Significance
Mutations in LRRK2 are associated with Parkinson’s disease. We have recently shown that LRRK2 is recruited and activated on damaged lysosomes; however, the mechanism underlying this process remains unclear. Here, we observe that lysosomal positioning regulates the ability of LRRK2 to phosphorylate and recruit Rab10 but not Rab12 on lysosomes. pRab10 is present almost exclusively at perinuclear LRRK2+ lysosomes, which also regulates LYTL (lysosomal tubulation/sorting driven by LRRK2) by recruiting its effector, JIP4. Manipulation of lysosomal positioning by promoting anterograde transport reduces pRab10 and JIP4 on lysosomes, while induction of retrograde transport has the opposite effect. This finding provides insight into the mechanism of LRRK2-dependent lysosomal damage regulation and supports future study of the role of LRRK2 in lysosomal biology.
Keywords: Parkinson's disease, JIP4, kinase, LLOMe, LYTL
Abstract
Genetic variation at the leucine-rich repeat kinase 2 (LRRK2) locus contributes to an enhanced risk of familial and sporadic Parkinson’s disease. Previous data have demonstrated that recruitment to various membranes of the endolysosomal system results in LRRK2 activation. However, the mechanism(s) underlying LRRK2 activation at endolysosomal membranes and the cellular consequences of these events are still poorly understood. Here, we directed LRRK2 to lysosomes and early endosomes, triggering both LRRK2 autophosphorylation and phosphorylation of the direct LRRK2 substrates Rab10 and Rab12. However, when directed to the lysosomal membrane, pRab10 was restricted to perinuclear lysosomes, whereas pRab12 was visualized on both peripheral and perinuclear LRRK2+ lysosomes, suggesting that lysosomal positioning provides additional regulation of LRRK2-dependent Rab phosphorylation. Anterograde transport of lysosomes to the cell periphery by increasing the expression of ARL8B and SKIP or by knockdown of JIP4 blocked the recruitment and phosphorylation of Rab10 by LRRK2. The absence of pRab10 from the lysosomal membrane prevented the formation of a lysosomal tubulation and sorting process we previously named LYTL. Conversely, overexpression of RILP resulted in lysosomal clustering within the perinuclear area and increased LRRK2-dependent Rab10 recruitment and phosphorylation. The regulation of Rab10 phosphorylation in the perinuclear area depends on counteracting phosphatases, as the knockdown of phosphatase PPM1H significantly increased pRab10 signal and lysosomal tubulation in the perinuclear region. Our findings suggest that LRRK2 can be activated at multiple cellular membranes, including lysosomes, and that lysosomal positioning further provides the regulation of some Rab substrates likely via differential phosphatase activity or effector protein presence in nearby cellular compartments.
Coding mutations in the leucine-rich repeat kinase 2 (LRRK2) gene can cause familial Parkinson’s disease (PD) (1, 2), and noncoding variants in the promoter of the same gene act as risk factors for sporadic PD (3). LRRK2 encodes LRRK2, a large protein consisting of dual enzymatic domains flanked by protein–protein interaction scaffold domains (4). Known LRRK2 pathogenic mutations are located in the Ras-of-complex/guanosine triphosphatase (ROC/GTPase), C-terminal of Roc, and kinase domains, and produce a toxic hyperactive protein (5, 6).
LRRK2 phosphorylates itself at serine 1292 (7) as well as serine/threonine residues in a conserved region of the switch II domain of a subset of Rab GTPases, leading to differential interactions with effector proteins and thus linking LRRK2 to vesicle-mediated transport (8, 9). Thus, LRRK2 has been nominated to play important roles at various endomembranes (4, 10–12). We and others have previously shown that membrane damage within the endolysosomal system results in LRRK2 activation as measured by Rab phosphorylation (13–15). LRRK2-dependent Rab10 phosphorylation at lysosomes results in the recruitment of c-Jun N-terminal kinase interacting protein 4 (JIP4), a motor adaptor protein, that initiates the formation of tubules from the lysosomal surface (lysosomal tubulation/sorting driven by LRRK2 [LYTL]) (13). We have shown that these tubules can resolve into vesicles that make contact with other lysosomes that are cathepsin B+ (CTSB+) (13). LYTL is likely a response to lysosomal membrane damage in which cargo from these lysosomes is delivered to active lysosomes for degradation. LRRK2 may also play an important role in the regulation of lysosomes in vivo, as lysosomal proteins are dysregulated in LRRK2 knockout mice (16) or after chronic LRRK2 kinase inhibition (17).
Although these prior data imply that LRRK2 is part of the cellular response to membrane damage, the mechanism(s) by which LRRK2 becomes activated at membranes are not fully understood. One study using a mitochondrially tagged Rab29 protein showed that directing LRRK2 to the mitochondrial membrane is sufficient to induce its activation and downstream Rab10 phosphorylation (18), suggesting that specific membrane identity is unimportant in LRRK2 activation. We have recently confirmed this finding at other membranes, in which directing LRRK2 to early, late, and recycling endosomes, plasma membrane, Golgi, and lysosomes was sufficient to activate LRRK2 kinase (19). However, how Rab phosphorylation patterns vary across different sub-cellular compartments remains to be defined.
Here, we designed two independent methods to deliver LRRK2 to early endosomes and lysosomes to investigate membrane-dependent patterns of direct LRRK2 substrates Rab10 and Rab12 at sites T73 and S106, respectively. We found that LRRK2 becomes kinase active at either membrane, as indicated by autophosphorylation at site S1292 and was able to phosphorylate both Rab substrates. However, only a subset of perinuclear LRRK2+ lysosomes could recruit pRab10, whereas pRab12 was shown at all LRRK2+ lysosomes throughout the cell. These patterns were not observed when directing LRRK2 to early endosomes. After manipulating the lysosomal positioning, we found that peripherally targeted LRRK2+ lysosomes were unable to recruit pRab10, and that clustering lysosomes into the perinuclear area significantly increased pRab10 signal and colocalization. Interestingly, pRab12 was not sensitive to lysosomal position, suggesting that additional signaling events may control the phosphorylation of specific Rabs on subsets of lysosomes. To address a possible mechanism for this observation, we knocked down PPM1H (protein phosphatase, Mg2+/Mn2+-dependent 1H), a phosphatase known to dephosphorylate T73 Rab10 (20). PPM1H knockdown resulted in a significant increase in pRab10 presence as well as frequency of pRab10/JIP4+ lysosomal tubules within the perinuclear area. These findings suggest that LRRK2-dependent recruitment and subsequent phosphorylation of Rab10 at lysosomes are in part affected by lysosome position, and that PPM1H plays a role as a limiting factor in pRab10-dependent tubulation at perinuclear lysosomes. As expected, the presence of pRab10 in lysosomes is heavily correlated with its effector JIP4. Therefore, by regulating the presence of pRab10 and JIP4 in lysosomes, lysosomal positioning is also regulating LYTL.
