Significance
An increase of invasive fungal infections is observed worldwide, whereby ABC transporters are known to play an important role in their drug resistance mediation. The purpose of this study was to contribute to the deeper understanding of drug recognition of fungal ABC-B transporters. We present a high-resolution structure of an ABC-B full-length transporter in complex with verapamil and observed local structural changes facilitating drug binding. Further, the structures of STE6-2p from Pichia pastoris revealed a sterol interaction site. Future studies may focus on the function of the sterol in the protein transport mechanism.
Keywords: ABC transporter, multidrug resistance, verapamil, protein–sterol interaction, yeast transport assay
Abstract
Adenosine triphosphate (ATP)-binding cassette (ABC) transporters are multidomain transmembrane proteins, which facilitate the transport of various substances across cell membranes using energy derived from ATP hydrolysis. They are important drug targets since they mediate decreased drug susceptibility during pharmacological treatments. For the methylotrophic yeast Pichia pastoris, a model organism that is a widely used host for protein expression, the role and function of its ABC transporters is unexplored. In this work, we investigated the Pichia ABC-B transporter STE6-2p. Functional investigations revealed that STE6-2p is capable of transporting rhodamines in vivo and is active in the presence of verapamil and triazoles in vitro. A phylogenetic analysis displays homology among multidrug resistance (MDR) transporters from pathogenic fungi to human ABC-B transporters. Further, we present high-resolution single-particle electron cryomicroscopy structures of an ABC transporter from P. pastoris in the apo conformation (3.1 Å) and in complex with verapamil and adenylyl imidodiphosphate (AMP-PNP) (3.2 Å). An unknown density between transmembrane helices 4, 5, and 6 in both structures suggests the presence of a sterol-binding site of unknown function.
One of the largest superfamilies of transmembrane proteins are the adenosine triphosphate (ATP)-binding cassette (ABC) transporters that can be found in all kingdoms of life. They transport chemically diverse substances across cellular membranes using ATP as an energy source (1). One of the first identified ABC transporters was human P-glycoprotein (P-gp/MDR1/ABC-B1), which is mostly known for its multidrug resistance (MDR) mediation during cancer treatment (2).
ABC transporters of yeast, some of them being homologous to P-gp, are of increasing medical relevance. The drug efflux pumps of fungi show their strong medical importance in the treatment of superficial and life-threatening invasive fungal diseases. Examples of individuals who are highly susceptible to invasive fungal infections (IFIs) are neonates or patients suffering from cancer, HIV, or other severe illnesses (3–7). It is important to note that during the current pandemic, Aspergillus caused SARS-CoV-2–associated pulmonary aspergillosis, an invasive infection that severely increased the mortality rate of COVID-19 intensive care unit patients (8). Important pathogenic yeasts causing severe IFIs are Candida albicans and Candida glabrata (9, 10). In the spotlight of emerging pathogens, special attention should be drawn to the new multidrug-resistant pathogenic yeast Candida auris. It was identified just a few years ago and is hypothesized to have evolved under human-induced environmental changes (11, 12). For the treatment of yeast infections, a limited set of antimycotic therapeutics is available with few more in the pipeline of clinical trials (13). Infectious yeast species have developed multiple ways to overcome drug sensitivity; one of them is the increased drug extrusion via ABC efflux pumps (14–16). Therefore, the dramatic rise of antifungal resistances underscores the necessity to investigate yeasts on a deeper molecular level (17).
Nonpathogenic yeasts are widely used as hosts for heterologous and homologous protein expression. Some of them are established systems, for which complete genome sequences and various molecular biology techniques are available, highlighting them as ideal model systems in many respects. One well-known host that is used as an expression system in both research and industry is the methylotrophic yeast Pichia pastoris. About 10% of the genome of this yeast encodes proteins that are either transporters or otherwise involved in transport activities (18, 19). By studying ABC transporters of nonpathogenic yeasts, we aim to help elucidate the fundamental mechanisms involved in substrate binding as well as the underlying differences in transport mechanisms of eukaryotic ABC transporters.
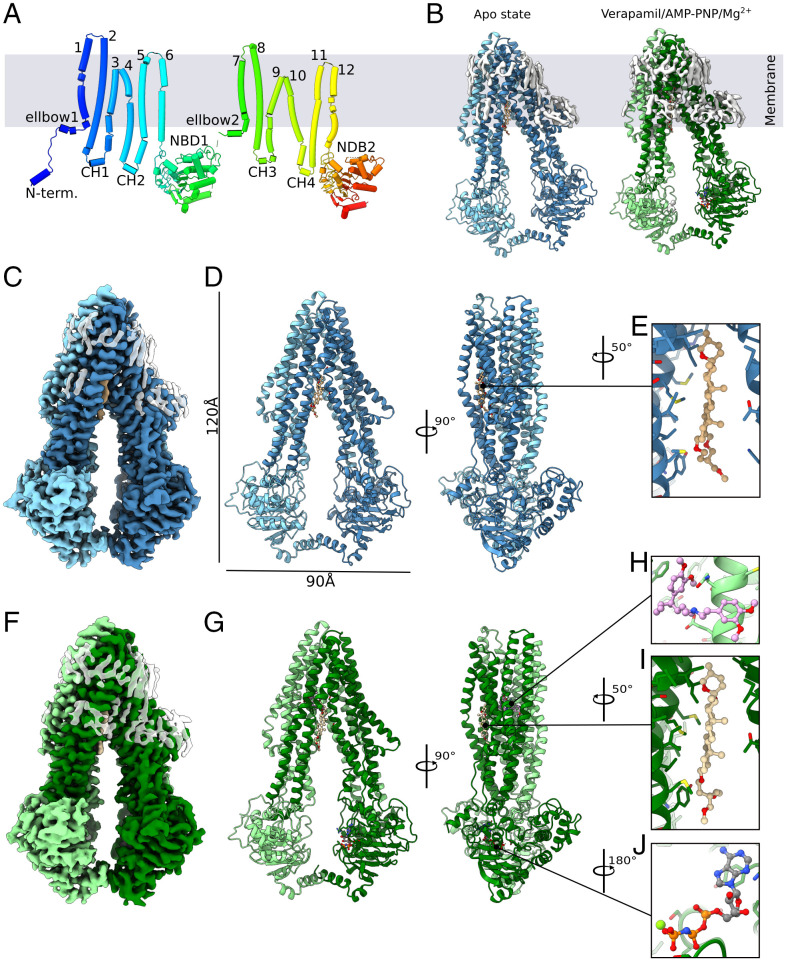
Eukaryotic ABC proteins are divided into eight subfamilies (A to H) based on their domain topology of transmembrane domains (TMDs) and nucleotide-binding domains (NBDs) (20, 21). Multidrug resistance is often mediated by members of subfamilies B and G. This study focuses on STE6-2p, a member of subfamily B. Transporters of this class follow a general TMD–NBD–TMD–NBD topology, whereby the first and second TMD–NBD repeats are connected via a long linker (Fig. 1A). Each TMD is the sum of one elbow helix, six transmembrane helices (TMs) with connecting extracellular loops and two cytoplasmic coupling helices (CHs). The full protein arranges the two TMD–NBD domain halves in a head-to-tail fashion exhibiting pseudosymmetry. Since TM4 and TM5 domain swap into the other functional transporter half, CH2 interacts with the NBD of the second transporter half and vice versa. Through these interprotein joints, TMDs and NBDs communicate ATP binding and protein motion (22, 23). The cytoplasmic NBDs contain highly conserved sequences across all phyla: Walker A and Walker B, the ABC signature motive, as well as the A, Q, D, and H loops. They are the hallmark of ABC proteins and are responsible for ATP binding and hydrolysis (24).
Fig. 1.
