Abstract
Introduction
Proviral HIV DNA integrated within CD4 T-cells maintains an archive of viral variants that replicate during the course of the infection, including variants with reduced drug susceptibility. We considered studies that investigated archived drug resistance, with a focus on virologically suppressed patients and highlighted interpretative caveats and gaps in knowledge.
Results
Either Sanger or deep sequencing can be used to investigate resistance-associated mutations (RAMs) in HIV DNA recovered from peripheral blood. Neither technique is free of limitations. Furthermore, evidence regarding the establishment, maintenance, expression and clinical significance of archived drug-resistant variants is conflicting. This in part reflects the complexity of the HIV proviral landscape and its dynamics during therapy. Clinically, detection of RAMs in cellular HIV DNA has a variable impact on treatment outcomes, modulated by the drugs affected, treatment duration and additional determinants of virological failure, including those leading to suboptimal drug exposure.
Conclusions
Sequencing cellular HIV DNA can provide helpful complementary information in treatment-experienced patients with suppressed plasma HIV RNA who require a change of regimen. However, care should be taken when interpreting the results. Presence of RAMs is not necessarily a barrier to treatment success. Conversely, even the most sensitive sequencing techniques will fail to provide a comprehensive view of the HIV DNA archive. To inform treatment decisions appropriately, the overall clinical and treatment history of a patient must always be considered alongside the results of resistance testing. Prospective controlled studies are needed to validate the utility of drug resistance testing using cellular HIV DNA.
Keywords: Archive, HIV DNA, Mutation, Resistance, Sequencing
Key Summary Points
| As a result of archiving within integrated HIV DNA, drug-resistant variants, either acquired at the time of infection or emerged during treatment, are postulated to retain life-long significance in people living with HIV. |
| There is growing interest in exploring how archived drug resistance-associated mutations (RAMs) modulate antiretroviral treatment outcomes and therefore how sequencing of cellular HIV DNA may aid treatment decisions. The main application is in virologically suppressed patients where sequencing plasma HIV RNA is not feasible. |
| HIV DNA sequencing may fail to reveal RAMs that had been previously detected in plasma HIV RNA or, when applied longitudinally, may suggest that certain RAMs disappear from the archive over time. More research is needed to understand the dynamics of the resistance archive. |
| Available evidence indicates that resistance test results obtained by HIV DNA sequencing should be interpreted in the context of the overall treatment history of a patient, considering any previous known or suspected resistance, keeping in mind the technical limitations of the methodology and the significant gaps in knowledge, and integrating the resistance data with the multiple additional factors that modulate antiretroviral treatment outcomes. |
Introduction
Infection with HIV-1 has become a manageable, chronic condition because of the availability of multiple antiretrovirals (ARVs) that safely and effectively suppress viraemia and restore and maintain CD4 cell counts [1, 2]. As all currently licensed ARVs are effective only if the virus is replicating and have no impact on the HIV DNA reservoir, lifetime treatment is required to maintain virus suppression. Ensuring patients’ physical and psychological wellbeing long term requires tailored treatment optimisation. Drug resistance testing using plasma HIV RNA, either before treatment initiation or in case of suboptimal treatment responses, continues to play a key role in informing therapeutic choices for viraemic patients [1, 2]. In recent years, there has been growing interest in exploring how drug resistance testing using cellular HIV DNA may also assist decision making. Recent US guidelines state “HIV-1 proviral DNA resistance assays may be useful in patients with HIV RNA below the limit of detection or with low-level viremia” (rated CIII—optional recommendation based on expert opinion) [1]. One potential application is when treated patients who are virologically suppressed require a switch of regimen to address issues such as pill burden, compliance, safety and tolerability, or drug-drug interactions [3]. Both conventional Sanger sequencing and deep sequencing (typically performed using the MiSeq platform [Illumina, San Diego, CA, USA]) can be applied to the study of resistance-associated mutations (RAMs) in the HIV DNA of CD4 T-cells within peripheral blood mononuclear cells (PBMC). However, there is conflicting evidence regarding the significance of RAMs in HIV DNA and to what extent detection (or lack of detection) should inform treatment choices. Determining the influence of archived RAMs on treatment outcomes, including understanding how they can inform safely switching regimens in virologically suppressed patients, is a clear research goal, with potential clinical implications. Our aim was to consider methodological and clinical studies that investigated RAMs in cellular HIV DNA, with a focus on treated patients with suppressed plasma HIV RNA, and to highlight interpretative caveats and gaps in knowledge.
Methods
A published literature search of MEDLINE and EMBASE was performed. The core search terms were “HIV”, “archived resistance”, “proviral DNA”, “DNA”, “minor”, “variant”, “unexpressed”, “resistance” and “RNA”. Key papers recommended by experts in the field were also reviewed. Care was taken to ensure published work was considered accurately and fairly and to exclude any work that raised ethics questions. This article is based on previously conducted studies and does not contain any new studies with human participants or animals performed by any of the authors.
