Significance
Interleukin-15 (IL-15) is considered essential for memory CD8+ T cell maintenance and self-renewal. However, IL-15–independent memory CD8+ T cells have now been described, notably including several tissue-resident subsets. This could indicate that during differentiation and tissue adaption, some memory CD8+ T cells lose the capacity to respond to IL-15. While IL-15 dependence is not shared between CD8+ T cell memory subsets, we demonstrate that IL-15 sensitivity is a conserved feature across populations, albeit with subset- and tissue-specific differences. Furthermore, IL-15 sensitivity can be exploited with IL-15 therapy to drive antigen-independent proliferation of recirculating and tissue-resident memory CD8+ T cell subsets across tissues in mice, with potential therapeutic applications.
Keywords: cytokine, CD8+ T cell memory, tissue-resident memory
Abstract
Interleukin-15 (IL-15) is often considered a central regulator of memory CD8+ T cells, based primarily on studies of recirculating subsets. However, recent work identified IL-15–independent CD8+ T cell memory populations, including tissue-resident memory CD8+ T cells (TRM) in some nonlymphoid tissues (NLTs). Whether this reflects the existence of IL-15–insensitive memory CD8+ T cells is unclear. We report that IL-15 complexes (IL-15c) stimulate rapid proliferation and expansion of both tissue-resident and circulating memory CD8+ T cell subsets across lymphoid and nonlymphoid tissues with varying magnitude by tissue and memory subset, in some sites correlating with differing levels of the IL-2Rβ. This was conserved for memory CD8+ T cells recognizing distinct antigens and elicited by different pathogens. Following IL-15c–induced expansion, divided cells contracted to baseline numbers and only slowly returned to basal proliferation, suggesting a mechanism to transiently amplify memory populations. Through parabiosis, we showed that IL-15c drive local proliferation of TRM, with a degree of recruitment of circulating cells to some NLTs. Hence, irrespective of homeostatic IL-15 dependence, IL-15 sensitivity is a defining feature of memory CD8+ T cell populations, with therapeutic potential for expansion of TRM and other memory subsets in an antigen-agnostic and temporally controlled fashion.
Memory CD8+ T cells can be divided into distinct subsets based on their functional and trafficking properties. Recirculating populations include “central” and various subpopulations of “effector” memory T cells, while tissue-resident memory CD8+ T cells (TRM) are locally maintained in diverse lymphoid tissue (LT) and nonlymphoid tissues (NLT). Some memory CD8+ T cell populations are maintained long-term by different combinations of individual cell longevity and basal or homeostatic proliferation (1–6). The common gamma chain (γC) family cytokine interleukin-15 (IL-15) was identified as a key player in maintenance of circulating memory CD8+ T cells, with evidence that it supports basal proliferation and that IL-15 deficiency leads to partial loss of the memory population (7–17). Building on these and other studies, IL-15 is now considered a key regulator of memory CD8+ T cells. However, the impact of IL-15 deficiency is not uniform across all circulating memory CD8+ T cell subsets. Cells with the phenotype of an effector memory subset, termed long-lived effector cells (LLECs), showed heightened sensitivity to IL-15 deficiency (17), as did a subset of memory CD8+ T cells that express Ly49 molecules (13). On the other hand, later studies identified settings where circulating memory CD8+ T cells became IL-15 independent, with seemingly normal maintenance and basal proliferation of circulating CD8+ T cell memory in the absence of IL-15 (1, 18).
Heterogeneity in IL-15 dependency is even more marked for tissue-resident memory CD8+ T cells. In the lung, kidney, salivary gland (SG), and skin of mice, the formation and maintenance of CD8+ TRM typically show considerable reliance on IL-15 (1, 19–21), while TRM in some other NLTs, such as the small intestine, pancreas, and female reproductive tract (FRT), were found to be IL-15 independent (1). The basis for these differences in IL-15 dependency are unclear. Expression of the IL-15 receptor component IL-2Rβ (CD122) in TRM depends on low levels of Tbet, which must be repressed for tissue residency, hinting that some TRM may have impaired IL-15 sensitivity (21, 22). Indeed, CD122 has been shown to be low on intraepithelial memory CD8+ T cells compared to memory CD8+ T cells from LTs (23, 24). Tumor antigen-specific CD8+ T cells can exhibit IL-15 insensitivity within tumors, in contrast to CD8+ T cells in other tissue sites, suggesting context-dependent loss of IL-15 sensitivity (25). As innate-like lymphocytes including natural killer cells and intraepithelial γδ T cells also rely on IL-15 (8, 10, 26), competition for IL-15 could potentially promote memory CD8+ T cell maintenance by other cytokines or tissue factors.
Such findings raise the question of whether sensitivity to IL-15 is entirely lost in some memory CD8+ T cell subpopulations or differs in degree. Graded expression of CD122 between different memory subsets could potentially act as a mechanism to limit homeostatic effects of IL-15 while permitting responsiveness to elevated IL-15 levels. Proliferation of circulating memory phenotype CD8+ T cells has been observed due to IL-15 production elicited by type 1 interferon during viral infection (7, 27). One approach to address this is comparing the efficacy of cytokine therapy on different populations of memory CD8+ T cells. Numerous groups have shown that various γC family cytokines can be used to induce proliferation and expansion of the circulating memory CD8+ T cell pool and that this response can be maximized using cytokine complexes (composed of cytokines bound to antibodies or cytokine receptor chains) (15, 16, 28, 29). IL-15 signals through IL-2Rβ and γC, while the high affinity receptor for IL-15, IL-15Rα, is not required by cells responding to IL-15; instead, IL-15Rα is used to “transpresent” IL-15 from the IL-15–synthesizing cell to another and can also form soluble complexes with IL-15 that increase during inflammation (30–36). Hence, IL-15/IL-15Rα-Fc complexes (IL-15c), as well as IL-2 complexes and IL-2 muteins designed to selectively engage IL-2Rβ/γC, have been used to drive expansion of memory CD8+ T cells (15, 16, 28, 37, 38), with potential therapeutic applications in the control of infectious diseases and cancer (39–42). However, these studies have typically limited their focus to recirculating memory CD8+ T cells: it is currently unknown whether cytokine complexes effectively act on CD8+ TRM in NLT and whether sensitivity to IL-15 therapy corresponds to IL-15 dependency during normal homeostasis. While TRM are known to locally expand in response to antigen challenge (43, 44), to our knowledge generalizable methods to elicit their proliferation without identifying antigens have not been demonstrated. We therefore set out to address whether IL-15 sensitivity is variable or conserved across memory CD8+ T cell subsets.
