Significance
RAS is mutated in nearly 20% of human cancers and has few direct, efficacious inhibitors. Here, we developed a synthetic protein, JAM20, to discover potential regions on RAS that may be used for inhibition. JAM20 robustly inhibited signaling mediated by multiple oncogenic KRAS, HRAS, and NRAS mutants. It bound to an epitope in the Switch I/II pocket that overlaps with previously developed small-molecule inhibitors whose efficacy in a mouse model has not been established. These findings validate targeting the Switch I/II region as an approach to inhibiting tumors driven by diverse oncogenic RAS mutants and demonstrate the utility of synthetic binding proteins as tools for drug discovery against difficult targets.
Keywords: synthetic binding protein, tool biologic, drug discovery, target validation
Abstract
RAS mutants are major therapeutic targets in oncology with few efficacious direct inhibitors available. The identification of a shallow pocket near the Switch II region on RAS has led to the development of small-molecule drugs that target this site and inhibit KRAS(G12C) and KRAS(G12D). To discover other regions on RAS that may be targeted for inhibition, we have employed small synthetic binding proteins termed monobodies that have a strong propensity to bind to functional sites on a target protein. Here, we report a pan-RAS monobody, termed JAM20, that bound to all RAS isoforms with nanomolar affinity and demonstrated limited nucleotide-state specificity. Upon intracellular expression, JAM20 potently inhibited signaling mediated by all RAS isoforms and reduced oncogenic RAS-mediated tumorigenesis in vivo. NMR and mutation analysis determined that JAM20 bound to a pocket between Switch I and II, which is similarly targeted by low-affinity, small-molecule inhibitors, such as BI-2852, whose in vivo efficacy has not been demonstrated. Furthermore, JAM20 directly competed with both the RAF(RBD) and BI-2852. These results provide direct validation of targeting the Switch I/II pocket for inhibiting RAS-driven tumorigenesis. More generally, these results demonstrate the utility of tool biologics as probes for discovering and validating druggable sites on challenging targets.
Mutated in almost 20% of human cancers, the RAS GTPase is a major drug target in oncology (1). Its isoforms (K, H, and NRAS) are responsible for regulating cell growth and other critical cellular processes through their function as GTPases. To regulate signal activation, wild-type (WT) RAS is converted to the active (GTP-bound) state via a Guanine Nucleotide Exchange Factor and then converted to the inactive (GDP-bound) state with the help of a GTPase Accelerating Protein (GAP) (2–5). Active, GTP-bound RAS interacts with diverse proteins termed effectors to promote signaling (6–10). Although this activation mechanism is properly regulated under normal circumstances, oncogenic RAS tends to exhibit reduced sensitivity to GAP-mediated GTP hydrolysis, leading to extended signal activation (11, 12).
Because of its prevalence in human cancers, there have been many attempts to inhibit oncogenic RAS-mediated signaling. Whereas attempts to directly inhibit RAS have been largely unsuccessful, the recent discovery of a pocket near the Switch II region (SII) and advancements in small-molecule chemistry have led to the successful development of mutant-specific RAS inhibitors that target this pocket. The SII pocket was only discovered when the RAS structure was determined with a tethered tool compound (13). Because of its dynamic nature, this pocket had remained undetected until this approach was used. This and other studies underscore the importance of developing tool ligands to discover new druggable sites on targets of interest (14).
Since the discovery of the SII pocket, the development of small-molecule inhibitors, such as sotorasib, adagrasib, and MRTX1133, have demonstrated that it is possible to engage the Switch region and achieve high selectivity for mutant over WT RAS (13, 15–17). Although these breakthroughs demonstrate the efficacy of targeting the RAS SII pocket for inhibition, it is unclear whether these molecules can be further derivatized for targeting other RAS mutants (18–20). Thus, there remains a substantial need to identify additional sites on RAS that can be targeted for direct inhibition.
Unlike small-molecule inhibitors, protein-based ligands are less reliant on deep pockets and can target relatively flat surfaces. Therefore, the development of peptides and synthetic proteins against RAS is generally less challenging than developing small molecules. Such reagents can be used as functional probes to identify sites on a target for inhibition. For example, synthetic proteins, such as monobodies, have been successfully developed against a diverse array of targets, including RAS, and tend to bind to underappreciated functional sites (21–28). Whereas inhibitors like sotorasib, adagrasib, and MRTX1133 bind near the Switch region of RAS, the monobody (Mb) NS1 marked the first inhibitor to bind to RAS outside of the Switch region (27). NS1 binds to the α4–β6–α5 region and inhibits signaling mediated by both KRAS and HRAS, demonstrating the feasibility of effectively inhibiting RAS-mediated signaling without targeting the Switch region. In addition to Mbs, other synthetic binding proteins, such as Designed Ankyrin Repeat Proteins (DARPins), have also been developed to identify regions on RAS for inhibition. KRAS-selective DARPins K13 and K19 have established the α3–loop 7–α4 region of KRAS as another site for RAS inhibition (29). These results demonstrate the utility of synthetic binding proteins to identify new sites on RAS that may be directly targeted for inhibition, ultimately with small molecules or via intracellular delivery of proteins.
In this study, we set out to address whether other regions exist on RAS that may be targeted for inhibition. We report a pan-RAS Mb termed JAM20 that bound to RAS in both nucleotide states. Furthermore, it robustly inhibited RAS-mediated signaling and cell transformation both in vitro and in vivo. Despite its low sensitivity to the bound nucleotide, JAM20 targeted an epitope within the Switch I (SI)/SII region of RAS and inhibited RAS–RAF(RBD) interaction. JAM20 bound to a similar region as low-affinity, small-molecule inhibitors as well as other synthetic binding proteins, whose efficacy in vivo has not been established (30, 31), making intracellularly expressed JAM20 a surrogate for these molecules. Our findings offer additional support for targeting the SI/SII pocket and its vicinity for future therapeutic development.
Results
Development of a Pan-RAS monobody, JAM20.
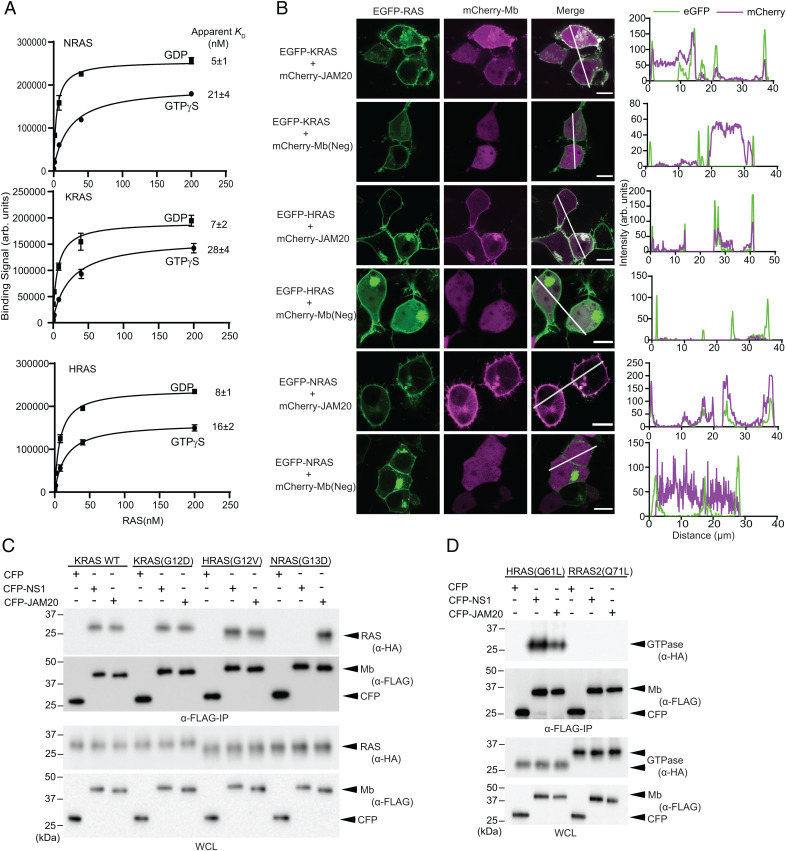
To identify regions on RAS that may be targeted for direct inhibition, we set out to develop Mbs that bind to a region other than those targeted by known ligands of RAS, such as RAF(RBD) or NS1. Natural effectors such as RAF bind to the Switch region and show strong nucleotide-state specificity, but little isoform specificity (8). Conversely, proteins that bind outside of the Switch region demonstrate distinct properties. For example, NS1 binds to both KRAS and HRAS with no nucleotide-state specificity, but does not bind to NRAS (27). To identify Mbs with properties distinct from these RAS-binding molecules—i.e., Mbs that are minimally dependent on the nucleotide state and cross-react with all isoforms—we performed library selection by alternating KRAS and NRAS as the target in either a GDP- or GTPγS-bound state over the course of successive rounds. We used the 4B isoform for KRAS, KRAS4B, throughout this study, unless otherwise stated. Additionally, the concentration of RAS was reduced over the course of the selection process to enrich for Mbs with high affinity. We identified a Mb termed JAM20 that bound to each RAS isoform in either the GTP- or GDP-bound state as measured by using JAM20 expressed on the surface of yeast (32, 33) (Fig. 1A and SI Appendix, Table S1). JAM20 exhibited higher affinity for the GDP-bound over the GTPγS-bound state of the RAS isoforms (Fig. 1A). Despite this preference, all apparent KD values were below 30 nM, indicating that we had successfully developed a pan-RAS Mb that exhibited limited nucleotide-state preference.
