Significance
Heterochromatin condensation during meiotic prophase I facilitates homologous chromosome interactions and ensures their accurate segregation, thereby maintaining genome integrity. However, the regulatory mechanisms governing meiotic heterochromatin condensation are poorly understood, especially in plants. Here, we show that the Arabidopsis catalytic subunit (POL2A) of DNA polymerase epsilon is required for meiotic heterochromatin formation. Two distinct domains in POL2A promote heterochromatin condensation, a C-terminal zinc finger domain (ZF1) that binds to the heterochromatin-enriched histone variant H3.1-H4 and the N terminus, which recruits the GHKL adenosine triphosphatase (ATPase) MORC1. We also demonstrate that mouse POL2A ZF1 binds to H3.1-H4, suggesting that the mechanism identified here appears to be conserved. These results expand the role of POL ε beyond DNA replication.
Keywords: meiosis, heterochromatin condensation, DNA polymerase epsilon, MORC1, H3.1-H4
Abstract
Heterochromatin is essential for genomic integrity and stability in eukaryotes. The mechanisms that regulate meiotic heterochromatin formation remain largely undefined. Here, we show that the catalytic subunit (POL2A) of Arabidopsis DNA polymerase epsilon (POL ε) is required for proper formation of meiotic heterochromatin. The POL2A N terminus interacts with the GHKL adenosine triphosphatase (ATPase) MORC1 (Microrchidia 1), and POL2A is required for MORC1’s localization on meiotic heterochromatin. Mutations affecting the POL2A N terminus cause aberrant morphology of meiotic heterochromatin, which is also observed in morc1. Moreover, the POL2A C-terminal zinc finger domain (ZF1) specifically binds to histone H3.1-H4 dimer or tetramer and is important for meiotic heterochromatin condensation. Interestingly, we also found similar H3.1-binding specificity for the mouse counterpart. Together, our results show that two distinct domains of POL2A, ZF1 and N terminus bind H3.1-H4 and recruit MORC1, respectively, to induce a continuous process of meiotic heterochromatin organization. These activities expand the functional repertoire of POL ε beyond its classic role in DNA replication and appear to be conserved in animals and plants.
In eukaryotes, DNA is complexed with proteins, primarily histones, to form chromatin. Canonical nucleosomes consist of DNA and two copies each of histones H2A, H2B, H3, and H4, but multiple variant histones can also be incorporated (1). In addition, the N-terminal tails of the histones can be posttranscriptionally modified with marks that convey epigenetic information. Chromatin can be classified as euchromatin or heterochromatin based on nucleosome density and transcriptional status. Euchromatin has lower nucleosome density and is enriched with actively transcribed genes, while heterochromatin has higher nucleosome density and often contains silenced repetitive sequences and transposable elements (TEs) (2). Proper establishment of both euchromatin and heterochromatin is essential for maintaining genomic stability, regulating transcription, and ensuring accurate chromosome segregation (3, 4). In contrast to extensively studied mitotic chromatin (5), much less is known about the assembly and function of chromatin during meiosis.
In Arabidopsis, histone H3 variant H3.1 is primarily deposited in newly synthesized chromatin during DNA replication by the CHROMATIN ASSEMBLY FACTOR 1 (CAF1) complex (6, 7). The H3.3 variant is associated with transcription by replacing the H3.1 variant in euchromatin throughout the cell cycle in a DNA replication–independent manner and mainly correlates with active genes (8–10). The Arabidopsis CAF1 complex has three subunits, FASCIATA1 (FAS1), FASCIATA2 (FAS2), and MULTICOPY SUPRESSOR OF IRA1 (MSI1), and deficiency in them disrupts H3.1 deposition, derepresses TEs, and disturbs the formation of constitutive heterochromatin in mitosis (6, 11, 12). In addition, the plant-specific H2A variant H2A.W (13), linker histone H1 (14), two members of the conserved microrchidia (MORC) adenosine triphosphatase (ATPase) family, MORC1 and MORC6 (15), and DNA methylation (16) also participate in heterochromatin condensation in mitosis.
Furthermore, specific N-terminal histone tail modifications are enriched in heterochromatin, including histone H3 lysine 9 di-methylation (H3K9me2) and H3 lysine 27 monomethylation (H3K27me1), which are enriched in chromatin that also includes the H3.1 variant (7, 10, 17). H3K27 methylation is usually present in transcriptionally repressed regions, and H3K27me1 is enriched in constitutive heterochromatin, whereas H3K27me3 is associated with facultative heterochromatin, primarily in the transcribed regions of individual genes in plants (18, 19). In animals, Polycomb Repressive Complex 2 (PRC2) catalyzes all three types of H3K27 methylation (20, 21). Intriguingly, plant PRC2 is primarily responsible for H3K27me2/3, while ARABIDOPSIS TRITHORAX-RELATED PROTEINs 5 and 6 (ATXR5 and ATXR6) specifically catalyze H3K27me1 of H3.1 at the replication fork and are critical for heterochromatin condensation (17, 22). In Arabidopsis, H3K9me2 is catalyzed by SU(VAR)3–9 HOMOLOG 4 (SUVH4), SUVH5, and SUVH6 and is preferentially localized to constitutive heterochromatin (23, 24). Arabidopsis H3K9me2 also participates in a positive-feedback loop with CHG DNA methylation (H is equal to A, T, or C) (25). Abolishing both H3K9me2 and H3K27me1 results in heterochromatin decondensation and reactivation of TEs (25, 26). However, whether any of these factors are also required for formation of meiotic heterochromatin is unknown.
DNA replication and chromatin establishment are highly orchestrated processes that ensure genetic inheritance and epigenetic memory through cell lineage (27). Loading of H3.1 and H3K27me1 takes place during replication and requires PCNA (proliferating cell nuclear antigen) (7, 28–30), while monomethylation of lysine 27 on H3.1 is essential for maintaining H3K27me3 to promote silencing memory (29). Both Arabidopsis DNA polymerase α (POL α) and POL ε are required for H3K27me3 maintenance at specific loci, including those that regulate flowering (31–34). Moreover, the largest catalytic subunit of POL ε (POL2A) physically interacts with LHP1 (LIKE HETEROCHROMATIN PROTEIN 1) and PRC2 components including MSI1, a shared component of both CAF1 and PRC2 complexes. Additionally, POL2A genetically interacts with CAF1 (32). More recently, POL2A has been reported to be required for preventing replicative stress to maintain heterochromatin structure and enforce genome-wide transcriptional silencing in mitotic cells (35). Together, these results suggest that POL ε acts in DNA replication–coupled heterochromatin organization and reestablishment of epigenetic marks, at least at specific loci.
Chromosome compaction is required for proper segregation during both mitosis and meiosis. During mitotic prophase to metaphase, chromatin fibers rapidly condense to form rod-shaped chromosomes (36). Meiosis has a protracted prophase I in which homologous chromosome interactions such as pairing, synapsis, and recombination occur (37). During this period, chromosomes continue to condense, forming thread-like structures (leptotene) and eventually highly compacted rod-shaped bivalents (pairs of homologs). In yeast, animal, and plant, heterochromatic associations at the centromeres facilitate stable pairing and precise segregation of homologs (38–42). However, the mechanistic basis for regulating meiotic heterochromatin condensation is unclear. Here, we show that Arabidopsis POL2A is required for the formation of meiotic heterochromatin. The N terminus of POL2A interacts with MORC1, which is known to mediate mitotic heterochromatin condensation (15). The morc1 single and morc1/2/6 triple mutants have defects in meiotic heterochromatin condensation similar to those of pol2a. POL2A’s C-terminal zinc finger (ZF) domain (ZF1) specifically recognizes histone variant H3.1-H4, which is enriched in heterochromatin following DNA replication (7). We also show that POL2A can associate with FAS1 in vivo, and our genetic analyses show that POL2A ZF1 is required for H3.1-H4 binding and heterochromatin formation in meiosis. These results reveal that POL2A ensures meiotic heterochromatin formation via distinct domains that couple H3.1 deposition and MORC1 recruitment. We also hypothesize that the mechanism identified here might be conserved among eukaryotes.
Results
AtPOL2A Is Required for Meiotic Heterochromatin Condensation.
