Significance
We illustrated a distinct molecular architecture of an endoplasmic reticulum (ER)–autophagosomal membrane contact site (EACS) in regulating plant autophagy, which involves the lipid-binding protein ORP2A, the ER residential protein VAP27-1, and the ubiquitin-like protein ATG8e. We further demonstrated that ORP2A functions in autophagosome formation, while knocking down of ORP2A affects phosphatidylinositol 3-phosphate (PI3P) redistribution.
Keywords: autophagy, oxysterol-binding protein–related protein, membrane contact site, PI3P
Abstract
Autophagy is an intracellular degradation system for cytoplasmic constituents which is mediated by the formation of a double-membrane organelle termed the autophagosome and its subsequent fusion with the lysosome/vacuole. The formation of the autophagosome requires membrane from the endoplasmic reticulum (ER) and is tightly regulated by a series of autophagy-related (ATG) proteins and lipids. However, how the ER contacts autophagosomes and regulates autophagy remain elusive in plants. In this study, we identified and demonstrated the roles of Arabidopsis oxysterol-binding protein–related protein 2A (ORP2A) in mediating ER–autophagosomal membrane contacts and autophagosome biogenesis. We showed that ORP2A localizes to both ER–plasma membrane contact sites (EPCSs) and autophagosomes, and that ORP2A interacts with both the ER-localized VAMP-associated protein (VAP) 27-1 and ATG8e on the autophagosomes to mediate the membrane contact sites (MCSs). In ORP2A artificial microRNA knockdown (KD) plants, seedlings display retarded growth and impaired autophagy levels. Both ATG1a and ATG8e accumulated and associated with the ER membrane in ORP2A KD lines. Moreover, ORP2A binds multiple phospholipids and shows colocalization with phosphatidylinositol 3-phosphate (PI3P) in vivo. Taken together, ORP2A mediates ER–autophagosomal MCSs and regulates autophagy through PI3P redistribution.
Macroautophagy (hereafter “autophagy”) is an evolutionarily conserved degradation mechanism which plays an important role in development, pathogen defense, stress response, and subcellular quality control, especially for plants when faced with various environmental cues (1, 2). A long-standing question in autophagy studies is how the cell controls precisely the biogenesis of autophagosomes, a double membrane–bound organelle formed during autophagy. Our mechanistic understanding of the autophagy process is based on the assembling of a series of autophagy-related (ATG) gene products, which are mostly conserved among eukaryotes. Numerous studies have reported that the endoplasmic reticulum (ER) is one of the most important endomembrane sources for autophagosome formation under stress conditions (3–5). In mammalian cells, autophagosome biogenesis is initiated at the ER from which a domain with an omega-shaped membrane (the omegasome) serves as a platform for the nucleation of the isolation membrane (IM, the precursor of the autophagosome) (6). The conserved machinery for autophagosome initiation contains two major complexes: the ULK1/ATG1 (unc-51–like autophagy activating kinase 1, known as Atg1 in yeast) complex and class III phosphatidylinositol 3-kinase (PI3K) complex I (7). Nucleation of the IM is led by the activity of the ULK1 complex, the PI3K complex, and ATG9, a singular conserved transmembrane protein in the core machinery (8, 9). The autophagy process in plants is highly conserved but has unique and plant-specific characteristics. For example, the Arabidopsis autophagy initiation protein ATG1 has four isoforms of likely functional redundancy (10, 11), whereas Arabidopsis ATG9 was shown to function in regulating autophagosome progression from the ER (12). In addition, a novel regulator, SH3 domain–containing protein 2 (SH3P2), was identified to be concentrated in phosphatidylinositol 3-phosphate (PI3P)–containing membranes in regulating autophagy in Arabidopsis (13).
In recruiting the core autophagy machinery proteins, PI3P plays a central role during the autophagy process (14). In mammalian cells, once the PI3P pools at the ER are established by the PI3K complex, various PI3P‐binding proteins such as double FYVE domain–containing protein 1 (DFCP1) and WD-repeat domain phosphoinositide-interacting protein 2 (WIPI2, Atg18 in yeast) are subsequently recruited (15–17). These proteins further recruit downstream regulators controlling almost all steps of autophagy including autophagosome expansion, ATG8 lipidation, and fusion with the lysosome/vacuole (18). Compared with mammals and yeast, plants have a number of conserved PI3P effectors and some plant-unique ones. For example, a plant-unique autophagy regulator, Arabidopsis SH3P2, was shown to be essential for seedling development and to function in regulating autophagosome formation at the ER (13). As a PI3P effector, SH3P2 can be detected in an autophagosomal membrane binding to PI3P and ATG8 (13). The function of SH3P2 at the autophagosomal membrane remains unclear, but it may promote membrane bending during phagophore expansion because SH3P2 is a BAR domain–containing protein for regulating membrane curvature (13). However, how PI3P is regulated at the forming autophagosome remains unknown.
Lipid transport is one of the functions of a membrane contact site (MCS), which provides a fast and nonvesicular way for material exchange. MCSs have a functional proteomic and/or lipidomic composition which is required for maintenance and function of the architecture (19, 20). In mammalian cells, the ER-localized proteins VAMP-associated protein (VAP) A and B have been shown to establish MCSs with multiple organelles including the Golgi, endosomes, and peroxisomes (21). In Arabidopsis, 10 VAP homologs termed VAP27s have been identified, of which VAP27-1 and 27-3 were shown to localize to both the ER and the ER–PM (plasma membrane) contact site (EPCS) (22). Recent studies have also revealed the function of VAP27-positive MCSs in endocytosis, autophagy, and lipid droplet biogenesis in Arabidopsis (23–25).
In mediating the MCSs with VAPs, one of the most important counterparts is the oxysterol-binding protein–related protein (ORP) family. The ORPs constitute a large family of lipid transfer proteins (LTPs) that bind oxysterols, cholesterol, and phospholipids within a conserved hydrophobic binding domain termed the oxysterol binding–related domain (ORD) (26). Besides the core lipid-binding ORD, a typical mammalian ORP has a tailed pleckstrin homology (PH) domain, which binds to specific lipids in a non-ER membrane (26). The Arabidopsis ORP family consists of 12 ORP members which are divided into four subgroups from a phylogenetic analysis (27). A PH domain and an FFAT (two phenylalanines [FF] in an acidic tract)-like (FFATL) motif have been identified in ORP1A, 1C, and 1D from subgroup 1 and in ORP2A and 2B from subgroup 2 (27, 28). However, little is known about the functions of the 12 ORPs in plants.
In this study, we identified and demonstrated that ORP2A mediates ER–autophagosomal MCSs which is crucial for autophagosome formation and progression in Arabidopsis. Live-cell imaging and biochemical analysis identified the molecular architecture of VAP–ORP–ATG8 interaction to mediate ER–autophagosomal MCSs in plant cells. In ORP2A artificial microRNA (amiRNA) knockdown (KD) mutant lines, autophagosomal formation was impaired while the ATG1 complex accumulated in the ATG8-positive autophagosomal structures associating with the ER membrane, indicating the roles of ORP2A in regulating autophagy initiation and progression from the ER. Further lipid-binding analysis showed that ORP2A binds to multiple phospholipids and modulates autophagy via regulating PI3P at the ER membrane. Thus, this study unveils a mechanism and function of ORP2A underlying the formation and maintenance of ER–autophagosomal MCSs in plants.
Results
ORP2A Is Involved in the Autophagic Pathway.
Previous studies have identified 12 ORPs in the Arabidopsis genome but with little information about their functions in plants (27, 28). Recent studies of ORPs in animal cells showed their involvement in autophagosome maturation (29), and thus we hypothesized that selective members of Arabidopsis ORPs may play roles in plant autophagy. To identify potential candidates of ORPs in the plant autophagic pathway, we first carried out subcellular localization studies on the 12 Arabidopsis ORP–XFP fusions upon their transient coexpression with the autophagosomal marker yellow fluorescent protein (YFP)–ATG8e in Arabidopsis plant system biology dark-type culture (PSBD) protoplasts. Indeed, red fluorescent protein (RFP)–tagged ORP2A showed localization with YFP-ATG8e in ring-like punctae upon their coexpression in PSBD protoplasts (SI Appendix, Fig. S1A). In addition, ORP2A-GFP (green fluorescent protein) also showed colocalization with two other autophagy-related proteins, SH3P2-CFP (cyan fluorescent protein) (13) and ATG9-CFP (12) (SI Appendix, Fig. S1B), suggesting that ORP2A may localize to different stages of autophagosomal membranes including the PI3P-positive isolation membrane (SH3P2-positive) and ATG9-positive membrane.
