Significance
Smith–Magenis syndrome (SMS) is a neurodevelopmental disorder associated with autism and epileptic seizures. SMS is caused by losing one copy of the gene encoding retinoic acid induced 1 (RAI1), a ubiquitously expressed transcriptional regulator. To pinpoint brain regions and cell types contributing to neuronal hyperexcitability in SMS, we combined electrophysiology and three-dimensional imaging of Fos expression in the intact mouse brain. We found that Rai1-deficient hippocampal dentate gyrus granule cells (dGCs) show increased intrinsic excitability and enhanced glutamatergic synaptic transmission. Our findings indicate that Rai1 safeguards the hippocampal network from hyperexcitability and could help explain abnormal brain activity in SMS.
Keywords: Smith–Magenis syndrome, neuronal excitability, dentate gyrus, whole-brain clearing and imaging, autism spectrum disorders
Abstract
Hyperexcitability of brain circuits is a common feature of autism spectrum disorders (ASDs). Genetic deletion of a chromatin-binding protein, retinoic acid induced 1 (RAI1), causes Smith–Magenis syndrome (SMS). SMS is a syndromic ASD associated with intellectual disability, autistic features, maladaptive behaviors, overt seizures, and abnormal electroencephalogram (EEG) patterns. The molecular and neural mechanisms underlying abnormal brain activity in SMS remain unclear. Here we show that panneural Rai1 deletions in mice result in increased seizure susceptibility and prolonged hippocampal seizure duration in vivo and increased dentate gyrus population spikes ex vivo. Brain-wide mapping of neuronal activity pinpointed selective cell types within the limbic system, including the hippocampal dentate gyrus granule cells (dGCs) that are hyperactivated by chemoconvulsant administration or sensory experience in Rai1-deficient brains. Deletion of Rai1 from glutamatergic neurons, but not from gamma-aminobutyric acidergic (GABAergic) neurons, was responsible for increased seizure susceptibility. Deleting Rai1 from the Emx1Cre-lineage glutamatergic neurons resulted in abnormal dGC properties, including increased excitatory synaptic transmission and increased intrinsic excitability. Our work uncovers the mechanism of neuronal hyperexcitability in SMS by identifying Rai1 as a negative regulator of dGC intrinsic and synaptic excitability.
Neuronal hyperexcitability, as indicated by increased seizure prevalence and epileptiform electroencephalogram (EEG) activity, is a common comorbidity of autism spectrum disorders (ASDs) (1–3). Many genes associated with ASDs encode key regulators of synaptic strength, intrinsic membrane excitability, or neurotransmitter release probability (2, 4–6). Although many of these ASD risk genes are widely expressed throughout the brain, neural circuits are differentially affected in each type of ASD (7). Therefore, developing mechanism-based interventions for neuronal hyperexcitability requires elucidating the mechanisms by which ASD risk genes regulate neural circuit activity in a brain region– and cell type–specific fashion.
Smith–Magenis syndrome (SMS) is a syndromic ASD characterized by intellectual disability, motor dysfunction, obesity, seizure, sleep disturbance, and autistic features (8–10). SMS is commonly caused by deletion of a 3.7-Mb genomic region on chromosome 17p11.2 that contains >70 genes (9). Importantly, 23% of SMS patients do not carry 17p11.2 deletions and instead have point mutations in retinoic acid induced 1 (RAI1) (11), which encodes a broadly expressed chromatin-interacting protein involved in transcriptional regulation (12). SMS patients with 17p11.2 deletions and RAI1 point mutations share highly overlapping neurocognitive symptoms, highlighting an essential role for RAI1 in brain function (13).
We previously showed that SMS-like obesity and neurobehavioral phenotypes in mice can be induced by subcortical glutamatergic neuron-specific Rai1 deletion (14–16). Although the function of Rai1 at the behavioral level is relatively well defined, it remains unclear how Rai1 regulates neuronal activity. Abnormal EEG patterns are found in 50% of SMS patients, of which 87% show epileptiform activity, with comparable prevalence among male and female patients (17). Both generalized and focal epileptiform EEG patterns are observed in SMS (17). Consistent with clinical findings, Rai1+/− and Rai1−/− mice display abnormal EEG patterns, with the latter developing symptoms that show greater severity and earlier onset (18, 19). Furthermore, spontaneous seizures have been detected in 30% of Rai1−/− mice and SMS patients (17, 19). In vitro studies have also found that short-hairpin RNA-mediated Rai1 knockdown increases synaptic efficacy and prevents synaptic upscaling (20).
Based on these findings, we hypothesize that Rai1 is involved in regulating neuronal excitability in vivo. To test this hypothesis, we combined the use of genetic mouse models with three-dimensional (3D) brain clearing and imaging, as well as in vivo and ex vivo electrophysiological approaches to study the function of Rai1 at the molecular, cellular, synaptic, and whole-brain levels. We found that loss of Rai1 induced hippocampal circuit hyperexcitability, accompanied by increased seizure susceptibility associated with prolonged epileptiform EEG patterns upon chemoconvulsant challenge. Loss of Rai1 also induced overexpression of ion channels, including Cav3.1, increased intrinsic dentate gyrus granule cell (dGC) excitability, and excitatory synaptic excitability onto dGCs. High-frequency neuronal firing in Rai1-deficient dGCs was rescued by applying NNC 55-0396 dihydrochloride, a selective T-type calcium channel blocker. Our findings, which identify Rai1-dependent neuronal subtypes, brain regions, and cellular/molecular mechanisms responsible for neuronal hyperexcitability, advance our understanding of the neurobiology of Rai1 and suggest potential therapeutic candidates for treating increased seizure susceptibility in SMS.
Results
Panneural Rai1 Loss Induces Increased Seizure Susceptibility Associated with Abnormal Brain Activity.
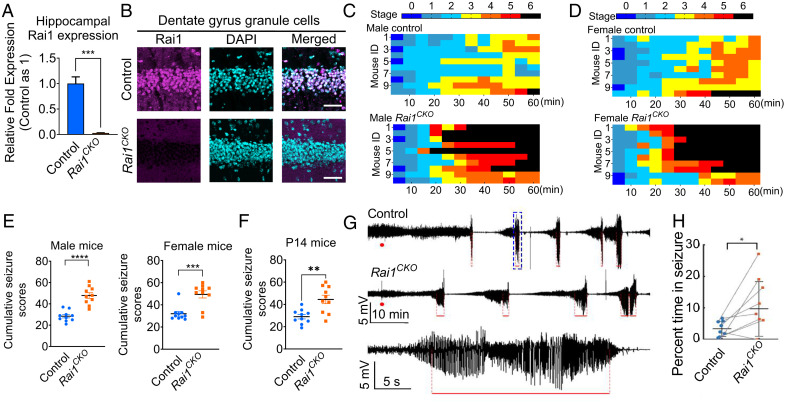
To determine how Rai1 regulates neural circuit function in vivo, we investigated whether deleting Rai1 alters the ability of neurons to respond to changes in activity. We used the panneural NestinCre driver and the Rai1flox allele to generate control (NestinCre/+) and whole-brain Rai1 homozygous knockout (KO) mice (NestinCre/+;Rai1flox/flox, which we referred to as Rai1CKO below) (21, 22). This strategy allowed us to bypass the high incidence of embryonic lethality (∼95%) caused by germline homozygous Rai1 deletion (12). Using qRT-PCR (Fig. 1A) and immunofluorescent staining (Fig. 1B), we confirmed a near complete loss of Rai1 mRNA and protein from the Rai1CKO brain.
Fig. 1.
Panneural Rai1 deletions increase seizure sensitivity and abnormal EEG activity. (A) qRT-PCR showing a near-complete loss of Rai1 mRNA expression in the hippocampus of NestinCre/+;Rai1flox/flox (Rai1CKO) mice when compared to their NestinCre/+ (control) littermates (n = 3, t test, ***P < 0.001). (B) Representative images of hippocampal sections from control and Rai1CKO mice stained with an anti-Rai1 antibody (magenta) and counterstained with DAPI (cyan). The Rai1 signal is reduced in Rai1CKO mice. (Scale bars: 50 µm.) (C) Time course of seizure severity following KA treatment (35 mg/kg, i.p.) in 6-wk-old male controls (Top, n = 10) and Rai1CKO mice (Bottom, n = 10). Each animal’s maximum seizure score was measured every 5 min over a 1-h period, with the timescale in one box being 5 min. (D) Time course of seizure severity following KA treatment (30 mg/kg, i.p.) in 6-wk-old female controls (Top, n = 10) and Rai1CKO mice (Bottom, n = 10). (E) Cumulative seizure scores showing that both male (Left) and female (Right) Rai1CKO mice are more susceptible to seizures than their control littermates (each dot represents one mouse, unpaired t test, males, ****P < 0.0001; females, ***P < 0.001). (F) P14 Rai1CKO mice show significantly increased seizure sensitivity when compared to control littermates (unpaired t test, **P < 0.01). (G) Hippocampal depth EEG recordings showing prolonged oscillatory events in the dentate gyrus of Rai1CKO mice upon KA injection (red dots); red lines indicate seizures. The Bottom panel illustrates an example of a detected seizure pattern (event in the blue dotted box in the Top panel with timescale expanded). (H) Quantification showing that Rai1CKO mice (n = 8) spend significantly more time in seizure than their paired controls (n = 8). *P = 0.02, Wilcoxon rank-sum nonparametric test for paired data after Bonferroni correction for multiple comparisons.
To monitor behavioral seizures within a defined timeframe, we adopted the well-established paradigm of injecting kainic acid (KA, a chemoconvulsant) to induce gamma frequency network oscillations in vitro and in vivo (23), as well as behavioral seizures in vivo (24). We then scored behavioral seizures according to a modified Racine scale (0, no symptoms; 5, whole-body in clonus; 6, death) (25). Over a 1-h period, 8-wk-old male Rai1CKO mice displayed more rapid onset of behavioral seizures and higher seizure severity than control littermates, with 80% of the Rai1CKO mice dying of clonic–tonic seizures during the observation period (35 mg/kg, Fig. 1 C and E). Similarly, 8-wk-old female Rai1CKO mice also showed an increased susceptibility to seizures compared to control littermates (30 mg/kg, Fig. 1 D and E). Importantly, the enhanced seizure susceptibility was detected as early as postnatal day 14 (P14) in Rai1CKO mice (Fig. 1F). This suggests that brain hyperexcitability in SMS is an early-onset feature.