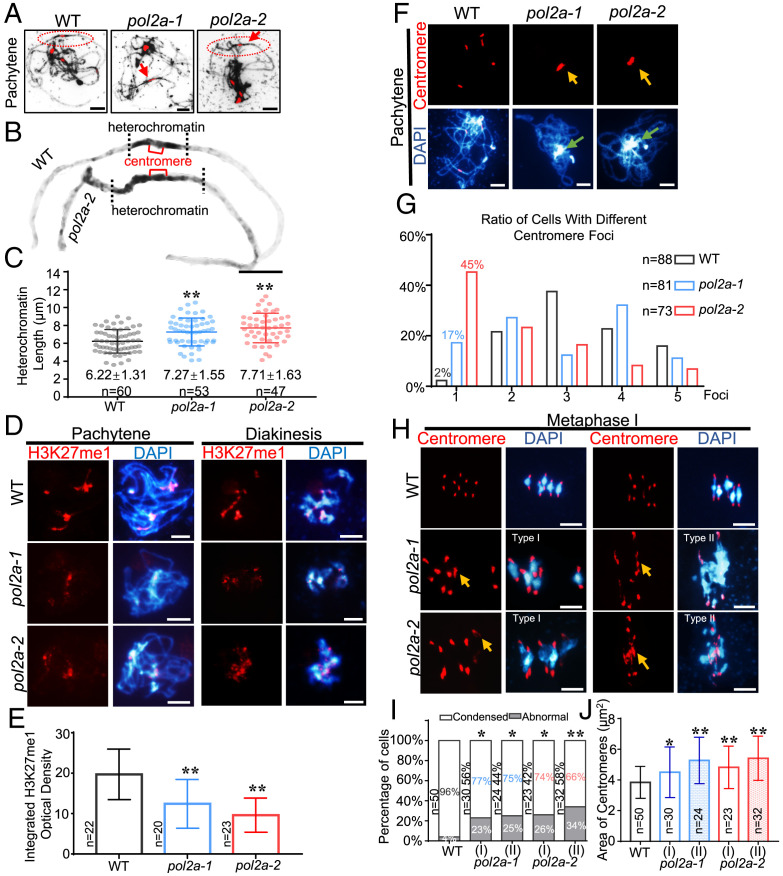
Arabidopsis POL2A, the catalytic subunit of POL ε, is essential for DNA replication and plant development (34, 43). We previously identified two AtPOL2A hypomorphic alleles, pol2a-1 with an N-terminal Transfer-DNA (T-DNA) insertion (12th intron) and pol2a-2 with a point mutation near its EXO (exonuclease) domain (SI Appendix, Fig. S1A). Both alleles have compromised meiotic recombination (44). Quantitative analysis of 4′,6-diamidino-2-phenylindole (DAPI)–stained pachytene chromosome spreads showed that pol2a-1 and pol2a-2 had longer heterochromatin regions compared to wild-type (WT) (Fig. 1 A–C). Moreover, WT had a weaker DAPI-staining centromeric core region flanked by heavier staining pericentromeric regions, while pol2a had uniformly heavy staining across the pericentromeric/centromeric region (Fig. 1B), indicative of a defect in appropriately defining heterochromatin domains.
Fig. 1.
pol2a has defective meiotic heterochromatin condensation. (A) Chromosome morphology with centromere FISH of WT, pol2a-1, and pol2a-2 at pachytene. Red arrows indicate longer heterochromatin than WT. (B) Enlarged images of chromosomes from A marked by red ellipses show abnormal pericentromeric/centromeric heterochromatin in pol2a compared to WT. (C) The average length of pachytene heterochromatin in WT (gray), pol2a-1 (blue), and pol2a-2 (red). (D) Immunofluorescence of H3K27me1 in WT, pol2a-1, and pol2a-2 chromosome spreads at pachytene and diakinesis. (E) Histogram showing the integrated density of H3K27me1 at pachytene. (F) Chromosome and centromere phenotypes of WT, pol2a-1, and pol2a-2 at pachytene. Yellow arrows indicate the enlarged and associated centromere signals. Green arrows indicate the abnormal association of heterochromatin of nonhomologs. Error bars indicate the standard deviations of the samples. (G) Percentage of cells in WT, pol2a-1, and pol2a-2 with different numbers of centromere foci at pachytene. (H) Chromosome and centromere phenotypes at metaphase I by FISH. Type I spreads five bivalents with enlarged centromeres; type II has multivalents (three or more associated chromosomes) with abnormal centromere associations. Yellow arrows indicate the enlarged and associated centromere signals. (I) Percentage of cells showing type I (five bivalents) and II (multivalents) with abnormal centromeres in pol2a-1 and pol2a-2 (Fisher’s exact test). The ratios near the bar indicate the percentage of type I or II in pol2a mutants. Cells with centromeric areas larger than 1.5 × 3.8 μm2 (the average area of centromere fluorescence in WT is 3.8 μm2) were defined as “Abnormal.” (J) Centromere signal area at metaphase I. Error bars indicate the standard deviations of the samples. *, P < 0.05; **, P < 0.01; two-tailed Student's t test. Scale bars, 5 μm.
To more fully characterize the pol2a heterochromatin defects, we examined the immunolocalization of modified histone H3K27me1 and H3K9me2, which are enriched in the heterochromatic regions of Arabidopsis somatic cells (22, 23). In WT male meiocytes, co-immunofluorescence using an anti-H3K27me1 antibody together with a centromeric fluorescence in situ hybridization (FISH) probe showed overlapping signals from leptotene to zygotene (SI Appendix, Fig. S1 B and C), indicating an enrichment of H3K27me1 on heterochromatin in meiocytes. These heterochromatic H3K27me1 signals were significantly reduced in both pol2a-1 and -2 compared to WT (Fig. 1 D and E and SI Appendix, Fig. S1 D and E). H3K9me2 was also more dispersed and slightly reduced in both pol2a mutants relative to WT (SI Appendix, Fig. S2 A and C), providing additional evidence to support a heterochromatin defect in pol2a. In contrast to these heterochromatic phenotypes, there was no obvious difference in the intensity or distribution of the euchromatic H3K4me3 mark between WT and pol2a male meiocytes (SI Appendix, Fig. S2 B and D). The milder pol2a-1 phenotype may reflect residual, low-level expression of full-length POL2A (SI Appendix, Fig. S8B), consistent with our previous findings (44), while the mutation in pol2a-2 may be more disruptive to meiotic function.
In addition to the heterochromatin phenotype, we found that pol2a centromeres tend to cluster in a single mass at pachytene at a significantly higher frequency relative to WT (Fig. 1 F and G). Analysis of preprophase male meiocytes with a centromeric FISH probe showed 10 foci in WT (SI Appendix, Fig. S3A) that were gradually reduced to three to five foci from leptotene through pachytene (SI Appendix, Fig. S3 A and D). The pol2a centromere signals were more diffuse at preprophase and leptotene (SI Appendix, Fig. S3 B and C), and then from zygotene to pachytene they frequently formed into an aberrant single cluster, which persisted through diakinesis (Fig. 1F and SI Appendix, Fig. S3E). At metaphase I, pol2a centromeres were more diffuse than WT, and centromere bridges were observed in some cells (Fig. 1 H–J and SI Appendix, Fig. S3E). To quantify the pericentromeric heterochromatin changes in pol2a, we measured the centromeric areas on highly condensed metaphase I chromosomes in WT and pol2a. We subcategorized pol2a metaphase I chromosome spreads into two morphological types: type I had the five bivalents typically observed in Arabidopsis; type II had aberrant multivalents (Fig. 1H) (44). In both subtypes, the centromere signals were significantly more diffuse in pol2a compared to WT (Fig. 1 H–J), further supporting a meiotic heterochromatin condensation defect in pol2a. Because POL2A is also required for the repair of SPO11-1 (SPORULATION 11–1)–mediated meiotic double-strand breaks (DSBs) (44), we examined spo11-1 single and pol2a-1 spo11-1 double mutants and found that pol2a-1 spo11-1 mutants have centromere clustering and decondensed heterochromatin phenotypes like pol2a-1 (SI Appendix, Fig. S3 E and G), but these phenotypes were not observed in spo11-1 (SI Appendix, Fig. S3F). These results suggest that the pol2a centromere and heterochromatin defects are DSB independent. We also analyzed mutants of RAD51 (45), a RecA recombinase homolog that functions upstream of POL2A during meiotic recombination. Centromere structure appeared normal, although the abnormal multivalents were typically seen in rad51 (SI Appendix, Fig. S3H), indicating that the pol2a centromeric defects are also independent of the early steps in meiotic recombination.
The POL2A N Terminus Interacts with MORC1 ATPase.
