Significance
Disruption of lung epithelial cell proteostasis causes activation of multiple unfolded protein response pathways, including inositol-requiring enzyme 1 (IRE1α) signaling. Hypothesized to be a proximal event in the development of pulmonary fibrosis, the mechanism by which proteostasis disruption causes disease is largely unknown. Utilizing a preclinical disease model of mice expressing a pulmonary fibrosis–associated mutation in the alveolar epithelial restricted surfactant protein C gene, we provide a mechanistic connection between disrupted epithelial proteostasis, the IRE1α signaling pathway, its major product XBP-1, and a cell-autonomous transcriptional reprogramming of distal lung epithelium identified in human pulmonary fibrosis. Our data support a therapeutic paradigm focused on lung epithelial cell proteostasis as an upstream target for intervention in pulmonary fibrosis pathogenesis.
Keywords: quality control, ER stress, surfactant protein C, unfolded protein response, transitional cell
Abstract
Disruption of alveolar type 2 cell (AEC2) protein quality control has been implicated in chronic lung diseases, including pulmonary fibrosis (PF). We previously reported the in vivo modeling of a clinical surfactant protein C (SP-C) mutation that led to AEC2 endoplasmic reticulum (ER) stress and spontaneous lung fibrosis, providing proof of concept for disruption to proteostasis as a proximal driver of PF. Using two clinical SP-C mutation models, we have now discovered that AEC2s experiencing significant ER stress lose quintessential AEC2 features and develop a reprogrammed cell state that heretofore has been seen only as a response to lung injury. Using single-cell RNA sequencing in vivo and organoid-based modeling, we show that this state arises de novo from intrinsic AEC2 dysfunction. The cell-autonomous AEC2 reprogramming can be attenuated through inhibition of inositol-requiring enzyme 1 (IRE1α) signaling as the use of an IRE1α inhibitor reduced the development of the reprogrammed cell state and also diminished AEC2-driven recruitment of granulocytes, alveolitis, and lung injury. These findings identify AEC2 proteostasis, and specifically IRE1α signaling through its major product XBP-1, as a driver of a key AEC2 phenotypic change that has been identified in lung fibrosis.
The alveolar epithelial type 2 cell (AEC2) plays a critical role in homeostasis of the distal adult lung, producing pulmonary surfactant to reduce alveolar surface tension and acting as a critical progenitor cell following injury (1, 2). With a high synthetic and secretory burden, the AEC2 is prone to cell quality control–related stress and maintains a proteostasis network for handling protein folding, misfolding, and degradation (3). Activation of the unfolded protein response (UPR) and the resultant endoplasmic reticulum (ER) stress is the hallmark of disruption of proteostasis and is a phenotypic feature of the AEC2 in chronic lung diseases like chronic obstructive pulmonary disease and pulmonary fibrosis (PF) (4–6). The genesis of AEC2 ER stress is thought to be from exogenous insults, such as cigarette smoke and viral infection (7, 8), but also can occur intrinsically with mutations in pulmonary surfactant proteins (9). Mutations in the AEC2-restricted SFTPC have been identified in pediatric patients with childhood interstitial lung disease (ChILD) and adults with PF and perhaps offer the best clinical evidence for the proximal role of AEC2 proteostasis dysfunction in lung disease (9–11). Recent in vivo murine modeling of both clinical SFTPC mutations and genetic knockout of the protein chaperone Bip in AEC2s has provided proof of concept for the role of AEC2 proteostasis as an upstream driver of PF (12–14).
While the UPR signaling pathways provide an adaptive response to protein misfolding by increasing proteostasis capacity with induction of chaperone genes and enhancing ubiquitin-proteasome system (UPS) degradation function, under prolonged stress, the response becomes maladaptive. AEC2 under terminal UPR activation undergoes differentiation, recruits immune cells, and becomes senescent and apoptotic (14–16). There are three distinct ER transmembrane sensors for misfolding proteins: inositol-requiring enzyme 1 (IRE1α), PKR-like ER kinase (PERK), and activating transcription factor 6 (ATF6). The IRE1α and PERK arms of the UPR are most associated with activation in the AEC2 in chronic lung disease, especially PF, and inhibition of each signaling pathway has been associated with improvement in the bleomycin model of murine lung injury and fibrotic remodeling (17–19). However, the precise mechanism by which attenuation of these UPR arms is protective in this model remains unanswered.
Recently, multiple groups have identified in murine models of severe lung injury and fibrotic repair that AEC2s undergo a transcriptional reprogramming to an alveolar epithelial cell state between AEC2 and the squamous alveolar epithelial type 1 cell (AEC1), defined by the marker genes Krt8 and Cldn4 (20–23). Similar alveolar epithelial reprogramming has been discovered in the lungs of patients with PF through trajectory inference analysis (19, 20). Both the IRE1α and PERK arms of the UPR have been implicated in a persistence of this reprogrammed state in the mouse lung (18, 19); however, unanswered is whether AEC2s enter this cell state through intrinsic UPR signaling. Given AEC2 UPR signaling has been identified in the lungs of patients with preclinical PF (24), defining the molecular and cellular mechanisms by which AEC2 reprogramming occurs may have implications for defining and intervening in the early pathogenesis of the human disease.
Uncovering the role of AEC2 UPR signaling as it relates to the maintenance of alveolar niche homeostasis and AEC2 cell fate in vivo has been limited by the lack of an adequate murine model to study the consequences of intrinsic disruption of AEC2 proteostasis. To overcome this, the current study leverages models expressing either of two PF-associated clinical Sftpc folding mutations. Utilizing these models, we show that disrupted proteostasis and UPR signaling drives intrinsic AEC2 dysfunction and the development of the reprogrammed AEC2 cell state. Through organoid modeling, in vitro activation of IRE1α ribonuclease (RNase) activity, and in vivo treatment with an IREIα inhibitor, our results suggest that the path to emergence of this reprogrammed cell state can be cell autonomous, independent of an exogenous distal lung epithelial injury, and mediated through the IRE1α-Xbp1 axis.
Results
Expression of a Clinical SP-C Mutation Challenges AEC2 Proteostasis.
Based on a clinical case of ChILD with a suspected pathogenic SFTPC missense variant in a SFTPC cysteine (C189) (SI Appendix, Fig. S1A) (25), we generated plasmids encoding the human SFTPC gene that contained a GFP tag and point mutations with a glycine substitution of the distal cysteine residues in the SFTPC BRICHOS domain (C121; C120; C148; C189). Western blotting performed 24 h after transfection of AEC2-like mouse lung epithelial MLE12 cells demonstrated proximal retention of both the GFP-tagged SP-CC121G and SP-CC189G mutations with failure to generate smaller intermediates (SI Appendix, Fig. S1B), consistent with the proposed role of each as critical cysteines for proper proSP-C trafficking and processing (26).
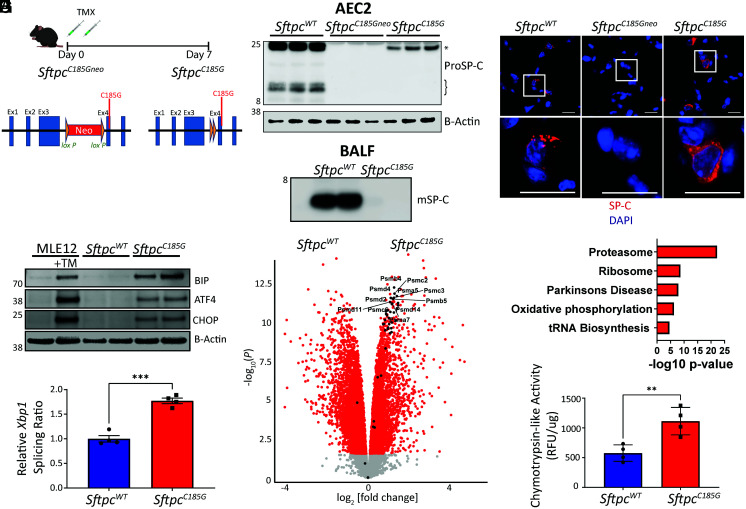
Given the equipoise from the in vitro expression studies, we next used a previously published embryonic stem cell–based strategy for modifying the Sftpc locus (14) to generate a murine model of the distal SFTPC (C189) mutation. The resultant founder line (SftpcC185Gneo) contained a glycine substitution for the homologous murine cysteine residue (C185G) and was functionally hypomorphic for Sftpc, owing to an intronic insertion of a neomycin cassette flanked by LoxP sites (Fig. 1A). When the founder line was crossed to the Rosa26ERT2cre line, administration of intraperitoneal (IP) tamoxifen (TMX) (450 mg/kg total delivered over two consecutive days) to the resultant SftpcC185Gneo/C185Gneo; Rosa26ERT2cre mice caused excision of the neomycin cassette and expression of the mutation (SftpcC185G). Western blotting of AEC2 lysates prepared 7 d after IP TMX demonstrated an expected absence of proSP-C protein in the hypomorphic SftpcC185Gneo founder line, while the SftpcC185G mice expressed but failed to process the primary proSP-C translation product to intermediate isoforms (Fig. 1B), a finding that conforms with other SP-C BRICHOS class mutations modeled in vitro and in vivo (12, 14, 27–29). Further, when compared with SftpcWT mice, immunostaining of SftpcC185G lung sections at 7 d showed abnormal patterning of mutant C185G proSP-C with ER retention and a failure to develop the cytosolic punctate appearance expected of proSP-C, indicative of normal anterograde trafficking and processing (Fig. 1C). Confirming this failure to process the mutant C185G proSP-C to the mature secreted peptide, Western blotting of the large-aggregate surfactant fraction of the bronchoalveolar lavage fluid (BALF) showed an absence of the mature SP-C (mSP-C) protein (Fig. 1B) in SftpcC185G mice.
