Significance
We found that evolutionary conflicts between the cytoplasmic and nuclear genomes have persistent effects on citrus genetics and breeding, based on an analysis of a citrus pan-mitogenome and population genomics. This conflict conferred a prominent divergence and increased the genetic load on the mitochondrial genome during diversification and domestication. Deficiencies in inheritance, the levels of heteroplasmy that occurred from paternal leakage, and mitochondrial–nuclear interactions were examined in hybrids. The data are consistent with two candidate chimeric open reading frames (ORFs) serving as species-specific cytoplasmic male sterility/restorer-of-fertility (CMS/Rf) genes. These findings on the cytoplasmic genomes expand our understanding of crop breeding.
Keywords: population genomics, domestication, cytonuclear, hybridization, citrus
Abstract
Although interactions between the cytoplasmic and nuclear genomes occurred during diversification of many plants, the evolutionary conflicts due to cytonuclear interactions are poorly understood in crop breeding. Here, we constructed a pan-mitogenome and identified chimeric open reading frames (ORFs) generated by extensive structural variations (SVs). Meanwhile, short reads from 184 accessions of citrus species were combined to construct three variation maps for the nuclear, mitochondrial, and chloroplast genomes. The population genomic data showed discordant topologies between the cytoplasmic and nuclear genomes because of differences in mutation rates and levels of heteroplasmy from paternal leakage. An analysis of species-specific SVs indicated that mitochondrial heteroplasmy was common and that chloroplast heteroplasmy was undetectable. Interestingly, we found a prominent divergence in the mitogenomes and the highest genetic load in the, which may provide the basis for cytoplasmic male sterility (CMS) and thus influence the reshuffling of the cytoplasmic and nuclear genomes during hybridization. Using cytoplasmic replacement experiments, we identified a type of species-specific CMS in mandarin related to two chimeric mitochondrial genes. Our analyses indicate that cytoplasmic genomes from mandarin have rarely been maintained in hybrids and that paternal leakage produced very low levels of mitochondrial heteroplasmy in mandarin. A genome-wide association study (GWAS) provided evidence for three nuclear genes that encode pentatricopeptide repeat (PPR) proteins contributing to the cytonuclear interactions in the Citrus genus. Our study demonstrates the occurrence of evolutionary conflicts between cytoplasmic and nuclear genomes in citrus and has important implications for genetics and breeding.
Cytoplasmic genomes are a hallmark feature of eukaryotic cells. Their evolutionary processes are distinct from those of nuclear genomes (1). The reshuffling of nuclear and cytoplasmic genomes that occurs during outbreeding is widespread and frequent, which could reshape the landscape and interactions between nuclear and cytoplasmic genomes (2, 3). Rearrangements, mutations, and inheritance modes affect interactions between nuclear and cytoplasmic genomes and lead to evolutionary conflicts that are expected to have profound effects on diversification, domestication, and hybridization. Nonetheless, the evolution of the interactions between nuclear and cytoplasmic genomes in crops is barely known, especially at the population level. The evolutionary conflicts between nuclear and cytoplasmic genomes are driven by at least three issues.
First, the extent of structural variations (SVs) differs substantially between the cytoplasmic and nuclear genomes. SVs occur at a very low frequency in chloroplast genomes (4). In contrast, repeat-mediated recombination generates extensive SVs in mitochondrial genomes (5). Indeed, high rates of genomic rearrangements occur in plant mitochondrial genomes because of frequent recombination at both the intraspecific and individual levels (6). These rearrangements in the mitochondrial genomes might affect the fusion of noncoding transcripts and conserved genes by shuffling the start and stop codons or by producing chimeric open reading frames (ORFs) in intergenic regions that are considered to be the cause of cytoplasmic male sterility (CMS) because they alter floral development (e.g., because they lead to the loss of the male gametophyte) (7–9). Nuclear restorer-of-fertility (Rf) genes suppress this type of CMS (10, 11). The selection patterns on Rf genes provide insight into the evolutionary conflict between the nuclear and cytoplasmic genomes.
Second, the mutation rate was estimated to be lower in cytoplasmic genomes than in nuclear genomes (12), which provides another source of evolutionary conflict. The differences in substitution rates between the nuclear and cytoplasmic genomes often lead to different estimates for phylogenetic relationships (13). In addition to differences in the substitution rates, decreases in the effective population size (Ne) could reduce the efficiency of selection for cytoplasmic genomes relative to nuclear genomes (4, 14). Because of the small Ne, genetic drift may lead to deleterious mutations (i.e., the genetic load) accumulating in cytoplasmic genomes (15–17). Crops have undergone population bottlenecks associated with decreases in genetic diversity during domestication (18, 19). Therefore, from our perspective, the patterns of genetic diversity that arose from crop domestication in the cytoplasmic genomes of crops might differ from the patterns observed in nuclear genomes.
Third, although the cytoplasmic genomes are inherited predominantly through the maternal lineage, the nuclear genome is biparentally transmitted in most plants, which may lead to evolutionary conflicts during hybridization (20). Our understanding of plant cytoplasmic genome evolution and diversity have expanded over the last two decades (21). First, massively deep resequencing has revealed an extraordinary level of cytoplasmic genome diversity among populations (22). Second, mixed populations of cytoplasmic genomes within individuals (i.e., heteroplasmy) have been observed at the cellular and organelle levels (23). Third, some degree of bipaternal inheritance (i.e., paternal leakage) allows for the possibility of cytoplasmic heteroplasmy (20, 24). In plants, the different mechanisms of transmission for nuclear and cytoplasmic genomes could reshape the levels of heteroplasmy and hybridization patterns (23). Heteroplasmy has been demonstrated to alter the expression of particular nuclear genes that perhaps in turn influence the level of heteroplasmy or increase the rearrangement response to the CMS/Rf system (25). In addition, new combinations of nuclear and cytoplasmic genomes are created during hybridization and introgression (26, 27). There is some evidence for cytonuclear hybrid incompatibility, which indicates that cytoplasmic inheritance may be affected by the admixed nuclear genomes that are produced during hybridization (28, 29).
Citrus has a complex evolutionary history (30–33). The term “citrus” includes seven basic groups—atalantia (Atalantia buxifolia), poncirus (Poncirus trifoliata), pummelo (Citrus maxima), citron (Citrus medica), mandarin (Citrus reticulata and Citrus unshiu), papeda (Citrus ichangensis), and kumquat (Fortunella spp.)—and four groups derived from admixtures—lemon (Citrus limon), sweet orange (Citrus sinensis), sour orange (Citrus aurantium), and grapefruit (Citrus paradisi) (34, 35). The nuclear and chloroplast genome sequences from citrus species indicate that sweet orange, sour orange, and grapefruit are admixtures derived from pummelo and mandarin and that the nuclear genome of lemon is an admixture derived from citron, pummelo, and mandarin (30, 32). Mandarin cultivars (Citrus unshiu) have been used to breed for citrus that produce seedless fruit because they provide male sterility based on the CMS/Rf system (36). Recent evidence proposes that a combination of dysfunctional mitochondrial genes and interacting nuclear restorer genes suppress anther formation in mandarin (37).
Here, we used citrus as a genetic system to investigate the evolutionary conflicts between cytoplasmic and nuclear genomes that occurred during diversification, domestication, and hybridization. We constructed a citrus pan-mitogenome based on 11 long read assemblies and one short read assembly. We also identified global rearrangement patterns and chimeric ORFs. The variation maps from the nuclear, mitochondrial, and chloroplast genomes from 184 accessions of citrus were used for population genomics analyses. The activities of the chimeric ORFs related to CMS were examined via cytoplasmic replacement experiments and expression analyses. Furthermore, mitochondrial heteroplasmy was estimated from species-specific SVs from a pan-mitogenome, and putative genetic regulators associated with cytonuclear interactions were identified through a genome-wide association study (GWAS). Our study aimed to address the following questions. How prevalent are SVs and chimeric ORFs in the citrus pan-mitogenome? What are the crucial SVs in mitogenomes that contribute to the CMS/Rf system in mandarin? Do cytonuclear interactions affect the inheritance of the cytoplasmic genome or influence paternal leakage during hybridization? Do the genomic data provide evidence of incongruence between the nuclear, mitochondrial, and chloroplast genomes or provide complementary interpretations for the evolutionary history of citrus?
Results
Extensive SVs in the Citrus Super Pan-Mitogenome.
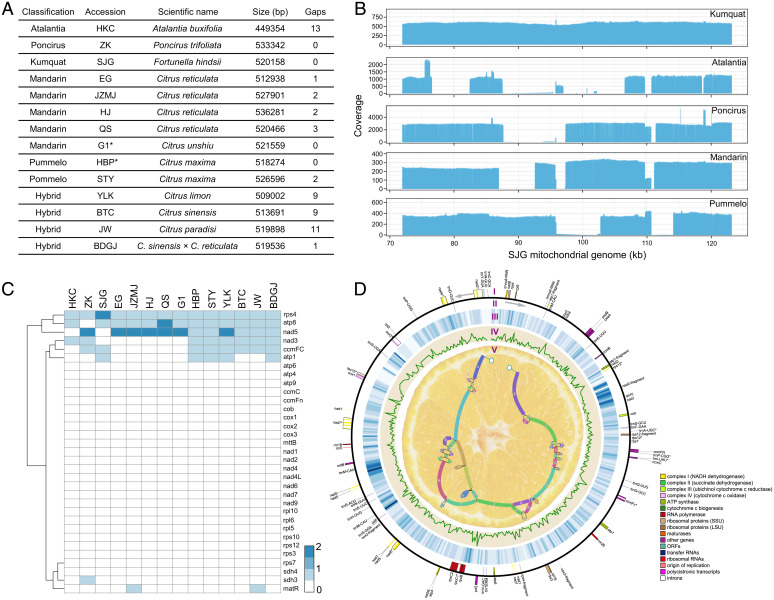
To investigate SVs and their associations in generating chimeric ORFs in citrus mitochondrial genomes, we started with the de novo assembly of mitochondrial genomes from 12 accessions (11 PacBio/Nanopore long read assemblies and one Illumina short read assembly) that represent a broad range of genetic diversity of mitochondrial genomes in citrus (Fig. 1A and SI Appendix, Tables 1 and 2). The sizes of our 12 mitochondrial assemblies ranged from 449.4 to 536.2 kb. With two additional published mitochondrial genomes from pummelo HBP and mandarin G1 (14 mitogenomes in total), we built an integrated graph-based genome (i.e., a super pan-mitogenome) containing 46 continuous segments from the kumquat SJG mitochondrial genome as a standard reference genome (no gap) (Fig. 1D). We identified an average of five chimeric ORFs (>300 bp) in the intergenic region that were derived from identifiable and conserved protein-coding gene fragments (i.e., transfer RNA [tRNA] and ribosomal RNA genes were excluded from the analysis), which is consistent with previous findings indicating that new ORFs are frequently generated in plant mitogenomes (38). The results showed that nad3 and nad5 from the nad family and atp1 and atp8 from the atp family formed prominent chimeric ORFs in all assemblies. We identified two chimeric ORFs in mandarin that contained fragments from nad5 (Fig. 1C and SI Appendix, Table 4).