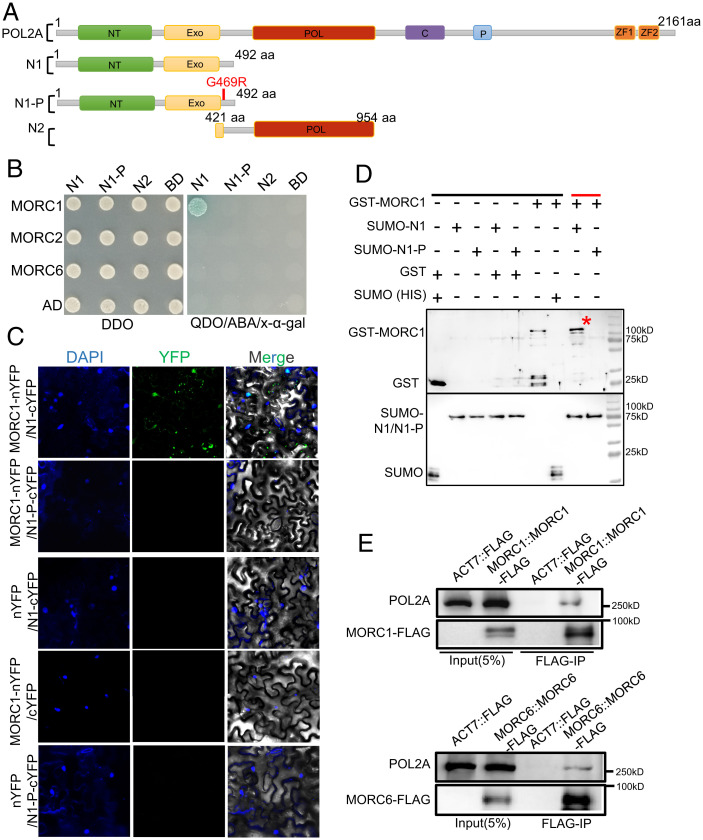
To identify proteins that interact with POL2A, we used the yeast two-hybrid (Y2H) assay to screen an Arabidopsis inflorescence complementary DNA library using the N terminus of POL2A (N1) as a bait (Fig. 2A) and identified the ATPase MORC1 as an interacting partner (Fig. 2B), which is required for mitotic heterochromatin condensation (15). The interaction was validated by affinity purification pull-down assays and bimolecular fluorescence complementation (BiFC) (Fig. 2 C and D). Arabidopsis has seven homologs, MORC1–MORC7, from which MORC1 and MORC2 have redundant gene silencing functions and form mutually exclusive heterodimers with MORC6 in somatic cells (46). The interactions of MORC1–MORC6 and MORC2–MORC6 were also validated in our study (SI Appendix, Fig. S4D). We used Y2H assays to test for interactions between POL2A and MORC2/6, but found no direct interaction (Fig. 2B). Interestingly, we also observed that POL2A-N1 colocalizes with MORC6 but not MORC2 by BiFC (SI Appendix, Fig. S4E). To assess whether full-length POL2A interacts with MORC1/6, we raised a polyclonal antibody against POL2A and performed coimmunoprecipitation (co-IP) of proteins extracted from MORC1/6-FLAG transgenic plant inflorescences using an anti-FLAG antibody. POL2A coprecipitated with FLAG-tagged MORC1 and MORC6 (Fig. 2E), supporting the in vivo interaction between them. Importantly, we did not observe interaction between MORC1 and the N terminus of POL2A engineered to include the pol2a-2 mutation (N1-P) (Fig. 2 B–D and SI Appendix, Fig. S4C), indicating that the conserved glycine near the EXO domain is essential for their interaction. Furthermore, we used Y2H assays to show that the POL2A NT domain and the MORC1 N terminus directly interact (SI Appendix, Fig. S4 A–C), although the G469R mutation of POL2A disrupts their interaction.
Fig. 2.
N terminus of POL2A interacts with ATPase MORC1. (A) Diagrams showing the full-length, truncated forms and point mutation of AtPOL2A. NT, N terminus; EXO, 3‘-5′ exonuclease; POL, 5‘-3′ polymerase; C, central; P, proliferating cell nuclear antigen interaction. (B) Y2H assay showing N-terminal (N1) POL2A interaction with MORC1, but not MORC2 or MORC6. N1-P is the G469R point mutation adjacent to the EXO-domain of POL2A indicated in A. DDO indicates SD/–Leu/–Trp medium, whereas QDO refers to SD/–Ade/–His/–Leu/–Trp medium. Blue dot refers to positive interaction. (C) Verification of POL2A and MORC1 interaction by BiFC assay. (D) Interaction between POL2A and MORC1 demonstrated using an affinity purification pull-down assay. MORC1::GST and N1-POL2A::SUMO-HIS were precipitated using Ni-NTA beads. Black line indicates input and negative control, and red line indicates the experimental group. Red asterisk indicates the GST-MORC1 band. (E) Co-IP of POL2A with MORC1-FLAG/MORC6-FLAG plants overexpressing FLAG as controls. aa, amino acid; ABA, Aureobasidin A; YFP: Yellow Fluorescent Protein; n/cYFP: n/c terminus of YFP; GST, glutathione S-transferase; SUMO (HIS), Small Ubiquitin-like Modifier-polyhistidine fusion tags.
Meiotic Heterochromatin Defects Are Similar in morc1 and pol2a.
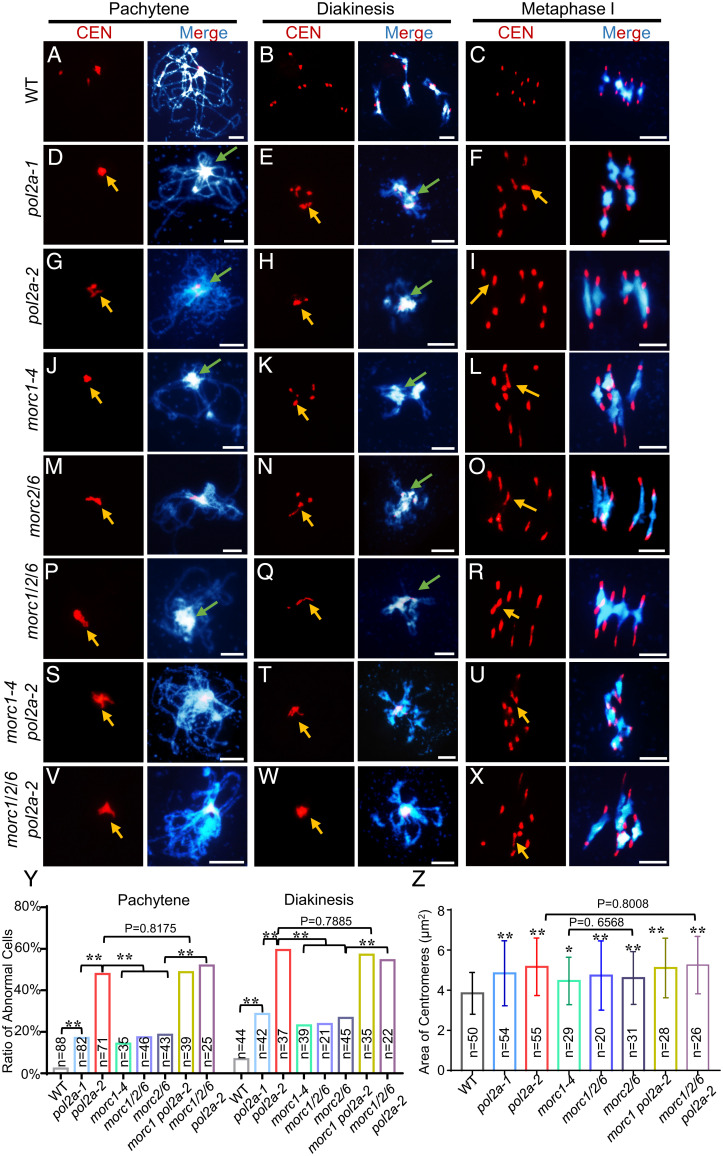
In mouse, Morc1 is required for spermatogenesis and meiosis (47, 48). Arabidopsis MORC1/MORC2 form heterodimers with MORC6, which function in mitotic heterochromatin condensation (15, 46). To test whether MORC1 has a similar function in meiosis, we examined morc1-4 single, morc2/6 double, and morc1/2/6 triple mutants (49), all of which have centromere clustering and heterochromatin condensation defects similar to pol2a (Fig. 3 J–R). Interestingly, no significant difference was found between morc1-4, morc2/6, and morc1/2/6 (Fig. 3 Y and Z), suggesting that MORC1 and MORC6 may function together in meiosis. This result is consistent with POL2A’s physical interaction with MORC1 and colocalization with MORC6 (Fig. 2B and SI Appendix, Fig. S4E). The abnormal centromere associations in morc1-4, morc2/6, and morc1/2/6 are similar to, but less severe than, those in pol2a-2 at pachytene and diakinesis (Fig. 3 G–R and Y). Similarly, at metaphase I, both pol2a and morc mutants had decondensed centromeres, but the former had more severe centromere bridging (Fig. 3 F, I, L, O, R, and Z). We also observed a reduction of both H3K27me1 and H3K9me2 signals in morc1, morc2/6, and morc1/2/6 mutants compared to WT (SI Appendix, Figs. S5 and S6). Taken together, the similarity of centromere clustering, heterochromatin condensation, and epigenetic patterning phenotypes seen in pol2a and morc mutants supports the idea that POL2A and MORC1 may have related functions. To test this hypothesis, we generated double morc1-4 pol2a-2 and quadruple morc1/2/6 pol2a-2 mutants and compared them with the single and triple mutants. The heterochromatin condensation and centromere clustering phenotypes in morc1 pol2a-2 were indistinguishable from pol2a-2 (Fig. 3 S–X and SI Appendix, Figs. S5 and S6). These results suggest that POL2A is epistatic to MORC1, and they might work in the same pathway to form or maintain meiotic heterochromatin.
Fig. 3.
The meiotic centromere phenotypes of morc mutants are similar to pol2a. (A–X) Chromosome and centromere phenotypes of WT, po12a-1, pol2a-2, morc1-4, morc2/6, morc1/2/6, morc1-4 pol2a-2, and morc1/2/6 pol2a-2 at pachytene, diakinesis, and metaphase I by centromere (CEN) FISH. Yellow arrows indicate clustered and associated centromere signals. Green arrows indicate the abnormal association of heterochromatin of nonhomologs. Scale bars, 5 μm. (Y) Histogram showing the ratio of cells with centromere defects from WT and mutants at pachytene and diakinesis, respectively (Fisher’s exact test). (Z) Area of centromere signals at metaphase I in each mutant (two-tailed Student’s t test). P values refer to the comparison between WT and mutants unless specified. Error bars indicate the standard deviations of the samples. *, P < 0.05; **, P < 0 0.01.
POL2A Is Required for Normal Localization of MORC1/6 on Chromosomes.