Fig. 1.
JAM20 binds to all RAS isoforms. (A) Binding measurements evaluated by using flow cytometry of JAM20 in yeast display against WT RAS isoforms bound to either GDP or GTPγS. The mean and SD of KD values from three technical replicates are plotted. (B) Colocalization of transiently expressed mCherry-tagged JAM20 or negative control Mb, Mb(Neg) (pseudocolored in magenta), with transiently expressed K, H, or NRAS(WT) (pseudocolored in green) in HEK293T cells. (Scale bars: 10 µm.) The graphs in B, Right show the fluorescence intensity profiles under the white lines across the microscopy images after subtracting a background signal. (C and D) JAM20 specifically binds RAS isoforms. Various HA-tagged GTPases were coexpressed along with CFP–FLAG–JAM20. CFP–NS1 and CFP–FLAG were used as controls. The bottom two panels show expression of HA-tagged GTPases and FLAG-tagged CFP-Mbs in whole cell lysates (WCL). Arb., arbitrary.
We next tested whether JAM20 was sufficiently specific to engage with RAS in cells by evaluating colocalization of fluorescent protein-tagged constructs expressed in HEK293T cells. eGFP-tagged RAS and mCherry-tagged JAM20 colocalized on the plasma membrane and within intracellular vesicles, whereas the negative control Mb, Mb(Neg), did not (Fig. 1B). Additionally, we evaluated whether JAM20 captured RAS in a coimmunoprecipitation (Co-IP) assay. HEK293 cells were cotransfected with CFP–FLAG-tagged Mbs and either HA-tagged oncogenic RAS or RAS-related protein (RRAS2) (Fig. 1 C and D). JAM20 captured various oncogenic forms of HRAS, KRAS, and NRAS, but not RRAS2(Q71L). In contrast, NS1 captured HRAS and KRAS, but not NRAS or RRAS2, as expected from its binding specificity (27). Although the colocalization experiment cannot exclude the possibility that JAM20 weakly interacts with other cytoplasmic proteins, these results demonstrate that JAM20 interacted with all RAS isoforms in the cellular environment.
JAM20 Inhibits RAS-Mediated Signaling.
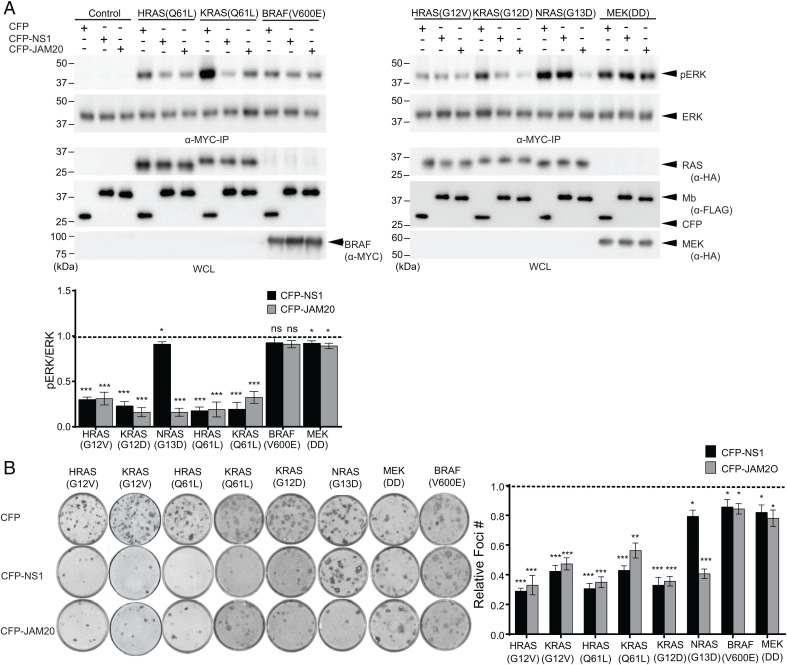
Next, we evaluated whether JAM20 inhibited RAS-mediated signaling in cells using a transient cotransfection assay, as described (27). CFP–FLAG-tagged JAM20 (hereafter referred to as JAM20) inhibited ERK activation by all oncogenic RAS mutants, whereas CFP–FLAG-NS1 (referred to as NS1) selectively inhibited only KRAS and HRAS (Fig. 2A). Both JAM20 and NS1 also inhibited EGF-induced activation of ERK mediated by endogenous RAS (SI Appendix, Fig. S1). Additionally, both Mbs did not impact ERK activation by either oncogenic BRAF(V600E) or MEK(DD), which are downstream components of the RAS–RAF–MEK–ERK pathway (34) (Fig. 2A). Taken together, these results show that JAM20 robustly and selectively inhibited the activation of MAPK signaling driven by all RAS isoforms.
Fig. 2.
JAM20 selectively inhibits RAS-mediated signaling in cells. (A) Effects of JAM20 on ERK activation mediated by oncogenic RAS mutants and downstream kinases. CFP, CFP-Mbs (NS1 or JAM20), and MYC-tagged ERK were coexpressed in HEK cells, and phosphorylation of MYC-tagged ERK was determined following IP and Western blot with phosphor-specific ERK antibodies. For each oncoprotein, the pERK level normalized to the total ERK level was determined. The resulting value in the presence of the indicated Mb was divided by the value for CFP alone. The dotted line at 1 represents the normalized pERK level without inhibition by an Mb. (B) Effects of JAM20 on foci formation of NIH/3T3 cells transfected with the indicated RAS mutants or downstream oncogenic kinases along with CFP or CFP-Mb. Relative foci numbers, the ratio of foci number in the presence of CFP-Mb to CFP alone, are shown in B, Right. The mean and SD (n = 3) are shown. P values were established via a Student’s t test between CFP and CFP-Mb for each oncogene. *P < 0.05; **P < 0.01; ***P < 0.001.
We also evaluated JAM20’s impact on RAS-driven cell transformation using the NIH/3T3 focus formation assay (Fig. 2B) (35). Both JAM20 and NS1 significantly reduced foci formation for oncogenic KRAS and HRAS (P < 0.001). JAM20 also significantly reduced foci formation for oncogenic NRAS (P < 0.001). Furthermore, JAM20 and NS1 did not inhibit foci formation driven by either oncogenic MEK(DD) or BRAF(V600E). Thus, JAM20 specifically disrupted RAS-mediated biological transformation in vitro.
JAM20 Inhibits RAS-Mediated Signaling and Tumorigenesis In Vivo.
