Significance
Most legumes benefit from nodulation, an ecologically and agriculturally important symbiosis with nitrogen-fixing bacteria. This endosymbiosis requires the formation of unique transcellular structures that mediate bacterial entry into root cells. Comparisons have been drawn between the subcellular structures that form during infection and those that precede cell division, but the evidence remains circumstantial. Here, we functionally link a deeply conserved mitotic module consisting of Aurora kinase, its regulators, and its targets to the symbiotic infection process. We propose that the recruitment of cell cycle machinery to nodulation is to promote the transcellular deposition of cell wall material, a feature common to infection and cell division. This progress provides a crucial entry point toward understanding the mechanisms that underlie endosymbiotic infection.
Keywords: cell cycle, Aurora kinase, rhizobial infection, microtubule, Medicago truncatula
Abstract
The subcellular events occurring in cells of legume plants as they form transcellular symbiotic-infection structures have been compared with those occurring in premitotic cells. Here, we demonstrate that Aurora kinase 1 (AUR1), a highly conserved mitotic regulator, is required for intracellular infection by rhizobia in Medicago truncatula. AUR1 interacts with microtubule-associated proteins of the TPXL and MAP65 families, which, respectively, activate and are phosphorylated by AUR1, and localizes with them within preinfection structures. MYB3R1, a rhizobia-induced mitotic transcription factor, directly regulates AUR1 through two closely spaced, mitosis-specific activator cis elements. Our data are consistent with a model in which the MYB3R1-AUR1 regulatory module serves to properly orient preinfection structures to direct the transcellular deposition of cell wall material for the growing infection thread, analogous to its role in cell plate formation. Our findings indicate that the eukaryotically conserved MYB3R1-TPXL-AUR1-MAP65 mitotic module was conscripted to support endosymbiotic infection in legumes.
Legumes form specialized lateral organs on their roots called nodules, which are colonized by rhizobia, a process referred to as nodulation (1). Nodulation entails the use of energy derived from the breakdown of photosynthetic carbon to reduce atmospheric dinitrogen to ammonia by rhizobial nitrogenase for direct use by the plant, which is of great ecological and economic importance (2). To establish functional nodules in most legumes, rhizobia must traverse the cells of the root epidermis and root cortex through unique intracellular structures called infection threads. Infection threads act as transcellular conduits for the rhizobia, providing them access to developing nodule cells where they are endocytosed, forming organelle-like structures called symbiosomes where they carry out nitrogen fixation (3–5). Infection threads are thin tubular invaginations of the host cell wall and plasma membrane that anticlinally traverse the length of the root hair cell, where they fuse to the distal cell wall and open into the apoplast, then sequentially form in underlying cortical cells, forming a network of aligned cell bridges. Although the root hairs and outer cortical cells infected by rhizobia do not divide, it has been noted that the subcellular structure that forms prior to infection thread formation, referred to as the preinfection thread, resembles the phragmosome which forms in premitotic cells (6–9). In each case, a thick, transcellular cytoplasmic bridge forms that predicts the path of cell wall deposition. These transcellular bridges are endoplasmic reticulum (ER)–rich and contain abundant actin and microtubules (3, 6). Our earlier study reported the increased expression of several cell cycle–related genes in root hairs after inoculation by rhizobia or in response to Nod factors (the key rhizobia-to-host signal), including several cyclins and the mitotic kinase Aurora1 (AUR1) (10).
The cell cycle comprises a highly ordered set of events that results in cellular fission to produce two daughter cells (11, 12). An event crucial for cell division is formation of the cell division plane, which involves dramatic changes in the organization of the cytoskeleton which involves Aurora kinases (AURs). AURs play multifaceted roles in cell cycle processes and their importance in mitosis is conserved across eukaryotes (13, 14). These roles, which include positional regulation of cytoskeletal rearrangements and cytokinesis, occur through interactions with microtubule-associated proteins (e.g., members of Targeting protein for Xenopus kinesin-like protein 2 [TPX2] and 65-kDa Microtubule-associated protein [MAP65] families) (15, 16). While extensively studied in mammals and yeast because of their role in cytokinesis and involvement in human cancers, the role of AURs in plants is less clear (17, 18). Plant AURs comprise a small gene family of two groups, with α-AURs functioning in early mitotic events and bipolar spindle formation and β-AURs having apparent roles in anaphase chromosome separation (18–20). Studies of AURs in plants have been mostly limited to the model plant Arabidopsis thaliana, where α-AUR Ataur1 aur2 knockdown mutants were shown to have a defective cytokinesis phenotype which featured randomly orientated cell plates (20). The increase of α-AUR1 expression during symbiotic interactions with nitrogen-fixing soil bacteria suggests the direct engagement of cell cycle machinery in the infection process (10).
Here, we show the requirement for the AUR1 Aurora kinase in symbiotic infection, and determine, in part, its biochemical role and how it is regulated. This study suggests that selected components of the fundamentally conserved cell division program were recruited to enable the transcellular deposition of cell wall material necessary for legume infection-thread formation.
Results
Cell Cycle Genes Are Induced in Symbiotic Tissues.
We began by investigating spatial and temporal expression patterns of several cell cycle–related genes previously reported to be induced in root hairs of Medicago truncatula seedlings after rhizobial inoculation, including the A-type cyclin CYCA3;1; D-type cyclin CYCD1;1; Cyclin-dependent kinases CDKB2;2 and CDKC2;1; and the mitotic kinase AUR1 (10). Data from public gene-expression databases indicate that these genes are highly expressed in the apices of shoots, roots, and nodules, consistent with their predicted roles in cell division (SI Appendix, Fig. S1A). The promoters of these genes were used to drive the expression of β-glucuronidase (GUS) in Agrobacterium rhizogenes–induced transgenic hairy roots of M. truncatula inoculated with Sinorhizobium meliloti (Rm2011), and reporter activity was characterized at different stages of nodulation. Results from GUS staining confirmed the expression of these genes in dividing cells, primarily lateral root primordia and root tips, nodule primordia, and the nodule apex/infection zone of mature root nodules (Fig. 1 B–D and SI Appendix, Fig. S1 B–O). In addition to being active in dividing cells, these reporters showed elevated expression in rhizobia-infected root hair cells (Fig. 1A and SI Appendix, Fig. S1 B–O).
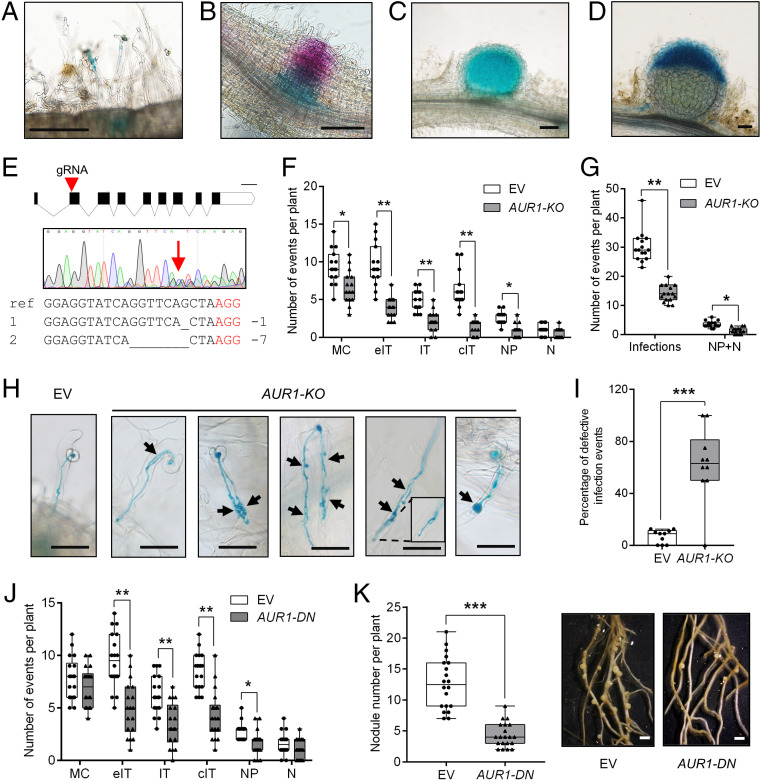
Fig. 1.
