: AB_142628 |
| Donkey anti-goat IgG (H + L) Antibody, Alexa Fluor 647 Conjugated | Thermo Fisher Scientific | Cat# A-21447 RRID: AB_141844 |
| Donkey anti-rat IgG (H + L) Antibody, Alexa Fluor 594 Conjugated | Thermo Fisher Scientific | Cat# A-21209 RRID: AB_2535795 |
| Alexa Fluor 647-AffiniPure Donkey Anti-Rat IgG (H + L) | Jackson ImmunoResearch Labs | Cat# 712-605-153 RRID: AB_2340694 |
| CD11b Monoclonal Antibody (M1/70), APC, eBioscience | Thermo Fisher Scientific | Cat# 17-0112-82 RRID: AB_469343 |
| CD45 Monoclonal Antibody (30-F11), PE-Cyanine7, eBioscience | Thermo Fisher Scientific | Cat# 25-0451082 RRID: AB_2734986 |
| Brilliant Violet 510 anti-mouse TCR beta chain | BioLegened | Cat# 109234 RRID: AB_2562350 |
| F4/80 Monoclonal Antibody (BM8), APC, eBioscience | Thermo Fisher Scientific | Cat# 17-4801082 RRID: AB_2784648 |
| Ly-6G Monoclonal Antibody (1A8-Ly6g), FITC, eBioscience | Thermo Fisher Scientific | Cat# 11-9668-80 RRID: AB_2572531 |
| CD4 Monoclonal Antibody (RM4-5), FITC, eBioscience | Thermo Fisher Scientific | Cat# 11-0042-82 RRID: AB_464896 |
| CD8a Monoclonal Antibody (53-6.7), Alexa Fluor 700, eBioscience | Thermo Fisher Scientific | Cat# 56-0081-82 RRID: AB_494005 |
| CD80 (B7-1) Monoclonal Antibody (16-10A1), APC, eBioscience | Thermo Fisher Scientific | Cat# 17-0801-82 RRID: AB_469417 |
| CD11c Monoclonal Antibody (N418), PE, eBioscience | Thermo Fisher Scientific | Cat# 12-0114-83 RRID: AB_465553 |
| GM-CSF Monoclonal Antibody (MP1-22 × 109), PE, eBioscience | Thermo Fisher Scientific | Cat# 12-7331-82 RRID: AB_466205 |
| IFN gamma Monoclonal Antibody (XMG1.2), APC, eBioscience | Thermo Fisher Scientific | Cat# 17-7311-82 RRID: AB_469504 |
| IL-17A Monoclonal Antibody (eBio17B7), eFluor 450, eBioscience | Thermo Fisher Scientific | Cat# 48-7177-80; RRID: 11149677 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| Mycobacterium tuberculosis | Becton, Dickinson, & Company | Cat# 231141 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| MOG Peptide | Bio-Synthesis | Cat# 12668-01 |
| Complete Freund’s Adjuvant | Sigma-Aldrich | Cat# F5881 |
| Pertussis toxin | List Biological Laboratories | Cat# 180 |
| Cuprizone | Sigma-Aldrich | Cat# 14690 |
| cOmplete Protease Inhibitor Cocktail | Roche | Cat# 11697498001 |
| PhosSTOP | Roche | Cat# 4906845001 |
| Ponceau S stain | Sigma-Aldrich | Cat# P7170 |
| Tissue-Plus O.C.T. Compound | Fisher Scientific | Cat# 23-730-571 |
| T-PER Tissue Protein Extraction Reagent | Thermo Fisher Scientific | Cat# 78510 |
| Sucrose | Sigma-Aldrich | Cat# S0389 |
| Triton X-100 | Sigma-Aldrich | Cat# 93418 |
| DAPI | Sigma-Aldrich | Cat# D9542 |
| ThioflavinS | AAT Bioquest | Cat# 23059 |
| ProLongGold Antifade Mountant | Invitrogen | Cat# P36930 |
| Methoxy-X04 | ApexBio | Cat# B5769 |
| Dimethyl Sulfoxide Anhydrous (DMSO) | Sigma-Aldrich | Cat# 276855 |
| Hanks Buffer Saline Solution (HBSS) | Thermo Fisher Scientific | Cat# 14025092 |
| DNase I, Grade II, from bovine pancreas | Sigma-Aldrich | Cat# 10104159001 |
| Papain, Suspension | Worthington Biochemical Corporation | Cat# LS003124 |
| DMEM/F-12, no glutamine | Thermo Fisher Scientific | Cat# 21331020 |
| Fetal Bovine Serum (FBS) | Thermo Fisher Scientific | Cat# 10082147 |
| Antibiotic-Antimycotic | Thermo Fisher Scientific | Cat# 15240096 |
| Glutamax | Thermo Fisher Scientific | Cat# 35050061 |
| Percoll | Cytvia | Cat# 17-0891-02 |
| MACS BSA Stock Solution | Miltenyi Biotec | Cat# 130-0910376 |
| Beta Amyloid (1-42), human | California peptide | Cat# 641-15 |
| Hexafluoroisopropanol (HFIP) | Sigma-Aldrich | Cat# 52517 |
| CypHer5E-NHS ester | Cytvia | Cat# PA15401 |
| IMDM | Thermo Fisher Scientific | Cat# 12440053 |
| Penicillin/Streptomycin | Thermo Fisher Scientific | Cat# 15140163 |
| Recombinant Murine M-CSF | PeproT ech | Cat# 315-02 |
| Tideglusib | Selleck Chemicals | Cat# S2823 |
| DMEM | Thermo Fisher Scientific | Cat# 11885-084 |
| CD11b microbeads (microglia) | Miltenyi Biotec | Cat# 130-093-634 |
| CD90.2 microbeads | Miltenyi Biotec | Cat# 130-121-278 |
| CD11b beads (monocytes) | Miltenyi Biotec | Cat# 130-049-601 |
| Fixable Viability Dye | eBioscience | Cat# 65-0866-14 |
| BODIPY 581/591 C11 (Lipid Peroxidation Sensor) | Fisher Scientific | Cat# D3861 |
| Fc Block | eBiosciene | Cat# 14-0161-86 |
| L-glutamine | Thermo Fisher Scientific | Cat# 25030-081 |
| Beta-mercaptoethanol | Thermo Fisher Scientific | Cat# 21985-023 |
| PMA | Sigma-Aldrich | Cat# P1585 |
| Ionomycin | Sigma-Aldrich | Cat# I19657 |
| Monensin | eBioscience | Cat# 00-4505-51 |
| IC Fixation Buffer | eBioscience | Cat# 00-8222-49 |
| Permeabilization Buffer | eBioscience | Cat# 00-8333-56 |
| 10% Formalin | Azer Scientific | Cat# NBF |
| Luxol Fast Blue | Thermo Fisher Scientific | Cat# 212170250 |
| Lithium Carbonate | Thermo Fisher Scientific | Cat# 446322500 |
| Hematoxylin | Sigma-Aldrich | Cat# HHS128 |
| ACK Lysis Buffer | Quality Biological | Cat# 118-156-101 |
| TRIzol | Life Technologies | Cat# 15596018 |
| Chloroform | Fisher Scientific | Cat# BP1145-1 |
| Isopropanol | Sigma-Aldrich | Cat# I9516 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Pierce 660nm Protein Assay Kit | Thermo Fisher Scientific | Cat# 22660 |
| Human/Mouse AKT Pathway Phosphorylation Array C1 | RayBiotech | Cat# AAH-AKT-1-8 |
| In Situ Cell Death Detection Kit, Fluorescein | Roche | Cat# 11684795910 |
| Amyloid beta 40 Mouse ELISA Kit | Thermo Fisher Scientific | Cat# KMB3481 |
| Amyloid beta 42 Mouse ELISA Kit | Thermo Fisher Scientific | Cat# KMB3441 |
| CellROX Deep Red Flow Cytometry Assay Kit | Thermo Fisher Scientific | Cat# C10491 |
| Bio-Rad Bio-Plex Pro Reagent Kit | Bio-Rad | Cat # 171-304070M |
| RNeasy Micro Kit | Qiagen | Cat# 74004 |
| Sensifast cDNA Synthesis Kit | Bioline | Cat# BIO-65054 |
| Sensifast Probe No-ROX Kit | Bioline | Cat# BIO-86005 |
|
| ||
| Deposited data | ||
|
| ||
| Gene expression data for replication analysis | This paper | GEO: GSE212310 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE212310 |
| Original code | This paper | Zenodo:https://zenodo.org/record/7026051#.YwkJKi-B3_Q |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: 5xFAD; B6SJL-Tg(APPSwFlLon, PSEN 1*M146L*L286V)6799Vas/Mmjax | The Jackson Laboratory | Jax Stock no. 034840-JAX |
| Mouse: B6.129P2-Syktm1.2Tara/J | The Jackson Laboratory | Jax Stock no. 017309-JAX |
| Mouse: B6.129P2(C)-Cx3cr1tm2.1(cre/ERT2)Jung/J | The Jackson Laboratory | Jax stock no. 020940-JAX |
| Mouse: B6.Cg-Gt(ROSA)26Sortm6(CAG-Zsgreen1)Hze/J | The Jackson Laboratory | Jax stock no. 007906-JAX |
|
| ||
| Oligonucleotides | ||
|
| ||
| qPCR primer: Sykb | Thermo Fisher Scientific | Mm01333035_m1 |
| qPCR primer: Gapdh | Thermo Fisher Scientific | Mm99999915_g1 |
|
| ||
| Software and algorithms | ||
|
| ||
| GraphPad Prism 9 | GraphPad | RRID: SCR_002798 |
| Imaris 9.5.1 | Imaris | RRID: SCR_007370 |
| Adobe Photoshop | Adobe | https://www.adobe.com/products/photoshop/ |
| BioRender | BioRender | RRID: SCR_018361 |
| LAS AF Software | Leica | https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/ |
| Fiji/ImageJ Software | Fiji | RRID: SCR_002285 |
| Noldus Ethovision XT Software | Noldus | https://www.noldus.com/ethovision-xt |
| Kaluza Acquisition Software | Beckman Coulter | https://www.beckman.com/flow-cytometry/software/kaluza |
| FlowJo Software | FloJo | RRID:SCR_008520 |
| Bio-Plex Manager software | Bio-Rad | https://www.bio-rad.com/en-us/product/bio-plex-200-systems |
| R Studio (V4.0.5) | https://rstudio.com/ | RRID:SCR_001905 |
| R | https://www.r-project.org | RRID:SCR_001905 |
| DESeq2 (v1.32.0) | N/A | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Cell Ranger (V1.3.1) | N/A | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/installation |
| Seurat (v4.0.2) | https://www.r-project.org/ | RRID:SCR_016341 |
| ToppCluster | Cincinnati Children's | https://toppcluster.cchmc.org |
| Monocle (v0.2.3.0) | N/A | N/A |
|
| ||
| Other | ||
|
| ||
| Leica TCS SP8 Confocal Microscope | Leica Microsystems | N/A |
| Mini-PROTEAN TGX Stain-Free Protein Gel | Bio-Rad | Cat# 4568093 |
| Mini-PROTEAN Tetra Cell | Bio-Rad | Cat# 1620264 |
| Trans-Blot Turbo Transfer System | Bio-Rad | Cat# 1704150 |
| Bio-Spin P-6 gel Columns, Tris Buffer | Bio-Rad | Cat# 7326227 |
| Mouse stereotaxic Frame | Stoelting | Cat# 51730U |
| Nanoliter Injector | World Precision Instruments | Cat# NL2010MC2T |
| LS columns | Miltenyi Biotec | Cat# 130-042-401 |
| QuadroMACS magnet | Miltenyi Biotec | Cat# 130-091-051 |
| Gallios Flow Cytometer - Navios System | Beckman Coulter | Cat# B83535 |
| Bio-Plex 200 System | Bio-Rad | https://www.bio-rad.com/en-us/product/bio-plex-200-systems |
| Keyence BZ-X810 Microscope | Keyence | https://www.keyence.com/landing/lpc/all-in-one-fluorescence-microscope.jsp |
| Cellometer Auto 2000 | Nexelcom Bioscience | https://www.nexcelom.com/nexcelom-products/cellometer-fluorescent-viability-cell-counters/cellometer-auto-2000/ |
| NanoDrop 2000 Spectrophotometer | Thermo Fisher Scientific | https://www.thermofisher.com/order/catalog/product/ND-2000 |
| CFX384 Real-Time PCR System | Bio-Rad | Cat# 1855484 |