Dynamics of HIV Drug Resistance
Integrated (“proviral”) HIV DNA within memory CD4 T-cell is an archive of viral variants that have replicated throughout the course of the infection, some of which may carry RAMs. Such RAMs may be acquired at the time of transmission and, in this case, through the ‘founder virus’ effect they are expected to seed the viral reservoir widely. Due to the error-prone nature of HIV replication, RAMs also occur spontaneously while the virus replicates, and once therapy is introduced, they become enriched by drug selective pressure if suppression of viral replication is incomplete. With ongoing viral replication during therapy, the variants continue to evolve accumulating further mutations that increase resistance and restore viral fitness [4]. Such variants can integrate as proviral DNA, thus seeding the viral reservoir [5, 6]. Proliferation of CD4 T-cells harbouring integrated virus, either homeostatic or as a result of antigenic stimulation, is thought to maintain the size of the HIV reservoir through many years of virologically suppressive ART [7–10]. Archived RAMs can re-emerge if virus production resumes, facilitated by lapses in adherence or treatment interruption, retaining potential clinical significance long term.
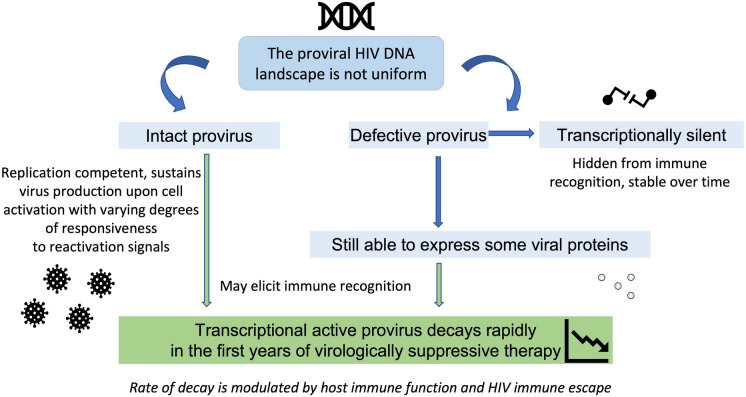
The HIV proviral landscape cannot be regarded as uniform [11–17]. In patients receiving virologically suppressive ART, a large proportion of HIV proviruses is known to be defective. Defective proviruses are either fully or partially transcriptionally silent and are unable to sustain virus production (Fig. 1). Even intact proviruses appear to show varying degrees of transcriptional silence, ranging from very deep latent and difficult-to-reactivate proviruses to viral sequences with high responsiveness to reactivation signals [13, 16]. These different HIV proviruses have a different distribution within memory CD4 T-cell subsets, may exert different effects on host genes and may undergo different selection pressures. Proviruses that have greater expression of viral antigens may be more sensitive to adaptive and innate mechanisms of immune clearance and may be more prone to causing cytopathic effects and, as a result, may decline over several years of virologically suppressive ART [13–16]. Host immune function, HIV ability to escape immune responses (e.g., via expression of the nef protein) and the intrinsic proliferative activity of CD4 T-cells modulate these dynamics [18]. In this complex context, further studies are clearly needed to understand how resistant variants are archived, maintained over time and potentially expressed.
Fig. 1.
The diverse HIV proviral landscape
HIV DNA Sequencing Methodologies
While drug resistance testing is routinely performed by sequencing plasma HIV RNA, the analysis of RAMs held within cellular HIV DNA may allow the identification of drug resistance when plasma viral load is undetectable. To this end, sensitive, scalable and affordable assays are a topic of ongoing research and development. Population Sanger sequencing of viral genes encoding ARV targets (principally protease, reverse transcriptase and integrase) is the conventional method for HIV drug resistance testing (Table 1). It is well established, reproducible and clinically validated. The technique is estimated to be able to detect a viral subpopulation within a sample when it represents at least 15–20% of the total viral population in that sample [19]. Sanger sequencing reveals the dominant variants present at time of testing and provides a composite “consensus” sequence for all the dominant variants in the sample. The method is performed in a ‘hands on’ fashion and is labour intensive and time consuming. Deep sequencing techniques, such as those that employ the Illumina platforms, offer an alternative to Sanger sequencing (Table 1). Features include the ability to detect minority or low abundance viral variants and to provide a quantitative estimate of the frequency of each variant in the sample down to a frequency level of ~ 2%, below which the technique becomes less reliable because of sequencing errors, amplification biases and other artefacts [19–23]. This technology permits high throughput and low cost per sequence. Complex bioinformatic methods are required to compose an informative report from the thousands of sequence reads obtained from each sample. Both Sanger and deep sequencings (mainly with Illumina MiSeq) have been applied to explore the detection of RAMs in cellular HIV DNA [21, 24–29]. Several studies comparing results of Sanger and deep sequencing showed a high correlation between the two methods, although, predictably, some major and minor RAMs were uniquely observed by deep sequencing. There is uncertainty as to the significance of RAMs that occur only as minority variants in a sample [30, 31]. When considering a comparison of plasma HIV RNA and cellular HIV DNA, consistency is greater when sequencing paired samples collected from patients with ongoing virus replication and viraemia [26], as newly transcribed HIV DNA is abundant within cells and mirrors plasma HIV RNA. Consistency is lower when comparing data from historical plasma HIV RNA and current HIV DNA in virologically suppressed patients [24, 25]. Fewer RAMs have generally been found in HIV DNA compared with historic plasma HIV RNA, indicating that some RAMs are either not archived or are not detectable in the HIV DNA reservoir of circulating cells. Population of archived variants may have been initially small, may be smaller in peripheral blood relative to tissue reservoirs or may have decayed below detection thresholds [32].