Structure of STE6-2p from P. pastoris. (A) Schematic 2D view of the inward-facing apo state. (B) Ribbon representation of both solved structures: apo state (blue) and substrate-bound (aquamarine). Unmodeled densities of residual micelles are shown (white). (C) EM density map (blue) with unmodeled residual micelle (transparent white) and (D) structure model of apo STE6-2p in ribbon representation (blue) with (E) GDN steroid fragment (pale brown) intercalated in transmembrane domains. (F) EM density map (green) with unmodeled residual micelle (transparent white) and (G) structure model of substrate bound STE6-2p in ribbon representation (green) with (H) bound verapamil (plum), (I) steroid fragment of GDN (wheat), and (J) AMP-PNP (gray).
Taking a closer look at ABC-B/MDR transporters known to mediate drug resistance in yeast, MDR1 (Cryprococcus neoformans) and PMD1 (Schizosaccharomyces pombe) have been characterized so far. Other full transporters of this subfamily are MAM1 (S. pombe), STE6 (Saccharomyces cerevisiae), and HST6 (C. albicans), all of which export yeast pheromones. ABC-B half transporters are associated with conjugated metal or peptide transport and are localized at inner cellular membranes (25).
We started the journey of studying P. pastoris ABC transporters with the putative ABC-B transporter STE6-2p. After the optimization of the expression construct, we succeeded in homologously overexpressing and purifying this protein. Consistent with a previous study, we demonstrated that STE6-2p does not function as an a-factor exporter (26). Our findings demonstrate that STE6-2p exhibits multidrug transport function. Using single-particle cryogenic electron microscopy (cryo-EM), it was possible to solve the structure in inward-facing conformations at high resolution in the absence and presence of the substrate and nucleotide at 3.1-Å and 3.2-Å resolutions, respectively. To our surprise, an additional density for an intercalated sterol derivative was found, which indicates the presence of a sterol-binding site.
Results
Purification Yields and Protein Quality.
The gene STE6-2 with N-terminal ramping sequence and C-terminal 10× His-tag was cloned into the pPICZ expression vector and transformed into P. pastoris strain X33. After homologous protein production, the protein was solubilized from yeast membranes and purified via a multistep procedure. Impurities and aggregates seen after immobilized metal affinity chromatography (IMAC) were removed by ion exchange chromatography (IEX) and subsequent size exclusion chromatography (SEC). The purified protein STE6-2p appears as a single sharp, monodisperse peak in SEC, indicating homogeneity of the sample (SI Appendix, Fig. S1). The single major band of ∼140 kDa on a sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel corresponds to the protein’s theoretical molecular weight and confirms the sample purity. The homogeneity of the purified protein was further verified by negative stain electron microscopy using an FEI Tecnai Spirit (BioTwin) transmission electron microscope (TEM). The yield of SEC-purified STE6-2p was ∼168 µg per liter expression culture (∼0.04% of total membrane protein).
Activity Measurements.
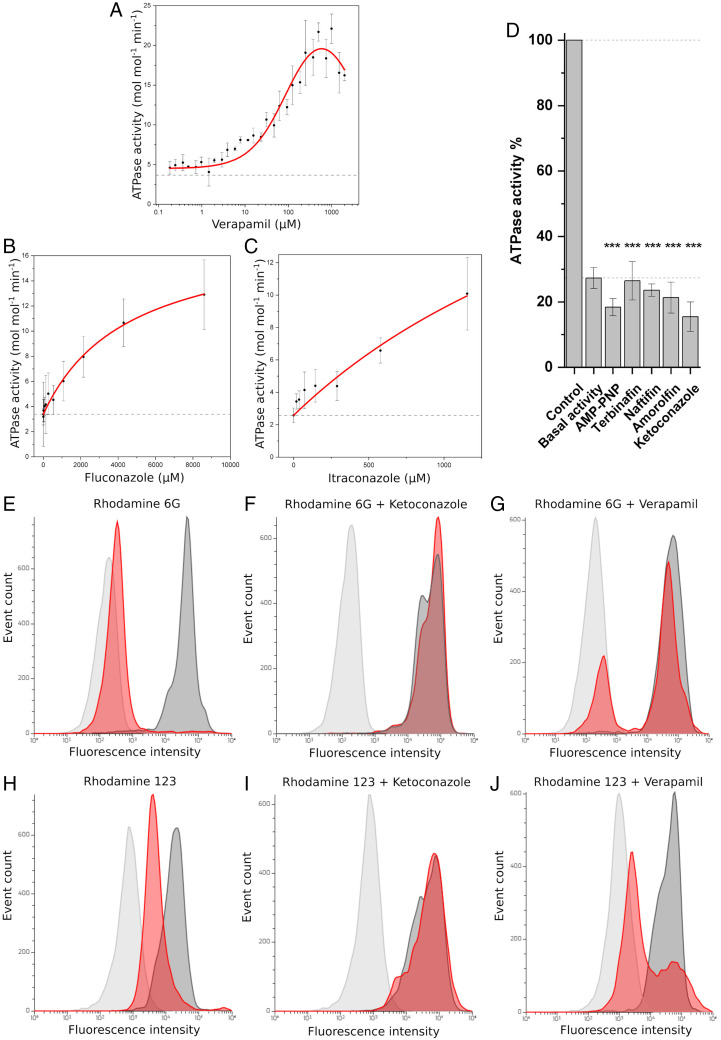
To find substrates for the Pichia ABC-B transporter STE6-2p, in vitro and in vivo experiments were performed. For a start, known ABC-B1 substrates, later yeast pheromones and antimycotics, as well as membrane components ergosterol, 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS), and total yeast lipid extract were tested for ATPase activity stimulation of purified protein. Except for lipids and pheromones, all other substances were dissolved in dimethyl sulfoxide (DMSO) and tested over a concentration range of up to 104 µM. Lipids were taken up in detergent buffer and screened at 0.1 mM or 0.1 mg/mL. Neither of the yeast pheromones, a-factor nor alpha-factor, stimulated ATP hydrolysis of STE6-2p (SI Appendix, Fig. S2). Among the other 16 tested compounds, verapamil, fluconazole, and itraconazole were identified to induce ATPase activity of STE6-2p (Fig. 2). For the well-known calcium channel blocker and P-gp modulator verapamil, a Km of 99.17 ± 19.50 µM and a Vmax of 20.18 ± 1.78 mol generated phosphate per mole STE6-2p and minute (mol·mol−1·min−1) was measured (Fig. 2A). As observed for the human ABC-B1 transporter (27) verapamil also exhibits an inhibitory effect on ATP hydrolysis at concentrations above 600 µM for STE6-2p.
Fig. 2.
Multidrug resistance activity of STE6-2p. (A–C) Stimulation of ATP hydrolysis indicates multidrug transport capacity (n = 9): (A) The anticancer drug verapamil with Vmax = 20.18 ± 1.78 (SD) mol·mol−1·min−1 and Km = 99.17 ± 19.50 (SD) µM and antimycotics (B) fluconazole and (C) itraconazole. (D) ATPase activity with verapamil can be reduced below base-line activity with AMP-PNP, terbinafine, naftifine, amorolfine, and ketoconazole. Significance value: ***P < 0.001 compared to control. (n = 9) (E–J) In vivo rhodamine 123 and rhodamine 6G accumulation and transport assay. STE6-2p overexpressing yeast (red) and X33 control cells (dark gray) after rhodamine treatment; yeast cell autofluorescence (light gray). (E) Cell fluorescence after incubation with rhodamine 6G. (F) Accumulation of rhodamine 6G by inhibiting the transport activity of STE6-2p by the presence of 500 µM ketoconazole. (G) A total of 500 µM verapamil largely abolishes the transport of rhodamine 6G. (H) Cell fluorescence after incubation with rhodamine 123. In the presence of (I) 500 µM ketoconazole, rhodamine 123 accumulates by inhibiting the transport activity of STE6-2p. (J) A total of 500 µM verapamil diminishes the extrusion of rhodamine 123 by STE6-2p. The figure presents one result from each of three independent measurements.