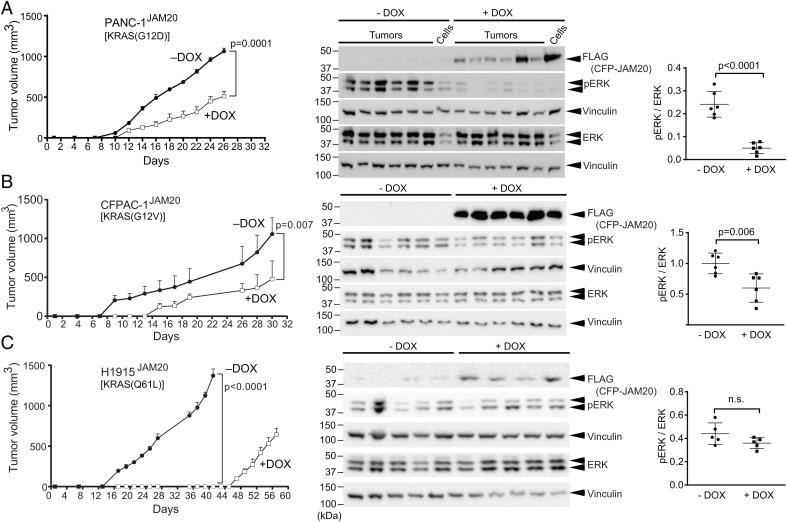
Because JAM20 effectively inhibited RAS-mediated signaling and transformation in cells in vitro, we next examined whether JAM20 also inhibited endogenous RAS-mediated signaling and tumorigenesis in vivo using RAS-mutant human tumor xenograft models (SI Appendix, Table S2 and Figs. S2 and S3). Doxycycline (DOX)-induced expression of JAM20 reduced ERK activation in human tumor cells driven by mutant KRAS, HRAS, or NRAS and reduced the anchorage-independent growth of these cell lines (SI Appendix, Figs. S2 and S3). Although HEC-1A and H1944 cell lines did not show a reduction of pERK level upon JAM20 induction in two-dimensional (2D) culture, their colony-formation activity was inhibited by JAM20 expression (SI Appendix, Fig. S3), consistent with published studies (36–38). JAM20 did not impair the MAPK signaling in A375 melanoma cells harboring the BRAF(V600E) mutant, as expected (SI Appendix, Fig. S4). These results demonstrate JAM20’s selectivity to RAS in cancer cell lines, consistent with its effects on model cellular systems, such as HEK293 and NIH/3T3 cells (Fig. 2). Furthermore, DOX-induced JAM20 expression reduced the tumor growth of PANC-1JAM20, CFPAC-1JAM20, and H1915JAM20 in vivo compared with uninduced (−DOX) cells (Fig. 3). Western blot analysis of tumor lysates confirmed JAM20 expression upon DOX addition (Fig. 3). JAM20 expression (+DOX) reduced the levels of pERK compared with −DOX controls for both PANC-1JAM20 and CFPAC-1JAM20 (Fig. 3 A and B). In contrast to the effects on RAS-MAPK signaling, the effects of JAM20 on AKT varied between 2D tissue culture conditions vs. three-dimensional tumor growth conditions. Expression of JAM20 increased pAKT in 2D tissue culture conditions, but significantly decreased the pAKT in tumor lysates (SI Appendix, Figs. S5 and S6). We have seen similar effects on RAS-mediated PI3K–AKT signaling with NS1 (37). Overall, these results demonstrated that JAM20 effectively inhibited endogenous oncogenic RAS-mediated signaling and tumorigenesis in vivo.
Fig. 3.
JAM20 inhibits signaling and tumorigenesis mediated by endogenous, oncogenic RAS mutants in vivo. Athymic nude mice were injected s.c. in the flanks with either PANC-1JAM20 (A), CFPAC-1JAM20 (B), or H1915JAM20 (C) cells. Mice were divided into two cohorts treated without (−) or with (+) DOX and monitored for tumor development. (Left) Average tumor volume at the indicated time point and analysis of average tumor volume at the end point of the experiment. (Center) Tumors harvested at end points (day 26 for PANC-1JAM20, day 30 for CFPAC-1JAM20, day 40 for H1915JAM20 −DOX, and day 57 H1915JAM20 +DOX) were used to make lysates, which were probed for pERK. Vinculin was used as a loading control. The two lanes at the end of −DOX and +DOX cohorts for PANC-1JAM20 represent lysates from parental PANC1JAM20 cells grown in culture and treated ± DOX. (Right) pERK/ERK ratio for each set of tumor lysates. P values were calculated via an unpaired Student’s t test. n.s., not significant.
Assessment of JAM20’s Mechanism of Inhibition.
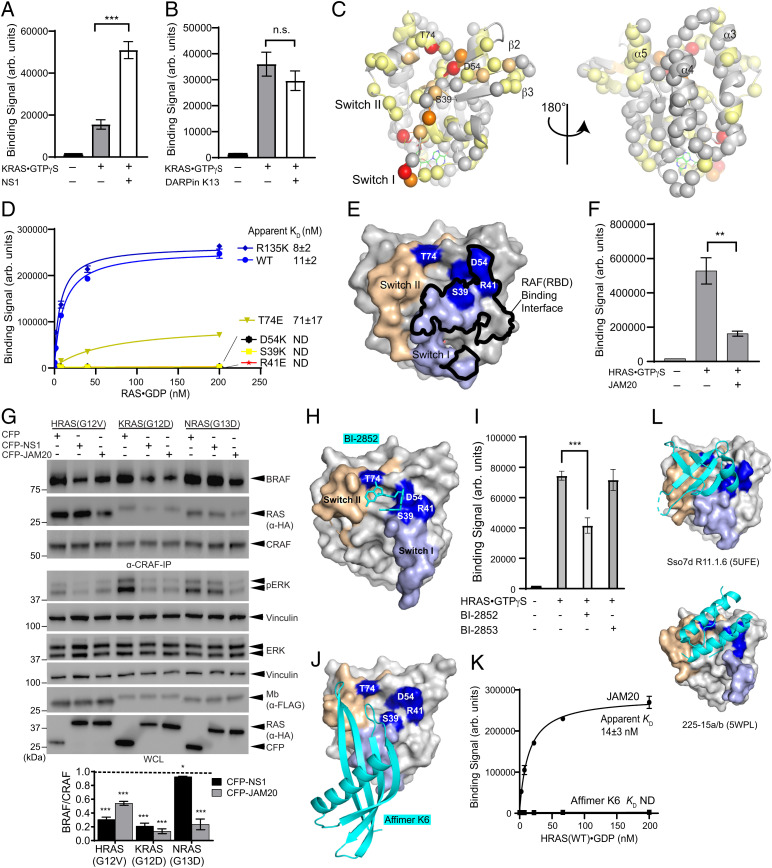
After determining that JAM20 inhibited RAS both in vitro and in vivo, we performed a series of experiments to define its mechanism of action. First, we tested whether JAM20 indeed binds to a region outside the epitopes of other RAS-targeting synthetic binding proteins. Competition by NS1 or DARPin K13 was evaluated by using JAM20 displayed on the surface of yeast. Neither NS1 nor DARPin K13 inhibited the binding of JAM20 to KRAS⋅GTPγS (Fig. 4 A and B and SI Appendix, Fig. S7 A and B). Interestingly, we observed a significant increase in signal intensity upon binding to KRAS⋅GTPγS complexed with NS1 (Fig. 4A). We speculate that JAM20 may interact with NS1 and/or NS1 may allosterically enhance JAM20 binding to RAS. As a control, we also tested competition between NS1 and DARPin K13 (SI Appendix, Fig. S7 A and B). As expected, yeast-displayed NS1 exhibited no binding to KRAS⋅GTPγS complexed with purified NS1. Furthermore, NS1 did not bind to RAS when complexed with DARPin K13, indicating that NS1 competed with the DARPin. The crystal structures of DARPin K13 and NS1 bound to RAS revealed that binding of these two molecules to RAS is mutually exclusive due to a steric clash, which we confirmed experimentally (SI Appendix, Fig. S7C) (27, 29). These results demonstrated that JAM20 did not interfere with either NS1 or DARPin K13 binding to RAS and confirmed that JAM20 bound to a region distinct from either the NS1 or DARPin K13 epitopes.
Fig. 4.