To determine how Rai1 deletion affects neuronal excitability in vivo, we performed simultaneous recordings of cortical EEG and hippocampal local field potentials (depth EEG) in eight pairs of control and Rai1CKO mice (12 to 16 wk old; see SI Appendix, Fig. S1A for electrode placement). We recorded 30 min of background cortical and hippocampal population activity in freely behaving mice and then injected a lower dose of KA (15 mg/kg) to induce brain activity without triggering clonic–tonic seizures and lethality. We found that although background brain activity remained similar between genotypes, Rai1CKO mice showed a significant increase in total seizure duration upon KA injection compared to controls, as measured by hippocampal depth EEG (Fig. 1 G and H). The latency to increased gamma power (a preseizure marker), the latency and number of seizures within the first hour after the initial KA dose, and the KA seizure threshold did not differ between genotypes (SI Appendix, Fig. S1B). However, cortical EEG patterns remained relatively normal in Rai1CKO mice (SI Appendix, Fig. S1C). Together, these data suggest that Rai1 deletion induces hippocampal hyperexcitability and increased seizure sensitivity in vivo.
Neuroanatomical and Glucose Metabolism Analyses in Control and Rai1CKO Mice.
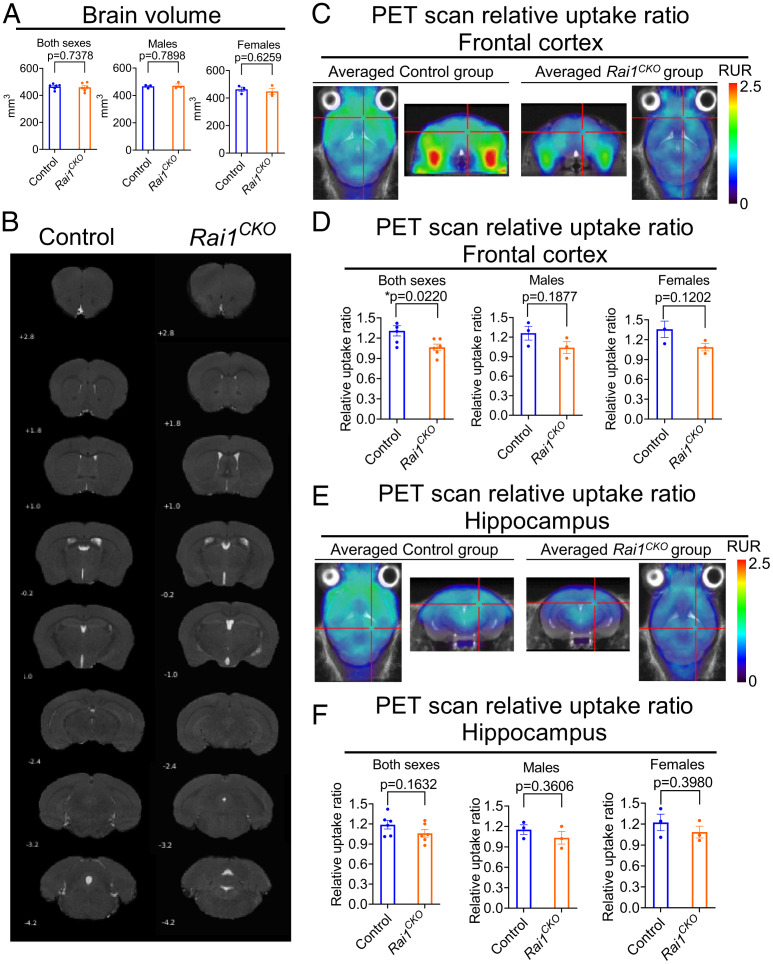
To determine whether panneural Rai1 deletions induce severe brain malformation that could underlie brain hyperexcitability, we performed 7-Tesla (7T) magnetic resonance imaging (MRI) in Rai1CKO mice and their control littermates (three males and three females for each genotype). Because Rai1CKO mice are underweight at birth and become overweight in adulthood (15), our brain imaging studies focused on 6-wk-old mice with similar body weight across groups (SI Appendix, Table S1). We found that whole-brain volume (Fig. 2A) did not differ between Rai1CKO mice and their control littermates. Furthermore, MRI showed that gross neuroanatomical structures remained intact in Rai1CKO mice (Fig. 2B). Although our results do not exclude the possibility that Rai1CKO mice could exhibit microstructural deficits in the brain, they are consistent with clinical evidence suggesting that macrostructural brain malformation is not a common feature in SMS patients (26).
Fig. 2.
MRI and 18F-FDG-PET imaging in control and Rai1CKO mice. (A) Quantification of whole-brain volume in 6-wk-old control and Rai1CKO mice. (B) Representative coronal MRI images in 6-wk-old control and Rai1CKO mice, with the position relative to Bregma indicated for each slide. (C) Averaged PET scan images showing the relative uptake ratio (RUR) of frontal cortex in 6-wk-old control and Rai1CKO mice (n = 6 per genotype) in 18F-FDG-PET imaging. The reticles indicate the regions of interest used for quantification. The images at the ends are in the transverse planes, and the images in the middle are in the coronal planes. (D) Quantification indicating that 6-wk-old Rai1CKO mice (males and females combined) show decreased RUR in the frontal cortex when compared to controls in 18F-FDG-PET imaging. *P < 0.05, unpaired t test. (E) Averaged PET scan images show comparable hippocampal RUR in 6-wk-old control and Rai1CKO mice (n = 6 per genotype) in 18F-FDG-PET imaging. The reticles indicate the regions of interest used for quantification. (F) Quantification indicating that 6-wk-old control and Rai1CKO mice (males and females combined) show similar hippocampal RUR in 18F-FDG-PET imaging. *P < 0.05, unpaired t test.
Synaptic activity is tightly coupled to glucose metabolism (27). Therefore, we quantified brain glucose metabolism in the resting state in vivo with 18Fluoro-2-deoxyglucose (18FDG) positron-emission tomography/computed tomography (PET/CT). First, we analyzed the standard uptake value (SUV) (SI Appendix, Table S2) and found similar SUVs in the whole-brain (SI Appendix, Fig. S2A), entorhinal cortex (SI Appendix, Fig. S2B), somatosensory cortex (SI Appendix, Fig. S2C), cingulate cortex (SI Appendix, Fig. S2D), hippocampus (SI Appendix, Fig. S2E), hypothalamus (SI Appendix, Fig. S2F), and frontal cortex (SI Appendix, Fig. S2G, showing averaged images of color-coded SUVs of control and Rai1CKO mice normalized by body weight). Given that the SUVs of the cerebellum are not significantly different between genotypes (SI Appendix, Table S2), we then analyzed the relative uptake ratio by normalizing radioactivity in each brain region of interest by cerebellar SUVs, and found that the frontal cortex showed a significant reduction of 18FDG uptake in Rai1CKO mice (male + female, P = 0.022, unpaired t test, Fig. 2 C and D). Subsequently, we analyzed the relative uptake ratios in other brain regions and found that the hippocampus (Fig. 2 E and F), entorhinal cortex (SI Appendix, Fig. S3A), cingulate cortex (SI Appendix, Fig. S3B), and hypothalamus (SI Appendix, Fig. S3C) did not show differential glucose uptake (see also SI Appendix, Table S3). By contrast, the motor cortex (SI Appendix, Fig. S3D) and somatosensory cortex (SI Appendix, Fig. S3E) showed reductions of the glucose uptake ratio in Rai1CKO mice (male + female). These observations suggest that, at least in the anesthetized state, selective brain regions in Rai1CKO mice have reduced metabolic activity.
Whole-Brain Clearing, Activity Mapping, and Imaging Identify Brain Regions Showing Differential Neuronal Activity upon Rai1 Deletion.
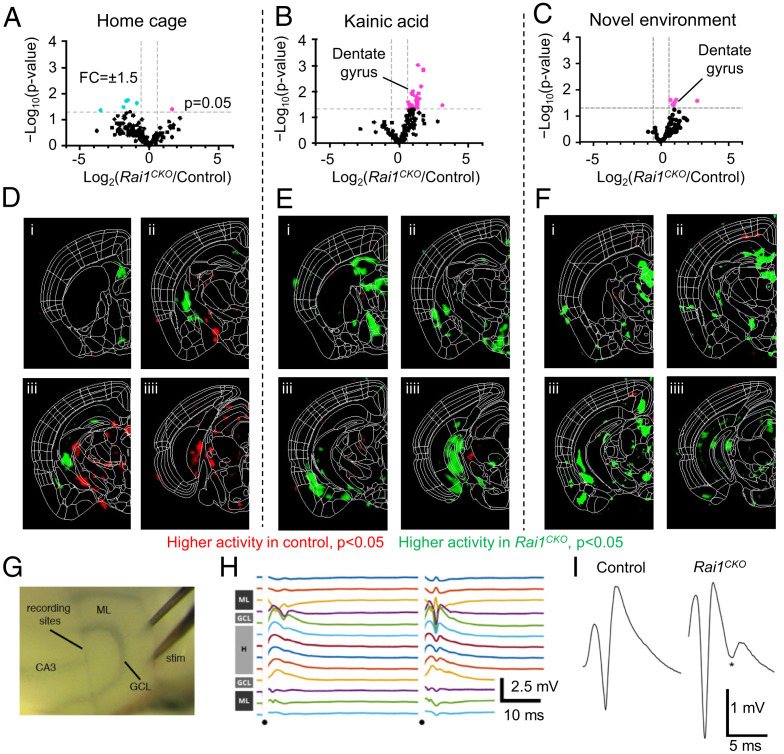
Although MRI provides neuroanatomical information and 18FDG-PET/CT scans are useful for measuring glucose metabolism in different brain regions, these methods currently lack single-cell resolution and are commonly limited to use in anesthetized mice. To address these limitations, we first performed iDISCO+ whole-brain clearing and activity mapping using expression of the Fos protein as a proxy for neuronal activity and searched for brain activity signatures of Rai1CKO mice at single-cell resolution in a high-throughput and unbiased manner (28). We then performed 3D light-sheet imaging, followed by automated cell mapping using the open source ClearMap program (28). We found that in the home cage (baseline condition), Fos expression patterns in control and Rai1CKO mice were largely comparable (Fig. 3A), with the exception of reduced activity in the midbrain, presubiculum, and parasubiculum of Rai1CKO mice (SI Appendix, Fig. S4A and Table S4). This is in line with the reduced 18FDG uptake in several brain regions of anesthetized Rai1CKO mice (Fig. 2).
Fig. 3.