ATP hydrolysis was measured at substrate concentrations of up to 8.60 mM fluconazole and 1.15 mM itraconazole with activity rates of 12.91 ± 2.76 and 10.09 ± 2.24 mol·mol−1·min−1, respectively, at the highest measured concentration (Fig. 2 B and C). The activity did not reach saturation in the measured concentration range and it was impossible to specify the interaction further due to the limited solubility of azole derivatives. We found that the presence of 500 µM terbinafine, naftifine, ketoconazole, or amorolfine in the in vitro assays reduced ATP hydrolysis below the protein baseline activity, similar to the ATP ortholog adenylyl-imidodiphosphate (AMP-PNP) (Fig. 2D). It is important to note that substrates not always lead to an increase of the ABC transporter ATPase activity (28). The in vitro negative data indicating nonsubstrates all scatter around the basal ATPase activity of STE6-2p, similar to the pheromones (SI Appendix, Fig. S2). Nevertheless, the in vitro experiments performed here are leading the way for the identification of the function of STE6-2p. Thus, the transport function of STE6-2p was further investigated in vivo. We measured the retention of MDR model substrates rhodamine 123 and rhodamine 6G in yeast cells by fluorescence-activated cell sorting (FACS) (Fig. 2 E–J). While both rhodamines accumulate in the control cells (P. pastoris strain X33), a significant decrease in fluorescence intensity is observed in the cells overexpressing STE6-2 (Fig. 2 E and H). Thus, STE6-2p transports both rhodamines. The addition of ketoconazole completely inhibited the efflux of rhodamine 6G as demonstrated by the high fluorescence intensity of the STE6-2p–producing cells, which is congruent with the intensity of the control cells (Fig. 2F). Including verapamil in the FACS studies as a potential inhibitor of STE6-2p decreases rhodamine extrusion in the majority of the cells (Fig. 2G). Similar effects were observed for rhodamine 123: Ketoconazole abolished rhodamine 123 extrusion completely (Fig. 2I) and verapamil partially (Fig. 2J).
Overall Architecture.
In this study, two high-resolution cryo-EM structures of STE6-2p from P. pastoris were solved. The first structure is of an ATP-free apo state at a resolution of 3.1 Å (Fig. 1 C–E). The second structure shows the protein in complex with verapamil and AMP-PNP at a resolution of 3.2 Å (Fig. 1 F–J). Both structures feature a sterol-binding site inhabited by the steroid group of glyco-diosgenin (GDN) (Fig. 1 E and I). Further, both structures exhibit inward-facing conformations that are near identical with each other (rmsd: 1.904 Å).
STE6-2p starts with a helical N-terminal extension, followed by an elbow helix and six TM helices (Fig. 1A). The transmembrane helices are connected via three extracellular loops and two intracellular coupling helices (CHs). The unit of elbow helix and all six TM helices is one transmembrane domain (TMD). This TMD is followed by a nucleotide-binding domain (NBD). A linker peptide connects this first transporter half to a second TMD–NBD domain repeat. Thus, STE6-2p comprises the architecture of an ABC-B full transporter with the domain order TMD–NBD–TMD–NBD. Both pseudosymmetrical transporter halves arrange in a head-to-tail fashion. Comparison to known eukaryotic ABC-B1 structures demonstrates a similar protein fold.
The linker between the two transporter halves is not visible in the EM map of neither solved structure. Also, a peripheral loop in NBD2 is not resolved. Hence, these peptides were excluded from the models. Both structures of STE6-2p comprise 1199 of the total 1288 residues with residues 32 to 645, 697 to 1144, and 1152 to 1288 in the apo structure, and 32 to 645, 697 to 1143, and 1151 to 1288 in the model of STE6-2p in complex with verapamil and AMP-PNP. An overview of the map resolutions and map-to-model statistics is given in SI Appendix, Figs. S3 and S4 and Table S1. Further, the EM density maps indicate the presence of multiple lipid molecules around the TMDs indicating a tilted orientation in the membrane (Fig. 1B).
Surprisingly, the N-terminal extension is partly visible in both EM densities. Residues P33 to R41 form a single helix and associate with NBD1 (SI Appendix, Fig. S5B). To our knowledge, this has not been observed for ABC-B transporters before. In the crystal structure of P-gp from Caenorhabditis elegans (Protein Data Bank [PDB]: 4F4C) the helical extension was found between the transmembrane helices (29). The N terminus of human ABC-B11 was observed to be associated within the TMDs at a different position and is hypothesized to stabilize the protein (30). Otherwise, this helix is widely missing in other known experimental ABC-B structures.
Nucleotide-Binding Domains.
Both NBDs show a typical fold, containing a RecA-type core with the conserved motives of Walker A, Q loop, Walker B, D loop, and H switch as well as the A loop and the ABC signature motif (SI Appendix, Fig. S5 A and B). The presence of all amino residues necessary for ATP coordination and hydrolysis indicates that both NBDs are probably catalytically active.
The substrate-bound structure coordinates AMP-PNP, a nonhydrolyzable ATP analog, in NBD1 (SI Appendix, Fig. S5C). Upon AMP-PNP binding, the nucleoside is in pi-stacking with Y402, which is stabilized by Y445. The triphosphate is held in place by Walker A, whereby the β- and γ-phosphates coordinate a magnesium ion together with D565 of Walker B and S435 of Walker A. The H switch flips away from the ATP analog. The weak EM density of the nucleoside in NBD2 suggests partial binding and was not used for AMP-PNP placement.
In the inward-open conformations observed here, the NBDs overlap with their C-terminal helices, whereby the interface is lined with hydrophobic residues such as isoleucine and leucine. In comparison to other ABC-B apo structures derived from detergent, the cytosolic domains of STE6-2p are much closer to each other. Short distances between NBDs are found in transporters that are reconstituted into native environment mimics like lipid vesicles or nanodiscs and are considered to be representative for the physiological state. Both structures reported here align with the observed NBD distance range of ABC transporter turnover experiments in nanodiscs (31).
Verapamil Binding of STE6-2p.
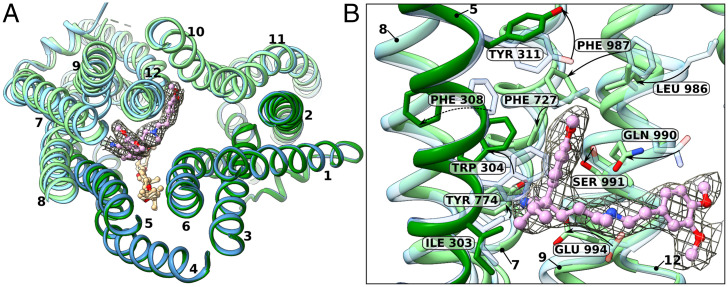
Verapamil binding is mainly coordinated by residues from TMs 5, 7, 8, 9, and 12 (Fig. 3). Comparison of our substrate-bound inward-facing structure with the ATP-free state gives an insight into the changes the protein undergoes upon substrate binding (Fig. 3). One of the aromatic rings of verapamil fills the position of W304, which flips outwards, followed by F308. The nitrile group of verapamil is in close proximity to Y774, suggesting hydrogen bond formation to the residue’s hydroxyl group. As the pKa of verapamil is 8.92, its tertiary amine will be protonated at the used experimental pH. Thus, it is possible for the carboxyamide group of Q990 to establish a hydrogen bond to N8 of verapamil. This residue is recognized to play an important role for substrate binding in human ABC-B1 (32). TM12 rolls over, placing F987 at the position of F727, which itself is flipped over. Y331 is pushed upwards. Verapamils second benzole ring with its methoxy groups interacts primarily with helical backbone of TM10 and TM12, with residues F996, V880, G876 and L873 in relative proximity. Some density noise was found next to this benzole ring, which was left unassigned.
Fig. 3.
Structural changes of STE6-2p induced by verapamil binding, comparison of apo state (blue) and substrate-bound state (green) showing (R)-verapamil fitted in the single-particle analysis cryo-EM density. (A) Cut Top view of STE6-2p in licorice representation with verapamil (plum) and GDN (burly wood/wheat). (R)-verapamil binding induces an outward shift of TM7 and TM9; TM8 shifts inward and TM12 rolls over. GDN does not change its position during verapamil binding. (B) Residue rearrangement undergone by verapamil binding.