Fig. 1.
SVs and chimeric ORFs in the citrus super pan-mitogenome. (A) Summary of the 12 de novo mitochondrial assemblies and 2 previously published mitochondrial genomes. The previously published sequences are highlighted with asterisks. (B) 70- to 120-kb regions identified based on the coverage depth of long-read mapping containing >5-kb deletions from five groups. The mitogenomic regions are shown on the x-axis. The different levels of coverage are indicated on the y-axis. (C) Chimeric ORFs related to conserved genes in the intergenic regions of 14 citrus mitochondrial genomes. The heatmap includes two previously reported mitochondrial genomes, mandarin G1 and pummelo HBP. (D) Circular representation of the mitochondrial pan-genome. The conserved gene structure was annotated for the forward strand (I) and the reverse strand (II). The heatmap indicates the distribution of SNPs and indels among the 184 accessions of citrus, with the mitochondrial genome from kumquat used as a reference (III). The diversity of SNPs and indels in the mitochondrial genomes in 184 citrus accessions is indicated (IV). The mitochondrial pan-genome is indicated with a schematic drawing (V).
We hypothesized that the evolution of the citrus mitogenome structure may be facilitated by the accumulation of large SVs. Our analysis combined with the 14 assembly alignments and 11 long read alignments to the kumquat SJG mitochondrial reference genome allowed us to identify 17 insertions (INSs), 12 deletions (DELs), 42 inversions (INVs), and 4 duplications (DUPs). We found three to nine SVs in each assembly (SI Appendix, Table 5). For example, we used a 50-kb window scan and found different sizes of deletions, ranging from 1.7 kb to 22 kb in five groups that excluded the hybrids (Fig. 1B and SI Appendix, Figs. 1 and 2). In addition to identifying SVs based on mitochondrial assemblies, we collected Illumina paired reads from 184 accessions (∼35.4× coverage) that included seven species (atalantia, papeda, poncirus, pummelo, mandarin, kumquat, and citron) and four admixture groups of Citrus (lemon, grapefruit, sweet orange, and sour orange) (SI Appendix, Table 6). Those reads were mapped to the kumquat SJG mitochondrial reference genome. We identified 14 DELs, 23 INVs, and 1 DUP (SI Appendix, Figs. 3–6 and Table 7). The results indicate extensive rearrangements in different populations, even at the individual level (e.g., pummelo cultivars STY and HBP; mandarin cultivars EG, JZMJ, HJ, and QS) (SI Appendix, Fig. 7). In addition, we built nuclear, mitochondrial, and chloroplast variation maps that contain 10,184,491 single-nucleotide polymorphisms (SNPs), 10,046 SNPs, and 6,445 SNPs, respectively.
Prominent Divergence in the Mitogenome of Mandarin.
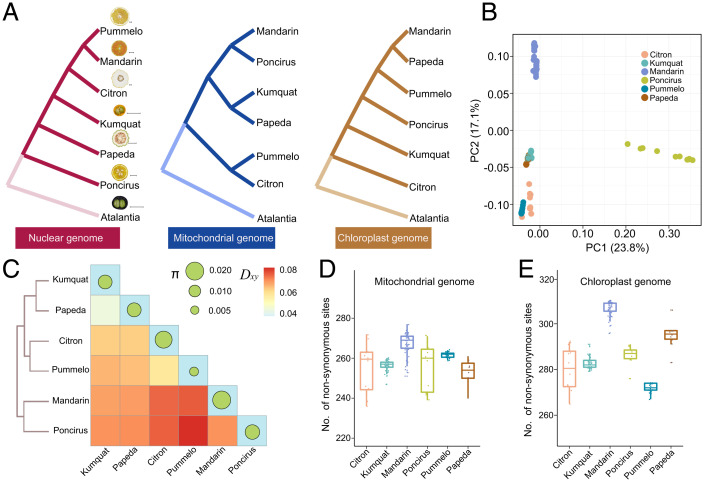
To better understand the evolutionary conflicts between the cytoplasmic and nuclear genomes that occurred in citrus during diversification, we estimated the phylogeny and the population structures of the six populations based on variants of the nuclear, mitochondrial, and chloroplast genomes (see Methods, SI Appendix, Figs. 8–12). Primitive citrus Atalantia buxifolia was used as an outgroup, and hybrid populations were excluded. It is notable that the topologies of the nuclear and cytoplasmic genomes among the different genera (Poncirus, Fortunella, and Citrus) were incongruent (Fig. 2A). Based on these three phylogenies, we elucidated two features of the Citrus genus (1). Mandarin and pummelo/citron were not clustered together in the phylogenetic analysis of the mitochondrial genomes but were clustered together in the chloroplast and nuclear genomes (2). Our phylogenetic analysis of the mitochondrial and nuclear genomes indicates that citron and pummelo are close, which conflicts with the result from the chloroplast genome. Furthermore, the ancestry composition estimation (SI Appendix, Figs. 10 and 12) and principal component analysis (PCA) (PC1, 23.8%; PC2, 17.1%) were performed with the cytoplasmic genomic dataset and corroborated the phylogenetic analysis (Fig. 2B and SI Appendix, Fig. 13).
Fig. 2.
Nonsynchronous genetic divergence of mitochondrial genomes in mandarin during citrus evolution. (A) Consensus phylogeny of nuclear genomes (Left), mitochondrial genomes (Middle), and chloroplast genomes (Right) from seven species of citrus. The phylogenetic diagram was constructed from the population dataset. The atalantia population (Atalantia buxifolia) was used as an outgroup. Scale bar, 2 cm. (B) PCA of mitochondrial genomes from six populations of citrus. The outgroup was excluded. PC1 (23.8%) and PC2 (17.1%) are shown. (C) Divergence (Dxy) and genetic diversity (π) analysis of mitochondrial genomes from six species of citrus. The size of the circle is proportional to the genetic diversity. The amount of divergence is indicated with a heatmap. (D and E) Evaluation of genetic load in mitochondrial and chloroplast genomes. The number of nonsynonymous mutations in predicted ORFs from six species is indicated on the y-axis.
To investigate the genetic features of the cytoplasmic genomes in six populations, we measured the divergence (Dxy) and genetic diversity (π) of the mitogenomes and attempted to evaluate the genetic load in cytoplasmic genomes by determining the number of nonsynonymous mutations in predicted ORFs (see Methods). The pairwise comparisons of divergence indicated that a dramatic differentiation of the mitogenomes occurred within the Citrus genus (e.g., the Dxy between pummelo and mandarin was 0.0666 ± 0.0116) that was even striking relative to the differentiation of mitogenomes in different genera (e.g., the Dxy between pummelo and poncirus was 0.0754 ± 0.0124) (Fig. 2C). At the same time, we found that the highest genetic diversity in the mitogenomes from the wild and cultivated mandarins (Citrus reticulata, π = 0.0195 ± 0.0019) (Fig. 2C). We used atalantia population as an outgroup to reduce reference bias, and interestingly, we found the highest genetic load in both the plastome and mitogenome of mandarin (Fig. 2 D and E and SI Appendix, Fig. 14). Overall, these analyses uncovered striking patterns of divergence and genetic load in the cytoplasmic genomes of mandarin. But whether the features of the cytoplasmic genomes that we observed influence phenotype remained an open question.
Domestication Affects the Genetic Features of the Cytoplasmic Genomes.