Fig. 1.
Expression of a clinical Sftpc mutation challenges AEC2 proteostasis and activates the UPR. (A) Schematic of SftpcC185Gneo founder line with LoxP-flanked inhibitory neomycin cassette and post-TMX induced Cre recombination to the active SftpcC185G. (B) (Top) Western blotting of AEC2 cell lysate 7 d post-TMX for SP-C shows primary translation product (*) in SftpcWT and SftpcC185G samples, an absence of SP-C in the SftpcC185Gneo founder line, and a failure of SftpcC185G SP-C to undergo proteolytic cleavage to intermediate isoforms (}). (Bottom) Western blotting of BALF large-aggregate surfactant fractions for mature SP-C (mSP-C) shows absence of mSP-C in alveolar compartment of SftpcC185G mice (n = 2 per genotype). (C) Representative immunostaining of lung section for SP-C shows punctae appearance in SftpcWT, an absence of SP-C in the SftpcC185Gneo founder line, or a reticular pattern to the SftpcC185G SP-C (60× magnification, scale bars = 20 μM). (D) Western blotting analysis for BIP, ATF4, and CHOP in MLE12 cells ± 500 nm tunicamycin (TM) and SftpcWT and SftpcC185G AEC2 lysates 7 d post-TMX. (E) Ratio of spliced to unspliced Xbp1 as assessed by qRT-PCR of SftpcWT and SftpcC185G AEC2s. (F) Volcano plot showing the differences in gene expression in SftpcWT versus SftpcC185G AEC2s (red = >1.5 fold change, adj P value < 0.05). Genes annotated to the proteasome are indicated and show enrichment in SftpcC185G AEC2s. (G) Kyoto Encyclopedia of Genes and Genomes pathway analysis of differentially expressed genes shows enrichment of the proteasome pathway in SftpcC185G AEC2s. (H) Proteasome activity assessed as chymotrypsin-like activity in cell lysates of AEC2 isolated 7 d post-TMX. **p < 0.005, ***p < 0.0005 by two-way t test.
Having demonstrated the mutant proSP-C failed to undergo normal proteolytic processing, we next assessed the cellular consequences of this proximally retained protein on the AEC2. Based on previous work that showed SP-C BRICHOS mutations activated the UPR when expressed in vivo (14, 30), we sorted AEC2s from SftpcC185G and SftpcWT mice at 7 d after IP TMX and subjected them to western blotting for UPR signaling. Using MLE12 cells treated with the UPR-activating agent tunicamycin as a positive control, we identified an increase in the general UPR marker, BIP (Fig. 1D and SI Appendix, Fig. S2A). Additionally, both the PERK arm of the UPR, as shown by both increases in CHOP and ATF4 (Fig. 1D and SI Appendix, Fig. S2A), and the IRE1α arm, as demonstrated by splicing of Xbp1 (Fig. 1E) by qRT-PCR, were activated by SftpcC185G.
To further understand the consequence of mutant proSP-C protein expression on AEC2s in this model, we performed bulk RNA sequencing on AEC2s purified at 7 d after IP TMX and demonstrated that SftpcC185G AEC2s transcriptionally segregate from both the SftpcWT and hypomorphic SftpcC185GGneo AEC2s (SI Appendix, Fig. S3 A and B). Analysis of the differentially expressed genes between SftpcC185G and SftpcWT AEC2s revealed enrichment of genes associated with the proteasome (Fig. 1F), and Kyoto Encyclopedia of Genes and Genomes pathway analysis of the differentially expressed genes confirmed enrichment of the proteasome pathway (Fig. 1G). SftpcC185G AEC2 lysates showed an increase in both 20a and 26s proteasome mass (SI Appendix, Fig. S2B), as well as a twofold increase in proteasome degradation capacity as defined by measurement of ex vivo chymotrypsin-like activity (Fig. 1H). Despite this increase in potential UPS capacity, the mutant SftpcC185G cells showed accumulation of polyubiquitinated proteins (SI Appendix, Fig. S2B). Together, these data show that, despite transcriptomic enrichment of proteasome components in response to mutant SftpcC185G expression, AEC2s failed to maintain functional proteostasis with resultant induction of the UPR.
Disrupted AEC2 Proteostasis Induces Acute Lung Injury in Sftpc Mutant Mice.
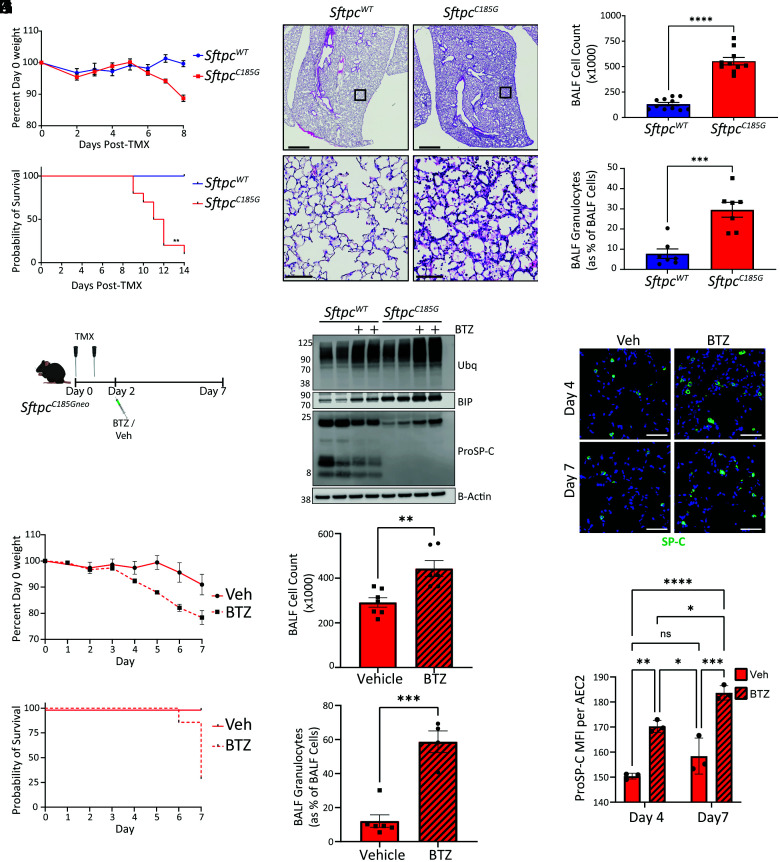
We next assessed whether the disruption on AEC2 proteostasis imposed by the mutant proSP-C resulted in a lung phenotype. We found that cohorts of mice expressing the SftpcC185G mutation developed weight loss beginning 7 d after receiving IP TMX (Fig. 2A) and developed fatal respiratory failure beginning at day 9 (Fig. 2B). At 7 d, the lung histology of SftpcC185G mice was marked by an acute lung injury pattern (Fig. 2C) and analysis of BALF revealed a granulocytic alveolitis (Fig. 2 D and E) with an accumulation of lung neutrophils (SI Appendix, Fig. S4A). A significant protein leak into the BALF (SI Appendix, Fig. S4B) was also consistent with acute lung injury. Analysis of BALF identified increases in granulocyte recruitment–associated cytokines CXCL1, CCL11, and CCL17, with commensurate enrichment of gene expression of these cytokines in SftpcC185G AEC2s (SI Appendix, Fig. S4C), showing the AEC2 challenged with the mutated proSP-C contribute to the lung injury phenotype.
Fig. 2.
