Significance
Genetic loci identified by genome-wide association studies for susceptibility to Alzheimer’s disease are robust; however, the underlying pathways are not well understood. By integrating genomics and metabolomics data in humans and mice models, we provide evidence that ABCA7, one of the main genes known for Alzheimer’s disease, is acting, at least in part, through alteration in sphingolipid metabolism and ceramides, which may offer new therapeutic opportunities for Alzheimer’s disease.
Keywords: genome-wide association study, Alzheimer’s disease, ABCA7, metabolomics, ceramide
Abstract
Genome-wide association studies (GWASs) have identified genetic loci associated with the risk of Alzheimer’s disease (AD), but the molecular mechanisms by which they confer risk are largely unknown. We conducted a metabolome-wide association study (MWAS) of AD-associated loci from GWASs using untargeted metabolic profiling (metabolomics) by ultraperformance liquid chromatography–mass spectrometry (UPLC-MS). We identified an association of lactosylceramides (LacCer) with AD-related single-nucleotide polymorphisms (SNPs) in ABCA7 (P = 5.0 × 10−5 to 1.3 × 10−44). We showed that plasma LacCer concentrations are associated with cognitive performance and genetically modified levels of LacCer are associated with AD risk. We then showed that concentrations of sphingomyelins, ceramides, and hexosylceramides were altered in brain tissue from Abca7 knockout mice, compared with wild type (WT) (P = 0.049–1.4 × 10−5), but not in a mouse model of amyloidosis. Furthermore, activation of microglia increases intracellular concentrations of hexosylceramides in part through induction in the expression of sphingosine kinase, an enzyme with a high control coefficient for sphingolipid and ceramide synthesis. Our work suggests that the risk for AD arising from functional variations in ABCA7 is mediated at least in part through ceramides. Modulation of their metabolism or downstream signaling may offer new therapeutic opportunities for AD.
Genome-wide association studies (GWASs) have robustly identified thousands of genetic loci for complex traits and disorders. Genetic loci have been used to identify pathways affected in the early stages of the disease and may differentiate potential causal pathways from effects secondary to the disease process. GWASs have so far identified 47 unique genetic loci for Alzheimer’s disease (AD) (1–3). However, the underlying pathway(s) mediating risk are poorly understood or unknown for most AD-associated genes.
Many of the genes associated with AD, including ABCA7, APOE, and TREM2, are associated with altered lipid metabolism, suggesting this may be a common feature of AD. Untargeted metabolomics measures metabolites, such as lipids and other small molecules, in biological samples and provides an objective means to investigate the metabolic features that may contribute to the disease risk (4–8). However, studying the association of metabolites with diseases in an observational setting is prone to well-characterized biases, such as confounding and reverse causation. Here, we conducted a metabolome-wide genetic association study (MWAS) to detect metabolic features in peripheral blood associated with genetic variants for AD identified in prior GWASs (1–3). Both GWASs and MWASs adopt an agnostic approach that is not biased according to prior knowledge and is more likely to discover novel pathways. The approach aims to uncover metabolic pathways conferring susceptibility to AD before disease onset and therefore provide mechanistic insight into the etiopathogenesis of AD.
We conducted MWASs on genetic variants reported by the largest GWAS on AD (1). We used data from two large epidemiologic cohorts—the Airwave Health Monitoring Study (Airwave) (9) and the Rotterdam Study (10)—with the metabolome measured by ultraperformance liquid chromatography–mass spectrometry (UPLC-MS). Among all genetic loci associated with AD in GWASs, we found the strongest associations between genetic variants in ABCA7 and lipid signals in the hexosylceramide pathway. We therefore focused further analysis on metabolites associated with ABCA7 and tested them against cognitive measures in the Airwave study, the Rotterdam Study, and the Finnish Geriatric Intervention Study to Prevent Cognitive Impairment and Disability (FINGER) trial (11). We then applied Mendelian randomization (12) to assess whether the identified metabolites may be on the causal pathway linking ABCA7 to AD and used a bioinformatic approach to investigate the extent to which ABCA7 was expressed in the brain. Then, taking a reverse translational approach from human studies to functional experiments, we performed targeted lipidomics and the analysis of transcriptomics datasets in brain tissue from the Abca7 knockout (KO) mouse (13) to investigate whether the Abca7 pathway influences lipid metabolism in the brain and whether this pathway also exists in mouse models of amyloidosis. Finally, we show that activation of microglia induces changes in hexosylceramides consistent with these classes of lipids playing a role in neuroinflammation.
Results
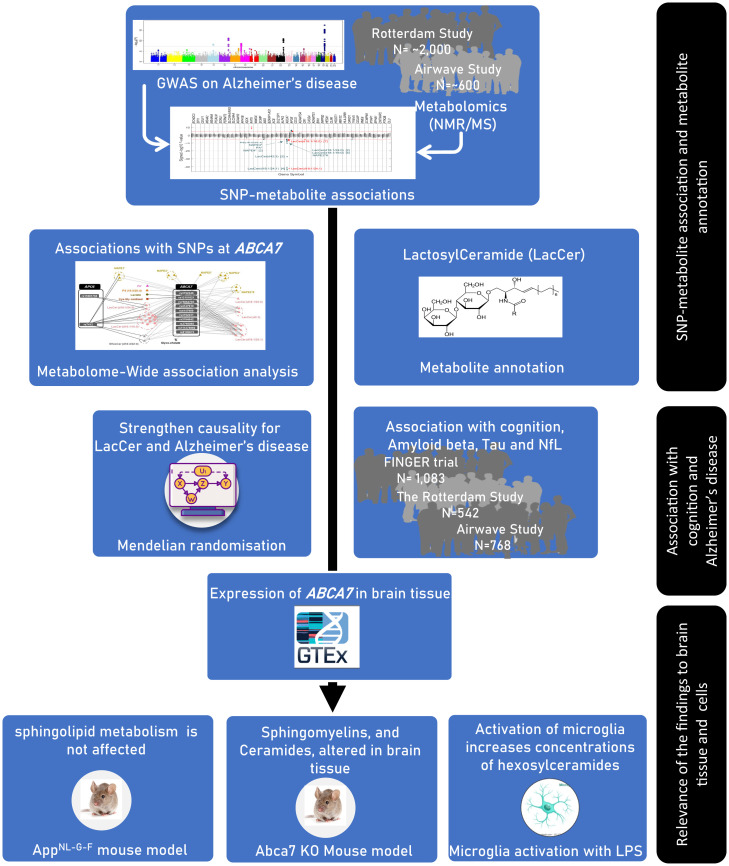
The study workflow is summarized in Fig. 1.
Fig. 1.
The overall design of the study.
MWAS on AD-Related Single-Nucleotide Polymorphisms (SNPs) Identifies Associations between ABCA7 and Lactosylceramides (LacCers).
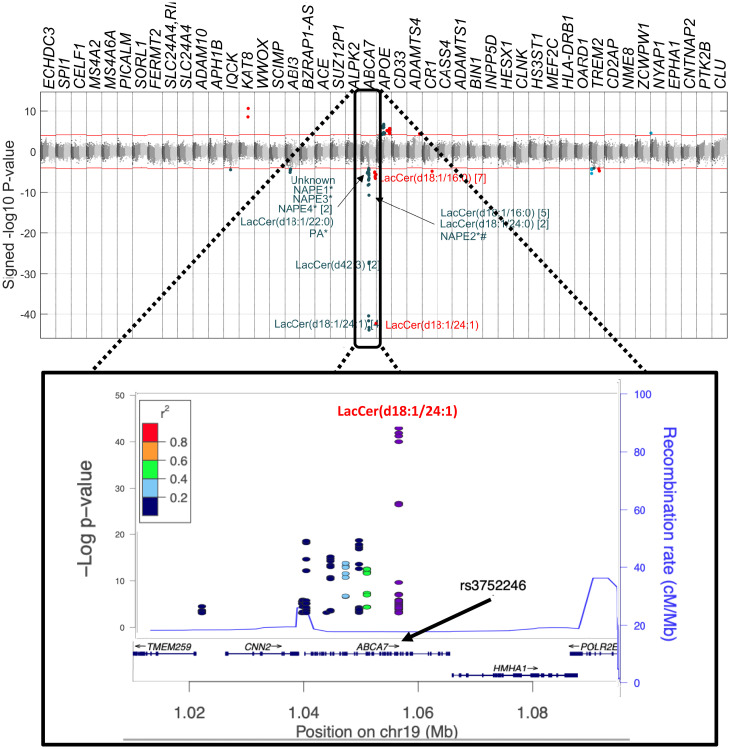
In our UPLC-MS analysis of the Airwave and Rotterdam Study data, we measured 5,199 mass spectral features (the unique mass to charge ratio–retention time pairing [m/z-RT]) and annotated 1,815 of them (Dataset S1). The 47 AD-related SNPs associated with metabolic features at a metabolome-wide significance level (MWSL) (SI Appendix, SI Methods) are shown in the Manhattan plot in Fig. 2. We observed the strongest association (Pearson correlation coefficient = −0.53, P = 7.16 × 10−44) between rs3752246 in ABCA7 and a feature with m/z = 1,068.696 and RT = 7.84 min in the negative ionization mode spectrum (abbreviated as LNEG, 1,068.6958_7.8401), which corresponds to LacCer(d18:1/24:1) (Fig. 2 and SI Appendix, Fig. S1 and Dataset S2). Overall, we identified 121 associations with 10 AD-related SNPs at ABCA7 that surpassed the 5% family-wise error rate (FWER) significance threshold (Dataset S2). These features corresponded to 41 unique mass spectrometry features (MWSL α = 0.05) mapped to 16 metabolites (Fig. 3 and Dataset S2 and SI Appendix, SI Methods). Ten of the 41 metabolic features were from LacCers or sulfatide hexosylceramides (SHexCers). In subsequent analyses where a single feature is used to represent a given metabolite, we have chosen either the dominant feature that represents the parent ion or the major adduct of the parent ion, whichever was the greater in relative intensity.