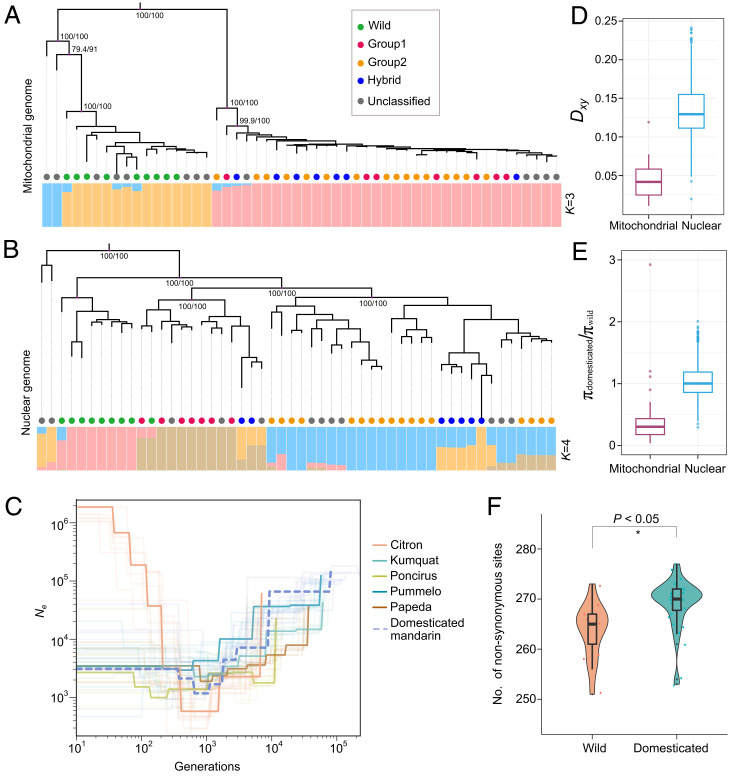
Previous studies have provided evidence that mandarins came from two independent domestication events (39). Therefore, we analyzed the cytoplasmic and nuclear genomes of wild species and two groups that may have been domesticated independently (MD1 and MD2) and additional accessions of mandarin, for a total of 53 samples (SI Appendix, Table 8). We found that the phylogenetic trees using mitochondrial and chloroplast genome sequences supported a separation event between the wild and domesticated mandarins and a mixed pattern of MD1 and MD2 groups (Fig. 3A and SI Appendix, Figs. 10 and 12). The phylogeny of nuclear genome sequences could separate the wild mandarins from the domesticated mandarins and from the MD1 and MD2 groups in the domesticated clade (Fig. 3B). The patterns of the population structures from the mitochondrial genome (best K = 3) and the nuclear genome (best K = 4) were similar (SI Appendix, Figs. 15 and 16). Furthermore, the demography estimated from the nuclear genomic data is consistent with a strong bottleneck occurring in the domesticated mandarin at 1,000 generations before the present (Fig. 3C).
Fig. 3.
Domestication history of mandarin inferred from mitochondrial genomes. (A) Population structure analysis based on the mitochondrial genomic dataset. The colored circles represent the classification of mandarin and are identical to those of a previous study. The ancestral composition (best K = 3) and bootstrap values are indicated. (B) Population structure analysis based on the nuclear genomic dataset. The ancestral composition (best K = 4) is presented. (C) Estimation of demography from nuclear genomes from six populations. The numbers of generations are indicated on the x-axis. The effective population size is indicated on the y-axis. (D) Comparison of the divergence statistic (Dxy) from the wild population and domesticated populations based on the nuclear and mitochondrial genomic datasets. The domesticated and wild groups were classified based on mitochondrial genomic datasets. (E) Ratio of genetic diversity (πdomesticated/πwild) for mandarin. (F) Evaluation of genetic load in mitochondrial genomes between wild and domesticated groups.
To test whether domestication had a similar influence on the nuclear and cytoplasmic genomes of mandarin, we compared the mitochondrial and nuclear genomes by using two statistics: Dxy in the domesticated and wild populations and the ratio of π from the domesticated population to π from the wild population. We found that Dxy was significantly lower (P < 0.001, Student’s t test) in the mitochondrial genome relative to the nuclear genome (Fig. 3D). We also found a 60.9% reduction in the genetic diversity of the mitochondrial genome during the domestication of mandarin (wild, π = 0.0105; domesticated, π = 0.0041). In contrast, we found that 103.5% of the genetic diversity was retained in the nuclear genome, which is consistent with previous work on the domestication of other perennial crops (Fig. 3E) (40, 41).
Furthermore, we compared the genetic load in the cytoplasmic genomes of wild and domesticated mandarin populations. We found a significantly higher genetic load in the cytoplasmic genomes of the domesticated population relative to the wild population (P < 0.05, Student’s t test; Fig. 3F and SI Appendix, Fig. 17). The dramatic reduction of Ne could increase the genetic load in the cytoplasmic genomes of domesticated mandarin through genetic drift.
Characterization of a Species-Specific CMS/Rf System in Mandarin.
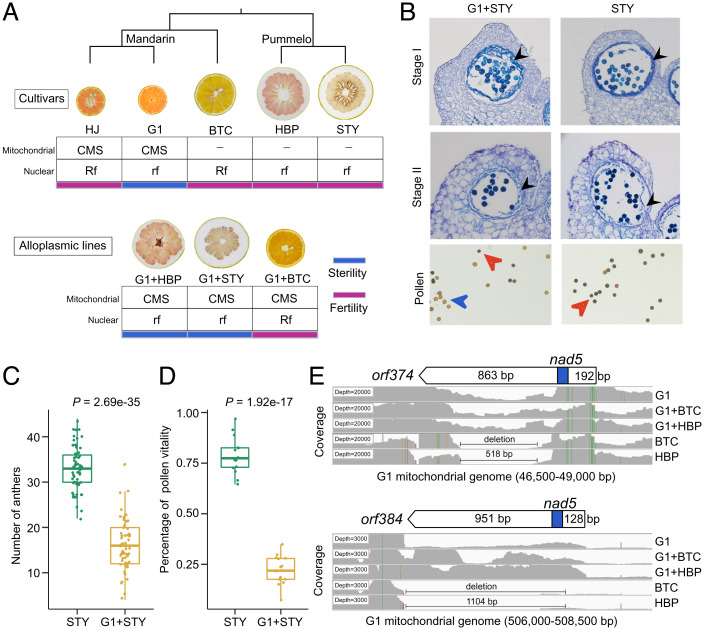
To examine the establishment of the species-specific CMS/Rf system in citrus, we combined three independent cytoplasmic replacement experiments via protoplast fusion (i.e., cell–cell fusions; see Methods), which produced three different groups of alloplasmic lines (pummelo HBP and STY; one sweet orange BTC) (42, 43) (Fig. 4A). The male sterility phenotypes were observed in the alloplasmic lines G1+HBP and G1+STY associated with seedless fruit (SI Appendix, Fig. 18). The alloplasmic line G1+HBP developed smaller petals and anther-like structures that did not develop to the mature stage (37). The tapetum contributes to the formation of pollen and is degraded during the process of pollen maturation (44). Our cytological observations of the alloplasmic line G1+STY indicate that the early stage (stage I) of tapetum development was tortuous, the tapetum was not degraded at the mature stage (stage II), and the anthers were indehiscent (Fig. 4B). The alloplasmic line G1+STY was used to investigate the number of anthers and to examine the viability of the pollen. We found that the number of anthers in the alloplasmic line G1+STY was significantly less than in the pummelo cultivar STY (P = 2.69e-35, Student’s t test) (Fig. 4C). Results from pollen staining experiments provide evidence that the vitality of pollen was reduced in the alloplasmic line G1+STY (P = 1.92e-17, Student’s t test) (Fig. 4D). Therefore, our study revealed that the alloplasmic lines that were produced by combining the cytoplasm from the mandarin G1 line with nuclei from different pummelo lines could lead to male sterility. The sweet orange cultivar BTC has an admixed nuclear genome from mandarin and pummelo (33). Both the sweet orange cultivar BTC and the alloplasmic line G1+BTC (group 3) produced normal anthers and seeds (Fig. 4A and SI Appendix, Fig. 19), which might be due to restorer genes in the nuclear genome of the sweet orange cultivar. Overall, our results demonstrate a species-specific CMS/Rf system in mandarin.
Fig. 4.
Analysis of the CMS/Rf system in mandarin with cytoplasmic replacement experiments. (A) Phenotypes and proposed mitonuclear genotypes in citrus cultivars and somatic cybrids. The male fertile mandarin cultivar HJ (Citrus reticulata), male sterile mandarin cultivar G1 (Citrus unshiu), male fertile sweet orange cultivar BTC, male fertile pummelo cultivar HBP, and STY are shown in the upper row. The male sterile alloplasmic line G1+HBP, male sterile alloplasmic line G1+STY, and male fertile alloplasmic line G1+BTC are shown in the lower row. The sweet orange BTC was derived from the hybridization of mandarin and pummelo. (B) Cytological observation of anthers during two different developmental stages and the staining of pollen from pummelo STY and the alloplasmic line G1+STY. Each micrograph is from a 40× field of view. In the pummelo cultivar STY, the tapetum was formed in stage I and was degraded in stage II. The tapetum is indicated with a black arrow. Activated pollen is indicated with an orange arrow. Inactivated pollen is indicated with a blue arrow. The bar is 100 μm. (C and D) Statistics for (C) the anther (100 flowers with three replications) and (D) pollen vitality (10 observations with three replications) in the pummelo STY and the alloplasmic line G1+STY. (E) RNA-seq reads mapped to the mandarin G1 mitochondrial genome (windows in IGV) and SVs for two chimeric ORFs (orf374 and orf384) in five samples including three cultivars (mandarin G1, sweet orange BTC, and pummelo HBP) and two alloplasmic lines (G1+BTC and G1+HBP). The x-axis indicates mitochondrial regions. The y-axis indicates coverage.
To identify candidate ORFs that respond to the species-specific CMS in mandarin, we collected RNA sequencing (RNA-seq) data (noncoding RNA libraries) from pummelo HBP, mandarin G1, sweet orange BTC, the alloplasmic line G1+BTC, and the alloplasmic line G1+HBP (45) and mapped the data to the G1 mitochondrial genome. In the mandarin cultivar G1, we identified five chimeric ORFs in the intergenic region that were derived from three conserved mitochondrial genes (rps4, atp8, and nad5) (Fig. 1C and SI Appendix, Fig. 20). Two chimeric ORFs (orf374 and orf384) that have 67-bp and 73-bp fragments, respectively, from nad5 are not capable of trans-splicing (SI Appendix, Table 4). Two deletion variants (518 bp and 1,104 bp) are responsible for the lack of orf374 and orf384 in the mitochondrial genomes of two pummelo cultivars (HBP and STY) (Fig. 4E). We found an individual-specific pattern in orf374 only in the mitogenome of mandarin cultivar G1 (SI Appendix, Fig. 21). In contrast, orf384 was found in the mitogenomes of all four mandarin cultivars with genome assemblies (SI Appendix, Fig. 22). Subsequently, we found that these two chimeric ORFs were highly expressed in the mandarin cultivar G1, alloplasmic line G1+BTC (male fertile), and alloplasmic line G1+HBP (Fig. 4E). Additionally, we found that orf384 was expressed at different levels in the mandarin cultivar G1 relative to the alloplasmic lines. This pattern might be related to the severe defect we observed in the alloplasmic line G1+HBP, which has a nuclear genome that is deficient in multiple restorer genes.