| Influx Cell Sorter | BD | https://med.virginia.edu/flow-cytometry-facility/equipment/influx-cell-sorter/ |
| GENEWIZ Next Generation Sequencing | Azenta | https://www.genewiz.com/en/Public/Services/Sanger-Sequencing |
| ChemiDoc MP Imaging System | Bio-Rad | Cat# 12003154 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All mouse experiments were performed in accordance with the relevant guidelines and regulations of the University of Virginia and approved by the University of Virginia Animal Care and Use Committee. 5xFAD mice (Stock #34848-JAX), Sykfl/fl mice (Stock #017309) and Cx3cr1ERT2cre mice (Stock #020940) were obtained from The Jackson Laboratory and were crossed to generate Sykfl/fl (denoted as Sykcon), Sykfl/fl Cx3cr1ERT2cre (denoted as SykΔMG), 5xFAD Sykfl/fl (denoted as 5xFAD), and 5xFAD Sykfl/fl Cx3cr1ERT2cre (denoted as 5xFAD SykΔMG) experimental mice. Upon weaning, female Sykcon and SykΔMG, 5xFAD, and 5xFAD SykΔMG littermates were fed tamoxifen diet (Envigo Teklad #TD.130858) ad libitum for two weeks and then returned to normal chow. Ai6-ZsGreen (Stock #007906-JAX) reporter mice were generously provided by Dr. Tajie Harris and crossed with Syk+/+ Cx3cr1ERT2Cre and Sykfl/fl Cx3cr1ERT2Cre mice. In EAE experiments, Sykcon, SykΔMG, Sykcon–Ai6, and SykΔMG–Ai6 mice were fed tamoxifen diet for two weeks upon weaning and then returned to normal chow. In cuprizone experiments, Sykcon and SykΔMG were fed tamoxifen diet for two weeks upon weaning and then returned to normal chow. Mice were housed for AD and demyelinating experiments in specific pathogen-free conditions under standard 12-h light/dark cycle conditions in rooms equipped with control for temperature (21 ± 1.5°C) and humidity (50 ± 10%).
Experimental autoimmune encephalomyelitis (EAE) induction and scoring
On the day of immunization (Day 0), mice were injected subcutaneously over each shoulder with 100 μL of an emulsion containing 0.5 mg/mL MOG35–55 peptide (Bio-Synthesis, 12–668–01) and 1.25 mg/mL heat-killed Mycobacterium tuberculosis (Becton, Dickinson, & Company, 231–141) in Complete Freund’s Adjuvant (Sigma Aldrich, F5881). Mice were intraperitoneally injected with 200 ng of Pertussis toxin (List Biological Laboratories, 180) on days 0 and 2 post-immunization.
Beginning at approximately day 7 post-immunization, mice were monitored daily for onset of hindlimb paralysis and scored for EAE severity using the following 5-point clinical scoring system: 0 = normal tail; 0.5 = limp at tip of tail; 1 = completely limp tail; 1.5 = partial hindlimb weakness/mouse can be flipped onto its back; 2 = complete hindlimb weakness with abnormal gait; 3 = partial hindlimb paralysis; 3.5 = complete hindlimb paralysis in both legs; 4 = hind- and fore-limb paralysis; 5 = moribund/dead.
Cuprizone model
For the cuprizone model, adult mice were fed regular chow mixed with 0.3% cuprizone (Sigma, 14,690) ad libitum for 5 weeks to induce demyelination.
METHOD DETAILS
Western blotting
MACS-sorted microglia, monocytes, and T cells were resuspended in Western blot lysis buffer [dH2O, RIPA, cOmplete Protease Inhibitor Cocktail (Roche), and PhosSTOP (Roche)]. Protein concentration was measured using Pierce 660 nm Protein Assay Reagent (Thermo Scientific, 22–660). 4X SDS loading dye was added to protein lysates and incubated at 95°C for 3 min. For each sample, 25 ug of protein was loaded per lane of a 4–20% Mini-PROTEAN TGX Stain-Free Protein Gel (BioRad, 4–568–093) and run at 120 V for 1.5 h using Mini-PROTEAN Tetra Cell (BioRad, 1–658–004). Proteins were transferred to an Immun-Blot Low Fluorescence PVDF membrane (BioRad, 1–620–264) using Trans-Blot Turbo transfer system (BioRad, 1–704–150) set to 2 mini gels and mixed MW for 21 min. SYK protein was probed using anti-SYK (D3Z1E) XP Rabbit mAb (Cell Signaling Technologies, 13,198, 1:1000 overnight at 4°C) and goat anti-rabbit IgG StarBright Blue 700 secondary antibody (BioRad, 12–004–161, 1:1000 for 2 h at room temperature). The stain-free gel and blotted membrane were imaged using ChemiDoc MP Imaging System (BioRad, 12–003–154). Total protein loaded was quantified using Image lab touch software (BioRad), Beta-Actin (Cell Signaling, 12–620), Ponceau S stain (Sigma-Aldrich, P7170) and SYK protein were quantified using Fiji. Samples underwent the same preparation for the AKT pathway phosphorylation kit (RayBiotech, AAH-AKT-1–8). The AKT assay was performed in accordance to manual instructions and pixel density was analyzed using Adobe Photoshop.
Brain sample preparation
Mice were euthanized using CO2 asphyxiation and transcardially perfused with 20 mL of 1xPBS. For AD experiments, brains were dissected out with the left hemisphere drop-fixed in 4% paraformaldehyde over night at 4°C and the right hemisphere flash-frozen and stored at −80°C. Drop-fixed samples were transferred to 30% sucrose for 48 h and then mounted and frozen in Tissue-Plus OCT compound (Fisher Scientific). These brains were then sectioned at 50 μm in thickness using a cryostat (Leica) and stored in PBS +0.05% sodium azide at 4°C for downstream staining and imaging. The flash-frozen brains were thawed for RNA and protein extraction and mechanically homogenized in 500 μL of Tissue Protein Extraction Reagent T-PER (Thermo Fisher, 78–510) containing phosphatase inhibitor cocktail PhosSTOP (Roche, 04–906–845–001) and protease inhibitor cocktail cOmplete (Roche, 11–873–580–001). Following homogenization, 50 μL of the brain sample was diluted in 500 μL Trizol for future RNA extraction and stored at −80°C. The stock brain samples were then spun down at 16,000 rpm for 10 min and the supernatant and pellet were isolated for soluble and insoluble amyloid beta analyses, respectively. For EAE experiments, brain tissue was dissected and immersion fixed in 4% paraformaldehyde for 48 h, followed by dehydration in 30% sucrose and freezing in OCT. Free-floating cryosections were cut (40 μm) and collected in PBS containing 0.02% sodium azide and stored at 4°C until further analysis.
ELISA
Brain samples underwent guanidine-extraction in which pelleted brain samples were incubated 1:6 in 5 M guanidine HCL/50 mM tris, pH = 8.0 at room temperature for 3 h, then diluted 1:5 in PBS containing protease inhibitor cocktail cOmplete (Roche, 11–873–580–001), centrifuged at 16,000 g for 20 min at 4°C, and the supernatant was collected and stored at −80°C pending ELISA. Triton X-extraction was performed by diluting the pelleted brain samples 1:5 in 1% Triton X-100 in T-PER buffer and sonicating the samples for 30 min at room temperature, then spun down at 16,000 g for 20 min at 4°C and the supernatant stored at −80°C until used for Aβ measurement by ELISA. Amyloid beta 40 or 42 Mouse ELISA kits were utilized (Thermo Fisher, KMB3481, KMB3441) on samples obtained with the soluble fraction (T-PER extracted supernatant) diluted 1:10, Triton X- fraction diluted 1:40, and the guanidine fraction diluted 1:200 following manufacturer’s instructions.
Immunofluorescence microscopy
Floating brains sectioned stored in PBS +0.05% sodium azide were blocked with 2% donkey serum, 1% BSA, 0.1% Triton-X, 0.05% Tween20 in PBS for 1 h at room temperature before applying the primary antibody master mix diluted in this block overnight at 4°C. Samples were stained with anti-Aβ (D54D2, Cell Signaling, 1:300; or 6e10, BioLegend, 1:1000) to label plaques. To study microglial morphology and numbers, sections were stained with Iba1 (ab5076, Abcam, 1:300). To further characterize microglia, we labeled with Ki67-EF660 (SoIA15, Thermo Fisher, 1:100), anti-Clec7a (R1–8g7, Invivogen, 1:30), Tmem119 (ab209064, Abcam, 1:300), and anti-CD68 (FA-11, BioRad, 1:1000). Neuronal health was probed by staining for anti-APP (Y188, ab32136, Abcam, 1:750), anti-phospho-tau (AT8, Thermo Fisher, 1:500), and anti-NeuN (MAB377, Millipore Sigma, 1:500). For cuprizone experiments, sections were stained with PDGFRα (AF1062, R&D Systems, 1:200), CC1 (OP80, 1:200, Millipore), Olig2 (AB9610, 1:500, Millipore), dMBP (AB5864, Millipore Sigma, 1:2000), and MBP (ab7349, Abcam, 1:1000). Sections were then washed 3 times for 10 min at room temperature in PBS and 0.05% tween 20, then incubated in matched donkey Alexa Fluor 488, 594, 647 anti-rabbit, -goat, -rat, -streptavidin, and -mouse (Thermo Fisher, 1:1000 dilution) at room temperature for 2 h. Samples were washed again 3 times for 10 min at room temperature and incubated with DAPI (1:1000) for 10 min at room temperature, or stained for plaques with ThioflavinS (Sigma-Aldrich, 2 mg/10mL) for 8 min followed by three 2-min washes with 50% ethanol at room temperature. To investigate cell death, sections were stained by TUNEL (Millipore Sigma, 11–684–795–910) following the manufacturer’s protocol. All tissue sections were then transferred to wells containing PBS before being mounted to glass slides with ProLongGold antifade reagent (Invitrogen, P36930) and coverslips. Mounted slides were stored at 4°C until imaged using LAS AF software (Leica Microsystems) on a Leica TCS SP8 confocal microscope. Quantification of images were performed using Fiji software or Imaris software (9.5.1).