Fig. 2.
Manhattan plot on the association of metabolites detected by mass spectrometry with AD-related SNPs. Features associated with ABCA7 SNPs are annotated. For some metabolites, more than one metabolic feature was found to be associated with AD-related SNPs, and the No. of metabolic features corresponding to the same metabolites is described in brackets. LacCer: lactosylceramide, NAPE: N-acyl phosphatidylethanolamine, PA: phosphatidic acid. Red features are assayed by negative ionization mode lipidomics and blue features by positive ionization mode lipidomics.
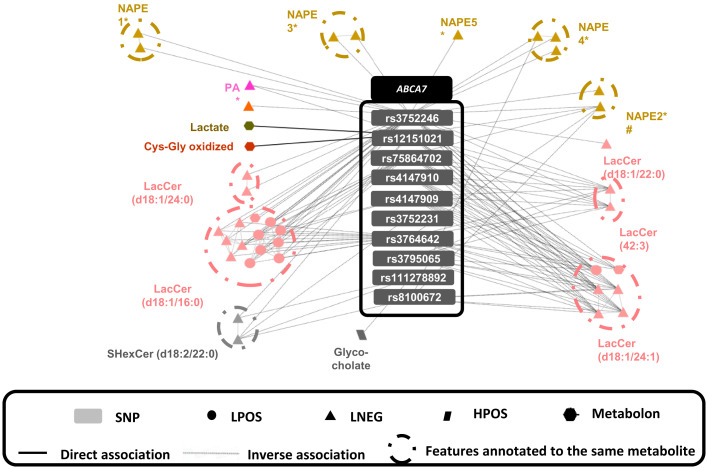
Fig. 3.
Metabolites associated with AD-related SNPs at ABCA7 (dark gray) and their association with AD-related SNPs at APOE (light gray). A circle surrounds all the multiplatform features annotated to the same metabolite, and at least one of those features is associated with the connected SNP. Colors indicate metabolite class, and symbols represent the analytical platform used. The features PA*, NAPE4*, and NAPE5* contain the carbon side chain (20:4).
Association with Cognitive Performance.
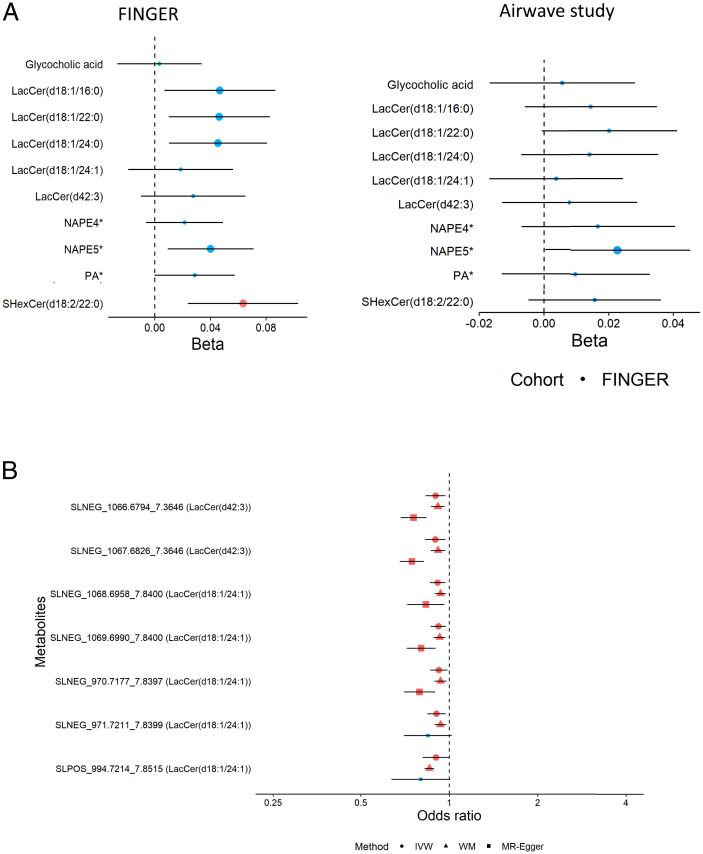
The association of 10 SNPs at ABCA7 with cognition is presented in Dataset S3. Of the 16 metabolomic features associated with ABCA7, 14 were available to test against individual cognitive performance variation in Airwave, 13 in the Rotterdam Study, and 11 in the FINGER trial data, a lifestyle intervention trial following individuals selected based on having a higher risk of late-life cognitive impairment (mean age 69.2 ± 4.7 y). Using mixed-effect regression models, five of the metabolites (including three LacCer species) were positively associated (P < 0.05/11, Bonferroni threshold) with a composite cognitive score (Fig. 4A) based on an extended version of the neuropsychological test battery (14). We then tested for their association with cognition in the Airwave cohort, a younger population (mean age 40.0 ± 9.2 y) unselected for risk of dementia. None of the metabolic features was associated with cognitive performance, suggesting that the association may depend on age and other risk factors (Fig. 4A). We also tested for associations between changes in these 14 metabolic features in plasma and changes in cognitive scores in the FINGER trial, comparing baseline and 2-y follow-up data, and found that a higher level of an N-acylphosphatidylethanolamine annotated as NAPE(P-18:1/20:4/18:1) or NAPE(O-18:0/20:4/18:1) (Dataset S2 for annotation details), and referred to here as NAPE5*, was associated with slower cognitive decline (SI Appendix, Fig. S2). Finally, we examined the association of the metabolic features with three cognitive domains, including working memory, executive function, and processing speed, in the Airwave study, FINGER trial, and the Rotterdam Study. Metabolic features annotated to NAPE5* and PA(O-18:1/20:4) and/or PA(P-18:0/20:4) were nominally associated with working memory and executive function. Moreover, LacCer(d18:1/22:0) was nominally associated with executive function and processing speed and SHexCer(d18:2/22:0) was associated with executive function (Dataset S4).
Fig. 4.
(A) The association of ABCA7-associated metabolites with the cognitive score in the FINGER trial and Airwave. For FINGER, the association between metabolite concentrations and cognition was tested using a linear mixed model adjusted for fixed effects of age at baseline, sex, intervention, visit (binary), and domain of participant groups and for random effects of within-subject variation. For Airwave, the association between metabolite concentrations and cognition at baseline was tested using a linear mixed model adjusted for fixed effects of age at baseline, sex, visit (binary) of participant groups, and random effects of within-subject variation. We obtained the z score for both metabolite levels and cognition values before analysis. Chemical species are represented by their molecular ion or most-abundant adduct ion. Larger dots indicate 95% CI, not including the null value; red dots indicate associations with P < 0.05/11 for FINGER and P < 0.05/14 for Airwave (Bonferroni correction for 11 and 14 features, respectively). (B) Associations of LacCer(d18:1/24:1) and LacCer(d42:3) with the risk of AD based on Mendelian randomization. Dots indicate inverse variance weighted (IVW) estimates, triangles indicate weighted median (WM), and squares indicate Egger estimates. Red and larger shapes indicate significant associations with P < 0.05.
Association with Mild Cognitive Impairment and Dementia.
We examined the association of the metabolic features with the incidence of dementia in the Rotterdam Study. We observed a nominally significant association between a metabolic feature annotated to PA(O-18:1/20:4) and incident dementia (SI Appendix, Tables S5 and S6).
Mendelian Randomization Analysis.
Mendelian randomization is an instrumental variable analysis in which genetic variants are used as proxies for specific exposures (in this case, metabolites) to overcome unmeasured confounding and reverse causation in observational studies (15). From GWAS on Airwave data, we identified genetic instruments (SNPs) for Mendelian randomization analysis (1) for 32 of the 41 mass spectral features related to the 10 AD-related SNPs at ABCA7.
The list of genetic variants for each metabolic feature is presented in Dataset S7. A two-sample Mendelian randomization analysis (16) was then performed using the genetic variants as instruments for the associated metabolites and testing these variants against the risk of AD in GWAS data. Using the inverse variance weighted method and after applying a false discovery rate of 0.05, we found supporting genetic evidence for inverse associations of LacCer(d18:1/24:1) and LacCer(d42:3) with AD (Fig. 4B and Dataset S8). To assess possible violation of assumptions underlying Mendelian randomization analysis, we carried out weighted median and Mendelian randomization (MR)-Egger methods as sensitivity analyses (see Materials and Methods). The findings were consistent with the inverse variance weighted estimates.
Association with Pathologic Biomarkers of AD.
Using data from the Rotterdam Study, we examined the association between 35 metabolic features associated with ABCA7 with pathologic biomarkers of AD. Metabolic features annotated to LacCer(d18:1/24:0) and LacCer(d18:1/24:1) showed nominally significant associations with the circulatory levels of amyloid beta 40, amyloid beta 42, and neurofilament light chain (NfL), but not Tau (SI Appendix, Tables S9–S12).
