Abstract
GBHs are the most widely used herbicides for weed control worldwide that potentially affect microorganisms, but the role of their sublethal exposure in the development of antibiotic resistance of Pseudomonas aeruginosa is still not fully investigated. Here, the effects of glyphosate acid (GLY), five glyphosate-based herbicides (GBHs), and POE(15), a formerly used co-formulant, on susceptibility to imipenem, a potent carbapenem-type antibiotic, in one clinical and four non-clinical environmental P. aeruginosa isolates were studied. Both pre-exposure in broth culture and co-exposure in solid media of the examined P. aeruginosa strains with 0.5% GBHs resulted in a decreased susceptibility to imipenem, while other carbapenems (doripenem and meropenem) retained their effectiveness. Additionally, the microdilution chequerboard method was used to examine additive/antagonistic/synergistic effects between GLY/POE(15)/GBHs and imipenem by determining the fractional inhibitory concentration (FIC) indexes. Based on the FIC index values, glyphosate acid and Total demonstrated a potent antagonistic effect in all P. aeruginosa strains. Dominator Extra 608 SL and Fozat 480 reduced the activity of imipenem in only one strain (ATCC10145), while POE(15) and three other GBHs did not have any effect on susceptibility to imipenem. Considering the simultaneous presence of GBHs and imipenem in various environmental niches, the detected interactions between these chemicals may affect microbial communities. The mechanisms of the glyphosate and GBH-induced imipenem resistance in P. aeruginosa are yet to be investigated.
Subject terms: Applied microbiology, Environmental impact, Outcomes research
Introduction
Glyphosate acid (GLY), usually formulated in form of salts1, is the most widely used active ingredient in glyphosate-based herbicides (GBH) with an annual volume of 700,000 tons2. GLY interrupts the metabolic pathway of shikimic acid, the precursor for the biosynthesis of aromatic amino acids in plants, by inhibiting the enzyme that catalyzes the formation of its central intermediate, 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS)3.
GLY and GBH use has been associated with adverse effects (cytotoxicity, carcinogenicity, teratogenicity, endocrine disruption, metabolic alterations) below the environmentally relevant concentrations or the recommended agricultural/horticultural usage doses4–6. Still, numerous GBHs are available on the worldwide pesticide market7 that may affect non-target organisms in aquatic-, and terrestrial ecosystems or threaten human health.
The shikimate pathway is present not only in the plants, but in bacteria and fungi as well, therefore, GLY can be considered as an antimicrobial agent4. GLY sensitivity partially depends on the class of EPSPS enzyme (class I, or class II) harbored by the microorganism: some Proteobacteria harboring the Class I EPSPS variant (E. coli, Salmonella, and species of Pseudomonas) may have higher GLY sensitivity8. Novel studies demonstrate the adverse effects of GLY and GBHs on environmental communities leading to changes in freshwater microbial communities9, influencing their activities in biogeochemistry cycles10, disturbing earthworms' gut microbiome11, and inducing microbiota dysbiosis in humans12,13 and in male Sprague–Dawley rats14. When it comes to assessing the influence of GLY, many studies focused on changes in soil and aquatic microbial community15–21 and verified that various microorganisms (especially bacteria) can use GLY as a sole source of phosphorus, carbon and nitrogen22.
Antibiotic resistance is a global, emerging problem that threatens human and animal health and raises environmental health concerns23 defined as an inherited ability of microorganisms to grow at high concentrations of an antibiotic, irrespective of the duration of treatment24. Over the last decades, the evolution of antibiotic resistance in pathogenic microorganisms correlated not only with the excessive antibiotic consumption but with the usage of biocides and commercial formulations of pesticides (e.g., 2,4-dichlorophenoxyacetic acid, dicamba, GLY)25–27. The association between antibiotic resistance of microorganisms and their GLY resistance was suggested by several authors28,29 and under the exposure to GLY isopropylamine (IPA) salt and Roundup, minimum inhibitory concentrations (MICs) of antibiotics in Escherichia coli and Salmonella enterica serovar Typhimurium isolates altered leading to antibiotic resistance phenotypes26. Exposure to sublethal concentrations of GLY may affect antibiotic susceptibility in E. coli and S. enterica as well30. Moreover, GLY can promote horizontal plasmid-mediated conjugative gene transfer of antibiotic resistance genes (ARGs) at 10 and 20 mg/L concentrations due to the increased cell membrane permeability in intragenic (E. coli) and intergenic (from E. coli to Bacillus oleronius) species31.
The correlation between the emergence of antibiotic-resistant bacteria and worldwide GLY use was suggested by several authors4,27. However, there is still a lack of knowledge about the effects of GLY and GBH exposure on the antibiotic resistance of Pseudomonas aeruginosa, a bacterium often found in locations associated with human activity32. P. aeruginosa is a frequent healthcare-associated pathogen with an outstanding capacity for antimicrobial resistance33, leading to the highest fatality rate of all opportunistic pathogen Gram-negative bacteria34. Due to their exceptional metabolic plasticity, P. aeruginosa strains are detectable in diverse ecological niches35 and can dominate contaminated sites36 with drug resistance, virulence, and biofilm-forming ability similar to their clinical counterparts37–39. P. aeruginosa can tolerate27 and degrade40 GLY, but the effects of sublethal exposure on its antibiotic resistance profile have not been investigated. To fill this gap, the initial aim of our research was to expose environmental and clinical isolates of opportunistic pathogen P. aeruginosa to sublethal concentrations of GLY and commercially available GBHs and to evaluate the induced alterations in the phenotypically detectable antibiotic resistance profiles. Based on the initial findings, our additional aim was to reveal the types of relationship (antagonism, synergism) between five chosen GBH formulations with different GLY salts and the formerly used co-formulant POE(15) (polyoxyethylene (15) tallow amine) with imipenem, a potent, carbapenem-type cell wall synthesis inhibitor41.