Behavior
All behavior experiments were performed between 8 am and 12 pm in a blinded fashion. All mice were 4 months old at the time of the assay. Mice were transported from their home vivarium room to the behavior core and allowed 30 min to habituate before beginning each test.
Morris water maze
The MWM was done as described previously (Da Mesquita et al., 2018). In brief, the test involved four days of training consisting of four trials and one day of probe consisting of one trial. Mice were alternately placed facing different visual cues for each trial in a 23°C pool made opaque with white paint. The hidden platform was placed 1 cm below the water surface. Each trail lasted 60 s, and the mouse was placed on the hidden platform for 5 s if unable to locate it within the 60 s trial. All trials were tracked and scored using video tracking software (Noldus Ethovision XT).
Elevated plus maze
EPM was used to investigate anxiety in mice and was performed as described previously (Lammert et al., 2020). The maze has two open arms (35 × 6 cm2) and two closed arms (35 × 6 cm2) with 20 cm-tall black plexiglass walls elevated 121 cm from the floor. The mice were placed in the center square connecting the open and closed arms and allowed to explore during a 5 min trial. Activity was monitored and scored using video tracking software (Noldus Ethovision XT).
In vivo Aβ phagocytosis assay
5xFAD, 5xFAD SykΔMG, and littermate controls were intraperitoneally injected with 10 mg/kg methoxy-X04 (ApexBio, B5769) in a 1:1 ratio of PBS and DMSO. A brain harvest was completed 3 h after injection in which mice were euthanized using CO2 asphyxiation before being intracardially perfused with 20 mL of PBS + Na heparin (5 units/mL). The brains were placed in a Hanks buffer saline solution (HBSS) (Thermo Fisher, 14–025–092) with DNAse I (50 U/mL) (Sigma-Aldrich, 10–104–159–001) and papain (2 mg/mL) (Worthington, LS003124) and homogenized using a 10 mL serological pipette. The brain homogenates were then incubated at 37°C for 15 min. This process was repeated for a total of 3 times with the final two homogenizations accomplished using a 5 mL serological pipette. The brain homogenates were then passed through a 70 μm strainer to create a single-cell suspension. The cell suspension was then placed in 20 mL of DMEM/F12 buffer (21–331–020, Thermo Fisher) with 10% fetal bovine serum (FBS) (Thermo Fisher, 10–082–147), 1% Anti-anti (Thermo Fisher, 15–240–096), and 1% Glutamax (Thermo Fisher, 35–050–061) and spun down with a slow brake at 300 g for 10 min. The cell pellet was then resuspended in 13 mL of 37% Percoll (Cytvia, 17–0891–02) and spun down with no brake at 2000 rpm for 12 min. Myelin was removed and cells were resuspended in MACS buffer (Miltenyi Biotec, 130–0910376) to wash. Flow cytometry for microglia was then performed (as described below) with additional gating for methoxy-X04.
CypHer5E-Aβ preparation
Monomerization of Aβ (1–42) (641–15, California peptide) was achieved using a previously published protocol (Stine et al., 2011), using hexafluoroisopropanol (HFIP) (52–517, Sigma-Aldrich). 5 mM monomeric Aβ samples were incubated for 24 h at 4°C in F12 media to make a 200 μM stock of oligomeric Aβ. Samples were then incubated with CypHer5E-NHS ester (PA15401, Cytvia) diluted in 0.1 M sodium bicarbonate for 30 min covered and at room temperature. Following incubation, Biospin columns (7–326–227, Bio-Rad) were used to quench unbound dye. CypHer5E-tagged Aβ oligomers were stored at 4°C prior to BMDM treatment.
In vitro Aβ phagocytosis assay
Bone marrow-derived macrophages (BMDMs) were isolated from the hindlimbs of WT and Sykfl/fl LysMCre mice. Marrow-containing bones were sprayed with 70% ethanol before being placed in IMDM (Thermo Fisher, 12–440–053) containing penicillin/streptomycin (P/S) (Thermo Fisher, 15–140–163). A 25-gauge needle was used to flush marrow from the bones using 20 mL of IMDM containing P/S. An 18-gauge needle was then used to triturate flushed bone marrow 5 times to make a single-cell suspension. Samples were spun down at 1500 rpm for 5 min at 4°C. Cell pellets were resuspended in BMDM media containing IMDM, 10% FBS, 1% non-essential amino acids, 1% P/S, and 50 ng/mL M-CSF (PeproTech, 315–02). Cell plating was performed using 150 × 25 mm culture dishes (430–597, Thomas Scientific). After three days, 5 mL of BMDM media was added to each dish. Six days post initial cell plating, media was aspirated from dishes and 10 mL of PBS was added to each plate and incubated for 10 min at 4°C. BMDMs were removed from the dish using a scraper and transferred to a conical tube, spun down, and resuspended in BMDM media. 100,000 cells/well were pipetted in into a flat bottom 96 well plate. The next day, BMDMs were treated with vehicle or 10 μM of Tideglusib (Selleck Chemicals, S2823) 1 h prior to treatment with 10 μM of oligomeric Aβ tagged with CypHer5E. 24 h post Aβ treatment, BMDMs were then collected and flow cytometry was used to assess CypHer5E fluorescence.
Intrahippocampal injection
5xFAD and 5xFAD SykΔMG mice were anesthetized before receiving a bilateral hippocampal injection of 2 μL of vehicle or 2 μg pustulan into the right and left hemisphere of the hippocampus (at ±2 mm lateral, −2 mm posterior, and −2 mm ventral relative to the intersection of the coronal and sagittal suture (bregma) at a rate of 200 nL/min) using a stereotaxic frame (51730U, Stoelting) and nanoliter injector (NL2010MC2T, World Precision Instruments). Seven days post injection, mice were euthanized using CO2 and transcardially perfused before preparing brains for immunofluorescent staining to evaluate Aβ clearance in the hippocampus. Images were analyzed using Fiji software and Imaris software (9.5.1).
MACS isolation of microglia, T cells, and monocytes
Mice were euthanized with CO2 and immediately transcardially perfused with 20 mL 1X PBS. Brains were collected and their meninges and choroid plexus were removed before using a previously described MACS-sorting protocol (Norris et al., 2018) to isolate microglia. Spleens from these mice were collected and kept on ice in DMEM (Thermo Fisher, 11–885–084) with penicillin/streptomycin (Thermo Fisher, 15–140–163) and homogenized by gently mashing through a 70 μm cell strainer. Spleen homogenates were then centrifuged at 1500 rpm for 5 min then resuspended in 2 mL ACK lysis buffer (Quality Biological,118–156–101) and incubated at room temperature for 3 min to lyse red blood cells. Finally, the MACS-sorting protocol was used to isolate T cells and monocytes. Spinal cords were dissected and kept on ice in DMEM (Thermo Fisher, 11–885–084) with penicillin/streptomycin (Thermo Fisher, 15–140–163). Tissues were homogenized by gently mashing through a 70 μm cell strainer. Homogenates were centrifuged at 1500 rpm for 5 min then resuspended in 13 mL 37% isotonic Percoll (GE Healthcare, 17–0891–01) in 1X PBS. Samples were centrifuged at 2000 rpm for 12 min at room temperature with no brake. After centrifugation, the top myelin layer and supernatant were aspirated and the cell pellet was resuspended in MACS buffer before using the MACS-sorting protocol to isolate microglia (Miltenyi Biotec, 130–0910376). We proceeded with column purification of microglia using CD11b microbeads (Miltenyi, 130–093–634), purification of T cells using CD90.2 microbeads (Miltenyi, 130–121–278), and purification of monocytes using CD11b microbeads (Miltenyi, 130–049–601). We performed sorting utilizing LS columns and a QuadroMACS magnet (Miltenyi, 130–042–401 and 130–091–051) according to manufacturer’s instructions. Column-bound cells were analyzed for purity by flow cytometry, probed for SYK expression by qPCR and Western blotting, or submitted for RNA-seq.
Flow cytometry
For MACS-sort validation, an aliquot of the microglia-positive and -negative fractions were transferred to a 96-well round bottom plate, then washed with 1X PBS and spun down at 1600 rpm for 5 min. The cells were then stained with fixable viability dye (eBioscience, 65–0866–14) at 1:1000 for 30 min at 4°C. Following incubation, cells were then washed with FACS buffer (pH 7.4; 0.1 M PBS; 1 mM EDTA, and 1% BSA). Cells were then stained 1:200 with CD11b (APC), CD45 (PE-Cy7), and TCR β chain (Brilliant Violet 510) flow antibodies (all from eBioscience) in FACS buffer (pH 7.4; 0.1 M PBS; 1 mM EDTA, and 1% BSA) for 15 min at 4°C. The cell pellets were then washed with FACS buffer then resuspended in 100 μL of FACS buffer. Microglia were identified as the CD45int and CD11bhi after gating for single and live cells.
For lipid-droplet-accumulation and reactive oxygen species (ROS) assessment in microglia, mice were euthanized with CO2 at 8 months of age and immediately transcardially perfused with 20 mL 1X PBS. Brains were collected and their meninges and choroid plexus removed and prepped as a single cell suspension as described in (Norris et al., 2018). Brain homogenates were centrifuged at 1500 rpm for 5 min then resuspended in 13 mL 37% isotonic Percoll (GE Healthcare, 17–0891–01) in 1X PBS. Samples were centrifuged at 2000 rpm for 12 min at room temperature with no brake. After centrifugation, the top myelin layer and supernatant were aspirated and the cell pellet was washed with PBS. Cells were then stained either 1:500 with CellROX (Thermo Fisher, C10491) for 30 min or 1:2000 with BODIPY (Invitrogen, D3861) for 10 min diluted in PBS at 37°C. Cells were spun down and washed with FACS buffer. Cells were then stained 1:200 with CD11b (APC), CD45 (PE-Cy7), and TCR β chain (Brilliant Violet 510) flow antibodies (all from eBioscience) in FACS buffer for 15 min at 4°C. The cell pellets were then washed with FACS buffer then resuspended 1:5000 with DAPI in 100 μL of FACS buffer. Microglia were identified as the CD45int and CD11bhi after gating for single and live cells.
For flow cytometry staining of BMDMs, cells were washed with FACS buffer and centrifuged at 1500 rpm for 5 min, and resuspended in 100 μL of FACS buffer with fluorescently labeled antibodies (all from eBioscience) specific for CD11b (clone M1/70) and F4/80 (clone BM8) diluted 1:200. Cells were incubated in the dark for 20 min at room temperature, washed with FACS buffer, and resuspended 1:5000 with DAPI in 100 μL of FACS buffer. BMDMs were identified as the CD11bhi and F4/80hi after gating for single and live cells.