Results
LRRK2 Is Activated at Early Endosomal or Lysosomal Membranes Using Two Orthogonal Methods of Relocalization.
LRRK2 has been reported to play a role in various compartments of the endolysosomal system (12). To study the activation of LRRK2 in this context, we designed two orthogonal methods to direct LRRK2 to lysosomal or early endosomal membranes. We chose these two distinct membranes as they are spatially and functionally distinct portions of the endolysosomal pathway. First, we used previously defined traps that use FKBP and FKBP-rapamycin-binding (FRB) rapamycin-binding domains from FK506 binding protein 12 (FKBP12) and mammalian target of rapamycin, respectively (21). Briefly, in the presence of rapamycin, these domains form a heterodimer, and rapidly and irreversibly direct a target protein to a target membrane (21). We therefore tagged LRRK2 with the FKBP sequence (3xFLAG-FKBP-LRRK2) and fused the FRB sequence to the lysosomal and early endosomal membrane markers lysosomal-associated membrane protein 1 (LAMP1) and Rab5, respectively, which we have previously characterized (19). Second, we cloned two chimera-LRRK2 constructs by tagging the N terminus of LRRK2 with a 39–amino acid (aa) transmembrane sequence from LAMTOR1 for lysosomal targeting and membrane-associated double zinc finger and coiled-coil domains FYVE-CC2 from hepatocyte growth factor (HRS) protein for early endosome targeting (22, 23).
We confirmed the efficiency of the translocation of LRRK2 to the lysosomal membrane using the LAMTOR1(1–39 aa)-LRRK2 chimera (referred to as LYSO-LRRK2 to denote a lysosomal chimera) in comparison to the FKBP/FRB lyso-trap. Cotransfection of FKBP-LRRK2 and LAMP1-FRB-Cyan Fluorescent Protein (CFP) constructs into HEK293FT cells resulted in strong colocalization of LRRK2 to the LAMP1 construct in the presence of rapamycin (Fig. 1A). Similarly, the LYSO-LRRK2 chimera colocalized with endogenous LAMP1, LAMTOR4, and CTSD (Fig. 1A and SI Appendix, Fig. S1 A–C). Having established correct localization of our constructs, we next evaluated whether the direction of LRRK2 to lysosomes was sufficient to activate the kinase activity of LRRK2 in both systems. Cells transfected with the FKBP/FRB constructs and treated with rapamycin showed enhanced LRRK2 activity compared to untreated control cells, as measured by a significant increase in the autophosphorylation of LRRK2 at site S1292 as well as phosphorylation of T73 Rab10 and S106 Rab12 (Fig. 1 B–E). A similar magnitude of LRRK2 activation was also seen with the LYSO-LRRK2 targeting construct compared to a LRRK2 construct without an additional targeting motif that remains cytosolic (referred to as NT-LRRK2 for “nontargeting”) (Fig. 1 B–E). To control for rapamycin treatment, NT-LRRK2-transfected cells were also treated with rapamycin, in which no significant difference in phosphorylation was observed compared to untreated cells, confirming that rapamycin alone does not activate LRRK2 (Fig. 1 B–E). Thus, directing LRRK2 to lysosomes is sufficient to increase kinase activity, even in the absence of lysosomal-damaging agents previously described to activate LRRK2 on lysosomes (13–15).
Fig. 1.
Activation of LRRK2 and subsequent Rab phosphorylation is achieved using FKBP/FRB complex and chimera-LRRK2 constructs regardless of membrane identity. HEK293FT cells transfected with either LAMP1-FRB-CFP and FKBP-LRRK2 plasmids (specified as “lyso-trap”; Top) or LYSO-LRRK2 plasmid (Bottom). Endogenous LAMP1 is also shown in the bottom image of LYSO-LRRK2. Construct designs are shown to the left of the respective ICC images in (A). Scale bar, 10 μm. (B–E) Western blot analyses of LRRK2 S1292 autophosphorylation, Rab10 and Rab12 phosphorylation under conditions of NT-LRRK2 or lyso-trap–expressing cells with or without rapamycin treatment, or LYSO-LRRK2 chimera only. (F) Cells transfected with iRFP-FRB-RAB5 and FKBP-LRRK2 plasmids treated with rapamycin (specified as “EE-trap”; Top) or the EE-LRRK2 chimera (Bottom). Endogenous EEA1 is also shown in the bottom image of EE-LRRK2. Construct designs are shown on the left of the images. Scale bar, 10 μm. DAPI is shown in blue. (G–J) Western blot analysis from densitometry measurements of LRRK2 S1292 autophosphorylation, Rab10 and Rab12 phosphorylation under conditions of NT-LRRK2, or EE-trap–expressing cells with and without rapamycin treatment or the EE-LRRK2 chimera alone. (C–E, H–J) One-way ANOVA with Tukey’s post hoc; n = 3; SD bars shown; (C) F(4, 10) = 19.67, (D) F(4, 10) = 16.87, (E) F(4, 10) = 81.71, (H) F(4, 10) = 25.86, (I) F(4, 10) = 25.22, and (J) F(4, 10) = 16.00. ns = not significant.
We similarly directed LRRK2 to early endosomal membranes using FKBP-LRRK2 and near-infrared fluorescent protein (iRFP)-FRB-Rab5 constructs and a C-terminal FYVE (Fab1, YOTB, Vac1, and EEA1) and CC2 (coiled-coil-2)-LRRK2 chimera (referred to as EE-LRRK2 to reflect an early endosomal chimera). Both methods resulted in LRRK2 colocalization with early endosome markers Rab5 and EEA1, respectively (Fig. 1F). Endogenous vacuolar protein sorting 35 (VPS35) was also stained as a secondary marker of early endosomes, and colocalization with the EE-LRRK2 chimera was quantified (SI Appendix, Fig. S1 D and F). The FYVE domain binds to phosphatidylinositol 3-phosphate (PI3P) on the surface of a subset of endosomes, with particular accumulation on early endosomes (24, 25). As expected, the overlap of EE-LRRK2 with the general early endosome marker EEA1 and VPS35+ endosomes (26) resulted in strong correlations, with an estimated average of 0.59 and 0.58 colocalization of total fluorescence in the region of interest, respectively (SI Appendix, Fig. S1F). Since PI3P can be present on other endosomal populations, we further quantified colocalization of EE-LRRK2 and endogenous Rab7, a marker of late endosomes. On average, the estimated fraction of colocalized pixels over total fluorescence was 0.18 when measuring EE-LRRK2 colocalization with Rab7, showing that EE-LRRK2 predominantly sequesters onto early endosomes, although there is some overlap with late endosomes (SI Appendix, Fig. S1 E and F). Cotransfection of FKBP/FRB-tagged plasmids significantly increased S1292 LRRK2 autophosphorylation and phosphorylation of Rab10 and Rab12 when cells were treated with rapamycin compared to untreated transfected cells (Fig. 1 G–J). The EE-LRRK2 chimera activated LRRK2 similarly to the rapamycin-treated FKBP/FRB method, whereas NT-LRRK2 expressing did not, even after rapamycin exposure (Fig. 1 H–J).