AUR1 contributes to rhizobial infection and nodulation. (A–D) Images showing AUR1pro:GUS activity in rhizobia-infected root hairs and the underlying cortical cells (A), in a nodule primordium (B), in a young nodule (C), and in an elongated nodule at 5 dpi (D). The blue color indicates GUS staining, and the magenta color in B indicates LacZ staining of S. meliloti Rm2011. Scale bars, 200 μm. (E) CRISPR/Cas9 mediated tissue-specific KO of AUR1 (AUR1-KO, ChOMT3pro:Cas9) in hairy roots. The guide RNA target position in AUR1 exons is indicated by the red triangle. Sequences having multiple peaks around the guide-RNA target are indicated by the red arrow, which suggests gene editing. The wild-type sequence (ref) at the targeting site and two primary edited forms are aligned where the deleted bases are indicated by underscores, and the red letters indicate the protospacer-adjacent motif (PAM) site. Scale bar, 200 bp. (F) Quantification of infection events at different developmental stages (including both normal and aberrant infections) and nodules/nodule primordia in the empty vector (EV) and tissue-specific AUR1-KO at 5 dpi. Infection-event categories are described in detail in (SI Appendix, Fig. S2A). MC, rhizobia microcolony; eIT, elongating infection-thread in root hair; IT, infection thread that fully traversed the root hair; cIT, infection thread that traversed the root hair and ramified into the underlying cortical cell; NP, nodule primordium; N, nodule. (G) Quantification of total infection events and nodules/nodule primordia from (F). (H) Representative infection phenotypes of tissue-specific KO of AUR1 at 5 dpi. Black arrowheads indicate defective infection threads. Scale bars, 50 μm. (I) Quantification of defective infection events in the EV control and tissue-specific AUR1-KO plants. (J) Root hair–infection phenotypes of tissue-specific expression of AUR1-DN (ChOMT3pro:AUR1-DN) at 7 dpi. (K) Nodulation phenotypes of tissue-specific expression of AUR1-DN at 14 dpi. Representative hairy roots expressing EV or AUR1-DN are shown on the Right. Scale bars, 1 mm. Means were compared using Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001. Boxes show the first quartile, median, and third quartile; whiskers show minimum and maximum values; dots show data points. Experiments were carried out two (J and K) or three times (A–I) with similar results.
A Role for AUR1 in Rhizobial Infection.
The specific induction of AUR1 in infected root hairs indicates its potential role in infection-thread development. Recent studies in Arabidopsis showed that ubiquitous overexpression of AtAUR1 leads to growth defects, and that Ataur1 aur2 double-knockdown mutants are severely dwarfed (18, 20, 21), suggesting that AUR1 expression has to be maintained within certain boundaries for normal growth and development. AUR1 is the only α-AUR gene in M. truncatula (SI Appendix, Fig. S1P), suggesting that aur1 mutants may have growth defects that would complicate interpretation of their phenotypes. Nonetheless, we attempted to generate AUR1 knockout (KO) roots but were unable to isolate biallelic mutations of AUR1 in M. truncatula hairy roots using a CRISPR/Cas9 KO strategy. Thus, we used pharmacological (SI Appendix, Fig. S2B) and alternative functional genetic approaches to study its function in nodulation.
To avoid defects in root growth and development, we designed a CRISPR/Cas9–based, tissue-specific KO strategy in which Cas9 is expressed from the M. truncatula chalcone O-methyltransferase 3 (ChOMT3) promoter, which is preferentially expressed in rhizobia-infected cells (SI Appendix, Fig. S2C) (22). A similar tissue-specific KO strategy has been successfully applied in Arabidopsis and M. truncatula (23, 24).
To evaluate the efficacy of gene editing in the roots transformed with the AUR1-KO construct, the roots were inoculated with rhizobia and DNA was extracted from the 2-mm root section corresponding to the infection zone of the positively transformed hairy roots indicated by Discosoma sp. red fluorescent protein (DsRed) fluorescence. Amplification and sequencing of the DNA fragments containing the targeted sequences revealed that AUR1 was edited in 9 of 10 root sections, having either double or multiple peaks in the vicinity of the predicted protospacer-adjacent motif sites (Fig. 1E). PCR products from three independently edited roots were pooled and subcloned; 23 of 30 clones sequenced contained mutations, indicating the root sections are composed of mixtures of wild-type and edited cells (SI Appendix, Fig. S2E). To further test if editing occurred specifically in rhizobia-infected cells of AUR1-KO roots, DNA was isolated from a single root hair containing an infection thread and from a single uninfected root hair of the same root, using laser capture microdissection. Sequencing of target-site amplicons from these samples indicated that mutation of AUR1 only occurred in the rhizobia-infected root hair, not in the uninfected cell (SI Appendix, Fig. S2F).
Quantification of infection events showed they were greatly reduced in the AUR1-KO roots compared with the control at 5 d postinoculation (dpi) with S. meliloti, and most of the infection threads that formed had abnormal morphologies (Fig. 1 F–I), including the infection threads that branched several times, those that branched and then later merged, and others that formed balloon-shaped structures (Fig. 1H). We further examined the infection events at 7 and 10 dpi and found that the number of infection threads of AUR1-KO roots was reduced, but the nodule number was similar to that of the controls (SI Appendix, Fig. S2G). These results suggest involvement of AUR1 in rhizobial infection and demonstrate the utility of tissue-specific KOs to study the symbiosis.
To further determine the role of AUR1 in nodulation of M. truncatula, we created a dominant negative version, AUR1-DN, by introducing a mutation in the ATP binding site (Lys66 to Arg), based on examples reported in other plant and animal species (21, 25). A construct for expressing AUR1-DN from the ChOMT3 promoter (ChOMT3pro:AUR1-DN) was introduced into M. truncatula by hairy-root transformation. Quantitative real-time PCR analysis of AUR1 transcripts in transgenic roots at 7 and 14 dpi showed approximately two-fold and four-fold increases of AUR1 transcripts vs. controls, respectively (SI Appendix, Fig. S2H). Since the expression of AUR1 was driven by the infected root hair–specific ChOMT3 promoter, the increase of AUR1 expression in these cells is likely much higher than from the quantification using from whole-root samples. The reduction of infection threads forming on AUR1-DN lines at 7 dpi was similar to what was observed for AUR1-KO roots (Fig. 1J), while nodule number at 14 dpi was strongly reduced (Fig. 1K). Expression of nodulation marker genes, including Nodule inception (NIN), Vapyrin (VPY), Early nodulin 11 (ENOD11), ENOD40, and Ethylene response factor required for nodulation 1 (ERN1), was reduced in the AUR1-DN transgenic roots at 14 dpi (SI Appendix, Fig. S2I). These results are consistent with a role for AUR1 in formation of infection structures and in nodule organogenesis.
AUR1, TUB6, and MAP65-1 Colocalize with ER in Rhizobia-Infected Root Hairs.
α-AURs have been implicated in the regulation of cytoskeletal rearrangements, which can be partly explained through their phosphorylation of MAP65s, a family of microtubule-binding proteins which have been implicated in tubulin cross-linking and cytokinesis in animals, fungi, and plants (15, 26). Analysis of publicly available data and promoter GUS indicates that the expression of MAP65-1 (Medtr5g093860) and MAP65-9 (Medtr6g061690) are induced in root hairs by rhizobia and Nod factors, respectively (SI Appendix, Fig. S4 A–G). The symbiotic induction of AUR1, MAP65-1, and MAP65-9 expression prompted us to further investigate their interaction and subcellular localization.
Potential interactions between MAP65s and AUR1 were tested using the yeast two-hybrid (Y2H) assay, and the results showed that AUR1 can bind to both MAP65-1 and MAP65-9 (SI Appendix, Fig. S4 H and I). Their interactions were further studied using coimmunoprecipitation (co-IP) and split luciferase complementation assays in detached leaves of Nicotiana benthamiana, which confirmed that MAP65-1 and MAP65-9 can both interact with AUR1 (Fig. 2 E and F and SI Appendix, Fig. S4 J and K).
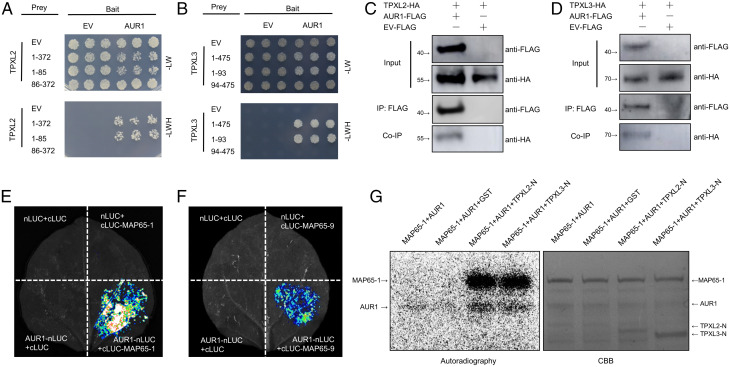
Fig. 2.