Results
IL-15 Complexes Stimulate Antigen-Independent Proliferation of Memory CD8+ T Cells across the Body.
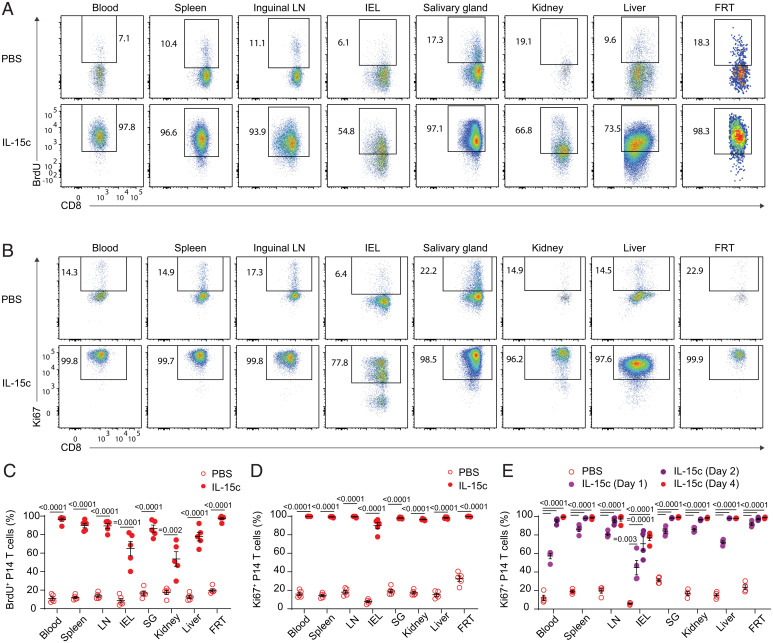
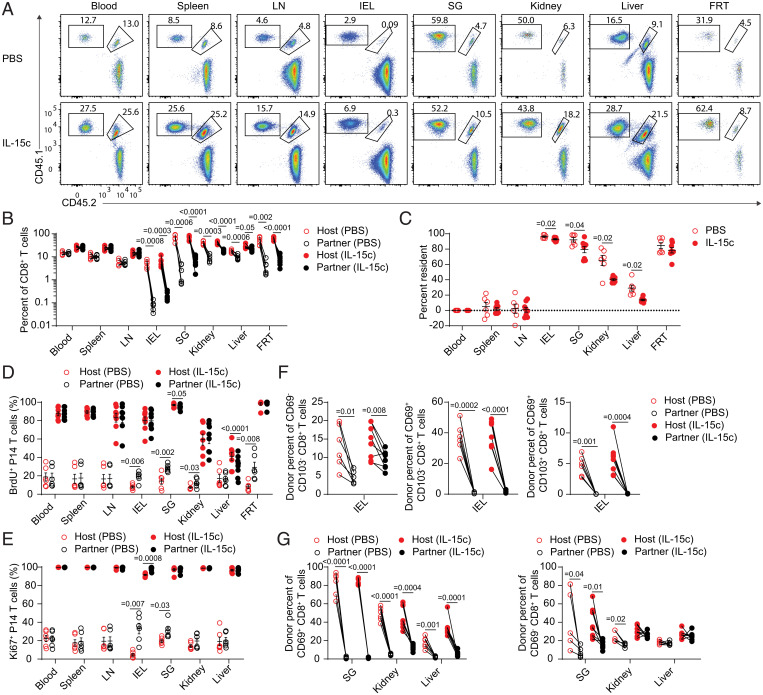
It is unclear whether the range in IL-15 dependency of memory CD8+ T cell populations predicts sensitivity or insensitivity to exogenous IL-15. To generate a defined and abundant CD8+ T cell memory population, we transferred T cell receptor (TCR) transgenic P14 CD8+ T cells, which recognize the gp33 epitope of LCMV, to naive recipient mice, which were then acutely infected with lymphocytic choriomeningitis virus (LCMV) Armstrong. After at least 4 wk to establish memory, these animals were treated with IL-15c and, in some experiments, the nucleotide analog bromodeoxyuridine (BrdU) to mark proliferating cells actively synthesizing DNA. To label intravascular cells in tissues, we intravenously (i.v.) injected a fluorochrome-conjugated antibody to CD8α, gating on unlabeled (“i.v.-negative”) cells as representing the parenchymal population (45, 46). As expected, IL-15c treatment induced marked proliferation of memory P14 populations in the blood and LTs, as indicated by both BrdU incorporation and expression of the cell cycling marker Ki67 (Fig. 1 A–D). Similar responses were observed for memory P14 T cells in the parenchyma of all NLTs studied (Fig. 1 A–D): notably, this applied to P14 T cells in the small intestine intraepithelial lymphocyte (IEL) fraction and FRT, which had previously been shown to be independent of IL-15 for normal homeostasis, as well as P14 T cells in the SG, kidney, and liver—sites in which normal homeostasis is IL-15 dependent (1). Furthermore, all P14 T cell populations responded rapidly and with similar kinetics, with marked Ki67 up-regulation by 24 h before plateauing around 48 h (Fig. 1E).
Fig. 1.
IL-15c are sufficient to induce proliferation of antigen-specific memory CD8+ T cells in the circulation and tissue sites. Naive B6 mice received congenically distinct P14 T cells, followed by LCMV-Armstrong challenge. Animals were then rested for at least 28 d. At days 0 and 2 of treatment, animals received phosphate buffered saline (PBS) or IL-15c. At day 0, all mice received intraperitoneal (i.p.) BrdU, followed by BrdU labeling via drinking water to mark proliferating cells. At day 4, mice were killed. (A and B) Flow cytometry for (A) BrdU incorporation and (B) Ki67 expression of donor P14 T cells in the blood, spleen, inguinal LN, IEL, SG, kidney, liver, and FRT from PBS- and IL-15c–treated mice. (C) Quantitation of the percentage of BrdU+ donor P14 T cells, gated as in A. (D and E) Quantitation of the percentage of Ki67+ donor P14 T cells in PBS- and IL-15c–treated mice at (D) Day 4 and (E) Days 1, 2, and 4 after start of treatment, gated as in B. (A, C, and D) Data are representative/pooled from two experiments with five mice per group. (B) Data are representative of six experiments with 12 to 14 mice per group. (E) Data are pooled from two experiments with four mice per group. Unpaired two-sided Student’s t tests. Error bars are ± standard error of the mean (S.E.M.).