Collectively, these results indicate that directing LRRK2 to a given membrane results in activation and phosphorylation of multiple Rab proteins, irrespective of membrane identity. Since both methods of LRRK2 translocation were sufficient for downstream phosphorylation of Rab GTPases, we chose to move forward with the lysosome and early endosome chimeric constructs so as to avoid unnecessary cotransfections and treatments (e.g., rapamycin) for subsequent experiments.
LRRK2 and Phosphorylated Rab10 Colocalization Patterns Differ at the Lysosomal and Early Endosomal Membranes.
We next examined the cellular distribution of two LRRK2 substrates, pT73 Rab10 and pS106 Rab12, at endogenous levels using immunostaining. When cells were transfected with LYSO-LRRK2, we noted a significant increase in pT73 Rab10 staining that colocalized with endogenous LAMP2 compared to cells transfected with NT-LRRK2 (Fig. 2 A and B). In addition, when treated with MLi-2, a LRRK2-specific kinase inhibitor, pRab10 signal was completely abolished (Fig. 2 A and B). Signal intensity of pS106 Rab12 was also significantly increased when LRRK2 was directed to lysosomes via the LYSO-LRRK2 construct compared to NT-LRRK2, and MLi-2 significantly decreased this signal (Fig. 2 C and D).
Fig. 2.
Rab phosphorylation is LRRK2 dependent and pRab colocalization patterns differ between Rab10 and Rab12 at the lysosomal membrane. HEK293FT cells were transfected with either NT-LRRK2 or LYSO-LRRK2 chimera, and pT73 Rab10 and pS106 Rab12 integrative densities were measured (A–D). MLi-2 treatment was compared to untreated cells expressing LYSO-LRRK2 (A–D). Scale bar, 10 μm. (E–I) Cells were transfected with either NT-LRRK2 or EE-LRRK2 chimera, and pT73 Rab10 and pS106 Rab12 integrative densities were measured (A–D). MLi-2 treatment was compared to untreated cells expressing EE-LRRK2 (E–I). Scale bar, 10 μm. In all merged images, “n” denotes the nucleus. (B, D, F, and H) One-way ANOVA with Tukey’s post hoc; (I) two-way ANOVA with Tukey’s post hoc; n = 3, n = 13 cells; SD bars shown; (B) F(2, 32) = 33.11, (D) F(2, 32) = 34.92, (F) F(2, 32) = 26.67, (H) F(2, 32) = 52.36, and (I) chimera: F(1, 36) = 93.49, P < 0.0001, Rab: F(1, 36) = 73.37, P < 0.0001.
When cells were transfected with the EE-LRRK2 construct, phosphorylation of both Rab10 and Rab12 were significantly increased and colocalized with endogenous EEA1 compared to the transfection of the untagged LRRK2 construct, and the addition of MLi-2 caused a significant reduction in signals for both pRabs (Fig. 2 E–H). These data suggest that Rab10 and Rab12 are phosphorylated by LRRK2 chimeras and can be recruited to and accumulated by these membranes where LRRK2 kinase is active, likely due to a lack of removal by GDI (Rab GDP dissociation inhibitor), as previously suggested (18).
Inspection of the same images suggested that not all LRRK2+ lysosomes were also pRab10+ (Fig. 2A). Therefore, we next measured LRRK2:pRab colocalization at LRRK2+ lysosomes and LRRK2+ early endosomes using Mander’s coefficient. Quantitatively, we found that only a small portion of LRRK2+ lysosomes colocalized to pT73 Rab10, whereas most LRRK2+ lysosomes showed colocalization with pS106 Rab12 (Fig. 2I). We also observed that this cluster of pRab10 lysosomes was situated in the perinuclear region, and not observed at distal lysosomes (SI Appendix, Fig. S2A). When measuring the signal intensity of pRab10 against distance from the nucleus, we observed that the majority of the pRab10 signal was situated at approximately 4 µm from the nucleus of cells (SI Appendix, Fig. S2B). In addition, we stained HEK293FT and U2OS cells expressing LYSO-LRRK2 with pericentrin, a marker for centrosomes located within the perinuclear region of cells, and observed that pRab10+ lysosomes were most often adjacent to pericentrin, further validating that the location of this subset of lysosomes is perinuclear (SI Appendix, Fig. S2 C and D). Since HEK293FT cells have relatively small lysosomes, we also used U2OS cells expressing LYSO-LRRK2 to improve our ability to analyze pRab10+ structures. Using superresolution microscopy, we were able to confirm the colocalization of pRab10 with LYSO-LRRK2, as well as endogenous lysosomal markers LAMP1, LAMP2, and a tagged version of TMEM192 (SI Appendix, Fig. S2E). In addition, with LRRK2 localized to lysosomes, spontaneous pRab10+ tubules were observed (SI Appendix, Fig. S2F). In contrast, nearly all of the LRRK2+ early endosomes colocalized with both pRab10 and pRab12, suggesting that the differing patterns between LRRK2-mediated phosphorylation of Rab10 and Rab12 at lysosomes is indeed lysosomal specific (Fig. 2I). Staining for total Rab10 on cells transfected with LYSO-LRRK2 revealed a pattern similar to that of pRab10, in which the total protein colocalized only to a subset of perinuclear LRRK2+ lysosomes (SI Appendix, Fig. S3 A and B). In addition, we stained for JIP4, a motor adaptor protein recruited by pRab10 to the lysosomal membrane (13, 27) and found it to cluster with the subset of LRRK2+ lysosomes on cells expressing LYSO-LRRK2 compared to NT-LRRK2 (SI Appendix, Fig. S3 C and D). Staining of JIP4 in U2OS cells also revealed JIP4+ tubules similar to those observed with pRab10 accumulation (SI Appendix, Fig. S3E). These data suggest that while Rab10 is retained at LRRK2+ lysosomes after phosphorylation, there are additional conditions that determine the presence of Rab10 at only a subset of lysosomes in the perinuclear region of the cell. To ensure that LYSO-LRRK2 itself did not affect lysosomal positioning, we transfected cells with either NT-LRRK2 or LYSO-LRRK2 and measured the mean lysosomal distance to the nucleus via endogenous LAMP2 staining (SI Appendix, Fig. S3 F and G). Our data show no significant difference between conditions, suggesting that the cluster of pRab10+ lysosomes situated in the perinuclear region are not caused by any alteration in lysosomes that have LYSO-LRRK2 present.
Expression of Motor Proteins ARL8B and SKIP Promote Dispersion of LRRK2+ Lysosomes to the Cell Periphery.