For flow cytometry staining in EAE experiments, cells were plated (100 μL of resuspended spinal cord or 1×106 splenocytes) in a 96-well plate and washed with FACS buffer. After centrifugation at 1500 rpm for 5 min and removal of supernatants, cells were resuspended in 100 μL 1X PBS with 1:1000 fixable viability dye (eBioscience, 65–0866–14) and 1:1000 Fc Block (eBioscience, 14–0161–86). Cells were incubated at 4°C for 30 min. Cells were then washed with FACS buffer, centrifuged at 1500 rpm for 5 min, and resuspended in 100 μL of FACS buffer with fluorescently labeled antibodies (all from eBioscience) specific for CD45 (clone 30-F11), CD11b (clone M1/70), Gr-1 (clone RB6–8C5), MHC-II (clone M5/114.15.2), TCRβ (clone H57–597), CD4 (clone RM4–5), CD8 (clone 53–6.7), CD11c (clone N418), and CD80 (clone 16–10A1) diluted 1:200. Cells were incubated in the dark for 20 min at room temperature, washed twice with FACS buffer, and fixed with 1% paraformaldehyde in FACS buffer.
For intracellular cytokine staining, cells were plated (100 μL of resuspended spinal cord or 1×106 splenocytes) in a 96-well plate in IMDM stimulation media (Iscove’s Modified Dulbecco’s Media) (Thermo Fisher, 12–440–053), penicillin/streptomycin (Thermo Fisher, 15–140–163), 10% heat-inactivated fetal bovine serum (Thermo Fisher, 10–082–147), 1% L-glutamine (Thermo Fisher, 25–030–081), and 50 μM beta-mercaptoethanol (Thermo Fisher, 21–985–023)] with 20 ng/mL PMA (Sigma-Aldrich, P1585), 1 μg/mL ionomycin (Sigma-Aldrich, I9657), and 1:1000 monensin (eBioscience, 00–4505–51). Cells were incubated for 5 h at 37°C with 5% CO2, then washed with 1X PBS prior to proceeding with surface staining for flow cytometry as described above. Cells were fixed and permeabilized using IC fixation buffer (eBioscience, 00–8222–49) and permeabilization buffer (eBioscience, 00–8333–56) following manufacturer’s instructions. Cells were then stained with 100 μL fluorescently labeled antibodies (all from Thermo Fisher) for GM-CSF (clone MP1–22 × 109), IFN-γ (clone XMG1.2), and IL-17A (clone eBio17B7) diluted 1:200 in 1X permeabilization buffer for 20 min at room temperature. Cells were washed twice with 1X permeabilization buffer, then twice with FACS buffer.
Sample data were acquired within a few days of fixation using a Gallios flow cytometer (10 colors, 3 lasers, B5-R1-V2 Configuration with Kaluza Acquisition; Beckman Coulter) and analyzed using FlowJo software (Becton, Dickinson, & Company).
Multiplex cytokine assay
Immune cells isolated from spleens were plated in a 96-well plate at up to 2×105 cells/well and stimulated with 30 μg/mL MOG35–55 peptide in IMDM stimulation media for 48 h at 37°C with 5% CO2. After incubation, cells were centrifuged at 1600 rpm for 5 min and supernatants were collected for storage at −80°C.
Supernatants were assayed for concentrations of various cytokines using Bio-Rad Bio-Plex Pro reagent kit (Bio-Rad, 171–304070M) and Bio-Plex Pro Mouse Cytokine 23-Plex Group I magnetic beads and detection antibodies for IL-1α, IL-1β, IL-2, IL-4, IL-6, IL-10, IL-17, G-CSF, GM-CSF, IFN-γ, KC, and TNF-α according to manufacturers’ instructions (Bio-Rad). Sample data for Bio-Plex Pro assays were acquired with a Bio-Plex 200 System (Bio-Rad) and analyzed using Bio-Plex Manager software (Bio-Rad).
Histopathological analysis of EAE spinal cords
Mice were euthanized with CO2 and immediately transcardially perfused with 20 mL 1X PBS followed by 20 mL 10% neutral buffered formalin (NBF; Azer Scientific). Spinal columns were dissected and immersion fixed in 10% NBF for at least 48 h. The thoracic region of the spinal cord was carefully dissected from the column, embedded in paraffin, sectioned coronally from the rostral end of each piece, and mounted on slides. Sections were stained with Luxol Fast Blue (LFB; Thermo Fisher, 212–170-250) to label myelin. In brief, after deparaffinization, sections were incubated at 60°C for 16–24 h in 0.1% Luxol Fast Blue in 95% ethanol +0.05% acetic acid. Excess stain was removed with 95% ethanol and slides were dipped in 0.05% lithium carbonate (Thermo Fisher, 446–322–500) in dH2O for 10–20 s then in 70% ethanol to remove LFB staining from gray matter but not from myelinated white matter. Slides were washed with dH2O then counterstained with hematoxylin (Sigma Aldrich, HHS128) to label nuclei. Brightfield images were acquired on a Keyence BZ-X810 microscope at 4X and 40X magnification.
Isolation of immune cells
Mice were euthanized with CO2 and immediately transcardially perfused with 20 mL 1X PBS. Spinal cords and spleens were dissected and kept on ice in DMEM (Thermo Fisher, 11–885–084) with penicillin/streptomycin (Thermo Fisher, 15–140–163). Tissues were homogenized by gently mashing through a 70 μm cell strainer. For spinal cords, homogenates were centrifuged at 1500 rpm for 5 min then resuspended in 13 mL 37% isotonic Percoll (GE Healthcare, 17–0891–01) in 1X PBS. Samples were centrifuged at 2000 rpm for 12 min at room temperature with no brake. After centrifugation, the top myelin layer and supernatant were aspirated and the cell pellet was resuspended in 500 μL DMEM with penicillin/streptomycin. For spleens, homogenates were centrifuged at 1500 rpm for 5 min then resuspended in 2 mL ACK lysis buffer (Quality Biological, 118–156–101) and incubated at room temperature for 3 min to lyse red blood cells. Cells were washed and resuspended in DMEM with penicillin/streptomycin. Cells were counted using a Cellometer Auto 2000 (Nexelcom Bioscience).
RNA isolation, cDNA synthesis, qPCR
RNA was isolated from the left hemisphere of the brain of 5xFAD and 5xFAD SykΔMG mice. 50 μL of brain homogenate (described in the brain sample preparation section) was added to 500 μL of TRIzol (Life Technologies, 15,596,018). Following vortexing of these samples, 200 μL of chloroform (Fisher Scientific, BP1145–1) was added and incubated for 5 min before being spun down at 14,000 rpm at 4°C for 15 min. The top aqueous fraction was transferred to a new tube and an equal volume of isopropanol (Sigma-Aldrich, I9516) was added then vortexed. The samples were incubated at room temperature for 10 min and then spun down at 12,000 rpm at 4°C for 5 min. The pellet was then washed with 1 mL 70% ethanol 2 times, then allowed to air dry for 15 min before resuspending the RNA pellet in 100 μL of DNAse/RNAse free water. RNA was isolated from MACS-sorted microglia in the brain and spinal cord using an RNeasy Micro kit (Qiagen, 74–004) according to manufacturer’s instructions. Sample quality and quantity for AD and EAE samples was evaluated using NanoDrop 2000 Spectrophotometer (Thermo Fisher). The RNA was then converted to cDNA using a Sensifast cDNA Synthesis kit (Bioline, BIO-65054). Levels of Sykb (Mm01333035_m1) and Gapdh (Mm99999915_g1) mRNA were determined using Taqman Gene Expression Assay primer/probe mix (Thermo Fisher), Sensifast Probe No-ROX kit (Bioline, BIO-86005), and a CFX384 Real-Time PCR System (BioRad, 1–855–484). All reagents were used according to manufacturer’s instructions.
FACS sorting for RNA sequencing
Ai6-ZsGreen reporter mice possess a LoxP-flanked STOP cassette that prevents the expression of green fluorescent protein variant ZsGreen1 until the stop site is excised after tamoxifen induction of Cre-recombinase activity. Following the withdrawal of tamoxifen, short-lived Ai6-ZsGreen expressing cells in the periphery are promptly replaced by newly derived cells that lack Ai6-ZsGreen expression. In contrast, the long-lived nature of microglia allows them retain their Ai6-ZsGreen signal for months post-tamoxifen withdrawal (Goldmann et al., 2013). Sykcon–Ai6 and SykΔMG–Ai6 mice were euthanized on EAE Day 35 with CO2 and immediately transcardially perfused with 20 mL ice-cold 1X PBS. Spinal cords were dissected and placed on ice in DMEM +10% FBS. Tissues were gently homogenized by mashing through a cell strainer then centrifuged at 1500 rpm for 5 min. Pellets were resuspended in 13 mL 37% isotonic Percoll (GE Healthcare, 17–0891–01) in 1X PBS. Samples were centrifuged at 2000 rpm for 12 min at room temperature with no brake. After centrifugation, the top myelin layer and supernatant were aspirated and the cell pellet was resuspended in FACS buffer. Cell suspensions were incubated with Fc Block and antibodies for CD45, CD11b, and Gr-1. After two washes with FACS buffer, cells were resuspended in FACS buffer +0.2 mg/mL DAPI and sorted on DAPI− CD45+ ZsGreen+ cells using an Influx Cell Sorter (BD) in the University of Virginia Flow Cytometry Core Facility.
RNA sequencing data analysis
AD microglia RNA-Seq
MACS-sorted microglia were sent to GENEWIZ Next Generation Sequencing. The raw sequencing reads (FASTQ files) were aligned to the UCSC mm10 mouse genome build using the splice-aware read aligner HISAT2. Samtools was used for quality control filtering. Reads were sorted into feature counts with HTSeq. DESeq2 (v1.32.0) was used to normalize the raw counts based on read depth, perform principal component analysis, and conduct differential expression analysis. The p values were corrected with the Benjamini-Hochberg procedure to limit false positives arising from multiple testing. The significantly repressed and enhanced genes were put into GProfiler to gather the KEGG terms. The analysis itself was performed using the Seq2Pathway, fgsea, tidyverse, and dplyr software packages. Heatmaps were generated using the pheatmap R package [https://github.com/raivokolde/pheatmap] while other plots were made with the lattice (https://lattice.r-forge.r-project.org/) or ggplot2 [https://ggplot2.tidyverse.org] packages.