Materials and methods
Microorganisms
The examined five P. aeruginosa strains were chosen to represent various clinical and environmental sources (Table 1). Type strains were obtained from the National Collection of Agricultural and Industrial Microorganisms (NCAIM), Hungary, environmental strains were chosen from the strain collection of the Department of Environmental Safety (MATE), Hungary. All examined strains represented antibiotic sensitive phenotypes (Table S1). Species level identification performed with the PCR targeted the 16S rDNA variable regions V2 and V8 as it was described previously42.
Table 1.
P. aeruginosa strains used in this study.
| Strain | Species | Source |
|---|---|---|
| HF234 (this study) | Pseudomonas aeruginosa | Surface water, Hungary |
| P6637 | Pseudomonas aeruginosa | Hydrocarbon contaminated groundwater, Hungary |
| ATCC 2785343 | Pseudomonas aeruginosa | Clinical (isolated from blood) |
| ATCC 1014544 | Pseudomonas aeruginosa | Type strain, unknown source |
| ATCC 1544245 | Pseudomonas aeruginosa | Water bottle in animal room |
Glyphosate, POE(15) and GBH formulations
The stock solution of GLY [(N-(phosphonomethyl) glycine) with chemical formula: C3H8NO5P, CAS 1071-83-6] was prepared from Pestanal analytical standard (Merck Ltd., Germany). POE(15), with an average of 15 ethylene oxide groups (chemical formula: R-N(CH2CH2O)m-H (CH2CH2O)n-H, CAS 61791-26-2) with 100% purity was purchased from Greyhound Chromatography and Allied Chemicals. The examined commercially available GBHs, their declared GLY concentrations, and co-formulants are summarized in Table 2.
Table 2.
Chemical properties of the examined water soluble concentrates of glyphosate-based herbicides (GBHs) examined in this study based on their Material Safety Data Sheets (MSDS).
| Formulation | Declared glyphosate concentration (g/L) | Type of glyphosate salt | Co-formulant(s) % (w/w) | Expiration date of approval in Hungary | |
|---|---|---|---|---|---|
| Glyphosate acid (active substance) | Glyphosate salt | ||||
| Dominator Extra 608 SL | 480 | 608 | Dimethylamine (DMA) salt |
d-Glucopyranose, oligomers, decyl octyl glycosides (< 5%), disodium cocoamphodipropionate (< 5%) Methyl alcohol (< 1%) |
31.12.2025 |
| Fozat 480 | 360 | 480 | Isopropylamine (IPA) salt | Non-declared | 31.12.2023 |
| Gladiator 480 SL | 360 | 486 | Isopropylamine (IPA) salt | Polyethoxylated (15) tallow amine [POE(15)] (13–18%) | 30.11.2016 |
| Roundup Mega | 450 | 551 | Potassium (P) salt | Ethoxylated ether alkylamine (7%) | 31.12.2025 |
| Total | 360 | 486 | Isopropylamine (IPA) salt | Non-declared (inert substance) | 31.12.2025 |
Preliminary screening assay
Preliminary experiments aimed to examine the effects of five commercially available GBHs (Table 2) on the growth and antibiotic resistance profile of a representative isolate of P. aeruginosa (HF234) originating from surface water, Hungary (Table 1). 45 mL Luria–Bertani (LB) medium (10.0 g tryptone, 5.0 g yeast extract, and 9.0 g NaCl in 1000 mL distilled water) was inoculated with 5.0 mL overnight bacterial suspension calibrated to an optical density of OD600 = 0.6 ± 0.02. The medium was supplemented with GBHs to reach a final GBH concentration of 0.5% (v/v), equivalent to 1.8–2.8 g/L GLY acid depending on the type of formulation. This concentration falls within the recommended dilution range of GBHs [0.2–3.5% (v/v)] for agricultural and household use and similar to those found in water after agricultural practices46. The experiment was performed in triplicates with an incubation period of 72 h at 28 °C on a horizontal shaker. During incubation, growth curves were determined based on regularly measured OD600 values to verify the sublethal effect of the chosen concentration. The antibiotic resistance assay was performed at the start (0 h) and endpoints (72 h) of the experiment using GBHs free Mueller–Hinton agar (Merck Ltd., Germany). Antibiotic resistance was examined with Liofilchem MIC test strips to determine Minimal Inhibitory Concentrations (MICs) according to the recommendations of the European Committee on Antimicrobial Susceptibility Testing (24 h incubation at 35 °C)47. The tested antibiotic agents represented six groups of antibiotics (Table S1). MIC differences between non-treated and pre-exposed cultures were analysed with a Two-way ANOVA multiple comparison (Tukey) test.
To evaluate the effect of co-exposure, preliminary assay was repeated with three GBHs (Roundup Mega, Dominator Extra 608 SL, Gladiator 480 SL) that significantly modified MICs of imipenem after 72 h pre-exposure. The overnight cultures of five environmental and clinical reference strains of P. aeruginosa (listed in Table 1) were involved in this setting. Bacterial suspensions were used without pre-exposition and were spread directly onto Mueller–Hinton agar containing 0.5% (v/v) GBHs. Antibiotic resistance test was performed as it was described above.
Microplate dilution assay
Based on the results of the preliminary assays, the imipenem-resistance inducing effect of GBHs, GLY, and its formerly used co-formulant POE(15) was examined further by the microdilution chequerboard method 48 with slight modifications. Assays were performed on 96-well, clear, U-shaped PS microplates (Greiner Bio-One GmbH, Austria) using two-fold dilution series of 2 g/L imipenem (CAS 74431-23-5, Supelco, Sigma-Aldrich Ltd.), 10.0 g/L GLY, 5 g/L POE(15) and 50% (v/v) GBH stock solutions prepared in sterile distilled water. Test materials were co-added to the x-, and y-axes across the plate to reach 0–64 mg/L imipenem (x-axis) and 0–800 mg/L GLY, 0–4 mg/L POE(15), 0–4% (v/v) GBH (y-axis) final concentrations, respectively. Controls containing only GLY/POE/GBH or imipenem have been carried out. Concentration ranges considered the water-solubility of the examined compounds.