To delineate the mechanism underlying the recruitment and phosphorylation of Rab10 to a subset of lysosomes, we took a spatial approach by manipulating lysosomal positioning. The small Arf-like GTPase ARL8B and kinesin-1 adaptor protein SKIP have been shown to direct lysosomes to the cell periphery when overexpressed (28, 29). Briefly, ARL8B binds to both the lysosomal membrane and SKIP, which contains two light-chain kinesin-binding domains to promote anterograde movement along microtubules (Fig. 3A). Interestingly, we found considerable morphological changes to HEK293FT cells with the addition of the ARL8B and SKIP proteins (mCherry-ARL8B and 2xMyc-SKIP constructs), in which cells formed long processes with a large accumulation of lysosomes situated at the tips (Fig. 3 B and C). When measuring the number of LRRK2+ lysosomes at the periphery versus total lysosome count, a significant increase was observed in the number of LRRK2+ lysosomes situated at the periphery when expressing ARL8B and SKIP together (Fig. 3 C and D).
Fig. 3.
ARL8B- and SKIP-expressing cells promote lysosomal repositioning to the periphery of cells. Representative schematic describing lysosomal translocation to the cell periphery via expression of ARL8B and SKIP proteins along the plus end of microtubules (A). Airyscan microscopy showing peripheral LYSO-LRRK2 lysosomes, colocalizing with ARL8B and SKIP (B). Cell edges are outlined. Scale bars, 10 μm. LYSO-LRRK2 was transfected in cells alone or cotransfected with ARL8B and SKIP (C). Scale bars, 10 μm. Quantification of the ratio of peripheral to total LRRK2+ lysosomes such as those represented in (C) are shown in (D). Peripheral fluorescence refers to the presence of LRRK2 within 2 μm from the cell vertices. Horizontal lines indicate the mean ± SD from 3 independent experiments.
Phosphorylated Rab10 Does Not Colocalize to Peripheral LRRK2+ Lysosomes while Lysosomal Clustering toward the Perinuclear Area Increases Colocalization.
We next cotransfected LYSO-LRRK2, ARL8B, and SKIP and stained for pT73 Rab10 to determine whether lysosomes situated at the periphery would be able to phosphorylate and retain Rab10. Both pRab10 levels and colocalization with LRRK2 were significantly decreased when cells were transfected with ARL8B and SKIP compared to cells transfected with LYSO-LRRK2 alone (Fig. 4 A–C). This pattern was also observed when staining for total Rab10 levels (SI Appendix, Fig. S3 H and I). Taken together, these data show that lysosomes situated within the perinuclear region have specific properties that allow recruitment and phosphorylation of Rab10 by LRRK2 that are not shared with peripheral lysosomes.
Fig. 4.
ARL8B- and SKIP-expressing cells prevent pRab10 colocalization on peripherally positioned LRRK2+ lysosomes. Representative confocal microscopy images show pT73 Rab10 signal, LYSO-LRRK2 with endogenous LAMP2 (Top), or ARL8B and SKIP (Bottom) (A). Integrative density and Mander’s correlation coefficient were used to measure pRab10 staining intensity (B) and LRRK2:pRab10 colocalization (C). (D) Cells with and without ARL8B/SKIP transfection were treated with 1 mM LLOMe for 2 h. Integrative density and Mander’s correlation coefficient were used to measure pRab10 staining intensity (E) and LRRK2:pRab10 colocalization (F). LYSO-LRRK2 expressing cells with and without ARL8B/SKIP cotransfection and LLOMe treatment were probed for S1292 LRRK2 and pT73 Rab10 for western blot densitometry analysis (G–J). In all merged images, “n” denotes the nucleus. (B, C, E, and F) Two-tailed unpaired t test; n = 13 cells across 2 independent experiments; error bars represent SDs; (B) ***P = 0.0004, (C) **P = 0.0071, (E) ****P < 0.0001, and (F) ****P < 0.0001. (H–J) One-way ANOVA with Tukey’s post hoc; n = 3 to 4; SD bars shown. (H) F(3, 12) = 59.39, (I) F(3, 12) = 4.624, and (J) F(3, 12) = 19.42. Scale bars, 10 μm. ns = not significant.
Previously, we have shown that the addition of the lysosomotropic agent l-leucyl-l-leucine methyl ester (LLOMe) induces LRRK2 localization to the lysosomal membrane and recruitment of Rab10 to initiate a lysosomal tubulation process (LYTL) via JIP4 (13). We thus conjectured that adding LLOMe to cells transfected with SKIP and ARL8B would push Rab10 to LRRK2+ lysosomes despite their location at the periphery. However, acute treatment with LLOMe did not affect the recruitment nor subsequent phosphorylation of Rab10 in cells transfected with SKIP and ARL8B compared to cells expressing LYSO-LRRK2 alone (Fig. 4 D–F). Western blot analysis supported these results further, in that LLOMe treatment significantly increases phosphorylation of Rab10 in cells transfected only with LYSO-LRRK2, but this effect was counteracted by cotransfection with ARL8B and SKIP (Fig. 4 G and H). Interestingly, we do not observe any impact of LRRK2 autophosphorylation at S1292 upon treatment with LLOMe (Fig. 4 G and I), suggesting either that LLOMe induces LRRK2 in a manner that is distinct from LRRK2 mutations that increase pS1292 or that LLOMe increases the proximity between LRRK2 and Rabs to provide more opportunity for LRRK2-dependent Rab phosphorylation without increasing LRRK2 kinase activity itself (30). In addition, it should be noted that LRRK2 transfection efficiency decreased when cotransfected with SKIP and ARL8B plasmids (Fig. 4 G and J), although a potential effect of SKIP/ARL8B overexpression leading to degradation of LRRK2 cannot be ruled out.
When staining for pRab12 in cells transiently expressing ARL8B and SKIP, we found strong colocalization between pRab12 and LRRK2+ peripheral lysosomes regardless of lysosomal position, and acute treatment with LLOMe significantly increased pRab12 in cells transfected with LYSO-LRRK2 alone or with ARL8B and SKIP (Fig. 5 A–D). Interestingly, pRab12 levels were slightly reduced in the ARL8B- and SKIP-expressing cells treated with LLOMe compared to LLOMe alone, suggesting that the accumulation of phosphorylated Rabs in general may be the most optimal in the perinuclear region. However, comparing the relatively stronger effect of positioning on pRab10 levels (Fig. 4 G and H) with the smaller effect for pRab12 levels (Fig. 5 C and D) suggests that there are differing mechanisms at play for the phosphorylation of Rab10 and Rab12 at damaged lysosomes by activated LRRK2.
Fig. 5.
Phosphorylated Rab12 is found at peripheral lysosomes in cells transiently transfected with ARL8B and SKIP plasmids. A superresolution confocal microscopy image shows pS106 Rab12 staining at LRRK2+ lysosomes when ARL8B and SKIP are coexpressed (A). In the merged image, “n” denotes the nucleus. Scale bar, 10 μm. Mander’s coefficient was used to measure LRRK2:pRab12 colocalization in conditions in which ARL8B and SKIP are coexpressed with LYSO-LRRK2 as well as when only LYSO-LRRK2 is expressed alone (B). Western blot analysis from densitometry measurements of Rab12 phosphorylation are shown (C and D). (B) Unpaired, two-tailed t test; n = 3, n = 12 cells each; SD error bars are shown; P = 0.6045. (D–F) One-way ANOVA with Tukey’s post hoc; n = 3; SD bars shown. (D) F(3, 8) = 87.73.