The largest backbone change is undergone by TM9. In the apo state, it poses a broken helix, with the inward-bend cytosolic part offset relative to its transmembrane part by a variable region. This region rearranges to form a more uniform helix upon verapamil binding and the helix appears to move outwards (Fig. 4A). TM7 shifts outwards, too, whereas TM8 shifts inwards.
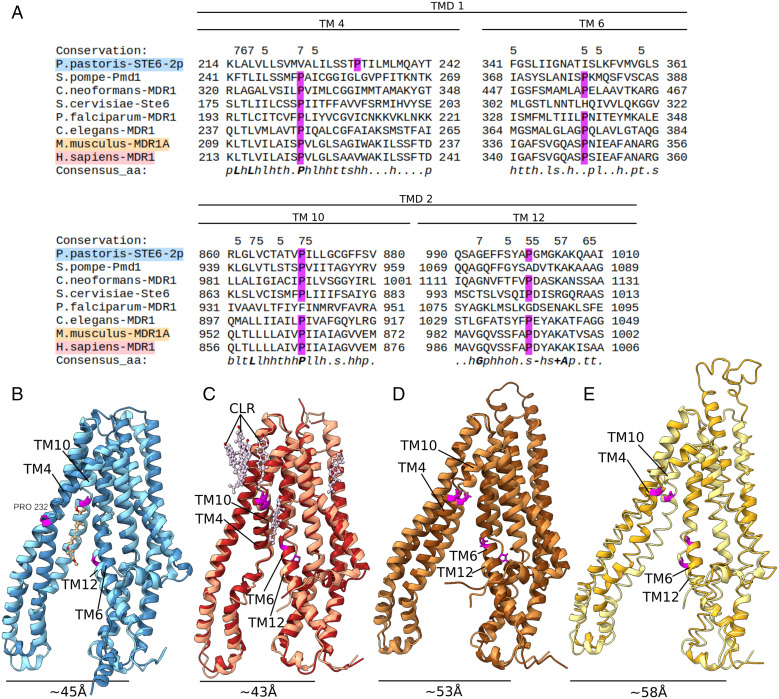
Fig. 4.
Multiple sequence alignment and TMD superimposition of several ABC-B transporters demonstrate shift of conserved proline in STE6-2p. (A) Multiple alignment generated using Promals3D server (73) of STE6-2p with PMD1 from S. pombe, Ste6 from S. cervisiae and MDR1 sequences from C. neoformans var. grubii, C. elegans, mouse, and human. Transmembrane helices 4, 6, 10, and 12 contain conserved prolines, which are shifted or missing in several yeast homologs of STE6-2p, but not in vertebrate homologs. The shift of the proline position in TM4 of STE6-2p allows the accommodation of GDN, which poses a possible sterol-binding site. (B–E) Superimposition of TMDs of ABC-B full transporters in the apo state pronounce the different bending of TM4 and TM10. (B) STE6-2p: TMD1 (steel blue) with bound GDN aligned with TMD2 (pale blue) emphasizes the strong conformational differences between TM4 and TM10 of this structure (rmsd all atoms: 4.123). The helical bending of TM4 is more similar to the ABC-B1 outward-facing state; TM10 represents the common apo conformation as seen in (C) human ABC-B1 (PDB: 7A65, rmsd: 3.065) TMD1 in red, TMD2 in rose. TM4 and TM10 are kinked toward the protein core, which is necessary to bring the NBDs in closer proximity. Many accessory cholesterols are present (CLR, pale violet), none of which are intercalated between helices. (D) Mouse MDR1 apo conformation (PDB: 5KPI chain A): TMD1 (dark brown) aligned with TMD2 (brown) (rmsd all atoms: 3.237). (E) Human ABC-B11 (PDB: 6LR0, rmsd: 19.518) with TMD1 (goldenrod) and TMD2 (pale yellow) has the same inward-bend TM4 and TM10.
A Sterol Interaction Site.
Interestingly, the EM maps show an unknown density sandwiched between TMs 4, 5, and 6 outside the binding pocket (SI Appendix, Fig. S6A). No ligand has been observed at this position in any ABC-B structure published so far. The nature of this additional density directed us toward a sterol derivative and both the yeast sterol ergosterol as well as the lipophilic steroid of GDN were found to be equally good fits (SI Appendix, Fig. S6 B and C). It is probable that STE6-2p binds ergosterol, which gets successively replaced by GDN during purification. Hence, for scientific accuracy, we decided to fit GDN into the deposited model but designate the position as a potential sterol-binding site. It is characterized by isoleucine and other small aliphatic amino acids, hence formed by the tertiary not primary protein structure (33). The ligand is placed at the height of the inner membrane leaflet (SI Appendix, Fig. S6D). K354 provides a positive charge close to the potential sterol hydroxyl group.
Sequence alignment of ABC-B from yeast, nematodes, rodents, and primates reveals conserved prolines in TMs 4, 6, 10, and 12 (Fig. 4). Superimpositions of TMD1 and TMD2 of mouse ABC-B1 (PDB: 5KPI) (34), human ABC-B1 (PDB: 7A65) (35), and human ABC-B11 (PDB: 6LR0) (30) apo structures show that the conserved prolines in TM4 and TM10 appear to be necessary to bend the helix toward the protein core.
The conserved prolines of TM10 and TM12 are both present in STE6-2p (P870, P1000), whereby TM10 bends toward the protein core, similar to the compared ABC-B structures. TM4 of STE6-2p differs from the aligned homologs as it has a valine (V224) at the position of the conserved proline (Fig. 4A). This mutation allows the formation of a small cavity between the helices in such a way that there is room for the hydrophobic ligand. A proline bending the membrane can be found C terminal at a short distance (P232). Superimposition of TMD1 (residues 32 to 379) and TMD2 (residues 697 to 1026) of STE6-2p emphasizes the difference between TM4 and TM10 within the pseudosymmetry (Fig. 4B). The bound ligand in STE6-2p takes the place of TM4 and pushes it outwards, adapting a conformation similar to the outward facing state of ABC-B transporters. Additionally, the asymmetrical arrangement of TM4 and TM10 leads to a shift in CH2 position relative to CH4. Although STE6-2p lacks the conserved proline in TM6, no structural difference to the compared structures is visible. It is possible that higher helical flexibility is necessary close to the steroid-binding site.
The impact of sterols on STE6-2p function still has to be investigated in the future.
Homology.
In total, 97 protein sequences met the selection criteria. The closest verified homologs are STE6-2p of Neosartorya fumigata (Aspergillus fumigatus) and the sophorolipid transporter from Starmerella bombicola (Candida bombicola) with 46% and 45.6% protein sequence identity, respectively. As shown in SI Appendix, Fig. S7, P. pastoris STE6-2p (“Komagataella phaffii_STE6-2p”) clusters with ABC-B transporters of the fungi kingdom. Evaluating the assigned functions of this clade reveals that most transporters are associated with multidrug resistance competencies. This fact points toward the hypothesis that STE6-2p is able to exhibit multidrug transport function, which was proven here in vivo and in vitro. Highlighting the full transporters with available experimental structures makes it obvious that the two structures reported here stand alone outside the vertebrate clade.