50 µL of overnight P. aeruginosa bacterial suspensions with optical density of OD600 = 0.6 ± 0.02 were added to each well. The final volume was adjusted to 250 µL with LB broth medium. 200 µL LB supplemented with 50 µL bacterial suspension was used as negative control.
Assays were carried out in triplicates using freshly prepared solutions and bacterial suspensions. Plates were incubated at 28 °C and at a speed of 350 rpm in a microplate shaker thermostat (PST-60HL-4, BioSan, Latvia). Absorbance was measured by an ELx800 microplate reader at 550 nm at the beginning of the incubation (0 h) and after 24 h of exposure.
Data analysis
Statistical data analysis and visualization were performed using GraphPad Prism 7 software, version 7.00 (GraphPad Software Inc., San Diego, USA). Replicate measurements of microplate assays were averaged prior to statistical evaluation. Absorbance data were expressed in mean values for heatmap visualization. To examine the additive/antagonistic/synergistic effects between imipenem and the examined glyphosate related chemicals, fractional inhibitory concentration index (FICI) was calculated using the following mathematical expression49:
MIC is the minimal inhibitory concentration; FIC is the fractional inhibitory concentration; FICI is the fractional inhibitory concentration index.
FIC index of < 0.5 indicates synergism, 0.5–4.0 means indifference, and ≥ 4.0 is considered as antagonism50.
To calculate the differences between GLY/POE(15)/GBHs and imipenem, the average absorbance values of the examined five strains under co-exposition and their respective imipenem control (samples containing bacterial suspension and imipenem) were compared. The analysis of variance was performed with a two-way ANOVA, followed by Dunnett’s multiple comparisons tests. Differences with p ≤ 0.05 were considered significant.
Results
Preliminary screening assay: pre-exposure with GBHs increases MIC of imipenem
According to our preliminary screening results, the HF234 isolate of P. aeruginosa could tolerate 0.5% (v/v) of Roundup Mega, Dominator Extra 608 SL, Gladiator 480 SL, Total, and Fozat 480 without any significant inhibitory effect (Fig. 1A). The applied 0.5% GBH pre-exposure caused a 1.5–2 fold increase in MIC values of piperacillin and gentamicin, but the detected changes were not statistically significant (Fig. 1B). At the same time, MIC values of imipenem showed a 2–32-fold increase (Fig. 1B,C), while the other examined carbapenems (meropenem and doripenem) retained their effectiveness. According to Tukey’s multiple comparison test, Dominator Extra 608 SL Gladiator 480 SL and Roundup Mega pre-exposure induced a significantly higher level of resistance against imipenem.
Figure 1.

Preliminary liquid broth assay of P. aeruginosa HF234 with 0.5% (v/v) GBH pre-exposure (averaged values of three technical replicates). (A) Optical density (OD600) of broth cultures during the 72 h pre-exposure with GBHs. (B) MIC values of HF234 cultures after 72 h of pre-exposure with GBHs, plated on GBHs-free agar plates. (C) Inhibition zones of HF234 cultures after 72 h of pre-exposure with GBHs, plated on GBHs-free agar plates. *,**Significantly different from the non-treated strain (p = 0.0332–0.0023).
Preliminary screening assay: co-exposure with GBHs increases MIC of imipenem
The repeated imipenem assay, extended to five P. aeruginosa strains (listed in Table 1.), verified, that Dominator Extra 608 SL and Gladiator 480 SL induce similar levels of MIC alterations in the case of imipenem without pre-exposure if 0.5% (v/v) of GBHs are mixed directly into the Mueller–Hinton medium (MIC values ranged between 16 and 32 mg/L) (Table S2).
Drug interactions in FIC assay: GBHs have an antagonistic effect on imipenem
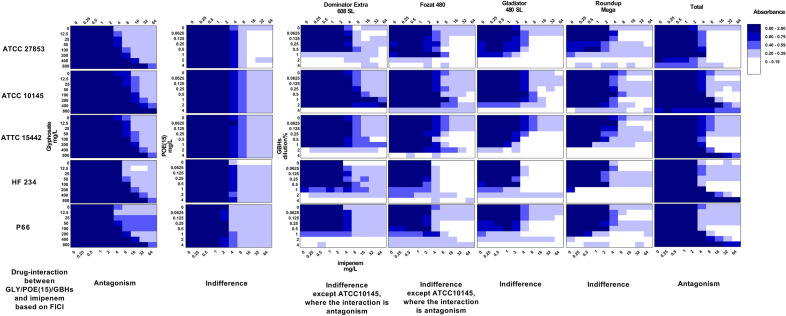
Averaged values of absorbance determined by microplate chequerboard test of the examined five P. aeruginosa strains exposed to both imipenem and GLY/POE(15)/GBHs are summarized in Fig. 2.
Figure 2.
Heatmap of averaged absorbance values of the examined P. aeruginosa strains under co-exposure with different concentrations of GLY/POE(15)/GBHs and imipenem. X-axis: imipenem concentrations (0–64 mg/L), y-axis: concentrations of test materials (GLY: 0–800 mg/L, POE(15): 0–4 mg/L, GBHs: 0–4% (v/v)). Blue bars indicate relative fold growth 51. Drug-interactions between GLY/POE(15)/GBHs and imipenem were determined by the FIC index calculated as described in the methods (see details in Table 3).
Results of FIC index determinations are available in Table 3. Summarized average percental differences in absorbance of P. aeruginosa strains after co-exposition to GLY/POE(15)/GBHs and imipenem compared to solo imipenem exposition are presented in Fig. 3.
Table 3.