Table 1.
Methodologies for the detection of HIV drug resistance-associated mutations
| Advantages | Disadvantages | |
|---|---|---|
| Sanger or population sequencing | Well-established, standardised and scientifically validated; highly reproducible; widely available | Inability to detect minority viral variants occurring at a frequency < 15–20% in a patient’s sample; labour intensive and time consuming |
| Next-generation ultradeep sequencinga | Able to detect low abundance viral variants occurring at a frequency between ~ 2% and 15%; capable of quantifying the relative frequency of each variant; high throughput and low cost per sample | Financial, infrastructural and logistical challenges impede widespread adoption; generates large amount of data and complex bioinformatic methodology is required for interpretation; uncertain clinical significance of variants detected at low frequency |
aTypically using the Illumina sequencing technology
A number of technical considerations apply when discussing detection of RAMs within HIV DNA. Studies commonly report on RAMs within buffy coats, whole blood, PBMC or less commonly isolated CD4 T-cells. The methodology will detect mutations within all forms of cell-associated HIV DNA and not exclusively within the archive of integrated provirus. In virologically suppressed patients, most but not all HIV DNA in PBMC is thought to represent integrated provirus [7]. Furthermore, in the setting of virological suppression, the size of the HIV DNA input into the sequencing test will be small, which can make RAM detection stochastic. The frequency of cells containing HIV DNA declines in the first 1–4 years of ART (with faster decay of cells carrying transcriptional active provirus) and remains relatively stable thereafter within the range of 1–3 per 10,000 CD4 cells during suppressive therapy [33]. Of note, these estimates are influenced by the interval between infection and commencement of ART.
Lambert-Niclot et al. investigated a cohort of virologically suppressed patients with or without previous virological failure who switched to a single tablet regimen of rilpivirine/emtricitabine/tenofovir disoproxil fumarate [27]. When comparing results obtained by Sanger sequencing of current cellular HIV DNA with the historical data from plasma HIV RNA, concordance was good in patients without prior virological failure, but less so among those with prior failure. Concordance was higher with a higher plasma viral load at the time of HIV RNA sequencing and a shorter mean time between the historical HIV RNA and the current HIV DNA sample, suggesting an effect of time on the ability to detect RAMs in HIV DNA. Nouchi et al. described 25 patients with historical detection of RAMs to lamivudine/emtricitabine, etravirine and rilpivirine in plasma HIV RNA who were currently virologically suppressed on regimens that did not exert selective pressure on those RAMs [28]. Using deep sequencing (Illumina MiSeq) of sequential PBMC samples, over 5 years, RAMs became no longer detectable in 72% (18/25) of patients. Based on the observation that more than half of the patients had residual viraemia at levels below 50 copies/ml, the authors proposed that ongoing virus replication may lead to the gradual replacement and evolution of cellular HIV DNA. This scenario may apply to a subset of treated patients with residual viraemia, although the bulk of evidence indicates that ongoing viral replication is unlikely at levels of residual viraemia below 10 copies/ml [34]. The data must also be placed into the context of the dynamics of HIV provirus described above.