Disruption of AEC2 proteostasis by SftpcC185G promotes early lung injury in mice. (A) Weight change following IP TMX to SftpcWT and SftpcC185G mice (n = 10 and 7, respectively). (B) Kaplan–Meier survival curve SftpcWT and SftpcC185G mice (n = 10 and 6, respectively), P = 0.0012 by log-rank (Mantel–Cox) test. (C) Representative hematoxylin and eosin (H&E) staining of lung section 7 d after TMX shows alveolitis in SftpcC185G lungs (top 4× magnification, scale bars = 1,000 μM; bottom 10× magnification, scale bars = 100 μM). (D and E) BALF cell counts (D) and percentage of BALF cells (E) that are granulocytes by Giemsa staining in SftpcWT and SftpcC185G mice at 7 d after TMX. ***p < 0.0005, ****p < 0.0001 by two-way t test. (F) Schematic for IP bortezomib (BTZ) in vivo proteasome inhibition studies (vehicle = 0.1% dimethyl sulfoxide [DMSO] in saline) in SftpcWT and SftpcC185G mice receiving OG TMX. (G) Western blotting for polyubiquitin (Ubq), BIP, and ProSP-C in AEC2 lysates isolated 6 h after in vivo BTZ (or vehicle) treatment. (H) (Top) Representative Immunostaining of lung section from SftpcC185G lungs at 4 and 7 d after BTZ or vehicle for proSP-C (20× magnification scale bars = 50 μM) and (Bottom) AEC2 mean fluorescence intensity (MFI) for proSP-C as measured in 40 most fluorescent AEC2s per 20× field per mouse (three 20× fields assessed per mouse). Not significant (ns), *p < 0.05, **p < 0.005, ***p < 0.0005, ****p < 0.0001 by one-way ANOVA with Tukey’s multiple comparisons test. (I) Mean weight change from baseline following OG TMX to SftpcC185G mice followed by BTZ or vehicle treatment. Data are shown as mean ± SEM (vehicle n = 5, BTZ n = 7). (J) Kaplan–Meier survival curve of SftpcC185G mice with OG TMX followed by BTZ or vehicle treatment (vehicle n = 5, BTZ n = 7) P = 0.0082 by log-rank (Mantel–Cox) test. (K and L) BALF cell counts (K) and percentage of BALF cells (L) that are granulocytes by manually counting Giemsa-stained cytospins of SftpcC185G mice at 7 d after OG TMX followed by BTZ or vehicle treatment. **p < 0.005, ***p < 0.0005 by two-way t test.
To further strengthen the connection between disrupted AEC2 proteostasis and alveolitis, we next constrained AEC2 proteasome function pharmacologically with the proteasome inhibitor bortezomib (BTZ). SftpcC185G mice were given 225 mg/kg oral gavage (OG) TMX to induce mutation expression, a TMX delivery modality associated with a less-severe lung phenotype than IP dosing (SI Appendix, Fig. S4 D–F), followed by IP treatment with 1 mg/kg BTZ or vehicle at day 2 after TMX (Fig. 2F). Analysis of AEC2 lysates from SftpcWT and SftpcC185G mice isolated 6 h following BTZ (or vehicle) exposure revealed in both genotypes an anticipated increase in polyubiquitinated proteins and BIP, signifying successful inhibition of the proteasome and compensatory UPR (Fig. 2G). We also found an increase in the mutant proSP-C isoform levels with proteasome inhibition and an increase in the fluorescence intensity of proSP-C-expressing cells at 4 and 7 d following TMX (Fig. 2 G and H), confirming previously published in vitro studies in transfected cell lines that had identified that AEC2s utilize the proteasome to manage misfolding SP-C mutations (31). BTZ inhibition of the proteasome accelerated the acute lung injury phenotype observed in the SftpcC185G, with BTZ-treated SftpcC185G mice showing accelerated weight loss and mortality (Fig. 2 I and J) and increases in both BALF cell No. and the proportion of granulocytes present in BALF at 7 d compared with vehicle-treated mice (Fig. 2 K and L). These experiments showed SftpcC185G mice have escalation of their lung injury phenotype with inhibition of the proteasome and strengthened a link between disruption of AEC2 proteostasis and AEC2-driven lung injury.
Disruption of AEC2 Proteostasis Accelerates Exogenous Bleomycin Lung Injury and Disrupts AEC2 Surfactant Homeostasis.
Despite an amplification of the alveolitis induced by SftpcC185G with proteasome inhibition, BTZ did not induce lung injury in SftpcWT mice (SI Appendix, Fig. S5 A–D). We hypothesized that SftpcWT mice challenged with extrinsic dysfunction to AEC2s from the fibrosing agent bleomycin (Bleo) would be susceptible to BTZ-induced proteasome inhibition similarly to the SftpcC185G mice with their intrinsic AEC2 dysfunction. Lung injury was induced in SftpcWT mice via intratracheal Bleo (3 U/kg) administered concurrently with IP BTZ (1 mg/kg) or vehicle (SI Appendix, Fig. S5E). SftpcWT mice given intratracheal saline with concurrent IP BTZ (1 mg/kg) or vehicle served as further controls. Two days following Bleo, mice receiving BTZ developed weight loss and began to succumb to respiratory failure (SI Appendix, Fig. S5 F and G). Analysis of the BALF from Bleo-treated mice at day 3 demonstrated no difference in the magnitude of elevation of BALF cell Nos. or granulocyte percent between BTZ and vehicle treatment; however, the BTZ-treated cohort did show an increase in BALF protein content (SI Appendix, Fig. S5 H–J), suggestive of accelerated lung injury.
Among the vital roles AEC2 has during lung inflammation is maintenance of pulmonary surfactant homeostasis, which reduces alveolar surface tension and prevents enhanced lung injury from repeated alveolar collapse and expansion (9). We next hypothesized that AEC2s injured from Bleo and challenged with proteasome inhibition from BTZ developed a disruption in their production of surfactant proteins. AEC2s sorted from the lungs of BTZ- and vehicle-treated mice at 3 d following Bleo showed a decrease in the mature, secreted, surfactant protein B (mSP-B) and mSP-C proteins (SI Appendix, Fig. S5L). Consistent with this AEC2 defect, Western blotting of the BALF large-aggregate surfactant fractions showed decreases in mSP-B and mSP-C in the alveolar compartment (SI Appendix, Fig. S5L). We next hypothesized that the proteostatically challenged AEC2s from the Bleo lung were developing a recently described AEC2 reprogrammed cell state that emerges following exogenous lung injury (20–23) defined by expression of Krt8. Analysis by qRT-PCR of sorted AEC2s showed increased expression of marker genes (Krt8, Cldn4, Sprr1a, and Lgals3) for this reprogrammed cell state in the BTZ-treated mice (SI Appendix, Fig. S5M). Together, these experiments demonstrated that coupling extrinsic Bleo AEC2 injury with concomitant pharmacological proteasome inhibition causes a disruption in AEC2 surfactant homeostasis, acceleration of lung injury, and promotion of a reprogrammed AEC2 cell state.
AEC2s Experiencing Endogenous Proteostatic Challenge Undergo Cell-Autonomous Reprogramming.
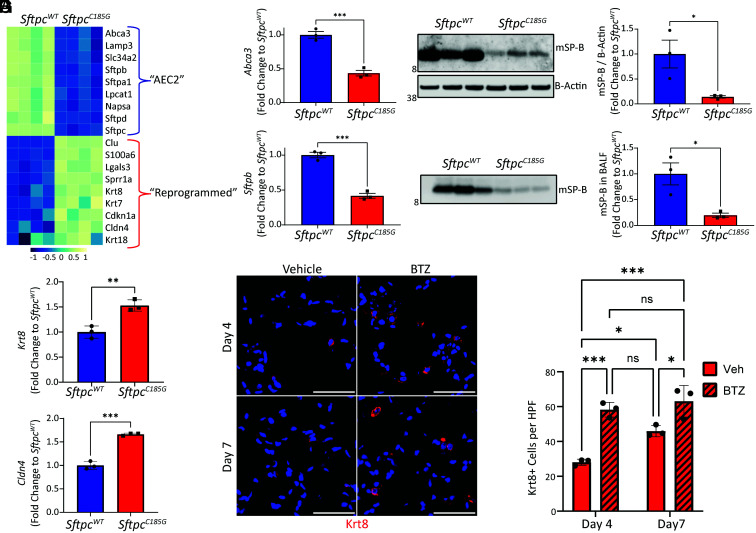
We next hypothesized that the SftpcC185G AEC2s, which have intrinsic disruption of proteostasis, lose their prototypical AEC2 program and develop the reprogrammed phenotype we observed in the AEC2s from the BTZ-treated Bleo injured mice. Further interrogation of the SftpcC185G AEC2 bulk RNA sequencing data (Fig. 1F) and confirmatory qRT-PCR revealed that the quintessential genes associated with pulmonary surfactant homeostasis, including Sftpb and Abca3, were all suppressed in the SftpcC185G AEC2s (Fig. 3 A and B). Western blotting of AEC2 lysates for SP-B identified a decreased mSP-B in the SftpcC185G AEC2 compared with SftpcWT (Fig. 3C), with a resultant decrease in mSP-B in the BALF large-aggregate surfactant fraction of SftpcC185G mice (Fig. 3D). This loss of surfactant in SftpcC185G AEC2s was coupled with transcriptomic enrichment in genes associated with the aforementioned reprogrammed cell state, including marker genes Krt8 and Cldn4 (Fig. 3 A and E and SI Appendix, Fig. S6A). Western blotting of AEC2 lysates for Krt8 confirmed the bulk transcriptional data (SI Appendix, Fig. S6B). Immunohistochemical analysis of lung sections from SftpcC185G mice treated with BTZ at 4 and 7 d post-TMX revealed increased Krt8+ cells compared with vehicle-treated SftpcC185G mice, further suggesting that challenges to proteostasis can drive AEC2 to a reprogrammed cell state (Fig. 3 F and G).
Fig. 3.