Brain-wide activity mapping uncovers elevated dentate network excitability in Rai1CKO mice. (A–C) Volcano plots showing Fos+ cells in each brain region of control (n = 6) and Rai1CKO (n = 6) mice in the home cage (A), 1 h after KA treatment (B, n = 8 control mice and n = 7 Rai1CKO mice), or 1 h after exposure to a novel environment (C, n = 6 control mice and n = 6 Rai1CKO mice). Each dot indicates a brain region. Magenta and cyan dots indicate regions with higher Fos counts in Rai1CKO mice and those with higher Fos counts in control mice, respectively. The x axis: relative Fos+ cells, threshold: fold change (FC) ±1.5 fold, indicated by dotted lines. The y axis: −Log10(P value), threshold: P < 0.05. (D–F) Coronal sections of the P value of voxel maps representing the densities of Fos+ cells in the home cage (D), 1 h after KA treatment (E), or 1 h after exposure to a novel environment (F). Red: brain areas with higher Fos activity in control mice. Green: brain areas with higher Fos activity in Rai1CKO mice. (G) Placement of a 16-channel recording probe and bipolar stimulating electrode (stim) in an acute horizontal hippocampal slice. GCL, granule cell layer; ML, molecular layer. (H) Multichannel recordings sample field potentials across the dentate gyrus following a paired-pulse facilitation stimulation paradigm. Dots indicate timing of stimulating pulses. Gray bars at Left show anatomical location of each electrode within the dentate gyrus. In this example, stimulation mainly activates one blade of the dentate gyrus (Upper traces). H, hilus. (I) Representative traces for population spike activity in control and Rai1CKO hippocampal slices after perforant path stimulation. The asterisk denotes a second population spike, indicative of a hyperexcitable dentate gyrus circuit. Number of slices with multiple population spikes: control = 1/13; Rai1CKO = 8/15. P < 0.05, t test, n = 6 per genotype.
We then increased brain activity by administering a single dose of KA (15 mg/kg) and harvested mouse brains 1 h after KA injection (Fig. 3B). Rai1CKO mice showed more robust Fos activation than control mice in the hippocampal dentate gyrus, as well as in hypothalamic regions, including the medial preoptic area, periventricular hypothalamus, medial and lateral zones, and dorsomedial nucleus (Fig. 3 B and E and SI Appendix, Fig. S4B and Table S5). Interestingly, although several brain regions were hypoactive in Rai1CKO mice in the home cage condition (Fig. 3 A and D), many brain regions became hyperactive in Rai1CKO mice when brain activity was elevated (Fig. 3 B and E). To determine whether increasing neuronal activity with a more physiological stimulus could induce brain hyperexcitability in Rai1CKO mice, we exposed control and Rai1CKO mice to a novel environment (NE) known to induce brain activity and Fos expression (29). One hour after exploration, mouse brains were harvested for Fos staining and processed using the iDISCO+/ClearMap pipeline. We found that several brain regions, including the hippocampal dentate gyrus, became hyperactive in Rai1CKO mice (Fig. 3 C and F and SI Appendix, Fig. S4C and Table S6; see SI Appendix, Fig. S5 for representative images). Our data suggest that although Rai1 is widely expressed in the brain, only specific brain regions, notably parts of the limbic system, become hyperactive upon Rai1 deletion when brain activity is elevated.
We then focused on the hippocampal dGCs, which consistently exhibited increased Fos activity in both the novel environment and KA injection paradigms. To verify that increased Fos activity reflects hyperexcitability of the dGC network, we performed ex vivo local field potential (LFP) recordings using acute hippocampal slices from 3-wk-old control and Rai1CKO mice. A 16-channel recording probe and bipolar stimulating electrode were placed in a horizontal slice (Fig. 3G). The recording electrodes were placed across the dentate gyrus, and the stimulating electrode was placed in the outer molecular layer to stimulate the perforant path (Fig. 3H), the major input onto dGCs. We found that stimulation was significantly more likely to evoke multiple population spikes in Rai1CKO slices, whereas it could only reliably evoke a single population spike in control slices (Fig. 3I; slices with multiple population spikes: control = 1/13; Rai1CKO = 8/15; P < 0.05, Fisher’s exact test, n = 6 per genotype). Therefore, Rai1 deletion results in abnormal repetitive dGC firing in response to incoming neural activity. Together, the whole-brain activity mapping and LFP experiments revealed that panneural Rai1 deletions induce neural hyperexcitability in selective cell types, including dGCs, a cell population that gates hippocampal excitability and has long been implicated in temporal lobe epilepsy (30, 31).
Cell Numbers, Dendritic Spine Density, and Axonal Projection Pattern of dGCs Are Rai1 Independent.
Loss of dGCs has been shown to exacerbate hippocampal hyperexcitability (32). To determine whether cell autonomous Rai1 deletion affects the generation or survival of dGCs, we performed clonal analysis using the mosaic analysis with double markers (MADM) technique (33–35) and generated wild-type (WT)-MADM (NestinCre/+;MADM11TG/GT) and Rai1 KO-MADM (NestinCre/+;MADM11TG/GT, Rai1-flox, SI Appendix, Fig. S6A) mice. In this scheme, both red and green neurons in WT-MADM mice were Rai1+/+ (SI Appendix, Fig. S6B), whereas in Rai1 KO-MADM mice, red neurons were Rai1−/− and green neurons were Rai1+/+ (SI Appendix, Fig. S6B). Quantification showed that the red-to-green ratio was similar between adolescent (3 wk old) WT-MADM and Rai1 KO-MADM mice (SI Appendix, Fig. S6C), and that this pattern persisted into adulthood when the mice were 6 mo old. Therefore, loss of Rai1 did not affect the neurogenesis or survival of dGCs.
Remodeling of dendritic spine density is associated with hippocampal hyperexcitability (36, 37). Our recent work found that Rai1 haploinsufficiency induces reduced spine density in the medial prefrontal cortex (14). Therefore, we investigated whether Rai1 loss in dGC alters dendritic spine density. By quantifying dGC spine density in the distal dendritic segments of dGCs, we found that WT-MADM and Rai1 KO-MADM dGCs show similar dendritic spine density (SI Appendix, Fig. S6D). Collateral sprouting of dentate mossy fibers, which leads to the formation of recurrent excitatory circuits, has been associated with temporal lobe epilepsy (38, 39). We immunolabeled mossy fibers with zinc transporter 3 (ZnT3) and found normal mossy fiber axonal terminal fields restricted to the hilus of both control and Rai1CKO mice (SI Appendix, Fig. S6 E and F), suggesting that Rai1 loss did not affect gross axonal targeting of dGCs. These data suggest that abnormal dGC activity does not appear to be contributed by dGC neuronal number, dendritic spine density, or gross axonal targeting; rather, it likely originates from alteration of cellular properties, such as synaptic function and membrane excitability.
Glutamatergic but Not GABAergic Rai1 Loss Enhances Seizure Susceptibility.
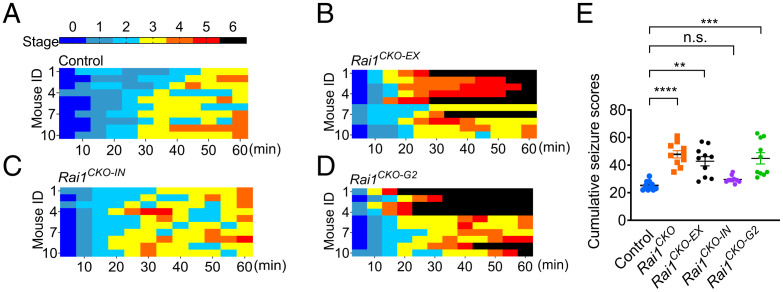
Rai1 is highly enriched in both glutamatergic and GABAergic neurons across multiple brain regions in mice (14, 15). To identify the neuronal populations that underlie seizure hypersensitivity associated with Rai1 loss, we conditionally deleted Rai1 from glutamatergic neurons (using Emx1Cre) or GABAergic neurons (using VgatCre) (40). Immunofluorescent staining confirmed the loss of Rai1 protein from cortical and hippocampal glutamatergic neurons in Emx1Cre/+;Rai1flox/flox (Rai1CKO-EX for conditional knockout in excitatory neurons) mice and from GABAergic neurons in VgatCre/+;Rai1flox/flox (Rai1CKO-IN for conditional knockout in inhibitory neurons) mice (SI Appendix, Fig. S7 A–C). Latencies to seizure following an initial exposure to KA were scored using the Racine scale. We found that Rai1CKO-EX mice showed a cumulative Racine score similar to that of Rai1CKO mice (Fig. 4 A, B, and E). By contrast, Rai1CKO-IN mice showed a seizure susceptibility comparable to that of control mice (Fig. 4 C and E). To independently confirm the contribution of glutamatergic neurons, we used Vglut2Cre to selectively delete Rai1 from cortical and subcortical excitatory neurons (SI Appendix, Fig. S7D) (40). We found that Vglut2Cre/+;Rai1flox/flox (Rai1CKO-G2) mice also showed a significantly increased seizure severity score (Fig. 4 D and E). Together, these data suggest that the loss of Rai1 in glutamatergic but not GABAergic neurons enhances epileptogenesis.
Fig. 4.
Rai1 loss in glutamatergic but not GABAergic neurons drives neural hyperexcitability in vivo. (A–D) Racine scale quantification for seizure severity following kainic acid treatment in control (A), Rai1CKO-EX (B), Rai1CKO-IN (C), and V Rai1CKO-G2 (D) mice. Each row represents a mouse (n = 10 per genotype). (E) Cumulative seizure scores in control and cell type–specific Rai1 conditional knockout mice (n = 10 per genotype) show that excitatory but not inhibitory neurons are responsible for heightened seizure sensitivity. n.s., not significantly different, **P < 0.01, ***P < 0.001, ****P < 0.0001, Tukey’s post hoc test.
Glutamatergic Rai1 Loss Induced Increased Excitatory Synaptic Transmission onto the dGCs without Altering Inhibitory Synaptic Transmission.
The dGCs constitute a group of glutamatergic neurons that normally exhibit low excitability owing to their hyperpolarized resting membrane potential, low input resistance, and high firing threshold (30, 31, 41). Therefore, the dGC circuitry has been postulated as a gate that prevents epileptogenesis by limiting the flow of excitatory input into the hippocampus (30, 31). Rai1 deletion could increase dGC excitability through a number of different mechanisms, including increased excitatory input, decreased inhibitory input, and/or increased intrinsic neuronal excitability. To distinguish between these possibilities, we performed whole-cell patch-clamp recording in acute hippocampal slices from 3- to 4-wk-old control (Emx1Cre/+) and Rai1CKO-EX (Emx1Cre;Rai1flox/flox) mice.