EAE microglia scRNA-Seq
FACS-sorted single cell suspensions were submitted to the University of Virginia Genome Analysis and Technology Core for single-cell RNA sequencing library preparation. The raw sequencing reads (FASTQ files) were aligned to the UCSC mm10 mouse genome build using Cell Ranger (v1.3.1) which performs alignment, filtering, barcode counting and unique molecular identifier (UMI) counting. R studio (v4.0.5) was used for all downstream analyses and Seurat (v.4.0.2) was used for filtering out low-quality cells, normalization of the data, determination of cluster defining markers and graphing of the data on UMAP. Low-quality cells were excluded in an initial quality-control (QC) step by removing genes expressed in fewer than three cells, cells with fewer than 150 genes expressed, and cells expressing more than 5000 genes. Cells with more than 20% of mitochondrial-associated genes and cells with more than 5% hemoglobin among their expressed genes were also removed. Genes with high variance were selected using the FindVariableGenes function, then the dimensionality of the data was reduced by principal component analysis (PCA) and identified by random sampling 20 significant principal components (PCs) for each sample with the PCElbowPlot function. Cells were clustered with Seurat’s FindClusters function. Differential gene expression analysis was performed within clusters using the ZinBWave function and DESeq2 (v1.32.0). ToppCluster (Cincinnati Children’s) was used for network analyses to identify KEGG and GO terms in the DAM cluster. Data was organized and graphs were created using patchwork, dplyr, tidyverse, and Seurat. The frequency plot was created using Prism GraphPad. Pseudotime analysis was conducted using Monocle (v0.2.3.0).
QUANTIFICATION AND STATISTICAL ANALYSIS
Mean values, SEM values, Student’s t test (unpaired), one-way ANOVA, and two-way ANOVA were calculated using Prism software (GraphPad). Significance for pooled EAE experiments was performed by a Mann-Whitney test. p values less than 0.05 were considered significant. ns = not significant, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Supplementary Material
Highlights.
Disruption of microglial SYK signaling exacerbates disease in models of AD and MS
Microglial proliferation and association with Aβ plaques is coordinated by SYK
SYK is a pivotal regulator of microglial activation and AKT/GSK3β-signaling
Phagocytic clearance of neurotoxic material by microglia is dependent on SYK
ACKNOWLEDGMENTS
This work was supported by funding from the NIH (1RF1AG071996–01, R01NS106383), The Alzheimer’s Association (ADSF-21–816651), the Cure Alzheimer’s Fund, and The Owens Family Foundation to J.R.L.. H.E. and K.E.Z. were supported by an NIH T32 (T32GM008136). H.E., K.E.Z., A.C.B., and C.R.L. were supported by Wagner Fellowships. H.E. and C.H. were supported by a Double Hoo Award. E.L.F. was supported by a National MS Foundation Fellowship (FG-1707–28590). D.A.S. was supported by a Harrison Fellowship. C.H. was supported by a College Science Scholars Award. A.C.B. was supported by funding from the NIH (5T32GM007267–38, 5T32AI007496–25, F30AG069396–01). C.R.L. was supported by an NIH T32 (3T32GM008328). J.A.K. was supported by an NIH T32 (T32DK007646). T.K.U. was supported by the NIH (R01AG070973).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
REFERENCES
- Back SA, Tuohy TMF, Chen H, Wallingford N, Craig A, Struve J, Luo NL, Banine F, Liu Y, Chang A, et al. (2005). Hyaluronan accumulates in demyelinated lesions and inhibits oligodendrocyte progenitor maturation. Nat Med 11, 966–972. 10.1038/nm1279. [DOI] [PubMed] [Google Scholar]
- Batista SJ, Still KM, Johanson D, Thompson JA, O’Brien CA, Lukens JR, and Harris TH (2020). Gasdermin-D-dependent IL-1α release from microglia promotes protective immunity during chronic Toxoplasma gondii infection. Nat. Commun. 11, 3687. 10.1038/s41467-020-17491-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemiller SM, McCray TJ, Allan K, Formica SV, Xu G, Wilson G, Kokiko-Cochran ON, Crish SD, Lasagna-Reeves CA, Ransohoff RM, et al. (2017). TREM2 deficiency exacerbates tau pathology through dysregulated kinase signaling in a mouse model of tauopathy. Mol. Neurodegener. 12, 74. 10.1186/s13024-017-0216-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MR, Radford SE, and Hewitt EW (2020). Modulation of beta-Amyloid Fibril Formation in Alzheimer’s Disease by Microglia and Infection. Front. Mol. Neurosci. 13, 609073. 10.3389/fnmol.2020.609073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu E, Mychasiuk R, Hibbs ML, and Semple BD (2021). Dysregulated phosphoinositide 3-kinase signaling in microglia: shaping chronic neuroinflammation. J. Neuroinflammation 18, 276. 10.1186/s12974-021-02325-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CG, Lee H, and Lee SB (2018). Mechanisms of protein toxicity in neurodegenerative diseases. Cell. Mol. Life Sci. 75, 3159–3180. 10.1007/s00018-018-2854-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung YH, Kim HY, Yoon BR, Kang YJ, and Lee WW (2019). Suppression of Syk activation by resveratrol inhibits MSU crystal-induced inflammation in human monocytes. J. Mol. Med. (Berl.) 97, 369–383. 10.1007/s00109-018-01736-y. [DOI] [PubMed] [Google Scholar]
- Cignarella F, Filipello F, Bollman B, Cantoni C, Locca A, Mikesell R, Manis M, Ibrahim A, Deng L, Benitez BA, et al. (2020). TREM2 activation on microglia promotes myelin debris clearance and remyelination in a model of multiple sclerosis. Acta Neuropathol. 140, 513–534. 10.1007/s00401-020-02193-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark EA, and Giltiay NV (2018). CD22: A Regulator of Innate and Adaptive B Cell Responses and Autoimmunity. Front. Immunol. 9, 2235. 10.3389/fimmu.2018.02235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colie S, Sarroca S, Palenzuela R, Garcia I, Matheu A, Corpas R, Dotti CG, Esteban JA, Sanfeliu C, and Nebreda AR (2017). Neuronal p38α mediates synaptic and cognitive dysfunction in an Alzheimer’s mouse model by controlling β-amyloid production. Sci. Rep. 7, 45306. 10.1038/srep45306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condello C, Yuan P, and Grutzendler J (2018). Microglia-Mediated Neuroprotection, TREM2, and Alzheimer’s Disease: Evidence From Optical Imaging. Biol Psychiatry 83, 377–387. 10.1016/j.biopsych.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condello C, Yuan P, Schain A, and Grutzendler J (2015). Microglia constitute a barrier that prevents neurotoxic protofibrillar Aβ42 hotspots around plaques. Nat. Commun. 6, 6176. 10.1038/ncomms7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinescu CS, Farooqi N, O’Brien K, and Gran B (2011). Experimental autoimmune encephalomyelitis (EAE) as a model for multiple sclerosis (MS). Br. J. Pharmacol. 164, 1079–1106. 10.1111/j.1476-5381.2011.01302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper-Knock J, Green C, Altschuler G, Wei W, Bury JJ, Heath PR, Wyles M, Gelsthorpe C, Highley JR, Lorente-Pons A, et al. (2017). A data-driven approach links microglia to pathology and prognosis in amyotrophic lateral sclerosis. Acta Neuropathol Commun 5, 23. 10.1186/s40478-017-0424-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornall RJ, Cheng AM, Pawson T, and Goodnow CC (2000). Role of Syk in B-cell development and antigen-receptor signaling. Proc. Natl. Acad. Sci. USA. 97, 1713–1718. 10.1073/pnas.97.4.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Mesquita S, Louveau A, Vaccari A, Smirnov I, Cornelison RC, Kingsmore KM, Contarino C, Onengut-Gumuscu S, Farber E, Raper D, et al. (2018). Functional aspects of meningeal lymphatics in ageing and Alzheimer’s disease. Nature 560, 185–191. 10.1038/s41586-018-0368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DaRocha-Souto B, Coma M, Perez-Nievas BG, Scotton TC, Siao M, Sanchez-Ferrer P, Hashimoto T, Fan Z, Hudry E, Barroeta I, et al. (2012). Activation of glycogen synthase kinase-3 beta mediates beta-amyloid induced neuritic damage in Alzheimer’s disease. Neurobiol. Dis. 45, 425–437. 10.1016/j.nbd.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deczkowska A, Keren-Shaul H, Weiner A, Colonna M, Schwartz M, and Amit I (2018). Disease-Associated Microglia: A Universal Immune Sensor of Neurodegeneration. Cell 173, 1073–1081. 10.1016/j.cell.2018.05.003. [DOI] [PubMed] [Google Scholar]
- Doble BW, and Woodgett JR (2003). GSK-3: tricks of the trade for a multi-tasking kinase. J. Cell Sci. 116, 1175–1186. 10.1242/jcs.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond RA, Saijo S, Iwakura Y, and Brown GD (2011). The role of Syk/CARD9 coupled C-type lectins in antifungal immunity. Eur. J. Immunol. 41, 276–281. 10.1002/eji.201041252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efthymiou AG, and Goate AM (2017). Late onset Alzheimer’s disease genetics implicates microglial pathways in disease risk. Mol. Neurodegener. 12, 43. 10.1186/s13024-017-0184-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron TF, and Petrucelli L (2009). The role of tau in neurodegeneration. Mol. Neurodegener. 4, 13. 10.1186/1750-1326-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann T, Wieghofer P, Muller PF, Wolf Y, Varol D, Yona S, Brendecke SM, Kierdorf K, Staszewski O, Datta M, et al. (2013). A new type of microglia gene targeting shows TAK1 to be pivotal in CNS autoimmune inflammation. Nat. Neurosci. 16, 1618–1626. 10.1038/nn.3531. [DOI] [PubMed] [Google Scholar]