Sftpc mutant expression reprograms AEC2s. (A) Heatmap of selected AEC2-defining genes and markers genes for reprogrammed cell state in RNA-seq from AEC2 isolated 7 d after IP TMX administration to SftpcWT and SftpcC185G mice. (B) qRT-PCR for AEC2 genes Abca3 and Sftpb in AEC2 isolated 7 d after TMX from SftpcWT and SftpcC185G lungs. ***p < 0.0005 by two-way t test. (C) Western blotting (Left) and densitometry (Right) of AEC2 cell lysates 7 d after TMX for SP-B shows decrease in mature SP-B (mSP-B) in SftpcC185G AEC2s compared with SftpcWT. *p < 0.05 by two-way t test. (D) Western blotting (Left) and densitometry (Right) for mSP-B in BALF large-aggregate surfactant fractions showed decreased content in alveolar compartment in SftpcC185G mice. *p < 0.05 by two-way t test. (E) qRT-PCR for reprogrammed cell genes Krt8 and Cldn4 in AEC2 cells 7 d after TMX from SftpcWT and SftpcC185G lungs. *p < 0.05, **p < 0.005, ***p < 0.0005 by two-way t test. (F) Representative immunostaining for Krt8 of lung sections at 4 and 7 d after TMX in vehicle and BTZ-treated SftpcC185G mice (20× magnification, scale bars = 50 μM). (G) Quantification of Krt8+ cells per high-power field (HPF). *p < 0.05, ***p < 0.0005 by one-way ANOVA with Tukey’s multiple comparisons test.
We next analyzed our published model (SftpcC121G) of spontaneous lung injury and fibrotic lung remodeling with expression of a mutation in the other critical SP-C BRICHOS domain cysteine required for folding (C121) to affirm that the development of the reprogrammed phenotype is not limited to SftpcC185G AEC2s. Analysis of AEC2 bulk RNA sequencing data from SftpcC121G mice at 5 d after TMX (SI Appendix, Fig. S6C), a timepoint preceding lung injury, revealed a similar transcriptional change to that identified in SftpcC185G AEC2s, with loss of key genes in pulmonary surfactant homeostasis and enrichment in genes associated with the reprogrammed cell phenotype. Additionally, reanalysis of the published AEC2 transcriptome following extreme loss of proteostasis imposed by AEC2 knockout of the 26s proteasome assembly protein RPT3 identified a similar reprogrammed cell state (SI Appendix, Fig. S6D), suggesting the AEC2 phenotype change in the setting of disrupted proteostasis was not unique to the Sftpc mutation models (32). Together, these data show resultant loss of quintessential AEC2 function and development of a reprogrammed cell phenotype as a consequence of disruption of AEC2 proteostasis.
The AEC2 Reprogrammed Phenotype Develops Early after UPR Activation.
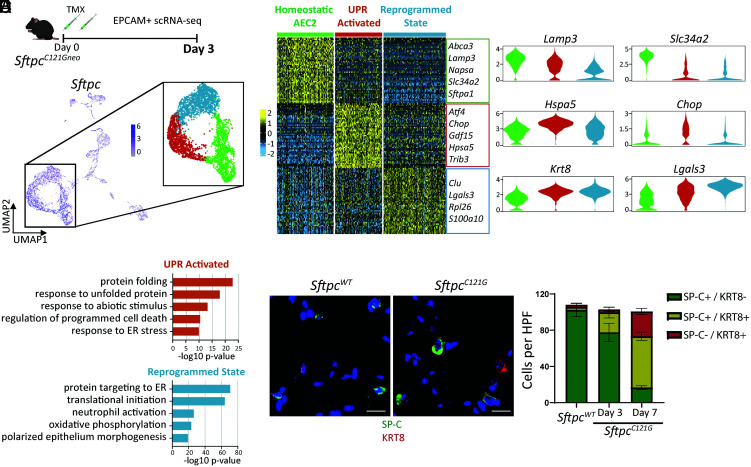
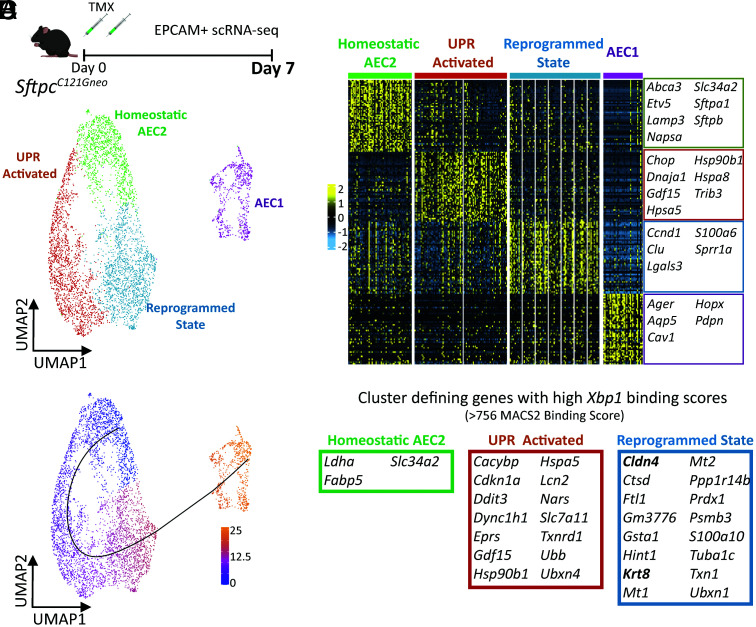
Based on our identification that AEC2 reprogramming precedes lung injury in the SftpcC121G model, we hypothesized that the development of this AEC2 phenotype arose as a response to intrinsic AEC2 stress signaling. To further understand the ontogeny of the reprogrammed AEC2 phenotype, we performed single-cell RNA sequencing (scRNA-seq) on epithelial (EPCAM+) enriched cell suspensions from the lungs of mice in the SftpcC121G model. EPCAM-enriched cell suspensions from two (one male and one female) non-TMX–treated SftpcC121G mice (day 0) and two (one male and one female) SftpcC121G mice prepared 3 d following IP TMX, a timepoint prior to appearance of an AEC2-driven lung injury phenotype, were subjected to scRNA-seq (SI Appendix, Fig. S7 A and C). Uniform Manifold Approximation and Projection (UMAP) projection and cluster annotation shows all expected lung epithelial populations in the lungs from both timepoints, with no single cluster dominated by Xist+ cells (signifying cells from the female sample) (SI Appendix, Fig. S7 B and D). Analysis of the SftpcC121G day 3 data revealed three clusters enriched in Sftpc (Fig. 4A). Analysis of the top differentially expressed genes across these clusters revealed a cluster enriched in quintessential AEC2 genes (homeostatic AEC2 cluster), a cluster enriched in UPR-associated genes (UPR activated cluster), and a cluster enriched in genes associated with the reprogrammed cell state (reprogrammed state cluster) (Fig. 4 B and C), with roughly a third of the SftpcC121G day 3 AEC2s segregating within each of the three clusters (34.1%, 28.4%, and 37.5%, respectively). Gene ontology (GO) pathway analysis on differentially expressed genes defining each cluster affirmed the UPR-activated AEC2 cluster was enriched in protein folding and response to cell stress pathways, whereas we found the reprogrammed state AEC2 cluster was enriched in pathways identified in Krt8+ cells in injury models (22), such as translational initiation, neutrophil activation, and oxidative phosphorylation (Fig. 4D). We next concatenated the SftpcC121G day 0 and SftpcC121G day 3 data for further analysis, which showed persistence of the three AEC2 clusters identified in the analysis of the SftpcC121G day 3 sample (SI Appendix, Fig. S7 E and F). The vast majority (96%) of AEC2s from the SftpcC121G day 0 control lungs grouped to the homeostatic AEC2 cluster, whereas the UPR-activated cluster and reprogrammed state cluster were almost entirely composed of SftpcC121G day 3 AEC2s (SI Appendix, Fig. S7 G and H). These analyses revealed the emergence of both an UPR-activated and a reprogrammed state in the SftpcC121G-expressing lung epithelia that preceded lung injury.
Fig. 4.
scRNA-seq of lung epithelial populations reveals early emergence of a reprogrammed AEC2 phenotype prior to lung injury. (A) (Top) Schematic for scRNA-seq of EPCAM+ sorted cells from SftpcC121G 3 d following TMX and (Bottom) UMAP projection annotated for Sftpc expression in the EPCAM+ populations. Inset shows the 3 Sftpc+ clusters that comprise the AEC2 states. (B) Heatmap of the top 50 cluster-defining genes by false discovery rate (FDR) for the three AEC2 cell states annotated with marker genes of each AEC2 state. (C) Violin plots of marker genes for homeostatic AEC2 state (Lamp3 and Slc34a2), UPR activation (Hspa5 and Chop), and reprogrammed state (Krt8 and Lgals3) in day 3 AEC2 clusters. (D) GO biological processes analysis of differentially expressed genes across the three AEC2 clusters shows pathways enriched in UPR-activated and reprogrammed states. (E) (Left) Representative immunostaining for SP-C (green) and Krt8 (red) expression in SftpcC121G and SftpcWT lungs 7 d post-TMX with coexpression of both (yellow) in AEC2 cells in SftpcC121G 7 d lungs (60× magnification, scale bars = 20 μM). (Right) Quantification of cells expressing one or both of the marker proteins counted in multiple 20× fields of sections acquired at days 3 and 7 after TMX in SftpcC121G and SftpcWT controls (n = 3 per genotype/timepoint).