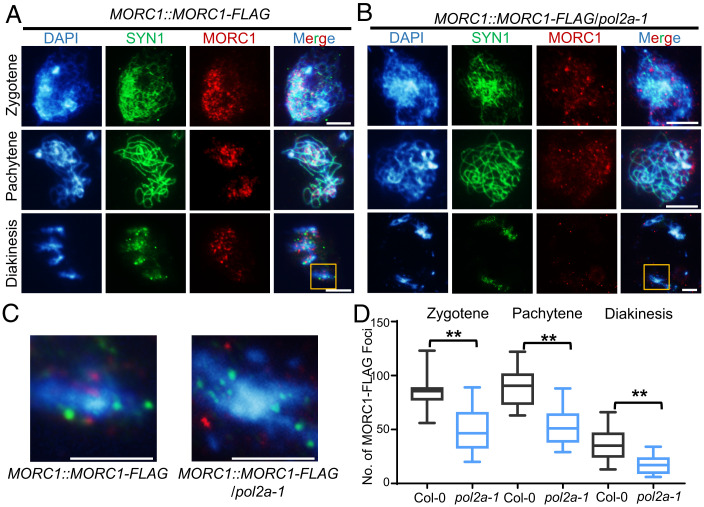
To test whether MORC1 and POL2A work together to facilitate meiotic heterochromatin formation and/or maintenance, we introduced a MORC1::MORC1-FLAG construct into WT and pol2a-1 backgrounds. Immunofluorescence of male meiocytes with anti-FLAG antibody showed that MORC1-FLAG signals often associate with chromosomes around the nucleolus at premeiosis in the WT (Col-0) background (SI Appendix, Fig. S7A). After entering meiosis, the MORC1-FLAG signals formed 84 ± 13 (n = 20) and 89 ± 18 (n = 20) foci at zygotene and pachytene, respectively (Fig. 4 A and D and SI Appendix, Fig. S7A). DAPI-bright heterochromatic regions often colocalized with these meiotic foci. Intriguingly, from zygotene to diakinesis MORC1-FLAG signals decreased and formed 35 ± 14 (n = 15) foci distributed along the highly condensed bivalents (Fig. 4 A, C, and D), which is consistent with a previous report that MORC1 and MORC6 form small nuclear bodies adjacent to chromocenters in somatic cells (15). In pol2a-1, MORC1-FLAG localization was compromised on chromosomes with 50 ± 18 (n = 20) foci at zygotene, 53 ± 16 (n = 20) at pachytene, and 18 ± 8 (n = 15) at diakinesis (Fig. 4 B and D). More importantly, MORC1-FLAG foci were less enriched near heterochromatin in pol2a-1 (Fig. 4 B and C). These results suggest that POL2A is required for normal distribution of MORC1 onto meiotic chromosomes, particularly to heterochromatin. We confirmed the decreased levels of MORC1-FLAG in pol2a-1 by probing Western blots of protein extracted from the nuclei of stage 1 to 14 inflorescences with anti-FLAG antibody (SI Appendix, Fig. S7B). In addition, we also examined the distribution of MORC6-FLAG in meiocytes and found that its localization is similar to that of MORC1 (SI Appendix, Fig. S7 B and C), implying that MORC1 and MORC6 may form heterodimers in meiosis as they do in mitosis.
Fig. 4.
MORC1 foci are significantly reduced in pol2a. (A and B) Anti-FLAG immunostaining showing localization of MORC1-FLAG at zygotene, pachytene, and diakinesis in WT and pol2a-1 background. In WT, MORC1-FLAG tends to form small bodies that are enriched in or near bright chromosome regions (heterochromatin) stained by DAPI at both zygotene and pachytene. In contrast, the MORC1 foci are significantly reduced in pol2a-1. SYN1, a meiosis-specific cohesin subunit, is used as control. (C) Enlarged regions in the yellow boxes from A and B. (D) Comparison of MORC1-FLAG foci counts in Col-0 and pol2a-1 at zygotene, pachytene, and diakinesis. Error bars indicate confidence interval. **, P < 0.01; two-tailed Student’s t test. Scale bars, 5 μm.
POL2A C-Terminal ZF Domain Is Essential for Meiotic Heterochromatin Condensation.
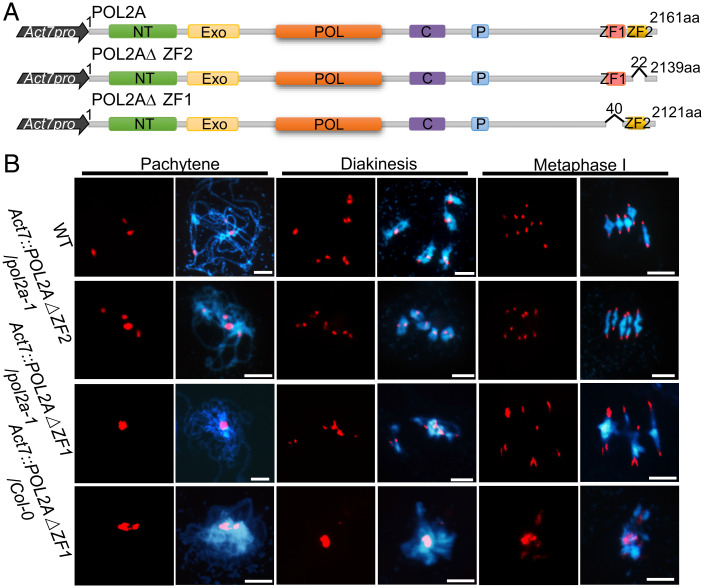
Mammalian MORC proteins have a CW-type ZF domain, which binds to H3K4me and plays important roles in regulating chromatin structure (50, 51). In contrast, the ZF domain is absent in plant MORCs including MORC1 (52). POL2A contains two conserved C-terminal ZF domains (ZF1 and ZF2; Fig. 2A). POL2A–MORC1 interaction and POL2A-dependent MORC1/6 localization raise the possibility that MORC1 deposition on meiotic chromosomes may rely on the POL2A ZF. If so, the POL2A ZFs could be essential for meiotic heterochromatin condensation. To test this, we generated transgenic plants expressing full-length POL2A and POL2AΔZF (truncated POL2A lacking both ZF domains) driven by the Actin7 promoter (Act7) (SI Appendix, Fig. S8A) (53) in a pol2a-1 background and obtained a total of 19 POL2A (Act7::POL2A/pol2a-1) and 35 POL2AΔZF (Act7::POL2AΔZF/pol2a-1) transformants. Analysis of Western blots probed with POL2A antibody showed higher POL2A expression in both transgenic lines compared to WT but lower expression in pol2a-1 (SI Appendix, Fig. S8B). Expression of full-length POL2A rescued the fertility and meiotic defects of pol2a, but expression of the truncated POL2AΔZF did not (SI Appendix, Fig. S8 C–F). To further test whether one or both of the ZFs are required for meiotic heterochromatin condensation in vivo, we made truncated POL2A lacking the ZF1 (Act7::POL2AΔZF1) and ZF2 (Act7::POL2AΔZF2) domains (Fig. 5A and SI Appendix, Fig. S9B) and transformed them into the pol2a-1 background. We found that POL2AΔZF2 but not POL2AΔZF1 could rescue the male meiotic heterochromatin defects of pol2a-1 partially (Fig. 5B). These results demonstrate that the ZF1 domain of POL2A is required for meiotic heterochromatin condensation.
Fig. 5.
ZF1 rather than ZF2 of POL2A is required for meiotic normal heterochromatin formation. (A) Illustration of synthetic constructs for whole-length POL2A and truncated POL2A without different ZFs. NT, N terminus; EXO, 3‘-5′ exonuclease; POL, 5‘-3′ polymerase; C, central; P, proliferating cell nuclear antigen interaction; aa, amino acid. (B) Chromosome phenotypes of WT, Act7::POL2AΔZF2/pol2a-1, Act7::POL2AΔZF1/pol2a-1, and Act7::POL2AΔZF1/Col-0 (DN-POL2AΔZF1) plants. Scale bars, 5 μm.
The POL2A ZF1 Domain Binds to H3.1.