Fractional inhibitory concentration index (FICI) determination of P. aeruginosa strains co-exposed to imipenem and glyphosate related chemicals (glyphosate acid, POE(15), glyphosate-based herbicides).
| Imipenem (mg/L) | Imipenem (mg/L) | Imipenem (mg/L) | Imipenem (mg/L) | Imipenem (mg/L) | Imipenem (mg/L) | Imipenem (mg/L) | |||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MICA | MICA | MICA | MICA | MICA | MICA | MICA | |||||||||||||||||||||||||||||
| Glyphosate acid (mg/L) | POE(15) (mg/L) | Dominator extra 608 SL, % (v/v) | Fozat 480, % (v/v) | Gladiator 480 SL, % (v/v) | Roundup mega, % (v/v) | Total, % (v/v) | |||||||||||||||||||||||||||||
| MICB | MICB | MICB | MICB | MICB | MICB | MICB | |||||||||||||||||||||||||||||
| ATCC27853 | ATCC10145 | ATCC15442 | HF234 | P66 | ATCC27853 | ATCC10145 | ATCC15442 | HF234 | P66 | ATCC27853 | ATCC10145 | ATCC15442 | HF234 | P66 | ATCC27853 | ATCC10145 | ATCC15442 | HF234 | P66 | ATCC27853 | ATCC10145 | ATCC15442 | HF234 | P66 | ATCC27853 | ATCC10145 | ATCC15442 | HF234 | P66 | ATCC27853 | ATCC10145 | ATCC15442 | HF234 | P66 | |
| MICA alone | 2 | 2 | 2 | 4 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 2 | 2 | 2 |
| MICA (in the presence of B) | 32 | > 64 | 32 | 32 | 32 | 2 | 2 | 4 | 2 | 1 | 4 | > 64 | 4 | 4 | 2 | 4 | 8 | 2 | 2 | 2 | 2 | 4 | 2 | 2 | 2 | 2 | 4 | 4 | 2 | 2 | 4 | 16 | 16 | > 64 | 16 |
| FICA | 16 | 32 | 16 | 8 | 16 | 1 | 1 | 2 | 1 | 0.5 | 2 | 32 | 2 | 2 | 1 | 2 | 4 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 4 | 8 | 8 | 32 | 8 |
| MICB alone | > 800 | > 800 | > 800 | > 800 | > 800 | > 4 | > 4 | > 4 | > 4 | > 4 | 0.5 | 2 | 1 | 0.5 | 0.5 | 0.5 | 2.0 | 2.0 | 0.5 | 0.25 | 0.125 | 1.0 | 1.0 | 0.25 | 0.125 | 0.125 | 2.0 | 1.0 | 0.5 | 1,0 | 2.0 | 2.0 | > 4.0 | > 4.0 | > 4.0 |
| MICB (in the presence of A) | > 800 | > 800 | > 800 | > 800 | > 800 | > 4 | > 4 | > 4 | > 4 | > 4 | 0.0625 | 1.0 | 0.0625 | 0.0625 | 0.0625 | 0.0625 | 0.0625 | 0.0625 | 0.0625 | 0.0625 | 0.0625 | 0.0625 | 0.0625 | 0.0625 | 0.0625 | 0.0625 | 0.0625 | 0.0625 | 0.0625 | 0.0625 | 0.0625 | 0.0625 | 0.0625 | 4.0 | 0.0625 |
| FICB | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.125 | 0.5 | 0.0625 | 0.125 | 0.125 | 0.125 | 0.0313 | 0.0313 | 0.125 | 0.25 | 0.5 | 0.0625 | 0.0625 | 0.25 | 0.5 | 0.5 | 0.0313 | 0.0625 | 0.125 | 0.0625 | 0.0313 | 0.0313 | 0.0156 | 1.0 | 0.0156 |
| FICI | 17.0 | 33.0 | 17.0 | 9.0 | 17.0 | 2.0 | 2.0 | 3.0 | 2.0 | 1.5 | 2.125 | 32.5 | 2.062 | 2.125 | 1.125 | 2.125 | 4.0313 | 1.0313 | 1.125 | 1.25 | 2.5 | 2.0625 | 1.0625 | 1.25 | 1.5 | 1.5 | 2.0313 | 2.0625 | 1.125 | 1.0625 | 4.0313 | 8.0313 | 8.0156 | 33.0 | 8.0156 |
| FICI (mean) | 18.6 | 2.1 | 1.9 (mean of four strains) | 1.9 (mean of four strains) | 1.6 | 1.5 | 12.8 | ||||||||||||||||||||||||||||
| FICI evaluation | Antagonism | Indifference |
Indifference Except ATCC10145, where FICI = 32.5–antagonism |
Indifference Except ATCC10145, where FICI = 4.03–antagonism |
Indifference | Indifference | Antagonism | ||||||||||||||||||||||||||||
FICI < 0.5—synergy; FICI = 0.5–4.0—indifference; FICI ≥ 4.0—antagonism.
Figure 3.
Summarized average differences of absorbance values (%) of all the examined five P. aeruginosa strains under co-exposition to imipenem with GLY, POE(15), and GBHs, compared with the absorbance values of solo imipenem exposition. X-axis: imipenem concentrations (0.25–64 mg/L) and seven concentrations of each test material (GLY: 12.5–800 mg/L, POE(15): 0.0625–4.0 mg/L, GBHs: 0.0625–4.0% (v/v)). Positive range—intensification; negative range—inhibition. Each bar represents the average absorbance value of five P. aeruginosa strains co-exposed to imipenem and test materials in three independent experiments. Two-way ANOVA and Dunnett’s multiple comparisons tests were used. *, **, *** and ****Significantly different from solo imipenem values (p < 0.0332, p < 0.0021, p < 0.0002 and p < 0.0001, respectively).
According to our results, glyphosate acid was not toxic to P. aeruginosa in the examined range (12.5–800 mg/L), but it had a strong antagonistic effect (FICImean = 18.6) on imipenem and induced a concentration-dependent resistance in all strains; MIC of imipenem increased from 1–3 mg/L up to 32–64 mg/L, respectively. The strongest antagonistic effect was detected in the case of P. aeruginosa ATTC 10145: the viability of the culture in the simultaneous presence of 800 mg/L GLY and 64 mg/L imipenem was identical to that of the untreated control (Fig. 2).
POE(15) exposition had no concentration-dependent effect on the growth and imipenem sensitivity of the examined P. aeruginosa strains, but a slight increase in absorbance (up to 50%) was observed during co-exposition with 4 mg/L imipenem.