Characterizing JAM20’s mechanism of inhibition. (A and B) Binding of JAM20 displayed on yeast to KRAS⋅GTPγS (70 nM) in the presence and absence of 1 μM NS1 or 1 μM DARPin K13. ***P = 0.0002. P values were determined by using a two-tailed unpaired t test. The mean and SD (n = 3) are plotted. Additional control experiments are shown in SI Appendix, Fig. S7. (C) Residues of HRAS⋅GDP exhibiting CSP in the HSQC spectrum upon JAM20(Y83S) binding mapped on the HRAS crystal structure (Protein Data Bank [PDB] ID code: 5E95). Spheres represent the backbone amides included in the analysis. Residues with a CSP > 0.3 are shown in red, those with CSP between 0.25 and 0.3 are in orange, those with CSP between 0.15 and 0.24 are in light orange, those with CSP between 0.045 and 0.14 are in pale yellow, and those with CSP < 0.045 are in gray. (D) Binding measurements of JAM20 in yeast display against HRAS mutants bound to GDP. The mean and SD (n = 3) of technical replicates are plotted. ND, not detectable. (E) Mutations affecting JAM20 binding mapped on the crystal structure of HRAS-WT bound to GppNHp (PDB ID code: 4G0N). SI and SII are highlighted in light blue and wheat, respectively. The surfaces for residues whose mutations disrupted JAM20 binding are highlighted in blue and labeled. The atoms within 4 Å of RAF(RBD) are outlined in black. (F) Bead-based binding measurements biotinylated RAF(RBD) immobilized on Dynabeads M280 streptavidin to HRAS⋅GTPγS (500 nM) in the presence and absence of JAM20 (1 μM). P values were determined by using a two-tailed unpaired t test. **P = 0.0012. The mean and SD (n = 3) are plotted. (G) Effect of JAM20 on heterodimerization of CRAF with RAS and BRAF. HEK cells were cotransfected with the indicated expression constructs. BRAF and RAS coimmunoprecipated with CRAF were quantified. The ratio of BRAF to CRAF was determined for each condition. The resulting value with an oncoprotein was divided by the value for CFP alone for each oncoprotein. Dotted line at 1 represents the normalized BRAF level without inhibition by an Mb. (H) The crystal structure of KRAS–G12D(C118S)–GDP in complex with BI-2852 (PDB ID code: 6ZL5). SI and SII are highlighted in light blue and wheat, respectively. The surfaces for residues disrupting JAM20 binding are highlighted blue and labeled. BI-2852 is colored cyan. (I) Binding of JAM20 displayed on yeast to HRAS⋅GTPγS (25 nM) in the presence of either 14.6 μM BI-2852 or BI-2853. Binding to RAS was evaluated in the presence of 0.75% DMSO. ***P = 0.0007. The mean and SD (n = 3) of three technical replicates are plotted. P values were determined by using a two-tailed unpaired t test. (J) The crystal structure of Affimer K6 bound to KRAS⋅GDP (PDB ID code: 6YR8). KRAS is presented in the same manner as in H and K6 as cartoon. (K) Binding measurements evaluated by using flow cytometry of both JAM20 and affimer K6 in yeast display against WT HRAS bound to GDP. The mean and SD values from three technical replicates are plotted on the graph and for KD values. (L) Crystal structures of two additional RAS-targeting proteins binding to the SI/SII pocket, presented in the same manner as in J, without residue labels for simplicity. Arb., arbitrary; n.s., not significant.
To gain structural insights into the interaction between JAM20 and RAS, we employed solution NMR spectroscopy using isotopically enriched HRAS and performed chemical-shift perturbation (CSP) analysis upon addition of Mb. We developed a JAM20 mutant with modestly improved solubility, JAM20(Y83S), that displayed binding characteristics to HRAS(WT)⋅GDP that were similar to the original JAM20 (Fig. 1A and SI Appendix, Fig. S8). 2D 1H/15N-heteronuclear single quantum coherence (1H/15N-HSQC) spectra were collected on [U-15N]-HRAS(WT)⋅GDP in the presence and absence of unlabeled JAM20(Y83S). Of the 122 assigned cross-peaks, we observed multiple CSPs for residues mostly near the Switch region and the β2–β3 strands of HRAS (Fig. 4C and SI Appendix, Fig. S9). By contrast, few perturbations were observed in the α3, α4, or α5 helices of RAS. These results suggest that JAM20 bound to a region within and/or near the Switch region of RAS.
To validate the epitope inferred from the NMR experiment, we tested the effects of mutating residues within the epitope inferred from the CSP data (Fig. 4D). For this purpose, we mutated select residues to either Arg, Glu, or Lys—i.e., amino acids with a bulky and charged side chain. Although Ala substitutions are more commonly used in scanning mutagenesis studies, we wanted to identify a mutation that disrupted the JAM20–RAS interaction instead of assessing the contribution of side chains to the binding energetics (39). These mutant HRAS proteins eluted from a size-exclusion chromatography column at the same volume as HRAS(WT) and bound equally well to NS1 displayed on the yeast surface, strongly suggesting that they are properly folded and monomeric (SI Appendix, Fig. S10 A and B). Of the mutants tested, mutants S39K, R41E, and D54K resulted in a complete loss of JAM20 binding, and T74E substantially reduced binding (Fig. 4C). R135K, a mutation that disrupts the HRAS–NS1 interaction and is located outside the predicted epitope for JAM20 (Fig. 4C) (27) had no effect on JAM20 binding, but eliminated binding of NS1, as expected (Fig. 4D and SI Appendix, Fig. S10B). These results further support JAM20’s epitope being in the SI/SII region, consistent with observations from solution NMR spectroscopy.
To understand how JAM20 disrupted RAS-mediated signaling, we also tested how the aforementioned HRAS mutants impacted the binding of the RAF(RBD) to RAS. Although RAF(RBD) maintained binding to the controls (WT, R135K, and T74E), its binding was severely reduced with the S39K, R41E, and D54K mutants (SI Appendix, Fig. S10C). In the crystal structure of the RAS:RAF(RBD) complex, S39 and R41 are located within 4 Å of the RAF(RBD) epitope, whereas D54 is more distal (Fig. 4E) (40). Despite its location, mutating D54 also resulted in reduced RAF(RBD) binding to RAS. Because the Switch region of RAS has been noted for its flexibility, the observed changes in RAF(RBD) binding to D54K may be a result of a local conformational change in the SI/SII region of RAS. Because both JAM20 and RAF(RBD) binding was disrupted by the same set of mutations and we expected that JAM20 binds to the same epitope in the GDP- and GTP-bound RAS, we also tested whether JAM20 competed with the RAF(RBD)–RAS interaction (Fig. 4F). JAM20 indeed inhibited the binding of RAF(RBD) to HRAS.
Furthermore, we evaluated whether JAM20 also disrupted RAF heterodimerization in cells. CFP-tagged Mbs were coexpressed with HA-tagged oncogenic RAS in HEK293 cells, and the amount of BRAF coimmunoprecipitated with CRAF was evaluated (Fig. 4G). Both CFP–JAM20 and NS1 disrupted RAF heterodimerization induced by oncogenic KRAS and HRAS. CFP–JAM20 also disrupted RAF heterodimerization for NRAS(G13D), whereas CFP–NS1 did not. These results are consistent with the inhibition of the interaction between an oncogenic RAS mutant and RAF by JAM20, the prerequisite for RAF dimerization.
JAM20 Bound to a Binding Hot Spot within the Switch Region of RAS.
Evaluating other reported molecules revealed that JAM20 may share an overlapping epitope with the small-molecule inhibitor BI-2852, a pan-RAS inhibitor that binds to a shallow pocket within the Switch region of RAS termed the SI/SII pocket (30). Further, BI-2852 interacts with several RAS residues, including S39, D54, and T74, which are three of the mutants that disrupted the JAM20–RAS interaction (Fig. 4 D and H) (30). Like JAM20, BI-2852 also binds to RAS in both nucleotide states, albeit with much weaker affinity with a KD value of 750 nM to KRAS(G12D) (30). Because of this close resemblance, we evaluated whether JAM20 competed with BI-2852 (Fig. 4I). The addition of BI-2852 resulted in a significant (P = 0.0007) reduction of binding of JAM20 to RAS. In contrast, a negative control compound, BI-2853, had no effect. Thus, these results strongly suggest that JAM20 bound to an epitope that overlapped with BI-2852.
A synthetic binding protein, termed affimer K6, was recently reported that appears to bind to a similar region as JAM20 and BI-2852 (Fig. 4J) (31). Affimer K6 showed no isoform specificity, and it was reported to have 0.6 µM half-maximal inhibitory concentration (IC50) for SOS1-mediated nucleotide exchange, consistent with its small binding interface observed in the crystal structure. Nevertheless, it was also reported to have a single-nanomolar KD value estimated from surface plasmon resonance, similar to JAM20. To examine whether affimer K6 has similar affinity toward HRAS as JAM20, we directly compared the two proteins for HRAS binding in the yeast display format (Fig. 4K). When binding was tested with monomeric HRAS, we only observed marginal binding of affimer K6 in the concentration range, in which JAM20 showed a robust binding signal. However, we were able to detect weak binding of affimer K6 when HRAS was tetramerized by first complexing with streptavidin, which dramatically enhances effective affinity due to multivalent effects (SI Appendix, Fig. S11). These results suggest that the affinity of affimer K6 is low, consistent with its reported IC50 value of 0.6 µM. More extensive survey identified two additional synthetic molecules that bind to similar regions of RAS (Fig. 4L) (41, 42), strongly suggesting that this region is a binding hot spot within RAS. Of note, none of these RAS ligands, except for JAM20, have been tested for their efficacy in inhibiting RAS-mediated tumorigenesis.