AUR1 interacts with multiple microtubule-associated proteins. (A) Y2H assays between AUR1 and TPXL2. The combinations of proteins expressed in either the prey vector (pGADT7) or the bait vector (pGBKT7) are indicated alongside the yeast colonies. The full-length of TPXL2 is indicated by 1-372, 1-85 indicates the N terminus of TPXL2, 86-372 indicates the C terminus of TPXL2. Yeast cells were plated onto SD-2/-Leu-Trp medium and SD-3/-Leu-Trp-His medium. (B) Y2H assays to test for interaction between AUR1 and TPXL3. The full-length of TPXL3 is indicated by 1-475, 1-93 indicates the N terminus of TPXL3, 94-475 indicates the C terminus of TPXL3. (C and D) Co-IP assays of AUR1-FLAG and TPXL2-HA (C) or TPXL3-HA (D) in N. benthamiana leaves. Proteins were immunoprecipitated (IP) with anti-FLAG-M2 beads and analyzed by Western blot using horseradish peroxidase–conjugated anti-FLAG or anti-HA antibody. (E and F) Split luciferase (LUC) complementation assays between AUR1 and MAP65-1 (E) or MAP65-9 (F). nLUC (N terminus of LUC)-tagged AUR1 was coinfiltrated into N. benthamiana leaves along with the cLUC (C terminus of LUC)-tagged MAP65-1 or MAP65-9. (G) Phosphorylation of recombinant MAP65-1 protein by AUR1 in the presence of TPXL2-N or TPXL3-N. An in vitro phosphorylation reaction was performed using purified proteins and 32P-labeled ATP and detected by autoradiography. The Upper band (Left panel) shows the phosphorylation of MAP65-1 by AUR1, and the bottom band shows AUR1 auto-phosphorylation. CBB, Coomassie brilliant blue staining. Experiments were carried out two (G) or three times (A–F) with similar results. EV, empty vector; HA, hemagglutinin.
We then investigated the subcellular localization of AUR1. AUR1 was fused with mCherry and driven by either the Lotus japonicus Ubiquitin (LjUBQ) promoter or its 1.4-kb version of the AUR1 promoter. No signal was observed when AUR1-mCherry was expressed from the AUR1 promoter, either in N. benthamiana leaves or in M. truncatula hairy roots. However, AUR1 fluorescent fusion proteins, when expressed from the LjUBQ promoter, could be reliably detected in M. truncatula roots and thus were used for subsequent experiments (SI Appendix, Fig. S3 A–C).
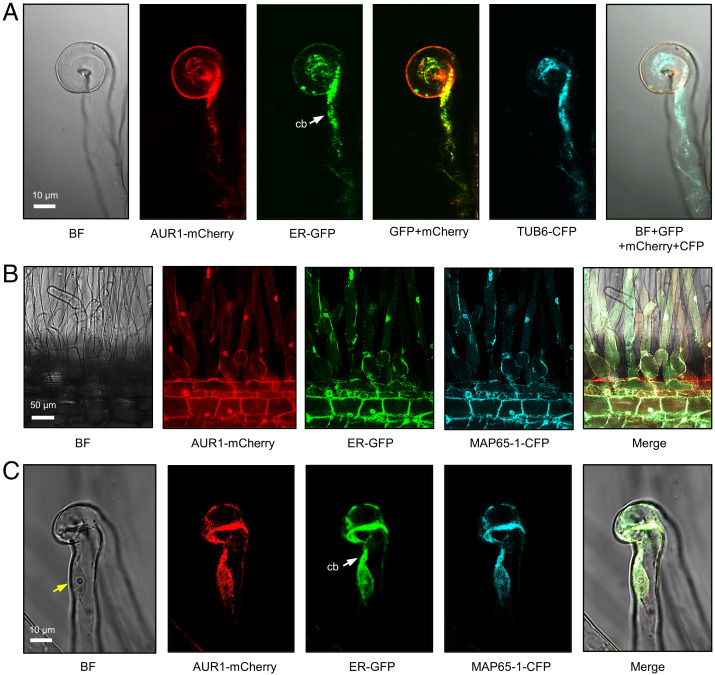
We then analyzed the subcellular localization of the symbiotically induced MAP65s in relation to AUR1 in M. truncatula roots. Both MAP65-1 and MAP65-9 fusion proteins were found to localize mainly in the ER of epidermal cells of the root tip and root hairs of composite transgenic plants (Fig. 3 B and C and SI Appendix, Fig. S3 D–G). We then made constructs for colocalization of AUR1 with either M. truncatula β-tubulin (TUB6) or MAP65-1, the first containing LjUBQpro:AUR1-mCherry and AtUBQpro:TUB6-CFP, and the second having LjUBQpro:AUR1-mCherry and 35Spro:MAP65-1-CFP. These vectors were transformed into the M. truncatula A2 line, which contains the ER marker GFP-HDEL expressed from the 35S promoter (6). AUR1-mCherry, TUB6-CFP, and MAP65-1-CFP all colocalized with the ER marker, both in normal root hairs and those with “shepherd’s crook” curls characteristic of rhizobial infection (Fig. 3 A–C). In addition, these markers colocalized with the ER signal in an infected root hair within the thick cytoplasmic bridge (i.e., preinfection structure) formed between the nucleus and curled root-hair tip (Fig. 3C). These findings are consistent with a potential role for AUR1 in the regulation of microtubule rearrangement within the ER-rich preinfection thread.
Fig. 3.
AUR1, TUB6, and MAP65-1 localize to the ER in rhizobia-infected root hairs. (A) Confocal images of a curled root hair showing subcellular localization of AUR1-mCherry, ER-GFP, and TUB6-CFP driven by constitutive promoters in M. truncatula A2 hairy roots. (B and C) Confocal images of AUR1-mCherry, ER-GFP, and MAP65-1-CFP in M. truncatula hairy roots. The white arrow indicates the cytoplasmic bridge (cb). The yellow arrow indicates the nucleus of the root hair. Scale bars, 10 μm (A and C) or 50 μm (B). Images were captured in A2 plants, which contain 35Spro:GFP-HDEL at 5 dpi with S. meliloti Rm2011. Experiments were carried out three times with similar results. BF, bright field.
AUR1 Interacts with Symbiotically Induced TPX2-Like Proteins.
In plants and animals, TPX2 family proteins directly activate α-AURs during cell division (27, 28) and can be grouped into TPX2 and TPX2-like (TPXL) proteins by their domain composition (29). TPX2 proteins contain an AUR binding domain, an importin domain, and a C-terminal microtubule-binding domain, while TPXLs can be divided into two groups, one having an AUR binding domain and a TPX2 importin domain (group A), and another with a TPX2 C-terminal domain (group B). We identified seven M. truncatula TPX2/TPXL homologs based on their AUR binding domains (SI Appendix, Fig. S5 A and B). Of these, two TPXLs (belonging to group A) were symbiotically induced in root hairs and expressed in nodules (SI Appendix, Fig. S5C); we named them TPXL2 (Medtr6g463310) and TPXL3 (Medtr4g101890). Promoter-GUS analysis revealed that both genes are expressed in infected root hairs and nodules (SI Appendix, Fig. S5 D–G).
We then used the Y2H assay to test whether TPXL2 and TPXL3 can interact with AUR1, using the TPX2 homolog Wave dampened-like (WDL) as a control (30). Results showed a positive interaction between AUR1 and TPXL2 or TPXL3, but not WDL (Fig. 2 A and B). Use of truncated versions of the TPXLs showed that their interaction with AUR1 depended on their N termini (Fig. 2 A and B and SI Appendix, Fig. S5 J and K), as previously demonstrated for TPX2 homologs in mammals and Arabidopsis (27, 28, 31). Similarly, in vivo co-IP and split luciferase complementation assays both confirmed that AUR1 can interact with TPXL2 and TPXL3 in N. benthamiana cells (Fig. 2 C and D and SI Appendix, Fig. S5 H–K).
Then we determined the intracellular localization of TPXL2 and TPXL3 with mCherry fused to their C termini. The genes encoding the fusion proteins were expressed from the LjUBQ promoter in roots of composite transgenic plants generated by hairy-root transformation. Confocal microscopy revealed that TPXL2-mCherry and TPXL3-mCherry localized to the ER and nuclei of root hairs of rhizobia-inoculated seedlings, as observed for AUR1 (SI Appendix, Fig. S3 H–K).
TPXLs Are Required for AUR1 Phosphorylation of MAP65-1.
During cell division in Arabidopsis, AtAUR1 phosphorylates microtubule-associated protein AtMAP65-1 to regulate microtubule dynamics (15), and AtTPXL3 was identified as the primary activator of AtAUR1 (28). We thus investigated whether TPXL2/3 can activate M. truncatula AUR1.
We used Escherichia coli to produce MAP65-1, AUR1, and TPXL2/3 fusion proteins with glutathione-S-transferase (GST) fused to their N termini. The MAP65-1 and AUR1 fusion proteins were incubated with 32P-labeled ATP with either GST alone (control) or with the TPXL2/3 fusion proteins. We found that the in vitro phosphorylation of MAP65-1 by AUR1 was almost entirely dependent on the presence of TPXL2 or TPXL3 (Fig. 2G). Collectively, these results suggest that a TPXL-AUR1-MAP65 module is active during rhizobial infection.