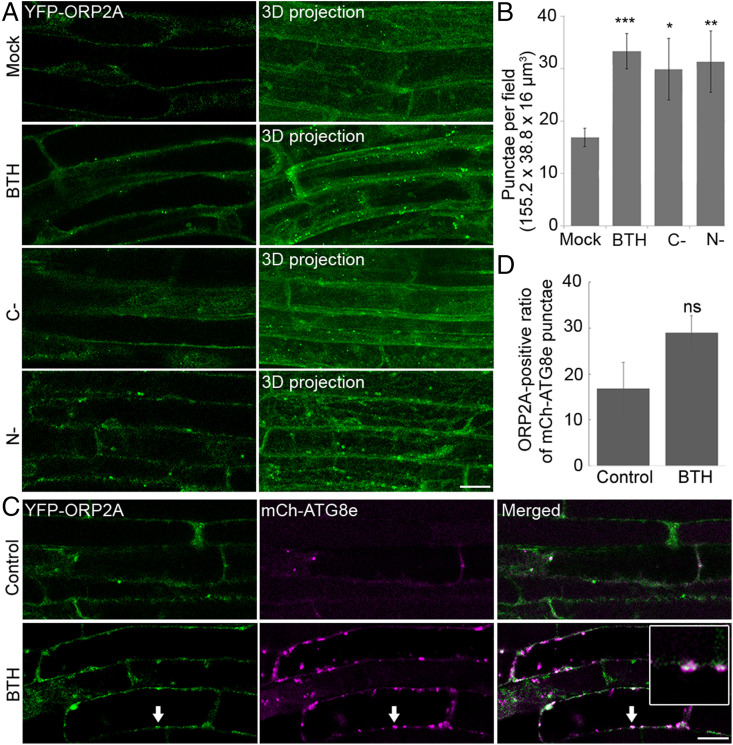
Since generation and transformation of protoplasts might be stresses that could induce autophagy, we next performed subcellular localization studies in plants. To confirm the autophagosomal localization of ORP2A observed in the protoplasts, we generated transgenic Arabidopsis plants expressing YFP-ORP2A under the control of a UBQ10 promoter. Consistent with results derived from transient expression in protoplasts (SI Appendix, Fig. S1A), transgenic Arabidopsis plants expressing YFP-ORP2A showed typical autophagic responses. As shown in Fig. 1, transgenic Arabidopsis root cells expressing YFP-ORP2A showed a mostly cytosolic pattern with a few of the punctae (mock) under normal conditions, but obviously changed to largely punctate patterns under three-dimensional (3D) confocal imaging projection upon autophagic induction by benzothiadiazole (BTH, a functional analog of salicylic acid) treatment, carbon starvation, and nitrogen depletion (Fig. 1A). Further quantification analysis supported this significant increase of YFP-ORP2A punctae upon these autophagic induction treatments (Fig. 1B). To further confirm the autophagosomal nature of these YFP-ORP2A–positive punctae, we also generated double-transgenic Arabidopsis plants coexpressing YFP-ORP2A and the autophagosome marker mCh-ATG8e. Indeed, the YFP-ORP2A–positive punctae showed partial colocalization with the mCh-ATG8e punctae upon various autophagy inductions by BTH treatment, carbon starvation, and nitrogen depletion (Fig. 1C and SI Appendix, Fig. S2C). In addition, treatment with concanamycin A (ConcA) greatly increased the mCh-ATG8e–positive autophagic bodies in the vacuoles, whereas the YFP-ORP2A punctae were not boosted nor colocalized with mCh-ATG8e in the vacuoles (SI Appendix, Fig. S2C), indicating that autophagy might not be the major pathway for YFP-ORP2A degradation. Although the YFP-ORP2A punctae showed correlated increases with the mCh-ATG8e punctae during autophagy induction, the colocalization rate (ratio of ORP2A-positive mCh-ATG8e punctae) showed no significant increase (Fig. 1D), implying that ORP2A may function at a certain stage of autophagosome formation. Indeed, time-lapse imaging analysis showed the dynamic association between the YFP-ORP2A punctae and the mCh-ATG8e–labeled autophagosomes (SI Appendix, Fig. S2). Interestingly, a fluorescent focus of mCh-ATG8e emerged at time 00:00 associated with the YFP-ORP2A signal but separated at 04:00, indicating that ORP2A might function at the autophagosome forming sites (SI Appendix, Fig. S2D).
Fig. 1.
YFP-ORP2A formed punctae upon autophagy induction. (A) Roots of 7-d-old transgenic Arabidopsis seedlings expressing YFP-ORP2A were subjected to various treatments of MS medium as control (mock), MS medium with 100 μM BTH for 6 h (BTH), carbon starvation (C-), and nitrogen deletion (N-), followed by confocal imaging and 3D projection. ΔZ = 0.8 μm, 20 stacks. (Scale bar, 10 μm.) (B) Quantification of the number of YFP-ORP2A–positive punctae per root section from different treatments shown in A. Error bars indicate ±SD; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, Student’s t test. (C) Roots of 7-d-old transgenic Arabidopsis seedlings coexpressing YFP-ORP2A/mCh-ATG8e were treated with BTH or without BTH (control) over a period of 6 h, followed by confocal imaging. Arrows indicate examples of colocalized punctae and enlargement in the box. (Scale bar, 10 μm.) (D) Quantification of the ORP2A-positive ratio of total mCh-ATG8e punctae from nine root sections (155.2 × 38.8 × 16 μm3) in treatments shown in C. Data represent the mean ± SD (standard deviation) of three independent experiments. ns, no significance.
To address the possible effect of overexpression using the UBQ10 promoter, we also generated transgenic Arabidopsis plants expressing ORP2A-eGFP (enhanced GFP) under the control of the native ORP2A promoter (ORP2Apro::OPP2A-eGFP) for further verification. Consistently, OPP2A-eGFP in transgenic ORP2Apro::OPP2A-eGFP lines showed similar responses to autophagic inductions by BTH treatment, carbon starvation, and nitrogen depletion (SI Appendix, Fig. S2 A and B). Taken together, these results indicated that YFP-ORP2A appears to be involved in the autophagic pathway in plants and that YFP-ORP2A under the control of either the UBQ10 promoter or the native ORP2A promoter showed similar autophagic responses in transgenic plants.
ORP2A Localizes to EPCSs.
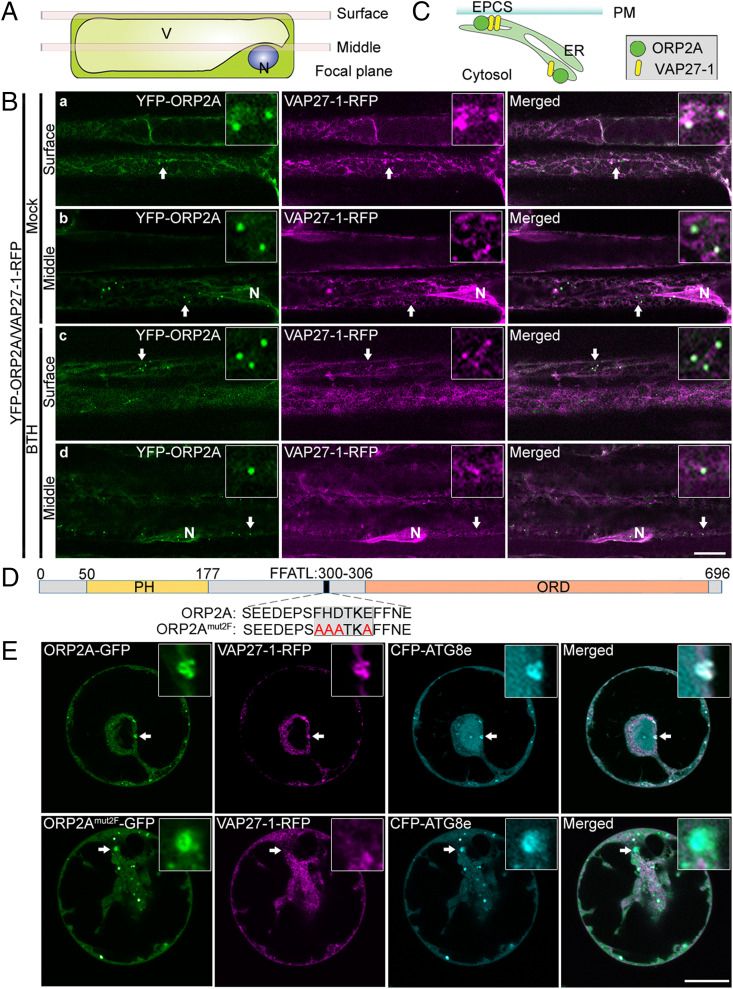
A working model from mammalian and yeast studies is that ORP works with VAP to mediate MCSs (21). A plant VAP homolog, VAP27-1, has been identified as an ER residential protein localizing to EPCSs and membrane junctions (25, 30). To test whether ORP2A is involved in MCSs in plants, we next generated transgenic Arabidopsis plants coexpressing YFP-ORP2A and VAP27-1-RFP. To observe the colocalization in cell cortical regions and equatorial planes, we adjusted the focal plane to the PM (surface) and perinuclear region (middle) (Fig. 2A). Consistent with previous studies (22, 30), VAP27-1-RFP showed largely punctae at the cell surface likely representing EPCSs (Fig. 2 B, a and c), but exhibited a slightly tubular pattern in the cytosol especially in the perinuclear region likely representing the ER (Fig. 2 B, b and d). Indeed, YFP-ORP2A showed colocalization with VAP27-1-RFP in punctae at the cell surface under normal (mock) or autophagy induction conditions by BTH treatment, indicating that YFP-ORP2A likely localized to EPCSs (Fig. 2 B, a and c and SI Appendix, Fig. S4 A, a and c) and associated with the VAP27-1–positive ER (Fig. 2C). Notably, the cytoplasmic YFP-ORP2A punctae largely colocalized with VAP27-1-RFP–positive signals (Fig. 2 B, b and d and SI Appendix, Fig. S4 A, b and d).
Fig. 2.
Dual localizations of ORP2A in MCSs. (A) Cartoon showing two axial positions of confocal imaging, plasma membrane (surface), and nucleic envelope (middle). N, nuclear; V, vacuole. (B) YFP-ORP2A punctae colocalized with VAP27-1-RFP. Roots of 7-d-old transgenic Arabidopsis seedlings coexpressing YFP-ORP2A and VAP27-1-RFP were treated with MS medium as control (a and b) or MS medium with BTH (c and d) for 6 h, followed by confocal imaging at the two focus positions, cell surface (surface) and midplane (middle), as illustrated in A. Quantification of colocalization is shown in SI Appendix, Fig. S4A. (Scale bar, 10 μm.) (C) Cartoon showing the dual localizations of ORP2A in EPCSs and VAP27-1–positive cytoplasmic ER. (D) Schematic diagram of ORP2A protein and mutagenesis of the FFATL motif. Numbers indicate the positions of amino acids (Top), while the WT construct (ORP2A) and the mutated construct ORP2Amut2F with mutated FFATL motif are shown below the diagram. (E) CFP-ATG8e and VAP27-1-RFP were transiently coexpressed together with ORP2A-GFP or ORP2Amut2F-GFP in Arabidopsis protoplasts, followed by confocal imaging. Arrows indicate examples of colocalized GFP/RFP punctae together with the autophagosomal marker CFP-ATG8e and their enlargement in the box. Quantification of colocalization is shown in SI Appendix, Fig. S4B. (Scale bar, 10 μm.)
The colocalization between VAP27-1-RFP and mCh-ATG8e (Figs. 1 and 2) indicated that YFP-ORP2A may locate to the ER/EPCSs and autophagosomes. To further test whether ORP2A and autophagosomes associated with the EPCSs, we transiently coexpressed the PM marker YFP-SCAMP (31) together with VAP27-1-YFP and CFP-ATG8e (SI Appendix, Fig. S3). To image the fluorescent signals from the PM and cytosol, we captured images from the cell surface and midplane, respectively (SI Appendix, Fig. S3A). As shown in SI Appendix, Fig. S3B, VAP27-1-YFP showed a punctate pattern representing the EPCSs at the cell-surface plane (22), whereas the CFP-ATG8e punctae were largely imaged in the midplane of the cell. Interestingly, a few ATG8e-positive punctae colocalizing with ORP2A were found proximately close to the PM, which may indicate EPCS-derived autophagosome formation (24). Taken together, these results indicated that the CFP-ATG8e–labeled autophagosomes were mostly cytosolic compartments with less association with the PM as well as the EPCSs. Further transient coexpressing of ORP2A-GFP, RFP-SCAMP, and CFP-ATG8e together in protoplasts confirmed the cytosolic localization of ORP2A-GFP and CFP-ATG8e with little association with the PM, demonstrating that the autophagosome-located (CFP-ATG8e–positive) ORP2A-GFP was largely distinct from the EPCSs (SI Appendix, Fig. S3C).