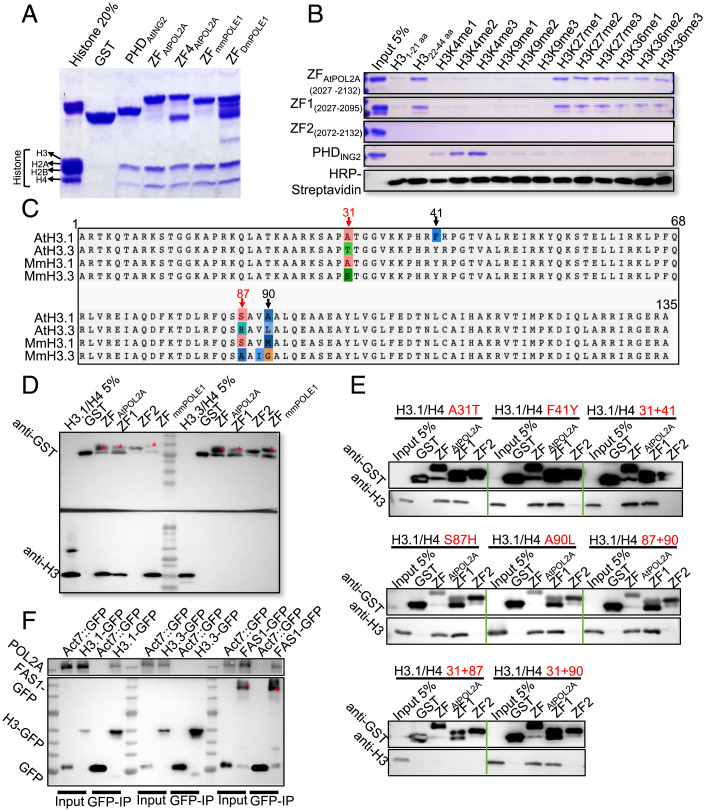
Mammalian MORC CW-type ZFs have four conserved cysteines and two or three position-conserved tryptophans and can specifically bind to H3K4me (54–56). Both ZF1 and ZF2 of POL2A are conserved across eukaryotes and contain four or five cysteines, and ZF1 harbors two additional conserved aromatic residues (one tryptophan and one tyrosine), which form an aromatic cage required for histone-binding activity in other proteins (57) (SI Appendix, Fig. S9A), suggesting that POL2A ZF might also bind histones. To test this hypothesis, we expressed and purified a GST–ZFPOL2A fusion protein containing both ZF1 and ZF2 domains and performed affinity purification pull-down assays with calf histones and human histone peptides (Fig. 6 A and B). The plant homeodomain (PHD) of AtING2 was used as a positive control, which specifically binds H3K4 methylated histone tails (58). We found that ZFPOL2A interacted with human unmodified H322–44, H3K27, and H3K36 methylated histone tails in vitro (Fig. 6B and SI Appendix, Fig. S9C). Interestingly, the H3K36me-binding ability of ZF1 was weaker than that of H3K27me. Similarly, modifications of residues neighboring K27, including H3K36me, which commonly promote transcription and are less abundant on H3.1 (9, 59), are detrimental to ATXR5/6 activity for binding nucleosomes and catalyzing monomethylation of K27 (9, 59, 60). Moreover, we also observed similar histone H3–binding specificity using mouse POLE1 (the ortholog of POL2A) ZF (Fig. 6A and SI Appendix, Fig. S9C), indicating conserved POLE1-ZF histone-binding activity in animals and plants. In contrast, POLD1 (the catalytic subunit of POL δ) also has two C-terminal ZF domains (ZFAtPOLD1) that do not have similar histone-binding activity (SI Appendix, Fig. S9C). To determine whether each AtPOL2A ZF can bind to histones, we individually expressed ZF1 and ZF2 with each fused to the alpha helix that separates them in the full-length protein (SI Appendix, Fig. S9A). ZF1 but not ZF2 binds to human unmodified H322–44 histone tails (Fig. 6B). These results agree with the in vivo meiotic functions of POL2A ZF1 we described earlier (Fig. 5 A and B).
Fig. 6.
ZF1 rather than ZF2 of POL2A binds to H3.1 both in vitro and in vivo. (A) Calf histone pull-down assays of glutathione S-transferase (GST)–fused ZFs of POL2A show that ZFPOL2A mainly binds to H3; the PHD of AtING2 is used as a positive control. ZFPOL2A (2027-2125) and ZF4POL2A (2034–2125) are truncated proteins harboring POL2A ZF1 and ZF2. (B) Biotinylated histone peptide pull-down assays of GST-fused ZFs, with AtING2 Plant Homeodomain (PHD) as a positive control. The biotin-labeled peptides were detected via Horseradish Peroxidase (HRP)-Streptavidin. (C) Alignment of H3.1 and H3.3 orthologs between Arabidopsis and mouse with four-residue difference marked above. (D) GST pull-down assay showing ZF-binding activity to H3.1/H4 and H3.3/H4 dimers. Red stars indicate the band corresponding to the predicted size of each protein. (E) GST pull-down assay showing ZF-binding activity to different mutated H3.1/H4 proteins corresponding to C. Red texts indicate the types of point mutation of H3.1. (F) POL2A coimmunoprecipitated with H3.1-GFP (HTR13::HTR13-GFP) and FAS1-GFP (Act7::FAS1-GFP), rather than H3.3-GFP (HTR5::HTR5-GFP), with Col-0 plants expressing empty GFP-tag vector as negative control. Red stars indicate the band corresponding to the predicted size of FAS1-GFP.
In Arabidopsis, histone H3 variant H3.1 localizes in heterochromatin, whereas H3.3 is usually enriched in euchromatin (7, 8, 10, 17). To explore whether POL2A ZF1 binds to H3.1-enriched heterochromatin rather than H3.3-enriched euchromatin, we synthesized Arabidopsis histone peptides H3.11–44 and H3.31–44, which differ at only two amino acids (17). To our surprise, affinity purification pull-down assays showed that POL2A ZF1 exhibits comparable binding affinity for both H3.11–44 and H3.31–44 (SI Appendix, Fig. S9D). Nucleosome assembly initiates with H3/H4 tetramer (two H3/H4 dimers) deposition onto DNA to form a nucleosome precursor in vivo (61, 62). To emulate the in vivo state, we expressed recombinant Arabidopsis H3.1/H4 and H3.3/H4 dimers using bacteria and performed affinity purification pull-down assays again with POL2A ZFs. Strikingly, POL2A ZF1 binds to the H3.1/H4 dimer but not the H3.3/H4 dimer, and ZF2 does not bind to either dimer (Fig. 6D). Moreover, we demonstrated that both mouse POLE1 ZF and Arabidopsis POL2A ZF1 have similar binding specificity with human H3.1-H4 or H3.3-H4 tetramers (Fig. 6D and SI Appendix, Fig. S9E). H3.1 and H3.3 only differ by four amino acids at positions 31, 41, 87, and 90 in plants (Fig. 6C) (63, 64). To investigate which of these residues contribute to ZF1-binding specificity, we introduced different point mutations into the H3.1/H4 protein dimer and found that mutating any single amino acid (AA) or any double mutants at AAs 87 and 90 or 37 and 41 did not disrupt the interaction with ZF1 (Fig. 6E). Interestingly, only simultaneous mutation of AAs 31 and 87 of H3.1/H4 disrupted binding with ZF of POL2A (Fig. 6E). It is noticeable that both AAs 31 and 87 of H3.1 were conserved between Arabidopsis and mouse, but AAs 41 and 90 were not (Fig. 6C). To validate these results with an independent approach, we obtained transgenic plants expressing H3.1–green fluorescent protein (GFP) and H3.3-GFP, conducted co-IP using an anti-GFP antibody, and confirmed that POL2A coprecipitates with H3.1 but not H3.3 in vivo (Fig. 6F). As expected, POL2A is also associated with FAS1, which is a CAF1 component (Fig. 6F). Taken together, our results suggest that POL2A may function in heterochromatin condensation by binding H3.1 with its ZF1 domain to facilitate the incorporation of H3.1-H4 into nucleosomes by the CAF1 complex.
POL2A Is Required for Heterochromatin H3 Deposition in Meiosis.
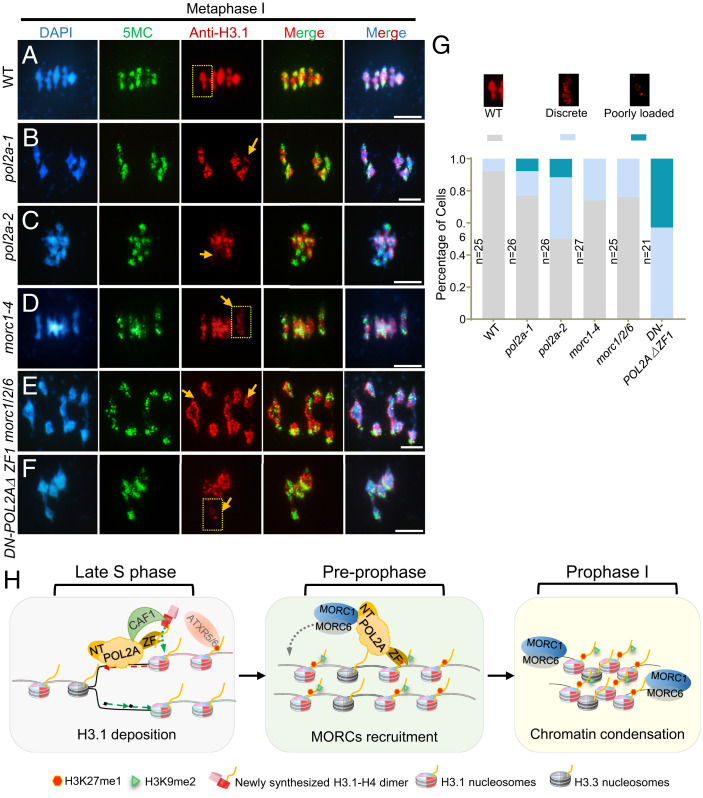
De novo deposition of H3.1 on mitotic heterochromatin is mediated by the CAF1 complex during DNA replication (7). Given that POL2A is the largest catalytic subunit of POL ε, which is required for leading-strand elongation (34), we hypothesized that POL2A might have a role in H3.1 deposition at the replication fork as well. Null alleles and C-terminal T-DNA insertion alleles of POL2A are lethal (32, 44), so we generated transgenic plants overexpressing POL2AΔZF1 driven by the ACTIN7 promoter and obtained 57 T1 plants (Act7::POL2AΔZF1/Col-0), most of which have aberrant meiotic centromere clustering and heterochromatin condensation defects even worse than Act7::POL2AΔZF1/pol2a-1 (Fig. 5B and SI Appendix, Fig. S9B). We then designated them as DN (dominant negative)–POL2AΔZF1 plants. Since H3.1 and H3.3 only differ by four or five amino acids between plants and animals, we used an antibody raised against mammalian histone H3.1 and found that anti-H3.1 recognized bacterially expressed H3.1 and H3.3 (SI Appendix, Fig. S10A). Immunolocalization signals using anti-H3.1 overlapped with DNA methylation (5MC, 5-methylcytosine) and colocalized with chromocenters in somatic cells (SI Appendix, Fig. S10B) and meiotic heterochromatin (SI Appendix, Fig. S10 C and D). In particular, in WT meiocytes, the H3.1 signals were associated with heterochromatic regions during meiotic prophase I (SI Appendix, Fig. S10 C and D) and overlapped with the highly condensed bivalents in metaphase I (Fig. 7A). To validate the specificity of anti-H3.1, we tested the signal examined by anti-H3.1 in fas1 and fas2 mutants, which have defective H3.1 loading during DNA replication (26, 65). The signals were significantly reduced in fas1/2 pachytene and metaphase I chromosomes compared with WT, especially on the less-compacted pachytene chromosomes (SI Appendix, Fig. S11 B, D, and E). Taken together, these results indicate that anti-H3.1 may recognize H3.1 in vivo and can at least reflect the density of H3 or the level of nucleosome condensation in heterochromatin. Interestingly, DN-POL2AΔZF1 male meiocytes also had dramatically decreased H3.1 density in heterochromatin, but normal 5MC signals compared to WT (SI Appendix, Fig. S10 C–E), indicating a potential role for POL2A in H3.1 deposition or maintenance in meiotic heterochromatin.