Using data from the FINGER trial, we found associations between metabolic features annotated to LacCer(d18:1/24:1), LacCer(d42:3), LacCer(d18:1/24:0), LacCer(d18:1/22:0), LacCer(d18:1/16:0), SHexCer(d18:2/22:0), NAPE(O-18:1/20:4/18:1)*, and NAPE(O-18:1/20:4/18:0)* with the total gray-matter volume as well as cortical thickness in AD signature regions (average of entorhinal, inferior temporal, middle temporal, and fusiform regions) (SI Appendix, Tables S13–S18) (17).
Ceramide Metabolism in Brain Tissue.
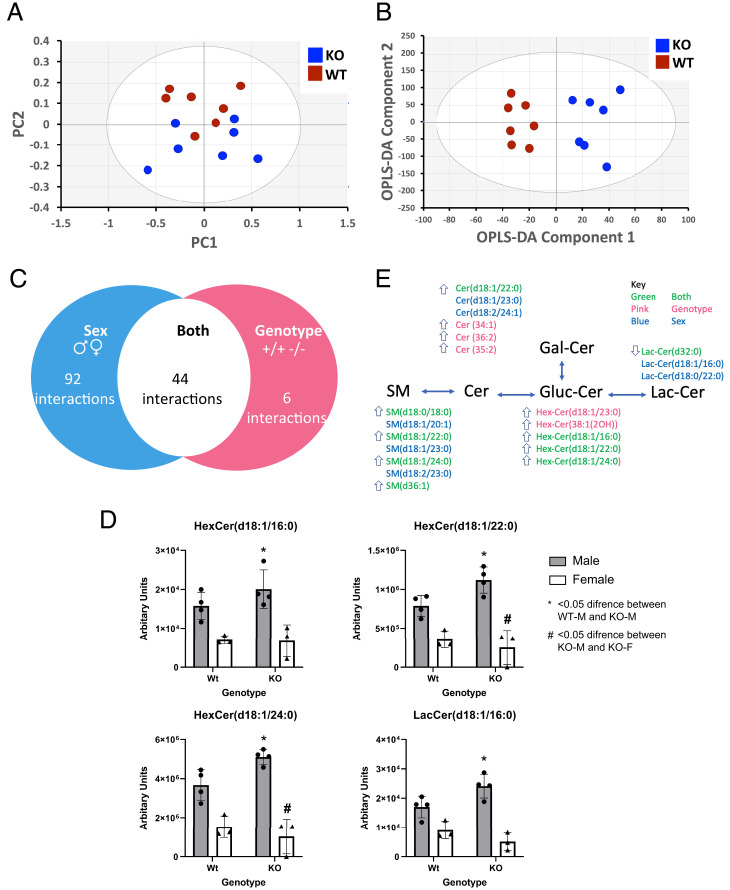
Using expression quantitative trait loci (eQTL) data from the Genotype-Tissue Expression project (18), we found that the 10 ABCA7 SNPs were associated with lower expression of ABCA7 in multiple human brain tissues, including the cerebellum and cerebral hemisphere (SI Appendix, Fig. S3). We then carried out metabolic profiling of brain tissue (left hemispheric cerebral cortex) from the Abca7 KO mouse versus wild type (WT) to test whether variations in lipid profiles related to ABCA7 in peripheral blood in the human studies above are also found in mouse brain tissue. We performed UPLC-MS in both positive and negative ionization modes on the lipid phase of chloroform/methanol/water extractions of the mouse cerebral cortex (Dataset S19) and then performed principal-component analysis (PCA) to visualize dominant patterns (Fig. 5). While the main variance in the positive ionization mode data was driven by sex (PC1 = 80.4%, PC2 = 8.7%), the principal factor discriminating samples in the negative ionization mode data was attributed to genotype (PC1 = 59%, PC2= 14.2%, PC3 = 6.9%).
Fig. 5.
Lipidomics analysis of the cortex of the Abca7-null mouse identifies alterations in ceramide metabolism in the brain. (A) Principal-component analysis (PCA) of the lipidomic data collected in negative ionization mode (variation represented by PC2 = 14.2%, PC3 = 6.9%). (B) PCA of the lipidomic data collected in positive ionization mode (variation represented by PC1 = 80.4%, PC2 = 8.7%). (C) Venn diagram of lipidomic changes detected in the dataset summarizing all sphingolipid changes associated with genotype (Abca7), sex, and their interaction by ANOVA. (D) Box plot of ANOVA results for hexosylceramide (HexCer) (d18:1/16:0), HexCer (d18:1/22:0), HexCer (d18:1/24:0), and lactosylceramide (LacCer) (d18:1/16:0). (E) Schematic of key changes detected in sphingomyelins (SMs), ceramides (Cers), HexCer, and LacCer detected in both the cortex of the Abca7-null mouse and the human MWAS analysis. Arrow indicates a significant increase or decrease in sphingolipid species by ANOVA in KO males relative to WT males (genotype). WT: wild type, KO: Abca7-null, m: male, f: female.
Based on associations of plasma LacCer concentrations with the ABCA7 locus in the human studies, we filtered the mouse lipidomic data specifically for sphingolipids (sphingomyelins, ceramides, and hexosylceramides). We performed a two-way ANOVA to investigate genotype–sex interactions. Nine sphingolipids detected in negative ionization mode differed between the Abca7 KO and WT mice (P < 0.05), with seven associated with genotype and two associated with sex (one passing the Bonferroni correction for multiple testing). Repeating this analysis for the positive ionization mode dataset, 244 differences were identified across the detected sphingolipids, of which 159 were associated with sex (two passing the Bonferroni correction for multiple testing) and 44 were associated with interactions between sex and Abca7 genotype (24 passing the Bonferroni correction for multiple testing). It was notable that, while the one common LacCer decreased in concentration in the brain tissue of Abca7 KO mice, the common sphingolipids, ceramides, and hexosylceramides all increased in concentration, suggesting Abca7 is associated with a failure to process sphingolipids into LacCer species. These data suggest a role for ABCA7 in regulating ceramide metabolism in the brain.
A previous study of Abca7 heterozygous mice failed to detect any changes in ceramide derivatives but did detect impaired microglia function following inflammatory stimuli (19). The lack of lipidomic changes may have been related to the small effect size in the heterozygous mice at a relatively early age (55–58 d), and so we examined the translation of enzymes involved in sphingolipid metabolism (sphingolipid metabolism, map 00600; sphingolipid signaling pathway, map 04071) and the ABCA transporters to test whether sphingolipid metabolism is altered in the brain of heterozygous mice (data from GSE 139592). The four groups in the dataset (WT and Abca+/−, with and without lipopolysaccharide [LPS] stimulation) were readily separated by orthogonal projections to latent structures discriminant analysis (OPLS-DA) of the mRNA levels of two pathways and Abca genes (SI Appendix, Fig. S4A; R2X = 91%, R2Y = 98%, Q2 = 47%; passing random permutation test). Abca+/− were associated with the increased translation of Abca15, Abca4, and Sptlc3 and decreased Abca9, Ugt8a, Hexb, Sgms1, and Sgpp2, while LPS treatment increased Sgms2, Acer2, and Abca1 and decreased Abca9 and 9130409I23Rik (ortholog delta 4-desaturase sphingolipid 1) (SI Appendix, Fig. S4B).
To test whether LacCers are common to all forms of AD, we examined the lipid profile of brain tissue (left cerebrum) from AppNL-G-F mouse (a model of amyloidosis with humanized APP with Swedish, Iberian, and Arctic familial mutation; n = 6 AppNL-G-F, n = 5 WT), performing LC-MS/MS and multivariate statistics to investigate perturbations in sphingolipid metabolism. While OPLS-DA could separate the samples from AppNL-G-F mouse brain from WT samples, these models failed to pass cross-validation (SI Appendix, Fig. S5). As these were young mice, we also examined a dataset from GEO examining the effects of aging in the APP/PS1 mouse (GEO GSE 86828), demonstrating that alterations in sphingolipid metabolism and Abca transporter expression in aged mice (7- vs. 18-mo-old mice) (SI Appendix, SI Results and Fig. S6). We also built an OPLS-DA model to examine sphingolipid metabolism in this dataset, and this also failed to pass cross-validation, indicating that any lipidomic changes associated with genotype had a relatively small effect size, particularly when compared with the Abca7 KO mouse model. For univariate analysis, no significant differences were detected for LacCer, suggesting that any alterations in sphingolipid metabolism in this mouse model of early-onset AD are much subtler than those associated with ABCA7 (SI Appendix, Fig. S6). Overall, these data do not suggest that sphingolipid metabolism is a shared underlying mechanism for early-onset as compared with late-onset AD.
Alterations in Ceramide Metabolism in Activated Microglia.
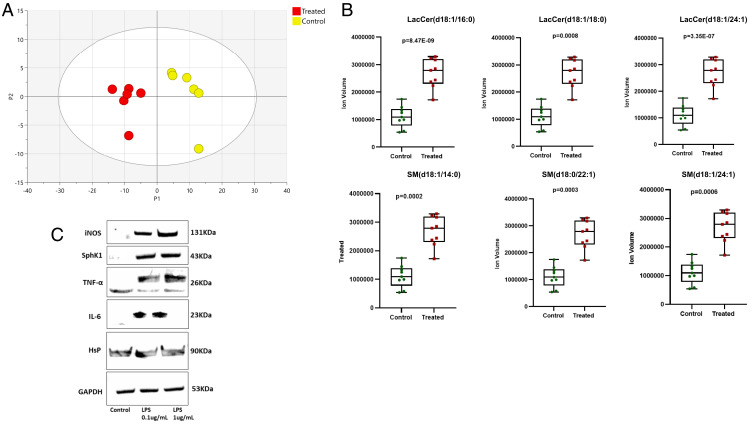
Given that inflammation was associated with alterations in sphingolipid metabolism and to further investigate the role of hexosylceramides in the brain, we examined lipid changes in neuroinflammation triggered by activation of microglia, a hallmark of early-stage AD. Human microglia (HMC3) was cultured in the presence of increasing concentrations (0.1 μg/mL; 1.5 μg/mL) of LPS to induce inflammation. Exposure to 1 μg/mL of LPS for 24 h caused a robust increase in a wide range of lipids as evidenced by PCA (Fig. 6A), including increases in LacCers (d18:1/16:0, d18:1/18:0, and d18:1/24:1) and sphingomyelins (SM(d18:1/14;0), SM(d18:0/22:1), and SM(d18:1/22:1)) (Fig. 6B). Activation of microglia was associated with an increase in expression of sphingosine kinase 1 (SphK1), as well as cytokines and other inflammatory markers (Fig. 6C).