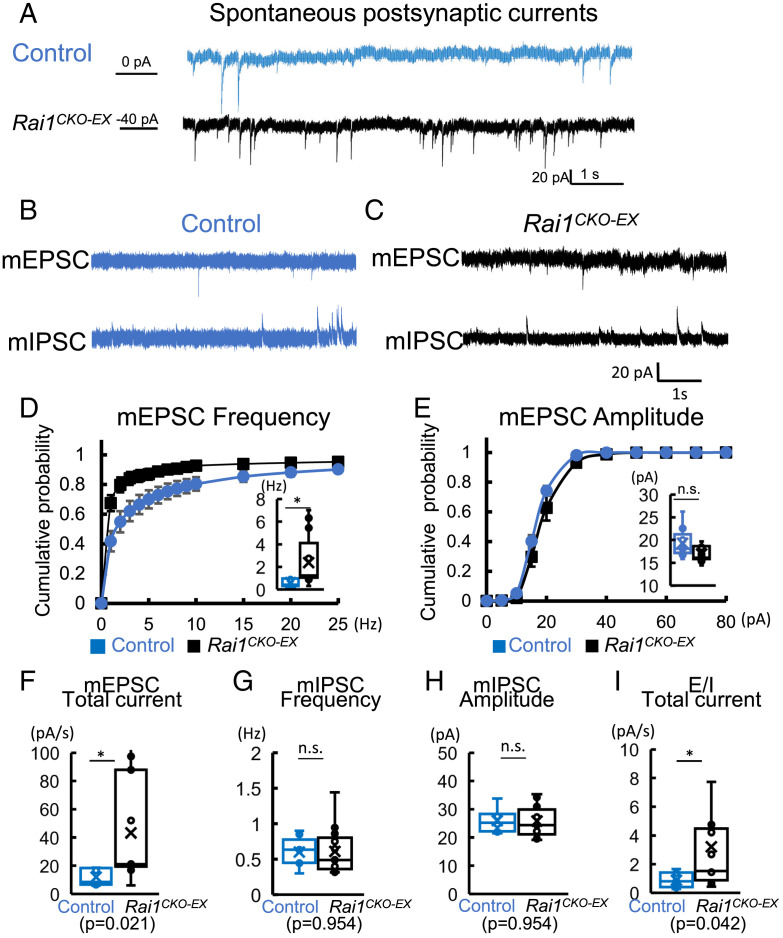
We measured spontaneous postsynaptic currents (sPSCs) from dGCs at a holding potential of −70 mV. Event frequencies in Rai1CKO-EX dGCs were higher than those in age-matched control neurons (Fig. 5A). This is consistent with the increased early adolescent seizure susceptibility we observed in P14 Rai1CKO mice (Fig. 1F). Although most inward events resulted from excitatory syanptic transmission at −70 mV, these events may contain both spontaneous spike-driven inward currents and quantal miniature excitatory postsynaptic currents (mEPSCs). Therefore, we isolated mEPSCs with 0.5 μM of tetrodotoxin (TTX, a Nav channel blocker) and Cs-gluconate–based internal solution to block K+ outflow at a holding potential of −70 mV (Top, Fig. 5 B and C). Rai1CKO-EX dGCs showed a significant increase in the frequency of mEPSCs (Fig. 5D) without significantly changing the averaged mEPSC amplitudes (Fig. 5E). This suggests potential increases in synapse number and/or alterations in release probability of vesicles onto dGCs. Moreover, the total current of mEPSCs showed a significant increase in Rai1CKO-EX dGCs (Fig. 5F). We also recorded miniature inhibitory postsynaptic currents (mIPSCs) to investigate whether loss of Rai1 from excitatory neurons changes inhibitory inputs onto dGCs. We found no significant changes in mIPSC frequency or peak amplitude in Rai1CKO-EX dGCs when compared to control neurons (Fig. 5 G and H).
Fig. 5.
Glutamatergic Rai1 loss enhances miniature EPSCs and E/I ratio. (A) Representative traces of spontaneous current under voltage clamp at a holding potential of −70 mV, from control (blue) and Rai1CKO-EX (black) dGCs in acute brain slices. (B) Representative traces of mEPSCs (Top, holding potential at −70 mV) and mIPSCs (Bottom, holding potential at 10 mV, recorded from the same cell) in control dGCs. (C) Representative traces of mEPSCs (Top, holding potential at −70 mV) and mIPSCs (Bottom, holding potential at +10 mV, recorded from the same cell) in Rai1CKO-EX dGCs. (D) When compared to controls (blue), Rai1CKO-EX (black) dGCs show an increase in comulative probability of mEPSC frequency (Inset: averaged mEPSC frequencies; control: n = 9 neurons from three mice; Rai1CKO-EX: 13 neurons from four mice). (E) The Rai1CKO-EX dGCs show similar mEPSC amplitude compared to those recorded from control dGCs (P = 0.068). (F) Rai1CKO-EX dGCs show significantly increased mEPSC total current (pA/s) when compared to controls (*P < 0.05, t test). (G) Rai1CKO-EX dGCs show a similar averaged mIPSC frequency to controls P = 0.954). (H) Rai1CKO-EX dGCs show similar averaged mIPSC amplitude when compared to controls (P = 0.954). n.s., not significantly different. (I) The E/I total current ratio is increased in Rai1CKO-EX mice (*P < 0.05, t test).
The disproportional increase in excitatory synaptic input may lead to an excitatory/inhibitory imbalance in dGC synaptic activity that contributes to the hyperexcitability phenotype. We estimated the mEPSC to mIPSC ratio (E/I ratio) and found an increase in E/I total current ratio in Rai1CKO-EX dGCs due to an increased mEPSC current (Fig. 5I). These results suggest that glutamatergic Rai1 loss results in alterations in E/I ratio in synaptic input onto dGCs that favor dGC excitation and, in turn, defective dentate gyrus gating, in Rai1CKO-EX mice.
Glutamatergic Rai1 Loss Also Induces Increased dGC Intrinsic Membrane Excitability.
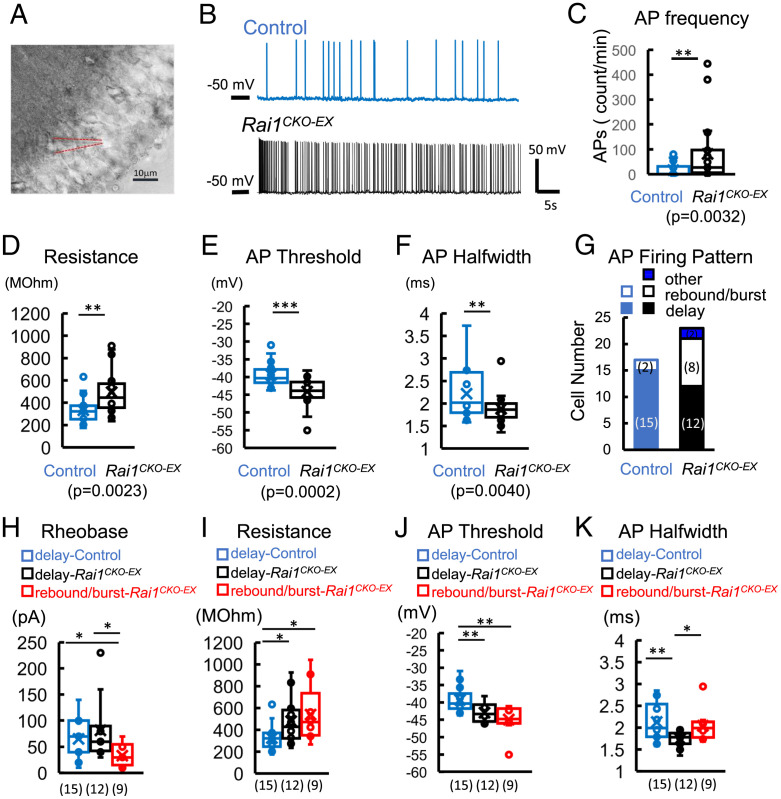
We further determined the possibility that glutamatergic Rai1 loss leads to alterations in the intrinsic neuronal properties of dGCs by measuring their membrane properties and action potentials (APs) in whole-cell recording (Fig. 6A). We found that in Rai1CKO-EX mice, dGCs fired APs more frequently at holding potentials of −60 to −50 mV (averaged AP frequency at a holding potential of −50 mV: control = 18.3 ± 7.0, Rai1CKO-EX = 81.6 ± 25.4; see Fig. 6 B and C and SI Appendix, Table S7). Rai1-deficient dGCs also showed significantly higher input resistance (58% increase), a lower AP firing threshold (12% decrease), and a narrower AP halfwidth (15% decrease) (Fig. 6 D–F), while displaying resting membrane potential and AP amplitude comparable to control neurons (SI Appendix, Table S7). Neuronal firing patterns such as burst and rebound firing are associated with neuronal excitability and gating, which could contribute to epileptogenesis (42). We classified dGCs based on their firing patterns into rebound-or-burst cluster, delay-firing cluster, and others (Fig. 6G and SI Appendix, Fig. S8 A and B) (43). We found that the number of rebound-or-burst neurons among dGCs increased in Rai1CKO-EX (39%) compared to control dGCs (12%). Rai1-deficient dGCs with rebound-or-burst firing patterns showed a lower rheobase (decreases of 59% and 49% compared to delay-firing Rai1CKO-EX and delay-firing control dGCs, respectively) and were able to fire APs in response to smaller depolarization step at a holding potential of −70 mV (Fig. 6H). Both rebound-or-burst and delay-firing Rai1-deficient dGCs contributed to increased input resistance (increases of 66% and 47%, respectively) and decreased AP threshold (decreases of 14% and 9%, respectively) (Fig. 6 I and J). Rai1-deficient delay-firing neurons displayed delayed AP initiation at rheobase and fired sharper APs (as evidenced by the decrease in AP halfwidth), which, in turn, led to an increase in AP firing compared to delay-firing control dGCs under a ramp test (Fig. 6K and SI Appendix, Fig. S8 C and D). These changes in intrinsic excitability support the notion that Rai1CKO-EX dGCs are hyperexcitable and fire more APs.
Fig. 6.