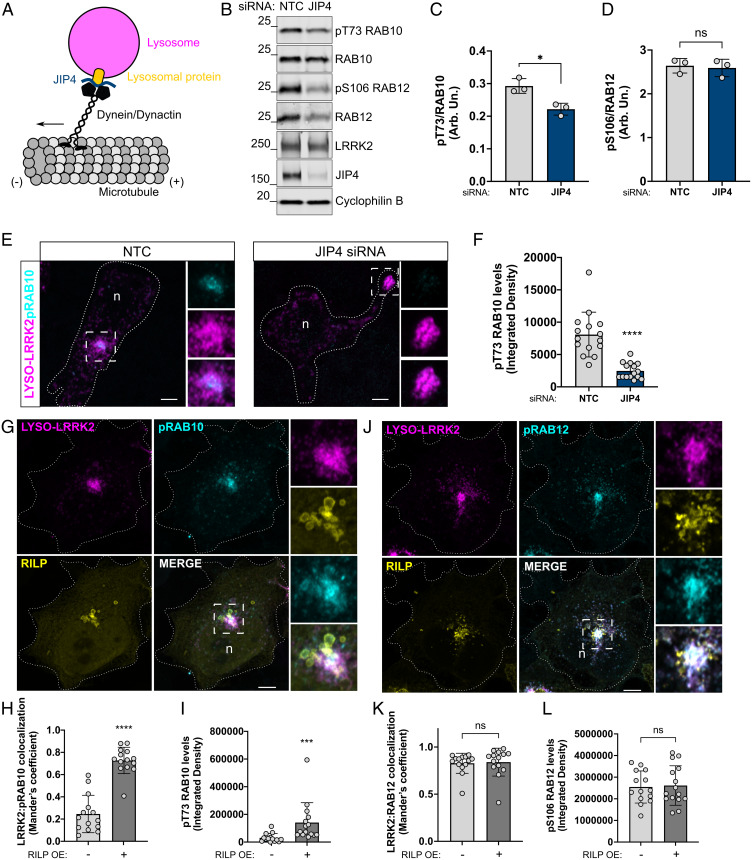
To further confirm these findings of position-specific Rab10 phosphorylation on LRRK2+ lysosomes, using an alternative method, we turned our attention to JIP4. Although we have previously shown that JIP4 binds to pRab10 to induce lysosomal tubulation, JIP4 is a motor adaptor protein that binds to the dynein/dynactin complex to promote the transport of lysosomes toward the minus end of microtubules (31) (Fig. 6A). Using small interfering RNA (siRNA), we knocked down JIP4 to prevent lysosomal retrograde transport to the perinuclear area. This was then followed by transient transfection of LYSO-LRRK2, and pRab10 levels were measured. Successful knockdown of JIP4 resulted in decreased Rab10 phosphorylation levels compared to cells transfected with nontargeting siRNA control (NTC) (Fig. 6 B and C). In addition, normalized Rab12 phosphorylation levels were unaffected by JIP4 knockdown, however, we found that JIP4 knockdown markedly reduced total Rab12 protein levels, suggesting a possible relationship between JIP4 and Rab12 (Fig. 6 B and D). Using immunocytochemistry (ICC), we confirmed that endogenous JIP4 knockdown had a similar effect on lysosomal segregation to the periphery as seen with ARL8B and SKIP overexpression where we measured a significant decrease in pRab10 signal compared to the nontargeting control siRNA condition (Fig. 6 E and F). This finding identifies JIP4 as an influencer of lysosomal positioning and thus LRRK2-dependent phosphorylation of Rab10. To make sure that the artificial expression of LYSO-LRRK2 was not contributing to this phenomenon, we also knocked down JIP4 in cells without additional transfections. This resulted in a significant increase in the average lysosome distance from the nucleus compared to NTC control cells (SI Appendix, Fig. S4 A and B), providing further evidence that JIP4 regulates the lysosome position independent of LRRK2.
Fig. 6.
Knockdown of endogenous JIP4 is sufficient to prevent peripheral lysosomes from returning to the perinuclear area, while RILP-expressing cells promote clustering of LRRK2+ lysosomes within the perinuclear region and significantly increase pRab10 signal. Simplified schematic of lysosomal movement into the perinuclear area via JIP4 adaptor binding to a lysosomal protein and the dynein/dynactin complex (A). Western blot analysis showing NTC (nontargeting control siRNA) versus JIP4 siRNA conditions in HEK293FT cells and densitometry measurements of pRab10 and pRab12 proteins (B–D). Representative confocal microscopy images of HEK293FT cells stained for LYSO-LRRK2 and pRab10 under conditions of NTC and JIP4 siRNA transfection (E), in which knockdown of JIP4 significantly reduces pRab10 signal (F). Cells expressing LYSO-LRRK2 and RILP were stained for pRab10 or pRab12 and colocalization with LRRK2 and integrated densities were measured, respectively (G–L). In all of the merged images, “n” denotes the nucleus. (C, D, F, H, I, K, and L) Two-tailed unpaired Student t test; SD error bars are shown. (C) P = 0.0132, n = 3, (F) P < 0.0001, n = 13 to 15 cells, (H) P < 0.0001, (I) P < 0.0001, (K) P = 0.6045, and (L) P = 0.8273, n = 12 to 15 cells.
LLOMe treatment not only works as a lysosomal membrane damaging reagent but it also promotes the clustering of lysosomes in the perinuclear region and consequently increases Rab10 phosphorylation in the perinuclear region. Therefore, we compared different stimuli to promote lysosomal retrograde transport in HEK293FT cells, including LLOMe treatment, Rab-interacting lysosomal protein (RILP) overexpression, and serum/aa starvation. RILP is a Rab7A effector that affects lysosomal positioning within the cell via the recruitment of dynein and dynactin proteins (32). Overexpressing RILP arrests Rab7A in its GTP-bound active form and sequesters the protein to late endosomal and lysosomal membranes and induces the recruitment of the dynein-dynactin complex, which promotes vesicle translocation along the minus end of microtubules toward the perinuclear region (33). In addition, serum/aa starvation has been shown to affect lysosomal positioning in that lysosomes cluster into the perinuclear region to facilitate autophagosome-lysosome fusion following nutrient deprivation in specific cell types (34). We found that both LLOMe treatment and RILP overexpression significantly increased the clustering of lysosomes into the perinuclear area, whereas starvation did not have a significant effect on lysosomal clustering as determined by measuring the mean distance of endogenous LAMP2 puncta to the nucleus in HEK293FT cells (SI Appendix, Fig. S4 C and D).