To further understand the relationship between UPR signaling and the development of the reprogrammed cell state and affirm these distinct cell states persist later in the model, we next performed scRNA-seq of SftpcC121G 7 d EPCAM+ cells (SI Appendix, Fig. S8A), a timepoint where the majority of proSP-C+ cells coexpressed Krt8 (Fig. 5E). UMAP projection and cluster annotation of these data, again, revealed expected lung epithelial populations, with no single cluster dominated by Xist+ cells (SI Appendix, Fig. S8 A and B). Analysis of the three clusters enriched in Sftpc revealed a persistence of the three AEC2 states identified at day 3 (SI Appendix, Fig. S8 C and D). Biplot analysis for coexpression of Sftpc and krt8 as well as Lamp3 coexpression with Cldn4 (SI Appendix, Fig. S8E) showed cells coexpressing markers of homeostatic AEC2 and reprogrammed cell state were located in the UPR-activated cluster. Immunostaining validated the emergence of alveolar cells coexpressing these marker proteins (SI Appendix, Fig. S8F), with the emergence of dual SP-C+/Krt8+ cells within 3 d of TMX-induced mutation expression with an increase in cells coexpressing these marker proteins over the 7 d (Fig. 4E). Thus, interrogation of the AEC2 pool by scRNA-seq at these early timepoints in the SftpcC121G model showed the development of both UPR active and reprogrammed cell state phenotypes, leading us to next hypothesize that the reprogrammed state arose through the UPR-activated state.
Fig. 5.
Alveolar epithelial cell analysis identifies trajectory from homeostasis to a reprogrammed state enriched in Xbp1 target genes. (A) Schematic (Top) for scRNA-seq of EPCAM+ sorted cells from SftpcC121G 7 d following and (Bottom) UMAP projection of alveolar epithelial cells (AEC2 states plus AEC1 cells). (B) Heatmap of the top 50 cluster-defining genes by FDR for the four alveolar epithelial cell clusters annotated with marker genes of each AEC2 state at AEC1 cells. (C) Trajectory analysis of the alveolar epithelium results in one putative lineage relationship; color represents cell localization along pseudotime. (D) Analysis of published mouse ChIP-seq dataset for XBP-1 binding domains reveals 14 and 16 genes with high binding scores (genes within top quartile for MACS2 binding score) within the top 50 cluster-defining genes for the UPR-activated and reprogrammed state clusters, respectively, compared with three genes with high XBP-1 binding scores in the homeostatic AEC2 cluster.
The IRE1α Arm of the UPR Mediates the AEC2 Reprogrammed State.
To further define the trajectory relationship between the three AEC2 states, we focused on the alveolar epithelial cell compartment at 7 d after TMX. We selected cells in the aforementioned three AEC2 clusters and AEC1 cells and reclustered these as alveolar epithelial cells (Fig. 5A), which now showed persistence of three AEC2 cell states and separation of the AEC1 from the AEC2 clusters as defined by marker genes for each cell state (Fig. 5B). Trajectory inference analysis using Slingshot starting from the homeostatic AEC2 cells cluster showed a pseudotemporal relationship (trajectory) that first went through the UPR-activated state into the reprogrammed state and terminated at the AEC1 clusters (Fig. 5C). This analysis suggests that the emergence of a reprogrammed cell state is through an AEC2 UPR-activated state and suggested some capacity for AEC2-to-AEC1 transition in this model.
Given recent reports implicating IRE1α signaling in the persistence of the reprogrammed AEC2 state in Bleo lung fibrosis (19), we hypothesized that IRE1α signaling and specifically the downstream transcription factor XBP-1 may mediate AEC2 reprogramming in the absence of exogenous injury. We interrogated the top 50 differentially expressed genes from each of the three AEC2 clusters for XBP-1 binding sites in promoter/enhancer regions using a publicly available chromatin immunoprecipitation sequencing (ChIP-seq) database (Signaling Pathways Project (33)) (Fig. 5D). Genes with high XBP-1 binding scores were defined as those genes with MACS2 binding scores in the top quartile in the XBP-1 ChIP-seq database (corresponding to a MACS2 binding score >756). This analysis revealed 3 of 50 cluster-defining genes in the homeostatic AEC2 cluster with high XBP-1 binding scores; 14 of 50 genes in the UPR-activated cluster, including known XBP-1 targets Cdkn1a and Hspa5; and 16 of 50 genes in the reprogrammed state cluster, including defining marker genes Krt8 and Cldn4. Thus, we found that IRE1α RNase activity, and specifically resultant downstream sXbp1, may promote the AEC2 reprogramming.
To further interrogate the role of IRE1α signaling in the mediation of the reprogrammed state, we next utilized the MLE12 distal lung epithelial cell line to test the hypothesis that IRE1α signaling, and specifically Xbp1 expression, is capable of participating in the reprogramming of the alveolar epithelium. MLE12 cells were treated with IXA4 (10 μM), a potent stimulator of IRE1α RNase activity (34), for 24 h and then harvested for gene-expression analysis. IXA4 treatment stimulated a marked increase in MLE12 expression of spliced Xbp1 and induced a twofold increase in Krt8 expression (SI Appendix, Fig. S9). Thus, in this lung epithelial cell line, IRE1α RNase activity was sufficient for stimulation of Krt8 expression.
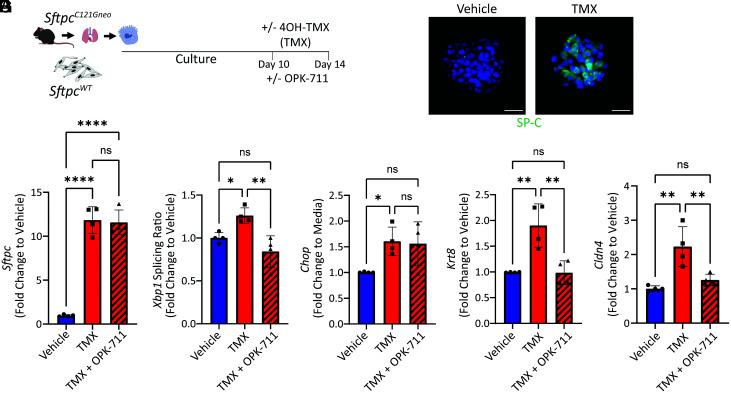
To further test whether the emergence of the reprogrammed cell state is a function of intrinsic AEC2 proteostatic stress that can be mediated through IRE1α signaling, we developed an ex vivo organoid culture of primary mouse AEC2s from our Sftpc mutation model. AEC2s from the inducible SftpcC121Gneo founder line were isolated and cultured in three-dimensional organoids with SftpcWT fibroblasts (Fig. 6A). Addition of 4OH-TMX (500 nM) to the culture media at 10 d (after organoids began to form) resulted in successful induction of the mutant proSP-C isoform by day 14 (Fig. 6B). At day 14, we then compared gene expression changes of the cultured cells between wells receiving vehicle, 4OH-TMX, and 4OH-TMX with the addition of an IRE1α inhibitor OPK-711 at a concentration of 200 nM (a dose we found inhibited IRE1α signaling in the MLE12 cells transfected with SP-CC121G plasmids, SI Appendix, Fig. S10). Expression of the mutant Sftpc in the organoid system caused activation of both the IRE1α and the PERK arms of the UPR marked by Xbp1 splicing and Chop expression, respectively (Fig. 6 D and E), with OPK-711 inhibiting Xbp1 splicing without affecting Chop expression. Expression of SftpcC121G also stimulated an increase in marker genes for the reprogrammed cell state (Cldn4 and Krt8), which was attenuated with the addition of OPK-711. In contrast, inhibition of the integrated stress response (ISR) using the pharmacological inhibitor ISRIB (200 nM) reduced expression of Chop but failed to resolve reprogrammed gene expression in this system (SI Appendix, Fig. S11 A–D). These data show that the reprogrammed cell state can emerge through intrinsic AEC2 signaling and is not dependent upon lung injury and that inhibition of IRE1α signaling can attenuate this reprogramming ex vivo.
Fig. 6.
Organoid modeling identifies that IRE1α signaling mediates the reprogrammed cell state. (A) Schematic for organoid experiments using uninduced SftpcC121G AEC2s and SftpcWT mouse lung fibroblasts with ex vivo induction of the Sftpc mutation using 1 μM 40H-TMX (vehicle = 0.1% EtOH) and treatment with 200 nM OPK-711 or vehicle (0.1% DMSO) for final 96 h of culture. (B) Representative immunostaining of day 14 organoids for SP-C shows presence of proSP-C in 4OH-TMX-treated culture (60× magnification scale bars = 20 μM). (C–G) Sftpc (C), Xbp1 splicing ratio (D), Chop (E), Krt8 (F), and Cldn4 (G) expression measured by qRT-PCR of day 14 organoid culture cells 96 h comparing combined vehicle (0.05% EtOH + 0.1% DMSO) and 4OH-TMX (in 0.05% EtOH) with 0.1% DMSO added to media and 4OH-TMX (0.05% EtOH) + OPK-711 (0.1% DMSO). n = 4 biological replicates. *p < 0.05, **p < 0.005, ***p < 0.0005 by one-way ANOVA with Tukey’s multiple comparisons test.
Disruption of IRE1α Signaling Attenuates AEC Reprogramming and Lung Injury In Vivo.