Glutamatergic Rai1 loss increases intrinsic excitability. (A) An image illustrating a whole-cell patch of a dGC in the dorsal blade of the dentate gyrus. (B) Representative traces of AP firing at a holding potential of −50 mV from control (blue) and Rai1CKO-EX (black) dGCs. (C) AP frequency is increased in Rai1CKO-EX dGCs (black open bar) compared to control dGCs (blue open bar). Spike numbers were quantified during the time of recording (shown as counts per minute). (D) Input resistance is increased in Rai1CKO-EX dGCs, as calculated from voltage clamp at a voltage close to AP firing. Rheobase was tested under current clamp, 500-ms steps from −110 mV in 10-pA increments. The properties of the first AP evoked at rheobase were measured. (E) AP threshold and (F) AP halfwidth are significantly decreased in Rai1CKO-EX dGCs (n = 17 and 22 from control and Rai1CKO-EX dGCs, respectively). (G) dGCs were classified into three clusters: a rebound-or-burst firing cluster (open bars); a delaying-firing cluster (closed bars); and others (stripe-patterned bar). The cell number in each cluster is shown in parentheses. The neuronal properties of delay-firing clusters from both genotypes and rebound-or-burst clusters from Rai1CKO-EX dGCs were compared. (H) The rheobase of rebound-or-burst Rai1CKO-EX dGCs is decreased significantly compared to that of delay-firing dGCs of either genotype. (I) Input resistance is increased significantly in delay-firing Rai1CKO-EX dGCs (black) and rebound-or-burst Rai1CKO-EX dGCs (red) compared to the control-delay cluster (blue). (J) The AP threshold of both delay-firing and rebound-or-burst Rai1CKO-EX dGCs is significantly decreased compared to controls. (K) The AP half width of delay-firing Rai1CKO-EX dGCs is decreased significantly compared to control and rebound-or-burst Rai1CKO-EX clusters.*P < 0.05, **P < 0.01, ***P < 0.001.
Glutamatergic Rai1 Loss Is Associated with Abnormal Expression of Ion Channels.
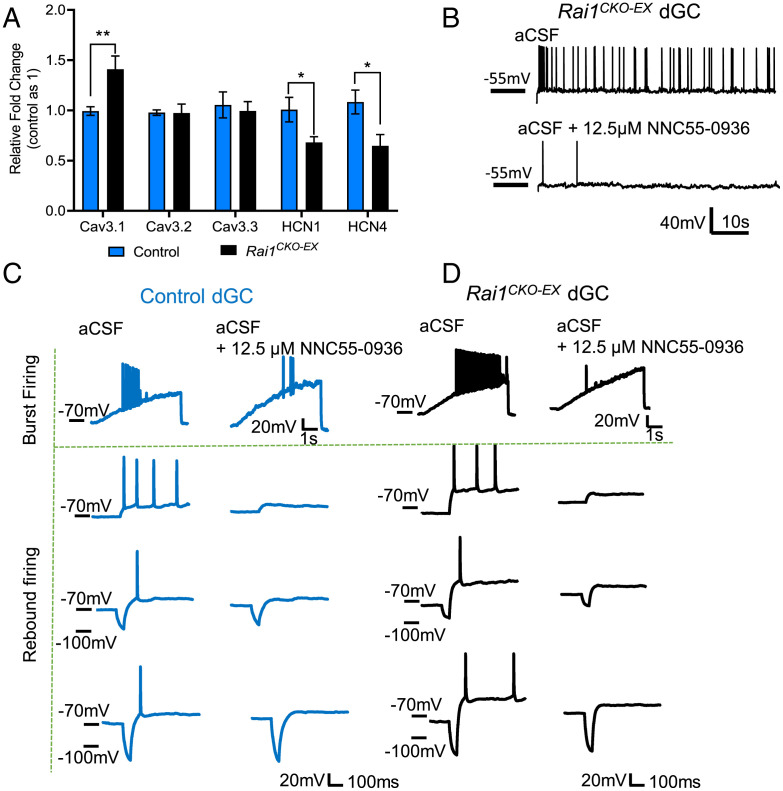
An alteration in intrinsic properties indicates potential changes in the ion channel composition of Rai1-deficient dGCs. Increased rebound-or-burst firing can be attributed to altered expression of voltage-gated T-type calcium channels (T-channels) and/or hyperpolarization-activated cyclic nucleotide-gated (HCN) channels. By performing qRT-PCR using manually dissected dentate gyrus tissues from control and Rai1CKO-EX mice, we found that the dentate gyrus isolated from Rai1CKO-EX mice showed a 40% increase in Cav3.1, a 30% reduction in HCN1, and a 35% reduction in HCN4 mRNA expression (Fig. 7A). By contrast, the mRNA levels of Cav3.2 and Cav3.3 remained unchanged (Fig. 7A).
Fig. 7.
Hyperexcitability in Rai1-deficient dGCs is associated with Cav3.1 overexpression. (A) qRT-PCR showing increased expression of Cav3.1 mRNA expression in the dentate gyrus dissected from Rai1CKO-EX mice when compared to control mice (n = 6 to 7 per genotype, unpaired two-tailed t test, *P < 0.05, **P < 0.01). (B) NNC 55-0396 dihydrochloride (12.5 mM) application reduces AP firing of a Rai1CKO-EX dGC at a holding potential of −55 mV. (C and D) NNC 55-0396 dihydrochloride (12.5 µM) inhibits long-current ramp (200 pA/5 s)-induced neuron bursting (Top rows) and hyperpolarzation-induced rebound firing (Bottom three rows) of a control dGC (C) and a Rai1CKO-EX dGC (D) at I-clamp. The AP firing thresholds were identified at I-ramp. To induce rebound firing, a 50-ms hyperpolarized step with progressively greater inject current was applied to the neuron following a 1.5-s step at sub-AP firing potentials. Similar results were found in five control and six Rai1CKO-EX rebound firing dGCs.
T-channels contribute to dGCs burst firing and can be activated at a more hyperpolarized membrane potential than the AP threshold of sodium channels. Therefore, T-channel overexpression could lead to neuronal hyperexcitability (44, 45), as evidenced by the increased propensity to develop epilepsy in mice that carry Cav3.1 overexpression transgenes and human patients that carry gain-of-function Cav3.1 mutations (46, 47). Conversely, CaV3.1-deficient mice show reduced seizure duration as detected by hippocampal depth EEG in response to KA injections (48, 49). To functionally test the involvement of T-type calcium channels in high frequency neuronal firing, we applied a pan-T-channel blocker, NNC 55-0396 dihydrochloride (IC50 = 6.7 μM), at a concentration that would not affect Na+ currents (12.5 μM), and found that AP firing of Rai1CKO-EX dGCs was decreased (Fig. 7B and SI Appendix, Fig. S8E). NNC 55-0396 perfusion also attenuated T-channel–dependent, depolarization-induced burst firing (Fig. 7 C and D, Top rows) and rebound firing at I-clamp (Fig. 7 C and D, Bottom three rows) in control (Fig. 7C) and Rai1CKO-EX (Fig. 7D) mice. These data confirmed the involvement of T-channels in dGC firing. Therefore, increased T-current due to Cav3.1 overexpression potentially contributes to dGC hyperexcitability in Rai1CKO-EX mice. Collectively, these data support the idea that glutamatergic loss of Rai1, a chromatin-binding protein, alters intrinsic membrane properties, AP firing, and dGC excitability.
Discussion
Abnormal EEG patterns and enhanced propensity for epileptogenesis are pervasive features in individuals afflicted with ASDs (50–52). In this study, we combined in vivo and ex vivo electrophysiological recordings, in vivo brain structural and metabolic imaging and whole-brain activity imaging/mapping to understand the neurobiology of Rai1, haploinsufficiency of which causes SMS. We made several discoveries. First, Rai1 regulates neuronal excitability in vivo without inducing gross neuroanatomical defects or neuronal survival. Second, specific brain regions, including the dentate gyrus become hyperactive in response to chemical or sensory stimuli upon Rai1 loss. Third, glutamatergic but not GABAergic neurons underlie enhanced seizure susceptibility in Rai1-deficient mice. Finally, glutamatergic Rai1 expression regulates dGC intrinsic excitability, excitatory (but not inhibitory) synaptic transmission, and ion channel expression. Together, this work uncovers molecular and neuronal mechanisms underlying brain hyperexcitability in SMS.
The complexity of genetic architecture poses a daunting challenge for understanding neuronal hyperexcitability in the ASD brain (53, 54), a task further complicated by the different mixtures of excitation and inhibition across varying timescales, cellular components, and microcircuits within multiple brain regions. Many ASD-risk genes, including Rai1, are widely expressed in multiple brain regions and cell types, making it challenging to pinpoint a specific circuit responsible for specific disease features. To assess neuronal activity patterns across the entire brain of Rai1 mutant mice in an unbiased manner, we took advantage of the compatibility of the iDISCO+/ClearMap pipeline with whole-brain clearing, Fos staining, volumetric light-sheet imaging, and automated brain region registration/quantification (28, 55). Fos expression patterns in control and Rai1CKO brains remained largely similar in the home cage, consistent with the subtle differences in resting-state glucose uptake detected by 18FDG-PET scans. By contrast, multiple brain regions in Rai1-deficient brains were hyperresponsive to a novel sensory experience and chemoconvulsant stimulation, consistent with the increased seizure susceptibility and abnormal EEG patterns we observed in vivo. Because immediate-early gene-based activity mapping provides a snapshot rather than the real-time dynamics of neuronal activity, we complemented this approach with the use of ex vivo acute brain slices and in vivo hippocampal depth EEG, which confirmed hippocampal hyperexcitability upon Rai1 loss. In comparison, cortical excitability was less affected by Rai1 loss, as demonstrated by our cortical EEG and iDISCO+ data. It is possible, however, that Rai1-dependent defects in cortical activity exist but were not detected by these methods. Together, our work suggests that the iDISCO+/Fos labeling pipeline can reliably uncover brain regions showing abnormal neuronal excitability in ASDs.
Apart from the hippocampus, our iDISCO+ analysis identified other brain regions that were hyperreactive upon Rai1 deletion, notably, the midbrain (i.e., superior colliculus and periaqueductal gray), as well as other limbic structures, including the bed nuclei of the stria terminalis, posterior hypothalamic nucleus, dorsomedial hypothalamic nucleus, and ventromedial hypothalamic nucleus. Furthermore, brain regions including CA1, CA3, and amygdala also show increased Fos expression in Rai1-deficient brains (though not all regions reach statistical significance). Therefore, the epilepsy phenotype likely originates from multiple sources, including hippocampal CA fields, neocortex, thalamus, and amygdala, which warrants future investigations. This also suggests that to develop effective therapies to treat altered brain excitability and behavioral problems in SMS, we will need to identify early interventions that uniformly restore Rai1 levels across multiple brain regions and cell types. In addition, our iDISCO+ and 18FDG-PET data indicate that some Rai1-deficient brain regions could be hypoactive in the home cage or anesthetized state—a topic that needs to be further studied.