Finally, in light of a recent publication reporting that binding of a fluorescent probe based on BI-2852 to KRAS(G12C) was inhibited by KRAS(G12C)-selective, covalent inhibitors binding to the SII pocket (43), we examined whether the JAM20–RAS interaction was inhibited by such an inhibitor. Interestingly, the treatment of KRAS(G12C)⋅GDP with AMG-510 (sotorasib) did not inhibit its binding to JAM20 and, rather, slightly increased the binding signal (SI Appendix, Fig. S12). We interpret this different effect of AMG-510 to be indicative of different modes of binding to RAS between JAM-20 and BI-2852. Notably, binding of the BI-2852–derived probe requires the formation of an RAS dimer, whereas JAM20 does not. The lack of inhibition by AMG-510 suggests the possibility of making molecules that occupy both the SII pocket and the SI/II pocket.
Discussion
The Mb reported here, JAM20, joins a growing family of intracellular biologics targeting RAS that can be used as genetically encoded reagents (SI Appendix, Table S3). This group of reagents exhibit at least 10 distinct profiles in function and epitope, providing powerful tools for interrogating aspects of RAS biology and testing effectiveness of targeting different isotypes, different mutants, and different surfaces of RAS in controlling RAS-mediated signaling and tumorigenesis.
JAM20 robustly inhibited RAS-driven tumor growth in vivo for all evaluated cell lines and reduced ERK activation in dissected tumors for both PANC-1JAM20 and CFPACJAM20 (Fig. 3 A and B). In contrast, it did not reduce ERK activation in H1915JAM20 tumors, despite reducing pERK levels in the same stable cell line in vitro (Fig. 3C and SI Appendix, Fig. S2C). This apparent discrepancy seems to be due to the low expression level of JAM20 in the remaining tumor cells following DOX-induced expression that emerged after a long lag phase following the initiation of DOX treatment (Fig. 3C). The low expression level of JAM20 may be caused by a loss of cells expressing JAM20 at high levels or suppression of JAM20 expression during tumor development. These results illustrate potential challenges in controlling the dosing of genetically encoded reagents over a long-term experiment. Advances in alternative methods of delivery, such as the delivery of messenger RNA using lipid nanoparticles (44, 45), may overcome these challenges.
We have successfully developed a reagent that validates a pan-RAS, pan-nucleotide state targeting approach in a mouse xenograft model. Pan-RAS Mb, JAM20, selectively bound to a region on RAS that was different from either the NS1 or DARPin K13 epitope (Fig. 1). Whereas NS1 and DARPin K13 are RAS isoform-selective, JAM20 bound to all RAS isoforms, demonstrating that it is distinct from either of these synthetic proteins. Additionally, JAM20 has distinct binding characteristics from natural RAS effectors, such as RAF(RBD), as JAM20 showed limited nucleotide state sensitivity, despite its epitope’s location within the SI/SII region (8). We were able to identify JAM20 without the use of direct competition with NS1 or DARPin K13. Our results indicate that a carefully designed library-sorting strategy can enrich for distinct Mbs without using competitors.
Directly inhibiting RAS remains a substantial challenge because it lacks obvious deep pockets for small-molecule inhibition. Indeed, BI-2852, which binds to the SI/SII region with low affinity, was reported in 2018 after decades of intense effort by the drug-discovery community. However, it has not been reported whether BI-2852 inhibits RAS-driven tumorigenesis in an animal model. Similarly, no efficacy data on tumorigenesis are available for other synthetic binding proteins targeting similar regions of RAS. In this work, we show that JAM20 bound to the SI/SII region and inhibited tumorigenesis driven by KRAS(G12D), KRAS(G12V), and HRAS(Q61L). These results provide strong support for the approach of inhibiting multiple oncogenic RAS mutants by targeting the SI/SII region, which, in turn, suggests potential antitumor efficacy of derivatives of BI-2852 with enhanced affinity. Future studies should also explore whether mutant-selective inhibitors can be developed that target this region.
Unlike small molecules, synthetic binding proteins do not depend on binding to a deep pocket to achieve high affinity. Thus, synthetic binding proteins are advantageous for rapidly identifying sites for inhibition on difficult targets such as RAS. This work is the second example of Mbs targeting essentially the identical region as small-molecule inhibitors. We previously developed a Mb targeting a peptide-binding site of the WDR5 subunit of the epigenetic regulatory complex, MLL (46). In both studies, we developed “drug-mimicking” Mbs without deliberately targeting a particular region of the target. As we reviewed previously (47), Mbs have strong propensity to bind to a functional site within a protein, presumably because such a site is predisposed to ligand interaction and offers an “easy” surface for a Mb to bind. Together, these studies demonstrate that synthetic proteins can be developed to target small-molecule binding sites and vice versa. We envision the utility of Mbs and other synthetic binding proteins as tool biologics for identifying and validating a potential binding site for small molecules and also for establishing drug-screening assays (25).
Materials and Methods
Protein Expression and Purification.
Proteins used for both Mb development and binding assays, including RAS constructs residues 1 to 174 [NRAS(WT), HRAS(WT), and KRAS4B either WT or mutants T74E, R41E, S39K, D54K, or R135K], Mbs, DARPins, and RAF-1 RBD (residues 51 to 131), were produced in the pHBT vector and had an N-terminal tag with 6×His, an Avi-tag, and a TEV protease recognition site (48). RAS and Mb mutants were made by using Kunkel Mutagenesis (49). Mb genes were cloned by using sticky-end PCR (50) into the pHBT vector. For RAS, Mb, and DARPin constructs, the DNA sequences were confirmed by using a T7 primer that anneals to the T7 promoter. All proteins were produced in BL21(DE3) Escherichia coli cells.
For the production of biotinylated proteins, BL21(DE3) E. coli cells containing a pBirA plasmid were transformed with the pHBT vector for the protein of interest. The cells was grown in medium that contained a final concentration of 50 μM biotin. The expressed proteins were purified via affinity chromatography using Ni–Sepharose columns (GE Healthcare), and dialyzed in Tris-buffered saline (TBS; 50 mM Tris⋅Cl, pH 7.5, with 150 mM NaCl). For RAS proteins, the dialysis buffer also contained 20 mM MgCl2 and 0.5 mM dithiothreitol (DTT). After dialysis, the samples were further purified by using size-exclusion chromatography using a Superdex 75 column on an AKTA Pure instrument (GE Healthcare). Nonbiotinylated proteins were produced by using the same workflow, but were expressed in medium lacking biotin.
For the production of 15N-labeled HRAS, the HRAS(WT) gene (residues 1 to 166) was cloned into the pHBT vector. BL21(DE3) E. coli cells harboring this vector were grown in minimum medium containing 15N-NH4Cl. The protein was purified as described above.
Both JAM20(WT) and JAM20(Y83S) were expressed in BL21(DE3) E. coli cells. The E. coli cells were lysed and centrifuged to separate the soluble and insoluble fractions. JAM20(WT) and JAM20(Y83S) were predominately found in the insoluble fraction. The insoluble fraction was then solubilized in a buffer composed of 20 mM Tris⋅Cl (pH 8) and 6 M GuHCl. After solubilization, the fraction was filtered through a 0.8-μm filter and applied to the Ni–Sepharose column (GE Healthcare). For refolding on the column, buffers composed of 20 mM Tris⋅Cl (pH 8), 500 mM NaCl (termed nickel column buffer), and 0.1% Triton X-100 or 5 mM beta-cyclodextrin were sequentially applied to the column. Last, the column was washed with the nickel column buffer containing 20 mM imidazole. Purified proteins were then eluted from the column by using nickel column buffer containing 500 mM imidazole. The purified protein was cleaved by using TEV protease and dialyzed in TBS with 20 mM MgCl2 and 0.5 mM DTT. The cleavage step was important for freezing and storing these proteins because the uncleaved proteins precipitated during freeze–thaw. The samples were applied to the Ni–Sepharose column, the column was washed with 20 mM Imidazole, and the flow-through fractions were collected and combined for dialysis in TBS with 20 mM MgCl2 and 0.5 mM DTT.