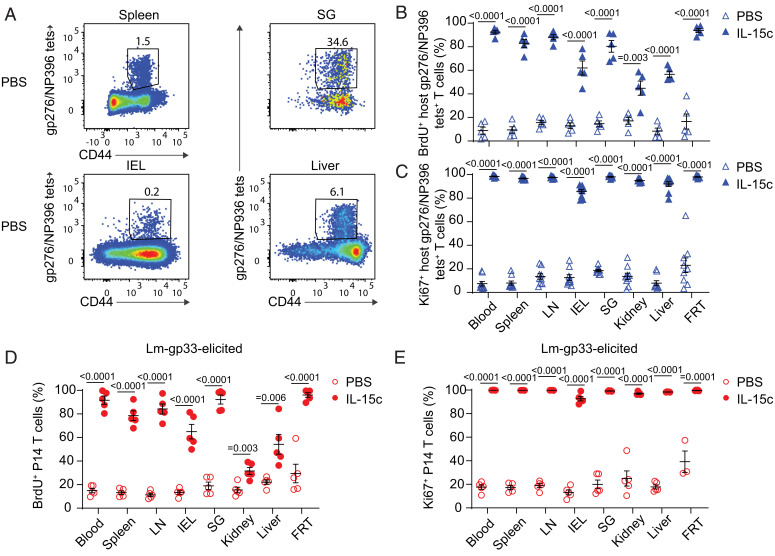
To exclude the possibility that the observed IL-15c–induced proliferation in diverse NLTs was unique to P14 TCR transgenic cells, we assessed host-derived LCMV-specific memory CD8+ T cell populations in these mice (gating on cells stained with either the LCMV glycoprotein 33 (gp33)/Db tetramer or a pool of gp276/Db and nucleoprotein 396 (NP396)/Db tetramers) (47). Like P14 T cells, endogenous antigen-specific memory CD8+ T cells in diverse lymphoid and nonlymphoid sites proliferated strongly in response to IL-15c (Fig. 2 A–C and SI Appendix, Fig. S1 A–C).
Fig. 2.
IL-15c induce proliferation of endogenous and TCR transgenic antigen-specific memory CD8+ T cells elicited by viral or bacterial challenge. (A) P14 memory mice treated as in Fig. 1 were analyzed for host gp276/NP396Db tet+ CD8+ T cells in the spleen, IEL, SG, and liver from PBS-treated mice. (B and C) Quantitation of the percentage of (B) BrdU+ and (C) Ki67+ host gp276/NP396/Db tet+ CD8+ T cells in the blood, spleen, LN, IEL, SG, kidney, liver, and FRT from PBS- and IL-15c–treated mice, gated as in A. (D and E) Intravenous challenge with 1*104 colony-forming units of Lm-gp33 was used as an alternative means to generate P14 memory in naive B6 recipients of congenically distinct P14 T cells. After establishment of memory, Lm-gp33–elicited P14 memory mice were treated with IL-15c and BrdU as in Fig. 1. Quantitation of the percentage of (D) BrdU+ and (E) Ki67+ donor P14 T cells in the blood, spleen, LN, IEL, SG, kidney, liver, and FRT from PBS- and IL-15c–treated mice. (A) Data are representative of seven experiments with 13 to 16 mice per group. (B) Data are pooled from two experiments with five mice per group. (C) Data are pooled from four experiments with eight or nine mice per group. (D and E) Data are pooled from two experiments with three to five mice per group. Unpaired two-sided Student’s t tests. Error bars are ±S.E.M.
We next considered whether this broad IL-15–induced proliferation across tissues was a phenomenon unique to LCMV-elicited memory. To address this, we transferred P14 T cells into naive recipients, which were then infected with Listeria monocytogenes expressing the gp33 peptide (Lm-gp33) (48) and rested to memory. When these animals were treated with IL-15c, we also observed strong IL-15–induced proliferation of memory P14 and gp33 tetramer-binding host CD8+ T cells (Fig. 2 D and E and SI Appendix, Fig. S1 D and E). Therefore, our data support conserved sensitivity to IL-15 and capacity for IL-15–induced proliferation in memory CD8+ T cells across tissues, antigen specificities, and eliciting pathogens.
IL-15c Elicits Prolonged but Reversible Expansion of Memory CD8+ T Cell Populations.
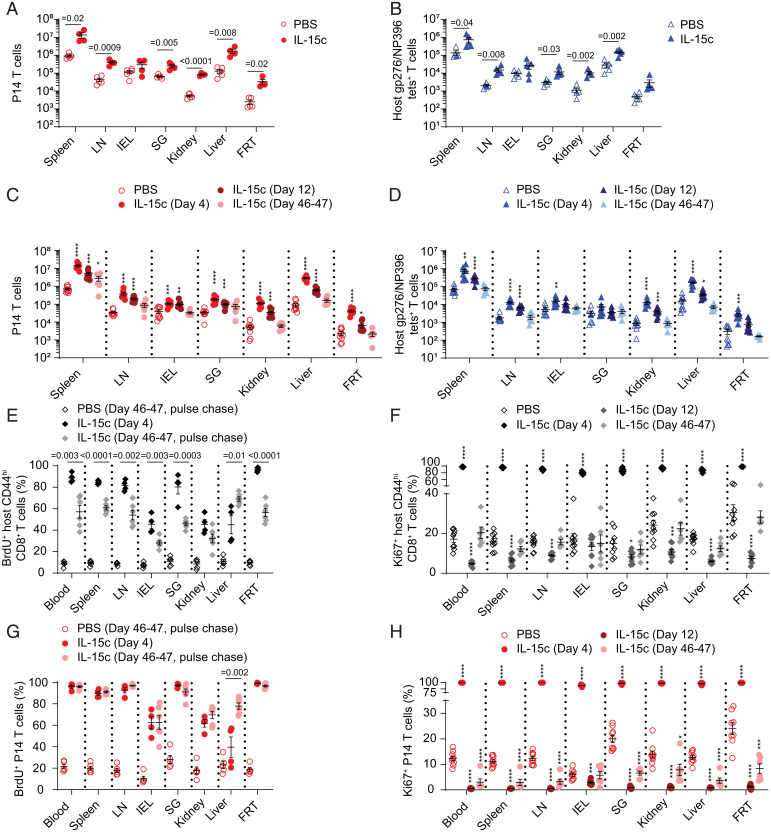
In light of the considerable proliferation induced by IL-15c, we assessed whether memory CD8+ T cell populations increased in number in IL-15c–treated compared to control-treated mice. For P14, gp33/Db tetramer-binding, and gp276/NP396/Db tetramer-binding memory CD8+ T cell populations elicited by LCMV, we saw increases in population numbers across most tissues (Fig. 3 A and B and SI Appendix, Fig. S1F). Similar outcomes were observed for P14 and gp33/Db tetramer-binding memory CD8+ T cell populations induced by Lm-gp33 infection (SI Appendix, Fig. S1 G and H). Interestingly, IL-15c–induced expansion was more muted for populations in the IEL—a site in which TRM homeostasis is IL-15 independent (1). We next assessed whether increases in antigen-specific memory populations were durable. We found declining but still-elevated numbers of most populations at 12 d after the start of IL-15c treatment (10 d after the final treatment) (Fig. 3 C and D and SI Appendix, Fig. S2A). By 46 to 47 d after IL-15c treatment, most populations had returned to baseline and were numerically similar to those of control-treated mice, with the possible exception of (despite substantial contraction) the spleen, the inguinal lymph node (LN), and potentially the SG (Fig. 3 C and D and SI Appendix, Fig. S2A). Taken together, IL-15c elicited broad expansion of antigen-specific memory CD8+ T cells across tissues, resulting in reversible increases that did not boost memory populations indefinitely.