The synaptic E/I ratio is temporally dynamic, a property that involves developmental compensation to stabilize circuit activity, and that varies in different brain regions, cell types, and synapses (6, 56). Guided by our iDISCO+ analysis, we focused on the hippocampal dentate gyrus as one of the main sources of neuronal hyperexcitability in Rai1-deficient mice. The dentate gate theory postulates that the dentate gyrus prevents hippocampal circuits from overactivation, and that its breakdown leads to increased propensity for epilepsy (30). Our findings suggest that Rai1 contributes to the stability of the hippocampal dentate gate. At the cellular level, Rai1 loss did not alter the survival, dendritic spine density, or gross axonal projection pattern of dGCs. However, glutamatergic neuron-specific Rai1 deletion induced synaptic dysfunction manifested by increased spontaneous and miniature excitatory synaptic transmission onto dGCs without altering inhibitory synaptic transmission, lending support to the E/I imbalance hypothesis in ASDs (1, 56).
Rai1 loss also affects dGC intrinsic excitability due to increased input resistance, decreased AP firing threshold, and increased rebound-or-burst firing cells. This is associated with abnormal expression of ion channels in dGCs, including Cav3.1, HCN1, and HCN4. Consistent with an important role of Cav3.1 in regulating neuronal firing, we found that using NNC 55-0396 dihydrochloride to block T-type calcium channels reduced the burst firing in Rai1-deficient dGCs. Because each subtype of T-type channel shows unique biophysical properties and spatial distributions, subtype-specific T-channel blockers are likely more effective in dampening the activity of overreactive T-channels and treating epilepsy (57, 58). Our study suggests that Cav3.1 overexpression potentially contributes to dGC hyperexcitability in SMS. Reduced HCN1 and HCN4 expression is of future interest because HCN channels are involved in maintaining resting membrane potentials (59). The average resting membrane potential is −73 mV for control dGCs. By contrast, the average resting membrane potential of rebound-or-burst dGCs is −67 mV, which allows the initiation of T-current. At this resting membrane potential, HCN channels could functionally interact with T-channels to facilitate T-current and increase neuronal excitability. In the future, the involvement of Cav3.1, HCN1, HCN4, and other ion channels should be evaluated as potential targets for treating neuronal hyperexcitability in SMS.
Does Rai1 prevent neuronal hyperexcitability in other brain regions through Cav3.1? Our previous work suggests that Rai1 function is highly context dependent and therefore it requires further investigations. For example, Rai1 loss in the cortex, striatum, and hypothalamus induces up- and down-regulation of vastly different genes, which do not include Cav3.1 (15). Furthermore, Rai1 only has a few common target genes in the developing and adult brains (16). Specifically, while embryonic Rai1 loss drives abnormal expression of neuronal wiring genes (15), adult Rai1 loss induces aberrant expression of genes involved in synaptic plasticity and cellular responses to hormone stimulus (16). Context-dependent Rai1 function could potentially explain the differences in synaptic transmission that we and others observed (20). Specifically, we found that genetic deletion of Rai1 from excitatory neurons increases mEPSC frequency without significant changes in mEPSC amplitude; Garay et al. found that reducing Rai1 increases mEPSC amplitude without impacting mEPSC frequency (20). Our current study genetically deletes Rai1 with Cre recombinase in vivo and measured synaptic transmission in acute hippocampal slices harvested from 3- to 4-wk-old mice. By contrast, Garay et al. performed acute Rai1 knockdown by applying short-hairpin RNA in rat hippocampal neurons cultured in neurobasal/B27 medium for 14 to 16 d (20). Therefore, discrepancy could be explained by differences in species, complexity of local circuits, loss-of-function approaches, and/or ages. Nevertheless, both studies found that at baseline level, reducing Rai1 in hippocampal neurons increases synaptic excitability (20). Further studies in different cell types and timepoints are needed to fully understand Rai1’s developmental and neuronal functions.
In summary, we found that loss of Rai1 (the causal gene for SMS) from excitatory neurons, but not inhibitory neurons, causes a failure to maintain hippocampal activity homeostasis and is associated with abnormal ion channel expression, increased dGC intrinsic excitability, and glutamatergic synaptic transmission. Protein-truncating variants of RAI1 have also been identified in other forms of ASD (60). Unlike many cases of ASD that show neuronal hyperexcitability due to reduced GABAergic inhibition (6), however, our data suggest that Rai1 expression in excitatory neurons maintains homeostatic neural network activity, and that its loss could underlie seizure disorders in individuals with SMS.
Materials and Methods
Mouse Models.
All procedures were performed in accordance with the guidelines of the Canadian Council on Animal Care, the Montreal General Hospital Facility Animal Care Committee, and Stanford University’s Administrative Panel on Laboratory Animal Care, with the appropriate approved protocols for animal use. Both male and female mice were used for the experiments. Mice were housed in groups on a 12-h light/12-h dark cycle with ad libitum access to food and water. The number of animals used for each experiment can be found in the text and figure legends. B6/CD1 F1 hybrid mice were used for all experiments. Rai1flox mice were generated by our group (15). MADM11, NestinCre, Emx1Cre, VgatCre, and Vglut2Cre mice were obtained from The Jackson Laboratory.
Mouse Genotyping.
Genotyping for NestinCre, Emx1Cre, VgatCre, and Vglut2Cre mice was performed using primers 5′- CACCCTGTTACGTATAGCCG-3′ and 5′-GAGTCATCCTTAGCGCCGTA-3′ for a 300-bp Cre band, and primers 5′-CCAATCTGCTCACACAGGATAGAGAGGGCAGG-3′ and 5′-CCTTGAGGCTGTCCAAGTGATTCAGGCCATCG-3′ for a 500-bp internal control band. Rai1-flox mice were identified using the following primers: 5′-CAGAGTCCAGATGGCACTACAGGGG-3′, 5′-GTGAGCTCCCGCTGAAATGGACAGT-3′, and 5′-GGAGGTCTGCGCTTCAGGGCTTAAT-3′. With these primers, the wild-type Rai1 allele produces a 396-bp band and the Rai1flox allele produces a 497-bp band. The mouse Rai1 gene is located on chromosome 11. The MADM11 TG cassette (H11TG) was detected using primers (5′-GGTACGTCCAGGAGCGCACCATCTTCTTCAAGG-3′ and 5′-GGACTGGGTGCTCAGGTAGTGGTTGTCG-3′) that produce an ∼326-bp band in the presence of H11TG. The MADM11 GT cassette (H11GT) was detected using primers (5′-CCAAGCTGAAGGTGACCAAG-3′ and 5′-TCTTCTTCTGCATTACGGGG-3′) that produce an ∼279-bp band in the presence of H11GT. To distinguish between wild-type (H11+/+), heterozygous (H11GT/+ or H11TG/+), and homozygous (H11GT/GT, H11TG/TG, or H11TG/GT) MADM11 alleles, an alternative set of primers (5′-TGGAGGAGGACAAACTGGTCAC-3′, 5′-TCAATGGGCGGGGGTCGTT-3′, and 5′-TTCCCTTTCTGCTTCATCTTGC-3′) was used; these primers amplify an ∼350-bp band from H11+ and an ∼230-bp band from H11TG and H11GT.
Immunostaining.
Immunostaining was performed as previously described (15, 61, 62). Briefly, mice were killed with isoflurane, perfused, and immersed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 24 h. The brains were then cryoprotected in 30% sucrose in PBS, immersed in optimal cutting temperature (OCT) compound (Thermo Fisher Scientific), and frozen by immersion in a dry ice/ethanol bath. Eight-micrometer-thick sections were mounted to Superfrost Plus slides and washed three times in PBS and then blocked for 2 h at room temperature in 10% normal donkey serum (NDS) in PBS. Slides were further incubated overnight at 4 °C with primary antibodies in 10% NDS in PBS, washed four times for 5 min in PBS, incubated for 2 to 3 h at room temperature with secondary antibodies in 10% NDS in PBS, washed four times for 5 min in PBS, incubated for 10 min in ∼1:30,000 dilution of 5 mg/mL DAPI in PBS, washed once for 5 min in PBS, and coverslipped in Fluoromount-G (SouthernBiotech). The following primary antibodies were used: rabbit anti-Rai1 (1:500, made in house), rabbit anti-ZnT3 (1:500, Synaptic Systems), rabbit anti-DsRed (1:500, Clontech), and chicken anti-EGFP (1:2,500, Aves Labs). Secondary antibodies conjugated to 488, Cy3, or Cy5 (Jackson Immunoresearch) were diluted 1:500 ∼ 1:2,000 from 50% glycerol stocks.
Dendritic Spine Analysis.
Confocal images of dGCs from WT-MADM and Rai1 KO-MADM mice were taken using a 63× (1.3 numerical aperture [NA]) oil objective and 5× zoom. Specifically, the distal ends of dendrite segments (20 to 30 µm in length) were imaged by 1-µm optical sections. A total of 20 cells/genotype from four mice were randomly selected for quantification. All protrusions, irrespective of their morphological characteristics, were counted as spines if they were in direct continuity with the dendritic shaft. All quantifications were done with the experimenter blind to the mouse genotypes. Statistical analysis was performed with Student’s t test.
qRT-PCR.
Total RNA was extracted by TRizol reagent (Thermo Fisher Scientific) and phenol-chloroform-isoamyl alcohol (Thermo Fisher Scientific) using dissected dentate gyrus or hippocampal tissues. The residual DNA was removed with on-column DNase digestion (Qiagen) for 30 min and RNA was further purified using the RNeasy Kit (Qiagen). Total RNA from three biological replicas of each genotype were used. The mRNA was reverse transcribed with the SuperScript III First-Strand Synthesis System (Thermo Fisher Scientific). Quantitative PCR reactions were conducted using SsoFast EvaGreen Supermix (Bio-Rad) on a Real-Time PCR Detection System (Bio-Rad).
Kainic Acid-Induced Seizure and Behavioral Seizure Scoring.