MYB3R1 Directly Activates the Expression of AUR1.
In addition to identifying the biochemical function of AUR1 in rhizobial infection, we investigated how AUR1 expression is induced in the symbiotic context. While the majority of rhizobia-induced genes require the master regulator NIN, AUR1 is an exception, with more than twofold higher expression in nin than in wild-type (10, 32). Analyses of AUR1 promoter sequences from 20 plant species highlighted the presence of mitosis-specific activator (MSA) cis elements (AACGG) near the predicted transcriptional initiation sites (most within 200 bp of the ATG; Fig. 4A and SI Appendix, Fig. S6A). Interestingly, most legume AUR1 promoters examined contain two closely spaced MSA elements, including M. truncatula, L. japonicus, Glycine max (soybean), Phaseolus vulgaris (common bean), Cicer arietinum (chickpea), and Pisum sativum (pea), while the promoters of the nonlegumes each contain a single MSA in this region (SI Appendix, Fig. S6A).
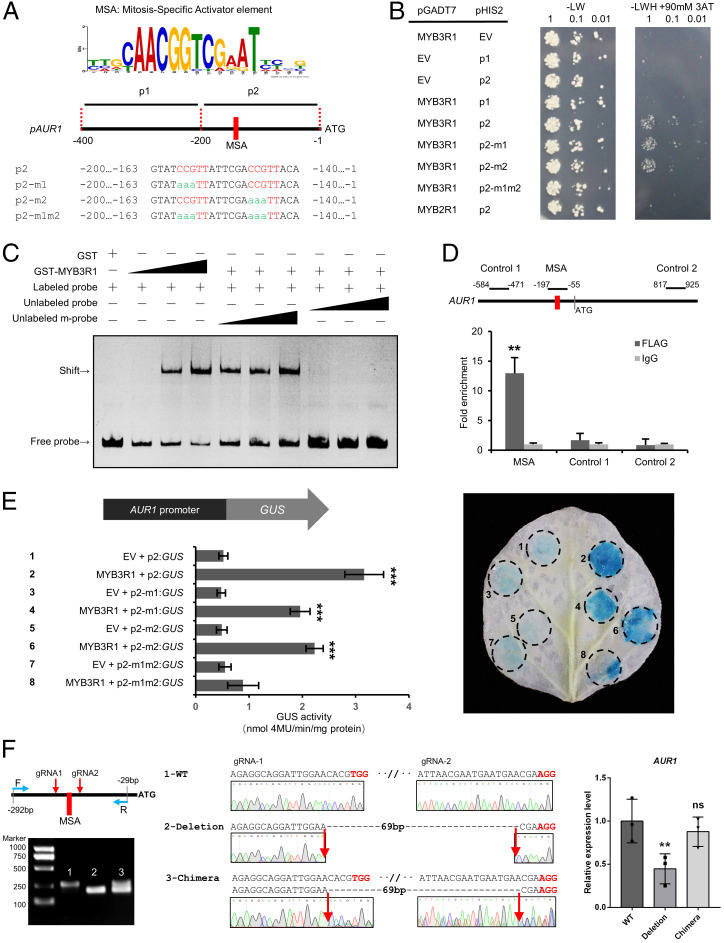
Fig. 4.
MYB3R1 directly activates the expression of AUR1. (A) Analysis of AUR1 promoters from 20 plant species conducted using MEME revealed consensus sites containing an MSA element (CCGTT/AACGG). Diagram of the M. truncatula AUR1 promoter sequences; these were used for further assays (B and E). (B) Interaction of MYB3R1 with the AUR1 promoter revealed by a Y1H assay. The AUR1 promoter sequence (refer to A) was used as bait (pHIS2) and MYB3R1 was used as a prey (pGADT7). Yeast cells were plated onto selection medium without Trp/Leu. In the absence of activation, the leaky expression of HIS was controlled by adding 90 mM 3-amino-1,2,4-triazole (3AT) to the selection medium without Trp/Leu/His. The combinations of MYB3R1 and versions of the AUR1 promoter are indicated alongside the yeast colonies. (C) Recombinant GST-MYB3R1 protein binds the AUR1 promoter in vitro. The probes covering the regions 197- and 90-bp upstream of the AUR1 coding sequence were incubated with increasing amounts of GST-tagged MYB3R1 fragment containing the DNA binding domain (0, 0.1, 0.3, and 0.9 μg); the corresponding EMSA results are shown in the first four lanes. Competition of GST-MYB3R1 binding with the unlabeled wild-type (WT) or mutant probes (both MSAs were mutated) are shown in the last six lanes, and this assay was performed with 0.3 μg of protein and 20-, 50-, and 100-fold excess amounts of unlabeled WT or mutant probe. A band shift indicates positive probe binding. (D) ChIP-qPCR analysis of MYB3R1 binding to the AUR1 promoter in M. truncatula. qPCR was performed using primers surrounding the MSAs or control region not containing the MSAs (Control 1/2). (E) Transactivation assays of the 200-bp AUR1pro:GUS reporter and MYB3R1 in N. benthamiana leaves. GUS expression driven by different AUR1 promoters (refer to A) coinfiltrated with MYB3R1. GUS activity was expressed in nmol 4-methylumbliferone min−1 mg−1 protein. Representative GUS staining is shown in the Right panel. (F) Relative expression (qRT-PCR) of AUR1 in M. truncatula hairy roots transformed with a CRISPR construct with two guide RNAs (gRNAs) flanking the MSA elements. WT indicates the roots were not edited (control band, 264 bp); Deletion indicates roots with biallelic mutations (195 bp); Chimera indicates the roots are not biallelic mutations (two bands). Expression levels were normalized against MtEF1, and data are reported as mean ± SD using three independent biological repeats. Means were compared using Student’s t test. **P < 0.01, ***P < 0.001. Experiments were carried out two (D–F) or three times (B and C) with similar results. EV, empty vector; ns, not significant.
MSA elements serve as targets for the MYB3R (R1R2R3-Myb) family of transcription factors, which are found across eukaryotes and have conserved roles in cell division (33, 34). MYB3Rs form a small distinct clade as part of the large MYB family in plants, with Arabidopsis and M. truncatula and each having five MYB3Rs (SI Appendix, Fig. S6B). In Arabidopsis, AtMYB3R1 and AtMYB3R4 bind MSAs to regulate the expression of cell cycle–related genes in a strict temporal sequence (34). One close homolog of Arabidopsis AtMYB3R1/4, designated here as MYB3R1 (Medtr3g110028), is induced in the M. truncatula root hairs during early infection (SI Appendix, Fig. S6 C and D). Transient expression of MYB3R1 fused with green fluorescent protein (GFP) in N. benthamiana leaves revealed its nuclear localization (SI Appendix, Fig. S6E). We then tested the ability of MYB3R1 to activate transcription when fused to the GAL4 DNA binding domain in yeast. Full-length MYB3R1 and a truncated version lacking the N-terminal MYB-DNA binding domain, but not a fragment containing only the MYB-DNA binding domain, activated transcription in yeast (SI Appendix, Fig. S6F), suggesting that MYB3R1 functions as a transcriptional activator.
We then tested whether MYB3R1 can directly bind to MSA sites found in the AUR1 promoter using the yeast one-hybrid (Y1H) system. We found that MYB3R1 was able to bind to the AUR1 promoter region containing two closely spaced MSAs or with one of the MSAs mutated but was unable to bind to the promoter fragment when both MSA sites were mutated (Fig. 4B). A MYB2R (R2R3-Myb) transcription factor (Medtr1g100667) failed to bind the promoter fragment, suggesting the interaction may be specific to MYB3Rs (Fig. 4B), consistent with earlier reports (35, 36). We further examined this interaction by performing an electrophoretic mobility shift assay (EMSA). This assay confirmed the binding of MYB3R1 to the AUR1 MSA-containing promoter fragment (Fig. 4C). Furthermore, the interaction could not be outcompeted with the addition of unlabeled DNA probe with mutations in both MSAs, while excess unlabeled probe successfully displaced the labeled promoter probes and eliminated the observed shift, indicating the specificity of the interaction (Fig. 4C). To determine whether the observed interaction occurs in vivo, chromatin immunoprecipitation (ChIP) was performed using MYB3R1-FLAG–transformed M. truncatula hairy roots. A monoclonal antibody against FLAG was used for ChIP and an IgG antibody was used as a negative control. Primers for qPCR were designed to amplify different AUR1 fragments. ChIP-qPCR results showed that the AUR1 promoter fragment containing the MSA elements was enriched, while the control fragments were not (Fig. 4D).