ORP2A Mediates ER–Autophagosomal Membrane Contact Sites.
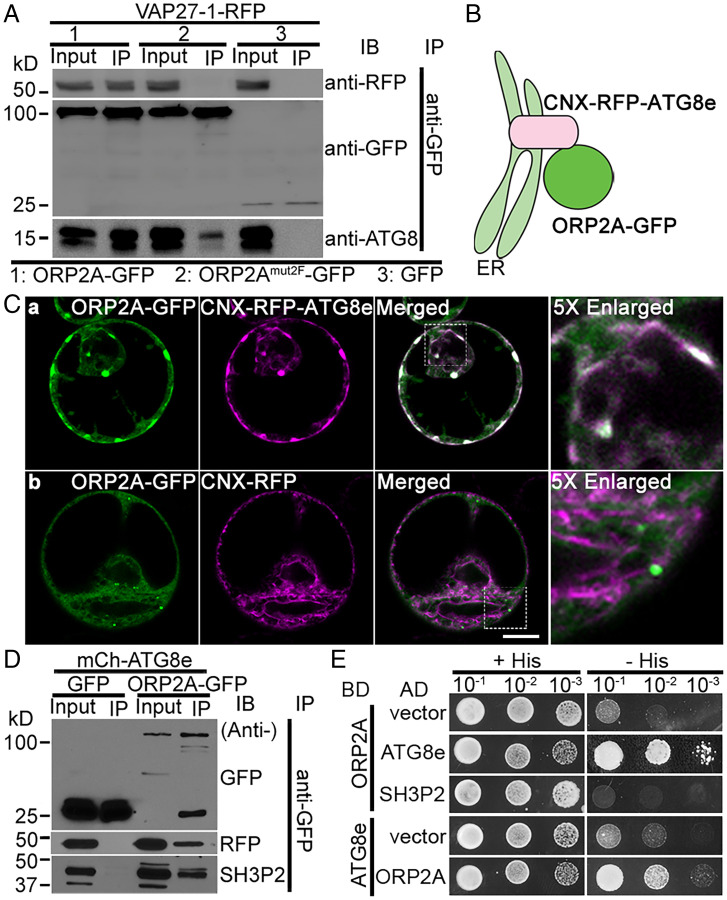
ORP2A contains an FFATL motif, which may be the potential interacting site with VAP proteins (32). To verify the importance of the FFATL motif for the colocalization of ORP2A with VAP27-1 in punctae, we generated an ORP2Amut2F-GFP fusion construct with a mutation on the FFATL motif (Fig. 2D) for subsequent coexpression analysis vs. ORP2A-GFP (Fig. 2E). Indeed, upon their transient coexpression with both VAP27-1-RFP and CFP-ATG8e in Arabidopsis protoplasts, the ORP2A-GFP punctae were concentrated on the sites containing both VAP27-1-RFP and CFP-ATG8e signals. In contrast, when ORP2Amut2F-GFP was coexpressed together with both VAP27-1-RFP and CFP-ATG8e in Arabidopsis protoplasts, ORP2Amut2F-GFP showed predominant colocalization with the CFP-ATG8e punctae lacking the VAP27-1-RFP signals (Fig. 2E). Since VAP27-1 and ATG8e have been known to localize to the ER/EPCS and autophagosomal membrane (phagophore/autophagosome), respectively, these results indicated that ORP2A may function in mediating ER–autophagosomal membrane contact sites (EACSs) by linking the ER-localized VAP27-1 together with the autophagosome-located ATG8e likely via VAP27–ORP2A–ATG8e interaction (SI Appendix, Fig. S5A), in which the VAP27–ORP2A interaction is achieved through the interaction of VAP27-1 with the FFATL motif of ORP2A (SI Appendix, Fig. S5B). Indeed, immunoprecipitation (IP) assays (SI Appendix, Supplementary Methods) showed that VAP27-1 interacts with ORP2A and ATG8 to form a protein complex but not with ORP2Amut2F (Fig. 3A). To test the possible role of VAP27-1 in forming and maintaining the MCSs, we expressed the amiRNA to knock down VAP27-1 in Arabidopsis protoplasts coexpressing ORP2A-GFP, VAP27-1-RFP, and CFP-ATG8e. Results showed that neither ORP2A-GFP localization nor CFP-ATG8e–positive autophagosome formation was obviously affected by the VAP27-1 KD (SI Appendix, Fig. S6A). The amiRNA KD efficiency was confirmed by immunoblot analysis of the expression level of VAP27-1-RFP (SI Appendix, Fig. S6B). To further test whether ORP2A can still bind to the ER without VAP27, we next used the VAP27-1 RNA interference (RNAi) line and the CRISPR line that knocked out both VAP27-1 and VAP27-3 (22). We coexpressed ORP2A-GFP together with an ER marker (RFP-CNX, calnexin) in leaf protoplasts of VAP27-1 RNAi KD or vap27-1/vap27-3 CRISPR knockout (KO) lines, showing similar ER association of ORP2A-GFP punctae in these VAP27-1 KD and KO mutants (SI Appendix, Fig. S6C). Thus, ORP2A can bind to membranes without VAP27-1. Taken together, ORP2A mediates EACSs likely by interacting with the ER-localized VAP27-1 via the FFATL motif.
Fig. 3.
ORP2A interacts directly with both ATG8e and VAP27-1. (A) ORP2A interacted with VAP27-1 in an IP assay. Cell lysates (input) prepared from Arabidopsis PSBD protoplasts transiently coexpressing VAP27-1-RFP with ORP2A-GFP/ORP2Amut2F-GFP/GFP (control) (Fig. 2B) were subjected to GFP-Trap assays and subsequent immunoblotting (IB) analysis using GFP, RFP, or ATG8 antibodies as indicated. (B) Working principle of the CNX recruitment assay. RFP-ATG8e was conjugated with the ER transmembrane protein CNX, resulting in ER localization of CNX-RFP-ATG8e with its C terminus facing the cytoplasm, thus recruiting ORP2A to the ER upon their coexpression because of ATG8e–ORP2A interaction. (C) ORP2A-GFP was recruited to the ER membrane by CNX-RFP-ATG8e. ORP2A-GFP was transiently coexpressed with CNX-RFP-ATG8e (a) or CNX-RFP (b, control), respectively, in Arabidopsis protoplasts, followed by confocal imaging. (Scale bar, 10 μm.) (D) ORP2A interacted with ATG8e and SH3P2 in IP assays. Cell lysates prepared from Arabidopsis PSBD protoplasts transiently coexpressing mCh-ATG8e/GFP (control) or mCh-ATG8e/ORP2A-GFP were subjected to GFP-Trap assays and subsequent IB analysis using GFP, RFP, or SH3P2 antibodies as indicated. (E) Yeast two-hybrid analysis of the binary interactions between ORP2A and ATG8e or SH3P2. AD, activation domain; BD, binding domain.
ORP2A Is Recruited to Autophagosomal Structures by Interacting with ATG8e.
To further understand the molecular links between ORP2A and the autophagy machinery, we next investigated the possible recruitment and interaction between ORP2A and ATG8e. We first designed and used a CNX recruitment assay to test the direct interaction between ATG8e and ORP2A in vivo, in which the ER membrane protein CNX was anchored to RFP-ATG8e, and thus CNX-RFP-ATG8e would show a tubular ER pattern along with its interacting proteins (e.g., ORP2A) upon their transient coexpression in Arabidopsis protoplasts (Fig. 3B). Indeed, ORP2A-GFP was specifically recruited by CNX-RFP-ATG8e to relocate to the ER upon their coexpression in Arabidopsis protoplasts (Fig. 3C). IP assays also showed a possible interaction of ORP2A with both ATG8e and SH3P2 (Fig. 3D). Further yeast two-hybrid assays (SI Appendix, Supplementary Methods) demonstrated that ORP2A directly interacts with ATG8e rather than SH3P2 (Fig. 3E). Taken together, ORP2A may mediate EACSs by interacting with the ER-localized VAP27-1 via the FFATL motif and the autophagosome-located ATG8e in plant cells.
Immunogold-Transmission Electron Microscopy and Electron Tomography Analysis of EACSs.