Discussion
In this study, we characterized the protein STE6-2p from P. pastoris. Previous automatic annotation of the amino acid sequence pointed to the two distinct functions of 1) a pheromone exporter or 2) a multidrug resistance protein. In this study, both hypotheses were addressed. With this first biochemical characterization of the purified protein, we could demonstrate drug-specific ATPase activity stimulation in the presence of the calcium channel blocker verapamil and the pharmacological triazole derivatives fluconazole and itraconazole (Fig. 2 A–C). Verapamil proves to be a modulator for this ABC transporter, similar to P-glycoprotein. The measured activity for STE6-2p with verapamil is fivefold lower than liposome reconstituted human ABC-B1 (27). FACS assays with rhodamine 123 and rhodamine 6G in cells overexpressing STE6-2 prove this ABC transporter to exhibit multidrug transport function (Fig. 2 E–J). Verapamil diminishes rhodamine extrusion of STE6-2 overexpressing cells. In previous fluorescence-based studies, verapamil served either as substrate or inhibitor of the studied ABC transporter (36–38). Our FACS results support the interaction of verapamil with STE6-2p in vivo, but do not allow further interpretation of the nature of the interaction. The transport of rhodamine, the model behavior of verapamil in vivo and in vitro, as well as its location in the cryo-EM density, plus the overall structural similarity of STE6-2p to known mammalian MDR transporters, substantiate the MDR hypothesis of this transporter. Together with previous in vivo studies (26), we conclude that STE6-2p is most likely a multidrug resistance protein and not—as hypothesized additionally— an a-factor exporter.
The first-generation azole therapeutic ketoconazole was identified as a potential inhibitor of STE6-2p. It is important to note that this antifungal drug possibly exhibits further effects on the yeast cells during the in vivo assay, influencing the retention of the rhodamines. Further, ATPase activity is inhibited in vitro by allylamines and amorolfine. The ATPase activity stimulation and rhodamine transport modulation of STE6-2p by antimycotics investigated in this study further support the hypothesis of MDR transport by STE6-2p. Nevertheless, more studies are needed in the future to unequivocally verify direct inhibition of STE6-2p function by these compounds.
STE6-2p has the overall structure of a typical ABC-B transporter (Fig. 1). Although the protein was imaged in a detergent environment, the structures exhibit a physiological state, indicated by the proximity of the NBDs (31). We assume that this condition was achieved by homologous expression and optimized purification, which preserved few annular lipids (Fig. 1B). The intercalation of a steroid between TMs 4, 5, and 6 was observed, to which we fitted ergosterol and the lipophilic steroid of GDN (Fig. 1 E and I and SI Appendix, Fig. S6). Notably, this ligand does not change its position upon ligand binding. The presence of this sterol derivative in between the TMs can be explained by the shift of a conserved proline in TM4 (Fig. 4A). It leads to an untypical outward bending of this helix. The pseudosymmetric second half of the transporter does not have a shift in the conserved prolines position of TM10, and thus, this TM is kinked toward the protein core. Comparison to other full-length ABC-B structures demonstrates that this conformational difference is in fact unusual (Fig. 4 B–E). This feature led us to the conclusion that the observation of an additional ligand at this position and the associated asymmetrical helix bending are thus far unique to STE6-2p.
The activity and specificity of human ABC-B1 can be modulated by different sterols (27, 39). Structures of ABC-B1 reconstituted into nanodiscs with high cholesterol content show that cholesterol binds to TMDs outer surface (35), but it hasn’t been observed to associate in such a way as in the case of STE6-2p reported here. It is possible that this sterol derivative represents an element needed for protein stability. Or it could place a steric hindrance in STE6-2p movement: The kinks of TM4 and TM10 of ABC-B1 are associated with an occluded conformation, which is necessary to bring the NBDs in close proximity (35). Locking TM4 in an outward-bent conformation decreases the possibility of reaching this transition state and would slow down if not abolish protein function. This was demonstrated in a recent study of lipid-flippase ABC-B4, where the lipid substrate hinders TM4 to adapt to the necessary kinked conformation (40). It is known that sterols exhibit different functions depending on the nature of protein interaction (41–45). The exact function relationship of STE6-2p with sterols has to be elucidated in the future.
Even though STE6-2p from P. pastoris is only a distant relative to its vertebrate homologs with relatively low sequence identity, the similarity of the protein fold is remarkable. Nevertheless, the importance of discovering the differences between ABC-B transporters of distinct species should not be underestimated. This statement holds true for the possible sterol-binding site discovered here. The shift or loss of the conserved proline in TM4 can be found in several fungal proteins of homologous proximity to STE6-2p, including pathogenic species (SI Appendix, Fig. S7). Hence, the observed differences in helical bending are probably found in several fungal ABC-B members.
We report an ABC-B transporter structure with bound verapamil, of which one molecule was identified in the binding pocket (Fig. 1H). Eight residues within a distance of 3.5 Å are identified to interact with verapamil, distributed over four TMs: I303 of TM5, Y774 of TM8, L872 and G876 of TM10, Q990, S991, F987, and C989 of TM12. They coordinate the ligand mainly by van der Waals interactions. A crucial residue appears to be Q990, which establishes a hydrogen bond with verapamil. Pi-stacking can be observed at a larger distance of ∼4.1 Å with W304, a residue that flips over during verapamil binding (Fig. 3). ABC-B1 is known to transport both verapamil enantiomers (46). In our study, we used a racemic mix and, regarding the cryo-EM density as well as the biochemical environment, modeled (R)-verapamil into the structure of STE6-2p. The nitrile at the enantiomeric center of (R)-verapamil establishes a hydrogen bond with Y774. The other side of the (R)-enantiomeric center of verapamil displays a propyl group. It faces toward I303, a nonpolar side chain. In (S)-verapamil, the propyl and nitrile groups are interchanged, thus, the propyl group of (S)-verapamil would face the polar Y774, and the nitrile would face the nonpolar I303. Therefore, we found it more appropriate to place (R)-verapamil into the structure of STE6-2p. Comparison of the human MDR1 structure (PDB: 7A65) reveals F770 at the spatial position of Y774. The lack of a hydroxyl group and thus lack of a possible hydrogen bond could explain P-gps lack of enantiomeric selectivity for verapamil.
In conclusion, our study demonstrates the molecular-binding mechanism of verapamil to STE6-2p, an MDR transporter. It provides a possible explanation for the lack of enantiomeric selectivity of human MDR1. Previous studies highlight verapamil to be a promising candidate to combat fungal pathogens such as C. albicans and A. fumigatus (47–49). Further, our experiments suggest that P. pastoris harbors an ABC transporter potentially capable of facilitating intrinsic azole resistance. It is possible that this fact applies to further opportunistic yeasts containing MDR1 homologs. Additionally, the retention and transport assay with rhodamine 123 and rhodamine 6G used by us support STE6-2p as MDR transporter and served as a method to highlight potential inhibitors. Nevertheless, to elucidate the specific interaction and prove the transport of azoles by STE6-2p, as well as further interpretation of the inhibitory effects of azoles on this transporter, requires additional comprehensive studies of antimycotics with the emerging model organism P. pastoris, and thus, calls for more research of this understudied subject of fungal multidrug resistance in the future. Finally, the observation of a sterol-binding site is thus far unique to STE6-2p. Its role in protein function is subject to further research.
Materials and Methods
Cloning.
Primers were designed based on the coding sequence of ABC-B transporter STE6-2p from P. pastoris (UniProt ID: C4R9D9 or F2QQK6; Gene ID: PAS_c034_0016 or PP7435_CHR1-1574). All primers are listed in SI Appendix, Table S2. Genomic DNA of P. pastoris strain X33, a K. phaffii CBS 7435 variant by Invitrogen (ATCC: 981350) was isolated using a QIAGEN DNeasy Blood and Tissue Kit. The coding sequence of STE6-2p was amplified using primer pair 1 and 2, and the reverse primer 2 was designed in a way that a 10× His-tag was introduced at the C terminus of the protein. The resulting DNA fragment was subcloned into the pPICZ vector using restriction-free cloning (50). Subsequently, a Flag-tag was added to the N terminus of STE6-2 using primer pair 3 and 4. Finally, the ribosomal translation ramp sequence ATGGTGAAAAACTAC (51) was inserted to further increase the protein yield using primer pair 5 and 6, resulting in the final expression vector pPICZ_STE6-2.