To further substantiate MYB3R1 transactivation of AUR1 in planta, we coexpressed LjUBQpro:MYB3R1 with various AUR1pro:GUS fusions in N. benthamiana leaves and measured GUS activity. MYB3R1 expression significantly increased GUS activity driven by the AUR1 promoter fragment containing the two MSA elements compared with the controls (Fig. 4E). Consistent with our in vitro binding assays, MYB3R1 was able to activate expression from the AUR1 promoter fragment lacking either of the two MSAs, but the activation level was lower than that of the WT fragment containing the two MSAs (Fig. 4E). Next, to evaluate the significance of the MSAs for AUR1 expression in M. truncatula, we deleted the cis elements in hairy roots, using CRISPR/Cas9. This resulted in strongly decreased expression of AUR1 (Fig. 4F). These results demonstrate the regulatory role of MYB3R1 in up-regulating AUR1 through the identified MSAs.
In addition, we overexpressed MYB3R1 in M. truncatula hairy roots. This resulted in higher levels of expression of AUR1, as well as several of other cell cycle–related genes which are induced during rhizobial infection in root hairs, including CYCA3;1, CYCD1;1, CDKB2;2, TPXL2, TPXL3, MAP65-1, MAP65-9, and KNOLLE (SI Appendix, Fig. S6H). Most of these genes contain more than one MSA in their promoters (SI Appendix, Fig. S6G), including KNOLLE, which is known to be a direct target of AtMYB3R1 in Arabidopsis (37). Altogether, our data demonstrate that MYB3R1 can bind the MSAs in the promoter of AUR1 to activate its expression and may similarly activate a suite of cell cycle–related genes during infection by rhizobia.
MYB3R1 Promotes Rhizobial Infection.
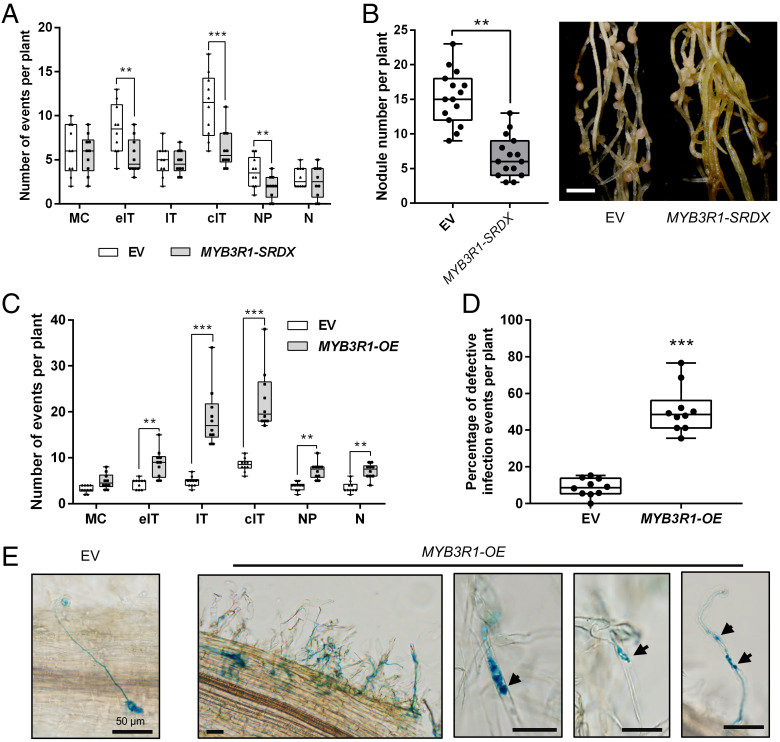
To investigate the role of MYB3R1 in legume nodulation, we used CRIPSR/Cas9 technology to knock out MYB3R1 in M. truncatula, but no abnormal phenotype was observed in the MYB3R1-KO hairy roots at 7 dpi (SI Appendix, Fig. S7A). We then investigated the possibility of functional redundancy. A recent study showed AtMYB3R1 and AtMYB3R4 physically interact to activate cell division in the A. thaliana shoot meristem (38). M. truncatula has five MYB3Rs, which include just two AtMYB3R1/4 orthologs, MYB3R1 and MYB3R2 (SI Appendix, Fig. S6B). Using a split-luciferase assay, we found that MYB3R1 can interact with MYB3R2 in planta (SI Appendix, Fig. S7B). To investigate whether these MYBs may act together to regulate nodulation, a construct with MYB3R1 fused to a plant-specific, ERF-associated amphiphilic repression domain (SRDX), which converts transcriptional activators into strong repressors (39), was used to transform M. truncatula seedlings. MYB3R1-SRDX hairy roots had impaired expression of AUR1 and developed fewer infection events and nodules, but otherwise grew normally (Fig. 5 A and B and SI Appendix, Fig. S7C). In contrast, overexpression of MYB3R1 (MYB3R1-OE) led to an increase of infection threads, nodule primordia, and nodules, but many of the infection threads that formed were defective (Fig. 5 C and D). In particular, a high frequency of branched infection threads was observed in the MYB3R1-OE roots (Fig. 5E).
Fig. 5.
MYB3R1 contributes to nodule symbiosis. (A) Rhizobial infection events, formation of nodule primordia, and nodules in the EV (empty vector) control and MYB3R1-SRDX plants at 7 dpi. MC, rhizobia microcolony; eIT, elongating infection thread in root hair; IT, infection thread that fully traversed the root hair; cIT, infection thread that traversed the root hair and ramified into the underlying cortical cell; NP, nodule primordium; N, nodule. (B) Expression of MYB3R1-SRDX resulted in reduced nodulation at 21 dpi. Representative hairy roots expressing EV or MYB3R1-SRDX are shown in the Right panel. Scale bar, 2 mm. (C) Quantification of infection events including normal and aberrant infections, nodule primordia, and nodules in the EV control and MYB3R1-OE plants at 7dpi. (D) Frequency of defective infection events in EV control and MYB3R1-OE plants. (E) A normal infection event in the EV control plants (Left panel). Typical defective infection events found in MYB3R1-OE plants. A cluster of infection events with examples of abnormal infection events is shown. Arrowheads indicate defective infection threads in root hairs. Scale bars, 50 μm. Boxes show the first quartile, median, and third quartile; whiskers show minimum and maximum values; dots show data points. Means were compared using Student’s t test. **P < 0.01, ***P < 0.001. Experiments were carried out twice with similar results.
To further test the effect of MYB3R1 on symbiotic gene expression, we expressed MYB3R1-OE and MYB3R1-SRDX in ENOD11pro:GUS transgenic plants (40). Introduction of the MYB3R1-OE construct enhanced GUS staining in both epidermal and cortical cells during nodulation, while MYB3R1-SRDX had the opposite effect (SI Appendix, Fig. S7D). These findings suggest that MYB3R1 plays a positive role in infection, like AUR1, but that its overexpression is deleterious. We propose that MYB3Rs act upstream of the TPXL-AUR1-MAP65 module to promote rhizobial infection.
Discussion
AUR1 Is Required for Rhizobial Infection.
In this study, we functionally link a mitotic module consisting of the regulatory kinase AUR1, its activator, and its microtubule-associated substrate to the symbiotic infection process (SI Appendix, Fig. S8). In A. thaliana, AtAUR1 exerts its effects on cytokinesis through inhibition of AtMAP65-1, which promotes microtubule bundling (15). The length and degree of crosslinking of microtubules, and the extent and orientation (parallel or antiparallel) of their bundling are important determinants of microtubule function. Microtubules play multiple roles in infection, including root-hair curling for entrapment of rhizobia and infection-thread growth. The number of microcolonies in AUR1-KO roots was decreased, suggesting that AUR1 contributes to events preceding the initiation of infection thread. However, root-hair curling, which involves the microtubule-dependent reorientation of root-hair growth induced by Nod factors (41), appeared normal in aur1 mutant roots (SI Appendix, Fig. S2J). This suggests that AUR1 could be involved in microcolony establishment within infection chambers. Additionally, the decreased expression of early-infection genes such as VPY, ERN1, and ENOD11 in aur1 roots suggests that AUR1 acts early in the infection process. Notably, both VPY and ERN1 are required for the progression, but not the initiation, of infection threads (42, 43). Like ERN1 and VPY, AUR1 is expressed not only in root hairs at the outset of infection but also in root hairs with partly finished infection threads in the cortical cells subtending infected root hairs, in nodule primordia, and in the infection zone of mature nodules (44, 45), and MYB3R1 shows a similar pattern of expression (SI Appendix, Fig. S6D). These findings suggest that, in addition to being important for microcolony establishment, the MYB3R1-AUR1 module is required for infection-thread progression. The only other microtubule-associated protein studied for its role in nodulation is Developmentally regulated plasma membrane polypeptide (DREPP), the loss of which resulted in fewer responsive (i.e., curled) root hairs and defective infection threads that were prematurely aborted or in which bacteria were released into the root hairs (46). Although mutations in AUR1 and DREPP result in distinct infection phenotypes, we cannot exclude that they may play related and/or overlapping roles.