Nucleotide Exchange of RAS.
Purified RAS proteins were diluted down to a final concentration of ∼2 μM using a buffer composed of 20 mM Tris⋅Cl (pH 7.5), 5 mM ethylenediaminetetraacetic acid, 0.1 mM DTT, and either 1 mM GDP or 1 mM GTPγS. Typically, the dilution factor was ∼25-fold. After incubating the diluted RAS protein at 30 °C for 30 min, MgCl2 was added to a final concentration of 20 mM, and the solution was incubated on ice for 5 min before use.
Monobody Development.
Detailed procedures for the development of Mbs have been described elsewhere (22, 48, 51, 52). Briefly, four rounds of phage display library sorting were performed by using the loop library (51, 53). In the first round, 0.2 nmol of KRAS⋅GTPγS and NRAS⋅GTPγS was first immobilized on streptavidin magnetic beads and then mixed with the phage display library in the total volume of 1 mL. In the subsequent rounds, enriched libraries were mixed with RAS proteins in solution, and then the RAS proteins with bound phages were captured with streptavidin magnetic beads. We used 100 nM KRAS⋅GDP in round 2, 50 nM NRAS⋅GTPγS in round 3, and 50 nM KRAS⋅GDP in round 4. Following phage display selection, the monobody genes from the enriched pool were transferred to a yeast display vector. A Bio-Rad S3e fluorescence-activated cell sorter was used for library sorting. The yeast display library was sorted via the following strategy: sort 1 enriched for Mbs with 400 nM NRAS⋅GTPγS, sort 2 with 75 nM KRAS(WT)⋅GTPγS, and sort 3 with 50 nM NRAS(WT)⋅GDP. Individual Mbs were then screened and selected based on whether they bound to all RAS isoforms in both nucleotide states.
Yeast Display Binding Assay.
Yeast cells were incubated with a biotinylated target and mouse anti-V5 (ThermoFisher, catalog no. MA5-15253; 1:75 for sorting ∼107 yeast cells and 1:300 for staining 105 yeast cells for analysis). Next, the cells were labeled with anti-mouse IgG conjugated to fluorescein isothiocyanate (Millipore Sigma, catalog no. F0257; 1:100; for detecting surface expression) and Neutravidin DyLight 650 was used to detect the binding of the biotinylated target. For streptavidin tetramerization, RAS was mixed in a 4:1 ratio with Streptavidin DyLight 650 and incubated on ice for 30 min before adding to yeast. For all titration experiments, both binding and labeling steps were performed at 4 °C, and all components were on ice prior to use. Samples were processed on an iQue flow cytometer (Sartorius). For data analysis, the median of the 75th- to 95th-percentile population in the fluorescence signal intensity from the red channel was used.
For testing effects of AMG-510, 32 nM KRAS(G12C)⋅GDP in TBS with 20 mM MgCl2 and 0.1% bovine serum albumin was reacted with 10.8 µM AMG510 at room temperature for 3 h. The residual DTT concentration in the reaction was no more than 0.8 µM. The AMG-510–reacted KRAS(G12C)⋅GDP was diluted to a final concentration of 8 nM for binding measurements with JAM20 displayed on yeast cells.
Bead Binding Assay.
A biotinylated protein, such as RAF-1 RBD, was bound to Dynabeads M-280 streptavidin (ThermoFisher) and diluted in a buffer containing biotin to block unoccupied biotin-binding sites. Next, the sample beads were incubated with a biotinylated protein of interest, such as KRAS(WT). Streptavidin DyLight650 was then used to detect the biotinylated protein of interest. To analyze binding, samples were processed by using an iQue flow cytometer (Sartorius). For data analysis, the median of the 75th- to 95th-percentile population in the fluorescence signal intensity from the red channel was used.
Solution NMR Spectroscopy.
For solution NMR experiments, proteins were produced as described above. The 15N-labeled HRAS(WT)⋅GDP and unlabeled JAM20(Y83S) were added in a 1:1 ratio with a final concentration of 107 μM. For 15N-labeled HRAS(WT)⋅GDP only, 107 μM protein was used. Both spectra were collected with samples in a buffer made up of TBS (pH 7.5), 20 mM MgCl2, 1 mM DTT, and 5% D2O. All NMR spectra were collected at 298 K on a Bruker 600 MHz (14.1 T) spectrometer, which was equipped with a triple-resonance TCI cryogenic probe. The 2D 1H/15N-HSQC experiments were recorded with a spectral width of 9,615.4 Hz (1H) and 2,189.7 Hz (15N). The acquisition (1H) and evolution times (15N) were set to 62.3 ms and 22.8 ms, respectively. The 2D 1H/15N-HSQC spectra were processed in NMRPipe (54) and analyzed by using NMRFAM-SPARKY (55). Assignments were based on a deposited assignment of HRAS(WT)⋅GDP (Biological Magnetic Resonance Bank [BMRB] ID: 18479) (56). CSPs were quantified with the equation below:
and are the 15N and 1H chemical-shift changes, respectively, between HRAS in the absence and presence of JAM20.
Transient Expression of RAS and Mb for Confocal Imaging.
One million HEK293T cells were cultured on a 6-cm glass-bottomed dish (MatTek Corp., catalog no. P35G-1.5-14-C). After 24 h, cells that were between 70% and 90% confluent were prepared for transfection by replacing the medium with antibiotic-free complete medium (Dulbecco’s modified Eagle medium [DMEM] supplemented with 10% fetal bovine serum [FBS]). The cells were then transfected based on the manufacturer’s recommended protocol using Lipofectamine 3000 (Thermo Fisher Scientific). pEGFP vectors containing either RAS(WT) with an N terminus eGFP tag and either JAM20 or Mb (neg) with an N terminus mCherry tag were cotransfected in a 2:1 ratio. After 24 h, the transfected cells were imaged by using a LSM710 confocal microscope (Ziess), and data were analyzed via FIJI for ImageJ (57).
Cell Culture and Transfections.
The effects of ectopic expression of JAM20 were determined by using HEK-293, HEK-293T, or NIH/3T3 cells. HEK293 and HEK293-T cells were cultured in DMEM with 10% FBS. NIH/3T3 cells were cultured in DMEM with 10% calf serum. RAS or RAF mutant cell lines were grown in the medium as per ATCC recommendations using 10% tetracycline-negative FBS. Polyethyleneimine (PEI) was used as a transfection reagent for all transient expressions. For HEK-293 and HEK-293T, 3 µL of 1 mg/mL PEI stock was used for each microgram of DNA in Opti-MEM reduced serum medium (Life Technologies). Briefly, Opti-MEM–PEI mixture was incubated for 10 min at room temperature. Next, DNA was added to the Opti-MEM:PEI mix and incubated for 20 min at room temperature. The Opti-MEM:PEI–DNA mixture was added to cells in serum-free medium and incubated not more than 3 h. The medium for transient transfections in HEK cells was replaced with complete medium after 3 h. Transfections in NIH/3T3 cells were done with same procedures, using 5 µL of 1 mg/mL PEI stock for each microgram of DNA, and medium was replenished after 5 h.
Cell-Signaling Assay.
Forty-eight hours after transient transfections or DOX administration, cells were serum-starved overnight (O/N). The following day, cell lysates were made by washing cells once in cold phosphate-buffered saline followed by lysis using PLC buffer [50 mM Hepes, pH 7.5, 150 mM NaCl, 10% glycerol, 1% Triton X-100, 1 mM ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid, 1.5 mM magnesium chloride, 100 mM sodium fluoride supplemented with 1 mM vanadate, 10 μg/mL leupeptin, and 10 μg/mL aprotinin). To generate tumor lysates, tumors were harvested, transferred to microfuge tubes, and snap-frozen by immersing in liquid nitrogen. Then, 40 to 50 mg of tissue was homogenized in ∼1 mL of ice-cold PLC buffer. Homogenate was passed through a 70-µm cell strainer and then centrifuged at 13,000 rpm for 20 min at 4 °C. Supernatant was collected and transferred to fresh tubes. The lysates were directly used for protein estimation and analysis or stored at −80 °C for later use. The following antibodies were used: monoclonal HA (clone 16B12; Biolegend catalog no. 90154), polyclonal rabbit HA (Poly9023; Biolegend catalog no. 923502), monoclonal FLAG (Clone M2; Sigma catalog no. F1804), polyclonal rabbit FLAG (Sigma catalog no. F7425), phospho-ERK (Thr202/Tyr204; CST catalog no. 9101), total ERK (CST catalog no. 9102), phospho-AKT (Ser473; CST catalog no. 9271), total AKT (CST catalog no. 9272S), phospho-AKT (T308; CST catalog no. 9275S), Vinculin (SC catalog no. 73614), anti-MYC (Clone A46; Millipore-Sigma catalog no. 05-724), CRAF (BD Biosciences catalog no. 610151), and BRAF (Santa Cruz catalog no. sc-9002).