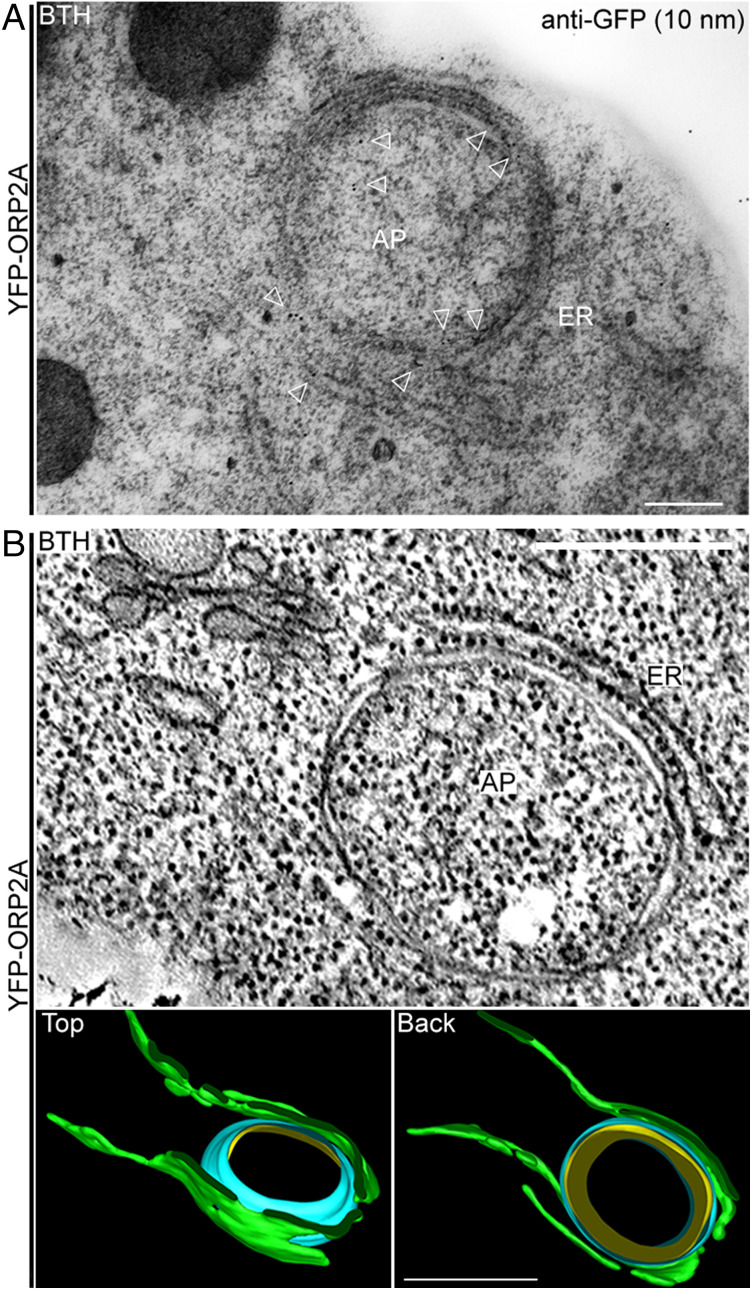
To further illustrate the ORP2A-positive EACSs at the ultrastructural level, we performed immunogold-TEM (transmission electron microscopy) and ET (electron tomography) analysis. Transgenic Arabidopsis seedlings expressing YFP-ORP2A were used for immunogold-TEM analysis with GFP antibodies to label YFP-ORP2A. Indeed, gold particles were specifically labeled alongside an autophagosome-like structure surrounded by the ER, likely representing EACSs (Fig. 4A). Three-dimensional ET analysis was also performed to have a close-up observation of an EACS (Fig. 4B). The reconstructed 3D model of the EACS showed that the ER (green) and the autophagosome outer membrane (blue) closely contacted each other without fusion (Fig. 4B).
Fig. 4.
Immunogold-TEM and ET analysis of EACSs. (A) Five-day-old transgenic Arabidopsis seedlings expressing YFP-ORP2A were treated with BTH for 6 h prior to HPFS and subsequent immunogold-TEM analysis using GFP antibodies. Arrowheads indicate examples of gold particle labeling. (Scale bar, 300 nm.) (B) Three-dimensional ET analysis of an EACS in Arabidopsis seedlings described in A. Two continuous 250-nm-thick sections were used for the ET analysis. (B, Top) Tomographic slice showing a representative example of the EACS in BTH-treated cells. (B, Bottom) The corresponding 3D tomographic model showing the profile of an EACS, which consists of ER (green), autophagosome outer membrane (blue), and inner membrane (yellow), viewed from the top and back. (Scale bars, 100 nm.) AP, autophagosome.
ORP2A Is Essential for Seedling Development and Autophagosome Formation.
We next used a loss-of-function approach to study the functional roles of ORP2A in plant autophagy. We failed to obtain T-DNA insertional KO mutants of ORP2A, likely due to the essential function of ORP2A in Arabidopsis as previously described for other genes (33). Alternatively, we therefore generated transgenic Arabidopsis plants expressing amiRNA targeting ORP2A under the control of the dexamethasone (DEX)-inducible promoter (DEX::amiR2A). Several transgenic lines with the three different microRNA targeting sites (SI Appendix, Fig. S7A) were obtained. When germinated and grown on DEX-containing Murashige and Skoog (MS) plates (SI Appendix, Supplementary Methods), the three DEX::amiR2A lines exhibited similar arrested growth phenotypes (SI Appendix, Fig. S7B). RT-PCR analysis of 7-d-old DEX::amiR2A seedlings pretreated with DEX showed that the ORP2A messenger RNA was properly knocked down (SI Appendix, Fig. S7C). As the microRNA targeting site of the DEX::amiR2A-1 line is on its 3′ untranslated region (UTR), the seedling phenotype could be expected to be rescued by introducing the full-length ORP2A coding DNA sequence. Indeed, overexpression of ORP2A-GFP (but not the GFP control) in DEX::amiR2A-1 (i.e., ORP2A-GFP/DEX::amiR2A-1) complemented the arrested growth seedling phenotype in the presence of DEX (SI Appendix, Fig. S7 D and E), demonstrating that ORP2A is required for seedling development and that the ORP2A-GFP fusion is fully functional in plants.
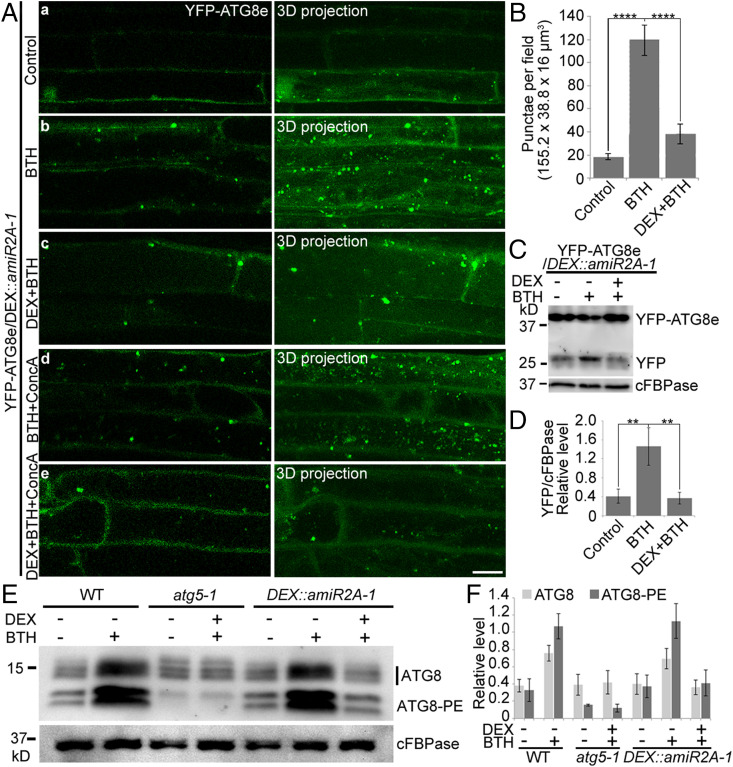
To further determine whether ORP2A KD can affect autophagy, we studied YFP-ATG8e–positive autophagosome formation in transgenic YFP-ATG8e/DEX::amiR2A-1 seedlings upon various treatments (Fig. 5A). As expected, BTH or BTH+ConcA treatments induced the visible accumulation of YFP-ATG8e punctae in the cytoplasm and vacuoles, respectively (Fig. 5 A, b and d). However, when similar treatments were performed in the presence of DEX with knocked-down ORP2A, the visible YFP-ATG8e punctae present in the cytoplasm or vacuoles were dramatically decreased (Fig. 5 A, c and e), which was further supported by quantification analysis (Fig. 5B). To further test the autophagic flux in ORP2A KD plants, we performed a fluorescent protein cleavage assay to analyze the YFP core accumulation in YFP-ATG8e/DEX::amiR2A-1 seedlings (34). Consistent with the confocal analysis, upon BTH treatment, an increased free YFP band was detected in control seedlings without DEX, whereas less free YFP was detected in the DEX-induced ORP2A KD line (Fig. 5C), which was further confirmed by quantification analysis of the relative free YFP level compared with control (cFBPase) (Fig. 5D). Further ATG8 lipidation assays were employed to examine the autophagy activity in ORP2A KD plants (34), together with the wild type (WT) and atg5-1 mutant (lipidation defect mutant) as controls. As shown in Fig. 5D, ATG8-PE (the lipidated form of ATG8) was increased in BTH-treated but not in DEX+BTH–treated DEX::amiR2A-1 seedlings, whereas the controls showed normal detection levels of ATG8-PE in WT with or without BTH treatments but hardly detected in the atg5-1 mutant as expected, respectively (Fig. 5 E and F). We also transformed the autophagy cargo receptor NBR1-RFP (neighbor of BRCA1) (35) into the ORP2A amiRNA KD to generate NBR1-RFP/DEX::amiR2A-1 plants to further study the autophagy defect of NBR1 in ORP2A KD (SI Appendix, Fig. S8). As expected, treatments of BTH and BTH+ConcA resulted in massive accumulation of NBR1-RFP punctae in the cytosol and vacuoles, respectively, compared with the control (SI Appendix, Fig. S8). However, in cells pretreated with DEX with knocked-down ORP2A, both BTH and BTH+ConcA treatments resulted in cytosolic accumulation of tubular NBR1-RFP structures in the cytosol but not in the vacuoles under BTH+ConcA treatment (DEX+BTH+ConcA) (SI Appendix, Fig. S8). These results indicated that normal autophagosome formation and likely subsequent autophagosome–vacuole fusion might be affected in ORP2A KD. To find out whether ORP2A KD plants exhibit autophagy-related phenotypes, we applied carbon starvation and nitrogen deprivation experiments (SI Appendix, Supplementary Methods) (36). Indeed, ORP2A KD plants displayed enhanced senescence-like symptoms compared with WT (Col-0) in carbon starvation and were hypersensitive to nitrogen deprivation, phenocopying atg5-1, a positive control which exhibited hypersensitivity to nutritional stress (SI Appendix, Fig. S9). These autophagy-related phenotypes of ORP2A KD could be fully reversed to WT phenotypes by complementation of expressing ORP2A-GFP (SI Appendix, Fig. S9). Taken together, these results indicated that ORP2A is required for seedling development and normal autophagosome formation during plant autophagy.
Fig. 5.