What role then, does AUR1 play at the molecular level? One possibility is that AUR1 is involved in formation of the preinfection thread, which contains longitudinally arranged microtubule bundles (47). The preinfection thread forecasts the path of the infection thread in the same manner as the phragmosome predicts the formation of the cell plate by the phragmoplast. Notably, repositioning of the phragmosome by mechanical means results in a corresponding change in the position of the cell wall that later forms (48). Intriguingly, formation of the phragmosome is prevented by the microtubule polymerization inhibitor oryzalin (49). Microtubules also feature prominently during infection-thread growth where longitudinally arranged microtubule bundles surround the growing infection thread and extend beyond to connect to the nucleus (47). Loss of AUR1 function resulted in defective infection threads that frequently formed multiple branches, suggesting misdirected cell wall deposition. This phenotype is reminiscent of the misorientation of cell plates observed in Arabidopsis Ataur1 aur2 mutants (20). Acquisition of a stable aur1 mutant with a partial loss of function associated with a less-severe phenotype would allow the possible involvement of AUR1 in preinfection-thread formation to be addressed.
Potential Roles for MAP65s in Infection-Thread Growth.
We show that the genes encoding AUR1, its activators TPXL2/3, and its targets MAP65-1/9 are expressed in rhizobia-infected root hairs. Most MAP65s, including AtMAP65-1 and AtMAP65-3, respective orthologs of the symbiotically induced MAP65-1 and MAP65-9, which can interact with AUR1, mediate microtubule bundling (50). This suggests that the function of the TPXL-AUR1-MAP65 module is to regulate microtubule functions to support infection-thread formation. AtMAP65-3 works redundantly with AtMAP65-4 and is required during formation of the cell plate, helping guide cell wall deposition, with mutants forming cell wall stubs and exhibiting misplaced or wavy phragmoplasts, the latter being the structure which supports formation of the cell plate during cytokinesis (51–53). Functional genetic analysis of MAP65-1 suggests that its phosphorylation during cytokinesis depends mainly on AUR1, but that this may occur partly through AUR1 phosphorylation of other kinases such as CDK1 or mitogen-activated protein kinase (15, 54, 55). Interestingly, loss of AtMAP65-3 causes defects in the formation of minicell walls, structures that are required for formation of giant cells induced by root knot nematodes, effectively blocking pathogenicity (51).
AtMAP65-1/3/5 have been shown to mediate antiparallel microtubule bundling both in vivo and in vitro (53, 56, 57). During mitosis, antiparallel microtubule bundles serve structural roles in establishment and maintenance of polarity and mechanical roles, participating in mitotic pole separation and chromosome segregation. In particular, AtMAP65-1/2/3 are essential for cell plate formation (15, 51, 58). AtMAP65-3 was shown to be required for recruitment of kinesins to the microtubule plus-ends in the midzone during anaphase. Kinesins are motor proteins that move along microtubule filaments, serving either in cargo transport along microtubule tracks or in the generation of pushing, pulling, or sliding forces (59, 60). In the moss Physcomitrella patens, kinesin-4 mediates shortening of the overlap regions of the antiparallel microtubules, and the kinesin-4 double mutant formed thick and irregular cell plates (61). It is also known that the antiparallel microtubule overlap region coincides with the position of the exocyst, which is involved in vesicle fusion at the cell plate (62). During rhizobial infection, the exocyst component EXO70H4 colocalizes and interacts with VPY in M. truncatula, and both components are required for infection-thread growth (43, 44). It is therefore possible that loss of AUR1 negatively impacts infection through affecting MAP65 kinesin recruitment, which could impact exocyst function. In this case, MAP65 may promote formation of antiparallel microtubules in infected root hairs to shape and maintain growth of the infection thread through directing cargo delivery by the exocyst complex at the infection-thread tip. Another possible, and nonexclusive, role for antiparallel microtubules could be in the “sliding” advancement of the rhizobia down growing infection threads, which would require a pulling motion to stretch the matrix in which the rhizobia are embedded (6, 60).
The Role of MYB3R1 in Symbiotic Infection.
Overexpression of MYB3R1 increased the expression of AUR1, TPXLs, MAP65s, and other genes involved in cell division and led to an increase in the number of rhizobial infection events on M. truncatula roots. Loss of MYB3R1 did not affect the symbiosis, while expression of MYB3R1-SRDX suppressed infection and the expression of AUR1 and other targets, suggesting MYB3R1 may act redundantly with homologs to promote expression of AUR1 during nodulation.
The two closest Arabidopsis homologs of Medicago MYB3R1, AtMYB3R1 and AtMYB3R4, were found to act together to promote cell division (38). Despite being close homologs, obvious growth phenotypes are only seen when both genes are mutated, suggesting that AtMYB3R1 and AtMYB3R4 are not functionally equivalent (34). This is further suggested by AtMYB3R4’s relatively restricted set of targets, ∼170 cell cycle genes, which composed a small subset of the >5,000 genes bound by AtMYB3R1 (38). In addition, AtMYB3R1 and AtMYB3R4 are differentially regulated; AtMYB3R4 accumulates in the nucleus at the G2-to-M transition in response to cytokinin, while AtMYB3R1, as we found for MtMYB3R1, always localizes within the nucleus. In addition, our sequence analysis revealed that MtMYB3R1 and AtMYB3R1 share an LCYEPP sequence that is conserved in a subset of MYB3Rs across plants but absent in AtMYB3R4 (SI Appendix, Fig. S6B). Thus, MtMYB3R1 and AtMYB3R1 share several features that distinguish them from AtMYB3R4. We speculate that the LCYEPP motif may define an MYB3R subtype and could be important for protein modification or recruitment of transcriptional activator(s) that confer a broader gene activation profile.
Our data suggest that AUR1 is a key target of MYB3R1 and that this control is conserved across plants via the presence of MSA elements in the proximal promoter regions of AUR1 orthologs. We identified a pair of MSA motifs in legume AUR1 promoters which confer higher expression than either MSA alone. Phylogenomic analysis suggests the second MSA arose in papilionoid legumes, and our experiments revealed that the additional element enhances the activation of AUR1 by MYB3R1, which may be part of the fine tuning of microtubule bundling dynamics required to support endosymbiotic infection in legumes.
Inoculation with rhizobia led to significant up-regulation of the expression of MYB3R1, TPXL, AUR1, and MAP65 in the root hairs. These cell cycle genes are also induced by Nod-factor treatment, suggesting that their expression in infected root-hair cells may require continuous secretion of Nod factors by rhizobia. However, the direct linkages between these cell cycle–related genes and the nodulation signaling pathway remain unknown. Notably, MYB3R1 expression is still induced in nin, ern1, or nuclear factor-ya 1 (nf-ya1) mutants, according to published transcriptomic data (SI Appendix, Fig. S7E) (10, 32). On this basis, we propose that the activation of cell cycle–related genes in root hairs may depend on components’ early symbiotic signaling, such as Doesn’t make infections 2 (DMI2), DMI3, Interacting protein of DMI3 (IPD3), Nodulation signaling pathway 1 (NSP1), or NSP2.
In conclusion, our results suggest that basic components required for cytokinesis were evolutionarily adapted to enable intracellular infection during nodulation. All the components studied, MYB3R1 (including its target MSA site), AUR1, TPXLs, and MAP65s, have homologs in other eukaryotes, reflecting their ancient origins and indispensability for cell division. Plants possess other strategies for creating cell wall invaginations, such as those employed to make transfer cells and xylem tracheary elements. However, the recruitment of mitotic machinery provides the advantages that it can mediate cell wall deposition that is transcellular, allowing bacterial passage across cells, and anticlinal, which serves to direct the infection inward. Perhaps most importantly, it creates an apoplastic compartment that bridges the cell, allowing for containment of the bacteria while allowing their transcellular passage. Numerous mitotic genes expressed during infection remain to be investigated and should reveal how and why this deeply ancient and fundamental program was co-opted into symbiosis.
Materials and Methods
Materials and Growth Conditions.
M. truncatula ecotype Jemalong A17 was used as the wild type in most experiments. For subcellular localization analysis, 35Spro:GFP-HDEL transgenic M. truncatula plants (A2 background) were used. ENOD11pro:GUS transgenic M. truncatula plants (A17 background) were used as a rhizobial infection markers for other gene functional analysis. N. benthamiana was used in this study for interaction analysis and subcellular localization analysis. M. truncatula seedlings were grown in a 3:1:1 mixture of vermiculite, peat soil, and perlite unless otherwise stated. N. benthamiana seeds were germinated and grown in a mixed soil containing a 3:1 ratio of peat soil and vermiculite. Plants were watered regularly as needed and kept in controlled environment chambers with 16 h of light, 8 h of dark at 22 °C with 65% relative humidity, and the light intensity was ∼220 μmol·m−2 s−1.