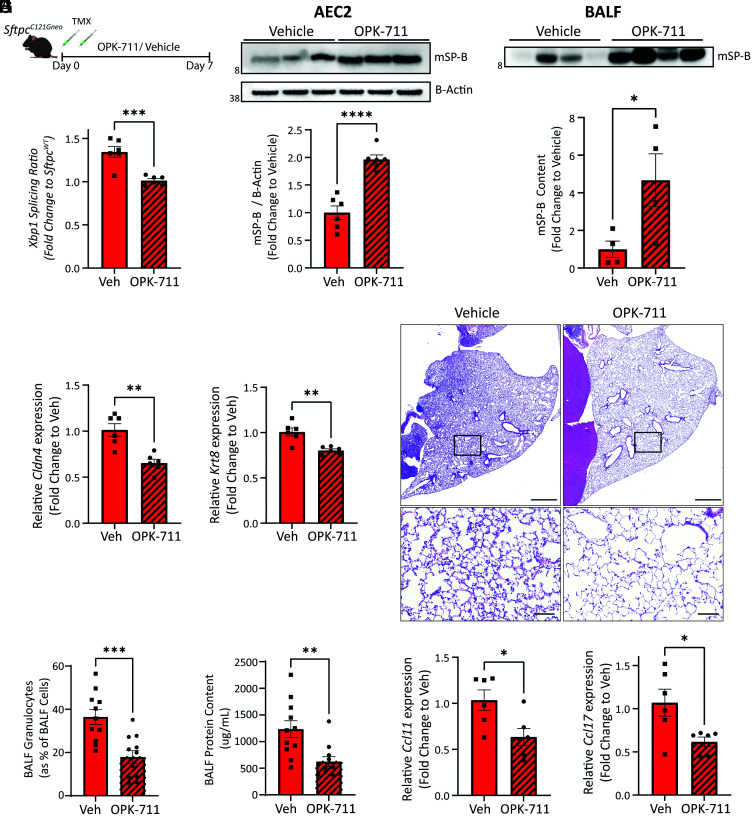
To test whether inhibition of IRE1α also attenuated the reprogramming of AEC2 in vivo, SftpcC121G mice were given daily 20 mg/kg OPK-711 by OG (or vehicle) for 7 d following TMX Sftpc mutation induction (Fig. 7A). Analysis of AEC2s at day 7 revealed OPK-711 reduced the splicing of Xbp1 (Fig. 7B) without affecting Atf4 or Chop expression (SI Appendix, Fig. S12 A–C). AEC2s in SftpcC121G mice treated with OPK-711 had increased production of mature, secreted, SP-B protein and subsequently showed increased SP-B content in the BALF large-aggregate surfactant fractions compared with vehicle-treated mice (Fig. 7 C and D, and SI Appendix, Fig. S12D). Consistent with AEC2 maintaining their homeostatic surfactant protein–producing state, AEC2 expression of both reprogrammed cell state marker genes Krt8 and Cldn4 was attenuated with IRE1α inhibition (Fig. 7E). Together, these analyses showed that inhibition of IRE1α signaling in vivo can attenuate reprogramming when AEC2s are confronted with intrinsic proteostatic stress.
Fig. 7.
IRE1α inhibition reduces the reprogrammed AEC2 cell state and AEC2 driven alveolitis in vivo. (A) Schematic for evaluating in vivo IRE1α inhibition in SftpcC121G mice using OPK-711 (20 mg/kg/day) or vehicle treatment. (B) Xbp1 splicing ratio as assessed by qRT-PCR at day 7 (n = 6 per group) referenced against SftpcWT. (C) Western blotting (Top) of AEC2 lysate for SP-B and densitometry analysis (Bottom) show increased mSP-B in AEC2 cells with OPK-711 IRE1α inhibition (see SI Appendix, Fig. S12C for additional Western blotting for densitometry). (D) Western blotting of BALF large-aggregate surfactant fractions (Top) and densitometry (Bottom) for mSP-B showed increased content in alveolar compartment with OPK-711 IRE1α inhibition. (E) Cldn4 (Left) and Krt8 (Right) expression determined by qRT-PCR of AEC2 cells at day 7 (n = 6 per group). (F) Representative H&E staining of lung sections shows reduced alveolitis in OPK-711-treated lungs (Top: 4× magnification scale bars = 1,000 μM, Bottom: 20× magnification, scale bars = 100 μM). (G and H) BALF granulocytes (G) quantified by manual counting of Giemsa-stained cytospins and expressed as a percentage of total BALF cells and BALF total protein (H) as assessed by Lowry assay in vehicle and OPK-711-treated SftpcC121G mice (n = 11 per group). (I) Ccl11 (Left) and Ccl17 (Right) expression determined by qRT-PCR of AEC2 cells at day 7 (n = 6 per group). *p < 0.05, **p < 0.005, ***p < 0.0005 by two-way t test.
We next assessed whether inhibition of IRE1α signaling improved the inflammatory lung phenotype observed in the model. Lung histology at 7 d demonstrated a reduced injury pattern with OPK-711 treatment (Fig. 7F), which was confirmed by BALF analysis, revealing a reduction in the percent and absolute No. of BALF granulocytes (Fig. 7G and SI Appendix, Fig. S12E) and reduced BALF protein content (Fig. 7H). Analysis for AEC2 expression of cytokine genes showed that two of the granulocyte recruitment–associated cytokines identified in the model, Ccl11 and Ccl17, were reduced (Fig. 7I) with IRE1α inhibition. Together, these data mechanistically link the IRE1α pathway in the reprogramming of AEC2s in this model of intrinsic AEC2 proteostatic dysfunction and also in the AEC2-driven inflammatory response to proteostatic stress.
Discussion
The AEC2 maintains lung homeostasis through the production of pulmonary surfactant to reduce alveolar surface tension and through acting as a facultative progenitor cell following injury. While loss of these critical cells through ablation causes spontaneous lung injury and fibrosis (35), the mechanisms by which intrinsic cell dysfunction challenges these AEC2 functions are largely unknown. Here, we identified that loss of AEC2 proteostasis was responsible for two additive pathological events, an AEC2-driven lung injury from immune cell recruitment and the reprogramming of AEC2s marked by loss of quintessential AEC2 surfactant protein production within the alveolar niche.
Disrupted alveolar epithelial proteostasis is implicated in the pathogenesis of chronic fibrotic lung disease, like PF (4, 36). Supporting the importance of AEC2 proteostasis in lung health and disease, loss of proteasome function through knockout of the proteasome regulatory unit in mouse AEC2s led to AEC2 apoptosis, loss of barrier function, and death (31). Less-extreme challenges to proteostasis through knockout of the key UPR chaperone BIP in AEC2 and through the expression of PF-associated SFTPC mutations have demonstrated a survivable spontaneous acute lung injury and subsequent fibrotic lung remodeling (13, 14). Our data provide a mechanistic link between loss of AEC2 proteostasis and lung injury. We identified that expression of the SftpcC185G mutation causes a cell-intrinsic disruption to proteostasis defined by the activation of UPR pathways. Further, we found that, through inhibition of the proteasome’s degradative capacity in the SftpcC185G AEC2s with the pharmacological proteasome inhibitor BTZ, we could stimulate a “second hit” to the proteostatic dysfunction and that this increased AEC2 UPR signaling and both magnified the AEC2-driven lung injury and the accelerated reprogramming of AEC2s. These studies provided a mechanistic link between intrinsic loss of AEC2 proteostasis from the expression of the misfolded proSP-C protein and a multifactorial (immune cell recruitment and loss of surfactant homeostasis) AEC2-driven lung injury.
There have been seemingly conflicting results in the literature of proteasome inhibition with BTZ in the Bleo model of lung injury and fibrosis. While several groups have shown an antifibrotic effect during the fibrotic lung remodeling phase (days 10–28) of the Bleo model, understood to occur through inhibition of the lung fibroblast proteasome, inhibition of the proteasome during the epithelial injury phase of the model (days 0–7) has shown a deleterious effect (37–39). Here, we have provided a mechanistic understanding for this adverse effect. We demonstrated that challenging AEC2 proteostasis with BTZ in the epithelial injury phase of the Bleo model amplifies lung injury and accelerates the loss of AEC2 surfactant protein production seen with AEC2 reprogramming. Speculatively, this may provide context for the pneumonitis observed in some patients receiving BTZ for the treatment of multiple myeloma (40, 41) and mechanistically connects our observation that intrinsic loss of proteostasis drives AEC2 reprogramming in the SftpcC185G model to the commonly used Bleo model of exogenous AEC2 injury, where a similar AEC2 reprogramming has been identified.