The expression vector was transformed into P. pastoris strain X33 following the lithium acetate method as described previously (52). In brief, yeast cells were grown in YPD medium (1% yeast extract, 2% peptone, 2% dextrose) overnight. Cells were permeabilized by a 30-min incubation in LiAc-buffer (10 mM Tris pH 7.5, 100 mM lithium acetate, 10 mM dithiothreitol [DTT], 600 mM sorbitol) at a ratio of 8 × 108 cells per transformation. Treated cells were washed with ice-cold 1 M sorbitol and mixed with 13 µg linearized plasmid DNA. Electroporation was done with 1.5 kV, 25 µF, and 200 Ω in a BIO-RAD Gene Pulser instrument. Transformed yeasts were selected on YDPS-agar plates (1% yeast extract, 2% peptone, 2% dextrose, 1 M sorbitol, 2% agar) containing various zeocin concentrations.
Expression and Purification of STE6-2p.
For protein production, cells from a frozen stock were plated on YPD agar and incubated at 30 °C for 2 d. From there, cells were grown in YPD medium at 29 °C for 24 h. The next evening, cells were precultured in BMGY medium (1% yeast extract, 2% peptone, 1.34% yeast nitrogen base without amino acid, 0.00004% biotin, 1% glycerol, 0.1 M phosphate buffer, pH 6) at 29 °C. Induction was achieved in BMMY medium (as BMGY with 1% methanol instead of glycerol) supplemented with casamino acids. Cultures were incubated at 29 °C for 2 d with methanol being adjusted to 1% after 24 h. After expression, cells were harvested at 1,500 × g for 15 min and cell pellets were resuspended in 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes), pH 7.4, 100 mM NaCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 5% glycerol and 1 mM phenylmethylsulfonyl fluoride (PMSF). Cells were broken surpassing a continuous-flow glass bead mill. The lysate was centrifuged at 1,500 × g and 4 °C for 10 min, followed by a high-spin centrifugation of the resulting supernatant at 100,000 × g and 4 °C for 1 h. Membrane pellets were homogenized at 0.1 g/mL in Hepes buffer.
Proteins were solubilized in 50 mM Hepes, pH 8, 120 mM NaCl, 10% glycerol, 0.5 mM EDTA, 1 mM β-mercaptoethanol, 0.5% lauryl maltose neopentyl glycol (LMNG), and 1 mM PMSF in a detergent:protein mass ratio of 1:1,000. The suspension was then centrifuged at 100,000 × g and 4 °C for 45 min, and the resulting supernatant was filtered. STE6-2p was purified over TOYOPEARL AF-chelate-650M resin (TOSOH Bioscience) and HiTrap Q FF (GE Healthcare) using ÄKTA systems. For cryo-EM analysis, protein was further purified over Superdex 200 Increase 3.2/300 (Cytiva) in 20 mM Hepes, pH 8, 100 mM NaCl, 0.0042% GDN, 0.25 mM Tris-(2-carboxyethyl)-phosphin (TCEP). Protein concentration was measured with a nanodrop photometer, using the theoretical extinction coefficient of 126,520 M−1·cm−1 at 280 nm generated by the ProtParam online tool (53). Additionally, quantitative amino acid analysis was used to accurately determine the concentration of purified STE6-2p at the Functional Genomics Center Zürich.
Activity Measurements.
The ATP-hydrolysis activity was measured using the malachite green colorimetric assay (54). A mix of ATP, MgCl2, and the putative substrate was added to the ion-exchange chromatography (IEX) purified protein, so that 0.5 µM protein was present in 25 µL reaction mix (20 mM Hepes, pH 8, 100 mM NaCl, 3 mM MgCl2, 2.5 mM ATP, 0.25 mM TCEP, 0.0042% GDN). Reactions were conducted at 28 °C for 10 min and stopped by the addition of 175 µL 20 mM H2SO4. A total of 175 µL of the resulting samples were mixed with 50 µL malachite-working solution (1.17 mg/mL malachite green, 1.5% Na2MoO4, 0.18% Tween, 14.2% H2SO4). After 8 min, samples were measured at 620 nm. The activity was calculated in mole protein per mole generated phosphate and minute (mol·mol−1·min−1). Inhibition of STE6-2p activity was measured with up to 500 µM putative inhibitor or 2.36 mM AMP-PNP in the presence of 250 µM verapamil. Each data point was measured in triplicate and in three independent assays, using protein from different preparations. The compounds tested for activity induction of STE6-2p were the a-factor (Zymo Research); the alpha-factor (Zymo Research); amorolfine (Arcos Organics); carbamazepine (Sigma); colchicine (Sigma); ergosterol (Tokyo Chemical Industry); fluconazole (Tokyo Chemical Industry); flucytosine (Sigma-Aldrich); itraconazole (Tokyo Chemical Industry); ketoconazole (Tokyo Chemical Industry); naftifine (Arcos Organics); terbinafine (Arcos Organics); verapamil (Sigma); voriconazole (Tokyo Chemical Industry); and POPC, POPE, POPS, and yeast total lipid extract (all from Avanti Polar Lipids).
Rhodamin 6G/123 Accumulation and Transport Assay.
For each sample, 1 × 107 yeast cells were centrifuged at 3,500 × g for 3 min and washed two times in 100 µL 20 mM Hepes, pH 7.5, 200 mM NaCl. The cell pellet was resuspended in 100 µL 20 mM Hepes, pH 7.5, 200 mM NaCl and 5 µM rhodamine 6G or 5 µM rhodamine 123 with and without 500 µM ketoconazole or 500 µM verapamil, respectively. After incubation in the dark at 22 °C for 2 h, cells were washed with 1 mL of 20 mM Hepes, pH 7.5, and 200 mM NaCl. The cell pellet was resuspended in 400 µL of the same buffer and flow cytometric analysis was performed using the S3e cell sorter (Bio-Rad). Instrument settings were as follows: Forward scatter at 303 V, side scatter at 323 V, fluorescence channel (FL) 1 at 580 V, FL2 at 550 V. Fluorescence was excited by the laser at 488 nm and analyzed by the 586/25 nm band-pass filter over the FL2 channel for rhodamine 6G or the 525/30 nm band-pass filter over the FL1 channel for rhodamine 123. A total of 10,000 cells were used for each measurement at a sampling rate of 1,000 events/s. Data were analyzed using ProSort v1.6.0.12 software and visualized using the online tool Floreada.io (https://floreada.io/analysis).
Cryo-EM Sample Preparation and Data Collection.
Before cryo-EM, purified STE6-2p was analyzed via negative staining using uranyl formate in a FEI Tecnai Spirit TEM.
C-Flat holey carbon copper grids 1.2/1.3 300 mesh were glow discharged twice with a PELCO easiGlow device at 15 mA for 45 s. 3 µL sample with a protein concentration of 2.8 mg/mL was applied on a grid and blotted with a Vitrobot Mk4 (Thermo Scientific) at 100% humidity, 4 °C for 7 s using a nominal blot force of -2. The grids were immediately plunged in liquid ethane and stored in liquid nitrogen until data acquisition. For substrate-bound STE6-2p, the protein sample was mixed with 1 mM AMP-PNP, 3 mM MgCl2 and 20-fold molar excess verapamil prior to vitrification.
Data were collected with a Titan Krios G3i (Thermo Scientific) 300 kV transmission electron microscope equipped with a Gatan BioQuantum imaging filter and K3 direct electron detector. The nominal magnification was 105,000× with a calibrated pixel size of 0.837 Å. Automated image collection was performed using EPU software (Thermo Fisher) in counted superresolution mode with aberration-free image shift (AFIS). Movies were recorded in 40 frames for 2 s at a total dose of 40 e−/Å2. A defocus range of −1.1 to −2.1 µm was applied.
Cryo-EM Image Processing.
The collected 8,218 images were corrected for electron beam–induced sample motion with MotionCor2 (55) in RELION 3.1 software (56) with dose weighting (1e−/Å2). Contrast transfer function (CTF) was estimated with Gctf estimation (57). Low-quality images were discarded and 1.24 M particles were picked with crYOLO picker (58). After two-dimensional (2D) classification, 612 k particles were further sorted in two rounds of three-dimensional (3D) classification using a previously generated initial model of STE6-2p (discussed below). After extensive refinement, CTF estimation, Bayesian polishing (59), and more particle sorting, the final refinement was performed using 89,358 particles reaching a resolution of 3.1 Å (Fourier shell correlation (FSC): 0.143). The final map was sharpened using the LocRes-function (60) applying a b-factor of −65. An overview of the workflow is shown in SI Appendix, Fig. S3 A–H.