Fig. 3.
IL-15c treatment induces reversible accumulation of memory CD8+ T cells. (A and B) P14 memory mice treated as in Fig. 1 were quantitated for (A) P14 donor and (B) host gp276/Db or NP396/Db tetramer-specific CD8+ T cells from the spleen, LN, IEL, SG, kidney, liver, and FRT from PBS- and IL-15c–treated mice. (C and D) P14 memory mice treated as in Fig. 1 were quantitated for (C) P14 donor and (D) host gp276/Db or NP396/Db tetramer-specific CD8+ T cells from the spleen, LN, IEL, SG, kidney, liver, and FRT from PBS- and IL-15c–treated mice on day 4, day 12, and day 46 to 47 after start of treatment. (E and F) Quantitation of the percentage of (E) BrdU+ and (F) Ki67+ host memory phenotype CD8+ T cells (gated as CD44hi in blood, spleen, and LN and on all host CD8+ T cells in NLT) from the blood, spleen, LN, IEL, SG, kidney, liver, and FRT from PBS- and IL-15c–treated mice on day 4, day 12 (Ki67 only), and day 46 to 47 after start of treatment, with BrdU labeling for the PBS and IL-15c–treated pulse-chased groups from day 0 to 6 only. (G and H) Quantitation of the percentage of (G) BrdU+ and (H) Ki67+ donor P14 T cells from the blood, spleen, LN, IEL, SG, kidney, liver, and FRT from PBS- and IL-15c–treated mice on day 4, day 12 (Ki67 only), and day 46 to 47 after start of treatment. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. (A and B) Data are pooled from two experiments with four or five mice per group. (C–H) Data are pooled from two to four experiments with four to nine mice per group. Unpaired two-sided Student’s t tests. Error bars are ±S.E.M.
It was previously shown that circulating memory phenotype CD44hi CD8+ T cells proliferate in response to interferon-induced IL-15 and remain highly labeled with BrdU for almost 2 mo (7, 27). In light of the contraction of IL-15c–expanded populations, we therefore assessed whether antigen-specific, BrdU-labeled memory CD8+ T cells that had undergone IL-15c–elicited cell division likewise persisted in order to determine whether they were long-lived or quickly lost across tissues. We set up a BrdU pulse-chase experiment, treating mice with IL-15c as usual (on days 0 and 2) while administering an extended 6-d course of BrdU to label proliferating cells until IL-15c–driven proliferation had ended, followed by cessation of BrdU treatment for the duration of the experiment. We first assessed memory phenotype CD8+ T cells (7, 27). IL-15c increased the frequency of memory phenotype circulating CD44hi CD8+ T cells, and interestingly, this population returned to baseline by 46 or 47 d (SI Appendix, Fig. S2 B and C). When we assessed proliferation of memory phenotype CD8+ T cells across tissues, we likewise observed long-term labeling of this population and a reduced frequency of BrdU+ cells over the chase period, corresponding with recovery to a normal proportion of Ki67+ proliferating cells (Fig. 3 E and F) (27).
In contrast, P14 T cells remained highly labeled with BrdU even after 46 to 47 d (Fig. 3G). We then asked whether ongoing proliferation of P14 memory cells (as assessed by Ki67) was affected long after IL-15c treatment. To our surprise, IL-15c–stimulated P14 T cells were still recovering toward the proportion of Ki67+ P14 T cells observed in PBS-treated controls at day 46 to 47, with P14 T cells in NLT sites potentially recovering more rapidly than recirculating populations (Fig. 3H). Intriguingly, IL-15–independent IEL populations seemed to respond least vigorously to IL-15c, which could be related to their more rapid recovery to normal levels of proliferation after IL-15c treatment. Taken together, these data indicate that memory P14 T cells that have experienced IL-15c–induced proliferation are not preferentially lost during contraction and can be long-lived. However, memory CD8+ T cells can exhibit reduced proliferation after IL-15c to varying degrees depending on the specific population. These results form a notable distinction from boosting with antigen, which can result in durable increases in antigen-specific memory populations (49), and suggest restraint on cytokine-elicited memory population expansion. This may allow short-term population expansion while avoiding permanent, nonspecific increases in memory cells after bursts of IL-15 like those seen during viral infection (7).
Memory CD8+ T Cell Subsets Share an Enhanced Sensitivity to IL-15.
Although IL-15c treatment affected memory CD8+ T cells in diverse tissues, it was unclear whether different memory subsets were equally responsive to IL-15 therapy. First, in agreement with previously published work (7), we found that host memory phenotype CD44hi CD8+ T cells proliferated to a far greater extent than naive CD44lo populations and increased considerably in frequency after IL-15c in both the LCMV-elicited and Lm-gp33–elicited memory models (SI Appendix, Fig. S2 B–I).