Influence of Reticulate Evolution on Mitochondrial Heteroplasmy.
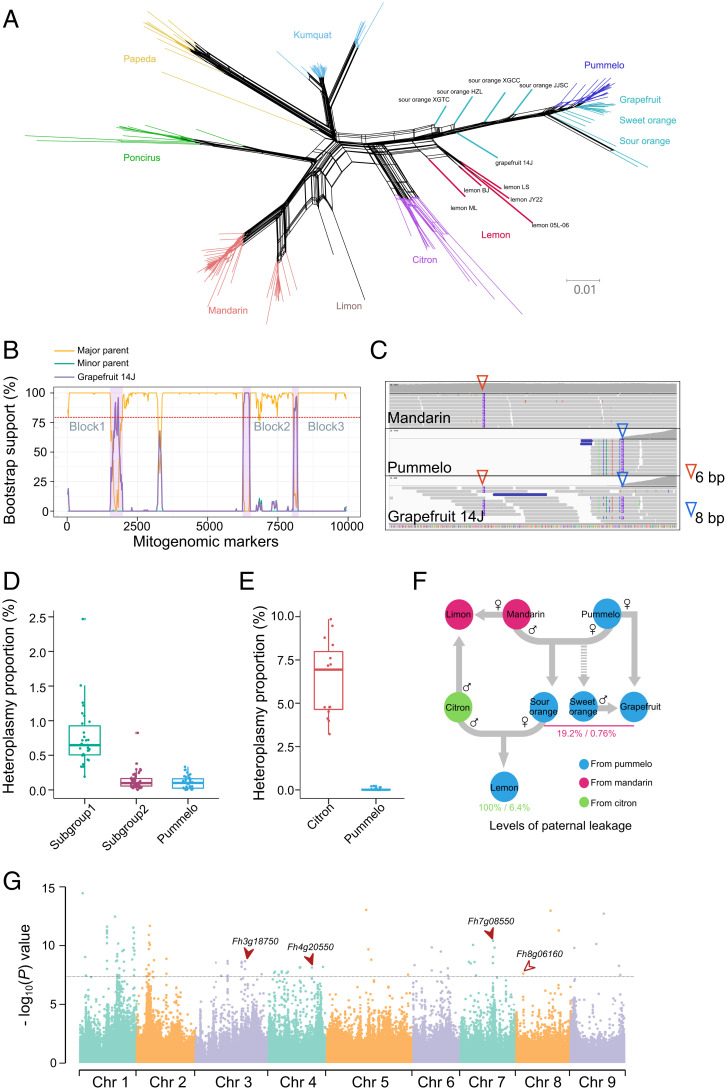
We found that a species-specific form of CMS is occurring in mandarin and that a restorer gene was lost from pummelo. These mutations may have affected the reshuffling of the cytoplasmic and nuclear genomes that occurred during hybridization in citrus. To clarify the incongruence between the cytoplasmic and nuclear genomes that occurred during hybridization, we used cytoplasmic genome variation datasets to construct the phylogenetic network (i.e., to perform a reticulate analysis) with 184 accessions including four hybrid populations (grapefruit, sweet orange, sour orange, and lemon). The split tree and the differentiation statistics make the case that the cytoplasmic genomes in grapefruit, sweet orange, sour orange, and lemon were prominently inherited from pummelo (Fig. 5A and SI Appendix, Fig. 23). Interestingly, we observed a long genetic distance to the pummelo cluster in the mitochondrial phylogenetic network for all lemon samples and five samples from sour orange and grapefruit (sour oranges XGTC, HZL, XGCC, and JJSC; grapefruit 14J) (Fig. 5A). In contrast, the pummelos and all hybrids were tightly clustered in the chloroplast phylogenetic network (SI Appendix, Fig. 24). For greater insight into the topology of the 10 hybrid individuals, we estimated the recombination breakpoints in the mitochondrial genome by using the aligned variations among the hybrid groups and their parental groups. Our recombination signal detection analysis demonstrated that all lemons and five individuals from grapefruit and sour orange have significant regions of recombination (SI Appendix, Figs. 25 and 26). For instance, there were three blocks with significant recombination signals (bootstrap support >80) in the grapefruit 14J mitochondrial genome (Fig. 5B). These results show that pummelo was a major parent of grapefruit 14J and that the three derived fragments (14–32 kb) were from mandarin, which was a minor parent (SI Appendix, Fig. 27).
Fig. 5.
Reticulate evolution and heteroplasmy in the mitochondrial genomes of citrus. (A) Network phylogeny of mitochondrial genomes in citrus. Four hybrid populations and two hybrid individuals (rangpur lime and rough lemon) were included in the analysis. The different colors indicate different species. Lemons are indicated with red. Sour oranges, sweet oranges, and grapefruits are indicated with light blue. (B) Bootstrap value (%) support for detected recombination. The yellow line represents the major parent (pummelo). The blue line represents the minor parent (mandarin). The purple line (bold) represents the individual grapefruit 14J (subgroup 1). Three blocks with higher bootstrap values (>80) are shaded. (C) Paternal leakage in the grapefruit 14J line. The IGV plot shows species-specific SVs with two different indel markers from pummelo and mandarin. (D) Proportions of heteroplasmy estimated by read coverage from species-specific SVs in mandarin and pummelo. Subgroup 1 contained five individuals (grapefruit 14J, sour orange XGTC, sour orange XGCC, sour orange HZL, and sour orange JJSC). Subgroup 2 contained the remaining individuals from grapefruit, sweet orange, and sour orange populations. (E) Proportions of heteroplasmy estimated in lemon populations (five of five individuals). (F) Inheritance of mitochondrial genomes for citrus hybrids and admixtures. The sample percentages and the heteroplasmy levels of paternal leakage are presented in different colors associated with the origin. (G) Candidate regions in nuclear genomes associated with cytonuclear interactions identified through a GWAS. The three solid arrows indicate the locations of three genes that encode PPR proteins (Fh3g18750, Fh4g20550, and Fh7g08550) that were expressed at significantly different levels. The hollow arrow indicates the location of a fourth gene that encodes a PPR protein (Fh8g06160) that was not expressed at significantly different levels.
Within the detected block regions, we found that there were large deletion variations in the pummelo mitogenome (i.e., regions without mapped reads) and reads from paternal leaks (citron or mandarin) that were mapped and that led to a recombination signal (SI Appendix, Fig. 28). For example, we identified a species-specific 6-bp deletion in mandarin that was located in the block 3 region (excluding the bias in homology mapping coverage) and a species-specific 8-bp deletion of pummelo that was located in the flanking region. Interestingly, we found both the 8-bp deletion and the 6-bp deletion in the five individuals that are hybrids of mandarin and pummelo (Fig. 5C). Accordingly, paternal leakage leads to the segregated cluster pattern in hybrids staying away from the material group in the phylogenetic network associated with the detectable recombination blocks in SVs.
Levels of Mitochondrial Heteroplasmy in Hybrids.
To clarify the levels of mitochondrial heteroplasmy in hybrids, we estimated the proportion of heteroplasmy that occurred from paternal leakage of mitochondrial genomes in each sample based on coverage depth in recombination blocks. Specifically, we quantified the prevalence of species-specific insertions and deletions that occurred from maternal and paternal transmissions based on the pan-mitogenome. We masked the homologous regions of the nuclear genome to exclude incorrectly mapped reads and estimated the proportion of heteroplasmy based on SV regions in whole mitogenomes to reduce the possibility of mapping errors. The results showed a significantly higher proportion of heteroplasmy (0.761 ± 0.435%, P < 4.3e-9) in the five hybrid individuals (four sour oranges and one grapefruit, subgroup 1), compared to the pummelo population (0.108 ± 0.092%) (Fig. 5D and SI Appendix, Fig. 29). At the same time, we found no significant difference (0.123 ± 0.141%, P = 0.22) in the proportion of heteroplasmy in the mitochondrial genomes from the rest of the hybrids: sour oranges, sweet oranges, and grapefruits (subgroup 2) (Fig. 5D). Although the cluster and phylogenic analysis revealed that the mitochondrial genome in lemon was derived from pummelo, the lemon group was separated from the subgroup 1 cluster (Fig. 5A). Lemon is different from hybrids, such as sweet orange, sour orange, and grapefruit. Indeed, lemons are admixtures derived from mandarin, pummelo, and citron. To avoid the influence of the mandarin mitochondrial genome, we estimated the proportion of heteroplasmy that depended on the species-specific insertions and deletions derived from pummelo and citron (i.e., excluding the SVs from pummelo and mandarin) (SI Appendix, Fig. 30). A much higher proportion of mitochondrial heteroplasmy (6.373 ± 2.106%, 8.4 times higher than that of subgroup 1) was found in all samples from the lemon population (Fig. 5E). Collectively, our analysis demonstrates that the mitochondrial genome from mandarin was maintained at an extremely low level as heteroplasmy derived from paternal leakage (Fig. 5F).