The dataset for the determination of STE6-2p in complex with verapamil and AMP-PNP comprised 9,471 movies. After movie alignment and Gctf estimation, low-quality images were discarded. A total of 2.49 M particles were picked with crYOLO. After 2D classification a particle stack of 988 k particles was sorted via 3D classification using a previously generated initial model (discussed below). Extensive refinement, particle sorting, Bayesian polishing, CTF refinement, and micelle subtraction resulted in a particle stack of 128,231 particles, generating a final map with a resolution of 3.2 Å (FSC: 0.143). It was sharpened using the LocRes-function with a b-factor of −75. An overview of the workflow is shown in SI Appendix, Fig. S4.
The initial model used for processing was generated from a separate dataset. Processing of 2,198 movies and particle picking was performed as described above. After intensive 2D classification a de novo initial model was generated in RELION (61). Subsequent 3D and 2D classifications of the initial particle stack and another de novo model generation led to the initial model used for processing. An overview of the workflow is given in SI Appendix, Fig. S3I.
Model Building, Refinement and Validation.
A homology model was prebuilt in SWISS-MODEL (62) online tool (reference model PDB: 5KPI) and fitted into the final cryo-EM density of STE6-2p ATP-free state using Chimera (63). The model was manually refined in Coot (64) and real-space refined using Phenix (65), whereby refinement results were manually corrected if necessary. Validation was done using Phenix comprehensive validation and the MolProbity online server (66). The substrate-bound structure of STE6-2p is based on the apo structure. Refinement and validation were performed as described above. Statistics of model parameters and cryo-EM maps are summarized in SI Appendix, Table S1. The final models were visualized with ChimeraX (67).
Homology.
The amino acid sequence of STE6-2p was searched with BLASTp (68) against the UniProt-KB/Swiss-Prot database (69). Results were filtered by e-value (<0.001), sequence coverage (>85%), and length (>1,000 amino acids). Subsequently, orthologs were identified manually by screening OrthoDB group “186078at2759” at Eukaryota level (70). Only protein sequences with a verified UniProt-ID were included. Homologous sequences, which were not found in the OrthoDB database group, were looked up in the InParanoid8 database (71). All homologous protein sequences were aligned with ClustalW and a maximum-likelihood tree (50× bootstrap) was generated utilizing MEGA software (72). The phylogenetic consensus tree was visualized in FigTree software.
Supplementary Material
Acknowledgments
We thank Gabriele Maul and Birgit Wolff for their support and assistance in the laboratory; the staff of the Central Electron Microscopy Facility at the Max Planck Institute of Biophysics, Dr. Susann Kaltwasser and Simone Prinz, for their support and assistance in data collection; Dr. Di Wu for guidance in cryo-EM data processing; and Dr. Uli Ermler for his support in structure building.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2202822119/-/DCSupplemental.
Data, Materials, and Software Availability
The cryo-EM maps used in this study have been deposited in the Electron Microscopy Data Bank (EMDB), where they can be found under accession nos. EMD-14050 (74) and EMD-14049 (75). Model files of STE6-2p in apo conformation and STE6-2p in substrate-bound conformation were deposited under accession nos. 7QKS (76) and 7QKR (77) in the RCSB PDB. All other data are included in the article and supporting information.
References
- 1.Ford R. C., Beis K., Learning the ABCs one at a time: Structure and mechanism of ABC transporters. Biochem. Soc. Trans. 47, 23–36 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Hodges L. M., et al. , Very important pharmacogene summary: ABCB1 (MDR1, P-glycoprotein). Pharmacogenet. Genomics 21, 152–161 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fridkin S. K., Kaufman D., Edwards J. R., Shetty S., Horan T., Changing incidence of Candida bloodstream infections among NICU patients in the United States: 1995-2004. Pediatrics 117, 1680–1687 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Pappas P. G., et al. , Invasive fungal infections among organ transplant recipients: Results of the Transplant-Associated Infection Surveillance Network (TRANSNET). Clin. Infect. Dis. 50, 1101–1111 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Armstrong-James D., Meintjes G., Brown G. D., A neglected epidemic: Fungal infections in HIV/AIDS. Trends Microbiol. 22, 120–127 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Charles P. E., et al. , Candida spp. colonization significance in critically ill medical patients: A prospective study. Intensive Care Med. 31, 393–400 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Miceli M. H., Díaz J. A., Lee S. A., Emerging opportunistic yeast infections. Lancet Infect. Dis. 11, 142–151 (2011). [DOI] [PubMed] [Google Scholar]
- 8.G. Dimopoulos, M.-P. Almyroudi, P. Myrianthefs, J. Rello, COVID-19-associated pulmonary Aspergillosis (CAPA). Intens. Med. 1, 71–80 (2021). [DOI] [PMC free article] [PubMed]
- 9.Kim J., Sudbery P., Candida albicans, a major human fungal pathogen. J. Microbiol. 49, 171–177 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Silva S., et al. , Candida glabrata, Candida parapsilosis and Candida tropicalis: Biology, epidemiology, pathogenicity and antifungal resistance. FEMS Microbiol. Rev. 36, 288–305 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Kean R., Brown J., Gulmez D., Ware A., Ramage G., . Candida auris: A decade of understanding of an enigmatic pathogenic yeast. JoF. 6, 30 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adams E., et al. ; Candida auris Investigation Workgroup, Candida auris in healthcare facilities, New York, USA, 2013-2017. Emerg. Infect. Dis. 24, 1816–1824 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seiler G. T., Ostrosky-Zeichner L., Investigational agents for the treatment of resistant yeasts and molds. Curr. Fungal Infect. Rep. 15, 104–115 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pemán J., Cantón E., Espinel-Ingroff A., Antifungal drug resistance mechanisms. Expert Rev. Anti Infect. Ther. 7, 453–460 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Healey K. R., Perlin D. S., . Fungal resistance to echinocandins and the MDR phenomenon in Candida glabrata. JoF. 4, 105 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfaller M. A., Antifungal drug resistance: Mechanisms, epidemiology, and consequences for treatment. Am. J. Med. 125(1, suppl.)S3–S13 (2012). [DOI] [PubMed] [Google Scholar]
- 17.Perlin D. S., Rautemaa-Richardson R., Alastruey-Izquierdo A., The global problem of antifungal resistance: Prevalence, mechanisms, and management. Lancet Infect. Dis. 17, e383–e392 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Karbalaei M., Rezaee S. A., Farsiani H., Pichia pastoris: A highly successful expression system for optimal synthesis of heterologous proteins. J. Cell. Physiol. 235, 5867–5881 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Schutter K., et al. , Genome sequence of the recombinant protein production host Pichia pastoris. Nat. Biotechnol. 27, 561–566 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Tweedie S., et al. , Genenames.org: The HGNC and VGNC resources in 2021. Nucleic Acids Res. 49 (D1), D939–D946 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kovalchuk A., Driessen A. J. M., Phylogenetic analysis of fungal ABC transporters. BMC Genomics 11, 177 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Locher K. P., Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 23, 487–493 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Thomas C., Tampé R., Structural and mechanistic principles of ABC transporters. Annu. Rev. Biochem. 89, 605–636 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Smith P. C., et al. , ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol. Cell 10, 139–149 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumari S., Kumar M., Gaur N. A., Prasad R., Multiple roles of ABC transporters in yeast. Fungal Genet. Biol. 150, 103550 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Heistinger L., et al. , Identification and characterization of the Komagataella phaffii mating pheromone genes. FEMS Yeast Res. 18, foy051 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimura Y., Kioka N., Kato H., Matsuo M., Ueda K., Modulation of drug-stimulated ATPase activity of human MDR1/P-glycoprotein by cholesterol. Biochem. J. 401, 597–605 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ernst R., et al. , A mutation of the H-loop selectively affects rhodamine transport by the yeast multidrug ABC transporter Pdr5. Proc. Natl. Acad. Sci. U.S.A. 105, 5069–5074 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin M. S., Oldham M. L., Zhang Q., Chen J., Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans. Nature 490, 566–569 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L., et al. , Cryo-EM structure of human bile salts exporter ABCB11. Cell Res. 30, 623–625 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zoghbi M. E., et al. , Substrate-induced conformational changes in the nucleotide-binding domains of lipid bilayer-associated P-glycoprotein during ATP hydrolysis. J. Biol. Chem. 292, 20412–20424 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alam A., Kowal J., Broude E., Roninson I., Locher K. P., Structural insight into substrate and inhibitor discrimination by human P-glycoprotein. Science 363, 753–756 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fantini J., Barrantes F. J., How cholesterol interacts with membrane proteins: An exploration of cholesterol-binding sites including CRAC, CARC, and tilted domains. Front. Physiol. 