ORP2A is required for plant autophagy. (A) KD of ORP2A affected autophagy. Seven-day-old YFP-ATG8e/DEX::amiR2A seedlings were treated with BTH (b), DEX+BTH (c), BTH+ConcA (d), and DEX+BTH+ConcA (e) for 6 h, followed by confocal imaging and 3D projection. ΔZ = 0.8 μm, 20 stacks. For DEX treatments, DEX was pretreated for 48 h prior to other treatments. (Scale bar, 10 μm.) (B) Quantification of YFP-ATG8e–positive punctae per root section in A. Error bars indicate ±SD; ****P ≤ 0.0001, Student’s t test. (C) Immunoblot detection of YFP-ATG8e turnover in transgenic YFP-ATG8e/DEX::amiR2A-1 plants. Seven-day-old seedlings of YFP-ATG8e/DEX::amiR2A-1 were subjected to treatments of DEX/BTH/ConcA as indicated, followed by protein extraction and immunoblot detection with GFP antibodies. cFBPase antibodies were used as a loading control. PE, phosphatidylethanolamine. (D) Quantification of free YFP normalized to cFBPase in C. At least three independent replicates; error bars indicate ±SD; **P ≤ 0.01, Student’s t test. (E) Immunoblot detection of the ATG8 lipidation level in WT, atg5-1, and DEX::amiR2A-1. Seven-day-old seedlings of WT, atg5-1, and DEX::amiR2A-1 were treated with/without BTH for 6 h and/or DEX for 48 h as indicated, followed by protein extraction and immunoblot analysis with ATG8 antibodies. cFBPase antibodies were used as a loading control. (F) Quantification of ATG8 upper (ATG8) and lower (ATG8-PE) bands normalized to cFBPase in E. At least three independent replicates; error bars indicate ±SD.
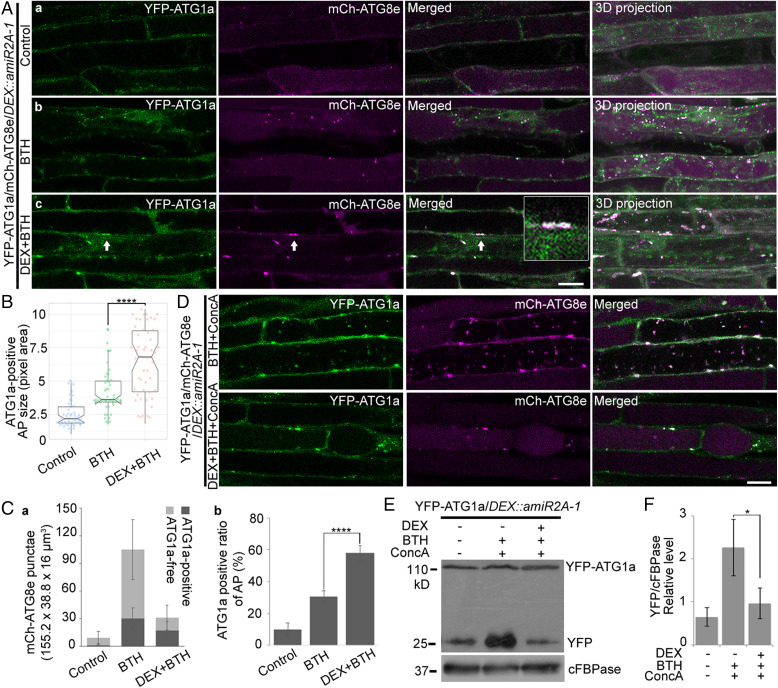
ORP2A KD Causes Accumulation of ATG1a in ATG8e-Positive Autophagosomal Structures.
In plants, the ATG1 kinase complex initiates the formation of the phagophore and responds to nutritional stresses (10). To find out if ORP2A KD would affect the early stage of autophagosome formation, we next monitored the fate of the ATG1 initiation complex in root cells of transgenic YFP-ATG1a/mCh-ATG8e/DEX::amiR2A-1 plants. As shown in Fig. 6A, upon autophagy induction by BTH treatment, YFP-ATG1a showed strong colocalization in the mCh-ATG8e–positive autophagosomal punctae as expected (Fig. 6 A, b). However, in DEX-induced ORP2A KD cells, YFP-ATG1a showed colocalization with mCh-ATG8e in abnormal tubular/enlarged autophagosomal structures (Fig. 6 A, c, arrows), and the sizes of the YFP-ATG1a–positive autophagosomal structures (colocalized YFP-ATG1a/mCh-ATG8e punctae) were significantly larger in cells of ORP2A KD plants than the WT (Fig. 6B). Further quantification analysis showed that the number of both ATG1a-free and ATG1a-positive autophagosomal structures was decreased in root cells of ORP2A KD plants upon autophagic induction (Fig. 6 C, a). Meanwhile, the quantification of the colocalization rate (ATG1a-positive ratio) showed that 57.71% of mCh-ATG8e punctae were colocalized with YFP-ATG1a upon autophagic induction in DEX-pretreated ORP2A KD plants, whereas the colocalization rates in control groups and BTH-treated groups were 10.14 and 30.46%, respectively (Fig. 6 C, b). Thus, both the ATG1a-positive initiation complexes and the ATG8e-positive autophagosomes showed higher colocalization rates in ORP2A KD plants than WT.
Fig. 6.
ATG1a recruited and accumulated on the enlarged autophagosomal structures in ORP2A KD plants. (A) Seven-day-old transgenic YFP-ATG1a/mCh-ATG8e/DEX::amiR2A-1 seedlings were treated with/without BTH for 6 h prior to confocal imaging and 3D projection (a and b). ΔZ = 0.8 μm, 20 stacks. For DEX+BTH treatment (c), DEX was pretreated for 48 h before BTH treatment, followed by confocal imaging and 3D projection. Arrows indicate examples of YFP-ATG1a–positive autophagic punctae containing mCh-ATG8e and enlargement in the box. (Scale bar, 10 μm.) (B) Quantification of the sizes of mCh-ATG8e–positive autophagosomes colocalized with YFP-ATG1a in A. Fifty punctae were measured using ImageJ from at least 20 root cells in each of the treatments shown in A. Error bars indicate ±SD; ****P ≤ 0.0001, Student’s t test. (C) Quantification of mCh-ATG8e punctae (a) and colocalization rate of ATG1a-positive mCh-ATG8e punctae (b) in A. (D) Seven-day-old transgenic YFP-ATG1a/mCh-ATG8e/DEX::amiR2A-1 seedlings were treated with BTH and ConcA with/without DEX pretreatment as indicated, followed by confocal imaging. (Scale bar, 10 μm.) (E) Immunoblot analysis of YFP-ATG1a protein turnover in YFP-ATG1a/DEX::amiR2A-1 seedlings. Seven-day-old transgenic YFP-ATG1a/DEX::amiR2A-1 seedlings were treated with/without BTH and ConcA for 6 h or pretreated with DEX for 48 h as indicated, followed by protein extraction and immunoblot detection with various antibodies. YFP-ATG1a and free YFP protein levels were detected by GFP antibodies. cFBPase antibodies were used as protein loading control. (F) Quantification of free YFP level normalized to cFBPase in E. Error bars indicate ±SD; *P ≤ 0.05, Student’s t test.
Under autophagic induction (e.g., nutrient-limiting conditions), ATG1a is degraded via an autophagic route (11). To monitor the fate of the ATG1 complex in the autophagic recycling process, we applied ConcA treatment to prevent vacuolar acidification to test the YFP-ATG1a degradation in ORP2A KD lines. As shown in Fig. 6D, YFP-ATG1a reached the vacuoles and associated with mCh-ATG8e in autophagic bodies in control cells, whereas vacuolar YFP-ATG1a and autophagic bodies were rarely seen in DEX-induced ORP2A KD lines, indicating that the autophagic recycling of ATG1a was blocked upon ORP2A KD. Further immunoblot analysis of YFP-ATG1a/DEX::amiR2A-1 seedlings under various treatments showed that the free YFP was accumulated in control cells upon BTH-induced autophagy, but such free YFP was significantly reduced upon DEX-induced ORP2A KD (Fig. 6E). Taken together, these results indicated that ATG1 proteins were likely trapped in an abnormal/enlarged autophagosomal structure and prevented their vacuolar delivery in ORP2A KD lines.
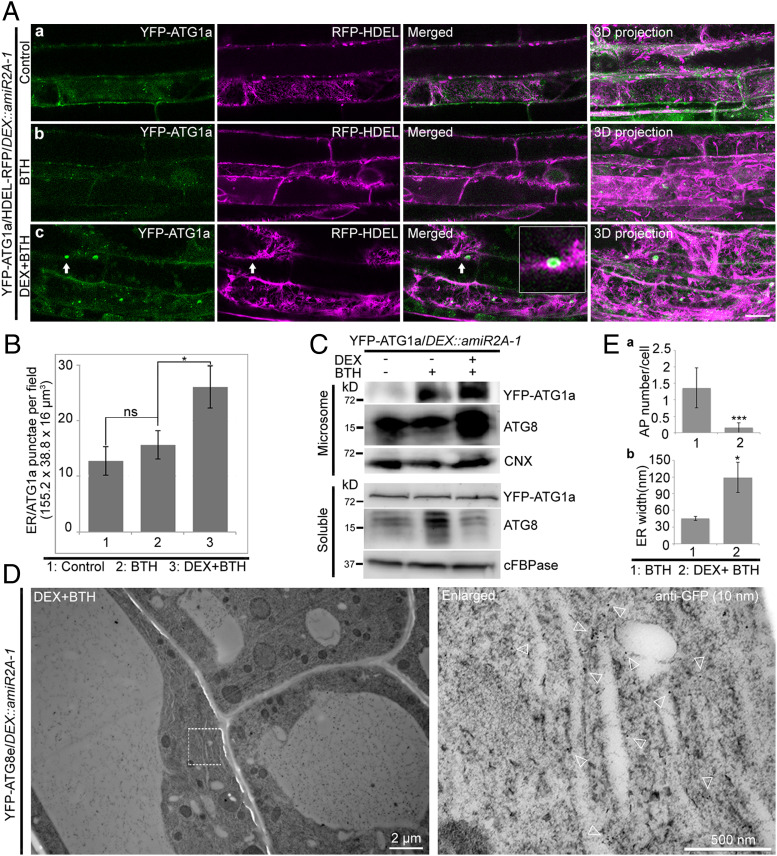
ATG1a/ATG8e Accumulates in ER Membranes during Autophagy in ORP2A KD Seedlings.