The nuclear genomes of modern Citrus cultivars are complex admixtures derived from hybridization and introgression in citron, mandarin, and pummelo (30). To test the hypothesis that cytonuclear interactions contribute to hybridization patterns or paternal leakage, we used a GWAS to identify candidate regions (nuclear linkage disequilibrium [LD] blocks) associated with the cytoplasmic genomes by using nuclear and mitochondrial variation maps constructed from 118 Citrus samples (see Methods, Fig. 5G and SI Appendix, Fig. 31). We found that a total of 168 candidate genes (Benjamini-Hochberg = 5%) were enriched, including three genes associated with the photosystem II process, a gene encoding mitochondrial transcription termination factor, a gene encoding mitochondrial import receptor subunit TOM22, and four genes that encode pentatricopeptide repeat (PPR) proteins (see SI Appendix, Table 9 for full list). PPR proteins often serve as restorer genes that respond to male sterility (46). Because cytonuclear interactions contribute to a complex trait that is influenced by both mitochondria–nuclear and chloroplast–nuclear interactions, narrowing down the crucial restorer genes by using a GWAS and annotations is not straightforward. Because of the precedent for PPR proteins serving as restorer genes, we focused on PPR genes associated with mitochondrial functions that are expressed in the anthers in mandarin and pummelo (SI Appendix, Fig. 32). Given the significant divergence between mandarin and pummelo, we collected RNA-seq data (complementary DNA libraries) from the anthers of eight pummelos and four mandarins and identified differences in expression after the normalization. We found that the expression of three PPR genes (Fh3g18750, Fh4g20550, and Fh7g08550) in mandarin was significantly higher than in pummelo (false discovery rate, adjusted P < 0.05) (Fig. 5G and SI Appendix, Fig. 33). Subsequently, the Gene Ontology analysis indicated that candidate genes located in the nuclear genome were significantly enriched (P < 0.01) in nucleotidyltransferase activity and membrane components and that these enrichments were not statistically significant after adjustment with the Benjamini–Hochberg method. We are cautious to not overinterpret these data, given the low power of GWAS to detect restorer genes in mandarin.
Discussion
In this study, our primary goal was to clarify the evolutionary conflicts between cytoplasmic and nuclear genomes and to determine the underlying evolutionary processes and their effects on crop breeding, using citrus as a genetic system. We built a super pan-mitogenome based on 12 assemblies to investigate the landscape of SVs and to identify chimeric ORFs. The population genetic analysis that used Illumina short reads from 184 accessions was used to investigate the conflicts in citrus during diversification, domestication, and hybridization. Our population genomic analyses describe the genetic features of mandarin mitogenomes, particularly their effects on domestication (Figs. 2 and 3). Our pan-mitogenomic data allowed us to identify two chimeric ORFs in the mandarin mitogenomes that possibly affect the production of pollen, as we demonstrated through cytoplasmic replacement experiments (Figs. 1 and 4). Our population genomics provides evidence for the prevalent paternal leakage of mitogenomes in citrus. In contrast, the plastome is strictly maternally inherited. Notably, the reshuffling of the cytoplasmic and nuclear genomes during hybridization could be selected by cytonuclear interactions and can lead to CMS (Fig. 5). Alternatively, selection for cytonuclear interactions and paternal leakage might partially explain the discordant phylogeny among nuclear, mitochondrial, and chloroplast genomes and the striking features of mandarin mitogenomes.
Comparative Population Genomics of cp, mt, and nu Genomes.
The integration of the three variation maps produced a better understanding of the evolutionary history of citrus. The phylogenies are incongruent in Citrinae, possibly due to introgressive hybridizations (47). There is evidence of extensive interspecific hybridization in citrus species, with Poncirus and Fortunella contributing to the gene pools of Citrus (34). As a possible explanation, the genetic information from extinct ancestors could be included in the modern species because of introgressive hybridizations that may have produced the incongruent topologies we observed between cytoplasmic and nuclear genomes. In this scenario, the CMS in mandarin would quickly lead to cytoplasmic capture even in the absence of significant nuclear exchange (27). This mechanism may have contributed to the prominent differences in the mitogenome of mandarin. Additionally, the chloroplast genome is of strict maternal inheritance, and the occasional paternal leakage of the mitochondrial genome may contribute to the mitochondrial haplotype variations (Fig. 5). The heteroplasmy and paternal leakage could contribute to the recombination of mitochondrial genomes and thus may help to explain the inconsistent phylogeny we observed between the mitochondrial and chloroplast genomes (Fig. 2) (20, 48).
Our population genomic analyses highlighted the mitogenome of mandarin for four reasons. First, the mitochondrial genome of mandarin is highly divergent relative to other genera and species of Citrus (Fig. 2). Second, domestication led to a dramatic reduction of genetic diversity and a higher genetic load in the mitochondrial genomes (Fig. 3). Third, mandarin has a persistent and species-specific CMS that could interfere with the production of pollen (Fig. 4). Fourth, the cytoplasmic genomes of mandarin were poorly inherited by hybrids, and extremely low levels of heteroplasmy occurred in the mitochondrial genomes through paternal leakage (Fig. 5). The cytonuclear interactions and paternal leakage may lead to the prominent divergence of mandarin mitogenomes, which, in turn, may provide the genetic basis for the species-specific CMS/Rf system.
Genetic Consequences of Cytoplasmic Genome Evolution.
Domestication may partially explain the increased genetic load. However, domestication probably does not explain the high genetic load of the cytoplasmic genomes that we found in both wild and domesticated mandarins. Another explanation for the genetic load of the cytoplasmic genomes is CMS. Balancing selection was proposed to influence CMS. Thus, the CMS/Rf system might influence the population history (i.e., the level of inbreeding and Ne) of mandarin that is associated with genetic load in mitochondrial genomes (49). It is also not possible to exclude the influence of different reproductive types in citrus. For example, facultative apomixis can reduce the selection efficiency associated with the higher genetic load in mandarin (50).
Furthermore, our findings indicate that different evolutionary processes and differences in the mode of transmission for mitochondrial genomes relative to nuclear genomes may produce an inconsistent explanation for the domestication of citrus. Previous analyses of nuclear genome sequences provide evidence that two independent domestication events that occurred in mandarin produced two different groups (39). Our mitochondrial genome sequence phylogeny reveals that a single domestication event may explain the domestication of mandarin (Fig. 3). Interestingly, a domestication bottleneck did not greatly affect the genetic diversity of the nuclear genome but reduced the genetic diversity of the mitochondrial genome (40). We propose that the mitochondrial genome can help improve our understanding of previous historical characteristics that became inconspicuous in the nuclear genome (51, 52).
Prevalence and Influence of Mitochondrial Heteroplasmy.
Contrary to the long-held view that most plants inherit mitochondrial genomes from the maternal genotype, our deep resequencing uncovered unanticipated and extreme genetic variation in the mitochondrial genomes at the individual level (Fig. 5). Limited by the methods available at the time and inherent awareness, few studies have examined mitochondrial heteroplasmy through paternal leakage (21, 48). Our genomic analysis makes the case that mitochondrial heteroplasmy may widely exist. The long-read sequencing provides for the accurate assembly of mitochondrial genomes and therefore allowed us to identity large SVs by using a pan-mitogenome (Fig. 1). The extensive SVs might affect cytonuclear interactions that occur during outcrossing. When hybridization occurs between parents with short divergent generations, variations in the cytoplasmic genomes may seem to be rare, and the interactions between the cytoplasmic and nuclear genomes are not affected (1).
The development of the female germline was associated with a genetic bottleneck that influences the segregation of mitochondrial genome heteroplasmy and may serve as a purifying selection mechanism that influences heteroplasmy levels in citrus during sexual reproduction (14). We found similar levels of heteroplasmy in different individuals (i.e., in subgroup 1 and the lemon group) derived from paternal leakage. However, there is no evidence for mitochondrial–nuclear interactions affecting the levels of mitochondrial heteroplasmy in these individuals (15). The amount of paternal leakage that occurs and persists in each generation remains an open question. The variations inherited from an ancient line probably contribute to this pattern. In addition, we found a significant difference in mitochondrial heteroplasmy in interspecific hybrids constructed from mandarin and pummelo (<1% in 5/26 interspecific hybrids) relative to the level of mitochondrial heteroplasmy derived from paternal leakage from citron in all lemons (>6%) (Fig. 5). The fitness reduction caused by heteroplasmic variations might be related to the unsafe threshold (i.e., the level of heteroplasmy) in mitochondrial DNA (20). This threshold is not known in citrus. Mitochondrial genome editing might be useful for studying mitochondrial heteroplasmy and for preventing conflicts between the nuclear and mitochondrial genomes (53).
Cytonuclear Interactions in Citrus Hybridization and Breeding.
Why are the mitochondrial genomes of hybrids from mandarin so deficient? One possibility is that mandarin is influenced by facultative apomixis, a kind of asexual reproduction that produces adventitious embryos derived from the nucellus that have a significant competitive advantage relative to embryos produced by sexual means (54). Thus, it is difficult for mandarin to serve as the female parent in natural hybridizations (55). Pummelo reproduces sexually, which is consistent with the mitochondria from pummelo contributing broadly to citrus hybrids (30, 31). However, our data provide alternative explanations that are supported by other observations that do not conform to previous assumptions. For example, the apomictic sour orange serves as a female parent when hybridized with sexually reproducing citron (30). Moreover, the frequency of leaky sexual reproduction in the facultative apomictic mandarin appeared to be higher than expected because of high genetic diversity in the mitochondrial and nuclear genomes of wild mandarin (39, 56). Our cytoplasmic replacement experiments led to male sterility (Fig. 4), and the GWAS analysis detected multiple loci that respond to species-specific cytonuclear interactions (Fig. 5), which suggests that interactions between cytoplasmic and nuclear genomes may influence the fate of hybrids and thus hybridization patterns in citrus. This hypothesis explains why the cytoplasmic genomes of the natural hybrids that we studied were typically derived from pummelo. These findings could expand our understanding of evolutionary conflicts involving mitochondria and provide a particularly attractive interpretation for the domestication, diversification, and breeding of crops.
Methods
Plant Materials and Sequencing.