Fig. 6.
Lipidomic changes in cultured human microglia following lipopolysaccharide (LPS)-induced activation. (A) PCA of the lipidomic profile of microglial cells with and without LPS activation. (B) Box plots of key changes in lactosylceramides and sphingomyelins following LPS activation. Control: no treatment. Treated: addition of LPS. P values are based on the Student’s t test. (C) Western blot analysis of protein changes induced by LPS activation in human microglia. iNOS: inducible nitric oxide synthase, Sphk1: sphingosine kinase 1, TNF-α: tumor necrosis factor-alpha, IL-6: interleukin-6, HsP: heat shock protein, GAPDH: glyceraldehyde-3-phosphate dehydrogenase.
Discussion
Our genomic and metabolomic data from human, animal, and in vitro studies provide evidence that LacCers play a potentially causal role in linking variants in ABCA7 with the risk of AD. Our MWAS indicated that, among genes so far identified for AD, ABCA7 had the strongest associations with metabolic traits in peripheral blood. Most of these features were annotated to LacCers; the strongest association was found between rs3752246 in ABCA7 and LacCer (d18:1/24:1). We confirmed that deletion of Abca7 in the mouse results in a range of alterations in sphingolipid metabolism, particularly for ceramides and hexosylceramides, which indicate a failure to process sphingolipids to LacCers. Reduced expression of Abca7 in heterozygous mice was also associated with alterations in sphingolipid metabolism. However, these associations were not seen in a mouse model of early-onset AD, although there was some evidence of altered sphingolipid metabolism in aging APP/PS1 mice, suggesting that the LacCers may preferentially be involved in the etiopathogenesis of late- as compared with early-onset AD. We also demonstrate that, during activation of microglia, as occurs in neuroinflammation, these hexosylceramides increase in intracellular concentration in part through induction in the expression of sphingosine kinase, an enzyme with a high control coefficient for sphingolipid and ceramide synthesis.
The genetic loci associated with AD can be separated into two broadly defined groups of genes. The first set includes a small No. of genes (APP, PSEN1, and PSEN2) with a strong and autosomal dominant hereditary effect and is associated with familial, early-onset AD. For these loci, potential underlying molecular mechanisms have been defined in some detail and underpin the “amyloid hypothesis” postulating that pathological amyloid-beta peptides are a primary cause of neurodegeneration (20–22). The second group of genes is more extensive and includes genes implicated in phagocytic, lipid, and inflammatory pathways (1) that may interact with comorbid disease or environmental risk factors in the development of late-onset AD (19, 23, 24). However, relative to the early-onset Mendelian forms, much less is known about primary causal mechanisms associated with late-onset AD and how mechanisms initiating the late- and early-onset forms of AD lead to a common pathology.
ABCA7 is part of the adenosine triphosphate–binding cassette (ABC) reporter family, important in regulating phospholipid and cholesterol transport across the cell membrane and within the cell between subcellular organelles (19). The transporter has also been shown to have a higher preference for transporting phosphatidylserine lipids relative to phosphatidylcholines out of the cell when compared with ABCA1 (25), suggesting the transporter has an important role in maintaining and regulating intracellular phosphatidylserine concentrations. ABCA7 is highly expressed in microglia in cell culture and neuronal cells, particularly in the hippocampus (26). From cell-culture experiments, ABCA7 expression is regulated by sterols through the SREBP2 pathway (27), and increased expression of ABCA7 has been shown to enrich HeLa cells with ceramides (28). Neuronal cells that lack ABCA7 show evidence for increased endoplasmic reticulum (ER) stress and amyloid beta 40 (Aβ40) and Aβ42 production. Sakae et al. (13) proposed that ABCA7 deficiency alters lipid metabolism in ways that secondarily increase SREBP2 and BACE1 expression and induce ER stress, contributing to the generation of pathological Aβ species, which accumulate and may lead to cognitive deficits prodromal to AD. Consistent with this model, genetic variants at ABCA7 are associated with relative atrophy of the cortex and hippocampus both in cognitively normal people and in those with mild cognitive impairment (29).
Our untargeted lipidomic assay of Abca7 KO versus WT mouse brains showed decreases in a wide range of phosphatidylcholines, phosphatidylserines, and phosphatidylglycerols and differences in the concentrations of more than 250 sphingolipids (including ceramides) in the KO model. These results support a role for ABCA7 in sphingolipid/ceramide metabolism, particularly in terms of the synthesis of hexosylceramides and LacCers. Consistent with this, Sakae and colleagues used a targeted lipidomics assay of 275 lipids to assess lipidomic changes in the brains of Abca7 KO mice (13). They found differences in 26 specific lipids: decreases in 12 species of phosphatidylethanolamines, three phosphatidylglycerols, one lysophosphatidylcholine, and two sphingomyelins and increases in three phosphatidylcholines, one ceramide, three sulfatides, and one cerebroside. Furthermore, we showed that activation of microglia using LPS caused an increase in intracellular hexosylceramides and sphingolipids, in part associated with an increase in expression with SphK1, a key enzyme in the synthesis of sphingolipids and ceramides. Alterations in sphingolipid metabolism were also apparent in the transcriptional profile of brain tissue of Abca7 heterozygous mice (19) and during aging in APP/PS1 mice at 18 mo, when plaque formation is most apparent (30).
One question raised by this research is how translatable the results are from the ABCA7 gene to other genetic causes of AD. We found no evidence of sphingolipid metabolism changes in the AppNL-G-F mouse of early-onset AD in a similarly powered mouse study, indicating that any alterations in lipid metabolism in this mouse model are subtler. However, at the transcriptional level, sphingolipid metabolism is perturbed as APP/PS1 mice age from 7 mo to 18 mo, over which time plaque formation progresses from mild to severe (30). As noted above, ABCA7 is associated with late-onset AD, along with several other genes associated with AD and altered lipid metabolism, including APOE and TREM2, suggesting sphingolipid metabolism may particularly be associated with late-onset AD compared with early-onset AD.
Previous human studies have shown inconsistent relationships of ceramides with cognitive impairment (31), memory impairment (32), and dementia field (33). The Women’s Health and Aging Study II and a clinic-based case-control study, both including small Nos. of individuals, reported inconsistent associations of plasma ceramides with memory loss, mild cognitive impairment, and risk of AD. A GWAS on 136 targeted metabolites in a Chinese population reported an inverse association between the risk allele of rs3752246 in ABCA7 and the ceramide d18:1/24:1 (34). However, that study was limited by the inclusion only of metabolites available in the targeted metabolomics platform, in comparison with our study, which had a broad representation of the metabolome through an untargeted metabolomics approach. Furthermore, our study was designed to investigate metabolic pathways associated with AD-related genetic loci, which provided greater specificity and statistical power than the genome-wide approach used in the above GWAS. In addition, we assessed possible causality by Mendelian randomization and applied a reverse translational approach to validate our findings in both animal and cellular models.
We used Mendelian randomization to test for potential causal relationships between LacCer concentrations in plasma and AD. We first performed GWAS on metabolic features in Airwave to generate genetic instruments for the target LacCers and then took advantage of the most-recent and largest AD GWAS data to test for associations of these genetic variants with AD (2). Using this approach, we found evidence of possible causal associations of decreased concentrations of plasma LacCer (d18:1/24:1) and LacCer (42:3) with increased risk of AD, suggesting that ABCA7 contributes to the regulation of systemic LacCer concentration in the genesis of AD.
Gene-set analyses have implicated pathways involving lipid metabolism in AD more generally, in addition to those for inflammation and amyloid processing (1). These observations suggest that the genesis of AD shares features with the metabolic disease–associated inflammation (“metainflammation”) that affects adipose and other peripheral tissues in the metabolic syndrome (35). In this context, we note that four metabolites, NAPE3*, NAPE4*, NAPE5*, and PA* (Dataset S2), and one unassigned metabolite corresponding to the feature SLNEG_782.4965 3.5812 associated with ABCA7 SNPs have arachidonic acid (20:4) side chains, while LacCers activate cPLA2-ɑ, a phospholipase that hydrolyses arachidonic acid from phospholipids (36). We further note that arachidonic acid, a substrate for cyclo-oxygenase and lipoxygenase, is a driver of macrophage and microglial proinflammatory pathways in the aging and AD (37).