Fig. 7.
POL2A is required for normal localization of H3 of heterochromatin and heterochromatin condensation in meiosis. (A–F) Immunofluorescence using anti-H3.1 at metaphase I in WT, pol2a-1, pol2a-2, morc1-4, morc1/2/6, and DN-POL2AΔZF1. 5-Methylcytosine (5MC) is used as a control to indicate the pericentromeric regions. Yellow arrows indicate bivalents with weak H3.1 signals. Scale bars, 5 μm. (G) Percentage of metaphase I cells showing poorly loaded (bivalent with only a few foci), discrete (several discontinuous foci along the length of the bivalent), and WT (bivalent with continuous signals) anti-H3.1 fluorescence in each line. The three examples above the diagram are enlarged regions of the yellow squares in A–F. (H) A proposed model showing the role of POL2A in meiotic heterochromatin establishment. At late S phase, POL2A catalyzes leading-strand DNA synthesis and may work with CAF1 complex to recruit H3.1/H4 dimers or recycling of parental histones by its ZF1. Assembly of nucleosomes that include H3.1 facilitates generation of H3.1K27me1 in heterochromatin. After S phase, at preprophase, POL2A may still associate with the chromosome by its ZF1 binding to H3.1. The POL2A N terminus interacts with MORC1/MORC6 heterodimer to mediate its association with chromatin in meiotic prophase I. MORC1/6 are responsible for condensation of chromosomes, especially on H3.1-enriched heterochromatic regions.
As A31 and S87 of H3.1/H4 are responsible for the binding specificity of POL2A, we further tested the function of the two amino acids in H3.1 deposition. H3.1 is encoded by five genes, HTR1, HTR2, HTR3, HTR9, and HTR13 in Arabidopsis. htr13 mutation was introduced into htr1 htr2 htr3 htr9 (htr1239) quadruple mutant by CRISPR-Cas9 to generate an h3.1 mutant (SI Appendix, Fig. S12A). Similar to the h3.1 knockdown mutants in the previous report (29), h3.1 mutants showed severe developmental defects, including abnormal tillers, fasciated stems, ectopic leaflets, enlarged inflorescence meristems, and reduced fertility (SI Appendix, Fig. S12C). To explore the function of A31 and S87, we generated transgenic lines expressing a series of amino acid substitutions of HTR1 in h3.1 background (SI Appendix, Fig. S12 B and C). Interestingly, single A31T or S87H mutation of H3.1 nearly rescued the developmental phenotypes of h3.1, but simultaneous mutation of A31 and S87 of H3.1 did not (SI Appendix, Fig. S12C). Furthermore, we observed H3.1 signals of the meiotic heterochromatin in each transgenic line. Expression of single A31T or S87H mutation of H3.1 mostly rescued the H3.1 signals on meiotic heterochromatin (SI Appendix, Fig. S12 D and E). Meanwhile, HTR1 A31T/S87H still showed much more serious defects (SI Appendix, Fig. S12 D and E), indicating that both sites of H3.1 are required for H3.1 deposition and function during meiosis.
To test whether H3 deposition in heterochromatin is mediated by POL2A-MORC1 in vivo, we performed immunostaining with anti-H3.1 in WT, pol2a-1, pol2a-2, and morc1 mutants. H3.1 signals were less contiguous in pol2a and morc1 mutants (Fig. 7 B–F). Furthermore, nearly 10% of bivalents had very little H3.1 signal but relatively normal DNA methylation levels (Fig. 7 B–D). We classified H3.1 localization into three patterns for phenotyping purposes (Fig. 7G). Using these categories, pol2a-2 had a more severe phenotype than morc1 (Fig. 7G), consistent with the difference in the severity of heterochromatin defects observed in pol2a and morc1 (Fig. 3). Given that POL2A coprecipitates with FAS1 in vivo, these results support the idea that POL2A may be required for H3.1 deposition by the CAF1 complex and formation of WT heterochromatin upon DNA replication and during meiosis.
Discussion
POL2A and MORC1 Are Required for Meiotic Heterochromatin Condensation.
POL2A, the largest catalytic subunit of POL ε, has multiple roles in mitotic and meiotic DNA repair, regulation of gene silencing, checkpoint signaling, and response to abscisic acid (34). Interestingly, a recent study isolated three strong hypomorphic pol2a mutants with mutations on DNA polymerase domain, which exhibited the release of heterochromatin silencing with compensating CHG hypermethylation and had little effect on H3K27me1, H2A.W, and small interfering RNA(35). However, the underlying molecular mechanism of how POL2A functions in heterochromatin organization is mysterious. Here, we have provided several lines of evidence that reveal a function of POL2A in meiotic heterochromatin condensation. First, two hypomorphic alleles of pol2a had defects in heterochromatin condensation, including longer heterochromatin regions, enlarged centromere signals (Fig. 1), and dispersed distribution of H3K27me1 and H3K9me2 heterochromatin marks (Fig. 1 and SI Appendix, Figs. S1 and S2). Second, we demonstrated that the N-terminal region of POL2A interacts with MORC1 both in vitro and in vivo (Fig. 2), while mutation of the POL2A N terminus disrupted this interaction (Fig. 2 B and D). Third, morc1 mutants had similar meiotic heterochromatin condensation defects compared to pol2a; and the pol2a morc1 double mutant resembled the pol2a single mutant. Together, these results support the notion that a POL2A-MORC1 module promotes heterochromatin condensation during meiosis.
Our results also reveal a meiotic function for MORC1 in plants. In both mice and Caenorhabditis elegans, MORC1 participates in germline silencing of heterochromatin-associated TEs (66, 67), and in Arabidopsis, it represses TEs and mediates heterochromatin condensation in mitotically dividing cells (15, 49). In mammals, MORCs contain a CW-type ZF domain that mainly binds to H3K4me (50, 51, 55). In contrast, plant MORC proteins do not have a ZF domain, which invokes a hypothesis that plant MORCs may need cofactors, such as POL2A, to associate with chromatin during meiosis. We have provided two lines of evidence to support the recruitment of MORC1 by POL2A. First, MORC1 directly interacted with POL2A both in vitro and in vivo (Fig. 2). Second, MORC1 localization on meiotic chromosomes was reduced in pol2a mutants (Fig. 4). This raises the question of how POL2A associates with chromatin after DNA replication. The catalytic subunits of three replicative DNA polymerases (POL ε, POL δ, and POL α) all have two conserved C-terminal ZF domains (two cysteine motifs) designated as ZF1 and ZF2 (68). The ZF2 domain of all three polymerases interacts with accessory subunits of the DNA replication machinery (69–71). In yeast, ZF1 of Pol3 (POL δ) mediates interaction with PCNA to maintain its processivity (69). Conversely, crystal structure analyses of human POL ε revealed that ZF1 neither directly participates in the interaction between subunits nor interacts with PCNA (70). We demonstrated that POL2A ZF1, but not ZF2, exclusively associates with a heterochromatin-enriched H3.1 (Fig. 6). In contrast, ZF1 of POL δ does not have similar histone-binding activity (SI Appendix, Fig. S9C). Furthermore, in vivo functional assays showed that POL2A ZF1 is crucial for heterochromatin condensation and H3.1 deposition during meiosis (Fig. 7 A–G and SI Appendix, Fig. S10 C and D). Therefore, we suggest a ZF-dependent POL ε function in coordinating deposition of H3.1 and promoting heterochromatin condensation by partnering with MORC1.