The ER is the most important membrane source for the forming autophagosome (37). To find out the possible link between the accumulated ATG1a/ATG8e–positive structures and the ER, we next generated double-transgenic lines of YFP-ATG1a/DEX::amiR2A-1 and mCh-ATG8e/DEX::amiR2A-1 coexpressing with the known ER markers of RFP-HDEL or GFP-HDEL. Indeed, in root cells of YFP-ATG1a/RFP-HDEL/DEX::amiR2A-1 seedlings, the accumulated ATG1a tubular/enlarged punctae largely associated with the ER marker RFP-HDEL upon BTH-induced autophagy and DEX-induced ORP2A KD (Fig. 7 A, c, arrows), in which the ER-located YFP-ATG1a punctae increased from 15.67 in BTH to 26.11 in DEX+BTH (Fig. 7B). Similar results were also obtained from the mCh-ATG8e/GFP-HDEL/DEX::amiR2A-1 lines, in which the accumulated ATG8e tubular/enlarged punctae were largely associated with the ER marker GFP-HDEL upon BTH+DEX treatments (SI Appendix, Fig. S10 A, c, arrows), in which the ER-associated autophagosomal structures increased from 9.72% in BTH to 65.12% in DEX+BTH treatments (SI Appendix, Fig. S10B).
Fig. 7.
ATG1a and ATG8e accumulated in and associated with the ER membrane during autophagy in ORP2A KD plants. (A) Seven-day-old transgenic YFP-ATG1a/RFP-HDEL/DEX::amiR2A-1 seedlings were treated without (a) or with BTH (b) for 6 h, followed by confocal imaging and 3D projection. ΔZ = 0.8 μm, 20 stacks. For DEX+BTH treatment (c), DEX pretreatment was performed for 48 h before BTH treatment, followed by confocal imaging and 3D projection. Arrows indicate examples of colocalized dots and the enlargement in the box. (Scale bar, 10 μm.) (B) Quantification of the numbers of RFP-HDEL–positive YFP-ATG1a (ER/ATG1a) punctae per root section in different treatments shown in A from three independent experiments. Error bars indicate ±SD; ns, no significance; *P ≤ 0.05. (C) Immunoblot analysis of YFP-ATG1a and ATG8 accumulation in microsomes of ORP2A KD plants. Seven-day-old transgenic YFP-ATG1a/amiR2A-1 seedlings were subjected to treatments as shown in A, followed by isolation of microsomes. Microsome and soluble fractions were analyzed by immunoblot detection using GFP, ATG8, and CNX (or cFBPase) antibodies as indicated. Protein loading was normalized by immunoblot detection of CNX (microsome) or cFBPase (soluble) using CNX/cFBPase antibodies. (D) Immunogold-TEM analysis of YFP-ATG8e accumulation in ORP2A KD plants. Five-day-old transgenic YFP-ATG8e/amiR2A-1 seedlings were pretreated with DEX for 48 h, followed by BTH treatment for 6 h and subsequent high-pressure frozen/freeze-substituted root cells. Ultrathin sections were then prepared for immunogold labeling using GFP antibodies. The white dashed-line box (Left) is enlarged (Right). Arrowheads indicate examples of gold particles. (E) Quantification of the numbers of autophagosomes per cell (a) and ER width (b) in F and in SI Appendix, Fig. S11 as control. (E, a) Data were collected from three independent experiments. Error bars indicate ±SD; ***P ≤ 0.001. (E, b) ER-labeled immunogold particles were measured. The ER in SI Appendix, Fig. S11 was used as control. ER width was measured by using ImageJ. Error bars indicate ±SD; *P ≤ 0.05.
To further confirm the association of the accumulated ATG1a/ATG8e proteins with the ER membrane, we isolated microsomes from YFP-ATG1a/DEX::amiR2A-1 seedlings upon various BTH and DEX treatments as indicated, followed by Western blot analysis using GFP/ATG8 antibodies to measure the YFP-ATG1a/ATG8e proteins, with CNX antibodies as loading control for normalization. Indeed, the amounts of YFP-ATG1a/ATG8e proteins presented in the microsomes were increased significantly upon BTH-induced autophagy and DEX-induced ORP2A KD as compared with the controls of mock and BTH treatment (Fig. 7C, microsome). As controls, the corresponding soluble fractions were also analyzed by immunoblot with GFP/ATG8 antibodies and normalized by cFBPase antibodies (Fig. 7C, soluble), in which ATG8 was significantly increased upon BTH treatment compared with the control and DEX+BTH treatment, whereas YFP-ATG1a showed no obvious change in these treatments (Fig. 7C, soluble).
To study the nature of ATG1a/ATG8e accumulation in ORP2A KD seedlings, we next performed immunogold-TEM analysis with GFP antibodies in root cells of YFP-ATG8e/DEX::amiR2A-1 seedlings upon DEX+BTH treatments. As shown, autophagosome-like structures were hardly observed (Fig. 7 D and E, a), whereas gold particles for YFP-ATG8e were labeled in the swollen ER membranes (Fig. 7 D and E, b). As controls, YFP-ATG8e/DEX::amiR2A-1 seedlings were treated with BTH only, followed by TEM analysis. As shown in SI Appendix, Fig. S11, autophagosome numbers in the control group were significantly higher than in ORP2A KD cells (Fig. 7 E, a). Meanwhile, ER luminal width in ORP2A KD root cells was significantly increased compared with the control (Fig. 7 E, b). Taken together, both ATG1a and ATG8e accumulated in the ER membrane in ORP2A KD plants during autophagy.
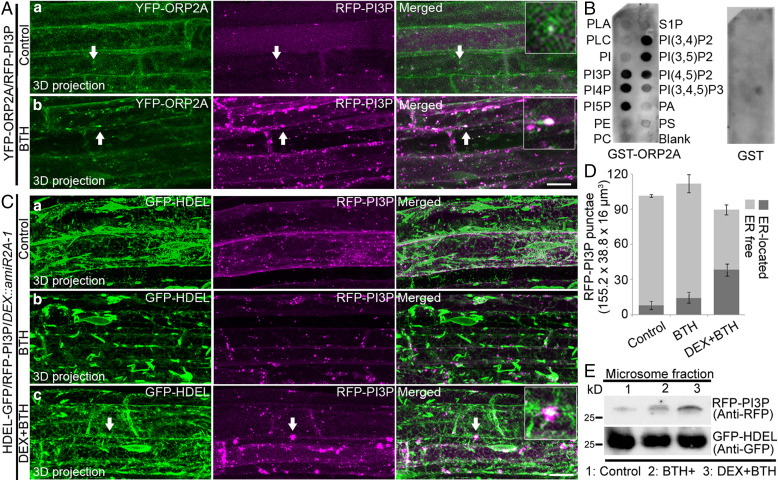
Down-Regulation of ORP2A Leads to PI3P Accumulation during Autophagy.
In mammalian cells and yeast, ORP/Osh binds multiple lipids and has a strong affinity for phosphoinositides (38), whereas the activation of ULK1/ATG1 results in PI3P synthesis (14), which takes place at autophagosome forming sites, presumably at the ER, recruiting a series of PI3P-binding autophagy effectors essential for autophagosome biogenesis (39). We thus hypothesized that ORP2A may function as a lipid regulator in plants and next generated double-transgenic plants coexpressing YFP-ATG8e with the PI3P reporter RFP-PI3P (RFP-2×FYVE) (40). Indeed, in root cells of transgenic YFP-ATG8e/RFP-PI3P seedlings, YFP-ATG8e largely colocalized with RFP-PI3P in punctae under both normal and autophagy induction conditions (SI Appendix, Fig. S12), indicating the autophagic activity and involvement of PI3P in plant autophagy. To further study the relationship between PI3P and ORP2A, we also generated double-transgenic YFP-ORP2A/RFP-PI3P plants. Indeed, YFP-ORP2A showed colocalization with RFP-PI3P in punctae in root cells of transgenic YFP-ORP2A/RFP-PI3P seedlings upon autophagy induction by BTH treatment (Fig. 8A). Quantification analysis showed that 26.4 and 35.9% of the RFP-PI3P punctae colocalized with YFP-ORP2A punctae before (control) and after BTH treatments, respectively (SI Appendix, Fig. S13). Further in vitro lipid-binding assays (SI Appendix, Supplementary Methods) showed that ORP2A binds multiple lipids including PI3P (Fig. 8B). To monitor PI3P behavior during autophagy in ORP2A KD lines, we next generated transgenic GFP-HDEL/RFP-PI3P/DEX::amiR2A-1 lines. Interestingly, the enlarged RFP-PI3P punctae were associated with the ER marker GFP-HDEL signals in cells upon BTH-induced autophagy and DEX-induced ORP2A KD (Fig. 8 C, c). Further quantification analysis showed that 43.4% of the RFP-PI3P punctae were associated with the ER in cells treated with DEX+BTH, whereas the numbers were 7.9 and 12.8% in mock and BTH-treated cells, respectively (Fig. 8D). Further immunoblot analysis using microsomes isolated from GFP-HDEL/RFP-PI3P/DEX::amiR2A-1 seedlings showed that the RFP-PI3P protein levels increased upon BTH-induced autophagy and DEX-induced ORP2A KD (Fig. 8E). Taken together, these results indicated that ORP2A KD caused enrichment of PI3P in the ER membrane.
Fig. 8.
PI3P accumulates in the ER membrane during autophagy in ORP2A KD plants. (A) Seven-day-old transgenic YFP-ORP2A/RFP-PI3P seedlings were treated without (a) or with BTH (b) for 6 h, prior to confocal imaging and 3D projection. ΔZ = 0.8 μm, 20 stacks. Arrows indicate examples of colocalized punctae and the enlargement in the box. (Scale bar, 10 μm.) (B) ORP2A binds to multiple phospholipids. Purified GST proteins or recombinant proteins (GST-ORP2A) were subjected to an in vitro lipid-binding assay, followed by immunoblot detection with GST antibodies. (C) Seven-day-old transgenic GFP-HDEL/RFP-PI3P/DEX::amiR2A-1 seedlings were treated without (a) or with BTH (b) for 6 h, followed by confocal imaging and 3D projection. ΔZ = 0.8 μm, 20 stacks. For DEX+BTH treatment (c), DEX pretreatment was performed for 48 h before BTH treatment (c), followed by confocal imaging and 3D projection. Arrows indicate examples of colocalized punctae and enlargement in the box. (Scale bar, 10 μm.) (D) Quantification of the numbers of ER-free and ER-located RFP-PI3P punctae per root section of different treatments shown in C. Error bars indicate ±SD. (E) Immunoblot detection of RFP-PI3P accumulation in GFP-HDEL/RFP-PI3P/DEX::amiR2A-1 plants shown in C. Seven-day-old YFP-ATG1a/DEX::amiR2A-1 seedlings were subjected to treatments as shown in C, followed by isolation of microsomes and subsequent immunoblot detection using RFP or GFP antibodies as indicated. Immunoblot detection of GFP-HDEL using anti-GFP was used as a loading control.