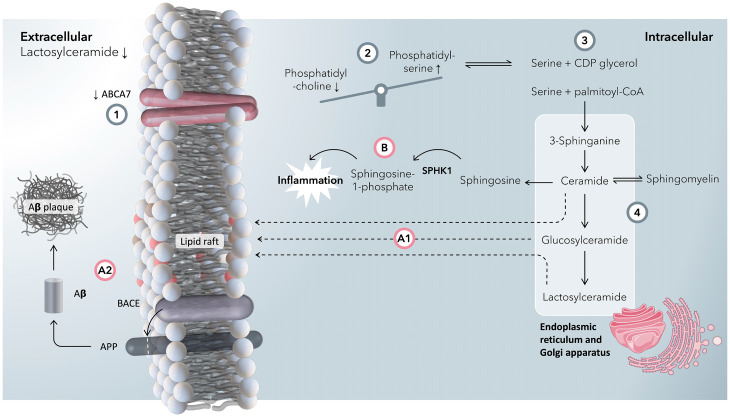
ABCA7 is widely expressed throughout the body, including the brain, particularly in myeloid cells (https://www.proteinatlas.org/). AD risk–raising SNPs were all associated with reduced ABCA7 activity, and previous work has shown that Abca7 KO increases soluble Aβ in an APP mouse model (38). Taken together, we propose a mechanism where impaired ABCA7 function results in alterations in sphingolipid metabolism in the brain, including both neuronal and microglial cells (Fig. 7). We postulate that the reduced concentration of LacCers in blood plasma associated with ABCA7 SNPs arises because of impaired efflux of phosphatidylserines impacting systemic metabolism. Increased intracellular concentrations of phosphatidylserines drive the production of sphingolipids, ceramides, and hexosylceramides as measured in the Abca7-null mouse. Ceramides and hexosylceramides have been postulated to increase lipid rafts in cell membranes (39), and in turn, it has been speculated that changes in cell-membrane fluidity drive increased processing of APP by BACE1 (13), increasing amyloid plaque formation.
Fig. 7.
A summary graphic of the proposed mechanism by which ABCA7 dysfunction is associated with an increased risk of developing Alzheimer’s disease. (1) A reduction in expression/activity of ABCA7 is associated with an increase in AD risk, as well as a reduction in blood-plasma concentrations of lactosylceramides. In our putative mechanism, we rationalize this with a reduction in the flux of lipids through ABCA7. (2) ABCA7 has more affinity to export phosphatidylserines (PSs) than phosphatidylcholines (PCs) across the plasma membrane compared with other ABCA lipid transporters (e.g., ABCA1) (25). With a reduction in the function of ABCA7, intracellular concentrations of PSs increase relative to PCs. (3) As PSs accumulate intracellularly, so does serine (PS is broken down to CDP-glycerol and serine). Serine and palmitoyl-coenzyme A (CoA) form 3-sphinganine at the start of the ceramide synthesis pathway. (4) In the ceramide pathway, 3-sphinganine is synthesized in the ER, which after some steps is converted into ceramides and, in subsequent steps, glucosylceramides and lactosylceramides in the Golgi apparatus. At this stage, there is a branch point. (A1) Ceramides, glucosylceramides, and lactosylceramides can all form lipid rafts (39) in the plasma membrane. (A2) Lipid rafts encourage interactions between β-site APP cleaving enzyme 1 (BACE1) and amyloid-β precursor protein (APP) to make amyloid-β, which ultimately forms plaques (13). (B) Ceramides are broken down to sphingosine in the ER, a precursor for sphingosine-1-phosphate via SPHK1, both of which are increased in inflammation as well as induce inflammation in a feedforward step. Because of disrupted PS metabolism and a failure in exporting PS to the extracellular space, blood concentrations of lactosylceramide decrease.
In addition, activation of microglia induces an increase in sphingolipid metabolism, in part through the induction of SphK1, which leads to an increase in intracellular hexosylceramides.
Thus, in summary, the observed reduction in blood serum lactose–ceramides detected in our cohort studies may reflect an impaired transport of these lipids out of cells in the brain, which subsequently accelerates the deposition of amyloid plaques and may also directly contribute to neuroinflammation. The same ceramides are also increased in the activation of microglia during inflammation and may contribute to lipid remodeling of cell membranes and the processing of APP to form amyloid plaques.
Our study has some limitations. First, GWAS signals may reflect indirect associations, which means that the causal variant may be a mutation in the vicinity of the top SNP and work through a gene other than the one closest to the SNP. Here, we present eQTL data supporting the sentinel SNP being associated with expression levels of the ABCA7 gene. Moreover, the mouse-model results indicate that the Abca7 gene is related to sphingolipid metabolism. Second, we discovered associations of LacCers with the ABCA7 locus using peripheral blood rather than directly in the brain. Thus, our results either imply that systemic biochemical pathology promotes the genesis of AD (e.g., via inflammation) or that there is a related lipid dyshomeostasis in the central nervous system. However, our observations of differences in concentrations of other lipids (particularly sphingolipids, ceramides, and hexosylceramides) in the Abca7 KO mouse brain relative to WT controls suggest the latter possibility. Third, rather than directly testing causal associations between concentrations of LacCers and AD, we carried out Mendelian randomization analyses in which genetic information was used to identify potential causal links between the LacCers and the risk of AD. This approach involves three key assumptions (15): (i) the genetic variants are associated with the risk factor (LacCer metabolites in this case); (ii) there are no unmeasured confounders between genetic variants and outcome (AD); and (iii) the genetic variants affect the outcome only through their effect on the risk factor (that is, there are no directional pleiotropic effects). We fulfilled the first assumption by setting strict criteria for selecting genetic variants (P < 5 × 10−8 and F-statistics > 10). It is not easy to verify the extent to which the second and third assumptions hold, but our findings were robust to sensitivity analyses, and the MR-Egger method did not indicate any potential directional pleiotropic effects. Finally, a heterozygote mouse model would have been preferential, as it could be a better representative of the lower ABCA7 expression in humans.
In conclusion, we report associations between SNPs at the ABCA7 locus and LacCers in peripheral blood. The SNPs were also associated with differences in cognitive function and have potential causal links with the risk of AD (40). Manipulation of sphingolipid metabolism is already being explored for augmenting the efficacy of anticancer and insulin resistance therapeutics (41). These may point to effective strategies that could be tested as “repurposed” treatments for AD.
Materials and Methods
Study Samples.
Airwave is an occupational cohort of 53,116 police officers and staff aged 18 y and over across Great Britain launched in June 2004 (9). Blood samples were obtained at the screening visit (with consent) and stored at −80 °C at the laboratory before being transferred to a biorepository facility and stored in vapor-phase liquid nitrogen. Metabolomic assays were done for ∼4,000 samples (Dataset S1). The samples were divided into two separate sample sets (referred to herein as Airwave 1 and Airwave 2). Airwave 1 utilized lithium heparin plasma samples, and Airwave 2 used ethylenediaminetetraacetic acid (EDTA) plasma samples. Both sets of samples were analyzed at the National Phenome Centre (Imperial College London, London, UK). In addition, a set of 2,250 samples was analyzed by Metabolon, Inc. (Morrisville, NC, USA), of which 1,000 were also included in Airwave 2.
The Rotterdam Study is a prospective cohort study of individuals in the Ommoord district of Rotterdam, the Netherlands (10). At baseline, between 1990 and 1993, 7,983 participants over 55 years old were interviewed at home and underwent extensive clinical examination at the research center. Serum samples were collected from 1997 to 1999, stored at −20 °C, and analyzed for metabolites by Metabometrix Ltd. (London, UK) using protocols adapted from the National Phenome Centre. The Airwave and Rotterdam sample and feature Nos. are described in Dataset S1.
FINGER is a randomized controlled trial of 1,260 individuals aged 60–77 y with increased risk for dementia based on the Cardiovascular Risk Factors, Aging, and Incidence of Dementia risk score and cognition at a mean level or slightly lower than expected for their age (42). The population was randomly assigned to a multidomain intervention (diet, exercise, cognitive training, and vascular risk monitoring) or a control group (general health advice) in a 1:1 ratio. We used metabolomics and cognitive data at baseline and 2-y follow-up across both the intervention and control groups. EDTA plasma samples were analyzed at the National Phenome Centre.
Genomics Data.
For both Airwave and the Rotterdam Study, DNA samples were extracted from leukocytes. Genotyping was done using Illumina Infinium HumanExome-12v1-1 BeadChip Array in Airwave and Illumina 550 K arrays in the Rotterdam Study. Both studies imputed their data to 1000 Genomes Phase 3 reference panel, and the imputed data were used throughout.
Metabolomics Data.
For each cohort (Airwave 1, Rotterdam Study, and FINGER), metabolomics data were acquired using UPLC-MS. Methods and quality control (QC) have been described previously (43–45). Briefly, a pooled study reference sample was prepared for each population. Serum and plasma long-term reference samples were prepared using commercial bulk serum and plasma (BioIVT [Seralab], West Sussex, UK). These QC samples were analyzed at regular intervals throughout data acquisition. Mixtures of authentic reference standards were added to the study reference, long-term reference, and study samples used in the UPLC-MS analysis to enable targeted data-quality monitoring during acquisition.
Plasma samples from FINGER were prepared and underwent UPLC-MS profiling analysis for lipids and small metabolites as previously described (43). The same procedures were used to analyze plasma samples from Airwave and serum samples from the Rotterdam Study except that 100 μL of samples was used without dilution before the addition of isopropanol for the lipidomics analyses. Briefly, batches of 80 samples were prepared into 96-well plates. Each sample was mixed with four parts of 4 °C isopropanol, incubated at 4 °C for 2 h in a plate shaker at 1,400 rpm, centrifuged, and the supernatant aliquoted into a 96-well plate.
All analyses were acquired on Acquity UPLC systems coupled to Xevo G2-S ToF mass spectrometers (Waters Corporation, Milford, MA, US). For these samples, three assays were employed: reversed-phase chromatography (RPC) for lipid profiling in positive ion mode (LPOS), RPC for LNEG, and hydrophilic interaction liquid chromatography for small molecule profiling in positive ion mode (HPOS).