HIV DNA Sequencing for the Detection of Drug Resistance During Virological Suppression
Ellis et al. found that it was possible to use HIV DNA sequencing to guide ART adjustments in virologically suppressed patients, allowing a switch to a regimen with reduced pill burden without a risk of virological rebound [35]. The study lacked a control group in whom therapy was not guided by HIV DNA sequencing. The authors suggested that HIV DNA sequencing may provide value for decision-making for selected patients, including those with multiple previous or foreseen ART adjustments due to comorbidities, medication interactions, difficulty accessing care, hesitancy regarding ART switches or other factors that could increase risk when switching therapy. Maybeck et al. also provided cohort data in support of HIV DNA sequencing to guide treatment decisions alongside historical data based on plasma HIV RNA sequencing [36]. Armenia et al. found that among virologically suppressed patients, detection of RAMs in HIV DNA, together with a low nadir CD4 cell count and a short duration of virological control, was predictive of virological rebound after switching therapy [37]. More recently, Cutrell et al. explored predictors of virological failure after switching virologically suppressed patients to long-acting cabotegravir and rilpivirine [38]. In a pooled analysis of Phase 3 clinical trials, the presence of rilpivirine RAMs in baseline HIV DNA contributed to predicting virological failure after switching, alongside HIV-1 subtype A6/A1, a higher body mass index and lower week 8 rilpivirine plasma trough concentrations; a combination of at least two of these factors significantly increased the risk of virological failure at 48 weeks. Data on the impact of archived rilpivirine RAMs are in line with older nevirapine-related data [39], illustrating how the barrier to resistance of the drugs affected by archived RAMs plays an important role in modulating their significance. There are general reasons why archived RAMs may not produce clinical impact: the variant population size may be too small for sufficient stochastic reactivation or may be too small or have insufficient levels of resistance or fitness to outcompete other variants during periods of viral rebound [40].
There is also evidence indicating that detection of RAMs within cellular HIV DNA may signal behavioural factors that impact on virological outcomes. A study investigated patients in Cameroon who had previously experienced failure of first-line therapy with two nucleos(t)ide reverse transcriptase inhibitors (NRTIs) and one non-nucleoside reverse transcriptase inhibitor (NNRTI) and were virologically suppressed on lopinavir/ritonavir plus two NRTIs [41]. Detection of NRTI and NNRTI RAMs in PBMC-associated HIV DNA predicted a reduced risk of virological rebound after a switch to monotherapy with darunavir/ritonavir and thus away from ARVs exerting selective pressure on the pre-existing RAMs. In this case, the presence of archived RAMs was taken to indicate a general predisposition towards a greater degree of adherence to treatment relative to patients that had previously experienced ART failure without accumulating drug resistance. Similar findings were reported about NRTI and NNRTI RAMs in plasma HIV RNA of cohorts in Africa starting second-line protease inhibitor-based ART after failure of NNRTI-based regimens [42].
The Unique Characteristics of M184V/I
One RAM of specific interest is the reverse transcriptase mutation M184V/I, which is common among treatment-experienced patients with virological failure and, when using deep sequencing (MiSeq) of plasma HIV RNA, is also found in ~ 10% of patients with a recent HIV infection [43]. The mutation reduces viral fitness, antagonises the emergence of resistance to the NRTIs zidovudine, stavudine and tenofovir, and increases susceptibility to the same NRTIs when occurring in combination with other NRTI RAMs, including thymidine analogue mutations and K65R [44–47]. In contrast, M184V/I directly contributes resistance to abacavir and didanosine. It should be noted that M184I (but not M184V) is part of the natural pool of mutations generated by APOBEC3G/F activity, a cellular defence mechanism that, independent of drug-selective pressure, induces hypermutations in viral genomes making them defective; finding M184I within HIV DNA should be interpreted with caution [48]. M184V is also described as a common inappropriate call by software, depending on settings for secondary peaks.
The available evidence suggests that in virologically suppressed patients, the presence of M184V/I, either documented in historical resistance tests performed with plasma HIV RNA or established with a test of current cellular HIV DNA, does not preclude continued virological response after switching to regimens with a high barrier to resistance. A pooled analysis of two switch studies assessed virological responses to the second-generation INSTI-based combination bictegravir, emtricitabine and tenofovir alafenamide (B/F/TAF) [3]. Pre-existing RAMs were assessed based on historical plasma HIV RNA sequencing data (documented resistance to study drugs was excluded at the time of inclusion in the trial) and by retrospective sequencing of cellular HIV DNA in PBMC collected at the time of switching. Overall, 40% of patients were found to have one or more pre-existing major RAM in protease, reverse transcriptase and/or integrase. Among patients treated with B/F/TAF, NRTI RAMs overall were detected in 16% (89/543), including M184V/I in 10% (54/543). At week 48 after switching to B/F/TAF, 98% (213/217) of patients with any pre-existing resistance and 96% (52/54) of those with M184V/I had HIV RNA < 50 copies/ml. Similar findings have been reported for triple ART regimens based on dolutegravir [49, 50]. Since pre-existing resistance may be detected but cannot be excluded based on HIV DNA sequencing alone, studies that demonstrate responses to high barrier triple ART regimens despite limited pre-existing resistance provide reassurance for clinicians in instances where HIV DNA sequencing is not available prior to a switch. Both clinical trial and real-world data also report a negligible impact of archived M184V/I on the dual combination of dolutegravir/lamivudine (DTG/3TC) [51, 52]. In the prospective cohort study DOLULAM, collating historical Sanger sequencing data obtained with plasma HIV RNA and performing deep sequencing of cellular HIV DNA revealed reverse transcriptase and integrase RAMs in 45% (10/22) and 21% (4/19) of virologically suppressed treatment-experienced patients, respectively [51]. M184I/V was detected in 63% (17/27) of patients; despite presence of M184V/I, however, all 17 patients maintained virological suppression 2 years after switching to dual therapy with DTG/3TC. One interesting observation from the LAMRES study was that patients with an archived M184V/I switching to DTG/3TC (n = 37) were significantly more likely to experience virological failure if the mutation was detected in the 5 years prior to the treatment switch; proportions with virological failure among those with M184V/I detected < 5 years vs. > 5 years were 20% vs. 0% at 1 year and 20% vs. 5% at 2 years after the switch [53]. These data again point to a temporally declining effect of archived RAMs, although the data require further confirmation, including a more detailed assessment of the characteristics of patients with older vs. more recent evidence of M184V/I. Observations on the clinical significance of archived M184V/I and other NRTI RAMs should also be placed into the context of emerging data on the impact of these same mutations when detected in plasma HIV RNA of patients with viraemia. In trials conducted in African populations, such RAMs do not appear to negatively affect the activity of regimens containing zidovudine or tenofovir plus lamivudine in combination with a high barrier third agent, either a boosted protease inhibitor [42] or dolutegravir [54].