A. rhizogenes Arqua1 (AC1060L, Weidi) was used in this study for M. truncatula hairy-root transformation. A. tumefaciens GV3101 (AC1001M, Weidi) was used for transformation of N. benthamiana leaves. Saccharomyces cerevisiae AH109 (YC1010M, Weidi) was used for the Y2H assay, and Y187 (YC1020S, Weidi) was used for Y1H assay. E. coli DH5α was used for vector construction, and E. coli Rosetta DE3 (EC1010, Weidi) was used for recombinant protein expression. S. meliloti Rm2011 (hemA:lacZ) was used for M. truncatula rhizobium inoculation.
Gene Cloning.
M. truncatula coding DNA sequences (CDS) and promoters used in this study were amplified from Jemalong A17 cDNA and genomic DNA, respectively. The CDS or promoters of each gene were amplified using KOD FX (KFX-101, TOYOBO) and then cloned into the pENTR vector and/or the Golden Gate L0 vectors using recombination reactions (NR005-01A, Novoprotein). Then these vectors were used to generate different final vectors, as described below. The integrity of each construct was confirmed by restriction digestion and Sanger sequencing. The sequences of the primers used for vector construction are listed in SI Appendix, Table S1. The accession numbers for genes mentioned in this study are listed in SI Appendix, Table S2.
Hairy-Root Transformation.
M. truncatula seedlings were cut slantwise above the hypocotyl in sterile conditions, and then the wounded area was dipped in a culture of A. rhizogenes Arqua1 (AC1060L, Weidi) transformed with a given binary vector for 1 min. The seedlings were then placed on Fåhraeus medium for 1 wk (22 °C, 16-h light/8-h dark photoperiod). Then all roots were removed, and the seedlings were transferred to FP medium containing 0.5 mM KNO3 and grown for 2 to 3 wk. Then positive hairy roots (dsRed+) were identified under a fluorescent microscope (SZX7, Olympus), and any nontransgenic roots were removed. For nodulation assays, transgenic composite plants were transferred to pots containing a 3:1 ratio of vermiculite to perlite, grown for 3 d in controlled environment chambers with 16 h of light, 8 h of dark at 22 °C with 65% relative humidity, and then inoculated with a 1-mL suspension of S. meliloti Rm2011 at an optical density at 600 nm (OD600nm) of 0.04∼0.08. At different time points after inoculation, roots were harvested and histochemically stained, and infection threads and nodules were assayed.
CRISPR/Cas9 KO.
CRISPR/Cas9 vector pKGSD401 and guide RNA-scaffold vector pG-DT1T2 were used for gene KO (63, 64). We used a website tool (CRISPR-P 2.0) for guide RNA design (65). Two specific primers were synthesized, gRNA-T1-F and gRNA-T2-R, and pG-DT1T2 was used as template for amplification of the guide RNA model, which was digested with BsaI (NEB) and ligated using T4 DNA ligase (NEB) in one reaction (4 min at 37 °C, 3 min at 16 °C, for 40 cycles) to make the CRISPR target vector pKGSD401-KO. For CRISPR/Cas9-mediated AUR1 promoter editing, two specific guide RNAs were designed and inserted into the vector pKGSD401. The binary vector was introduced into A. rhizogenes strain Arqua1 (AC1060L, Weidi) that was then used to transform M. truncatula A17 roots. More than 100 DsRed+ roots were used for DNA extraction and genotyping (T5 direct PCR kit, TSE011, TsingKe); the homozygous deletion of AUR1 promoter editions were chosen for testing the expression of AUR1 compared with controls. For tissue-specific CRISPR/Cas9 KO, the CaMV 35S promoter upstream of Cas9 was removed from pKGSD401 by BstEII, XbaI double digestion; a fragment containing ChOMT3 promoter (1.8 kb) was amplified, then ligated into pKGSD401-BstEII-XbaI by using recombination assembly (NR005-01A, Novoprotein), it was renamed as pKGCD401. Two specific guide RNAs were designed and inserted into the vector pKGCD401, as previously described.
Confocal Microscopy.
For subcellular localization analyses, constructs to express AUR1 fused to mCherry, TUB6, or MAP65-1 fused to CFP (cyan fluorescent protein), TPXL2, TPXL3, MAP65-1, or MAP65-9 fused to mCherry were generated by Golden Gate cloning. The plasmids were transformed into A. rhizogenes Arqua1 (AC1060L, Weidi) for hairy-root transformation. Two to three weeks after transformation, composite plants were transferred to 10-cm square Petri dishes containing modified Fåhraeus medium with 3 mM MgSO4, 0.5% Phytagel (P8169-100G, Sigma-Aldrich), and 100 nM 2-aminoethoxyvinyl glycine (AVG; 55720–26-8, Cayman). The roots were covered with Lumox film (Sarstedt). Plants were then grown vertically in growth chambers at 22 °C with a 16 h/8 h light/dark photoperiod and 65% relative humidity. In each case, more than 20 composite plants were inoculated with Rm2011 (OD600 nm = 0.002), unless otherwise mentioned. Roots were imaged under a Leica SP8 confocal laser scanning microscope. The excitation wavelengths used for CFP, GFP, and mCherry were 457, 488, and 561 nm, respectively, and collection wavelengths used were 465 to 485 nm for CFP, 500 to 530 nm for GFP, and 600 to 630 nm for mCherry.
Details about additional methods are described in SI Appendix, including promoter GUS analysis; Y1H and Y2H assays; co-IP; split luciferase biomolecular complementation; RNA extraction and real-time PCR; protein expression; in vitro phosphorylation; EMSA; transactivation assay in yeast; transactivation assay in N. benthamiana and GUS quantification; ChIP analysis; and statistical analysis.
Supplementary Material
Acknowledgments
We are grateful to Fang Xie, Lin Xu, Ertao Wang, and Weibing Yang (Center for Excellence in Molecular Plant Sciences [CEMPS]) for discussions and suggestions. We thank Wen-Juan Cai and Shui-Ning Yin at the core facility center of CEMPS for technical assistance with the confocal microscope; Wei-Hua Tang, Xiaolin Li, and Gang Wang (CEMPS) for experimental support; and Joëlle Fournier (Université de Toulouse) for providing the GFP-HDEL Medicago truncatula seeds. We also thank the members in the J.D.M. laboratory for discussions and comments on the manuscript. J.-P.G. received support from Xiaoyan Shi. This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (grant XDB27040209), National Key Research & Development Program of China (grant 2019YFA0904703), CAS Project for Young Scientists in Basic Research (grant YSBR-011), National Natural Science Foundation of China (grants 32200208, 32150710527 and 32170250), Chinese Academy of Sciences (grant 153D31KYSB20160074), China Postdoctoral Science Foundation (grant 2022M713146), and Shanghai Postdoctoral Excellence Program (grant 2021388).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2202606119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Roy S., et al. , Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 32, 15–41 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oldroyd G. E. D., Leyser O., A plant’s diet, surviving in a variable nutrient environment. Science 368, eaba0196 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Oldroyd G. E., Murray J. D., Poole P. S., Downie J. A., The rules of engagement in the legume-rhizobial symbiosis. Annu. Rev. Genet. 45, 119–144 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Gage D. J., Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol. Mol. Biol. Rev. 68, 280–300 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brewin N. J., Plant cell wall remodelling in the Rhizobium-legume symbiosis. Crit. Rev. Plant Sci. 23, 293–316 (2004). [Google Scholar]
- 6.Fournier J., et al. , Mechanism of infection thread elongation in root hairs of Medicago truncatula and dynamic interplay with associated rhizobial colonization. Plant Physiol. 148, 1985–1995 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Brussel A. A., et al. , Induction of pre-infection thread structures in the leguminous host plant by mitogenic lipo-oligosaccharides of Rhizobium. Science 257, 70–72 (1992). [DOI] [PubMed] [Google Scholar]
- 8.Yang W.-C., et al. , Rhizobium nod factors reactivate the cell cycle during infection and nodule primordium formation, but the cycle is only completed in primordium formation. Plant Cell 6, 1415–1426 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rae A. L., Bonfantefasolo P., Brewin N. J., Structure and growth of infection threads in the legume symbiosis with Rhizobium leguminosarum. Plant J. 2, 385–395 (1992). [Google Scholar]
- 10.Breakspear A., et al. , The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for auxin signaling in rhizobial infection. Plant Cell 26, 4680–4701 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Veylder L., Beeckman T., Inzé D., The ins and outs of the plant cell cycle. Nat. Rev. Mol. Cell Biol. 8, 655–665 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Gutierrez C., 25 Years of cell cycle research: What’s ahead? Trends Plant Sci. 21, 823–833 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Glover D. M., Leibowitz M. H., McLean D. A., Parry H., Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell 81, 95–105 (1995). [DOI] [PubMed] [Google Scholar]