Based on these results, we coexpressed LYSO-LRRK2 and RILP and stained for pT73 Rab10. We were excited to find that RILP overexpression is sufficient to cluster LRRK2+ lysosomes into the perinuclear region, resulting in the recruitment and phosphorylation of Rab10, as demonstrated by LRRK2:pRab10 colocalization (Fig. 6 G and H). We also observed that phosphorylation levels of Rab10 were significantly increased compared to cells that were transfected with LYSO-LRRK2 alone (Fig. 6I). These results further demonstrate that lysosomes situated at the perinuclear area possess favorable characteristics for the recruitment and phosphorylation of Rab10 by LRRK2. Overexpression of RILP also clustered pRab12+ lysosomes; however, we observed no difference in colocalization with LRRK2 or levels of phosphorylated Rab12, suggesting, again, that pRab12 is unaffected by lysosomal position (Fig. 6 J–L).
Dispersion of Lysosomes to the Cell Periphery Is Sufficient to Block Recruitment of Rab10 in Untagged LRRK2 and Endogenous LRRK2 Models.
We next wanted to test whether the phenomenon of peripheral lysosomes being recalcitrant to pRab10 formation could be recapitulated using LLOMe treatment in nontargeted, wild-type LRRK2-expressing cells. NT-LRRK2 is primarily cytosolic, and treatment with LLOMe promotes colocalization to the lysosomal membrane (13, 14). Therefore, we treated cells transfected with NT-LRRK2 with LLOMe for 2 h. We observed that LRRK2 relocalized to the lysosomal membrane with or without the addition of ARL8B and SKIP overexpression, shown as colocalization with endogenous LAMP2 (Fig. 7 A and B). Interestingly, when cells were treated with LLOMe, colocalization of LRRK2 and pRab10 was found only in the subset of lysosomes within the perinuclear region, and this was blocked when lysosomes were pushed to the periphery upon ARL8B and SKIP overexpression (Fig. 7 C and D). Furthermore, JIP4 recruitment to LLOMe-treated lysosomes was also prevented by ARL8B and SKIP expression (Fig. 7 E and F), suggesting that LYTL cannot be activated at peripheral lysosomes. Western blot analysis confirmed that the addition of SKIP and ARL8B inhibits pRab10 in cells treated with LLOMe compared to those without ARL8B and SKIP overexpression (Fig. 7 G–I). In addition, we observed that pRab12 was recruited to LRRK2+ lysosomes after LLOMe treatment with and without cotransfection of ARL8B and SKIP proteins (SI Appendix, Fig. S5 A and B). This suggests that regardless of lysosomal position, LLOMe treatment is sufficient to bring cytosolic LRRK2 to the lysosomal membrane to recruit pRab12. Interestingly, the movement of most lysosomes to the periphery does not affect the ability of galectin-3 recruitment to damaged lysosomes after treatment with LLOMe, suggesting that lysosomal membrane damage also occurs in peripheral lysosomes (SI Appendix, Fig. S5C).
Fig. 7.
ARL8B- and SKIP- expressing cells treated with LLOMe recruit NT-LRRK2-expressing HEK293FT and endogenous LRRK2 in primary mouse astrocytes but not pRab10. Representative Airyscan microscopy images of NT-LRRK2 with and without ARL8B/SKIP coexpressing cells under conditions of DMSO or LLOMe treatment, and “n” denotes the nuclei. (A and B) LRRK2+ lysosomes are counted in each condition followed by pT73 Rab10 (C and D), and lastly, JIP4 (E and F). Scale bars, 10 μm. (G–I) Western blot analysis from densitometry measurements of pRab10 from LYSO-LRRK2-expressing cells ± ARL8B/SKIP and ± LLOMe. (J) Primary astrocytes were treated with DMSO or LLOMe and probed for endogenous LAMP1 and pRab10 at endogenous levels of LRRK2. In the image shown with ARL8B, “periph.” and “perinuc.” abbreviations are shown for the insets that are peripheral and perinuclear, respectively. (K) pRab10+ lysosomes were counted in these treatment conditions ± ARL8B/SKIP. Scale bars, 10 μm. (B, D, F, H, I, and K) One-way ANOVA with Tukey’s post hoc; (B, D, F, and K) n = 32 to 40 cells counted. (H and I) n = 3; SD error bars are shown. (B) F(2, 105) = 15.91, (D) F(2, 107) = 141.1, (F) F(2, 94) = 116.4, (H) F(3, 8) = 14.54, (I) F(3, 8) = 0.5329, and (K) F(2, 99) = 27.14.
We then decided to evaluate whether the same effects were seen with LRRK2 at endogenous levels and selected mouse primary astrocytes for their high levels of LRRK2 expression (13, 35). Acute LLOMe treatment for 4 h significantly increased phosphorylated Rab10 at lysosomes compared to those only treated with DMSO (Fig. 7 J and K), whereas ARL8B and SKIP overexpression resulted in a significant reduction in pRab10+ lysosomes, and minimal recruitment to peripheral lysosomes was observed (Fig.7 J and K). Of note, LLOMe treatment clusters lysosomes to the perinuclear region in mouse primary astrocytes in a similar way as that in HEK293FT cells (SI Appendix, Fig. S6 A and B). Taken together, our results indicate that LRRK2-dependent Rab10 recruitment to the lysosomal membrane is position specific and is recapitulated across multiple models, including at endogenous levels of LRRK2.
Knockdown of PPM1H Increases Phosphorylation of Rab10 at Perinuclear Lysosomes and Increases the Frequency of Lysosomal Tubulation via JIP4.
A recent siRNA screen identified that PPM1H phosphatase dephosphorylates T72 Rab8A and T73 Rab10, thus mitigating LRRK2 signaling (20). Of interest, exogenously expressed PPM1H is reported to be located at the Golgi, thus potentially having spatially restricted effects on cellular phosphorylation. We therefore hypothesized that PPM1H may counteract LRRK2-dependent Rab10 phosphorylation on lysosomes in the perinuclear area. Knockdown of PPM1H resulted in a significant increase in pRab10 signal on LYSO-LRRK2 lysosomes within the perinuclear region, but no increase in signal at peripheral lysosomes was observed (Fig. 8 A and B). This was verified by western blot, in which levels of pRab10 significantly increased in cells transfected with PPM1H siRNA compared to nontargeting siRNA control (NTC) control cells (Fig. 8 C and D). As expected, Rab12 phosphorylation levels remained unchanged, as PPM1H is not a Rab12 phosphatase (Fig. 8 C and E). A pattern similar to that of pRab10 was seen when quantifying JIP4 signal via ICC (Fig. 8 F and G). Upon PPM1H knockdown, we also found a significant increase in the frequency of pRab10+ and JIP4+ lysosomal tubules (Fig. 8 H–K). This finding demonstrates that PPM1H limits pRab10 phosphorylation at the perinuclear lysosomes and thereby counteracts LYTL.
Fig. 8.