Conclusions
Whilst controlled data are required, available observations point to the potential utility of HIV DNA sequencing to guide clinical practice. Several uncertainties remain. It is important to emphasise that results must be interpreted with caution, keeping in mind the technical properties of the methodology and placing the resistance data in the context of all available indicators that may interact to determine treatment outcomes. We highlight the importance of correctly interpreting the concept of archived resistance when considering how the data can guide the most appropriate drug choice. The existence of archived RAMs is not necessarily a barrier to treatment success with high barrier triple drug regimens and in the case of M184V/I may also have limited impact on dual DTG/3TC regimens. However, when considering a treatment switch, each case must be assessed on its merits. A careful risk/benefit assessment is required that considers the overall clinical and treatment history and avoids unnecessarily exposing patients to an increased risk of virological failure and treatment-emergent resistance.
Acknowledgements
Funding
No funding or sponsorship was received for publication of this article. The journal’s Rapid Service Fee was funded by Gilead Sciences Europe Ltd.
Medical Writing Assistance
Yvonne Adebola and Mark Davies of inScience Communications, Springer Healthcare Ltd, UK, provided medical writing support funded by Gilead Sciences Europe Ltd.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Author Contributions
Anna Maria Garetti wrote the bulk of the article and revised it after review. Jose Luis Blanco, Anne Genevieve Marcelin, Carlo Federico Perno, Hans Jurgen Stellbrink, Dan Turner and Tuba Zengin contributed extensively to the concept, critical review, and revision of all drafts of the article.
Disclosures
Anna Maria Garetti reports personal payment and grants (to the institution) from Roche Pharma Research & Early Development, personal fees and grants (to the institution) from ViiV Healthcare and Gilead, and personal fees from Janssen, Theratecnologies, and GSK. Jose Luis Blanco has received honoraria for lectures or advisory boards from Gilead, Janssen, and MSD. Carlo Federico Perno has received honoraria for advisory boards from Abbvie, Gilead, ViiV, Janssen, Merck and Theratechnologies, and research grants from Abbvie, Gilead, ViiV, Janssen, and Merck. Hans Jurgen Stellbrink reports personal fees from ViiV Healthcare during the study; personal fees from Gilead Sciences, Janssen-Cilag, Theratechnologies, and Merck Sharp & Dohme, outside of the submitted work. Tuba Zengin is an employee of Gilead Sciences. Anne Genevieve Marcelin and Dan Turner have nothing to declare.
Compliance with Ethics Guidelines
This article is based on previously conducted studies and does not contain any new studies with human participants or animals performed by any of the authors. Care was taken to ensure published work was considered accurately and fairly and to exclude any work that raised ethics questions.
Data Availability
Data sharing is not applicable to this article as no datasets were generated or analysed.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the use of antiretroviral agents in adults and adolescents with HIV. Department of Health and Human Services. Available at https://clinicalinfo.hiv.gov/sites/default/files/guidelines/documents/AdultandAdolescentGL.pdf. Accessed 7 Mar 2022.
- 2.European AIDS Clinical Society. Guidelines Version 11.0 October 2021. Available at https://www.eacsociety.org/media/final2021eacsguidelinesv11.0_oct2021.pdf. Accessed 7 Mar 2022.