POL ε Likely Facilitates H3.1 Deposition and Histone Modification during Meiotic DNA Replication.
Nucleosomes and relevant epigenetic marks need to be restored following each round of DNA replication. The newly synthesized H3.1/H4 histone dimers are thought to be chaperoned by CAF1 onto chromatin (61). In Arabidopsis, the largest subunit, FAS1, interacts with PCNA and is required for binding H3/H4 dimers (28, 29). FAS1 also interacts specifically with H3.1 in vivo and nonspecifically with both H3.1 or H3.3 in vitro (29). Interestingly, we found that POL2A not only specifically bound to the H3.1/H4 dimer via its ZF1 domain both in vivo and in vitro (Fig. 6 D–F) but also associated with FAS1 in vivo (Fig. 6F). Human POL ε colocalizes with PCNA for heterochromatin replication at late S phase (72), and Arabidopsis POL2A directly interacts with MSI1 of the CAF1 complex (31). These findings suggest that POL2A may associate with CAF1 to facilitate newly synthesized H3.1 deposition or recycling of parental H3.1-nucleosomes at the replication fork. This also agrees with previous findings that budding yeast DPB3 and DPB4, two nonessential subunits of POL ε, are also required for assembly of (H3-H4)2 tetramers onto leading strands and function in the inheritance of heterochromatin (73, 74). This hypothesis was supported by our observation that heterochromatin-associated H3 was significantly reduced from preprophase to metaphase I in pol2a mutants, especially in POL2AΔZF1-overexpressing plants (Fig. 7 A–G and SI Appendix, Fig. S10), which was also seen in fas mutants (SI Appendix, Fig. S11). Moreover, simultaneous mutation of A31 and S87 of H3.1 disrupted POL2A binding with H3.1-H4 (Fig. 6E), which appear to be essential for H3.1 deposition and function during meiosis (SI Appendix, Fig. S12). Integration of in vitro experiments (Fig. 6 and SI Appendix, Fig. S9), we speculate that POL2A-ZF has a binding specificity to the N terminus of H3.1 in H3.1-H4 dimer/tetramer, while both A31 and S87 mutation likely leads to the configuration change of the H3.1-H4 dimer, thus affecting its interaction with POL2A-ZF. Altogether, these results indicate that POL2A may be essential for H3.1 deposition during the S phase of both the mitotically and meiotically dividing cells.
Why are H3K27me1 signals reduced in pol2a? In plants, ATXR5 and ATXR6 methylate lysine-27 on histone H3. They also selectively modify variant histone H3.1 (H3.1K27me1) in a replication-dependent manner, while replication-independent H3.3 inhibits their activity (17). However, there is no evidence that POL2A can directly interact with ATXR5/6. One possibility is that in pol2a, heterochromatin condensation is compromised due to a decrease in H3.1 density, resulting in reduced H3.1K27me1 signal (Figs. 1 and 7 B and C). To test this hypothesis, we examined H3K27me1 immunolocalization in fas mutants and found that both fas1-4 and fas2-4 had decreased H3K27me1 signals in meiosis, which is consistent with the compromised H3.1 signals in the two mutants (SI Appendix, Fig. S11). This phenotype is similar to that of pol2a mutants. Another possibility is that PCNA interacts with both POL2A and ATXR5/6, tethering them to promote monomethylation on H3.1 at the replication fork (75). Interestingly, Schizosaccharomyces pombe uses this kind of mechanism, wherein its H3K9me histone methyltransferase and POL2A homolog share the same silencing complex to participate in mitotic heterochromatin condensation (74, 76). Thus, POL2A may play multiple roles in coordination of DNA synthesis, H3.1 deposition, and H3K27 or H3K9 modification during DNA replication. Moreover, a recent study using a strong allele of pol2a-12 identified that over 85% of up-regulated TEs are also activated in atxr5/6, accompanied by a modest reduction of H3K27me1 level (35). This result is slightly inconsistent with our data. We conjecture that this difference may be due to differences in the pol2a alleles used in each study or because of differences in cellular context.
A POL2A-Facilitated H3.1 Deposition and Meiotic Heterochromatin Condensation Model.
Based on our results and previous data, we present a model to illustrate POL ε–coupled H3.1 incorporation at DNA replication and heterochromatin condensation during meiosis. It has been reported that heterochromatin replication is delayed relative to euchromatin (77) and that human POL ε only colocalizes with PCNA for heterochromatin replication in the late S phase (72). During premeiotic DNA replication, we hypothesize that POL ε catalyzes DNA polymerization on the leading strand and recycles parental (H3.1-H4)2 tetramers (73) or recruits newly synthesized H3.1/H4 dimers using its ZF1 domain (Fig. 6) and associates with CAF1 via MSI1 (31) to facilitate H3.1 deposition and nucleosome assembly (Fig. 7H, Left). It is plausible that ATXR5/6 subsequently restores K27me1 on H3.1 (17). Our results suggest that POL2A may bind to H3.1 on heterochromatin and recruit other epigenetic factors, such as MORC1, to facilitate heterochromatin condensation and modification throughout the cell cycle, similar to HP1’s function in yeast and mammals (78). Following DNA replication, the ZF1 domain of POL2A may still be associated with H3.1 to maintain the association of POL2A with the chromosome and to facilitate the stabilization of H3.1K27me1 by POL2A. The POL2A N terminus interacts with MORC1/MORC6 heterodimers (Fig. 2 and SI Appendix, Fig. S4) and recruits them to the chromatin (Fig. 7H, Middle). Consequently, MORC1/6 facilitate chromatin condensation, particularly in H3.1-enriched heterochromatin regions (Fig. 7H, Right). We also have shown that POL ε ZF1 specifically binds to H3.1 in animals. These findings represent broad relevance for chromosome transmission, genome stability, and reproductive biology.
Materials and Methods
All pol2a and morc mutants were described previously (15, 44, 49). Details on the following are available in SI Appendix, SI Materials and Methods: plant materials, morphological and cytological analyses, constructs for plant transformation and Y2H assays, BiFC assay, affinity purification pull-down assay, co-IP, calf-histone, histone peptide and H3/H4 dimer-binding assays, and accession numbers. The primers used in this study are listed in SI Appendix, Table S1. Peptides used in this study are listed in SI Appendix, Table S2.
Supplementary Material
Acknowledgments
We thank Xinjian He (National Institute of Biological Sciences, Beijing, China) for the morc1/2/6 mutants. We thank Chaoyi Yu and Qiang Luo at Fudan University for advice on the histone-binding assay. This research was supported by grants from the National Natural Science Foundation of China (Grant Nos. 31925005, 31870293, and 32000246), the State Key Laboratory of Genetic Engineering, the China Postdoctoral Science Foundation, National Postdoctoral Program for Innovative Talents and Fudan University, and Guangdong Laboratory for Lingnan Modern Agriculture.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2213540119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J., Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Grewal S. I., Jia S., Heterochromatin revisited. Nat. Rev. Genet. 8, 35–46 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Allshire R. C., Madhani H. D., Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol. 19, 229–244 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng W., Michaels S. D., Accessing the inaccessible: The organization, transcription, replication, and repair of heterochromatin in plants. Annu. Rev. Genet. 49, 439–459 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Klemm S. L., Shipony Z., Greenleaf W. J., Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 20, 207–220 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Benoit M., et al. , Replication-coupled histone H3.1 deposition determines nucleosome composition and heterochromatin dynamics during Arabidopsis seedling development. New Phytol. 221, 385–398 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Tagami H., Ray-Gallet D., Almouzni G., Nakatani Y., Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116, 51–61 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Jiang D., Berger F., Histone variants in plant transcriptional regulation. Biochim. Biophys. Acta Gene Regul. Mech. 1860, 123–130 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Wollmann H., et al. , Dynamic deposition of histone variant H3.3 accompanies developmental remodeling of the Arabidopsis transcriptome. PLoS Genet. 8, e1002658 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stroud H., et al. , Genome-wide analysis of histone H3.1 and H3.3 variants in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 109, 5370–5375 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ono T., et al. , Chromatin assembly factor 1 ensures the stable maintenance of silent chromatin states in Arabidopsis. Genes Cells 11, 153–162 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Kirik A., Pecinka A., Wendeler E., Reiss B., The chromatin assembly factor subunit FASCIATA1 is involved in homologous recombination in plants. Plant Cell 18, 2431–2442 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yelagandula R., et al. , The histone variant H2A.W defines heterochromatin and promotes chromatin condensation in Arabidopsis. Cell 158, 98–109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi J., Lyons D. B., Kim M. Y., Moore J. D., Zilberman D., DNA methylation and histone H1 jointly repress transposable elements and aberrant intragenic transcripts. Mol. Cell 77, 310–323.e7 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Moissiard G., et al. , MORC family ATPases required for heterochromatin condensation and gene silencing. Science 336, 1448–1451 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H., Lang Z., Zhu J. K., Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 19, 489–506 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Jacob Y., et al. , Selective methylation of histone H3 variant H3.1 regulates heterochromatin replication. Science 343, 1249–1253 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X., et al. , Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 5, e129 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathieu O., Probst A. V., Paszkowski J., Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. EMBO J. 24, 2783–2791 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simon J. A., Kingston R. E., Mechanisms of polycomb gene silencing: Knowns and unknowns. Nat. Rev. Mol. Cell Biol. 10, 697–708 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Xiao J., Wagner D., Polycomb repression in the regulation of growth and development in Arabidopsis. Curr. Opin. Plant Biol. 23, 