Discussion
In this study, we propose a working model for the possible roles of ORP2A in plant autophagy as compared with those in mammals (SI Appendix, Fig. S14): Under normal conditions, ORP2A locates to EPCSs and interacts with VAP27-1 (SI Appendix, Fig. S14A); however, upon autophagic induction, ORP2A interacts with VAP27-1 and ATG8e as a complex to bridge the EACSs, whereas ORP2A may also work as a PI3P regulator to control local PI3P redistribution (SI Appendix, Fig. S14 B, b).
In mediating EPCSs, more than one conserved tether has been identified among species. In a mammalian model, one of the linkers mediating the EPCSs is ORP5/8, which lacks the ER targeting sequence FFAT motif but contains a single C-terminal transmembrane domain to anchor the protein onto the ER membrane, while an N-terminal PH domain binds the PI4P in the PM (41). In plants, continuing studies of Arabidopsis EPCSs are expanding our knowledge about their function in the cellular and developmental process (42–45). A well-established model for an EPCS in Arabidopsis is mediated by the cytoskeleton, NET3C, and VAP27-1 (SI Appendix, Fig. S14A) (30). In this study, we demonstrated that ORP2A interacts with VAP27-1 and localizes to the VAP27-1–labeled EPCSs in the cell cortical layer. As a potential LTP, ORP2A might represent a missing part of a functioning EPCS, because nonvesicular lipid transfer is one of the most important functions for an MCS.
EACSs play a pivotal role in autophagosome formation (46). In recent mammalian studies, two molecular linkers have been identified to mediate the contacts between ER and autophagosomal membranes: 1) the ATG2–WIPI complex, and 2) VAPs and the FFAT motif containing autophagic core machinery proteins (SI Appendix, Fig. S14B) (47, 48). In ATG2-mediated EACSs, ATG2 may function to transfer phospholipids from the ER to an expanding IM through an unknown mechanism (49–51); however, the identification of ATG2 as a multifunctional protein provides insights into the regulation of an expanding IM. In the other model for autophagosomal membrane tethering, the main mechanism involves the interaction of VAPs with varieties of autophagic core machineries including ULK1, FIP200, and WIPI2 (SI Appendix, Fig. S14B) (47). In plants, a close association between the ER and the autophagosomal membrane has been observed but the MCSs and the molecular tethering remain unknown. In this study, we first identified the molecular tethering of an EACS, which involves ORP2A, VAP27-1, and ATG8e (SI Appendix, Fig. S14 B, b). In this architecture, ORP2A directly interacts with ATG8e on the autophagosomes, while anchoring to the ER membrane by interacting with VAP27-1 via an FFATL motif. Interestingly, ORP2A tethers other membranes (i.e., autophagosomal membranes) by interacting with the autophagic protein ATG8e, rather than merely relying on lipid binding, which is unique in comparison with any other known mechanisms. Notably, ORP2A can bind to the ER membrane in the absence of VAP27-1 (SI Appendix, Fig. S6), indicating that there might be other tethers anchoring ORP2A onto the membranes. Since the VAP27 family has 10 members (22) and ORP2A binds to multiple lipids in vitro (Fig. 8B), the binding of ORP2A to the membrane could be achieved indirectly via interacting with other VAP27 members or directly via binding to lipids in the membranes. Future studies are needed to test these possibilities in plants. Interestingly, only two VAP27 homologs (VAPA and B) participated in forming MCSs in mammalian cells (26). It will be interesting in future studies to further explore how ORP2A is tethered onto membranes in plants.
ORPs show distinct functions among different organisms. For example, ORP family members in humans exhibit functional diversity (52); however, ORPs in yeast and Caenorhabditis elegans display compensatory functions because deletion of all ORPs but not an individual one in yeast and C. elegans resulted in a lethal phenotype (53, 54). In mammals, deletion of OSBP is lethal at early stages of embryogenesis (55). In plants, the function of ORPs in plant development is elusive. In this study, we employed an inducible amiRNA KD approach to study the function of ORP2A in plants. Interestingly, unlike most autophagy-related mutants in plants, ORP2A KD seedlings displayed a strong developmental defect (SI Appendix, Fig. S7), which could be explained by the strong association between ORP2A and PI3P/PI3K (Fig. 8), as shown in previous studies of the PI3K components (13, 56, 57). Since an ER–MCS protein shows a multiple lipid–binding ability, it remains elusive if ORP2A would be involved in other pathways in plants.
In mammalian cells, a multiple lipid–binding protein, ATG2, mediates the EACSs and transfers lipids between membranes (49, 58). Knocking out the mammalian ATG2s (ATG2A/B) resulted in the accumulation of autophagosome-associated proteins including LC3B-II, GL1, and p62 under full medium conditions (49). Furthermore, LC3B fails to undergo autophagy flux but accumulates at abnormally large immunofluorescent structures (49). Interestingly, such an autophagy phenotype can be reversed by expressing the mini-ATG2A only with lipid transport ability, indicating that a main function of ATG2A is lipid transport (49). In this study, KD of ORP2A caused impaired autophagy levels and ATG1a/ATG8e accumulation in the swollen ER membrane (Figs. 5, 6, and 7), indicating that ORP2A may play a role in the process of autophagosome initiation and progression from the ER. Meanwhile, PI3P enrichment can be detected in the ER membrane in Arabidopsis ORP2A KD plants (Fig. 8), indicating that an EACS is essential for proper regulation of lipids. Since ORP2A works as an MCS protein and may be involved in similar regulation of lipids, a working model for the possible effects of ORP2A KD in autophagosome formation is proposed: 1) Autophagy can be initiated by the nucleation of ATG1 and recruitment of ATG8; 2) in the subsequent elongation and maturation, autophagosomes fail to properly progress from the ER membrane, probably owing to the misredistribution of the lipids at the autophagosome forming site; and 3) autophagy-related proteins and lipids fail to undergo autophagosome progression but accumulate in the ER membrane, likely resulting in ER dilation (SI Appendix, Fig. S14C). Notably, in ORP2A KD plants, autophagy was not totally blocked and the ATG1a/ATG8e accumulation was not universally found in every cell, likely due to the DEX-induced KD efficiency. In Arabidopsis, the ATG1 complex initiates the formation of the autophagosome responding to stresses (10, 11). This study may provide insights in studying the plant stress response in the future.
In yeast and mammalian cells, ORP belongs to a large family of the LTPs and transfers multiple lipids between membranes at diverse MCSs (59). However, it remains unknown whether ORP functions similarly in plants. As the sole autophagy regulator from the ORP family to date, ORP2A possesses all the essential domains working with VAP27-1 as a typical ORP–VAP lipid transfer complex (SI Appendix, Fig. S14B). In this study, we showed that ORP2A bound to PI3P in vivo and PI3P was accumulated in ER membrane in ORP2A KD plants (Fig. 8). Although the exact mechanism underlying the lipid transfer remains elusive, this study on ORP2A provides evidence of a direct involvement of a PI3P-binding protein in autophagosome biogenesis. Future studies will be focused on understanding whether and which lipids are transferred between membranes by ORP2A.
In conclusion, plants have evolved and developed a sophisticated system of responding to environmental cues. Autophagy is one of the most important mechanisms in response to stresses. This study not only sheds light onto the regulation of plant autophagy but also provides leads for future studies on organelle interaction and communication through MCSs and lipid regulation in plants (60).
Materials and Methods
Plasmid Construction.
For the constructs used for Arabidopsis PSBD transient expression, pBI221 vectors were used with modification containing CFP, GFP, RFP, or mCherry tags under the control of the UBQ10 promoter. For transgenic plants, complementary DNAs (cDNAs) were cloned into pBI121 vectors under the UBQ10 or ORP2A native promoter as indicated in the figures. To generate amiRNA KD lines, amiRNAs were designed following the procedure described previously (61). Three amiRNA sequences were designed: amiR-1: 5′-TAAGACTTTACAGTGACGCTC-3′; amiR-2: 5′-TTGAAAACTCATGCTTGCCCG-3′; and amiR-3: 5′-TATAGTAAAGGCTATCACCCC-3′. The amiRNAs were introduced into the pTA7002 vector as described (61) under the control of the DEX-inducible promoter.
Transient Expression in Protoplasts and Confocal Imaging.
Arabidopsis PSBD protoplasts were used to perform transient expression as described previously (62). Confocal fluorescence images were acquired 16 h after protoplast transformation using a Leica SP8 system. For the generation of 3D image projection, 20 slices were collected with a step thickness of 0.8 μm, to ensure to image a whole cell from top to bottom. Projection was done by Leica SP8 software. Images were processed using Adobe Photoshop software. The number and size of dots were quantified by using ImageJ.
High-Pressure Freezing and Immunogold-TEM Analysis.
High-pressure freezing and freeze substitution (HPFS) were performed as described previously (63). Briefly, 5-d-old Arabidopsis root tips were frozen in a high-pressure freezer (Leica EM ICE). High-pressure frozen root tips were transferred to an AFS (Leica Microsystems) and freeze-substituted in dry acetone containing 0.1% uranyl acetate at −85 °C for 48 h. Infiltration with HM20, embedding, and ultraviolet polymerization were performed stepwise at −45 °C. Immunogold labeling was performed as described previously (64). GFP antibodies were diluted at 40 μg/mL in phosphate-buffered saline (PBS) with 1% bovine serum albumin (BSA), and gold-coupled rabbit secondary antibodies were at 1:40 dilution in PBS with 1% BSA.
ET Analysis.
The general procedures for ET analysis were performed as described previously (65). Briefly, HPFS samples sectioned at 250 nm were recorded using a Tecnai F20 electron microscope (Thermo Fisher) operated at 200 kV. For each grid, a tilt image stack (64 images) from +48° to −48° with 1.5° intervals was collected and the second tilt image stack was collected by rotating the grid 90°. Dual-axis tomograms were calculated from pairs of image stacks with the etomo program of the IMOD software package. For model generation, the contours were drawn and meshed with the 3dmod program in the IMOD software package.