Heretofore, the AEC2-reprogrammed Krt8+ cell state has been shown to emerge following severe and direct lung epithelial cell injury, triggering the need for alveolar epithelial regeneration, be it from viral infection, Bleo, or AEC1 ablation (20–23). In these lung injury models, senescence-associated TP53 signaling and UPR signaling have been implicated in the persistence of this cell state and a failure to efficiently differentiate to AEC1s (18–20). Specifically, recent work by Watanabe et al. and Auyeung et al., showing inhibition of the ISR and IRE1α signaling, respectively, can mediate the capacity of reprogrammed AEC2 to differentiate into AEC1 following injury, has implicated proteostatic networks in persistence of the reprogrammed state. However, less is known about the mechanistic ontogeny of this cell state. Choi et al. (21) identified interleukin-1B signaling from alveolar macrophages during lung injury as mediating AEC2 reprogramming, implicating epithelial-immune cell signaling in the reprogramming. Here, we show through identification that this reprogrammed state develops before epithelial injury in vivo and through organoid modeling that the development of this cell state does not require a lung injury response or immune cell signaling but rather can be an AEC2-autonomous process through intrinsic proteostatic stress. Given that AEC2 UPR signaling has been identified in the lungs of patients with preclinical PF (24), defining molecular and cellular mechanisms by which AEC2 reprogramming occurs has implications for the human disease.
When interrogation of our single-cell dataset of the alveolar epithelial compartment showed a pseudotemporal relationship that went from AEC2 homeostasis through the UPR-activated state to the reprogrammed state, we further hypothesized that UPR signaling may mediate this cell-autonomous reprogramming. We then focused on the IRE1α signaling arm of the UPR, given its implication to the reprogramming of distal epithelial cells in human PF (19). Activation of IRE1α stimulates both kinase and RNase activity, which causes the splicing, and subsequent activation, of the transcription factor XBP-1. Based on our identification that many of the genes that defined the AEC2 reprogrammed state, and specifically Krt8 and Cldn4, noted high binding scores of XBP-1 to promoter/enhancer regions, we went on to both show that pharmacological stimulation IRE1α RNase activity and subsequent increase in sXbp1 enhanced Krt8 expression in a lung epithelial cell line. We then used an IRE1α inhibitor, OPK-711, for in vitro and in vivo studies that demonstrated that inhibition of IRE1α signaling led to maintenance of AEC2 surfactant homeostasis, attenuated AEC2 reprogramming, and improved AEC2-driven lung injury. While UPR signaling and ER stress have been implicated in cell differentiation in the gastrointestinal epithelium (42) and ER stress in lung epithelial cell culture induces an epithelial–mesenchymal transition in vitro (15), the implication that IRE1α signaling, and specifically the transcription factor XBP-1, can induce AEC2 differentiation to a reprogrammed state represents a finding with therapeutic implications.
Our study has important limitations. Our in vivo BTZ and OPK-711 studies cannot exclude the effect of proteasome inhibition (BTZ) and IRE1α inhibition (OPK-711) on other resident and immune cells in the lung. In the future, genetic-based interventions targeting AEC2-specific programming could overcome that limitation. Second, unanswered in this current study is the capacity of the AEC2 when under proteostatic stress to differentiate fully to AEC1s. Our trajectory inference analysis suggests a lineage relationship between AEC2 and AEC1 in the Sftpc mutation model, but a limitation of the biallelic Sftpc mutation model is that it does not allow for the use of currently available AEC2-specific genetic lineage–tracing reagents (owing to the SftpcERT2Cre being the lone AEC2-specific cre-driver). We therefore cannot assess in this model the capacity of AEC2s entering the reprogrammed state to fully differentiate into AEC1 cells. Future lineage-tracing studies with novel AEC2-specific cre-drivers would be required to define the ability and capacity of AEC2s entering the UPR-activated state to fully differentiate into AEC1 cells. Finally, while we demonstrated using organoid modeling that inhibition of ISR signaling did not attenuate AEC2 expression of reprogramming genes, no comparative in vivo studies of PERK or ATF6 inhibition were conducted to fully define the role of the full UPR signaling repertoire in AEC2 autonomous programming when confronted with proteostatic stress.
In conclusion, we have shown that AEC2s confronted with intrinsic proteostatic stress undergo cell-autonomous reprogramming and lose quintessential AEC2 function. Inhibition of IRE1α signaling can attenuate this reprogramming and appears to be a key mediator of an AEC2 phenotypic change that has been implicated in human PF. Targeting epithelial proteostatic networks is a promising therapeutic paradigm in PF and may permit interventions at a more-proximal point in disease pathogenesis.
Materials and Methods
Mice.
The SftpcC185Gneo and SftpcC121Gneo founder lines were commercially produced (Genoway, Inc. Lyon, France) as previously described (14). For TMX-inducible removal of PGK-neo cassettes, the founder lines were crossed to the Rosa26ERT2Cre (strain B6.129-Gt(ROSA)26Sortm1(cre/ERT2)Tyj/J; stock No. 008463; The Jackson Laboratory, Bar Harbor, ME). The progeny was backcrossed to homozygosity of the Sftpc mutant allele and the Rosa26ERT2Cre allele and IP or OG TMX (dissolved in corn oil at a concentration of 20 mg/mL) at 225/mg/kg/day × 2 d was used to induce Sftpc mutation expression. SftpcWT mice harboring homozygous Rosa26ERT2Cre allele treated with TMX as above were used as controls. Both male and female animals (aged 8–12 wk) were utilized, and all mouse strains and genotypes generated for these studies were congenic with C57/B6/J. See SI Appendix for detailed methods on Bleo instillation and pharmacological interventions with BTZ and OPK-711. All experimental and breeding protocols were approved by the Institutional Animal Care and Use Committee of the Perelman School of Medicine at the University of Pennsylvania.
BALF Analysis and Lung Histology.
After euthanasia, the trachea was cannulated with a blunt needle, and BALF was collected by five sequential normal-saline 1-mL lavages and analyzed for cell content, cell differential cell counts, protein content, cytokine measurements, and surfactant protein content as detailed in SI Appendix. Following lung lavage, lungs were fixed or removed en bloc or prepared for multichannel flow cytometry (additional methods can be seen in SI Appendix).
Immunofluorescence Staining, Imaging, and Quantification.
Immunofluorescence staining of lung sections was performed with combinations of primary and secondary antibodies outlined in detail in the SI Appendix. Imaging was performed on a Nikon Eclipse Ni at 20× and 60× magnification using the NIS-Elements AR 5.02 software and captured in the manufacturer’s fluorescein isothiocyanate , Cy5, and Dapi channels. Images were analyzed on Fiji (ImageJ, v.1.8.0) with quantification methods detailed in the SI Appendix.
AEC2 Isolation and Analysis.
Mouse AEC2s were isolated as previously described (14). Briefly, a single-cell suspension was obtained by instilling Dispase (Sigma Aldrich, DLW354235) into perfused lungs, followed by mechanical dissociation and treatment with 20 μg/mL DNase I (Sigma Aldrich, D5025-150KU). Differential adherence on plastic culture dishes negatively selected mesenchymal cells. CD45+ cells were depleted by negative selection using Dynabeads untouched mouse T cell kit (No. 11413D) and Dynabeads mouse DC enrichment kit (No. 11429D). AEC2 protein lysate was collected and subjected to western blot analysis, proteasome activity assay, RT-qPCR, and RNA sequencing (see additional detailed methods in SI Appendix).
EPCAM-Enriched scRNA-Seq and Analysis.
EPCAM+ cells from the SftpcC121G model were prepared from SftpcC121G mice at day 0 (SftpcC121Gneo), day 3, and day 7 as described in the SI Appendix and loaded individual GemCode instrument (10X Genomics), one for each of the three timepoints. Single-cell barcoded droplets were produced using 10X Single Cell 3′ v3 chemistry. Libraries generated were sequenced using an Illumina HiSeq2500 instrument in a high output mode. See SI Appendix for detailed methods on processing, integration, and downstream analysis using SeuratV3, trajectory analysis using the R slingshot package, and GO analysis.
XBP-1 Binds Scores Using Signaling Pathways Project ChIP-Seq Analysis.
Cluster-defining gene lists were prepared by comparing differential gene expression between the three AEC2 clusters in the alveolar clusters subset from the SftpcC121G day 7 analysis. The top 50 cluster-defining genes were identified as those with highest differential expression (with a minimum of 50% of cells within a cluster expressing the gene of interest). These three gene lists were queried for XBP-1 MACS2 binding scores on The Signaling Pathways Project database (www.signalingpathways.org/), which utilized the published mouse XBP-1 ChIP-seq analysis from Argemí et al. (43). Genes with high MACS2 binding scores were defined as genes with top quartile MACS2 binding scores in the full Argemí et al. dataset, corresponding to a score >756.
MLE12 and Primary Mouse AEC2 Organoid Culture.
MLE12 cells were obtained from American Type Culture Collection (CRL-2110) and cultured in Dulbecco’s modified Eagle’s medium + 10% fetal bovine serum that included 1% penicillin-streptomycin and were incubated at 37 °C and 5% CO2. See SI Appendix for detailed methods on SP-C construct transfections and treatment with tunicamycin and IXAV4. Organoid culture assays were performed as previously described (44). Inducible SftpcC121Gneo AEC2 were isolated from mouse lungs and cocultured with primary mouse lung fibroblasts. At day 10 of culture, media were changed to include 4OH-TMX, OPK-711, ISRIB, or vehicle, and at day 14, the organoids were analyzed (see SI Appendix for detailed methods).
Supplementary Material
Acknowledgments
This study was supported by funding from the National Institutes of Health by grants K08 HL150226 to JBK, R01 HL132999, R01 HL087825, R01 HL152194, and U01 HL134745 to EEM, R01 HL145408 and U01 HL152970 to MFB, and funding support from the Pulmonary Fibrosis Foundation and Parker B Francis Foundation to JBK.