To determine how ancestral states of mitogenomes have retained or diverged during citrus domestication and hybridization, we collected 12 samples from six groups.
We obtained and sequenced eight distinct types of germplasm, including four mandarins (EG, JZMJ, HJ, and QS), one F1 hybrid (BDGJ), one pummelo (STY), one lemon (YLK), and one grapefruit (JW) from the National Citrus Breeding Center at Huazhong Agricultural University (Wuhan, China) (SI Appendix, Table 1). Total DNA was isolated from young leaves. All plant material was immediately frozen in liquid nitrogen and ground into a powder. High molecular weight genomic DNA was extracted as described by Chin et al. (57). The concentration and quality of the DNA were determined with a NanoDrop 1000 spectrophotometer (Thermo Scientific, USA) and checked via pulsed-field gel electrophoresis, respectively. Different sequencing strategies were adopted. Approximately 4 Gb of single-molecule long reads from four mandarins, one F1 hybrid, one pummelo, and one lemon were generated on either the PacBio Sequel II platform (SMRTbell libraries) or Nanopore GridION (Ligation Sequencing Kit). In addition, mitochondria from grapefruit JW were also enriched via discontinuous sucrose gradient centrifugation from fresh 45-d-old etiolated seedlings. At least 2 μg of DNA from purified mitochondria was used for sequencing on the Illumina HiSEq 150 platform (AmpliSeq Library). After adapter sequences were removed, ∼1 Gb of Illumina short reads were obtained.
In addition to the new sequence data in this study, we collected the published Nanopore long reads from sweet orange (BTC) (58) and PacBio long reads from atalantia (HKC) (31), kumquat (SJG) (35), and poncirus (ZK) (59). The published mitochondrial genomes of mandarin (G1) and pummelo (HBP) were also collected to identify the SVs and to construct the mitochondrial pan-genome (sum to 12 samples) in citrus (45). In addition, Illumina paired reads from 184 accessions of citrus were collected from the National Center for Biotechnology Information (NCBI) to analyze the evolutionary conflicts of cytoplasmic and nuclear genomes in citrus (SI Appendix, Table 6) (30, 31, 56, 60).
We collected publicly available short read sequence data of 184 accessions from 11 types of citrus species (atalantia, poncirus, kumquat, papeda, pummelo, mandarin, citron, grapefruit, sweet orange, sour orange, lemon) and two hybrids (rangpur lime and rough lemon). Grapefruit, sweet orange, sour orange, and lemon are admixtures derived from pummelo, mandarin, and citron. These Illumina paired reads were used to identify the SVs and construct the nuclear, mitochondrial, and chloroplast variation maps. Among the 184 accessions, there were 53 mandarins including 32 samples (MD1 group and MD2 group) from a previous domestication analysis (39). Those samples were used for the phylogeny and the ancestry composition analysis of mandarin domestication.
Mitochondrial Genome Assembly and Variant Maps.
Mitochondrial genome assembly and annotation, structural variation identification, and variant maps construction were described according to methods (SI Appendix).
Phylogeny and Population Genomics Analyses.
To infer the diversification of seven species of citrus (atalantia, poncirus, papeda, kumquat, citron, mandarin, and pummelo), we constructed phylogenetic trees by using a maximum likelihood–based method with the three variation maps from 184 accessions. The LD pruned dataset (∼0.225 million variations) was used to construct the nuclear phylogenetic tree in IQ-TREE version 2.0 with 1,000 μLtrafast bootstrap replicates that yield support values for each node with the GTR + I+G model (61).
The reliable phylogenetic trees were constructed for the mitochondrial and chloroplast genomes with the IQ-TREE program and 184 accessions. In addition to performing the bootstrap test, we also estimated the Shimodaira–Hasegawa (SH)-like approximate likelihood ratio to examine the incongruence in monophyly mitochondrial, chloroplast, and nuclear topologies by using the parameter “SH-aLRT 1000.” The Bayesian-like transformation of aLRT (aBayes) was combined to validate the robustness of those three topologies (62). Simultaneously, we tested the consistency of phylogenetic topologies with or without homologous regions. Subsequently, we tested the consistency of phylogenetic topology based on cytoplasmic variation maps with different missing rate filtering, <40% missing genotypes, and <20% missing genotypes and without filtering (SI Appendix, Fig. 8). To reduce the sampling error, we estimated 1,000 phylogenies for seven species at an individual level. The induvial trees were used as consensus to interpret the probability of the species tree (SI Appendix, Fig. 9). Collectively, this study inferred strong and reliable cytoplasmic topologies for citrus species.
The population structure was directly analyzed through the mitochondrial and chloroplast variation maps. The ancestry components were estimated in Admixture with fivefold cross-validation (−cv = 5) and cluster number K ranging from 2 to 11 (63). Also, we conducted the PCA for cytoplasmic genomes by using PLINK v1.90b6.21 (64) and plotted the results with the R package. Overall, the samples were clustered into 11 populations (atalantia, poncirus, papeda, kumquat, citron, mandarin, pummelo, sweet orange, sour orange, grapefruit, and lemon) and examined with nuclear, mitochondrial, and chloroplast variation maps. The cytoplasmic genomes of the four hybrid populations were inherited from pummelo. Therefore, the population structure determined from cytoplasmic genome variations could not distinguish hybrids from pummelos (K < 11). When we estimated the population structure in the mandarin group, the ancestry components were estimated with fivefold cross-validation (−cv = 5) and cluster number K ranging from 2 to 6.
The nuclear variation dataset without filtering was used to infer the demographic history of six populations (papeda, poncirus, kumquat, citron, domesticated mandarin, and pummelo) in SMC++ v1.15.5 with a mutation rate of 2.2 × 10−8 per site per generation (31, 65). To increase reliability, genome regions were masked when the coverage depth was <15 or mapping quality <20. We split the phased VCF into nine chromosomes and estimated demographic history separately for each chromosome. The results from nine chromosomes were combined to infer the demography of the population. A jackknife procedure with 20 replicates was used to verify the results.
The genetic load for the mitochondrial and chloroplast genomes in six populations was estimated from the number of nonsynonymous mutations in the predicted ORFs based on the kumquat SJG reference mitochondrial genome. The nuclear and mitochondrial phylogenetic trees supported the atalantia population as an outgroup. We constructed an ancestry sequence statement based on the atalantia population and built the unfolded VCF file to reduce the bias of the reference and mutation rate by using custom scripts. First, the predicted ORFs from the Hong Kong kumquat mitochondrial and chloroplast genomes were used to construct the annotations. Second, the mitochondrial and chloroplast genomic datasets were annotated in SnpEff v5.1 (66). Finally, the numbers of nonsynonymous variations in the predicted ORFs were calculated to evaluate the genetic load. Because the cytoplasmic genomes of the four hybrid populations were inherited from pummelo, the estimate for hybrids was used as a negative control.
The statistics of divergence (Dxy), differentiation (Fst), and genetic diversity (π) were calculated based on the nuclear variation map and the mitochondrial variation map as recommended by Python scripts in genomics_general (https://github.com/simonhmartin/genomics_general) (67).
Analysis of Mitochondrial Heteroplasmy.
To quantify the mitochondrial heteroplasmy that occurs during hybridization in citrus, we analyzed the reticulate evolution, performed a Recombination Detection Program (RDP v5.0) analysis, and analyzed the species-specific SVs (i.e., deletions and insertions) (68). The cytoplasmic phylogenetic network was performed in SpliteTree v4 (69) based on 184 accessions. The input FASTA file was generated from the mitochondrial and chloroplast variation maps in vcf2phylip (https://github.com/edgardomortiz/vcf2phylip). Notably, the indel sites were filtered, and missing sites were replaced as “N.” Also, the FASTA files were aligned for the purpose of detecting potential recombination signals. For the RDP analysis, the FASTA files were separated into two groups. Group 1 contained mandarin, pummelo, and three admixtures (i.e., sweet orange, sour orange, and grapefruit). Group 2 contained citron, pummelo, mandarin, and lemon (i.e., the admixture derived from citron, pummelo, and mandarin). Seven methods integrated into the RDP software were used for the recombination signal analysis, including RDP, GENECONV, Bootscan, MaxChi, Chimaera, SiScan, and TOPAL DSS. All of the significant sites (bootstrap values >80) were tested via phylogeny analysis to compare the recombined regions to other regions and to identify the major parent and the minor parent. The species-specific SVs were used for quantifying mitochondrial heteroplasmy. We filtered the species-specific SVs among mandarin, pummelo, and citron. Two types of SVs were used to analyze mitochondrial heteroplasmy, including species-specific SVs between mandarin and pummelo that were used in the analysis of sweet orange, sour orange, and grapefruit and species-specific SVs in pummelo and citron that were used in the analysis of lemon, excluding the SVs in mandarin. The species-specific indels linked to the SVs were filtered to verify the mitochondrial heteroplasmy, and the species-specific SVs were plotted in IGV.
To estimate the proportion of mitochondrial heteroplasmy in citrus hybrids, we calculated the coverage depth of the species-specific insertion/deletion SVs and compared them to the flanking regions. The proportion of mitochondrial heteroplasmy through paternal leakage was calculated from a division of the low depth of coverage in the deletion regions to the coverage depth in the linked regions. To avoid mapping errors in the deleted regions, we masked the fragments that were homologous sequences from the nuclear genome and the chloroplast genome.
Phenotypic Characterization of Alloplasmic Lines.
We previously performed cytoplasmic replacement experiments including mandarin G1 + pummelo HBP (G1+HBP, group 1), mandarin G1 + pummelo STY (G1+STY, group 2), and mandarin G1 + sweet orange BTC (G1+BTC, group 3). The group 1 diploid cybrid individual G1+HBP was generated by Guo et al. (43), and the associated phenotype and RNA-seq data were already published (37, 45). For other groups, protoplast isolation, fusion, and cytoplasmic replacement experiments were performed and described by Cai et al. (42), and the phenotype and RNA-seq datasets of group 2 and group 3 were generated in this study.