- Griciuc A, Patel S, Federico AN, Choi SH, Innes BJ, Oram MK, Cereghetti G, McGinty D, Anselmo A, Sadreyev RI, et al. (2019). TREM2 Acts Downstream of CD33 in Modulating Microglial Pathology in Alzheimer’s Disease. Neuron 103, 820–835.e7. e827. 10.1016/j.neuron.2019.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudi V, Gingele S, Skripuletz T, and Stangel M (2014). Glial response during cuprizone-induced de- and remyelination in the CNS: lessons learned. Front. Cell. Neurosci. 8, 73. 10.3389/fncel.2014.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha J, Kim EJ, Lim S, Shin DW, Kang YJ, Bae SM, Yoon HK, and Oh KS (2012). Altered risk-aversion and risk-taking behaviour in patients with Alzheimer’s disease. Psychogeriatrics 12, 151–158. 10.1111/j.1479-8301.2011.00396.x. [DOI] [PubMed] [Google Scholar]
- Hadas S, Spira M, Hanisch UK, Reichert F, and Rotshenker S (2012). Complement receptor-3 negatively regulates the phagocytosis of degenerated myelin through tyrosine kinase Syk and cofilin. J. Neuroinflammation 9, 166. 10.1186/1742-2094-9-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez F, Lucas JJ, and Avila J (2012). GSK3 and tau: two convergence points in Alzheimer’s disease. J Alzheimers Dis 33 (Suppl 1), S141–S144. 10.3233/JAD-2012-129025. [DOI] [PubMed] [Google Scholar]
- Hickman S, Izzy S, Sen P, Morsett L, and El Khoury J (2018). Microglia in neurodegeneration. Nat. Neurosci. 21, 1359–1369. 10.1038/s41593-018-0242-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman SE, Allison EK, Coleman U, Kingery-Gallagher ND, and El Khoury J (2019). Heterozygous CX3CR1 Deficiency in Microglia Restores Neuronal beta-Amyloid Clearance Pathways and Slows Progression of Alzheimer’s Like-Disease in PS1-APP Mice. Front. Immunol. 10, 2780. 10.3389/fimmu.2019.02780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Happonen KE, Burrola PG, O’Connor C, Hah N, Huang L, Nimmerjahn A, and Lemke G (2021). Microglia use TAM receptors to detect and engulf amyloid beta plaques. Nat. Immunol. 22, 586–594. 10.1038/s41590-021-00913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado DE, Molina-Porcel L, Carroll JC, Macdonald C, Aboagye AK, Trojanowski JQ, and Lee VMY (2012). Selectively silencing GSK-3 isoforms reduces plaques and tangles in mouse models of Alzheimer’s disease. J. Neurosci. 32, 7392–7402. 10.1523/JNEUROSCI.0889-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Multiple Sclerosis Genetics Consortium, Baranzini SE, Santaniello A, Shoostari P, Cotsapas C, Wong G, Beecham AH, James T, Replogle J, Vlachos IS, McCabe C, et al. (2019). Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science 365, eaav7188. 10.1126/science.aav7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Multiple Sclerosis Genetics Consortium and The Wellcome Trust Case Control Consortium 2, Sawcer S, Hellenthal G, Pirinen M, Spencer CC, Patsopoulos NA, Moutsianas L, Dilthey A, Su Z, Freeman C, Hunt SE, et al. (2011). Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 476, 214–219. 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jawhar S, Trawicka A, Jenneckens C, Bayer TA, and Wirths O (2012). Motor deficits, neuron loss, and reduced anxiety coinciding with axonal degeneration and intraneuronal Aβ aggregation in the 5XFAD mouse model of Alzheimer’s disease. Neurobiol. Aging 33, 196.e29. e129–196.e40. 10.1016/j.neurobiolaging.2010.05.027. [DOI] [PubMed] [Google Scholar]
- Kanno T, Tsuchiya A, and Nishizaki T (2014). Hyperphosphorylation of Tau at Ser396 occurs in the much earlier stage than appearance of learning and memory disorders in 5XFAD mice. Behav. Brain Res. 274, 302–306. 10.1016/j.bbr.2014.08.034. [DOI] [PubMed] [Google Scholar]
- Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, David E, Baruch K, Lara-Astaiso D, Toth B, et al. (2017). A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 169, 1276–1290.e17. e1217. 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- Krasemann S, Madore C, Cialic R, Baufeld C, Calcagno N, El Fatimy R, Beckers L, O’Loughlin E, Xu Y, Fanek Z, et al. (2017). The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity 47, 566–581.e9. e569. 10.1016/j.immuni.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammert CR, Frost EL, Bellinger CE, Bolte AC, McKee CA, Hurt ME, Paysour MJ, Ennerfelt HE, and Lukens JR (2020). AIM2 inflammasome surveillance of DNA damage shapes neurodevelopment. Nature 580, 647–652. 10.1038/s41586-020-2174-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampron A, Larochelle A, Laflamme N, Prefontaine P, Plante MM, Sanchez MG, Yong VW, Stys PK, Tremblay ME, and Rivest S (2015). Inefficient clearance of myelin debris by microglia impairs remyelinating processes. J. Exp. Med. 212, 481–495. 10.1084/jem.20141656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latour S, Fournel M, and Veillette A (1997). Regulation of T-cell antigen receptor signalling by Syk tyrosine protein kinase. Mol. Cell Biol. 17, 4434–4441. 10.1128/MCB.17.8.4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor B, Segurado R, Kennelly S, Olde Rikkert MGM, Howard R, Pasquier F, Borjesson-Hanson A, Tsolaki M, Lucca U, Molloy DW, et al. (2018). Nilvadipine in mild to moderate Alzheimer disease: A randomised controlled trial. PLoS Med. 15, e1002660. 10.1371/journal.pmed.1002660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CD, Daggett A, Gu X, Jiang LL, Langfelder P, Li X, Wang N, Zhao Y, Park CS, Cooper Y, et al. (2018). Elevated TREM2 Gene Dosage Reprograms Microglia Responsivity and Ameliorates Pathological Phenotypes in Alzheimer’s Disease Models. Neuron 97, 1032–1048.e5. e1035. 10.1016/j.neuron.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionakis MS, Iliev ID, and Hohl TM (2017). Immunity against fungi. JCI Insight 2, 93156. 10.1172/jci.insight.93156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loving BA, Tang M, Neal MC, Gorkhali S, Murphy R, Eckel RH, and Bruce KD (2021). Lipoprotein Lipase Regulates Microglial Lipid Droplet Accumulation 10, 198. Cells. 10.3390/cells10020198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik A, Sharma D, Malireddi RS, Guy CS, Chang TC, Olsen SR, Neale G, Vogel P, and Kanneganti TD (2018). SYK-CARD9 Signaling Axis Promotes Gut Fungi-Mediated Inflammasome Activation to Restrict Colitis and Colon Cancer. Immunity 49, 515–530.e5. e515. 10.1016/j.immuni.2018.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik M, Simpson JF, Parikh I, Wilfred BR, Fardo DW, Nelson PT, and Estus S (2013). CD33 Alzheimer’s risk-altering polymorphism, CD33 expression, and exon 2 splicing. J. Neurosci. 33, 13320–13325. 10.1523/JNEUROSCI.1224-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschallinger J, Iram T, Zardeneta M, Lee SE, Lehallier B, Haney MS, Pluvinage JV, Mathur V, Hahn O, Morgens DW, et al. (2020). Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat. Neurosci. 23, 194–208. 10.1038/s41593-019-0566-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin LJ, Pardo CA, Cork LC, and Price DL (1994). Synaptic pathology and glial responses to neuronal injury precede the formation of senile plaques and amyloid deposits in the aging cerebral cortex. Am. J. Pathol. 145, 1358–1381. [PMC free article] [PubMed] [Google Scholar]
- Mateo I, Infante J, Llorca J, Rodriguez E, Berciano J, and Combarros O (2006). Association between Glycogen Synthase Kinase-3β Genetic Polymorphism and Late-Onset Alzheimer’s Disease. Dement Geriatr Cogn Disord 21, 228–232. 10.1159/000091044. [DOI] [PubMed] [Google Scholar]
- Mc Guire C, Wieghofer P, Elton L, Muylaert D, Prinz M, Beyaert R, and van Loo G (2013). Paracaspase MALT1 deficiency protects mice from autoimmune-mediated demyelination. J. Immunol. 190, 2896–2903. 10.4049/jimmunol.1201351. [DOI] [PubMed] [Google Scholar]
- Mocsai A, Ruland J, and Tybulewicz VLJ (2010). The SYK tyrosine kinase: a crucial player in diverse biological functions. Nat. Rev. Immunol. 10, 387–402. 10.1038/nri2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinero LL, Cubre A, Mora-Solano C, Wang Y, and Alegre ML (2012). T cell receptor/CARMA1/NF-κB signaling controls T-helper (Th) 17 differentiation. Proc. Natl. Acad. Sci. USA. 109, 18529–18534. 10.1073/pnas.1204557109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris GT, Smirnov I, Filiano AJ, Shadowen HM, Cody KR, Thompson JA, Harris TH, Gaultier A, Overall CC, and Kipnis J (2018). Neuronal integrity and complement control synaptic material clearance by microglia after CNS injury. J. Exp. Med. 215, 1789–1801. 10.1084/jem.20172244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaum RL, and Ellis CE (2003). Alzheimer’s disease and Parkinson’s disease. N. Engl. J. Med. 348, 1356–1364. 10.1056/NEJM2003ra020003. [DOI] [PubMed] [Google Scholar]
- Parakalan R, Jiang B, Nimmi B, Janani M, Jayapal M, Lu J, Tay SS, Ling EA, and Dheen ST (2012). Transcriptome analysis of amoeboid and ramified microglia isolated from the corpus callosum of rat brain. BMC Neurosci. 13, 64. 10.1186/1471-2202-13-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris D, Ait-Ghezala G, Bachmeier C, Laco G, Beaulieu-Abdelahad D, Lin Y, Jin C, Crawford F, and Mullan M (2014). The spleen tyrosine kinase (Syk) regulates Alzheimer amyloid-beta production and Tau hyperphosphorylation. J. Biol. Chem. 289, 33927–33944. 10.1074/jbc.M114.608091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plastini MJ, Desu HL, and Brambilla R (2020). Dynamic Responses of Microglia in Animal Models of Multiple Sclerosis. Front. Cell. Neurosci. 14, 269. 10.3389/fncel.2020.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluvinage JV, Haney MS, Smith BAH, Sun J, Iram T, Bonanno L, Li L, Lee DP, Morgens DW, Yang AC, et al. (2019). CD22 blockade restores homeostatic microglial phagocytosis in ageing brains. Nature 568, 187–192. 10.1038/s41586-019-1088-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramagopalan SV, Dobson R, Meier UC, and Giovannoni G (2010). Multiple sclerosis: risk factors, prodromes, and potential causal pathways. Lancet Neurol. 9, 727–739. 10.1016/S1474-4422(10)70094-6. [DOI] [PubMed] [Google Scholar]