- 3.Andreatta K, Willkom M, Martin R, et al. Switching to bictegravir/emtricitabine/tenofovir alafenamide maintained HIV-1 RNA suppression in participants with archived antiretroviral resistance including M184V/I. J Antimicrob Chemother. 2019;74:3555–3564. doi: 10.1093/jac/dkz347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung PK, Shahid A, Dong W, et al. Impact of combinations of clinically observed HIV integrase mutations on phenotypic resistance to integrase strand transfer inhibitors (INSTIs): a molecular study. J Antimicrob Chemother. 2022;77:979. doi: 10.1093/jac/dkab498. [DOI] [PubMed] [Google Scholar]
- 5.Noë A, Plum J, Verhofstede C. The latent HIV-1 reservoir in patients undergoing HAART: an archive of pre-HAART drug resistance. J Antimicrob Chemother. 2005;55:410–412. doi: 10.1093/jac/dki038. [DOI] [PubMed] [Google Scholar]
- 6.Wind-Rotolo M, Durand C, Cranmer L, et al. Identification of nevirapine-resistant HIV-1 in the latent reservoir after single-dose nevirapine to prevent mother-to-child transmission of HIV-1. J Infect Dis. 2009;199:1301–1309. doi: 10.1086/597759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chomont N, El-Far M, Ancuta P, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruggiero A, De Spiegelaere W, Cozzi-Lepri A, et al. During stably suppressive antiretroviral therapy integrated HIV-1 DNA load in peripheral blood is associated with the frequency of CD8 cells expressing HLA-DR/DP/DQ. EBioMedicine. 2015;2:1153–1159. doi: 10.1016/j.ebiom.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwon KJ, Siliciano RF. HIV persistence: clonal expansion of cells in the latent reservoir. J Clin Invest. 2017;127:2536–2538. doi: 10.1172/JCI95329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau CY, Adan MA, Maldarelli F. Why the HIV reservoir never runs dry: clonal expansion and the characteristics of HIV-infected cells challenge strategies to cure and control HIV infection. Viruses. 2021;13:2512. doi: 10.3390/v13122512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiener B, Horsburgh BA, Eden JS, et al. Identification of genetically intact HIV-1 proviruses in specific CD4(+) T cells from effectively treated participants. Cell Rep. 2017;21:813–822. doi: 10.1016/j.celrep.2017.09.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pinzone MR, VanBelzen DJ, Weissman S, et al. Longitudinal HIV sequencing reveals reservoir expression leading to decay which is obscured by clonal expansion. Nat Commun. 2019;10:728. doi: 10.1038/s41467-019-08431-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Einkauf KB, Lee GQ, Gao C, et al. Intact HIV-1 proviruses accumulate at distinct chromosomal positions during prolonged antiretroviral therapy. J Clin Invest. 2019;129:988–998. doi: 10.1172/JCI124291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peluso MJ, Bacchetti P, Ritter KD, et al. Differential decay of intact and defective proviral DNA in HIV-1-infected individuals on suppressive antiretroviral therapy. JCI Insight. 2020;5:e132997. doi: 10.1172/jci.insight.132997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gandhi RT, Cyktor JC, Bosch RJ, et al. Selective decay of intact HIV-1 proviral DNA on antiretroviral therapy. J Infect Dis. 2021;223:225–233. doi: 10.1093/infdis/jiaa532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Einkauf KB, Osborn MR, Gao C, et al. Parallel analysis of transcription, integration, and sequence of single HIV-1 proviruses. Cell. 2022;185:266–282. doi: 10.1016/j.cell.2021.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siliciano JD, Siliciano RF. In vivo dynamics of the latent reservoir for HIV-1: new insights and implications for cure. Annu Rev Pathol. 2022;17:271–294. doi: 10.1146/annurev-pathol-050520-112001. [DOI] [PubMed] [Google Scholar]
- 18.Duette G, Hiener B, Morgan H, et al. The HIV-1 proviral landscape reveals Nef contributes to HIV-1 persistence in effector memory CD4+ T-cells. J Clin Invest. 2022;132:e154422. doi: 10.1172/JCI154422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tzou PL, Ariyaratne P, Varghese V, et al. Comparison of an in vitro diagnostic next-generation sequencing assay with Sanger sequencing for HIV-1 genotypic resistance testing. J Clin Microbiol. 2018;56:e00105–e118. doi: 10.1128/JCM.00105-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parikh UM, McCormick K, van Zyl G, Mellors JW. Future technologies for monitoring HIV drug resistance and cure. Curr Opin HIV AIDS. 2017;12:182–189. doi: 10.1097/COH.0000000000000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alidjinou EK, Coulon P, Hallaert C, et al. Routine drug resistance testing in HIV-1 proviral DNA, using an automated next-generation sequencing assay. J Clin Virol. 2019;121:104207. doi: 10.1016/j.jcv.2019.104207. [DOI] [PubMed] [Google Scholar]