Of the five GBHs microplate assay verified that Total was not only non-toxic to the examined P. aeruginosa strains in the tested concentrations, but induced resistance towards imipenem: absorbance during co-exposition increased up to 192%, while MIC values of imipenem have significantly increased to 4–64 mg/L (Fig. 3, Table 3). FIC indexes verified the antagonistic effect between Total and imipenem (FICI was 12.8) in all strains. Dominator Extra 608 SL and Fozat 480 decreased susceptibility to imipenem in only strain ATCC10145 (FICIs were 32.5 and 4.03, respectively, indicating antagonism), while the interactions were indifferent in all the other strains (Table 3). The least pronounced effect of co-exposure was detected in the case of clinical reference strain ATTC 27853 with Total and P66 with Dominator Extra 608 SL, leading to only a slight increase (from 1 to 4 mg/L) in imipenem tolerance.
Gladiator 480 SL, Fozat 480, and Roundup Mega led to significant decreases in absorbance at higher [2–4% (v/v)] concentrations of GBHs, equivalent to 7.2–19.2 g/L glyphosate acid, verifying their cytotoxic effect on the examined strains (Figs. 2 and 3). At lower concentrations [0.0625–1.0% (v/v)], the above mentioned GBHs slightly increased imipenem tolerance compared to the solo exposure to the antibiotic, but the differences were significant in the case of Roundup Mega and Fozat 480 co-exposed with 4–8 mg/L imipenem only.
According to our results, the more pronounced antagonistic effect of Dominator Extra 608 SL and Total on imipenem can be related to their lower cytotoxicity to P. aeruginosa: their cytotoxicity was not significant even at higher concentrations [2.0–4.0% (v/v) equivalent to 7.2–14.4 g/L glyphosate acid]. Therefore, they could stimulate a more pronounced resistance to imipenem (up to 64 mg/L) in P. aeruginosa.
Discussion
GLY, the most frequently used herbicide worldwide, is the focus of scientific and public discussions, especially in the European Union, where the renewing of its approval is approaching (currently approved in the EU until 15 December 2022). Recently, besides examining the acute and chronic biological effects (cytotoxicity, carcinogenicity, teratogenicity, endocrine disruption) of GLY4–6 and some GBHs52,53, the focus has been on the occurrence of antibiotic resistance evoked by GLY in soil and bacterioplankton communities54,55, E. coli56, and Salmonella strains26,57. At the same time, other pathogenic bacteria like P. aeruginosa, with a high capacity to tolerate and utilize GLY as the sole P or N source leading to its complete degradation within 96h40, have not been investigated from the antibiotic resistance point of view27.
We demonstrated for the first time that GLY acid significantly decreases susceptibility to imipenem in P. aeruginosa strains: with microplate chequerboard method we proved that all five examined isolates had a higher level of imipenem resistance in a concentration-dependent manner when they were exposed to GLY, but not to POE(15). Regarding GBHs, the examined four environmental and one clinical P. aeruginosa strain had a GBH tolerance up to 1–4% (v/v) (equivalent to 3.6–14.4 g/L GLY acid depending on the type of formulation), which are within the range of the commonly recommended doses for agricultural and horticultural use. The phenotypic imipenem resistance of GBH co-exposed P. aeruginosa strains was detectable at a concentration range of 4–64 mg/L, while the MIC breakpoint of imipenem resistance is 4 mg/L58. Out of five examined GBHs, the freely available, POE(15)-free, IPA salt containing Total caused pronounced increase in MICs of all strains during co-exposure with imipenem, while Fozat 480 and Dominator Extra 608 SL (containing IPA and DMA salt, respectively) decreased the efficiency of imipenem in only type strain ATCC 10145. Therefore, the type of GLY salt used for formulation was not a determining factor of the detected alteration in susceptibility to imipenem.
The nonheritable resistance to antibiotics in E. coli induced by salicylates and other chemotactic repellents59, the kanamycin resistance of E. coli in the presence of GLY26 and the kanamycin/ciprofloxacin resistance of S. Typhimurium induced by the exposure to Roundup26 were previously reported. The imipenem resistance of P. aeruginosa induced by GLY and GBHs has a similar pattern and rapidity: therefore, it may be activated by an efflux or permeability-related mechanism as it was suggested in the case of E. coli26, or by the activation of general mechanisms against potential stressors, which does not necessarily require a change in the specifically targeted structures60.
In clinical settings, carbapenem resistance of P. aeruginosa has been divided into three phenotypes: imipenem resistant-meropenem susceptible type I (IRMS), meropenem resistant-imipenem susceptible type II (MRIS), and imipenem resistant-meropenem resistant type III (IRMR). It has been described that IRMS is primarily caused by the numerous different mutations observed across various loops in the oprD porin leading to the porin down regulation61. At the same time, MRIS is due to the over-expression of the mexAB-oprM efflux operon in clinical P. aeruginosa strains, while strains harbouring plasmid-mediated carbapenemase genes usually belong to the type III (IRMR) group61. In our study, we have discovered phenotypic imipenem resistance in parallel with meropenem and doripenem susceptibility in both clinical and environmental P. aeruginosa strains after co-exposure to antibiotics and GBHs, therefore, we presume, that the detected changes can be related to the porin regulation of oprD as it was described in the case of IRMS type strains61 or to other general mechanisms against stressors60. The molecular mechanisms of the glyphosate and GBH-induced imipenem resistance in P. aeruginosa are yet to be investigated.
Conclusions
Over the years, many studies have proved the harmful biological effects of GLY and GBHs or co-formulants used in GBHs. We report for the first time that GLY acid and freely available GBHs (containing a variety of co-formulants, but free of POE(15) induce significant, phenotypically detectable, discrepant imipenem resistance in clinical and environmental P. aeruginosa strains, while POE(15), the formerly used and banned co-formulant does not affect imipenem sensitivity. Considering the worldwide use of GLY and GBHs, and the simultaneous emergence of antibiotic-resistant bacteria in environmental matrices, the detected interactions between these chemicals may affect microbial communities and possess a potential environmental- and human health risk. Exploring the underlying mechanisms of this phenomenon is essential for further risk management.