- Reddy PH (2013). Amyloid beta-induced glycogen synthase kinase 3β phosphorylated VDAC1 in Alzheimer’s disease: Implications for synaptic dysfunction and neuronal damage. Biochim. Biophys. Acta 1832, 1913–1921. 10.1016/j.bbadis.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard BC, Kurdakova A, Baches S, Bayer TA, Weggen S, and Wirths O (2015). Gene Dosage Dependent Aggravation of the Neurological Phenotype in the 5XFAD Mouse Model of Alzheimer’s Disease. J Alzheimers Dis 45, 1223–1236. 10.3233/JAD-143120. [DOI] [PubMed] [Google Scholar]
- Schaffer BAJ, Bertram L, Miller BL, Mullin K, Weintraub S, Johnson N, Bigio EH, Mesulam M, Wiedau-Pazos M, Jackson GR, et al. (2008). Association of GSK3B with Alzheimer disease and frontotemporal dementia. Arch. Neurol. 65, 1368–1374. 10.1001/archneur.65.10.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweig JE, Yao H, Beaulieu-Abdelahad D, Ait-Ghezala G, Mouzon B, Crawford F, Mullan M, and Paris D (2017). Alzheimer’s disease pathological lesions activate the spleen tyrosine kinase. Acta Neuropathol Commun 5, 69. 10.1186/s40478-017-0472-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steen E, Terry BM, J Rivera E, Cannon JL, Neely TR, Tavares R, Xu XJ, Wands JR, and de la Monte SM (2005). Impaired insulin and insulin-like growth factor expression and signaling mechanisms in Alzheimer’s disease–is this type 3 diabetes? J Alzheimers Dis 7, 63–80. 10.3233/jad-2005-7107. [DOI] [PubMed] [Google Scholar]
- Stine WB, Jungbauer L, Yu C, and LaDu MJ (2011). Preparing Synthetic Aβ in Different Aggregation States. Methods Mol. Biol. 670, 13–32. 10.1007/978-1-60761-744-0_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Prinz M, Stagi M, Chechneva O, and Neumann H (2007). TREM2-transduced myeloid precursors mediate nervous tissue debris clearance and facilitate recovery in an animal model of multiple sclerosis. PLoS Med. 4, e124. 10.1371/journal.pmed.0040124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JP, Hardy J, and Fischbeck KH (2002). Toxic proteins in neurodegenerative disease. Science 296, 1991–1995. 10.1126/science.1067122. [DOI] [PubMed] [Google Scholar]
- Trapp BD, and Nave KA (2008). Multiple sclerosis: an immune or neurodegenerative disorder? Annu. Rev. Neurosci. 31, 247–269. 10.1146/annurev.neuro.30.051606.094313. [DOI] [PubMed] [Google Scholar]
- Ulland TK, Song WM, Huang SCC, Ulrich JD, Sergushichev A, Beatty WL, Loboda AA, Zhou Y, Cairns NJ, Kambal A, et al. (2017). TREM2 Maintains Microglial Metabolic Fitness in Alzheimer’s Disease. Cell 170, 649–663.e13. e613. 10.1016/j.cell.2017.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers JC, King AE, Woodhouse A, Kirkcaldie MT, Staal JA, McCormack GH, Blizzard CA, Musgrove RE, Mitew S, Liu Y, et al. (2009). Axonopathy and cytoskeletal disruption in degenerative diseases of the central nervous system. Brain Res. Bull. 80, 217–223. 10.1016/j.brainresbull.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Walker DG, and Lue LF (2015). Immune phenotypes of microglia in human neurodegenerative disease: challenges to detecting microglial polarization in human brains. Alzheimer’s Res. Ther. 7, 56. 10.1186/s13195-015-0139-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cella M, Mallinson K, Ulrich J, Young K, Robinette M, Gilfillan S, Krishnan G, Sudhakar S, Zinselmeyer B, et al. (2015). TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell 160, 1061–1071. 10.1016/j.cell.2015.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ulland TK, Ulrich JD, Song W, Tzaferis JA, Hole JT, Yuan P, Mahan TE, Shi Y, Gilfillan S, et al. (2016). TREM2-mediated early microglial response limits diffusion and toxicity of amyloid plaques. J. Exp. Med. 213, 667–675. 10.1084/jem.20151948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinger JG, Brosnan CF, Loudig O, Goldberg MF, Macian F, Arnett HA, Prieto AL, Tsiperson V, and Shafit-Zagardo B (2011). Loss of the receptor tyrosine kinase Axl leads to enhanced inflammation in the CNS and delayed removal of myelin debris during experimental autoimmune encephalomyelitis. J. Neuroinflammation 8, 49. 10.1186/1742-2094-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker Hawkins RF, Patenaude A, Dumas A, Jain R, Tesfagiorgis Y, Kerfoot S, Matsui T, Gunzer M, Poubelle PE, Larochelle C, et al. (2017). ICAM1+ neutrophils promote chronic inflammation via ASPRV1 in B cell-dependent autoimmune encephalomyelitis. JCI Insight 2, 96882. 10.1172/jci.insight.96882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wißfeld J, Nozaki I, Mathews M, Raschka T, Ebeling C, Hornung V, Brustle O, and Neumann H (2021). Deletion of Alzheimer’s disease-associated CD33 results in an inflammatory human microglia phenotype. Glia 69, 1393–1412. 10.1002/glia.23968. [DOI] [PubMed] [Google Scholar]
- Wu X, Saito T, Saido TC, Barron AM, and Ruedl C (2021). Microglia and CD206(+) border-associated mouse macrophages maintain their embryonic origin during Alzheimer’s disease. Elife 10, e71879. 10.7554/eLife.71879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Coppola K, Schweig JE, Crawford F, Mullan M, and Paris D (2019). Distinct Signaling Pathways Regulate TREM2 Phagocytic and NFκB Antagonistic Activities. Front. Cell. Neurosci. 13, 457. 10.3389/fncel.2019.00457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye XC, Hao Q, Ma WJ, Zhao QC, Wang WW, Yin HH, Zhang T, Wang M, Zan K, Yang XX, et al. (2020). Dectin-1/Syk signaling triggers neuroinflammation after ischemic stroke in mice. J. Neuroinflammation 17, 17. 10.1186/s12974-019-1693-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan J, Mann T, Joost S, Behrangi N, Frank M, and Kipp M (2020). The Cuprizone Model: Dos and Do Nots 9, 843. Cells. 10.3390/cells9040843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegenfuss JS, Biswas R, Avery MA, Hong K, Sheehan AE, Yeung YG, Stanley ER, and Freeman MR (2008). Draper-dependent glial phagocytic activity is mediated by Src and Syk family kinase signalling. Nature 453, 935–939. 10.1038/nature06901. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Bulk and Single-cell RNA-seq data have been deposited at GEO and are publicly available as of the date of publication (accession number: GEO: GSE212310) and can be found in the key resources table. All original code has been deposited at Zenodo and is publicly available as of the date of publication (Zenodo: https://doi.org/10.5281/zenodo.7026051) and is listed in the key resources table. Any additional information required to reanalyze the data reported in this paper will be shared by the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| SYK (D3Z1E) XP Rabbit Antibody | Cell Signaling Technology | Cat# 13198 RRID: AB_2687924 |
| StarBright Blue 700 Goat Anti-Rabbit IgG | Bio-Rad | Cat# 12004161 RRID: AB_2721073 |
| β-actin (D6A8) Rabbit mAb (HRP Conjugate) | Cell Signaling Technology | Cat# 12620 RRID: AB_2797972 |
| β-Amyloid (D54D2) XP Rabbit mAb | Cell Signaling Technology | Cat# 8243 RRID: AB_2797642 |
| Purified (azide-free) anti-beta-Amyloid, 1-16 | BioLegend | Cat# 803004 RRID: AB_2715854 |
| Iba1 antibody | Abcam | Cat# ab5076 RRID: AB_2224402 |
| Ki-67 Monoclonal Antibody (SolA15), Alexa Fluor 600, eBioscience | Thermo Fisher Scientific | Cat# 606-5698-80 RRID: AB_2896285 |
| Anti-mDectin-1-IgG | InvivoGen | Cat# mabg-mdect RRID: AB_2753143 |
| Anti-TMEM119 antibody [29–3] – Microglial marker | Abcam | Cat# ab209064 RRID: AB_2800343 |
| Rat Anti-Mouse CD68 Monoclonal antibody, Unconjugated, Clone FA-11 | Bio-Rad | Cat# MCA1957 RRID: AB_322219 |
| Amyloid beta precursor protein antibody [Y188] | Abcam | Cat# ab32136 RRID: AB_2289606 |
| Phospho-Tau (Ser202, Thr205) Monoclonal Antibody (AT8) | Thermo Fisher Scientific | Cat# MN1020 RRID: AB_223647 |
| Anti-NeuN | Millipore | Cat# MAB377 RRID: AB_2298772 |
| Mouse PDGF R alpha Antibody | R and D Systems | Cat# AF1062 RRID: AB_2236897 |
| Anti-APC (Ab-7) Mouse mAb (CC-1) | Millipore | Cat# OP80 RRID: AB_2057371 |
| Anti-Olig-2 Antibody | Millipore | Cat# AB9610 RRID: AB_570666 |
| Anti-Myelin Basic Protein | Millipore | Cat# AB5864 RRID: AB_2140351 |
| Rat Anti-Myelin Basic Protein Monoclonal Antibody, Unconjugated, Clone 12 | Abcam | Cat# ab7349 RRID: AB_305869 |
| Donkey anti-Rabbit IgG (H + L) Antibody, Alexa Fluor 488 Conjugated | Thermo Fisher Scientific | Cat# A-21206 RRID: AB_2535792 |
| Donkey anti-Rabbit IgG (H + L) Antibody, Alexa Fluor 594 Conjugated | Thermo Fisher Scientific | Cat# A-21207 RRID: AB_141637 |
| Donkey anti-Rabbit IgG (H + L) Antibody, Alexa Fluor 647 Conjugated | Thermo Fisher Scientific | Cat# A-31573 RRID: AB_2536183 |
| Donkey anti-mouse IgG (H + L) Antibody, Alexa Fluor 488 Conjugated | Thermo Fisher Scientific | Cat# A21202 RRID: AB_141607 |
| Donkey anti-mouse IgG (H + L) Antibody, Alexa Fluor 647 Conjugated | Thermo Fisher Scientific | Cat# A-31571 RRID: AB_162542 |
| Donkey anti-goat IgG (H + L) Antibody, Alexa Fluor 488 Conjugated | Thermo Fisher Scientific | Cat# A-11055 RRID: AB_2534102 |