Kainic acid (Abcam) dissolved in 0.9% saline was injected intraperitoneally (i.p.). Mice were then placed individually in cages for observation, with genotypes blinded to the experimenter. Behavioral seizures were scored using a modified Racine scale (score = 0, mice behaved normally; score = 1, mice displayed absence-like immobility; score = 2, mice displayed tail extension/rigid posture/bulged eyes; score = 3, mice displayed circling, head bobbing, and scratching; score = 4, mice displayed rearing, forelimb clonus, and falling; score = 5, mice displayed generalized tonic–clonic seizure; score = 6, death. The highest score in each 5-min bin was recorded for a total of 60 min.
In Vivo Cortical EEG, Hippocampal Depth EEG, and Data Analysis.
Eight pairs of control and Rai1CKO mice (12 to 16 wk old) were used. Four silver wire electrodes were implanted under the skull, just above the cortical surface, to record EEG. A bilateral pair of frontal electrodes was implanted 1 mm anterior and 1 mm lateral to Bregma, and a bilateral pair of parietal electrodes was implanted 1.5 mm posterior and 2.5 mm lateral to Bregma (SI Appendix, Fig. S1A). Ground and reference wires were implanted posteriorly, ∼4 mm posterior and 1 mm lateral to Bregma. An array of four tungsten electrodes was also inserted into the right hippocampal formation to record local field potentials (depth EEG, 1.6 mm posterior and 1 mm lateral to Bregma, spanning a depth of 1.5 to 2 mm). After a 30-min baseline recording period, KA (15 mg/kg) was administered subcutaneously, and simultaneous video and EEG were recorded. EEG recordings were conducted using OpenEphys, and EEG data were analyzed in MatLab. A common average reference was applied separately for cortical and hippocampal EEGs. Signals were then filtered from 0.5 to 10 Hz, and seizures were detected when these band-passed signals exceeded 5 to 10 times the SD of the background recording period. The 5 to 10 times threshold multiplier was set manually on a per-recording basis. Signals were band-pass filtered from 20 to 50 Hz, and the amplitude of the filtered signal was used to measure gamma power in the preseizure period.
MRI.
Animals (control and Rai1CKO mice, 6 wk old) were anesthetized using 4 to 5% isoflurane, moved to a 7T Bruker 70/30 MRI scanner, and placed in the supine position in an MRI-compatible bed. The animals were maintained at 37 °C using an MRI-compatible air-warming system (SA Instruments, Inc.). The anesthesia was maintained at 1.5 to 2.5% isoflurane for ∼1 h. Following completion of scanning, animals were given 0.5 mL of sterile, warmed saline subcutaneously, returned to the procedure suite, and monitored during recovery from anesthesia under a warming lamp.
PET and CT.
Animals (control and Rai1CKO mice, 6 wk old) were anesthetized with isoflurane (5% induction, 1.5 to 2% maintenance for ∼3 min) and radiotracer fluorodeoxyglucose (18F-FDG) was injected into the tail vein. Following a 35-min uptake period in the habituation cage, the animal was reanesthetized (isoflurane, 5% induction, 1.5 to 2% maintenance throughout the scan) and habituated for 10 min in a Mediso nanoScan PET machine in the prone position (spatial resolution 0.7 mm). Following completion of PET scanning, a CT scan was performed for anatomical localization and attenuation correction. Animals were fasted for at least 16 h prior to scanning to prevent blood glucose levels from affecting 18F-FDG uptake. Glucose levels were measured prior to 18F-FDG delivery, and the respiration rate and body temperature were continuously monitored, with the latter maintained at 37 °C using a feedback-regulated warming system during scanning. Following completion of a 30-min scan, animals were given 0.5 mL of sterile, warmed saline subcutaneously and monitored during recovery from anesthesia under a warming lamp.
iDISCO+ Whole-Brain Clearing, Fos Mapping, and Light-Sheet Imaging.
Mice in the 1) home cage, 2) KA injection (15 mg/kg), and 3) novel environment conditions were perfused transcardially with PBS followed by 4% paraformaldehyde. After a 24-h postfixation period, immunolabeling (Fos antibody from Synaptic Systems 226 003; Alexa Fluor 647 from Thermo Fisher Scientific), whole-brain clearing, and ClearMap analysis were performed as reported (28). Cleared samples were scanned by a blind experimenter on a light-sheet microscope (Ultramicroscope II, LaVision Biotec) equipped with a sCMOS camera (Andor Neo) and a 2×/0.5 NA objective lens (MVPLAPO 2×) equipped with a 6-mm working distance dipping cap. The samples were scanned with a 3-μm step size using the continuous light-sheet scanning method with a contrast adaptive algorithm for the 640-nm channel (20 acquisitions per plane), and without horizontal scanning for the 488-nm autofluorescence channel. Cell counts were calculated blind to experimental conditions using the ClearMap cell detection module, with detection parameters optimized and validated by two expert users based on the intensity and shape parameters of the immunolabeling profile for Fos. SI Appendix, Tables S4–S6 list the mean and SD of Fos+ cells in each brain region in all samples, with the criterion for statistical significance set at P < 0.05. Whole-brain Fos maps for P values, mean, and SD were uploaded to https://zenodo.org/record/7140061.
Local Field Potential Recordings from Acute Brain Slices.
Horizontal slices 400-µm thick were prepared, and recordings were performed in a humidified oxygenated interface chamber at 34 °C and superfused at a rate of 3 mL/min with oxygenated artificial cerebrospinal fluid (aCSF). A 16-channel recording probe was placed across the dentate gyrus, and a bipolar stimulating electrode was placed in the outer molecular layer. Evoked responses to paired electrical stimulations (0.1 ms in duration, 100-ms interspike interval) were measured over a range of voltages (50 to 500 µA in 50-µA steps, average of five trials per voltage level), and population spikes were quantified on the recording channel where they were most prominent. Population spikes were detected in MatLab using the findpeaks function.
Electrophysiology of Acute Brain Slices.
Mouse brains were removed and placed in ice-cold carbogenated slicing aCSF, which contained (in millimoles) 125 NaCl, 2.5 KCl, 1.25 NaH2PO4, 25 NaHCO3, 12.5 D-glucose, 0.5 CaCl2, and 5 MgCl2. Coronal sections (300 µm) were cut on a Leica vibratome. Slices were allowed to recover at 30 °C for 60 min and then at 23 to 25 °C for 30 min to 6 h. Slices were then placed in carbogenated recording aCSF (125 NaCl, 2.5 KCl, 25 NaHCO3, 1.25 NaH2PO4, 1 MgCl2, 2 CaCl2, and 25 D-glucose, in millimoles). Signals were recorded with a 5× gain, low-pass filtered at 2 kHz, digitized at 10 kHz (Molecular Devices Multiclamp 700B), and analyzed with pClamp 11 (Molecular Devices). Whole-cell recordings were made using 3 to 5 MΩ pipettes filled with an internal solution that contained (in millimoles) 131 potassium gluconate, 8 NaCl, 20 KCl, 2 ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid (EGTA), 10 Hepes, 2 MgATP, and 0.3 Na3GTP (pH 7.2, with KOH, 300 to 310 mOsm with sucrose). For miniature current recording, the internal solution contains 130 gluconate acid, 8 CsCl, 1 NaCl, 2 EGTA, 0.8 CsOH, 10 Hepes, 2 MgATP, and 0.3 Na3GTP (pH 7.2, with CsOH, 300 to 310 mOsm with sucrose). mEPSCs were recorded in the presence of 0.5 µM TTX at holding potential of −70 mV. mIPSCs were recorded at holding potential of +10 mV. The mEPSCs/mIPSCs were analyzed from the events of 3-min continuous recordings that contain >15 events/cell (Molecular Devices, Clampfit 11). Electrophysiology protocols for Fig. 6 and 7 are uploaded to https://zenodo.org/record/7140061.
Quantification and Statistical Analysis.
Additional calculations, statistical analysis, and plotting were performed in Microsoft Excel, MatLab, or Prism (GraphPad). Significance was defined as having a P value <0.05. The statistical tests used for each analysis (e.g., Student’s t test, Fisher’s exact test, analysis of variance [ANOVA]) are indicated in the text and legends.
Supplementary Material
Acknowledgments
This work was supported by the Canadian Institutes of Health Research to W.-H.H. We thank members of the W.-H.H. laboratory for feedback on this project and comments on the manuscript. L.L. is an investigator of the Howard Hughes Medical Institute. W.-H.H. is a member of the Azrieli Centre for Autism Research and supported by a salary award from the Fonds de Recherche du Québec–Santé.
Footnotes
Reviewers: J.L.R., University of California San Francisco; and S.B.N., Brandeis University.
Competing interest statement: Both corresponding authors have a joint paper with J.L.R. (a reviewer of this paper) in 2020. That was a special paper in which the principal investigator (Dr. Ann Marie Craig) identified problems in germline recombination of Cre driver lines and wanted to collect similar problems by soliciting from other laboratories. Our laboratory and J.L.R. laboratory (among many others) contributed some examples and hence became co-authors of the resulting paper (PMID 32027825).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2210122119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and supporting information, except the electrophysiology protocols and whole-brain Fos maps, which can be accessed at https://zenodo.org/record/7140061 (63).