The mandarin cultivar HJ, pummelo cultivars HBP and STY, sweet orange BTC, and the alloplasmic line G1+BTC are seedy (i.e., male fertile). The mandarin cultivar G1 and the alloplasmic lines G1+HBP and G1+STY are seedless (i.e., male sterile). We squeezed the anthers of the alloplasmic line G1+STY to facilitate the release of pollen so that we could examine the viability of the pollen. Our cytological observations on pollen development were performed with the pummelo STY and the alloplasmic line G1+STY. Paraffin section analysis was performed to determine the mature stage of pollen grain development. The anthers were fixed overnight in Formalin-Aceto-Alcohol mixed stationary liquid, dehydrated with an ascending series of ethanol from 30 to 70%, cleared in xylene, and gradually embedded in paraffin. A series of sections were prepared, stained with hematoxylin, and observed with an Olympus BX61 microscope (Olympus, Japan).
Expression of Mitochondrial Genes and GWAS Candidate Genes.
The RNA-seq data were obtained from noncoding RNA libraries from a mixture of tissues that included flowers, leaves, stems, seeds, roots, and fruits and were collected previously from three types of germplasm (mandarin G1, the alloplasmic line G1+HBP, and pummelo HBP) (45) but were reanalyzed in this study to clarify the expression of chimeric ORFs in the mitogenomes. In addition to these RNA-seq data, we obtained new RNA-seq data from noncoding RNA libraries that were derived from the anthers of the restored alloplasmic line G1+BTC and the sweet orange cultivar BTC. Each library contained ∼10 Gb of reads. These data were mapped to the mandarin G1 reference mitochondrial genome. We split the reference genome into 100-bp nonoverlapping bins to reduce the possibility that ORFs were overlooked during annotation and calculated the depth of RNA-seq reads in each window. The depth data were transformed into transcripts per million for the subsequent analysis. Meanwhile, the expression levels of the chimeric ORFs associated with the conserved gene fragments were compared between the three samples. We focused on chimeric ORFs that were expressed at high levels from the mandarin mitochondrial genomes in the mandarin cultivar G1, the alloplasmic line G1+HBP, and the alloplasmic line G1+BTC but not expressed at high levels from the pummelo mitochondrial genomes in the pummelo cultivar HBP and sweet orange cultivar BTC.
Previous studies revealed many different types of CMS systems within and between species. Common genetic features of CMS systems appear to be that CMS is associated with chimeric ORFs in the mitogenome, and the restoration of fertility is often associated with genes encoding PPR proteins. Indeed, we identified four genes that encode PPR proteins by performing a GWAS for CMS/Rf genes. Our underlying assumption is that restorer genes in the mandarin nuclear genome could repair the CMS caused by chimeric ORFs in the mitogenome. However, there are no obvious restorer genes in the nuclear genome of pummelo. Therefore, we reasoned that restorer genes would be highly or specifically expressed from the nuclear genome of mandarin. To identify potential restorer genes in mandarin, we collected RNA-seq data from complementary DNA libraries prepared from anthers from eight pummelos (two biological replicates per genotype) and four mandarins (three biological replicates per genotype) (60, 70, 71). Given the highly divergent genome-wide expression patterns of mandarin and pummelo, we normalized read counts in DEseq2 and focused on 168 candidate genes (72). First, the means of the counts for each gene <20 were filtered. Second, the eight pummelos were compared with the four mandarins with default parameters in the DESeqDataSetFromMatrix function. Third, the genome-wide expression matrix was normalized, and the P values were adjusted with the false discovery rate. The four PPR genes were compared via normalized read counts from the anthers of mandarins and pummelos.
Cytonuclear Interactions and the GWAS.
The domestication of Citrus was associated with wide admixtures between citron, mandarin, and pummelo. Cytonuclear interactions may influence the selection of nuclear genes in admixtures (e.g., the CMS system in mandarin). To evaluate interactions between the nuclear and cytoplasmic genomes, we performed a GWAS based on 118 samples within Citrus including mandarin, pummelo, citron, and particular admixtures: sweet orange, sour orange, grapefruit, and lemon. The haplotypes of the mitochondrial genome were consistent with the chloroplast genome and were calculated with a characteristic value (PC1 eigenvalue in PCA, 28.1%) that was based on a mitochondrial variation map constructed in PLINK. We constructed the LD blocks based on 10-kb nonoverlapping windows in PLINK with parameter “50 10 0.8” based on the nuclear variation dataset. The 10-kb window size was inferred based on the length of the LD decay, as recommended by Wang et al. (34). Those filtered variations were used for the GWAS analysis and depended on linear mixed models constructed in GEMMA (v0.98.5) (73). The kinship matrix was computed in GEMMA with the parameter “-gk 2.” The regression analysis was performed based on the kinship matrix with the parameter “-lmm 4.” The output (adjusted P value) was plotted with R package ggplot2 (74).
Supplementary Material
Acknowledgments
We thank the anonymous reviewers for helpful comments and suggestions. We also thank Dr. Sanwen Huang (Chinese Academy of Tropical Agricultural Sciences), Dr. Daniel Sloan (Colorado State University), and Dr. Jeffrey P. Mower (University of Nebraska–Lincoln) for discussions during the project. This research was financially supported by grants from the Ministry of Science and Technology of China (No. 2018YFD1000106), the National Natural Science Foundation of China (Nos. 31820103011 and 31530065), and the Foundation of Hubei Hongshan Laboratory (No. 2021hszd009).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. J.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2206076119/-/DCSupplemental.
Data, Materials, and Software Availability
Data supporting the findings of this work are available in the article and SI Appendix. Sequencing data are accessible through the National Center for Biotechnology Information (NCBI) associated with BioProject ID PRJNA807745 (75). Mitochondrial genome assembly and annotation of genome and variation maps were uploaded to Zenodo (https://zenodo.org/record/5826754) (76). Custom scripts and workflows are available on the repository in GitHub (https://github.com/wangnan9394/coevolution_mitochondrial_nuclear) (77). All genome sequencing data, genome assembly, and annotation of genome and variation maps, workflows, and scripts generated in this study have been deposited in NCBI/Zenodo/GitHub (75–77).
References
- 1.Mower J. P., Sloan D. B., Alverson A. J., Plant mitochondrial genome diversity: The genomics revolution. Plant Genome Diversity 1, 123–144 (2012). [Google Scholar]
- 2.Mackenzie S. A., The influence of mitochondrial genetics on crop breeding strategies. Plant Breeding Rev. 115–138 (2010). [Google Scholar]
- 3.Eyre-Walker A., Gaut R. L., Hilton H., Feldman D. L., Gaut B. S., Investigation of the bottleneck leading to the domestication of maize. Proc. Natl. Acad. Sci. U.S.A. 95, 4441–4446 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou Y., et al. , Importance of incomplete lineage sorting and introgression in the origin of shared genetic variation between two closely related pines with overlapping distributions. Heredity 118, 211–220 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole L. W., Guo W., Mower J. P., Palmer J. D., High and variable rates of repeat-mediated mitochondrial genome rearrangement in a genus of plants. Mol. Biol. Evol. 35, 2773–2785 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Zhu A., Guo W., Jain K., Mower J. P., Unprecedented heterogeneity in the synonymous substitution rate within a plant genome. Mol. Biol. Evol. 31, 1228–1236 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Chase C. D., Cytoplasmic male sterility: A window to the world of plant mitochondrial-nuclear interactions. Trends Genet. 23, 81–90 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Tuteja R., et al. , Cytoplasmic male sterility-associated chimeric open reading frames identified by mitochondrial genome sequencing of four Cajanus genotypes. DNA Res. 20, 485–495 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujii S., Kazama T., Yamada M., Toriyama K., Discovery of global genomic re-organization based on comparison of two newly sequenced rice mitochondrial genomes with cytoplasmic male sterility-related genes. BMC Genomics 11, 209 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Budar F., Touzet P., De Paepe R., The nucleo-mitochondrial conflict in cytoplasmic male sterilities revisited. Genetica 117, 3–16 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Fujii S., Bond C. S., Small I. D., Selection patterns on restorer-like genes reveal a conflict between nuclear and mitochondrial genomes throughout angiosperm evolution. Proc. Natl. Acad. Sci. U.S.A. 108, 1723–1728 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolfe K. H., Li W.-H., Sharp P. M., Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. U.S.A. 84, 9054–9058 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mower J. P., Touzet P., Gummow J. S., Delph L. F., Palmer J. D., Extensive variation in synonymous substitution rates in mitochondrial genes of seed plants. BMC Evol. Biol. 7, 135 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neiman M., Taylor D. R., The causes of mutation accumulation in mitochondrial genomes. Proc. Biol. Sci. 276, 1201–1209 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chinnery P. F., et al. , The inheritance of mitochondrial DNA heteroplasmy: Random drift, selection or both? Trends Genet. 16, 500–505 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Meiklejohn C. D., Montooth K. L., Rand D. M., Positive and negative selection on the mitochondrial genome. Trends Genet. 23, 259–263 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Ballard J. W. O., Whitlock M. C., The incomplete natural history of mitochondria. Mol. Ecol. 13, 729–744 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Doebley J. F., Gaut B. S., Smith B. D., The molecular genetics of crop domestication. Cell 127, 1309–1321 (2006). [DOI] [PubMed] [Google Scholar]
- 19.Gaut B. S., Seymour D. K., Liu Q., Zhou Y., Demography and its effects on genomic variation in crop domestication. Nat. Plants 4, 512–520 (2018). [DOI] [PubMed] [Google Scholar]