| Donkey anti-goat IgG (H + L) Antibody, Alexa Fluor 546 Conjugated | Thermo Fisher Scientific | Cat# A-11056 RRID: AB_142628 |
| Donkey anti-goat IgG (H + L) Antibody, Alexa Fluor 647 Conjugated | Thermo Fisher Scientific | Cat# A-21447 RRID: AB_141844 |
| Donkey anti-rat IgG (H + L) Antibody, Alexa Fluor 594 Conjugated | Thermo Fisher Scientific | Cat# A-21209 RRID: AB_2535795 |
| Alexa Fluor 647-AffiniPure Donkey Anti-Rat IgG (H + L) | Jackson ImmunoResearch Labs | Cat# 712-605-153 RRID: AB_2340694 |
| CD11b Monoclonal Antibody (M1/70), APC, eBioscience | Thermo Fisher Scientific | Cat# 17-0112-82 RRID: AB_469343 |
| CD45 Monoclonal Antibody (30-F11), PE-Cyanine7, eBioscience | Thermo Fisher Scientific | Cat# 25-0451082 RRID: AB_2734986 |
| Brilliant Violet 510 anti-mouse TCR beta chain | BioLegened | Cat# 109234 RRID: AB_2562350 |
| F4/80 Monoclonal Antibody (BM8), APC, eBioscience | Thermo Fisher Scientific | Cat# 17-4801082 RRID: AB_2784648 |
| Ly-6G Monoclonal Antibody (1A8-Ly6g), FITC, eBioscience | Thermo Fisher Scientific | Cat# 11-9668-80 RRID: AB_2572531 |
| CD4 Monoclonal Antibody (RM4-5), FITC, eBioscience | Thermo Fisher Scientific | Cat# 11-0042-82 RRID: AB_464896 |
| CD8a Monoclonal Antibody (53-6.7), Alexa Fluor 700, eBioscience | Thermo Fisher Scientific | Cat# 56-0081-82 RRID: AB_494005 |
| CD80 (B7-1) Monoclonal Antibody (16-10A1), APC, eBioscience | Thermo Fisher Scientific | Cat# 17-0801-82 RRID: AB_469417 |
| CD11c Monoclonal Antibody (N418), PE, eBioscience | Thermo Fisher Scientific | Cat# 12-0114-83 RRID: AB_465553 |
| GM-CSF Monoclonal Antibody (MP1-22 × 109), PE, eBioscience | Thermo Fisher Scientific | Cat# 12-7331-82 RRID: AB_466205 |
| IFN gamma Monoclonal Antibody (XMG1.2), APC, eBioscience | Thermo Fisher Scientific | Cat# 17-7311-82 RRID: AB_469504 |
| IL-17A Monoclonal Antibody (eBio17B7), eFluor 450, eBioscience | Thermo Fisher Scientific | Cat# 48-7177-80; RRID: 11149677 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| Mycobacterium tuberculosis | Becton, Dickinson, & Company | Cat# 231141 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| MOG Peptide | Bio-Synthesis | Cat# 12668-01 |
| Complete Freund’s Adjuvant | Sigma-Aldrich | Cat# F5881 |
| Pertussis toxin | List Biological Laboratories | Cat# 180 |
| Cuprizone | Sigma-Aldrich | Cat# 14690 |
| cOmplete Protease Inhibitor Cocktail | Roche | Cat# 11697498001 |
| PhosSTOP | Roche | Cat# 4906845001 |
| Ponceau S stain | Sigma-Aldrich | Cat# P7170 |
| Tissue-Plus O.C.T. Compound | Fisher Scientific | Cat# 23-730-571 |
| T-PER Tissue Protein Extraction Reagent | Thermo Fisher Scientific | Cat# 78510 |
| Sucrose | Sigma-Aldrich | Cat# S0389 |
| Triton X-100 | Sigma-Aldrich | Cat# 93418 |
| DAPI | Sigma-Aldrich | Cat# D9542 |
| ThioflavinS | AAT Bioquest | Cat# 23059 |
| ProLongGold Antifade Mountant | Invitrogen | Cat# P36930 |
| Methoxy-X04 | ApexBio | Cat# B5769 |
| Dimethyl Sulfoxide Anhydrous (DMSO) | Sigma-Aldrich | Cat# 276855 |
| Hanks Buffer Saline Solution (HBSS) | Thermo Fisher Scientific | Cat# 14025092 |
| DNase I, Grade II, from bovine pancreas | Sigma-Aldrich | Cat# 10104159001 |
| Papain, Suspension | Worthington Biochemical Corporation | Cat# LS003124 |
| DMEM/F-12, no glutamine | Thermo Fisher Scientific | Cat# 21331020 |
| Fetal Bovine Serum (FBS) | Thermo Fisher Scientific | Cat# 10082147 |
| Antibiotic-Antimycotic | Thermo Fisher Scientific | Cat# 15240096 |
| Glutamax | Thermo Fisher Scientific | Cat# 35050061 |
| Percoll | Cytvia | Cat# 17-0891-02 |
| MACS BSA Stock Solution | Miltenyi Biotec | Cat# 130-0910376 |
| Beta Amyloid (1-42), human | California peptide | Cat# 641-15 |
| Hexafluoroisopropanol (HFIP) | Sigma-Aldrich | Cat# 52517 |
| CypHer5E-NHS ester | Cytvia | Cat# PA15401 |
| IMDM | Thermo Fisher Scientific | Cat# 12440053 |
| Penicillin/Streptomycin | Thermo Fisher Scientific | Cat# 15140163 |
| Recombinant Murine M-CSF | PeproT ech | Cat# 315-02 |
| Tideglusib | Selleck Chemicals | Cat# S2823 |
| DMEM | Thermo Fisher Scientific | Cat# 11885-084 |
| CD11b microbeads (microglia) | Miltenyi Biotec | Cat# 130-093-634 |
| CD90.2 microbeads | Miltenyi Biotec | Cat# 130-121-278 |
| CD11b beads (monocytes) | Miltenyi Biotec | Cat# 130-049-601 |
| Fixable Viability Dye | eBioscience | Cat# 65-0866-14 |
| BODIPY 581/591 C11 (Lipid Peroxidation Sensor) | Fisher Scientific | Cat# D3861 |
| Fc Block | eBiosciene | Cat# 14-0161-86 |
| L-glutamine | Thermo Fisher Scientific | Cat# 25030-081 |
| Beta-mercaptoethanol | Thermo Fisher Scientific | Cat# 21985-023 |
| PMA | Sigma-Aldrich | Cat# P1585 |
| Ionomycin | Sigma-Aldrich | Cat# I19657 |
| Monensin | eBioscience | Cat# 00-4505-51 |
| IC Fixation Buffer | eBioscience | Cat# 00-8222-49 |
| Permeabilization Buffer | eBioscience | Cat# 00-8333-56 |
| 10% Formalin | Azer Scientific | Cat# NBF |
| Luxol Fast Blue | Thermo Fisher Scientific | Cat# 212170250 |
| Lithium Carbonate | Thermo Fisher Scientific | Cat# 446322500 |
| Hematoxylin | Sigma-Aldrich | Cat# HHS128 |
| ACK Lysis Buffer | Quality Biological | Cat# 118-156-101 |
| TRIzol | Life Technologies | Cat# 15596018 |
| Chloroform | Fisher Scientific | Cat# BP1145-1 |
| Isopropanol | Sigma-Aldrich | Cat# I9516 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Pierce 660nm Protein Assay Kit | Thermo Fisher Scientific | Cat# 22660 |
| Human/Mouse AKT Pathway Phosphorylation Array C1 | RayBiotech | Cat# AAH-AKT-1-8 |
| In Situ Cell Death Detection Kit, Fluorescein | Roche | Cat# 11684795910 |
| Amyloid beta 40 Mouse ELISA Kit | Thermo Fisher Scientific | Cat# KMB3481 |
| Amyloid beta 42 Mouse ELISA Kit | Thermo Fisher Scientific | Cat# KMB3441 |
| CellROX Deep Red Flow Cytometry Assay Kit | Thermo Fisher Scientific | Cat# C10491 |
| Bio-Rad Bio-Plex Pro Reagent Kit | Bio-Rad | Cat # 171-304070M |
| RNeasy Micro Kit | Qiagen | Cat# 74004 |
| Sensifast cDNA Synthesis Kit | Bioline | Cat# BIO-65054 |
| Sensifast Probe No-ROX Kit | Bioline | Cat# BIO-86005 |
|
| ||
| Deposited data | ||
|
| ||
| Gene expression data for replication analysis | This paper | GEO: GSE212310 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE212310 |
| Original code | This paper | Zenodo:https://zenodo.org/record/7026051#.YwkJKi-B3_Q |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: 5xFAD; B6SJL-Tg(APPSwFlLon, PSEN 1*M146L*L286V)6799Vas/Mmjax | The Jackson Laboratory | Jax Stock no. 034840-JAX |
| Mouse: B6.129P2-Syktm1.2Tara/J | The Jackson Laboratory | Jax Stock no. 017309-JAX |
| Mouse: B6.129P2(C)-Cx3cr1tm2.1(cre/ERT2)Jung/J | The Jackson Laboratory | Jax stock no. 020940-JAX |
| Mouse: B6.Cg-Gt(ROSA)26Sortm6(CAG-Zsgreen1)Hze/J | The Jackson Laboratory | Jax stock no. 007906-JAX |
|
| ||
| Oligonucleotides | ||
|
| ||
| qPCR primer: Sykb | Thermo Fisher Scientific | Mm01333035_m1 |
| qPCR primer: Gapdh | Thermo Fisher Scientific | Mm99999915_g1 |
|
| ||
| Software and algorithms | ||
|
| ||
| GraphPad Prism 9 | GraphPad | RRID: SCR_002798 |
| Imaris 9.5.1 | Imaris | RRID: SCR_007370 |
| Adobe Photoshop | Adobe | https://www.adobe.com/products/photoshop/ |
| BioRender | BioRender | RRID: SCR_018361 |
| LAS AF Software | Leica | https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/ |
| Fiji/ImageJ Software | Fiji | RRID: SCR_002285 |
| Noldus Ethovision XT Software | Noldus | https://www.noldus.com/ethovision-xt |
| Kaluza Acquisition Software | Beckman Coulter | https://www.beckman.com/flow-cytometry/software/kaluza |
| FlowJo Software | FloJo | RRID:SCR_008520 |
| Bio-Plex Manager software | Bio-Rad | https://www.bio-rad.com/en-us/product/bio-plex-200-systems |
| R Studio (V4.0.5) | https://rstudio.com/ | RRID:SCR_001905 |
| R | https://www.r-project.org | RRID:SCR_001905 |
| DESeq2 (v1.32.0) | N/A | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Cell Ranger (V1.3.1) | N/A | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/installation |
| Seurat (v4.0.2) | https://www.r-project.org/ | RRID:SCR_016341 |
| ToppCluster | Cincinnati Children's | https://toppcluster.cchmc.org |
| Monocle (v0.2.3.0) | N/A | N/A |
|
| ||
| Other | ||
|
| ||
| Leica TCS SP8 Confocal Microscope | Leica Microsystems | N/A |
| Mini-PROTEAN TGX Stain-Free Protein Gel | Bio-Rad | Cat# 4568093 |
| Mini-PROTEAN Tetra Cell | Bio-Rad | Cat# 1620264 |
| Trans-Blot Turbo Transfer System | Bio-Rad | Cat# 1704150 |
| Bio-Spin P-6 gel Columns, Tris Buffer | Bio-Rad | Cat# 7326227 |
| Mouse stereotaxic Frame | Stoelting | Cat# 51730U |
| Nanoliter Injector | World Precision Instruments | Cat# NL2010MC2T |
| LS columns | Miltenyi Biotec | Cat# 130-042-401 |
| QuadroMACS magnet | Miltenyi Biotec | Cat# 130-091-051 |
| Gallios Flow Cytometer - Navios System | Beckman Coulter | Cat# B83535 |
| Bio-Plex 200 System | Bio-Rad | https://www.bio-rad.com/en-us/product/bio-plex-200-systems |
| Keyence BZ-X810 Microscope | Keyence | https://www.keyence.com/landing/lpc/all-in-one-fluorescence-microscope.jsp |
| Cellometer Auto 2000 | Nexelcom Bioscience | https://www.nexcelom.com/nexcelom-products/cellometer-fluorescent-viability-cell-counters/cellometer-auto-2000/ |
| NanoDrop 2000 Spectrophotometer | Thermo Fisher Scientific | https://www.thermofisher.com/order/catalog/product/ND-2000 |
| CFX384 Real-Time PCR System | Bio-Rad | Cat# 1855484 |
| Influx Cell Sorter | BD | https://med.virginia.edu/flow-cytometry-facility/equipment/influx-cell-sorter/ |
| GENEWIZ Next Generation Sequencing | Azenta | https://www.genewiz.com/en/Public/Services/Sanger-Sequencing |
| ChemiDoc MP Imaging System | Bio-Rad | Cat# 12003154 |