- 22.Lee ER, Parkin N, Jennings C, et al. Performance comparison of next generation sequencing analysis pipelines for HIV-1 drug resistance testing. Sci Rep. 2020;10:1634. doi: 10.1038/s41598-020-58544-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zanini F, Brodin J, Albert J, Neher RA. Error rates, PCR recombination, and sampling depth in HIV-1 whole genome deep sequencing. Virus Res. 2017;239:106–114. doi: 10.1016/j.virusres.2016.12.009. [DOI] [PubMed] [Google Scholar]
- 24.Wirden M, Soulie C, Valantin MA, et al. Historical HIV-RNA resistance test results are more informative than proviral DNA genotyping in cases of suppressed or residual viraemia. J Antimicrob Chemother. 2011;66:709–712. doi: 10.1093/jac/dkq544. [DOI] [PubMed] [Google Scholar]
- 25.Delaugerre C, Braun J, Charreau I, ANRS 138-EASIER study group et al. Comparison of resistance mutation patterns in historical plasma HIV RNA genotypes with those in current proviral HIV DNA genotypes among extensively treated patients with suppressed replication. HIV Med. 2012;13:517–525. doi: 10.1111/j.1468-1293.2012.01002.x. [DOI] [PubMed] [Google Scholar]
- 26.Geretti AM, Conibear T, Hill A, SENSE Study Group et al. Sensitive testing of plasma HIV-1 RNA and Sanger sequencing of cellular HIV-1 DNA for the detection of drug resistance prior to starting first-line antiretroviral therapy with etravirine or efavirenz. J Antimicrob Chemother. 2014;69:1090–1097. doi: 10.1093/jac/dkt474. [DOI] [PubMed] [Google Scholar]
- 27.Lambert-Niclot S, Allavena C, Grude M, et al. Usefulness of an HIV DNA resistance genotypic test in patients who are candidates for a switch to the rilpivirine/emtricitabine/tenofovir disoproxil fumarate combination. J Antimicrob Chemother. 2016;71:2248–2251. doi: 10.1093/jac/dkw146. [DOI] [PubMed] [Google Scholar]
- 28.Nouchi A, Nguyen T, Valantin MA, et al. Dynamics of drug resistance-associated mutations in HIV-1 DNA reverse transcriptase sequence during effective ART. J Antimicrob Chemother. 2018;73:2141–2146. doi: 10.1093/jac/dky130. [DOI] [PubMed] [Google Scholar]
- 29.Millière L, Bocket L, Tinez C, et al. Assessment of intra-sample variability in HIV-1 DNA drug resistance genotyping. J Antimicrob Chemother. 2021;76:2143–2147. doi: 10.1093/jac/dkab149. [DOI] [PubMed] [Google Scholar]
- 30.Geretti AM, Paredes R, Kozal MJ. Transmission of HIV drug resistance: lessons from sensitive screening assays. Curr Opin Infect Dis. 2015;28:23–30. doi: 10.1097/QCO.0000000000000136. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen T, Fofana DB, Le MP, et al. Prevalence and clinical impact of minority resistant variants in patients failing an integrase inhibitor-based regimen by ultra-deep sequencing. J Antimicrob Chemother. 2018;73:2485–2492. doi: 10.1093/jac/dky198. [DOI] [PubMed] [Google Scholar]
- 32.Clarridge KE, Blazkova J, Einkauf K, et al. Effect of analytical treatment interruption and reinitiation of antiretroviral therapy on HIV reservoirs and immunologic parameters in infected individuals. PLoS Pathog. 2018;14:e1006792. doi: 10.1371/journal.ppat.1006792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson EM, Maldarelli F. The role of integration and clonal expansion in HIV infection: live long and prosper. Retrovirology. 2018;15:71. doi: 10.1186/s12977-018-0448-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doyle T, Geretti AM. Low-level viraemia on HAART: significance and management. Curr Opin Infect Dis. 2012;25:17–25. doi: 10.1097/QCO.0b013e32834ef5d9. [DOI] [PubMed] [Google Scholar]
- 35.Ellis KE, Nawas GT, Chan C, et al. Clinical outcomes following the use of archived proviral HIV-1 DNA genotype to guide antiretroviral therapy adjustment. Open Forum Infect Dis. 2020;7:ofz533. doi: 10.1093/ofid/ofz533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meybeck A, Alidjinou EK, Huleux T, et al. Virological outcome after choice of antiretroviral regimen guided by proviral HIV-1 DNA genotyping in a real-life cohort of HIV-infected patients. AIDS Patient Care STDS. 2020;34:51–58. doi: 10.1089/apc.2019.0198. [DOI] [PubMed] [Google Scholar]
- 37.Armenia D, Zaccarelli M, Borghi V, et al. Resistance detected in PBMCs predicts virological rebound in HIV-1 suppressed patients switching treatment. J Clin Virol. 2018;104:61–64. doi: 10.1016/j.jcv.2018.04.001. [DOI] [PubMed] [Google Scholar]