For the binding assays using Co-IP, FLAG-tagged CFP–JAM20 was cotransfected with the indicated GTPase mutant (RAS and RRAS2) into HEK293 cells. FLAG-tagged CFP alone and FLAG-tagged CFP–NS1 were used as negative and positive controls, respectively. At 48 h posttransfection, cells were replenished with fresh serum containing medium or serum-starved O/N, depending on the experimental purpose. Lysates were collected and run for determining the expression of RAS or RRAS2 and Mb. Following this, lysates were immunoprecipitated by using FLAG antibody, and immunoprecipitated samples were analyzed by Western blot for coprecipitation of the HA-tagged RAS or RRAS2 (TC21).
For transient MAPK cell-signaling assays, HEK293 cells were transfected with CFP–JAM20 or the indicated HA-tagged RAS mutant or non-RAS oncogenic kinases, MEKDD or BRAF(V600E), and MYC-tagged ERK. Forty-eight hours after transfection, cells were serum-starved O/N in DMEM alone and stimulated with EGF (100 ng/mL for 10 min) for WT endogenous RAS activation. Oncogene-transfected cells were not stimulated with growth factor. Cells were then lysed in PLC buffer and immunoprecipitated by using MYC antibody. The immunoprecipitated fractions were used for Western blots and analyzed for effects on MAPK signaling. Western blots were quantified with the software Image Studio Lite (version [v].5.2.5, LI-COR Biosciences) using the Analysis function. The ratio of active (pERK) divided by total ERK was determined for each condition. The resulting values were divided by value for CFP alone for each protein. The dotted line represents the level of ERK–MAPK activation by each RAS mutant in the presence of CFP and was arbitrarily set to one.
The effects of JAM20 on RAS interaction with CRAF and BRAF were evaluated by Co-IP. HEK293 cells were transfected with the indicated constructs. After immunoprecipitation (IP) of endogenous CRAF, samples were analyzed by Western blotting for coprecipitation of BRAF and HA-tagged RAS. The ratio for BRAF to CRAF was determined for each condition. The resulting value was divided by the value for CFP alone for each oncoprotein. The dotted line represents the level of BRAF/CRAF activation by each RAS mutant in the presence of CFP and was arbitrarily set to one. For determining the effects of JAM20 on RAS–RAF association, the ratio of IP fraction HA-RAS and CRAF was evaluated for each condition. Again, the values of each oncoprotein for NS1 and JAM20 were divided by CFP alone. HA-RAS/CRAF for each condition for CFP was arbitrarily set to one.
NIH/3T3 Focus-Formation Assay.
NIH/3T3 cells were freshly revived and passaged no more than two times. Cells were seeded in 60-mm2 dishes to a density of 2.5 × 105 cells in complete medium. Cells were cotransfected with the indicated RAS mutant or downstream oncogenic kinases, negative control CFP-alone vector, positive control CFP–NS1 vector, and test CFP–JAM20 vector using PEI. The medium was replenished every 2 d. Usually, the foci for oncogenic RAS mutant transformation begin to form approximately 10 d posttransfection, and the assay terminated by 2 wk, depending on the isoform/oncogenic mutant. However, the foci for downstream oncogenic kinases usually appear after 2 wk, and the assay was terminated after 3 wk. Foci were stained with 0.1% crystal violet and counted. All assays were performed in triplicate and repeated three times.
Stable Cell Generation of Inducible JAM20.
HEK-293T cells were used for the generation of the viral particles for DOX-inducible JAM20 expression. Briefly, the packaging HEK-293T cells were transfected with pCW57.1-CFP-JAM20 (transfer plasmid) along with a plasmid-encoding packaging plasmid (pCMVdR8.74) and the viral envelope (pMD2.G) in a 4:3:1 ratio using calcium phosphate. On the following day, cells were placed in fresh medium, and on day 2 posttransfection, conditioned medium from the HEK-293T cells were collected and filtered by using 0.45-µm syringe filters. The filtered supernatant was used to infect various RAS mutant cell lines, and cells were selected by using puromycin. The selected colonies were pooled to generate a polyclonal cell line that expresses JAM20 on DOX induction.
Soft Agar Colony-Formation Assay.
Soft agar colony-formation assays were performed essentially as described elsewhere (35). A base agar layer (0.5%) was allowed to solidify and topped with cell suspension in 0.33% soft agar. This was allowed to set at room temperature for 15 to 20 min. Cells were fed one to two times per week by careful dropwise addition of growth medium to the top layer. DOX was added to induce expression of Mb. Two to three weeks after platting, cells were stained by using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT); 100 μL of 2 mg/mL solution MTT was used for each well. NIH ImageJ was used for quantification of colonies.
Xenograft Tumor Assay.
The procedures described herein are in compliance with protocols approved by the Institutional Animal Care and Use Committee at the Medical University of South Carolina and the Ralph H. Johnson Veterans Affairs Medical Center. Five-week-old male or female athymic nude mice were purchased from Charles River Laboratories or Taconic Biosciences. The mice were acclimatized for 1 wk. About 100-µL suspension of 10 × 106 cells in a 1:1 solution (volume [vol]/vol) of serum-free RPM1/Matrigel basement membrane matrix were injected subcutaneously (s.c.) into the flanks of the nude mice. Two days postinjection, mice were randomly segregated to control or treatment cohorts. DOX was provided at a concentration of 2 mg/mL with water supplemented with sucrose. Tumor dimensions were noted thrice a week by using a digital caliper, and the tumor volume was estimated as V(mm3) = π/6(length × width2).
Supplementary Material
Acknowledgments
We thank Boehringer Ingelheim for generously providing BI-2852 and BI-2853 via its open-innovation platform (opnME). We acknowledge Takamitsu Hattori, Lorenzo Maso, and Padma Akkapeddi for their technical assistance and helpful discussion; Michael Cammer and Yan Deng of the Microscopy Laboratory at New York University (NYU) Grossman School of Medicine for assistance; and Mark Philips for the pEGFP-RAS(WT) vector. This work was supported by NIH Grants R01 AI108889 (to N.J.T.), R01 CA194864 (to S.K.), R01 CA212608 (to J.P.O. and S.K.), P30CA138313 (to J.P.O), and P20GM130457 (to J.P.O). J.P.O. was also supported in part by a grant from the Department of Veterans Affairs Biomedical Laboratory Research and Development Service MERIT Award 1I01BX002095. L.W. was supported in part by T32 GM 88118-10. K.W.T. was supported in part by NIH Fellowship F32 CA225131 and American Cancer Society Fellowship PF-18-180-01-TBE. The core facilities of the NYU School of Medicine were partially supported by Cancer Center Support Grant P30CA016087. The 600-MHz cryoprobe used for data collection at NYU was supported by NIH Grant S10 OD016343.
Footnotes
Competing interest statement: S.K. was an SAB member and received consulting fees from Black Diamond Therapeutics; is a co-founder and holds equity in Revalia Bio; and receives research funding from Black Diamond Therapeutics, Puretech Health, and Argenx BVBA. A.K. and S.K. are listed as inventors on issued and pending patents on the monobody technology filed by The University of Chicago (US Patent 9512199 B2 and related pending applications). The other authors declare no competing interests.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2204481119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and/or supporting information.