Data availability
The datasets generated during and/or analysed during the current study are available as Supplementary data. Further supplementary information is available from the corresponding author on reasonable request.
Supplementary Information
Author contributions
All authors contributed to the study conception and design. J.H.: Investigation, Methodology, Visualization, Writing—original draft, writing—reviewing and editing, B.K.: Funding acquisition, Project administration, Resources, Supervision, Reviewing and editing, G.T.: Investigation, Writing—original draft, writing—reviewing and editing, D.J.: Investigation, Data analysis, Writing—original draft, M.F.: Investigation, I.S.: Data curation, Supervision, B.G.: Investigation, B.U.: Funding acquisition, Resources, Supervision, S.S.: Conceptualization, Data curation, Funding acquisition, Resources, Supervision, E.K.: Conceptualization, Investigation, Methodology, Visualization, Writing—original draft, writing—reviewing and editing.
Funding
Open access funding provided by Hungarian University of Agriculture and Life Sciences. This research was supported by 2020-1.1.2-PIACI-KFI-2021-00239 provided by the Ministry of Innovation and Technology of Hungary from the National Research, Development and Innovation Fund, financed under the PIACI KFI funding scheme, by the Ministry of Innovation and Technology within the framework of the Thematic Excellence Programme 2020, National Challenges Subprogramme (TKP2020-NKA-16), and by the ÚNKP-21-4 (J. Háhn) New National Excellence Program of the Ministry for Innovation and Technology from the source of the National Research, Development and Innovation Fund. The scientific work of E. Kaszab was supported by the János Bolyai Research Grant of the Hungarian Academy of Sciences (BO/00236/20/4), D. Jiang was supported by the Stipendium Hungaricum scholarship.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Judit Háhn, Balázs Kriszt, Sándor Szoboszlay and Edit Kaszab.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-23117-9.
References
- 1.Székács, A. & Darvas, B. Forty years with glyphosate. in Herbicides—Properties,SynthesisandControlofWeeds. 247–284. (IntechOpen, 2012).
- 2.Maggi F, Tang FHM, la Cecilia D, McBratney A. PEST-CHEMGRIDS, global gridded maps of the top 20 crop-specific pesticide application rates from 2015 to 2025. Sci. Data. 2019;61(6):1–20. doi: 10.1038/s41597-019-0169-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orcaray L, Igal M, Marino D, Zabalza A, Royuela M. The possible role of quinate in the mode of action of glyphosate and acetolactate synthase inhibitors. Pest Manag. Sci. 2010;66:262–269. doi: 10.1002/ps.1868. [DOI] [PubMed] [Google Scholar]
- 4.Van Bruggen AHC, et al. Environmental and health effects of the herbicide glyphosate. Sci. Total Environ. 2018;616–617:255–268. doi: 10.1016/j.scitotenv.2017.10.309. [DOI] [PubMed] [Google Scholar]
- 5.Meftaul IM, et al. Controversies over human health and ecological impacts of glyphosate: Is it to be banned in modern agriculture? Environ. Pollut. 2020;263:114372. doi: 10.1016/j.envpol.2020.114372. [DOI] [PubMed] [Google Scholar]
- 6.Tóth G, et al. Cytotoxicity and hormonal activity of glyphosate-based herbicides. Environ. Pollut. 2020;265:115027. doi: 10.1016/j.envpol.2020.115027. [DOI] [PubMed] [Google Scholar]
- 7.Nagy K, Tessema RA, Budnik LT, Ádám B. Comparative cyto- and genotoxicity assessment of glyphosate and glyphosate-based herbicides in human peripheral white blood cells. Environ. Res. 2019;179:108851. doi: 10.1016/j.envres.2019.108851. [DOI] [PubMed] [Google Scholar]
- 8.Sørensen MT, Poulsen HD, Katholm CL, Højberg O. Review: Feed residues of glyphosate—Potential consequences for livestock health and productivity. Animal. 2021;15:100026. doi: 10.1016/j.animal.2020.100026. [DOI] [PubMed] [Google Scholar]
- 9.Sabio y García CA, et al. Rethinking the term “glyphosate effect” through the evaluation of different glyphosate-based herbicide effects over aquatic microbial communities. Environ. Pollut. 2022;292:118382. doi: 10.1016/j.envpol.2021.118382. [DOI] [PubMed] [Google Scholar]
- 10.Chávez-Ortiz P, Tapia-Torres Y, Larsen J, García-Oliva F. Glyphosate-based herbicides alter soil carbon and phosphorus dynamics and microbial activity. Appl. Soil Ecol. 2022;169:104256. [Google Scholar]
- 11.Owagboriaye F, et al. Impacts of a glyphosate-based herbicide on the gut microbiome of three earthworm species (Alma millsoni, Eudrilus eugeniae and Libyodrilus violaceus): A pilot study. Toxicol. Reports. 2021;8:753–758. doi: 10.1016/j.toxrep.2021.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mesnage R, Antoniou MN. Computational modelling provides insight into the effects of glyphosate on the shikimate pathway in the human gut microbiome. Curr. Res. Toxicol. 2020;1:25–33. doi: 10.1016/j.crtox.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maddalon A, Galbiati V, Colosio C, Mandić-Rajčević S, Corsini E. Glyphosate-based herbicides: Evidence of immune-endocrine alteration. Toxicology. 2021;459:152851. doi: 10.1016/j.tox.2021.152851. [DOI] [PubMed] [Google Scholar]
- 14.Lozano VL, et al. Sex-dependent impact of roundup on the rat gut microbiome. Toxicol. Reports. 2018;5:96–107. doi: 10.1016/j.toxrep.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wardle DA, Parkinson D. Influence of the herbicides 2,4-D and glyphosate on soil microbial biomass and activity: A field experiment. Soil Biol. Biochem. 1992;24:185–186. [Google Scholar]