The polyclonal CD44hi host CD8+ T cell pool encompasses both antigen-experienced memory (or AEM) cells and “virtual memory” (VM) cells, which are thought to be induced through homeostatic mechanisms (50, 51). The responsiveness of these populations to IL-15c has not been directly compared. VM cells can be distinguished from AEM cells by low expression of the marker CD49d (52), but it is unclear whether expression of CD49d on AEM cells might be altered by IL-15c treatment. Analysis of LCMV-primed P14 T cells confirmed that AEM cells remained CD49dhi rather than converting to a CD49dlo phenotype in response to IL-15c–elicited proliferation (SI Appendix, Fig. S3 A and B). Both AEM- and VM-phenotype host CD8+ T cells expanded in frequency in response to IL-15c (SI Appendix, Fig. S3 A–D). Interestingly, the proportional expansion of CD44hi compared to CD44lo host T cells was in large part due to expansion of VM cells. However, at late time points after IL-15c treatment, VM-phenotype CD8+ T cells returned to their resting frequency and number, while AEM-phenotype CD8+ T cells (like donor P14 T cells) remained slightly elevated (SI Appendix, Fig. S3 E and F). To ascertain whether this reflected total replacement of the VM-phenotype pool after IL-15c, we assessed the stability of pulse-chase BrdU labeling of AEM- and VM-phenotype CD8+ T cells. Both populations remained highly labeled long after the termination of IL-15c and BrdU treatment (SI Appendix, Fig. S3G), indicating that VM-phenotype populations proliferate at a similar rate to AEM-phenotype populations and neither is substantially replaced in the aftermath of IL-15c treatment. Like AEM CD8+ T cells, VM CD8+ T cells responded rapidly to IL-15c by up-regulating Ki67 (SI Appendix, Fig. S3H). These data suggest that IL-15 sensitivity is shared by VM CD8+ T cells but also indicate that VM and AEM populations differ in their behavior after the cessation of IL-15c treatment.
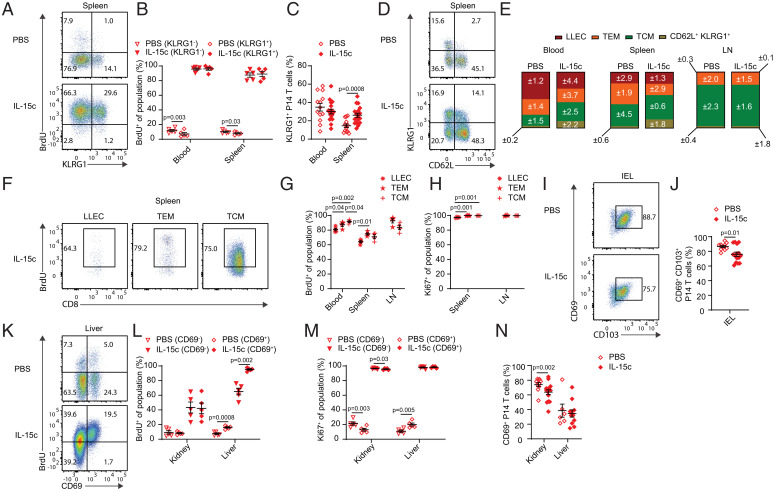
At memory phase, KLRG1 marks a subset of effector memory CD8+ T cells termed LLECs, which have been alternately described in different studies as terminally differentiated or still capable of proliferation (3, 4). Memory cells of this phenotype are highly dependent on IL-15 for generation and/or maintenance (17). Therefore, we assessed whether KLRG1+ P14 T cells could proliferate in response to IL-15c in both the LCMV-elicited and Lm-gp33–elicited memory models. When we compared proliferation of KLRG1+ versus KLRG1− P14 T cells, we observed similar proportions of BrdU+ and Ki67+ cells between the two populations, consistent with IL-15c–driven proliferation of LLECs (Fig. 4 A and B and SI Appendix, Fig. S4A). IL-15c induced a modest but significant increase in the frequency of KLRG1+ memory CD8+ T cells in the spleen (Fig. 4C and SI Appendix, Fig. S4B).
Fig. 4.
IL-15c stimulate proliferation of memory CD8+ T cell subsets. Analysis of donor P14 T cells from P14 memory mice treated as in Fig. 1. (A) Flow cytometry for BrdU incorporation of KLRG1+ and KLRG1− donor P14 T cells in the spleen from PBS- and IL-15c–treated mice. (B) Quantitation of the percentage of BrdU+ KLRG1+ (of KLRG1+ P14 T cells) and KLRG1− (of KLRG1− P14 T cells) donor P14 T cells in the blood and spleen, gated as in A. (C) Quantitation of the percentage of KLRG1+ donor P14 T cells (of all donor P14 T cells) in the blood and spleen from PBS- and IL-15c–treated mice. (D) Flow cytometry for P14 circulating memory subsets in the spleen from PBS- and IL-15c–treated mice. (E) Quantitation of the percentages of P14 circulating memory subsets in the blood, spleen, and LN, gated as in D (days 50 to 55 post-LCMV). Values represent SEM. (F) Flow cytometry for BrdU incorporation of LLEC, TEM, and TCM P14 T cells in the spleen from PBS- and IL-15c–treated mice. (G) Quantitation of the percentage of BrdU+ donor LLEC, TEM, and TCM P14 T cells in the blood, spleen, and LN (TEM and TCM only) from IL-15c–treated mice, gated as in F. (H) Quantitation of the percentage of Ki67+ donor LLEC, TEM, and TCM P14 T cells in the blood, spleen, and LN (TEM and TCM only) from IL-15c–treated mice. (I) Flow cytometry for CD69 and CD103 expression on donor IEL P14 T cells from PBS- and IL-15c–treated mice. (J) Quantitation of the percentage of CD69+ CD103+ donor IEL P14 T cells (of all donor P14 T cells) gated as in I. (K) Flow cytometry for BrdU incorporation of CD69+ and CD69− donor P14 T cells in the liver from PBS- and IL-15c–treated mice. (L and M) Quantitation of the percentage of (L) BrdU+ and (M) Ki67+ CD69+ (of CD69+ P14 T cells) and CD69− (of CD69− P14 T cells) donor P14 T cells in the kidney and liver from PBS- and IL-15c–treated mice. (N) Quantitation of the percentage of CD69+ donor P14 T cells (of all donor P14 T cells) in the kidney and liver from PBS- and IL-15c–treated mice. (A, D, F, I, and K) Data are representative of two to five experiments with 4 to 15 mice per group. (B, E, G, H, L, and M) Data are pooled from two experiments with four or five mice per group. (C, J, and N) Data are pooled from three to six experiments with PBS = 6 to 13 and IL-15c = 11 to 18 mice per group. (B, G, H, L, and M) Paired and (C, J, and N) unpaired two-sided Student’s t tests. Error bars are ±S.E.M.