PPM1H knockdown affects pRab10-mediated lysosomal tubulation. Representative confocal images showing LYSO-LRRK2 and pT73 Rab10 after siRNA knockdown of NTC or PPM1H, where “n” denotes the nuclei (A). The integrated density of pRab10 signal was measured in ICC (B) and western blot densitometry measurements, with additional probing for Rab12 (C–E). Representative confocal images of JIP4 are shown accompanied by integrated density measurements in cells transfected with NTC or PPM1H siRNA (F and G). Airyscan images are shown of pT73 Rab10+ and JIP4+ tubules, and the number of tubules was counted (H–K). (B, D, E, G, J, and K) Two-tailed unpaired t test; n = 16 (B and G); (D and E) n = 3; n = 35 (J and K); SD bars shown. (B) P = 0.0013, (D) P = 0.0141, (E) P = 0.6540, (G) P = 0.0066, (J) P = 0.0055, and (K) P = 0.0038.
Discussion
Understanding the patterns of LRRK2 kinase activity is of crucial interest in PD pathogenesis and future therapeutics targeted at this enzyme. Recent work from our laboratory and other laboratories has shown that LRRK2 can be targeted to different membranous compartments, where it can phosphorylate and recruit several Rab substrates under specific cell signaling conditions (13–15, 36–38). However, the dynamics underlying LRRK2 membrane presence, activation, and Rab phosphorylation remain unclear. In the present study, we have shown that LRRK2-dependent Rab10 phosphorylation is influenced by lysosomal positioning and the action of spatially restricted phosphatases.
In addition, we characterized pRab localization patterns after LRRK2 was translocated to the lysosomal and early endosomal membranes. We observed a striking difference in colocalization pattern between Rabs, with pT73 Rab10 showing strict perinuclear localization in a subset of lysosomes, whereas pS106 Rab12 was found on LRRK2+ lysosomes throughout the cell, suggesting that LRRK2 may have a more direct influence over Rab12 phosphorylation. Recently, we proposed that pRab12 more accurately than pRab10 in vivo reports the effects of hyperactivation as well as inhibition of LRRK2 kinase(17). Taken together, our current data suggest that Rab12 is spatially more readily available to LRRK2 mediation once LRRK2 is translocated to the lysosomal membrane, whereas Rab10 is regulated by mechanisms distinct from Rab12 that affect its phosphorylation by LRRK2.
We have identified a spatially dependent, lysosomal-specific mechanism through which LRRK2 recruits and phosphorylates Rab10. When LRRK2+ lysosomes are pushed to the periphery via ARL8B and SKIP overexpression, Rab10 can no longer be recruited, in contrast to Rab12. This phenomenon was also seen with the knockdown of endogenous JIP4, revealing a role for JIP4 in LRRK2-dependent phosphorylation of Rab10 at lysosomes positioned near the nucleus. We speculate that there may be a feedforward signaling pathway where lysosomal positioning can be promoted by JIP4 to the perinuclear region that would then localize JIP4 for the initiation of LYTL after the phosphorylation of Rab10. In contrast to the overexpression of ARL8B and SKIP, the use of JIP4 knockdown did not affect LRRK2 expression levels, confirming that a reduction in pRab10 is position dependent. Interestingly, however, our data also suggest that there may be a smaller effect on pRab12 levels in the perinuclear area in the context of activated LRRK2.
What remains to be identified are the key conditions that determine the spatial specificity of the phosphorylation of specific Rabs at lysosomes. Lysosomes are highly dynamic and transient organelles responsible for wide-ranging cellular functions (39), and clustering lysosomes into the organelle-dense perinuclear cloud has been observed under various conditions of cellular stress and compromise (34, 40). LYTL, exacerbated by LLOMe treatment, requires the recruitment and phosphorylation of Rab10 and its subsequent recruitment of JIP4 to induce tubulation, which we previously hypothesized was beneficial to sort undegraded cargo to other active lysosomes for proper disposal (13). Our data collectively suggest that at least two conditions are required for the presence of pRab10/JIP4 on lysosomes: (1) LRRK2 must be active and recruited to lysosomes, and (2) LRRK2+ lysosomes must be located near the nucleus. Thus, the current data suggest a complex regulation of signaling downstream of LRRK2 that involves multiple conditions and varies depending on which Rab is phosphorylated by LRRK2.
Recent work by Berndsen et al. identified T73 Rab10 as a substrate for PPM1H phosphatase (20). Here, we show that the knockdown of PPM1H significantly increased pRab10 signal as well as pRab10+ and JIP4+ tubules on perinuclear lysosomes, demonstrating that endogenous PPM1H is not restricted to Golgi membranes. By extension, this suggests that as-yet-unidentified phosphatases may influence Rab10 phosphorylation at peripheral lysosomes. Of note, Berndsen et al. also reported that pS106 Rab12 was not influenced by PPM1H. In addition, it is possible that there are Rab10 GTPase-activating proteins/guanine exchange factor proteins that are also spatially restricted that may drive Rab10 accumulation on perinuclear lysosomes. Taken together, it will be important to identify key proteins that contribute to this mechanism in follow-up studies.
Overall, our results suggest that the presence of LRRK2 in a membrane is sufficient to trigger its activation, Rab phosphorylation, and retention at membranes, and that Rab10-specific recruitment to lysosomes is, in part, controlled by lysosomal positioning. Further studies are needed to identify the key proteins involved in determining optimal conditions for LRRK2-dependent Rab10 recruitment at the lysosomal membrane, including the description of phosphatases that can act on Rab12 and on Rab10 at peripheral lysosomes. Our results suggest LRRK2 signaling pathways are complex and will function as a product of which Rab is phosphorylated and which phosphatases act to counteract or terminate that signal.
Materials and Methods
Reagents and Treatments.
For all of the experiments using the FKBP-FRB complex, rapamycin (Cayman Chemicals, cat no. 13346) was added at 200 nM for 15 min before fixation in 4% paraformaldehyde. MLi-2 (Tocris, cat. no. 5756) was used at 1 µM in dimethyl sulfoxide (DMSO) for 90 min, or LLOMe (Sigma-Aldrich, cat. no. L7393) was added at 1 mM in DMSO for 2 h before fixation or lysing cells for downstream analyses. Starvation experiments consisted of rinsing cells with phosphate-buffered saline, followed by incubation in Earle’s balanced salt solution for 3 h before fixation. Time points from 2 to 16 h of starvation were explored and all resulted in no significant lysosomal clustering in HEK293FT cells. The full experimental procedures and associated references can be found in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Institute on Aging (to M.R.C.), and the University of Reading. We thank every member of the Cookson laboratory for critical feedback. We also thank Rosa Puertollano for kindly providing the LAMP1-FRB-CFP construct and Juan S. Bonifacino for the gift of the mCherry-ARL8B and 2xMyc-SKIP constructs.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. M.S.M. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2205492119/-/DCSupplemental.
Data, Materials, and Software Availability
All of the study data are included in the article and/or supporting information.