Footnotes
Competing interest statement: T.P. and G.R. are employees of OptiKira, LLC. The remaining authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2123187119/-/DCSupplemental.
Data, Materials, and Software Availability
Sequencing data generated in this study has been deposited to GEO with accession No. GEO:GSE189480 (45).
All study data are included in the article and/or supporting information.
References
- 1.Beers M. F., Moodley Y., When is an alveolar type 2 cell an alveolar type 2 cell? A conundrum for lung stem cell biology and regenerative medicine. Am. J. Respir. Cell Mol. Biol. 57, 18–27 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barkauskas C. E., et al. , Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 123, 3025–3036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katzen J., Beers M. F., Contributions of alveolar epithelial cell quality control to pulmonary fibrosis. J. Clin. Invest. 130, 5088–5099 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lawson W. E., et al. , Endoplasmic reticulum stress in alveolar epithelial cells is prominent in IPF: Association with altered surfactant protein processing and herpesvirus infection. Am. J. Physiol. Lung Cell. Mol. Physiol. 294, L1119–L1126 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Malhotra D., et al. , Heightened endoplasmic reticulum stress in the lungs of patients with chronic obstructive pulmonary disease: The role of Nrf2-regulated proteasomal activity. Am. J. Respir. Crit. Care Med. 180, 1196–1207 (2009). Correction in: Am. J. Respir. Crit. Care Med. 193, 344 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Balch W. E., et al. , Malfolded protein structure and proteostasis in lung diseases. Am. J. Respir. Crit. Care Med. 189, 96–103 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torres-González E., et al. , Role of endoplasmic reticulum stress in age-related susceptibility to lung fibrosis. Am. J. Respir. Cell Mol. Biol. 46, 748–756 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelsen S. G., et al. , Cigarette smoke induces an unfolded protein response in the human lung: A proteomic approach. Am. J. Respir. Cell Mol. Biol. 38, 541–550 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Mulugeta S., Nureki S., Beers M. F., Lost after translation: Insights from pulmonary surfactant for understanding the role of alveolar epithelial dysfunction and cellular quality control in fibrotic lung disease. Am. J. Physiol. Lung Cell. Mol. Physiol. 309, L507–L525 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nogee L. M., et al. , A mutation in the surfactant protein C gene associated with familial interstitial lung disease. N. Engl. J. Med. 344, 573–579 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Thomas A. Q., et al. , Heterozygosity for a surfactant protein C gene mutation associated with usual interstitial pneumonitis and cellular nonspecific interstitial pneumonitis in one kindred. Am. J. Respir. Crit. Care Med. 165, 1322–1328 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Sitaraman S., et al. , Surfactant protein C mutation links postnatal type 2 cell dysfunction to adult disease. JCI Insight 6, e142501 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borok Z., et al. , Grp78 loss in epithelial progenitors reveals an age-linked role for endoplasmic reticulum stress in pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 201, 198–211 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katzen J., et al. , An SFTPC BRICHOS mutant links epithelial ER stress and spontaneous lung fibrosis. JCI Insight 4, e126125 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanjore H., et al. , Alveolar epithelial cells undergo epithelial-to-mesenchymal transition in response to endoplasmic reticulum stress. J. Biol. Chem. 286, 30972–30980 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maguire J. A., Mulugeta S., Beers M. F., Endoplasmic reticulum stress induced by surfactant protein C BRICHOS mutants promotes proinflammatory signaling by epithelial cells. Am. J. Respir. Cell Mol. Biol. 44, 404–414 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thamsen M., et al. , Small molecule inhibition of IRE1α kinase/RNase has anti-fibrotic effects in the lung. PLoS One 14, e0209824 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watanabe S., et al. , Resetting proteostasis with ISRIB promotes epithelial differentiation to attenuate pulmonary fibrosis. Proc. Natl. Acad. Sci. U.S.A. 118, e2101100118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Auyeung V. C., et al. , IRE1α drives lung epithelial progenitor dysfunction to establish a niche for pulmonary fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 322, L564–L580 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi Y., et al. , Persistence of a regeneration-associated, transitional alveolar epithelial cell state in pulmonary fibrosis. Nat. Cell Biol. 22, 934–946 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi J., et al. , Inflammatory signals induce AT2 cell-derived damage-associated transient progenitors that mediate alveolar regeneration. Cell Stem Cell 27, 366–382.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strunz M., et al. , Alveolar regeneration through a Krt8+ transitional stem cell state that persists in human lung fibrosis. Nat. Commun. 11, 3559 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang P., et al. , Ineffectual type 2-to-type 1 alveolar epithelial cell differentiation in idiopathic pulmonary fibrosis: Persistence of the KRT8hi transitional state. Am. J. Respir. Crit. Care Med. 201, 1443–1447 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kropski J. A., et al. , Extensive phenotyping of individuals at risk for familial interstitial pneumonia reveals clues to the pathogenesis of interstitial lung disease. Am. J. Respir. Crit. Care Med. 191, 417–426 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delestrain C., et al. , Methylprednisolone pulse treatment improves ProSP-C trafficking in twins with SFTPC mutation: An isoform story? Br. J. Clin. Pharmacol. 87, 2361–2373 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Willander H., et al. , High-resolution structure of a BRICHOS domain and its implications for anti-amyloid chaperone activity on lung surfactant protein C. Proc. Natl. Acad. Sci. U.S.A. 109, 2325–2329 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bridges J. P., Wert S. E., Nogee L. M., Weaver T. E., Expression of a human surfactant protein C mutation associated with interstitial lung disease disrupts lung development in transgenic mice. J. Biol. Chem. 278, 52739–52746 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Kabore A. F., Wang W. J., Russo S. J., Beers M. F., Biosynthesis of surfactant protein C: Characterization of aggresome formation by EGFP chimeras containing propeptide mutants lacking conserved cysteine residues. J. Cell Sci. 114, 293–302 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Mulugeta S., et al. , Misfolded BRICHOS SP-C mutant proteins induce apoptosis via caspase-4- and cytochrome c-related mechanisms. Am. J. Physiol. Lung Cell. Mol. Physiol. 293, L720–L729 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Lawson W. E., et al. , Endoplasmic reticulum stress enhances fibrotic remodeling in the lungs. Proc. Natl. Acad. Sci. U.S.A. 108, 10562–10567 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hawkins A., Gonzales L., Guttentag S., Mulugeta S., Beers M., Protein quality control in alveolar type 2 cells: The proteasome is essential for control of aggregation-prone SP-C mutations. FASEB J. 29, 1_supplement.1015.5 (2015).25561464 [Google Scholar]
- 32.Sitaraman S., et al. , Proteasome dysfunction in alveolar type 2 epithelial cells is associated with acute respiratory distress syndrome. Sci. Rep. 9, 12509 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ochsner S. A., et al. , The Signaling Pathways Project, an integrated 'omics knowledgebase for mammalian cellular signaling pathways. Sci. Data 6, 252 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grandjean J. M. D., et al. , Pharmacologic IRE1/XBP1s activation confers targeted ER proteostasis reprogramming. Nat. Chem. Biol. 16, 1052–1061 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sisson T. H., et al. , Targeted injury of type II alveolar epithelial cells induces pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 181, 254–263 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korfei M., et al. , Epithelial endoplasmic reticulum stress and apoptosis in sporadic idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 178, 838–846 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Penke L. R. K., Speth J., Wettlaufer S., Draijer C., Peters-Golden M., Bortezomib inhibits lung fibrosis and fibroblast activation without proteasome inhibition. Am. J. Respir. Cell Mol. Biol. 66, 23–37 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mutlu G. M., et al. , Proteasomal inhibition after injury prevents fibrosis by modulating TGF-β(1) signalling. Thorax 67, 139–146 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fineschi S., et al. , In vivo investigations on anti-fibrotic potential of proteasome inhibition in lung and skin fibrosis. Am. J. Respir. Cell Mol. Biol. 39, 458–465 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Saglam B., et al. , Bortezomib induced pulmonary toxicity: A case report and review of the literature. Am. J. Blood Res. 10, 407–415 (2020). [PMC free article] [PubMed] [Google Scholar]
- 41.Miyakoshi S., et al. , Severe pulmonary complications in Japanese patients after bortezomib treatment for refractory multiple myeloma. Blood 107, 3492–3494 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Heijmans J., et al. , ER stress causes rapid loss of intestinal epithelial stemness through activation of the unfolded protein response. Cell Rep. 3, 1128–1139 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Argemí J., et al. , X-box binding protein 1 regulates unfolded protein, acute-phase, and DNA damage responses during regeneration of mouse liver. Gastroenterology 152, 1203–1216.e15 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Liberti D. C., et al. , Alveolar epithelial cell fate is maintained in a spatially restricted manner to promote lung regeneration after acute injury. Cell Rep. 35, 109092 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.J. Katzen, M.F. Beers, A. Babu, SP-C BRICHOS mutation mouse model. NCBI Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE189480. Deposited 24 November 2021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data generated in this study has been deposited to GEO with accession No. GEO:GSE189480 (45).
All study data are included in the article and/or supporting information.