Data Processing.
Peak picking was completed using Bioconductor R-package XCMS (46). Drift correction was done using a method previously described (47). For FINGER, we used nPYc toolbox (48). Negative values were replaced with zeros, and the data were natural log transformed after adding one. We filtered the data based on RT to exclude nonretained and cleaning phase features—only features in the following RTs (in minutes) were accepted: HPOS (0.5–7), LNEG (0.3–9.5), and LPOS (0.45–12). We used PCA to identify samples that were outliers and excluded them (HPOS = 8, LNEG = 4, and LPOS = 1).
Further, we excluded values that were greater than five median absolute deviations (MADs) from the median (HPOS = 7,216, LNEG = 6,106, and LPOS = 8,285). We used ten principal components from genome-wide scans to adjust for population stratification. Finally, we transformed the data into z scores using median and MAD, so that we had comparable intensities across studies. We determined cross-study feature correspondence for the Airwave and the Rotterdam Study using an in-house algorithm that selects the matches that are the closest in RT and m/z ratio in the two prealigned datasets.
Metabolomic feature intensities were normalized across samples to be comparable across cohorts. In each cohort separately, the value of each variable was brought to zero, then one was added and was transformed using a natural log scale. Each feature was then subtracted from its median and divided by its MAD. After combining the data from all cohorts, the dataset was additionally subtracted from its grand median and divided by the MAD.
UPLC-MS Metabolite Annotation.
Lipid annotation was initially completed by matching accurate mass fragmentation measurements to reference spectra from online databases (LIPID MAPS (49), Metlin (50), and Human Metabolome Database (51)) and previous publications. Where chemical reference materials were commercially available (Avanti Polar Lipids, Sigma Aldrich, Cayman Scientific), they were used to generate definitive molecular identification by direct matching of chromatographic and spectral qualities (including accurate mass, MS/MS spectra, and isotopic distribution) to those observed in the profiling data. NAPE standards were synthesized in house using the protocol described in SI Appendix, Supplementary Methods. For FINGER data, annotations were transferred from the Airwave data after finding feature correspondence between the two datasets, using the same method as described above. Annotation confidence is reported according to guidance in the Metabolomics Standards Initiative (52) (details in Dataset S2 footnotes), any additional annotations were done as described above.
Selection of genetic variants.
We identified 47 unique genetic loci based on three recent GWASs on AD (1–3). The lead SNP from 45 genetic loci was available in our data and was analyzed in the exploratory-stage MWAS (see below). To focus on the pathways linking ABCA7 and the risk of AD, we selected SNPs with a P < 1 × 10−5 within ±500 kb from the lead SNP in ABCA7 using the most recent GWAS by the International Genomics of Alzheimer’s Project (IGAP) consortium (2) (discovery stage: Ncase/control = 21,982/41,944). For correlated SNPs with r2 ≥ 0.5, we kept the SNP with the smallest P value, resulting in 10 SNPs selected for the MWAS focusing on ABCA7.
MWAS.
We carried out linear regression to study the association of each SNP with all metabolomic features (Metabolon, HPOS, LNEG, and LPOS) with adjustment for age, sex, and cohort. We used a permutation-based method to estimate the MWAS significance level (53, 54) to account for multiple testing and the high degree of correlation in metabolomics datasets. A P value threshold giving a 5% FWER was computed for each SNP in each metabolomics platform.
Representative features.
For subsequent analysis, we used a single feature to represent each metabolite. The representative feature was chosen based on the major detected ion (either parent ion, closest adduct, or isotope that gave the largest intensity and was not confounded by isobaric species).
Association with Cognition.
We examined the association of ABCA7 SNPs with cognition using summary statistics data from the largest GWAS on cognition by Davies et al. (55). We observed a nominally significant association with rs3752246, which had the strongest association with LacCer(d18:1/24:1). We used data from the Airwave (n = 768), FINGER (n = 1,083), and the Rotterdam (n = 542) studies to examine the association of identified metabolites with three cognitive domains (executive function, processing speed, and working memory) as well as an overall score of cognitive performance.
In the Airwave study, cognition was measured using a version of the Cardiff Cognitive Battery (56) in Airwave and using standard neuropsychological tests (an extended version of the neuropsychological test battery) in FINGER (57). In the Airwave study, processing speed was assessed (after training) using 60 stimuli presented as a two-choice, reaction-time task where participants respond to one of two targets on the computer screen. Working memory was tested by a digit-span test where participants had to recall an increasing No. of digits, starting from one digit and ending with the first incorrect answer. Executive function was assessed by a verbal and numeric reasoning test including 12 items that gradually increased in difficulty.
In the Rotterdam Study, cognitive function was assessed using a neuropsychological test battery comprising the letter–digit substitution task (No. of correct digits in 1 min), the verbal fluency test (animal categories), the Stroop test (error-adjusted time in seconds), a 15-word learning test (immediate and delayed recall), and Purdue pegboard task (58). For each test, z scores were calculated by dividing the difference between the individual and mean test scores by the SD. We used the word-learning test to estimate working memory; Stroop reading, color-naming task, and letter–digit substitution task (weighted half) for information processing; and Stroop interference task, verbal fluency test, and letter–digit substitution task (weighted half) for executive function.
The analysis for FINGER was based on metabolomics and cognition data both available at baseline and the follow-up visit. In Airwave and the Rotterdam Study, however, the analysis was cross-sectional using the cognition and metabolomic data at the same visit.
Mendelian Randomization.
We performed two-sample Mendelian randomization analyses to investigate the potential causal relationship between metabolomic features associated with SNPs in ABCA7 and AD risk. We conducted GWAS on these metabolomic features in Airwave to identify genetic instruments for the Mendelian randomization analyses. For each feature, data points >5 MAD were deleted. GWAS was performed using PLINK2 (59). The analysis included Airwave participants separately genotyped using Affymetrix (n = 882), Illumina (n = 2,003), and Global Screening Array (n = 823) arrays and was adjusted for age and sex. Population stratification was controlled by adjusting for 10 principal components. Summary results for each metabolite feature were combined using inverse-variance–weighted meta-analysis using METAL (n = 3,708) (60).
For each metabolic feature, SNPs with P < 5 × 10−8 and F statistics >10 were selected as genetic instruments for the Mendelian randomization analysis. To mitigate weak instrument bias, we only included SNPs with a minor allele frequency greater than 5% and imputation quality greater than 0.7. We removed correlated SNPs (r2 > 0.1) by retaining the SNPs with the smallest P value. Mendelian randomization analyses were performed for metabolic features with more than three independent genetic instruments to allow further sensitivity analyses. The genetic association of AD was based on the most recent GWAS on AD by the IGAP Consortium (61).
We searched for genetic instruments for 36 mass spectral features. Three or more independent instruments were available for 32 features, annotated to 11 metabolites (Dataset S7). We performed Mendelian randomization analysis for each feature. We estimated the effect of metabolic features on the risk of AD using the inverse variance weighted (IVW) method (62). Potential pleiotropic effects were assessed using weighted median (WM) and MR-Egger regression as sensitivity analyses. Potential outlier SNPs were identified using MR-pleiotropy residual sum and outlier and were excluded from the analyses (63). We accounted for multiple testing using false discovery rate correction on causal estimates calculated using the IVW method.
Association with Neuroimaging Measurements.
We used data from the FINGER trial to examine the association of the metabolic features with neuroimaging measures. Selection criteria and imaging protocol for the FINGER neuroimaging exploratory substudies (n = 132 MRI scans and n = 48 Pittsburgh compound-B positron emission tomography [PiB-PET] scans conducted in connection to the baseline visit) have been previously described in detail (64, 65). In brief, brain MRI scans were conducted at four of the six study sites and PiB-PET scans at one site. The MRI/PET populations were not significantly different in demographic, clinical, and cognitive characteristics from the populations without MRI/PET at these sites. However, the MRI/PET populations were slightly older than the rest of the FINGER participants (mean 70.2 ± 4.6 vs. 69.2 ± 4.7 y for MRI and 70.8 ± 5 vs. 69.3 ± 4.7 y for PiB-PET).
Regional cortical thickness and volumes were measured using the Freesurfer image analysis suite (version 5.0.3). For the present study, we chose four MRI measurements with clear established links to dementia/AD: hippocampus volume, total gray matter (GM) volume, white matter lesion (WML) volume (all MRI volumes were divided by total intracranial volume), and a measure of cortical thickness in AD signature regions calculated as the average of cortical thickness in entorhinal, inferior temporal, middle temporal, and fusiform regions (17).
For PiB-PET, we used a visual rating of overall amyloid status (positive/negative) and a composite measure of amyloid deposition calculated as the average across the prefrontal, parietal, lateral temporal, precuneus, anterior cingulate, and posterior cingulate regions of interest as previously described (65).
After zero-skewness log transformation for all neuroimaging variables that were not normally distributed (hippocampus, total GM and WML volumes, and PIB composite), linear-regression models were used to assess the associations between each MRI measure or the PiB composite score (as dependent variables) and metabolic features (as independent variables). All models were adjusted for age and sex, and MRI models were additionally adjusted for the study site. All results are shown uncorrected for multiple testing. Similar logistic-regression models were conducted with positive/negative amyloid status on PiB-PET as the dependent variable.
Mouse Abca7 KO Model.