References
- 1.Paisán-Ruíz C., et al. , Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron 44, 595–600 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Zimprich A., et al. , The PARK8 locus in autosomal dominant parkinsonism: Confirmation of linkage and further delineation of the disease-containing interval. Am. J. Hum. Genet. 74, 11–19 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nalls M. A., et al. ; 23andMe Research Team; System Genomics of Parkinson’s Disease Consortium; International Parkinson’s Disease Genomics Consortium, Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet Neurol. 18, 1091–1102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roosen D. A., Cookson M. R., LRRK2 at the interface of autophagosomes, endosomes and lysosomes. Mol. Neurodegener. 11, 73 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greggio E., et al. , Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol. Dis. 23, 329–341 (2006). [DOI] [PubMed] [Google Scholar]
- 6.West A. B., et al. , Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc. Natl. Acad. Sci. U.S.A. 102, 16842–16847 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheng Z., et al. , Ser1292 autophosphorylation is an indicator of LRRK2 kinase activity and contributes to the cellular effects of PD mutations. Sci. Transl. Med. 4, 164ra161 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Steger M., et al. , Phosphoproteomics reveals that Parkinson’s disease kinase LRRK2 regulates a subset of Rab GTPases. eLife 5, e12813 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonet-Ponce L., Cookson M. R., The role of Rab GTPases in the pathobiology of Parkinson’ disease. Curr. Opin. Cell Biol. 59, 73–80 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hur E.-M., Jang E.-H., Jeong G. R., Lee B. D., LRRK2 and membrane trafficking: Nexus of Parkinson’s disease. BMB Rep. 52, 533–539 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yun H. J., et al. , An early endosome regulator, Rab5b, is an LRRK2 kinase substrate. J. Biochem. 157, 485–495 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Bonet-Ponce L., Cookson M. R., LRRK2 recruitment, activity, and function in organelles. FEBS J., 10.1111/febs.16099 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonet-Ponce L., et al. , LRRK2 mediates tubulation and vesicle sorting from lysosomes. Sci. Adv. 6, eabb2454 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herbst S., et al. , LRRK2 activation controls the repair of damaged endomembranes in macrophages. EMBO J. 39, e104494 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eguchi T., et al. , LRRK2 and its substrate Rab GTPases are sequentially targeted onto stressed lysosomes and maintain their homeostasis. Proc. Natl. Acad. Sci. U.S.A. 115, E9115–E9124 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pellegrini L., et al. , Proteomic analysis reveals co-ordinated alterations in protein synthesis and degradation pathways in LRRK2 knockout mice. Hum. Mol. Genet. 27, 3257–3271 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kluss J. H., et al. , Preclinical modeling of chronic inhibition of the Parkinson’s disease associated kinase LRRK2 reveals altered function of the endolysosomal system in vivo. Mol. Neurodegener. 16, 17 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gomez R. C., Wawro P., Lis P., Alessi D. R., Pfeffer S. R., Membrane association but not identity is required for LRRK2 activation and phosphorylation of Rab GTPases. J. Cell Biol. 218, 4157–4170 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kluss J. H., Bonet-Ponce L., Lewis P. A., Cookson M. R., Directing LRRK2 to membranes of the endolysosomal pathway triggers RAB phosphorylation and JIP4 recruitment. Neurobiol. Dis. 170, 105769 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berndsen K., et al. , PPM1H phosphatase counteracts LRRK2 signaling by selectively dephosphorylating Rab proteins. eLife 8, e50416 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson M. S., Sahlender D. A., Foster S. D., Rapid inactivation of proteins by rapamycin-induced rerouting to mitochondria. Dev. Cell 18, 324–331 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raiborg C., et al. , FYVE and coiled-coil domains determine the specific localisation of Hrs to early endosomes. J. Cell Sci. 114, 2255–2263 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Nada S., et al. , The novel lipid raft adaptor p18 controls endosome dynamics by anchoring the MEK-ERK pathway to late endosomes. EMBO J. 28, 477–489 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komada M., Soriano P., Hrs, a FYVE finger protein localized to early endosomes, is implicated in vesicular traffic and required for ventral folding morphogenesis. Genes Dev. 13, 1475–1485 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gillooly D. J., et al. , Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19, 4577–4588 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Follett J., Bugarcic A., Collins B. M., Teasdale R. D., Retromer’s role in endosomal trafficking and impaired function in neurodegenerative diseases. Curr. Protein Pept. Sci. 18, 687–701 (2017). [DOI] [PubMed] [Google Scholar]
- 27.Waschbüsch D., et al. , Structural basis for Rab8a recruitment of RILPL2 via LRRK2 phosphorylation of Switch 2. Structure 28, 406–417.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosa-Ferreira C., Munro S., Arl8 and SKIP act together to link lysosomes to kinesin-1. Dev. Cell 21, 1171–1178 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keren-Kaplan T., Bonifacino J. S., ARL8 relieves SKIP autoinhibition to enable coupling of lysosomes to kinesin-1. Curr. Biol. 31, 540–554.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuwahara T., et al. , Roles of lysosomotropic agents on LRRK2 activation and Rab10 phosphorylation. Neurobiol. Dis. 145, 105081 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Willett R., et al. , TFEB regulates lysosomal positioning by modulating TMEM55B expression and JIP4 recruitment to lysosomes. Nat. Commun. 8, 1580 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cantalupo G., Alifano P., Roberti V., Bruni C. B., Bucci C., Rab-interacting lysosomal protein (RILP): The Rab7 effector required for transport to lysosomes. EMBO J. 20, 683–693 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jordens I., et al. , The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr. Biol. 11, 1680–1685 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Korolchuk V. I., et al. , Lysosomal positioning coordinates cellular nutrient responses. Nat. Cell Biol. 13, 453–460 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beilina A., et al. , The Parkinson’s disease protein LRRK2 interacts with the GARP complex to promote retrograde transport to the trans-Golgi network. Cell Rep. 31, 107614 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beilina A., et al. ; International Parkinson’s Disease Genomics Consortium; North American Brain Expression Consortium, Unbiased screen for interactors of leucine-rich repeat kinase 2 supports a common pathway for sporadic and familial Parkinson disease. Proc. Natl. Acad. Sci. U.S.A. 111, 2626–2631 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Purlyte E., et al. , Rab29 activation of the Parkinson’s disease-associated LRRK2 kinase. EMBO J. 38, e101237 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee H., et al. , LRRK2 is recruited to phagosomes and co-recruits RAB8 and RAB10 in human pluripotent stem cell-derived macrophages. Stem Cell Reports 14, 940–955 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cabukusta B., Neefjes J., Mechanisms of lysosomal positioning and movement. Traffic 19, 761–769 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Martín Garrido N., Aylett C. H. S., Nutrient signaling and lysosome positioning crosstalk through a multifunctional protein, folliculin. Front. Cell Dev. Biol. 8, 108 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the study data are included in the article and/or supporting information.