Analysis of CD62L expression allowed us to resolve LLEC (CD62L− KLRG1+), T effector memory (TEM; CD62L− KLRG1−), and T central memory (TCM; CD62L+ KLRG1−) P14 subsets. The relative proportions of each subset remained mostly stable despite modest declines in effector-like memory populations in some tissues (Fig. 4 D and E). Likewise, following cessation of IL-15c treatment, the proportions of TCM, TEM, and LLEC largely remained similar to those of control animals (SI Appendix, Fig. S4C). Interestingly, while the overall proportions of BrdU-incorporating and Ki67-expressing cells were similar across subsets (Fig. 4 F–H), the pattern of BrdU-labeling differed between TEM/LLEC and TCM (SI Appendix, Fig. S4 D and E), suggesting subset-specific nuance to the IL-15c–driven proliferation of these populations, despite their shared homeostatic IL-15 dependence.
While our data were strongly suggestive of IL-15c–driven proliferation of tissue-resident memory CD8+ T cells, it was possible that the cycling cells were recirculating effector memory cells, not resident cells. As a first step to address this, we determined how the expression of markers associated with TRM was affected by IL-15c treatment in both the LCMV-elicited and Lm-gp33–elicited memory models. TRM in the IEL typically have a CD69+ CD103+ phenotype (2, 53), and antigen-specific CD8+ T cells with this phenotype were abundant in animals treated with IL-15c, although transiently reduced compared to controls (Fig. 4 I and J and SI Appendix, Fig. S3 F and G). TRM in other NLTs are typically identified using CD69 expression, though this is known to be an imperfect marker of resident cells (53–55). Examination of kidney and liver revealed similar levels of IL-15c–induced proliferation by CD69+ populations compared to CD69− populations after IL-15c, consistent with balanced effects on both resident and nonresident populations (Fig. 4 K–M and SI Appendix, Fig. S4 H and I). BrdU labeling of CD69+ P14 T cells was higher in the liver with or without IL-15c treatment despite equivalent Ki67 expression, suggesting that the kinetics of proliferation differed between CD69+ and CD69− cells in this tissue site. Nevertheless, the relative frequency of CD69+ P14 T cells was modestly decreased in the kidney and liver before recovering over time, implying somewhat increased representation of circulating cells after IL-15c (Fig. 4N and SI Appendix, Fig. S4 J and K).
To limit the possibility of memory P14 T cells proliferating in the circulation and then infiltrating into NLTs during the 4-d labeling period, we alternatively labeled with BrdU for the last 24 h rather than the entire course of IL-15c treatment. We still observed clear proliferation of memory P14 T cells (SI Appendix, Fig. S5 A–C). As a shorter BrdU-labeling period also limited the opportunity for memory cells to change phenotype after proliferation, we then assessed short-term IL-15c–elicited BrdU labeling of VM and AEM-phenotype CD8+ T cells; LLEC, TEM, and TCM P14 T cells; and CD69− and CD69+ P14 T cells. We observed generally similar behavior compared with longitudinal BrdU labeling, with slight trends toward lower BrdU incorporation by LLECs (compared to TEM and TCM) and minor differences between CD69+ and CD69− P14 T cells (SI Appendix, Fig. S5 D–F). However, with short-term labeling, a higher proportion of LT P14 T cells incorporated BrdU than for most NLT P14 populations after IL-15c, particularly when sacrificed at day 4 (with labeling from day 3) compared to day 2 (with labeling from day 1). IEL P14 T cell BrdU incorporation was especially reduced with 24-h labeling as opposed to complete longitudinal labeling during treatment (SI Appendix, Fig. S5 A–C and G). This suggested tissue-specific differences in the duration of IL-15c–induced proliferation or infiltration of proliferating circulating memory cells, which quickly acquired CD69 and CD103.
In light of different degrees of proliferation between tissue sites, we next assessed expression of the IL-2 receptor β chain (CD122), which is required for IL-15 signaling. Interestingly, differences in CD122 were observed even for circulating memory subsets. VM CD8+ T cells exhibited increased CD122 expression compared to AEM cells (SI Appendix, Fig. S6 A and B), as previously described (56). While all circulating P14 T cells expressed CD122, LLEC P14 T cells expressed lower levels of CD122 than TEM and TCM as previously seen (3, 4), particularly after IL-15c (SI Appendix, Fig. S6 C–F). IL-15c treatment resulted in notable up-regulation of CD122 on most circulating memory subsets, including VM cells (SI Appendix, Fig. S6 A–F). P14 T cells in NLT had generally lower expression of CD122 compared to LT P14 T cells, with IEL P14 T cells exhibiting the lowest CD122 expression (SI Appendix, Fig. S6 G–J), consistent with earlier reports (23, 24). Furthermore, NLT P14 T cells had only modest, if any, increase in CD122 upon IL-15c treatment (SI Appendix, Fig. S6 G–J). Based on the generally lower proliferation of IEL P14 T cells observed and the reduced short-term BrdU incorporation by NLT P14 T cells, our data suggest that reduced capacity to sense IL-15 does affect the degree of IL-15 responsiveness in this population. However, it is important to note that FRT P14 T cells, which (like IEL) were shown to be IL-15c independent for homeostatic maintenance (1), express CD122 at levels comparable to cells from other NLTs and responded rapidly and durably to IL-15c, showing a high degree of BrdU labeling even with short-term labeling (SI Appendix, Fig. S6 G and H). Furthermore, while circulating memory P14 subsets all express relatively high levels of CD122, they nonetheless exhibit differences in their pattern of proliferation. Therefore, while CD122 likely imposes constraints on IL-15c–elicited responsiveness, other factors also contribute to response patterns. Taken together, while all memory CD8+ T cell populations respond to IL-15c, the degree of sensitivity and responsiveness may differ between subsets.
Tissue-Resident Memory CD8+ T Cells Proliferate in Response to IL-15c.