- 16.Haney R, Senseman S, Hons F, Zuberer D. Effect of glyphosate on soil microbial activity and biomass | weed science | Cambridge core. Weed Sci. 2000;48:89–93. [Google Scholar]
- 17.Busse MD, Ratcliff AW, Shestak CJ, Powers RF. Glyphosate toxicity and the effects of long-term vegetation control on soil microbial communities. Soil Biol. Biochem. 2001;33:1777–1789. [Google Scholar]
- 18.Allegrini M, Zabaloy MC, del Gómez EV. Ecotoxicological assessment of soil microbial community tolerance to glyphosate. Sci. Total Environ. 2015;533:60–68. doi: 10.1016/j.scitotenv.2015.06.096. [DOI] [PubMed] [Google Scholar]
- 19.Newman MM, et al. Glyphosate effects on soil rhizosphere-associated bacterial communities. Sci. Total Environ. 2016;543:155–160. doi: 10.1016/j.scitotenv.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 20.Pérez GL, et al. Effects of the herbicide Roundup on freshwater microbial communities: A mesocosm study. Ecol. Appl. 2007;17:2310–2322. doi: 10.1890/07-0499.1. [DOI] [PubMed] [Google Scholar]
- 21.Lu T, et al. Understanding the influence of glyphosate on the structure and function of freshwater microbial community in a microcosm. Environ. Pollut. 2020;260:114012. doi: 10.1016/j.envpol.2020.114012. [DOI] [PubMed] [Google Scholar]
- 22.Zhan H, Feng Y, Fan X, Chen S. Recent advances in glyphosate biodegradation. Appl. Microbiol. Biotechnol. 2018;102:5033–5043. doi: 10.1007/s00253-018-9035-0. [DOI] [PubMed] [Google Scholar]
- 23.Robinson TP, et al. Antibiotic resistance is the quintessential One Health issue. Trans. R. Soc. Trop. Med. Hyg. 2016;110:377–380. doi: 10.1093/trstmh/trw048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brauner A, Fridman O, Gefen O, Balaban NQ. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 2016;14:320–330. doi: 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- 25.Whitehead RN, Overton TW, Kemp CL, Webber MA. Exposure of Salmonella enterica Serovar Typhimurium to high level biocide challenge can select multidrug resistant mutants in a single step. PLoS ONE. 2011;6:e22833. doi: 10.1371/journal.pone.0022833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurenbach B, et al. Sublethal exposure to commercial formulations of the herbicides dicamba, 2,4-dichlorophenoxyacetic acid, and Glyphosate cause changes in antibiotic susceptibility in Escherichia coli and Salmonella enterica serovar Typhimurium. MBio. 2015;6:00009. doi: 10.1128/mBio.00009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raoult D, Hadjadj L, Baron SA, Rolain JM. Role of glyphosate in the emergence of antimicrobial resistance in bacteria? J. Antimicrob. Chemother. 2021;76:1655–1657. doi: 10.1093/jac/dkab102. [DOI] [PubMed] [Google Scholar]
- 28.Bote K, Pöppe J, Merle R, Makarova O, Roesler U. Minimum inhibitory concentration of glyphosate and of a glyphosate-containing Herbicide formulation for Escherichia coli isolates—Differences between pathogenicand non-pathogenic isolates and between host species. Front. Microbiol. 2019;10:932. doi: 10.3389/fmicb.2019.00932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pöppe J, Bote K, Merle R, Makarova O, Roesler U. Minimum inhibitory concentration of glyphosate and a glyphosate-containing herbicide in Salmonella enterica isolates originating from different time periods, hosts, and serovars. Eur. J. Microbiol. Immunol. 2019;9:35–41. doi: 10.1556/1886.2019.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurenbach B, et al. Herbicide ingredients change Salmonella enterica sv. Typhimurium and Escherichia coli antibiotic responses. Microbiology. 2017;163:1791–1801. doi: 10.1099/mic.0.000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X, et al. Herbicide promotes the conjugative transfer of multi-resistance genes by facilitating cellular contact and plasmid transfer. J. Environ. Sci. 2022;115:363–373. doi: 10.1016/j.jes.2021.08.006. [DOI] [PubMed] [Google Scholar]
- 32.Crone S, et al. The environmental occurrence of Pseudomonas aeruginosa. APMIS. 2020;128:220–231. doi: 10.1111/apm.13010. [DOI] [PubMed] [Google Scholar]
- 33.Horcajada JP, et al. Epidemiology and Treatment of Multidrug-Resistant and Extensively Drug-Resistant Pseudomonas aeruginosa Infections. Clin. Microbiol. Rev. 2019;32:31. doi: 10.1128/CMR.00031-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aliaga L, Mediavilla JD, Cobo F. A clinical index predicting mortality with Pseudomonas aeruginosa bacteraemia. J. Med. Microbiol. 2002;51:615–619. doi: 10.1099/0022-1317-51-7-615. [DOI] [PubMed] [Google Scholar]
- 35.Lorè NI, et al. Cystic fibrosis-Niche adaptation of Pseudomonas aeruginosa reduces virulence in multiple infection hosts. PLoS ONE. 2012;7:35648. doi: 10.1371/journal.pone.0035648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Al-Saleh E, Akbar A. Occurrence of Pseudomonas aeruginosa in Kuwait soil. Chemosphere. 2015;120:100–107. doi: 10.1016/j.chemosphere.2014.06.031. [DOI] [PubMed] [Google Scholar]
- 37.Kaszab E, et al. The occurrence of multidrug-resistant Pseudomonas aeruginosa on hydrocarbon-contaminated sites. Microb. Ecol. 2010;59:37–45. doi: 10.1007/s00248-009-9551-7. [DOI] [PubMed] [Google Scholar]
- 38.Kaszab E, et al. Antibiotic resistance profiles and virulence markers of Pseudomonas aeruginosa strains isolated from composts. Bioresour. Technol. 2011;102:1543–1548. doi: 10.1016/j.biortech.2010.08.027. [DOI] [PubMed] [Google Scholar]
- 39.Radó J, et al. Characterization of environmental Pseudomonas aeruginosa using multilocus sequence typing scheme. J. Med. Microbiol. 2017;66:1457–1466. doi: 10.1099/jmm.0.000589. [DOI] [PubMed] [Google Scholar]