- 20.McCauley D. E., Paternal leakage, heteroplasmy, and the evolution of plant mitochondrial genomes. New Phytol. 200, 966–977 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Bentley K. E., Mandel J. R., McCauley D. E., Paternal leakage and heteroplasmy of mitochondrial genomes in Silene vulgaris: Evidence from experimental crosses. Genetics 185, 961–968 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Z., Waneka G., Sloan D. B., The tempo and mode of angiosperm mitochondrial genome divergence inferred from intraspecific variation in Arabidopsis thaliana. G3 (Bethesda) 10, 1077–1086 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCauley D. E., Bailey M. F., Sherman N. A., Darnell M. Z., Evidence for paternal transmission and heteroplasmy in the mitochondrial genome of Silene vulgaris, a gynodioecious plant. Heredity 95, 50–58 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Ellis J. R., Bentley K. E., McCauley D. E., Detection of rare paternal chloroplast inheritance in controlled crosses of the endangered sunflower Helianthus verticillatus. Heredity 100, 574–580 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Wade M. J., McCauley D. E., Paternal leakage sustains the cytoplasmic polymorphism underlying gynodioecy but remains invasible by nuclear restorers. Am. Nat. 166, 592–602 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Rieseberg L. H., Baird S. J. E., Gardner K. A., Hybridization, introgression, and linkage evolution. Plant Mol. Biol. 42, 205–224 (2000). [PubMed] [Google Scholar]
- 27.Rieseberg L. H., Soltis D., Phylogenetic consequences of cytoplasmic gene flow in plants. Evolutionary Trends in Plants 5, 65–84 (1991). [Google Scholar]
- 28.Sweigart A. L., Fishman L., Willis J. H., A simple genetic incompatibility causes hybrid male sterility in mimulus. Genetics 172, 2465–2479 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fishman L., Willis J. H., A cytonuclear incompatibility causes anther sterility in Mimulus hybrids. Evolution 60, 1372–1381 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Wu G. A., et al. , Genomics of the origin and evolution of Citrus. Nature 554, 311–316 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Wang X., et al. , Genomic analyses of primitive, wild and cultivated citrus provide insights into asexual reproduction. Nat. Genet. 49, 765–772 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Wu G. A., et al. , Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotechnol. 32, 656–662 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu Q., et al. , The draft genome of sweet orange (Citrus sinensis). Nat. Genet. 45, 59–66 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Wang N., et al. , Structural variation and parallel evolution of apomixis in citrus during domestication and diversification. Natl. Sci. Rev. 10.1093/nsr/nwac114 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu C., et al. , Genome sequencing and CRISPR/Cas9 gene editing of an early flowering Mini-Citrus (Fortunella hindsii). Plant Biotechnol. J. 17, 2199–2210 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goto S., et al. , QTL mapping of male sterility and transmission pattern in progeny of Satsuma mandarin. PLoS One 13, e0200844 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng B.-B., et al. , Comparative transcript profiling of a male sterile cybrid pummelo and its fertile type revealed altered gene expression related to flower development. PLoS One 7, e43758 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mower J. P., Case A. L., Floro E. R., Willis J. H., Evidence against equimolarity of large repeat arrangements and a predominant master circle structure of the mitochondrial genome from a monkeyflower (Mimulus guttatus) lineage with cryptic CMS. Genome Biol. Evol. 4, 670–686 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L., et al. , Genome of wild mandarin and domestication history of mandarin. Mol. Plant 11, 1024–1037 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Gaut B. S., Díez C. M., Morrell P. L., Genomics and the contrasting dynamics of annual and perennial domestication. Trends Genet. 31, 709–719 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Zhou Y., Massonnet M., Sanjak J. S., Cantu D., Gaut B. S., Evolutionary genomics of grape (Vitis vinifera ssp. vinifera) domestication. Proc. Natl. Acad. Sci. U.S.A. 114, 11715–11720 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cai X.-D., Fu J., Deng X.-X., Guo W.-W., Production and molecular characterization of potential seedless cybrid plants between pollen sterile Satsuma mandarin and two seedy Citrus cultivars. Plant Cell Tissue Organ Cult. 90, 275–283 (2007). [Google Scholar]
- 43.Guo W. W., et al. , Targeted cybridization in citrus: Transfer of Satsuma cytoplasm to seedy cultivars for potential seedlessness. Plant Cell Rep. 22, 752–758 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Seale M., Callose deposition during pollen development. Plant Physiol. 184, 564–565 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang S., et al. , Assembly of Satsuma mandarin mitochondrial genome and identification of cytoplasmic male sterility–specific ORFs in a somatic cybrid of pummelo. Tree Genet. Genomes 16, 1–13 (2020). [Google Scholar]
- 46.Dahan J., Mireau H., The Rf and Rf-like PPR in higher plants, a fast-evolving subclass of PPR genes. RNA Biol. 10, 1469–1476 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsutsui K., et al. , Incongruence among mitochondrial, chloroplast and nuclear gene trees in Pinus subgenus Strobus (Pinaceae). J. Plant Res. 122, 509–521 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Fontaine K. M., Cooley J. R., Simon C., Evidence for paternal leakage in hybrid periodical cicadas (Hemiptera: Magicicada spp.). PLoS One 2, e892 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Charlesworth D., Balancing selection and its effects on sequences in nearby genome regions. PLoS Genet. 2, e64 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Charlesworth B., Fundamental concepts in genetics: Effective population size and patterns of molecular evolution and variation. Nat. Rev. Genet. 10, 195–205 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Gaut B. S., Molecular Clocks and Nucleotide Substitution Rates in Higher Plants. Evolutionary Biology (Springer, 1998), pp. 93–120. [Google Scholar]
- 52.Gray M. W., Burger G., Lang B. F., Mitochondrial evolution. Science 283, 1476–1481 (1999). [DOI] [PubMed] [Google Scholar]
- 53.Aushev M., Herbert M., Mitochondrial genome editing gets precise. Nature 583, 521–522 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Koltunow A. M., Soltys K., Nito N., McClure S., Anther, ovule, seed, and nucellar embryo development in Citrus sinensis cv. Valencia. Can. J. Bot. 73, 1567–1582 (1995). [Google Scholar]
- 55.Xu Y., et al. , Regulation of nucellar embryony, a mode of sporophytic apomixis in Citrus resembling somatic embryogenesis. Curr. Opin. Plant Biol. 59, 101984 (2021). [DOI] [PubMed] [Google Scholar]
- 56.Wu G. A., et al. , Diversification of mandarin citrus by hybrid speciation and apomixis. Nat. Commun. 12, 4377 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chin C.-S., et al. , Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13, 1050–1054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang L., et al. , Somatic variations led to the selection of acidic and acidless orange cultivars. Nat. Plants 7, 954–965 (2021). [DOI] [PubMed] [Google Scholar]
- 59.Huang Y., et al. , Genome of a citrus rootstock and global DNA demethylation caused by heterografting. Hortic. Res. 8, 69 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liang M., et al. , Evolution of self-compatibility by a mutant Sm-RNase in citrus. Nat. Plants 6, 131–142 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kalyaanamoorthy S., Minh B. Q., Wong T. K. F., von Haeseler A., Jermiin L. S., ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anisimova M., Gil M., Dufayard J.-F., Dessimoz C., Gascuel O., Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst. Biol. 60, 685–699 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alexander D. H., Novembre J., Lange K., Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Purcell S., et al. , PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Terhorst J., Kamm J. A., Song Y. S., Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat. Genet. 49, 303–309 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cingolani P., et al. , A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martin S. H., Davey J. W., Jiggins C. D., Evaluating the use of ABBA-BABA statistics to locate introgressed loci. Mol. Biol. Evol. 32, 244–257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martin D. P., et al. , RDP5: A computer program for analyzing recombination in, and removing signals of recombination from, nucleotide sequence datasets. Virus Evolution 7, veaa087 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huson D. H., Bryant D., Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267 (2006). [DOI] [PubMed] [Google Scholar]
- 70.Zhang C., et al. , Seedless mutant ‘Wuzi Ougan’ (Citrus suavissima Hort. ex Tanaka ‘seedless’) and the wild type were compared by iTRAQ-based quantitative proteomics and integratedly analyzed with transcriptome to improve understanding of male sterility. BMC Genet. 19, 106 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ye L.-X., et al. , Comparative analysis of the transcriptome, methylome, and metabolome during pollen abortion of a seedless citrus mutant. Plant Mol. Biol. 104, 151–171 (2020). [DOI] [PubMed] [Google Scholar]
- 72.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zoubarev A., et al. , Gemma: A resource for the reuse, sharing and meta-analysis of expression profiling data. Bioinformatics 28, 2272–2273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wickham H., ggplot2: Elegant Graphics for Data Analysis (Springer, 2016). [Google Scholar]
- 75.Wang N., et al. , Pacbio and Illumina sequence reads for citrus mitogenome assemble. NCBI BioProject. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA807745. Deposited 16 February 2022.
- 76.Wang N., Genetic basis of cytonuclear conflicts in citrus hybridization, domestication, and diversification. Zenodo. https://zenodo.org/record/5826754. Deposited 3 October 2022. [DOI] [PMC free article] [PubMed]
- 77.Wang N., et al. , co-evolution_mitochondrial_nuclear. GitHub. https://github.com/wangnan9394/coevolution_mitochondrial_nuclear. Deposited 20 March 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this work are available in the article and SI Appendix. Sequencing data are accessible through the National Center for Biotechnology Information (NCBI) associated with BioProject ID PRJNA807745 (75). Mitochondrial genome assembly and annotation of genome and variation maps were uploaded to Zenodo (https://zenodo.org/record/5826754) (76). Custom scripts and workflows are available on the repository in GitHub (https://github.com/wangnan9394/coevolution_mitochondrial_nuclear) (77). All genome sequencing data, genome assembly, and annotation of genome and variation maps, workflows, and scripts generated in this study have been deposited in NCBI/Zenodo/GitHub (75–77).