Supplementary Material
Acknowledgments
We thank Prof. Pengwei Wang (Huazhong Agricultural University, China) for sharing the VAP27-1 RNAi KD and vap27-1/vap27-3 CRISPR KO seeds. This work was supported by grants from the National Natural Science Foundation of China (91854201) and the Research Grants Council of Hong Kong (AoE/M-05/12, CUHK14100818, CUHK14101219, C4033-19E, C4002-17G, C4002-20W, C4002-21EF, C2009-19G, C4041-18E, and R4005-18), The Chinese University of Hong Kong Research Committee, and The Chinese Academy of Sciences–Croucher Funding Scheme for Joint Laboratories (to L.J.). Portions of this work were developed from the doctoral dissertation completed at The Chinese University of Hong Kong by H.Y.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. D.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2205314119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Mizushima N., Autophagy: Process and function. Genes Dev. 21, 2861–2873 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Bozhkov P. V., Plant Autophagy: Mechanisms and Functions (Oxford University Press, Oxford, UK, 2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hollenstein D. M., Kraft C., Autophagosomes are formed at a distinct cellular structure. Curr. Opin. Cell Biol. 65, 50–57 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamasaki M., et al. , Autophagosomes form at ER-mitochondria contact sites. Nature 495, 389–393 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Zhu D., Zhang M., Gao C., Shen J., Protein trafficking in plant cells: Tools and markers. Sci. China Life Sci. 63, 343–363 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Hayashi-Nishino M., et al. , A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 11, 1433–1437 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Li W., Zhang L., “Regulation of ATG and autophagy initiation” in Autophagy: Biology and Diseases, Z.-H. Qin, Ed. (Springer, 2019), pp. 41–65. [DOI] [PubMed] [Google Scholar]
- 8.Hurley J. H., Young L. N., Mechanisms of autophagy initiation. Annu. Rev. Biochem. 86, 225–244 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karanasios E., et al. , Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles. Nat. Commun. 7, 12420 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suttangkakul A., Li F., Chung T., Vierstra R. D., The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23, 3761–3779 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang X., et al. , Genetic analyses of the Arabidopsis ATG1 kinase complex reveal both kinase-dependent and independent autophagic routes during fixed-carbon starvation. Plant Cell 31, 2973–2995 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhuang X., et al. , ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 114, E426–E435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhuang X., et al. , A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell 25, 4596–4615 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nascimbeni A. C., Codogno P., Morel E., Phosphatidylinositol-3-phosphate in the regulation of autophagy membrane dynamics. FEBS J. 284, 1267–1278 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Cheung P. C., Trinkle-Mulcahy L., Cohen P., Lucocq J. M., Characterization of a novel phosphatidylinositol 3-phosphate-binding protein containing two FYVE fingers in tandem that is targeted to the Golgi. Biochem. J. 355, 113–121 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baskaran S., Ragusa M. J., Boura E., Hurley J. H., Two-site recognition of phosphatidylinositol 3-phosphate by PROPPINs in autophagy. Mol. Cell 47, 339–348 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Obara K., Sekito T., Niimi K., Ohsumi Y., The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J. Biol. Chem. 283, 23972–23980 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizushima N., Yoshimori T., Ohsumi Y., The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Scorrano L., et al. , Coming together to define membrane contact sites. Nat. Commun. 10, 1287 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X., et al. , Plant multiscale networks: Charting plant connectivity by multi-level analysis and imaging techniques. Sci. China Life Sci. 64, 1392–1422 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Wu H., Carvalho P., Voeltz G. K., Here, there, and everywhere: The importance of ER membrane contact sites. Science 361, eaan5835 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang P., et al. , Plant VAP27 proteins: Domain characterization, intracellular localization and role in plant development. New Phytol. 210, 1311–1326 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Stefano G., et al. , Plant endocytosis requires the ER membrane-anchored proteins VAP27-1 and VAP27-3. Cell Rep. 23, 2299–2307 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Wang P., et al. , Plant AtEH/Pan1 proteins drive autophagosome formation at ER-PM contact sites with actin and endocytic machinery. Nat. Commun. 10, 5132 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greer M. S., et al. , SEIPIN isoforms interact with the membrane-tethering protein VAP27-1 for lipid droplet formation. Plant Cell 32, 2932–2950 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weber-Boyvat M., Kentala H., Peränen J., Olkkonen V. M., Ligand-dependent localization and function of ORP-VAP complexes at membrane contact sites. Cell. Mol. Life Sci. 72, 1967–1987 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skirpan A. L., et al. , Identification and characterization of PiORP1, a Petunia oxysterol-binding-protein related protein involved in receptor-kinase mediated signaling in pollen, and analysis of the ORP gene family in Arabidopsis. Plant Mol. Biol. 61, 553–565 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Umate P., Oxysterol binding proteins (OSBPs) and their encoding genes in Arabidopsis and rice. Steroids 76, 524–529 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Wijdeven R. H., et al. , Cholesterol and ORP1L-mediated ER contact sites control autophagosome transport and fusion with the endocytic pathway. Nat. Commun. 7, 11808 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang P., et al. , The plant cytoskeleton, NET3C, and VAP27 mediate the link between the plasma membrane and endoplasmic reticulum. Curr. Biol. 24, 1397–1405 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Lam S. K., et al. , Rice SCAMP1 defines clathrin-coated, trans-Golgi-located tubular-vesicular structures as an early endosome in tobacco BY-2 cells. Plant Cell 19, 296–319 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaiser S. E., et al. , Structural basis of FFAT motif-mediated ER targeting. Structure 13, 1035–1045 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Gao C., et al. , A unique plant ESCRT component, FREE1, regulates multivesicular body protein sorting and plant growth. Curr. Biol. 24, 2556–2563 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Klionsky D. J., et al. , Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 17, 1–382 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung H., et al. , Arabidopsis cargo receptor NBR1 mediates selective autophagy of defective proteins. J. Exp. Bot. 71, 73–89 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin Y., et al. , Plant Rho GTPase signaling promotes autophagy. Mol. Plant 14, 905–920 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Zhuang X., Chung K. P., Jiang L., Origin of the autophagosomal membrane in plants. Front. Plant Sci. 7, 1655 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong L. H., Čopič A., Levine T. P., Advances on the transfer of lipids by lipid transfer proteins. Trends Biochem. Sci. 42, 516–530 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ktistakis N. T., ER platforms mediating autophagosome generation. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1865, 158433 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Simon M. L. A., et al. , A multi-colour/multi-affinity marker set to visualize phosphoinositide dynamics in Arabidopsis. Plant J. 77, 322–337 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chung J., et al. , INTRACELLULAR TRANSPORT. PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER-plasma membrane contacts. Science 349, 428–432 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang P., Hawes C., Hussey P. J., Plant endoplasmic reticulum–plasma membrane contact sites. Trends Plant Sci. 22, 289–297 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Michaud M., Jouhet J., Lipid trafficking at membrane contact sites during plant development and stress response. Front. Plant Sci. 10, 2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zaman M. F., Nenadic A., Radojičić A., Rosado A., Beh C. T., Sticking with it: ER-PM membrane contact sites as a coordinating nexus for regulating lipids and proteins at the cell cortex. Front. Cell Dev. Biol. 8, 675 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruiz-Lopez N., et al. , Synaptotagmins at the endoplasmic reticulum-plasma membrane contact sites maintain diacylglycerol homeostasis during abiotic stress. Plant Cell 33, 2431–2453 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang H., Lipid transfer at ER-isolation membrane contacts. Nat. Rev. Mol. Cell Biol. 21, 121 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Zhao Y. G., et al. , The ER contact proteins VAPA/B interact with multiple autophagy proteins to modulate autophagosome biogenesis. Curr. Biol. 28, 1234–1245.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Maeda S., Otomo C., Otomo T., The autophagic membrane tether ATG2A transfers lipids between membranes. eLife 8, e45777 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valverde D. P., et al. , ATG2 transports lipids to promote autophagosome biogenesis. J. Cell Biol. 218, 1787–1798 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otomo T., Maeda S., ATG2A transfers lipids between membranes in vitro. Autophagy 15, 2031–2032 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Osawa T., Ishii Y., Noda N. N., Human ATG2B possesses a lipid transfer activity which is accelerated by negatively charged lipids and WIPI4. Genes Cells 25, 65–70 (2020). [DOI] [PubMed] [Google Scholar]
- 52.Kentala H., Pfisterer S. G., Olkkonen V. M., Weber-Boyvat M., Sterol liganding of OSBP-related proteins (ORPs) regulates the subcellular distribution of ORP-VAPA complexes and their impacts on organelle structure. Steroids 99 (Pt B), 248–258 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Beh C. T., Cool L., Phillips J., Rine J., Overlapping functions of the yeast oxysterol-binding protein homologues. Genetics 157, 1117–1140 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kobuna H., et al. , Multivesicular body formation requires OSBP-related proteins and cholesterol. PLoS Genet. 6, e1001055 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mesmin B., et al. , Sterol transfer, PI4P consumption, and control of membrane lipid order by endogenous OSBP. EMBO J. 36, 3156–3174 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee Y., et al. , The Arabidopsis phosphatidylinositol 3-kinase is important for pollen development. Plant Physiol. 147, 1886–1897 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harrison-Lowe N. J., Olsen L. J., Autophagy protein 6 (ATG6) is required for pollen germination in Arabidopsis thaliana. Autophagy 4, 339–348 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Osawa T., et al. , Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat. Struct. Mol. Biol. 26, 281–288 (2019). [DOI] [PubMed] [Google Scholar]
- 59.Pietrangelo A., Ridgway N. D., Bridging the molecular and biological functions of the oxysterol-binding protein family. Cell. Mol. Life Sci. 75, 3079–3098 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ye H., “Molecular characterization of AtORP2A in mediating autophagosome formation,” Doctoral dissertation, The Chinese University of Hong Kong, Hong Kong, China, (2021).
- 61.Schwab R., Ossowski S., Riester M., Warthmann N., Weigel D., Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18, 1121–1133 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Miao Y., Jiang L., Transient expression of fluorescent fusion proteins in protoplasts of suspension cultured cells. Nat. Protoc. 2, 2348–2353 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Kang B.-H., Electron microscopy and high-pressure freezing of Arabidopsis. Methods Cell Biol. 96, 259–283 (2010). [DOI] [PubMed] [Google Scholar]
- 64.Cui Y., et al. , A whole-cell electron tomography model of vacuole biogenesis in Arabidopsis root cells. Nat. Plants 5, 95–105 (2019). [DOI] [PubMed] [Google Scholar]
- 65.Toyooka K., Kang B.-H., “Reconstructing plant cells in 3D by serial section electron tomography” in Plant Cell Morphogenesis, V. Žárský, F. Cvrčková, Eds. (Springer, 2014), pp. 159–170. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.