While our findings were consistent with IL-15c inducing proliferation in established TRM populations, it was possible that IL-15c treatment induced massive recruitment and proliferation of recirculating cells, some of which quickly acquired a TRM phenotype in NLT. Indeed, short-term BrdU incorporation assays showed reduced labeling of NLT P14 compared to longitudinal BrdU labeling throughout IL-15c treatment: such data might indicate that cells labeled in the circulation subsequently acquire a TRM (CD69-positive) phenotype, but could also be explained by distinct kinetics of IL-15c–induced proliferation by distinct memory subsets. If IL-15–independent TRM populations were also IL-15 insensitive (IEL, FRT), these populations might be overrun by infiltrating circulating memory cells proliferating in response to IL-15c. To directly assess whether IL-15c treatment led to recruitment of cells from the circulation into the NLT TRM pool, we turned to parabiosis. Naive recipient B6 mice received either CD45.1/CD45.2 or CD45.1/CD45.1 P14 T cells and were then infected with LCMV to generate memory as before. Seven to eight weeks later, mice receiving congenically distinct donor P14 T cells were conjoined by parabiosis. After the establishment of a shared circulation (∼3 to 4 wk, confirmed by similar representation of both P14 cohorts in the blood), we treated both animals in a pair with phosphate-buffered saline (PBS) or IL-15c (i.e., PBS/PBS or IL-15c/IL-15c) and concurrently labeled with BrdU. In this manner, we could assess whether IL-15c treatment led to erosion of host tissue residency with concurrent, de novo establishment of tissue-resident partner cells, as well as the relative proliferation of resident and recirculating cells in diverse tissues. As expected, recirculating populations (in blood, spleen, and LNs) were in similar proportions regardless of IL-15c treatment (Fig. 5 A and B). In NLTs of PBS-treated mice, memory P14 T cells showed disequilibrium, indicating tissue residency in all sites, albeit to different degrees depending on the tissue (Fig. 5 A–C) (54). Notably, the degree of NLT tissue residency was generally similar in IL-15c–treated animals. However, while IL-15–independent sites (IEL, FRT) showed relatively little infiltration of additional partner-derived cells after IL-15c, IL-15–dependent sites (SG, kidney, liver) showed greater evidence of infiltration of partner-derived cells (Fig. 5 A–C). Following IL-15c treatment, the proportions of proliferating host cells and partner-derived cells were similar across tissues (Fig. 5 D and E), indicating that resident populations were genuinely capable of local, IL-15c–driven proliferation.
Fig. 5.
Tissue-resident memory CD8+ T cells proliferate in response to IL-15c. Parabiosed pairs of CD45.1/CD45.1 and CD45.1/CD45.2 P14 memory mice were treated with PBS or IL-15c, with both members of each pair receiving the same treatment course as in Fig. 1. (A) Flow cytometry for donor P14 T cells (in this example, CD45.1/CD45.1 P14 T cells were host derived and CD45.1/CD45.2 P14 T cells were partner derived). (B) Quantitation of the percentage of the host and partner P14 population in the blood, spleen, LN, IEL, SG, kidney, liver, and FRT from PBS- and IL-15c–treated parabiosed mice gated as in A. (C) Blood-adjusted percent resident donor P14 T cells in the blood, spleen, LN, IEL, SG, kidney, liver, and FRT from PBS- and IL-15c–treated parabiosed mice. (D and E) Quantitation of the percentage of (D) BrdU+ and (E) Ki67+ host and partner P14 T cells in the blood, spleen, LN, IEL, SG, kidney, liver, and FRT (BrdU only) from PBS- and IL-15c–treated parabiosed mice. (F) Quantitation of the percentage of P14 T cells within the CD69+ CD103+, CD69+ CD103−, and CD69−CD103− IEL CD8+ T cell populations from PBS- and IL-15c–treated parabiosed mice. (G) Quantitation of the percentage of CD69+ and CD69− P14 T cells within CD69+ and CD69− SG, kidney, and liver CD8+ T cell populations from PBS- and IL-15c–treated parabiosed mice. Data are representative of/pooled from three experiments with three or four parabiosed pairs per group. (B and D–G) Paired and (C) unpaired two-sided Student’s t tests. Error bars are ±S.E.M.
In IL-15c–treated mice, CD69+ CD103+ and CD69+ CD103− P14 T cells were predominantly derived from the host (Fig. 5 F and G and SI Appendix, Fig. S7). While a degree of increased infiltration into tissues was evident in the SG, kidney, and liver following IL-15c treatment, this consisted of CD69− CD103− partner (and host) P14 T cells, arguing against infiltrating cells rapidly acquiring a resident phenotype in response to IL-15c (Fig. 5 F and G and SI Appendix, Fig. S7). Therefore, IL-15c treatment does not erase preexisting residency and elicits similar proportions of proliferating resident and recirculating cells. Rather than driving enhanced infiltration of circulating memory cells into sites with IL-15–independent TRM maintenance, if anything IL-15c induced moderately greater recruitment of circulating cells into IL-15–dependent NLTs, though TRM populations remained abundant. Taken together, these data demonstrate that TRM are sensitive and proliferative in response to IL-15 signals.
Discussion
Previous studies have shown that dependency on IL-15 for CD8+ T cell memory is not fixed but varies among TRM in distinct NLTs. This raised the question of whether memory CD8+ T cell populations that are IL-15 independent are also IL-15 insensitive or, alternatively, whether responsiveness to IL-15 is a hallmark of all CD8+ T cell memory subpopulations. IL-15 independence could arise, for example, by IL-15–independent memory CD8+ T cells losing the ability to sense IL-15 in favor of responding to homeostatic cues. Instead, our findings establish a conserved capacity of IL-15c to drive the proliferation and (reversible) expansion of memory CD8+ T cell populations across various subsets, tissues, antigen specificities, and models of memory induction. However, intriguingly, we also observed some points of distinction between different subsets and in different tissues, including between IL-15–dependent circulating memory subsets. Our data suggest that cytokine therapy could be customized to promote expansion of memory subsets, including the possibility of increasing CD8+ TRM abundance in desired NLTs. The significance of these subset and tissue-specific nuances remains to be fully explored but is consistent with IL-15c stimulation operating within a framework imposed by context-specific programming, with the degree of IL-15c sensitivity being limited in part by expression of the IL-15β receptor chain, CD122. Building on previous work that established the existence of IL-15–independent memory CD8+ T cells (1, 18), our study reveals that while IL-15 is not always necessary, it is sufficient for potent effects on memory CD8+ T cell proliferation.