All animal procedures were approved by the Mayo Clinic’s Institutional Animal Care and Use Committee and were performed following the NIH’s Guide for the Care and Use of Laboratory Animals (66). Abca7 KO mice (Abca7−/−) (67) were crossbred with WT C57BL/6 inbred mice. Littermate male and female Abca7+/+ (WT) and Abca7−/− (KO) mice were killed at 50 wk of age (four WT male, three WT female, four KO male, and three KO female), and the left hemisphere of the brain was rapidly dissected and frozen on dry ice. The left cerebral cortex (20 mg of wet weight tissue) was extracted using chloroform/methanol/water biphasic extraction, and the lipid fraction was analyzed along with three pooled samples using UPLC-MS with reverse phase chromatography (LPOS and LNEG). Peak picking was performed using the R library XCMS after optimizing peaks extraction and RT correction parameters with the library IPO (68). Further data processing was completed using KniMet (69). Briefly, features were removed from further analysis if detected in less than 50% of the pooled samples with a relative SD higher than 20%. Annotations were made by the exact mass match with the LIPID MAPS database followed by a comparison with RTs, collision cross-section, and fragmentation patterns from previous analyses (Dataset S6). Missing values were either imputed using the R library “impute” (70) for multivariate statistical analysis or replaced with half of the minimum value found in the feature for univariate statistical analysis. In total, 10,837 features were detected in positive and negative ionization mode, with 5,503 of these features being assigned by a combination of exact mass, RT matching, and comparison with standards. Multivariate statistics comparing metabolic features detected in KO versus WT mice were performed on both negative and positive ionization mode data.
AppNL-G-F Mouse Model.
We examined the lipid profile of brain tissue (left cerebrum) from the AppNL-G-F mouse (n = 6 AppNL-G-F, n = 5 WT). All procedures were performed as described for the ABCA7 KO mouse model above.
Supplementary Material
Acknowledgments
The authors thank Prof. Takashi Saito, Nagoya City University Graduate School of Medical Sciences, Japan and Prof. Takomi Saido, RIKEN Centre for Brain Science, Japan for access to the AppNL-G-F mouse model. The authors also want to thank Dr. Jenny Hällqvist for helping to illustrate Fig. 7. This work is supported by the UK Dementia Research Institute at Imperial College, which receives its funding from UK Dementia Research Institute Ltd., funded by the UK Medical Research Council (MRC), Alzheimer’s Society, and Alzheimer’s Research UK. A.D. is funded by a Wellcome Trust seed award (206046/Z/17/Z). R.M. is funded by the President’s PhD Scholarship from Imperial College London. P.M.M. acknowledges generous personal and research support from the Edmond J. Safra Foundation and Lily Safra and a National Institute for Health Research (NIHR) Senior Investigator Award (to 2020). J.L.G. is funded by the MRC (MC_UP_A090_1006, MC_PC_13030, MR/P011705/1, MR/P01836X/1, and MR/S010483/1) and Wellcome Trust (MetaboFlow). M.K. is funded by the NIHR Imperial Biomedical Research Centre (BRC), Wallenberg Clinical Scholars, Academy of Finland, and Swedish Research Council. A.S. is funded by the Academy of Finland (287490, 294061, and 319318), European Research Council (804371), Alzheimerfonden, and Region Stockholm ALF (Sweden). This work was supported by the MRC and NIHR (grant No. MC_PC_12025). P.E. is the director of the MRC Centre for Environment and Health (MR/L01341X/1). He acknowledges support from the NIHR Health Protection Research Units in Chemical and Radiation Threats and Hazards and Health Impact of Environmental Hazards. P.E. is a codirector of the Health Data Research UK London site, which is supported, among others, by MRC, NIHR, Engineering and Physical Sciences Research Council, Economic and Social Research Council, Wellcome Trust, and British Heart Foundation (BHF). P.E. acknowledges support from the BHF Centre for Research Excellence at Imperial College. Infrastructure support for this research was provided by the NIHR Imperial BRC Science.
Footnotes
Competing interest statement: P.M.M. has received consultancy fees from Biogen, Roche, Celgene, and Ipsen Pharmaceuticals. He has received honoraria or speaker’s fees from Novartis, Biogen, Neurodiem, and Medscape and has received research or educational funds from Biogen, Novartis, and GlaxoSmithKline. J.L.G. has received research funds from Astra Zeneca and Unilever.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2206083119/-/DCSupplemental.
Data, Materials, and Software Availability
All summary statistics for association data are included in the manuscript and/or supporting information. All AIRWAVE metabolomic data sets are available for download here https://doi.org/10.14469/hpc/6945 (71). All other AIRWAVE data may be accessed upon application to the Dementias Platform UK Data portal (https://portal.dementiasplatform.uk/Apply/ApplicationProcess) (72). The FINGER data presented in this article are not readily available because data can be made available only for those fulfilling the requirements for viewing confidential data as required by Finnish law and the Finnish Institute for Health and Welfare. Moreover, the purpose of the research must be in alignment with the informed consent provided for this study and/or the FINGER study, with Finnish law and regulations at the Finnish Institute for Health and Welfare. Requests are to be submitted to the Finnish Institute for Health and Welfare: kirjaamo@thl.fi. Data for the Rotterdam Study can be obtained upon request. Requests should be directed toward the management team of the Rotterdam Study (datamanagement.ergo@erasmusmc.nl), which has a protocol for approving data requests. Because of restrictions based on privacy regulations and the informed consent of the participants, data cannot be made freely available in a public repository. Data for ABCA7 KO mice is available here https://www.ebi.ac.uk/metabolights/MTBLS6093 (73).
References
- 1.Jansen I. E., et al. , Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat. Genet. 51, 404–413 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kunkle B. W., et al. ; Alzheimer Disease Genetics Consortium (ADGC); European Alzheimer’s Disease Initiative (EADI); Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium (CHARGE); Genetic and Environmental Risk in AD/Defining Genetic, Polygenic and Environmental Risk for Alzheimer’s Disease Consortium (GERAD/PERADES), Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 51, 414–430 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lambert J. C., et al. ; European Alzheimer’s Disease Initiative (EADI); Genetic and Environmental Risk in Alzheimer’s Disease; Alzheimer’s Disease Genetic Consortium; Cohorts for Heart and Aging Research in Genomic Epidemiology, Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 45, 1452–1458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lotta L. A., et al. , Genetic predisposition to an impaired metabolism of the branched-chain amino acids and risk of type 2 diabetes: A Mendelian randomisation analysis. PLoS Med. 13, e1002179 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wittemans L. B. L., et al. , Assessing the causal association of glycine with risk of cardio-metabolic diseases. Nat. Commun. 10, 1060 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tzoulaki I., et al. , Serum metabolic signatures of coronary and carotid atherosclerosis and subsequent cardiovascular disease. Eur. Heart J. 40, 2883–2896 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holmes E., et al. , Human metabolic phenotype diversity and its association with diet and blood pressure. Nature 453, 396–400 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Illig T., et al. , A genome-wide perspective of genetic variation in human metabolism. Nat. Genet. 42, 137–141 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elliott P., et al. , The Airwave Health Monitoring Study of police officers and staff in Great Britain: Rationale, design and methods. Environ. Res. 134, 280–285 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Ikram M. A., et al. , Objectives, design and main findings until 2020 from the Rotterdam Study. Eur. J. Epidemiol. 35, 483–517 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marengoni A., et al. , The effect of a 2-year intervention consisting of diet, physical exercise, cognitive training, and monitoring of vascular risk on chronic morbidity-the FINGER randomized controlled trial. J. Am. Med. Dir. Assoc. 19, 355–360.e1 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Smith G. D., Ebrahim S., ‘Mendelian randomization’: Can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 32, 1–22 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Sakae N., et al. , ABCA7 deficiency accelerates amyloid-β generation and Alzheimer’s neuronal pathology. J. Neurosci. 36, 3848–3859 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrison J., et al. , A neuropsychological test battery for use in Alzheimer disease clinical trials. Arch. Neurol. 64, 1323–1329 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Davies N. M., Holmes M. V., Davey Smith G., Reading Mendelian randomisation studies: A guide, glossary, and checklist for clinicians. BMJ 362, k601 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burgess S., Scott R. A., Timpson N. J., Davey Smith G., Thompson S. G.; EPIC- InterAct Consortium, Using published data in Mendelian randomization: A blueprint for efficient identification of causal risk factors. Eur. J. Epidemiol. 30, 543–552 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jack C. R. Jr., et al. , Different definitions of neurodegeneration produce similar amyloid/neurodegeneration biomarker group findings. Brain 138, 3747–3759 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Consortium G. T.; GTEx Consortium, The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aikawa T., Holm M. L., Kanekiyo T., ABCA7 and pathogenic pathways of Alzheimer’s disease. Brain Sci. 8, 27–39 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Götz J., Chen F., van Dorpe J., Nitsch R. M., Formation of neurofibrillary tangles in P301l tau transgenic mice induced by Abeta 42 fibrils. Science 293, 1491–1495 (2001). [DOI] [PubMed] [Google Scholar]
- 21.LaFerla F. M., Tinkle B. T., Bieberich C. J., Haudenschild C. C., Jay G., The Alzheimer’s A beta peptide induces neurodegeneration and apoptotic cell death in transgenic mice. Nat. Genet. 9, 21–30 (1995). [DOI] [PubMed] [Google Scholar]
- 22.Oddo S., et al. , Triple-transgenic model of Alzheimer’s disease with plaques and tangles: Intracellular Abeta and synaptic dysfunction. Neuron 39, 409–421 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Nugent A. A., et al. , TREM2 regulates microglial cholesterol metabolism upon chronic phagocytic challenge. Neuron 105, 837–854.e9 (2020). [DOI] [PubMed] [Google Scholar]