- 38.Cutrell AG, Schapiro JM, Perno CF, et al. Exploring predictors of HIV-1 virologic failure to long-acting cabotegravir and rilpivirine: a multivariable analysis. AIDS. 2021;35:1333–1342. doi: 10.1097/QAD.0000000000002883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jourdain G, Wagner TA, Ngo-Giang-Huong N, et al. Association between detection of HIV-1 DNA resistance mutations by a sensitive assay at initiation of antiretroviral therapy and virologic failure. Clin Infect Dis. 2010;50:1397–1404. doi: 10.1086/652148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boltz VF, Shao W, Bale MJ, et al. Linked dual-class HIV resistance mutations are associated with treatment failure. JCI Insight. 2019;4:e130118. doi: 10.1172/jci.insight.130118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geretti AM, Abdullahi A, Mafotsing Fopoussi O, et al. An apparent paradox: resistance mutations in HIV-1 DNA predict improved virological responses to antiretroviral therapy. J Antimicrob Chemother. 2019;74:3011–3015. doi: 10.1093/jac/dkz264. [DOI] [PubMed] [Google Scholar]
- 42.Stockdale AJ, Saunders MJ, Boyd MA, et al. Effectiveness of protease inhibitor/nucleos(t)ide reverse transcriptase inhibitor-based second-line antiretroviral therapy for the treatment of Human Immunodeficiency Virus type 1 infection in sub-Saharan Africa: a systematic review and meta-analysis. Clin Infect Dis. 2018;66:1846–1857. doi: 10.1093/cid/cix1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mbisa JL, Ledesma J, Kirwan P, et al. Surveillance of HIV-1 transmitted integrase strand transfer inhibitor resistance in the UK. J Antimicrob Chemother. 2020;75:3311–3318. doi: 10.1093/jac/dkaa309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diallo K, Gotte M, Wainberg MA. Molecular impact of the M184V mutation in human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 2003;47:3377–3383. doi: 10.1128/AAC.47.11.3377-3383.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gallant JE. The M184V mutation: what it does, how to prevent it, and what to do with it when it's there. AIDS Read. 2006;16:556–559. [PubMed] [Google Scholar]
- 46.Stanford University HIV Drug Resistance Database. HIV resistance notes, https://hivdb.stanford.edu/dr-summary/resistance-notes/NRTI/. Accessed 7 Mar 2022.
- 47.Kuritzkes DR. New perspectives on the virologic consequences of M184V or I in Human Immunodeficiency Virus-1 reverse transcriptase. J Infect Dis. 2020;222:1067–1069. doi: 10.1093/infdis/jiz632. [DOI] [PubMed] [Google Scholar]
- 48.Fourati S, Malet I, Lambert S, et al. E138K and M184I mutations in HIV-1 reverse transcriptase coemerge as a result of APOBEC3 editing in the absence of drug exposure. AIDS. 2012;26:1619–1624. doi: 10.1097/QAD.0b013e3283560703. [DOI] [PubMed] [Google Scholar]
- 49.Acosta RK, Willkom M, Andreatta K, et al. Switching to Bictegravir / Emtricitabine / Tenofovir Alafenamide (B/F/TAF) from Dolutegravir (DTG)+F/TAF or DTG+F/Tenofovir Disoproxil Fumarate (TDF) in the presence of pre-existing NRTI resistance. J Acquir Immune Defic Syndr. 2020;85:363–371. doi: 10.1097/QAI.0000000000002454. [DOI] [PubMed] [Google Scholar]
- 50.Ndashimye E, Arts EJ. Dolutegravir response in antiretroviral therapy naive and experienced patients with M184V/I: Impact in low-and middle-income settings. Int J Infect Dis. 2021;105:298–303. doi: 10.1016/j.ijid.2021.03.018. [DOI] [PubMed] [Google Scholar]
- 51.Charpentier C, Montes B, Perrier M, Meftah N, Reynes J. HIV-1 DNA ultra-deep sequencing analysis at initiation of the dual therapy dolutegravir + lamivudine in the maintenance DOLULAM pilot study. J Antimicrob Chemother. 2017;72:2831–2836. doi: 10.1093/jac/dkx233. [DOI] [PubMed] [Google Scholar]
- 52.Baldin G, Ciccullo A, Rusconi S, et al. Long-term data on the efficacy and tolerability of lamivudine plus dolutegravir as a switch strategy in a multi-centre cohort of HIV-1-infected, virologically suppressed patients. Int J Antimicrob Agents. 2019;54:728–734. doi: 10.1016/j.ijantimicag.2019.09.002. [DOI] [PubMed] [Google Scholar]
- 53.Santoro MM, Armenia D, Teyssou E, et al. Impact of M184V on the virological efficay of switch to 3TC/DTG in real life. Topics Antiviral Med. 2021;29(154):P429. [Google Scholar]
- 54.Paton N, Musaazi J, Kityo C, et al. Nucleosides and darunavir/dolutegravir in Africa (NADIA) trial: outcomes at 96 weeks. In: 2022 CROI, February 12–16 and 22–24, 2022. Abstract 137.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analysed.