- 14.Nikonova A. S., Astsaturov I., Serebriiskii I. G., R. L. Dunbrack, Jr, Golemis E. A., Aurora A kinase (AURKA) in normal and pathological cell division. Cell. Mol. Life Sci. 70, 661–687 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boruc J., et al. , Phosphorylation of MAP65-1 by Arabidopsis Aurora kinases is required for efficient cell cycle progression. Plant Physiol. 173, 582–599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eyers P. A., Erikson E., Chen L. G., Maller J. L., A novel mechanism for activation of the protein kinase Aurora A. Curr. Biol. 13, 691–697 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Goldenson B., Crispino J. D., The aurora kinases in cell cycle and leukemia. Oncogene 34, 537–545 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weimer A. K., Demidov D., Lermontova I., Beeckman T., Van Damme D., Aurora kinases throughout plant development. Trends Plant Sci. 21, 69–79 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Demidov D., Van Damme D., Geelen D., Blattner F. R., Houben A., Identification and dynamics of two classes of aurora-like kinases in Arabidopsis and other plants. Plant Cell 17, 836–848 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Damme D., et al. , Arabidopsis α Aurora kinases function in formative cell division plane orientation. Plant Cell 23, 4013–4024 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demidov D., et al. , Altered expression of Aurora kinases in Arabidopsis results in aneu- and polyploidization. Plant J. 80, 449–461 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Chen D. S., et al. , Identification of a core set of rhizobial infection genes using data from single cell-types. Front Plant Sci. 6, 575 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Decaestecker W., et al. , CRISPR-TSKO: A technique for efficient mutagenesis in specific cell types, tissues, or organs in Arabidopsis. Plant Cell 31, 2868–2887 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang S., et al. , NIN-like protein transcription factors regulate leghemoglobin genes in legume nodules. Science 374, 625–628 (2021). [DOI] [PubMed] [Google Scholar]
- 25.Terada Y., et al. , AIM-1: A mammalian midbody-associated protein required for cytokinesis. EMBO J. 17, 667–676 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walczak C. E., Shaw S. L., A MAP for bundling microtubules. Cell 142, 364–367 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Bayliss R., Sardon T., Vernos I., Conti E., Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol. Cell 12, 851–862 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Boruc J., et al. , TPX2-LIKE PROTEIN3 is the primary activator of alpha-Aurora kinases and is essential for embryogenesis. Plant Physiol. 180, 1389–1405 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dvořák Tomaštíková E., et al. , Functional divergence of microtubule-associated TPX2 family members in Arabidopsis thaliana. Int. J. Mol. Sci. 21, 2183 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smertenko A., et al. , A guide to plant TPX2-like and WAVE-DAMPENED2-like proteins. J. Exp. Bot. 72, 1034–1045 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zorba A., et al. , Molecular mechanism of Aurora A kinase autophosphorylation and its allosteric activation by TPX2. eLife 3, e02667 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu C. W., et al. , NIN acts as a network hub controlling a growth module required for rhizobial infection. Plant Physiol. 179, 1704–1722 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ito M., Conservation and diversification of three-repeat Myb transcription factors in plants. J. Plant Res. 118, 61–69 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Haga N., et al. , Mutations in MYB3R1 and MYB3R4 cause pleiotropic developmental defects and preferential down-regulation of multiple G2/M-specific genes in Arabidopsis. Plant Physiol. 157, 706–717 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grotewold E., Drummond B. J., Bowen B., Peterson T., The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76, 543–553 (1994). [DOI] [PubMed] [Google Scholar]
- 36.Prouse M. B., Campbell M. M., The interaction between MYB proteins and their target DNA binding sites. Biochim. Biophys. Acta 1819, 67–77 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Haga N., et al. , R1R2R3-Myb proteins positively regulate cytokinesis through activation of KNOLLE transcription in Arabidopsis thaliana. Development 134, 1101–1110 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Yang W., et al. , Molecular mechanism of cytokinin-activated cell division in Arabidopsis. Science 371, 1350–1355 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M., Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34, 733–739 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Journet E. P., et al. , Medicago truncatula ENOD11: A novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol. Plant Microbe Interact. 14, 737–748 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Sieberer B. J., Timmers A. C., Emons A. M., Nod factors alter the microtubule cytoskeleton in Medicago truncatula root hairs to allow root hair reorientation. Mol. Plant Microbe Interact. 18, 1195–1204 (2005). [DOI] [PubMed] [Google Scholar]
- 42.Middleton P. H., et al. , An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19, 1221–1234 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murray J. D., et al. , Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant J. 65, 244–252 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Liu C.-W., et al. , A protein complex required for polar growth of rhizobial infection threads. Nat. Commun. 10, 2848 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cerri M. R., et al. , Medicago truncatula ERN transcription factors: Regulatory interplay with NSP1/NSP2 GRAS factors and expression dynamics throughout rhizobial infection. Plant Physiol. 160, 2155–2172 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Su C., et al. , The Medicago truncatula DREPP protein triggers microtubule fragmentation in membrane nanodomains during symbiotic infections. Plant Cell 32, 1689–1702 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perrine-Walker F. M., Lartaud M., Kouchi H., Ridge R. W., Microtubule array formation during root hair infection thread initiation and elongation in the Mesorhizobium-Lotus symbiosis. Protoplasma 251, 1099–1111 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Murata T., Wada M., Formation of a phragmosome-like structure in centrifuged protonemal cells of Adiantum capillus-veneris L. Planta 201, 273–280 (1997). [DOI] [PubMed] [Google Scholar]
- 49.Panteris E., Apostolakos P., Quader H., Galatis B., A cortical cytoplasmic ring predicts the division plane in vacuolated cells of Coleus: The role of actomyosin and microtubules in the establishment and function of the division site. New Phytol. 163, 271–286 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Chan J., Jensen C. G., Jensen L. C. W., Bush M., Lloyd C. W., The 65-kDa carrot microtubule-associated protein forms regularly arranged filamentous cross-bridges between microtubules. Proc. Natl. Acad. Sci. U.S.A. 96, 14931–14936 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caillaud M. C., et al. , MAP65-3 microtubule-associated protein is essential for nematode-induced giant cell ontogenesis in Arabidopsis. Plant Cell 20, 423–437 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H., et al. , Arabidopsis MAP65-4 plays a role in phragmoplast microtubule organization and marks the cortical cell division site. New Phytol. 215, 187–201 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Ho C. M. K., et al. , Interaction of antiparallel microtubules in the phragmoplast is mediated by the microtubule-associated protein MAP65-3 in Arabidopsis. Plant Cell 23, 2909–2923 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu C., Lau E., Schwarzenbacher R., Bossy-Wetzel E., Jiang W., Spatiotemporal control of spindle midzone formation by PRC1 in human cells. Proc. Natl. Acad. Sci. U.S.A. 103, 6196–6201 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sasabe M., et al. , Phosphorylation of NtMAP65-1 by a MAP kinase down-regulates its activity of microtubule bundling and stimulates progression of cytokinesis of tobacco cells. Genes Dev. 20, 1004–1014 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tulin A., McClerklin S., Huang Y., Dixit R., Single-molecule analysis of the microtubule cross-linking protein MAP65-1 reveals a molecular mechanism for contact-angle-dependent microtubule bundling. Biophys. J. 102, 802–809 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gaillard J., et al. , Two microtubule-associated proteins of Arabidopsis MAP65s promote antiparallel microtubule bundling. Mol. Biol. Cell 19, 4534–4544 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sasabe M., Kosetsu K., Hidaka M., Murase A., Machida Y., Arabidopsis thaliana MAP65-1 and MAP65-2 function redundantly with MAP65-3/PLEIADE in cytokinesis downstream of MPK4. Plant Signal. Behav. 6, 743–747 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nebenführ A., Dixit R., Kinesins and myosins: Molecular motors that coordinate cellular functions in plants. Annu. Rev. Plant Biol. 69, 329–361 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tolić-Nørrelykke I. M., Push-me-pull-you: How microtubules organize the cell interior. Eur. Biophys. J. 37, 1271–1278 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Keijzer J., Kieft H., Ketelaar T., Goshima G., Janson M. E., Shortening of microtubule overlap regions defines membrane delivery sites during plant cytokinesis. Curr. Biol. 27, 514–520 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Tang H., et al. , Exocyst subunit Sec6 is positioned by microtubule overlaps in the moss phragmoplast prior to cell plate membrane arrival. J. Cell Sci. 132, ••• (2019). [DOI] [PubMed] [Google Scholar]
- 63.Xing H.-L., et al. , A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 327 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gao J. P., et al. , Nod factor receptor complex phosphorylates GmGEF2 to stimulate ROP signaling during nodulation. Curr. Biol. 31, 3538–3550.e5 (2021). [DOI] [PubMed] [Google Scholar]
- 65.Liu H., et al. , CRISPR-P 2.0: An improved CRISPR-Cas9 tool for genome editing in plants. Mol. Plant 10, 530–532 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.