- 24.Fernandez C. G., Hamby M. E., McReynolds M. L., Ray W. J., The role of APOE4 in disrupting the homeostatic functions of astrocytes and microglia in aging and Alzheimer’s disease. Front. Aging Neurosci. 11, 14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Picataggi A., et al. , Specificity of ABCA7-mediated cell lipid efflux. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1867, 159157 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim W. S., Guillemin G. J., Glaros E. N., Lim C. K., Garner B., Quantitation of ATP-binding cassette subfamily-A transporter gene expression in primary human brain cells. Neuroreport 17, 891–896 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Iwamoto N., Abe-Dohmae S., Sato R., Yokoyama S., ABCA7 expression is regulated by cellular cholesterol through the SREBP2 pathway and associated with phagocytosis. J. Lipid Res. 47, 1915–1927 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Kielar D., et al. , Adenosine triphosphate binding cassette (ABC) transporters are expressed and regulated during terminal keratinocyte differentiation: A potential role for ABCA7 in epidermal lipid reorganization. J. Invest. Dermatol. 121, 465–474 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Ramirez L. M., et al. , Common variants in ABCA7 and MS4A6A are associated with cortical and hippocampal atrophy. Neurobiol. Aging 39, 82–89 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Yang J., et al. , Age- and nicotine-associated gene expression changes in the hippocampus of APP/PS1 Mice. J. Mol. Neurosci. 69, 608–622 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Mielke M. M., et al. , Plasma ceramides are altered in mild cognitive impairment and predict cognitive decline and hippocampal volume loss. Alzheimers Dement. 6, 378–385 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mielke M. M., et al. , Serum sphingomyelins and ceramides are early predictors of memory impairment. Neurobiol. Aging 31, 17–24 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mielke M. M., et al. , Serum ceramides increase the risk of Alzheimer disease: The Women’s Health and Aging Study II. Neurology 79, 633–641 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chai J. F., et al. , Associations with metabolites in Chinese suggest new metabolic roles in Alzheimer’s and Parkinson’s diseases. Hum. Mol. Genet. 29, 189–201 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Li C., et al. , Macrophage polarization and meta-inflammation. Transl. Res. 191, 29–44 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamura H., Murayama T., Role of sphingolipids in arachidonic acid metabolism. J. Pharmacol. Sci. 124, 307–312 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Wang P., et al. , Aggravation of Alzheimer’s disease due to the COX-2-mediated reciprocal regulation of IL-1β and Aβ between glial and neuron cells. Aging Cell 13, 605–615 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Satoh K., Abe-Dohmae S., Yokoyama S., St George-Hyslop P., Fraser P. E., ATP-binding cassette transporter A7 (ABCA7) loss of function alters Alzheimer amyloid processing. J. Biol. Chem. 290, 24152–24165 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moll T., et al. , Membrane lipid raft homeostasis is directly linked to neurodegeneration. Essays Biochem. 65, 999–1011 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis A. C., Wallington-Beddoe C. T., Powell J. A., Pitson S. M., Targeting sphingolipid metabolism as an approach for combination therapies in haematological malignancies. Cell Death Discov. 4, 72 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chaurasia B., et al. , Targeting a ceramide double bond improves insulin resistance and hepatic steatosis. Science 365, 386–392 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kivipelto M., et al. , The Finnish geriatric intervention study to prevent cognitive impairment and disability (FINGER): Study design and progress. Alzheimers Dement. 9, 657–665 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Izzi-Engbeaya C., et al. , The effects of kisspeptin on β-cell function, serum metabolites and appetite in humans. Diabetes Obes. Metab. 20, 2800–2810 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lewis M. R., et al. , Development and application of ultra-performance liquid chromatography-TOF MS for precision large Scale Urinary Metabolic Phenotyping. Anal. Chem. 88, 9004–9013 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Dona A. C., et al. , Precision high-throughput proton NMR spectroscopy of human urine, serum, and plasma for large-scale metabolic phenotyping. Anal. Chem. 86, 9887–9894 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Smith C. A., Want E. J., O’Maille G., Abagyan R., Siuzdak G., XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 78, 779–787 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Dunn W. B., et al. ; Human Serum Metabolome (HUSERMET) Consortium, Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 6, 1060–1083 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Sands C. J., et al. , The nPYc-Toolbox, a Python module for the pre-processing, quality-control and analysis of metabolic profiling datasets. Bioinformatics 35, 5359–5360 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fahy E., Sud M., Cotter D., Subramaniam S., LIPID MAPS online tools for lipid research. Nucleic Acids Res. 35, W606-12 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith C. A., et al. , METLIN: A metabolite mass spectral database. Ther. Drug Monit. 27, 747–751 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Wishart D. S., et al. , HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 46 (D1), D608–D617 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sumner L. W., et al. , Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 3, 211–221 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chadeau-Hyam M., et al. , Metabolic profiling and the metabolome-wide association study: Significance level for biomarker identification. J. Proteome Res. 9, 4620–4627 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Castagné R., et al. , Improving visualization and interpretation of metabolome-wide association studies: An application in a population-based cohort using untargeted 1H NMR metabolic profiling. J. Proteome Res. 16, 3623–3633 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davies G., et al. , Study of 300,486 individuals identifies 148 independent genetic loci influencing general cognitive function. Nat. Commun. 9, 2098 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gallacher J., et al. , A platform for the remote conduct of gene-environment interaction studies. PLoS One 8, e54331 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ngandu T., et al. , A 2 year multidomain intervention of diet, exercise, cognitive training, and vascular risk monitoring versus control to prevent cognitive decline in at-risk elderly people (FINGER): A randomised controlled trial. Lancet 385, 2255–2263 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Hoogendam Y. Y., Hofman A., van der Geest J. N., van der Lugt A., Ikram M. A., Patterns of cognitive function in aging: The Rotterdam Study. Eur. J. Epidemiol. 29, 133–140 (2014). [DOI] [PubMed] [Google Scholar]
- 59.Chang C. C., et al. , Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Willer C. J., Li Y., Abecasis G. R., METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bellenguez C., et al. ; EADB; GR@ACE; DEGESCO; EADI; GERAD; Demgene; FinnGen; ADGC; CHARGE, New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat. Genet. 54, 412–436 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burgess S., Thompson S. G., Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 32, 377–389 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Verbanck M., Chen C. Y., Neale B., Do R., Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stephen R., et al. , Associations of CAIDE dementia risk score with MRI, PIB-PET measures, and cognition. J. Alzheimers Dis. 59, 695–705 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kemppainen N., et al. , Brain amyloid load and its associations with cognition and vascular risk factors in FINGER Study. Neurology 90, e206–e213 (2018). [DOI] [PubMed] [Google Scholar]
- 66.National Research Council (U.S.), Committee for the Update of the Guide for the Care and Use of Laboratory Animals., Institute for Laboratory Animal Research (U.S.), National Academies Press (U.S.), Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, D.C.), pp xxv, 220 p. [Google Scholar]
- 67.Kim W. S., et al. , Abca7 null mice retain normal macrophage phosphatidylcholine and cholesterol efflux activity despite alterations in adipose mass and serum cholesterol levels. J. Biol. Chem. 280, 3989–3995 (2005). [DOI] [PubMed] [Google Scholar]
- 68.Libiseller G., et al. , IPO: A tool for automated optimization of XCMS parameters. BMC Bioinformatics 16, 118 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liggi S., et al. , KniMet: A pipeline for the processing of chromatography-mass spectrometry metabolomics data. Metabolomics 14, 52 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hastie T., Tibshirani R., Narasimhan B., Chu G., impute: Imputation for microarray data. https://bioconductor.org/packages/release/bioc/html/impute.html. (2016). Accessed 13 October 2022.
- 71.Robinson O., AIRWAVE Metabolomic data sets. Imperial College London. https://data.hpc.imperial.ac.uk/resolve/?doi=6945. Deposited 11 March 2020.
- 72.Dementias Platform UK. Application Process. Dementias Platform UK Data Portal. https://portal.dementiasplatform.uk/Apply/ApplicationProcess. Accessed 14 October 2022.
- 73.Dehghan A., et al. , Metabolome-wide association study on ABCA7 indicates a role of ceramide metabolism in Alzheimer’s disease. MetaboLights. https://www.ebi.ac.uk/metabolights/MTBLS6093. Deposited 17 October 2022. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All summary statistics for association data are included in the manuscript and/or supporting information. All AIRWAVE metabolomic data sets are available for download here https://doi.org/10.14469/hpc/6945 (71). All other AIRWAVE data may be accessed upon application to the Dementias Platform UK Data portal (https://portal.dementiasplatform.uk/Apply/ApplicationProcess) (72). The FINGER data presented in this article are not readily available because data can be made available only for those fulfilling the requirements for viewing confidential data as required by Finnish law and the Finnish Institute for Health and Welfare. Moreover, the purpose of the research must be in alignment with the informed consent provided for this study and/or the FINGER study, with Finnish law and regulations at the Finnish Institute for Health and Welfare. Requests are to be submitted to the Finnish Institute for Health and Welfare: kirjaamo@thl.fi. Data for the Rotterdam Study can be obtained upon request. Requests should be directed toward the management team of the Rotterdam Study (datamanagement.ergo@erasmusmc.nl), which has a protocol for approving data requests. Because of restrictions based on privacy regulations and the informed consent of the participants, data cannot be made freely available in a public repository. Data for ABCA7 KO mice is available here https://www.ebi.ac.uk/metabolights/MTBLS6093 (73).