References
- 1.Rubenstein J. L., Merzenich M. M., Model of autism: Increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 2, 255–267 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Contractor A., Klyachko V. A., Portera-Cailliau C., Altered neuronal and circuit excitability in fragile X syndrome. Neuron 87, 699–715 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richard A. E., Scheffer I. E., Wilson S. J., Features of the broader autism phenotype in people with epilepsy support shared mechanisms between epilepsy and autism spectrum disorder. Neurosci. Biobehav. Rev. 75, 203–233 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Bateup H. S., et al. , Excitatory/inhibitory synaptic imbalance leads to hippocampal hyperexcitability in mouse models of tuberous sclerosis. Neuron 78, 510–522 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu H., et al. , Loss and gain of MeCP2 cause similar hippocampal circuit dysfunction that is rescued by deep brain stimulation in a Rett Syndrome mouse model. Neuron 91, 739–747 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson S. B., Valakh V., Excitatory/inhibitory balance and circuit homeostasis in autism spectrum disorders. Neuron 87, 684–698 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sahin M., Sur M., Genes, circuits, and precision therapies for autism and related neurodevelopmental disorders. Science 350, aab3897 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laje G., et al. , Autism spectrum features in Smith-Magenis syndrome. Am. J. Med. Genet. C. Semin. Med. Genet. 154C, 456–462 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith A. C., et al. , Interstitial deletion of (17) (p11.2p11.2) in nine patients. Am. J. Med. Genet. 24, 393–414 (1986). [DOI] [PubMed] [Google Scholar]
- 10.Greenberg F., et al. , Molecular analysis of the Smith-Magenis syndrome: A possible contiguous-gene syndrome associated with del(17)(p11.2). Am. J. Hum. Genet. 49, 1207–1218 (1991). [PMC free article] [PubMed] [Google Scholar]
- 11.Boot E., Linders C. C., Tromp S. H., van den Boogaard M. J., van Eeghen A. M., Possible underreporting of pathogenic variants in RAI1 causing Smith-Magenis syndrome. Am. J. Med. Genet. A. 185, 3167–3169 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bi W., et al. , Inactivation of Rai1 in mice recapitulates phenotypes observed in chromosome engineered mouse models for Smith-Magenis syndrome. Hum. Mol. Genet. 14, 983–995 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Javed S., Selliah T., Lee Y. J., Huang W. H., Dosage-sensitive genes in autism spectrum disorders: From neurobiology to therapy. Neurosci. Biobehav. Rev. 118, 538–567 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Huang W. H., et al. , Early adolescent Rai1 reactivation reverses transcriptional and social interaction deficits in a mouse model of Smith-Magenis syndrome. Proc. Natl. Acad. Sci. U.S.A. 115, 10744–10749 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang W. H., et al. , Molecular and neural functions of Rai1, the causal gene for Smith-Magenis syndrome. Neuron 92, 392–406 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Javed S., Lee Y. J., Xu J., Huang W. H., Temporal dissection of Rai1 function reveals brain-derived neurotrophic factor as a potential therapeutic target for Smith-Magenis syndrome. Hum. Mol. Genet. 31, 275–288 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Goldman A. M., et al. , Epilepsy and chromosomal rearrangements in Smith-Magenis Syndrome [del(17)(p11.2p11.2)]. J. Child Neurol. 21, 93–98 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Diessler S., Kostic C., Arsenijevic Y., Kawasaki A., Franken P., Rai1 frees mice from the repression of active wake behaviors by light. eLife 6, e23292 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bi W., et al. , Rai1 deficiency in mice causes learning impairment and motor dysfunction, whereas Rai1 heterozygous mice display minimal behavioral phenotypes. Hum. Mol. Genet. 16, 1802–1813 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Garay P. M., et al. , RAI1 regulates activity-dependent nascent transcription and synaptic scaling. Cell Rep. 32, 108002 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tronche F., et al. , Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 23, 99–103 (1999). [DOI] [PubMed] [Google Scholar]
- 22.Luo L., et al. , Optimizing nervous system-specific gene targeting with Cre driver lines: Prevalence of germline recombination and influencing factors. Neuron 106, 37–65.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLeod F., et al. , Reduced seizure threshold and altered network oscillatory properties in a mouse model of Rett syndrome. Neuroscience 231, 195–205 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Sperk G., et al. , Kainic acid induced seizures: Neurochemical and histopathological changes. Neuroscience 10, 1301–1315 (1983). [DOI] [PubMed] [Google Scholar]
- 25.Racine R. J., Modification of seizure activity by electrical stimulation. II. Motor seizure. Electroencephalogr. Clin. Neurophysiol. 32, 281–294 (1972). [DOI] [PubMed] [Google Scholar]
- 26.Maya I., et al. , Abnormal brain magnetic resonance imaging in two patients with Smith-Magenis syndrome. Am. J. Med. Genet. A. 164A, 1940–1946 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Magistretti P. J., Allaman I., A cellular perspective on brain energy metabolism and functional imaging. Neuron 86, 883–901 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Renier N., et al. , Mapping of brain activity by automated volume analysis of immediate early genes. Cell 165, 1789–1802 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guenthner C. J., Miyamichi K., Yang H. H., Heller H. C., Luo L., Permanent genetic access to transiently active neurons via TRAP: Targeted recombination in active populations. Neuron 78, 773–784 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heinemann U., et al. , The dentate gyrus as a regulated gate for the propagation of epileptiform activity. Epilepsy Res. Suppl. 7, 273–280 (1992). [PubMed] [Google Scholar]
- 31.Krook-Magnuson E., et al. , In vivo evaluation of the dentate gate theory in epilepsy. J. Physiol. 593, 2379–2388 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bengzon J., et al. , Apoptosis and proliferation of dentate gyrus neurons after single and intermittent limbic seizures. Proc. Natl. Acad. Sci. U.S.A. 94, 10432–10437 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zong H., Espinosa J. S., Su H. H., Muzumdar M. D., Luo L., Mosaic analysis with double markers in mice. Cell 121, 479–492 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Hippenmeyer S., et al. , Genetic mosaic dissection of Lis1 and Ndel1 in neuronal migration. Neuron 68, 695–709 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Espinosa J. S., Wheeler D. G., Tsien R. W., Luo L., Uncoupling dendrite growth and patterning: Single-cell knockout analysis of NMDA receptor 2B. Neuron 62, 205–217 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Müller M., Gähwiler B. H., Rietschin L., Thompson S. M., Reversible loss of dendritic spines and altered excitability after chronic epilepsy in hippocampal slice cultures. Proc. Natl. Acad. Sci. U.S.A. 90, 257–261 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peebles C. L., et al. , Arc regulates spine morphology and maintains network stability in vivo. Proc. Natl. Acad. Sci. U.S.A. 107, 18173–18178 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dudek F. E., Sutula T. P., Epileptogenesis in the dentate gyrus: A critical perspective. Prog. Brain Res. 163, 755–773 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Luo W., et al. , Recurrent rewiring of the adult hippocampal mossy fiber system by a single transcriptional regulator, Id2. Proc. Natl. Acad. Sci. U.S.A. 118, e2108239118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vong L., et al. , Leptin action on GABAergic neurons prevents obesity and reduces inhibitory tone to POMC neurons. Neuron 71, 142–154 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amaral D. G., Scharfman H. E., Lavenex P., The dentate gyrus: Fundamental neuroanatomical organization (dentate gyrus for dummies). Prog. Brain Res. 163, 3–22 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huguenard J., Current controversy: Spikes, bursts, and synchrony in generalized absence epilepsy: Unresolved questions regarding thalamocortical synchrony in absence epilepsy. Epilepsy Curr. 19, 105–111 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Venkadesh S., Komendantov A. O., Wheeler D. W., Hamilton D. J., Ascoli G. A., Simple models of quantitative firing phenotypes in hippocampal neurons: Comprehensive coverage of intrinsic diversity. PLOS Comput. Biol. 15, e1007462 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheong E., Shin H. S., T-type Ca2+ channels in normal and abnormal brain functions. Physiol. Rev. 93, 961–992 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Dumenieu M., et al. , The low-threshold calcium channel Cav3.2 mediates burst firing of mature dentate granule cells. Cereb. Cortex 28, 2594–2609 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ernst W. L., Zhang Y., Yoo J. W., Ernst S. J., Noebels J. L., Genetic enhancement of thalamocortical network activity by elevating alpha 1g-mediated low-voltage-activated calcium current induces pure absence epilepsy. J. Neurosci. 29, 1615–1625 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chemin J., et al. , De novo mutation screening in childhood-onset cerebellar atrophy identifies gain-of-function mutations in the CACNA1G calcium channel gene. Brain 141, 1998–2013 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Kim C. H., Cav3.1 T-type calcium channel modulates the epileptogenicity of hippocampal seizures in the kainic acid-induced temporal lobe epilepsy model. Brain Res. 1622, 204–216 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Kim D., et al. , Lack of the burst firing of thalamocortical relay neurons and resistance to absence seizures in mice lacking alpha(1G) T-type Ca(2+) channels. Neuron 31, 35–45 (2001). [DOI] [PubMed] [Google Scholar]
- 50.Danielsson S., Gillberg I. C., Billstedt E., Gillberg C., Olsson I., Epilepsy in young adults with autism: A prospective population-based follow-up study of 120 individuals diagnosed in childhood. Epilepsia 46, 918–923 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Reilly C., et al. , Neurobehavioral comorbidities in children with active epilepsy: A population-based study. Pediatrics 133, e1586–e1593 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Krishnan V., et al. , Autism gene Ube3a and seizures impair sociability by repressing VTA Cbln1. Nature 543, 507–512 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eyring K. W., Geschwind D. H., Three decades of ASD genetics: Building a foundation for neurobiological understanding and treatment. Hum. Mol. Genet. 30, R236–R244 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choi L., An J. Y., Genetic architecture of autism spectrum disorder: Lessons from large-scale genomic studies. Neurosci. Biobehav. Rev. 128, 244–257 (2021). [DOI] [PubMed] [Google Scholar]
- 55.Friedmann D., et al. , Mapping mesoscale axonal projections in the mouse brain using a 3D convolutional network. Proc. Natl. Acad. Sci. U.S.A. 117, 11068–11075 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sohal V. S., Rubenstein J. L. R., Excitation-inhibition balance as a framework for investigating mechanisms in neuropsychiatric disorders. Mol. Psychiatry 24, 1248–1257 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simms B. A., Zamponi G. W., Neuronal voltage-gated calcium channels: Structure, function, and dysfunction. Neuron 82, 24–45 (2014). [DOI] [PubMed] [Google Scholar]
- 58.Nanou E., Catterall W. A., Calcium channels, synaptic plasticity, and neuropsychiatric disease. Neuron 98, 466–481 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Benarroch E. E., HCN channels: Function and clinical implications. Neurology 80, 304–310 (2013). [DOI] [PubMed] [Google Scholar]
- 60.Satterstrom F. K., et al. ; Autism Sequencing Consortium; iPSYCH-Broad Consortium, Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell 180, 568–584.e23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang W. H., et al. , Atoh1 governs the migration of postmitotic neurons that shape respiratory effectiveness at birth and chemoresponsiveness in adulthood. Neuron 75, 799–809 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang W. H., Performing single-cell clonal analysis in the mouse brain using mosaic analysis with double markers (MADM). Methods Mol. Biol. 2515, 59–74 (2022). [DOI] [PubMed] [Google Scholar]
- 63.Ya-Ting Chang et al., Loss of Rai1 enhances hippocampal excitability and epileptogenesis in mouse models of Smith–Magenis syndrome. Zenodo. https://zenodo.org/record/7140061#.Yzs2Y3bMLEZ. Deposited 3 October 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information, except the electrophysiology protocols and whole-brain Fos maps, which can be accessed at https://zenodo.org/record/7140061 (63).