References
- 1.Prior I. A., Hood F. E., Hartley J. L., The frequency of Ras mutations in cancer. Cancer Res. 80, 2969–2974 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boriack-Sjodin P. A., Margarit S. M., Bar-Sagi D., Kuriyan J., The structural basis of the activation of Ras by Sos. Nature 394, 337–343 (1998). [DOI] [PubMed] [Google Scholar]
- 3.Hall B. E., Yang S. S., Boriack-Sjodin P. A., Kuriyan J., Bar-Sagi D., Structure-based mutagenesis reveals distinct functions for Ras switch 1 and switch 2 in Sos-catalyzed guanine nucleotide exchange. J. Biol. Chem. 276, 27629–27637 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Rudack T., Xia F., Schlitter J., Kötting C., Gerwert K., Ras and GTPase-activating protein (GAP) drive GTP into a precatalytic state as revealed by combining FTIR and biomolecular simulations. Proc. Natl. Acad. Sci. U.S.A. 109, 15295–15300 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scheffzek K., Lautwein A., Kabsch W., Ahmadian M. R., Wittinghofer A., Crystal structure of the GTPase-activating domain of human p120GAP and implications for the interaction with Ras. Nature 384, 591–596 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Tran T. H., et al. , KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation. Nat. Commun. 12, 1176 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terrell E. M., Morrison D. K., Ras-mediated activation of the Raf family kinases. Cold Spring Harb. Perspect. Med. 9, a033746 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wittinghofer A., Herrmann C., Ras-effector interactions, the problem of specificity. FEBS Lett. 369, 52–56 (1995). [DOI] [PubMed] [Google Scholar]
- 9.Pacold M. E., et al. , Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell 103, 931–943 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Amendola C. R., et al. , KRAS4A directly regulates hexokinase 1. Nature 576, 482–486 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simanshu D. K., Nissley D. V., McCormick F., RAS proteins and their regulators in human disease. Cell 170, 17–33 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheffzek K., et al. , The Ras-RasGAP complex: Structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science 277, 333–338 (1997). [DOI] [PubMed] [Google Scholar]
- 13.Ostrem J. M., Peters U., Sos M. L., Wells J. A., Shokat K. M., K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503, 548–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thanos C. D., DeLano W. L., Wells J. A., Hot-spot mimicry of a cytokine receptor by a small molecule. Proc. Natl. Acad. Sci. U.S.A. 103, 15422–15427 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X., et al. , Identification of MRTX1133, a noncovalent, potent, and selective KRASG12D inhibitor. J. Med. Chem. 65, 3123–3133 (2022). [DOI] [PubMed] [Google Scholar]
- 16.Lanman B. A., et al. , Discovery of a covalent inhibitor of KRASG12C (AMG 510) for the treatment of solid tumors. J. Med. Chem. 63, 52–65 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Hallin J., et al. , The KRASG12C inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov. 10, 54–71 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Bryan J. P., Pharmacological targeting of RAS: Recent success with direct inhibitors. Pharmacol. Res. 139, 503–511 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore A. R., Rosenberg S. C., McCormick F., Malek S., RAS-targeted therapies: Is the undruggable drugged? Nat. Rev. Drug Discov. 19, 533–552 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunter J. C., et al. , Biochemical and structural analysis of common cancer-associated KRAS mutations. Mol. Cancer Res. 13, 1325–1335 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Tanaka S., et al. , Monobody-mediated alteration of enzyme specificity. Nat. Chem. Biol. 11, 762–764 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wojcik J., et al. , A potent and highly specific FN3 monobody inhibitor of the Abl SH2 domain. Nat. Struct. Mol. Biol. 17, 519–527 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wojcik J., et al. , Allosteric inhibition of Bcr-Abl kinase by high affinity monobody inhibitors directed to the Src homology 2 (SH2)-kinase interface. J. Biol. Chem. 291, 8836–8847 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilbreth R. N., et al. , Isoform-specific monobody inhibitors of small ubiquitin-related modifiers engineered using structure-guided library design. Proc. Natl. Acad. Sci. U.S.A. 108, 7751–7756 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akkapeddi P., Teng K. W., Koide S., Monobodies as tool biologics for accelerating target validation and druggable site discovery. RSC Med. Chem. 12, 1839–1853 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teng K. W., et al. , Selective and noncovalent targeting of RAS mutants for inhibition and degradation. Nat. Commun. 12, 2656 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spencer-Smith R., et al. , Inhibition of RAS function through targeting an allosteric regulatory site. Nat. Chem. Biol. 13, 62–68 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khan I., et al. , Identification of the nucleotide-free state as a therapeutic vulnerability for inhibition of selected oncogenic RAS mutants. Cell Rep. 38, 110322 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bery N., et al. , KRAS-specific inhibition using a DARPin binding to a site in the allosteric lobe. Nat. Commun. 10, 2607 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kessler D., et al. , Drugging an undruggable pocket on KRAS. Proc. Natl. Acad. Sci. U.S.A. 116, 15823–15829 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haza K. Z., et al. , RAS-inhibiting biologics identify and probe druggable pockets including an SII-α3 allosteric site. Nat. Commun. 12, 4045 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boder E. T., Wittrup K. D., Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15, 553–557 (1997). [DOI] [PubMed] [Google Scholar]
- 33.Gai S. A., Wittrup K. D., Yeast surface display for protein engineering and characterization. Curr. Opin. Struct. Biol. 17, 467–473 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samatar A. A., Poulikakos P. I., Targeting RAS-ERK signalling in cancer: Promises and challenges. Nat. Rev. Drug Discov. 13, 928–942 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Clark G. J., Cox A. D., Graham S. M., Der C. J., Biological assays for Ras transformation. Methods Enzymol. 255, 395–412 (1995). [DOI] [PubMed] [Google Scholar]
- 36.Janes M. R., et al. , Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell 172, 578–589.e17 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Khan I., Spencer-Smith R., O’Bryan J. P., Targeting the α4-α5 dimerization interface of K-RAS inhibits tumor formation in vivo. Oncogene 38, 2984–2993 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vartanian S., et al. , Identification of mutant K-Ras-dependent phenotypes using a panel of isogenic cell lines. J. Biol. Chem. 288, 2403–2413 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cunningham B. C., Wells J. A., High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science 244, 1081–1085 (1989). [DOI] [PubMed] [Google Scholar]
- 40.Fetics S. K., et al. , Allosteric effects of the oncogenic RasQ61L mutant on Raf-RBD. Structure 23, 505–516 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kauke M. J., et al. , An engineered protein antagonist of K-Ras/B-Raf interaction. Sci. Rep. 7, 5831 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McGee J. H., et al. , Exceptionally high-affinity Ras binders that remodel its effector domain. J. Biol. Chem. 293, 3265–3280 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vasta J. D., et al. , KRAS is vulnerable to reversible switch-II pocket engagement in cells. Nat. Chem. Biol. 18, 596–604 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lim S., et al. , Exquisitely specific anti-KRAS biodegraders inform on the cellular prevalence of nucleotide-loaded states. ACS Cent. Sci. 7, 274–291 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rurik J. G., et al. , CAR T cells produced in vivo to treat cardiac injury. Science 375, 91–96 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gupta A., et al. , Facile target validation in an animal model with intracellularly expressed monobodies. Nat. Chem. Biol. 14, 895–900 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sha F., Salzman G., Gupta A., Koide S., Monobodies and other synthetic binding proteins for expanding protein science. Protein Sci. 26, 910–924 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sha F., et al. , Dissection of the BCR-ABL signaling network using highly specific monobody inhibitors to the SHP2 SH2 domains. Proc. Natl. Acad. Sci. U.S.A. 110, 14924–14929 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kunkel T. A., Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. U.S.A. 82, 488–492 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeng G., Sticky-end PCR: New method for subcloning. Biotechniques 25, 206–208 (1998). [DOI] [PubMed] [Google Scholar]
- 51.Koide A., Wojcik J., Gilbreth R. N., Hoey R. J., Koide S., Teaching an old scaffold new tricks: Monobodies constructed using alternative surfaces of the FN3 scaffold. J. Mol. Biol. 415, 393–405 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zorba A., et al. , Allosteric modulation of a human protein kinase with monobodies. Proc. Natl. Acad. Sci. U.S.A. 116, 13937–13942 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swers J. S., Kellogg B. A., Wittrup K. D., Shuffled antibody libraries created by in vivo homologous recombination and yeast surface display. Nucleic Acids Res. 32, e36 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Delaglio F., et al. , NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995). [DOI] [PubMed] [Google Scholar]
- 55.Lee W., Tonelli M., Markley J. L., NMRFAM-SPARKY: Enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Connor C., Kovrigin E. L., Assignments of backbone 1H, 13C and 15N resonances in H-Ras (1-166) complexed with GppNHp at physiological pH. Biomol. NMR Assign. 6, 91–93 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.