15–24 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Jacob Y., et al. , ATXR5 and ATXR6 are H3K27 monomethyltransferases required for chromatin structure and gene silencing. Nat. Struct. Mol. Biol. 16, 763–768 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ebbs M. L., Bartee L., Bender J., H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases. Mol. Cell. Biol. 25, 10507–10515 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ebbs M. L., Bender J., Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell 18, 1166–1176 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du J., Johnson L. M., Jacobsen S. E., Patel D. J., DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 16, 519–532 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramirez-Parra E., Gutierrez C., The many faces of chromatin assembly factor 1. Trends Plant Sci. 12, 570–576 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Liu Q., Gong Z., The coupling of epigenome replication with DNA replication. Curr. Opin. Plant Biol. 14, 187–194 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Shibahara K., Stillman B., Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96, 575–585 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Jiang D., Berger F., DNA replication-coupled histone modification maintains Polycomb gene silencing in plants. Science 357, 1146–1149 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Jacob Y., et al. , Regulation of heterochromatic DNA replication by histone H3 lysine 27 methyltransferases. Nature 466, 987–991 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Del Olmo I., et al. , Arabidopsis DNA polymerase ϵ recruits components of Polycomb repressor complex to mediate epigenetic gene silencing. Nucleic Acids Res. 44, 5597–5614 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.del Olmo I., et al. , EARLY IN SHORT DAYS 7 (ESD7) encodes the catalytic subunit of DNA polymerase epsilon and is required for flowering repression through a mechanism involving epigenetic gene silencing. Plant J. 61, 623–636 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Hyun Y., et al. , The catalytic subunit of Arabidopsis DNA polymerase α ensures stable maintenance of histone modification. Development 140, 156–166 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Pedroza-Garcia J. A., De Veylder L., Raynaud C., Plant DNA polymerases. Int. J. Mol. Sci. 20, 4814 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bourguet P., et al. , DNA polymerase epsilon is required for heterochromatin maintenance in Arabidopsis. Genome Biol. 21, 283 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirano T., Chromosome dynamics during mitosis. Cold Spring Harb. Perspect. Biol. 7, a015792 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma H., A molecular portrait of Arabidopsis meiosis. Arabidopsis Book 4, e0095 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eyster C., Chuong H. H., Lee C. Y., Pezza R. J., Dawson D., The pericentromeric heterochromatin of homologous chromosomes remains associated after centromere pairing dissolves in mouse spermatocyte meiosis. Chromosoma 128, 355–367 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Da Ines O., Abe K., Goubely C., Gallego M. E., White C. I., Differing requirements for RAD51 and DMC1 in meiotic pairing of centromeres and chromosome arms in Arabidopsis thaliana. PLoS Genet. 8, e1002636 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dernburg A. F., Sedat J. W., Hawley R. S., Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell 86, 135–146 (1996). [DOI] [PubMed] [Google Scholar]
- 41.Griffiths S., et al. , Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439, 749–752 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Da Ines O., White C. I., Centromere associations in meiotic chromosome pairing. Annu. Rev. Genet. 49, 95–114 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Ronceret A., et al. , Genetic analysis of two Arabidopsis DNA polymerase epsilon subunits during early embryogenesis. Plant J. 44, 223–236 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Huang J., et al. , Formation of interference-sensitive meiotic cross-overs requires sufficient DNA leading-strand elongation. Proc. Natl. Acad. Sci. U.S.A. 112, 12534–12539 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li W., et al. , The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc. Natl. Acad. Sci. U.S.A. 101, 10596–10601 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moissiard G., et al. , Transcriptional gene silencing by Arabidopsis microrchidia homologues involves the formation of heteromers. Proc. Natl. Acad. Sci. U.S.A. 111, 7474–7479 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inoue N., et al. , New gene family defined by MORC, a nuclear protein required for mouse spermatogenesis. Hum. Mol. Genet. 8, 1201–1207 (1999). [DOI] [PubMed] [Google Scholar]
- 48.Watson M. L., et al. , Identification of morc (microrchidia), a mutation that results in arrest of spermatogenesis at an early meiotic stage in the mouse. Proc. Natl. Acad. Sci. U.S.A. 95, 14361–14366 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Z. W., et al. , Two components of the RNA-directed DNA methylation pathway associate with MORC6 and silence loci targeted by MORC6 in Arabidopsis. PLoS Genet. 12, e1006026 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoppmann V., et al. , The CW domain, a new histone recognition module in chromatin proteins. EMBO J. 30, 1939–1952 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y., et al. , MORC3 forms nuclear condensates through phase separation. iScience 17, 182–189 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koch A., et al. , MORC proteins: Novel players in plant and animal health. Front. Plant Sci. 8, 1720 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McDowell J. M., An Y. Q., Huang S., McKinney E. C., Meagher R. B., The Arabidopsis ACT7 actin gene is expressed in rapidly developing tissues and responds to several external stimuli. Plant Physiol. 111, 699–711 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li D. Q., Nair S. S., Kumar R., The MORC family: New epigenetic regulators of transcription and DNA damage response. Epigenetics 8, 685–693 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li S., et al. , Mouse MORC3 is a GHKL ATPase that localizes to H3K4me3 marked chromatin. Proc. Natl. Acad. Sci. U.S.A. 113, E5108–E5116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Y., et al. , Family-wide characterization of histone binding abilities of human CW domain-containing proteins. J. Biol. Chem. 291, 9000–9013 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu R., Li X., Chen W., Du J., Structure and mechanism of plant histone mark readers. Sci. China Life Sci. 61, 170–177 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Peña P. V., et al. , Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 442, 100–103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnson L., et al. , Mass spectrometry analysis of Arabidopsis histone H3 reveals distinct combinations of post-translational modifications. Nucleic Acids Res. 32, 6511–6518 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bergamin E., et al. , Molecular basis for the methylation specificity of ATXR5 for histone H3. Nucleic Acids Res. 45, 6375–6387 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grover P., Asa J. S., Campos E. I., H3-H4 histone chaperone pathways. Annu. Rev. Genet. 52, 109–130 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Smith S., Stillman B., Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 10, 971–980 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ingouff M., Berger F., Histone3 variants in plants. Chromosoma 119, 27–33 (2010). [DOI] [PubMed] [Google Scholar]
- 64.Lu L., Chen X., Qian S., Zhong X., The plant-specific histone residue Phe41 is important for genome-wide H3.1 distribution. Nat. Commun. 9, 630 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Otero S., Desvoyes B., Peiró R., Gutierrez C., Histone H3 dynamics reveal domains with distinct proliferation potential in the Arabidopsis root. Plant Cell 28, 1361–1371 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pastor W. A., et al. , Erratum: MORC1 represses transposable elements in the mouse male germline. Nat. Commun. 6, 7604 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weiser N. E., et al. , MORC-1 integrates nuclear RNAi and transgenerational chromatin architecture to promote germline immortality. Dev. Cell 41, 408–423.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sengupta S., van Deursen F., de Piccoli G., Labib K., Dpb2 integrates the leading-strand DNA polymerase into the eukaryotic replisome. Curr. Biol. 23, 543–552 (2013). [DOI] [PubMed] [Google Scholar]
- 69.Netz D. J., et al. , Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat. Chem. Biol. 8, 125–132 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baranovskiy A. G., et al. , Crystal structure of the human Polϵ B-subunit in complex with the C-terminal domain of the catalytic subunit. J. Biol. Chem. 292, 15717–15730 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suwa Y., et al. , Crystal structure of the human pol α B subunit in complex with the C-terminal domain of the catalytic subunit. J. Biol. Chem. 290, 14328–14337 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fuss J., Linn S., Human DNA polymerase epsilon colocalizes with proliferating cell nuclear antigen and DNA replication late, but not early, in S phase. J. Biol. Chem. 277, 8658–8666 (2002). [DOI] [PubMed] [Google Scholar]
- 73.Yu C., et al. , A mechanism for preventing asymmetric histone segregation onto replicating DNA strands. Science 361, 1386–1389 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.He H., et al. , Coordinated regulation of heterochromatin inheritance by Dpb3-Dpb4 complex. Proc. Natl. Acad. Sci. U.S.A. 114, 12524–12529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Davarinejad H., Joshi M., Ait-Hamou N., Munro K., Couture J. F., ATXR5/6 forms alternative protein complexes with PCNA and the nucleosome core particle. J. Mol. Biol. 431, 1370–1379 (2019). [DOI] [PubMed] [Google Scholar]
- 76.Li F., Martienssen R., Cande W. Z., Coordination of DNA replication and histone modification by the Rik1-Dos2 complex. Nature 475, 244–248 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wallace J. A., Orr-Weaver T. L., Replication of heterochromatin: Insights into mechanisms of epigenetic inheritance. Chromosoma 114, 389–402 (2005). [DOI] [PubMed] [Google Scholar]
- 78.Kwon S. H., Workman J. L., The heterochromatin protein 1 (HP1) family: Put away a bias toward HP1. Mol. Cells 26, 217–227 (2008). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.