4, 31 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Esser L., et al. , Structures of the multidrug transporter P-glycoprotein reveal asymmetric ATP binding and the mechanism of polyspecificity. J. Biol. Chem. 292, 446–461 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nosol K., et al. , Cryo-EM structures reveal distinct mechanisms of inhibition of the human multidrug transporter ABCB1. Proc. Natl. Acad. Sci. U.S.A. 117, 26245–26253 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sasitharan K., Iqbal H. A., Bifsa F., Olszewska A., Linton K. J., . ABCB1 does not require the side-chain hydrogen-bond donors Gln347, Gln725, Gln990 to confer cellular resistance to the anticancer drug taxol. IJMS. 22, 8561 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zolnerciks J. K., et al. , The Q loops of the human multidrug resistance transporter ABCB1 are necessary to couple drug binding to the ATP catalytic cycle. FASEB J. 28, 4335–4346 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Maliyakkal N., Appadath Beeran A., Udupa N., Nanoparticles of cisplatin augment drug accumulations and inhibit multidrug resistance transporters in human glioblastoma cells. Saudi Pharm. J. 29, 857–873 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clay A. T., Lu P., Sharom F. J., Interaction of the P-glycoprotein multidrug transporter with sterols. Biochemistry 54, 6586–6597 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Nosol K., et al. , Structures of ABCB4 provide insight into phosphatidylcholine translocation. Proc. Natl. Acad. Sci. U.S.A. 118, e2106702118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lemel L., et al. , The ligand-bound state of a G protein-coupled receptor stabilizes the interaction of functional cholesterol molecules. J. Lipid Res. 62, 100059 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laursen L., et al. , Cholesterol binding to a conserved site modulates the conformation, pharmacology, and transport kinetics of the human serotonin transporter. J. Biol. Chem. 293, 3510–3523 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levitan I., Singh D. K., Rosenhouse-Dantsker A., Cholesterol binding to ion channels. Front. Physiol. 5, 65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cosco J., et al. , ATP modulates SLC7A5 (LAT1) synergistically with cholesterol. Sci. Rep. 10, 16738 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeppelin T., Ladefoged L. K., Sinning S., Periole X., Schiøtt B., A direct interaction of cholesterol with the dopamine transporter prevents its out-to-inward transition. PLOS Comput. Biol. 14, e1005907 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luurtsema G., et al. , (R)- and (S)-[11C]verapamil as PET-tracers for measuring P-glycoprotein function: In vitro and in vivo evaluation. Nucl. Med. Biol. 30, 747–751 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Vega-Chacón Y., de Albuquerque M. C., Pavarina A. C., Goldman G. H., Mima E. G. O., Verapamil inhibits efflux pumps in Candida albicans, exhibits synergism with fluconazole, and increases survival of Galleria mellonella. Virulence 12, 231–243 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeng Q., et al. , In vitro and in vivo efficacy of a synergistic combination of itraconazole and verapamil against Aspergillus fumigatus. Front. Microbiol. 10, 1266 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nazik H., Choudhary V., Stevens D. A., Verapamil inhibits aspergillus biofilm, but antagonizes voriconazole. J. Fungi (Basel) 3, 50 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Unger T., Jacobovitch Y., Dantes A., Bernheim R., Peleg Y., Applications of the Restriction Free (RF) cloning procedure for molecular manipulations and protein expression. J. Struct. Biol. 172, 34–44 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Verma M., et al. , A short translational ramp determines the efficiency of protein synthesis. Nat. Commun. 10, 5774 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu S., Letchworth G. J., High efficiency transformation by electroporation of Pichia pastoris pretreated with lithium acetate and dithiothreitol. Biotechniques 36, 152–154 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Gasteiger E., et al. , “Protein identification and analysis tools on the expasy server” in The Proteomics Protocols Handbook, Walker J. M., Ed. (Humana Press, 2005), pp. 571–607. [Google Scholar]
- 54.Chifflet S., Torriglia A., Chiesa R., Tolosa S., A method for the determination of inorganic phosphate in the presence of labile organic phosphate and high concentrations of protein: Application to lens ATPases. Anal. Biochem. 168, 1–4 (1988). [DOI] [PubMed] [Google Scholar]
- 55.Zheng S., Palovcak E., Armache J.-P., Cheng Y., Agard D., Anisotropic correction of beam-induced motion for improved single-particle electron cryo-microscopy. BioRxiv [Preprint] (2016), 10.1101/061960. Accessed 10 August 2021. [DOI]
- 56.Scheres S. H. W., Amyloid structure determination in RELION-3.1. Acta Crystallogr. D Struct. Biol. 76, 94–101 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang K., Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wagner T., et al. , SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2, 218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scheres S. H. W., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kucukelbir A., Sigworth F. J., Tagare H. D., Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Waterhouse A., et al. , SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46 (W1), W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pettersen E. F., et al. , UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 64.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liebschner D., et al. , Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Williams C. J., et al. , MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pettersen E. F., et al. , UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Altschul S. F., et al. , Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.UniProt Consortium, UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 49 (D1), D480–D489 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kriventseva E. V., et al. , OrthoDB v10: Sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 47 (D1), D807–D811 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sonnhammer E. L. L., Östlund G., InParanoid 8: Orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res. 43, D234–D239 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kumar S., Stecher G., Li M., Knyaz C., Tamura K., MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pei J., Kim B.-H., Grishin N. V., PROMALS3D: A tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 36, 2295–2300 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schleker E. S. M., Reinhart C., Cryo-EM structure of ABC transporter STE6-2p from Pichia pastoris in apo conformation at 3.1 A resolution. EMDB. https://www.ebi.ac.uk/emdb/EMD-14050. Deposited 18 December 2021.
- 75.Schleker E. S. M., Reinhart C., Cryo-EM structure of ABC transporter STE6-2p from Pichia pastoris with Verapamil at 3.2 A resolution. EMDB. https://www.ebi.ac.uk/emdb/EMD-14049. Deposited 18 December 2021.
- 76.Schleker E. S. M., Reinhart C., Cryo-EM structure of ABC transporter STE6-2p from Pichia pastoris in apo conformation at 3.1 A resolution. RCSB PDB. https://www.rcsb.org/structure/7QKS. Deposited 18 December 2021.
- 77.Schleker E. S. M., Reinhart C., Cryo-EM structure of ABC transporter STE6-2p from Pichia pastoris with Verapamil at 3.2 A resolution. RCSB PDB. https://www.rcsb.org/structure/7QKR. Deposited 18 December 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM maps used in this study have been deposited in the Electron Microscopy Data Bank (EMDB), where they can be found under accession nos. EMD-14050 (74) and EMD-14049 (75). Model files of STE6-2p in apo conformation and STE6-2p in substrate-bound conformation were deposited under accession nos. 7QKS (76) and 7QKR (77) in the RCSB PDB. All other data are included in the article and supporting information.