- 40.Hoodaji M, Tahmourespour A, Partoazar M. The efficiency of glyphosate biodegradation by Pseudomonas (aeruginosa) Microbes Appl. Res. Curr. Adv. Challenges (Malaga, Spain, 14–16 Sept) 2012;2011:183–186. [Google Scholar]
- 41.Ameen N, Memon Z, Shaheen S, Fatima G, Ahmed F. Imipenem Resistant Pseudomonas aeruginosa: The fall of the final quarterback. Pak. J. Med. Sci. 2015;31:561. doi: 10.12669/pjms.313.7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Atzél B, Szoboszlay S, Mikuska Z, Kriszt B. Comparison of phenotypic and genotypic methods for the detection of environmental isolates of Pseudomonas aeruginosa. Int. J. Hyg. Environ. Health. 2008;211:143–155. doi: 10.1016/j.ijheh.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 43.Medeiros AA, O’Brien TF, Wacker WEC, Yulug NF. Effect of salt concentration on the apparent in-vitro susceptibility of Pseudomonas and other gram-negative Bacilli to gentamicin. J. Infect. Dis. 1971;124:S59–S64. doi: 10.1093/infdis/124.supplement_1.s59. [DOI] [PubMed] [Google Scholar]
- 44.Skerman, V. B. D., McGowan, V. & Sneath, P. H. A. Approved list of bacterial names, amended edition. Am.Soc.Microbiol.30, 72 (1989). [PubMed]
- 45.Wang Y, Li C, Gao C, Ma C, Xua P. Genome sequence of the nonpathogenic Pseudomonas aeruginosa strain ATCC 15442. Genome Announc. 2014;2:421. doi: 10.1128/genomeA.00421-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cassault-Meyer E, Gress S, Séralini GÉ, Galeraud-Denis I. An acute exposure to glyphosate-based herbicide alters aromatase levels in testis and sperm nuclear quality. Environ. Toxicol. Pharmacol. 2014;38:131–140. doi: 10.1016/j.etap.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 47.EUCAST:MICDetermination. https://www.eucast.org/ast_of_bacteria/mic_determination/?no_cache=1. Accessed 9 May 2022.
- 48.Fratini F, et al. A novel interpretation of the fractional inhibitory concentration index: The case Origanum vulgare L. and Leptospermum scoparium J.R. et G. Forst essential oils against Staphylococcus aureus strains. Microbiol. Res. 2017;195:11–17. doi: 10.1016/j.micres.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 49.Faleiro, M. L. & Miguel, M. G. Use of essential oils and their components against multidrug-resistant bacteria. inFightingMultidrugResistancewithHerbalExtracts,EssentialOilsandTheirComponents. 65–94. (Academic Press, 2013).
- 50.Odds FC. Synergy, antagonism, and what the chequerboard puts between them. J. Antimicrob. Chemother. 2003;52:1. doi: 10.1093/jac/dkg301. [DOI] [PubMed] [Google Scholar]
- 51.Ceballos-Garzon A, et al. Genotypic, proteomic, and phenotypic approaches to decipher the response to caspofungin and calcineurin inhibitors in clinical isolates of echinocandin-resistant Candida glabrata. J. Antimicrob. Chemother. 2022;77:585–597. doi: 10.1093/jac/dkab454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsui MTK, Chu LM. Aquatic toxicity of glyphosate-based formulations: comparison between different organisms and the effects of environmental factors. Chemosphere. 2003;52:1189–1197. doi: 10.1016/S0045-6535(03)00306-0. [DOI] [PubMed] [Google Scholar]
- 53.Novotny E. Glyphosate, roundup and the failures of regulatory assessment. Toxics. 2022;10:321. doi: 10.3390/toxics10060321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao H, et al. Herbicide selection promotes antibiotic resistance in soil microbiomes. Mol. Biol. Evol. 2021;38:2337–2350. doi: 10.1093/molbev/msab029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.da Costa NB, et al. A glyphosate-based herbicide cross-selects for antibiotic resistance genes in bacterioplankton communities. mSystems. 2022;7:1482. doi: 10.1128/msystems.01482-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Staub JM, Brand L, Tran M, Kong Y, Rogers SG. Bacterial glyphosate resistance conferred by overexpression of an E. coli membrane efflux transporter. J. Ind. Microbiol. Biotechnol. 2012;39:641–647. doi: 10.1007/s10295-011-1057-x. [DOI] [PubMed] [Google Scholar]
- 57.Kurenbach B, Hill AM, Godsoe W, Van Hamelsveld S, Heinemann JA. Agrichemicals and antibiotics in combination increase antibiotic resistance evolution. PeerJ. 2018;6:5801. doi: 10.7717/peerj.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.EUCAST:ClinicalBreakpointsandDosingofAntibiotics. https://www.eucast.org/clinical_breakpoints/. Accessed 9 May 2022 (2022).
- 59.Rosner JL. Nonheritable resistance to chloramphenicol and other antibiotics induced by salicylates and other chemotactic repellents in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA. 1985;82:8771–8774. doi: 10.1073/pnas.82.24.8771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pöppe J, et al. Selection for resistance to a glyphosate-containing Herbicide in Salmonella enterica does not result in a sustained activation of the tolerance response or increased cross-tolerance and cross-resistance to clinically important antibiotics. Appl. Environ. Microbiol. 2020;86:1204. doi: 10.1128/AEM.01204-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pragasam AK, Raghanivedha M, Anandan S, Veeraraghavan B. Characterization of Pseudomonas aeruginosa with discrepant carbapenem susceptibility profile. Ann. Clin. Microbiol. Antimicrob. 2016;15:1–4. doi: 10.1186/s12941-016-0127-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available as Supplementary data. Further supplementary information is available from the corresponding author on reasonable request.