How can memory CD8+ T cells be IL-15 independent during normal homeostasis, yet highly responsive to IL-15 therapy? One possibility is that IL-15–independent memory CD8+ T cells are maintained in sites with minimal IL-15; however, several studies have identified IL-15–independent CD8+ memory T cells in sites also housing IL-15–dependent populations (1, 8, 10, 18, 26). It is also possible that some memory CD8+ T cell populations display greater versatility in responding to alternate cytokines (or other factors) that permit maintenance in the absence of IL-15. Favoring this interpretation, we found that several NLT P14 populations exhibited low expression of CD122, suggesting that NLT P14 T cells may not be as sensitive to homeostatic IL-15 levels. The substantial IL-15c–driven rise of (CD122hi) VM-phenotype CD8+ T cells to occupy a much greater portion of the host CD8+ T cell compartment, followed by a precipitous decline, suggests that heightened IL-15 sensitivity possesses its own perils. Tuned expression of CD122, possibly related to repression of Tbet as part of the tissue-residency program (21, 22), may allow appropriate (but not exaggerated) responses of TRM to spikes in IL-15. IEL memory P14 T cells were particularly notable for their low expression of CD122, and this may account for their weaker response to IL-15c. Whether IL-15c treatment also mediates indirect effects remains to be determined. While IL-15Rα is typically dispensable on circulating memory CD8+ T cells during homeostasis (and is not required for IL-15 complex signaling) (30–35), it has been found on circulating memory CD8+ T cells and, when overexpressed in naive CD8+ T cells, promotes proliferation (14, 57). Whether T cell–associated IL-15Rα is required by memory CD8+ T cells in homeostatic or inflammatory contexts in specific tissues should be considered in future studies. Interpretation of previous studies using IL-15–deficient mice may also need to be tempered by the realization that loss of IL-15 affects numerous lymphocyte populations and may thereby reduce the competition for other resources by the remaining antigen-primed IEL CD8+ TRM.
Interestingly, memory cells that had undergone considerable IL-15c–elicited proliferation were still found weeks later in a state of reduced proliferation, and memory populations largely contracted back to their previous pre–IL-15c size. This suggests that IL-15c–induced population expansion does not erase the previous “set point” of a memory population and that slowed proliferation may contribute to contraction of memory populations to basal levels after IL-15c. Thus, IL-15 may act as a regulator to restore homeostatic levels of memory rather than inappropriately expanding memory populations to an excessive level. IL-15 could act as a proprietary cytokine to preserve preexisting memory CD8+ T cells during effector responses and particularly viral infections characterized by interferon-elicited IL-15 (7, 27), as well as to enhance bystander CD8+ T cell responses. Thus, while high (and likely superphysiological) doses of IL-15c were used in our study, our results are likely informative as to the response of memory CD8+ T cells to enhanced IL-15 levels during viral infection. Our findings highlight key differences in the outcomes of T cell proliferation induced by IL-15 therapy versus T cell receptor-dependent restimulation. While antigen reencounter enables a stable increase in memory T cell numbers and access of boosted cells into NLTs (2, 49), we show that IL-15c treatment resulted in transient amplification of the memory CD8+ T cell pool, which then contracted to pretreatment levels, and did not lead to expansive recruitment of circulating cells into NLTs.
Cytokine-based T cell immunotherapy approaches for cancer and some infectious diseases are making significant clinical progress. IL-2 therapy has substantial limitations due to toxicity and its ability to expand CD4+ regulatory T cells, but IL-15–based immunotherapies as well as engineered forms of IL-2 that act similarly to IL-15 are under active investigation for treatment of cancers and HIV (39–42). Our data suggest that targeted, local delivery of IL-15 (58, 59) could have potent effects on tissue-localized memory CD8+ T cells, especially pathogen-specific and also potentially tumor-infiltrating populations, though resistance to IL-15 has been described in this latter population (25, 60).
We propose that while IL-15 is not universally required to sustain the constellation of memory CD8+ T cell populations induced by different pathogens across the body, sensitivity to IL-15 is a defining feature of CD8+ T cell memory. While circulating and tissue-resident memory CD8+ T cells in diverse locations likely rely on varying supportive factors, they exhibit a shared responsiveness to IL-15 that permits therapy using this cytokine to promote proliferation and transient expansion of memory CD8+ T cell subsets in diverse tissues in an antigen-agnostic manner. Precise targeting of IL-15 to any tissue may hold the potential to expand local memory populations, with potential implications for CD8+ T cell vaccines and anti-tumor immunity.
Materials and Methods
Mice.
LCMV-DbGP33–specific P14 TCR transgenic mice were maintained on the C57BL/6N background and were previously crossed to B6.SJL animals to generate CD45.1/CD45.1 and CD45.1/CD45.2 offspring for tracking after cell transfer. CD45.1/CD45.1 or CD45.1/CD45.2 P14 mice of 6 to 14 wk of age were used as donors, and NCI C57BL/6NCr mice (strain 556) were purchased from Charles River Laboratories as recipients and received cell transfer between 6 and 14 wk of age. Animals were housed under specific pathogen-free conditions at the University of Minnesota, and viral and bacterial infections were performed in a BSL2 animal facility. All animal procedures were approved by the institutional animal care and use committee of the University of Minnesota.
CD8± T Cell Transfers.
CD8+ T cells were isolated from the spleens of donor P14 TCR transgenic mice with the Miltenyi Biotec CD8a+ T cell isolation kit, mouse (130-104-075) and Miltenyi Biotec LS columns (130-042-401). Cells were washed and counted; 50,000 were transferred to naive recipient mice by retro-orbital or tail vein i.v. injection.
Infections and Treatments.
One day after cell transfer, mice were infected with LCMV-Armstrong (2*105 plaque-forming units by intraperitoneal (i.p.) injection) or Lm-gp33 (1*104 colony-forming units by i.v. tail vein injection) to induce memory. Animals were bled (see below) after 4 wk to confirm the establishment of memory. Memory mice were used at least 4 wk after transfer.
For treatment with IL-15c, 7 μg IL-15Rα-Fc chimera (R&D Systems, 551-MR) at 0.2 mg/mL was combined with 0.75 μg murine IL-15 at 0.5 mg/mL (eBioscience, 14–8153-80 or Tonbo Biosciences, 21–8153-U500) in PBS (61). After incubation in a 37 °C water bath for 20 to 30 min, complexes were briefly placed on ice before injection i.p. Complexes were prepared fresh each day and given on day 0 and 2 of experiments.
For longitudinal BrdU labeling, mice were given 1 mg BrdU i.p. (Sigma-Aldrich, B5002) at the start of cytokine complex treatment. For 4 d, mice were given 0.8 mg/mL BrdU in 2% sucrose (Sigma-Aldrich, S1888) water ad libitum before harvest. For 24-h BrdU labeling, mice were given 1 mg BrdU i.p. 24 h before killing. For pulse-chase experiments, mice were given 1 mg BrdU i.p. at the start of cytokine complex treatment, followed by 6 d of 0.8 mg/mL BrdU in 2% sucrose water ad libitum. BrdU water was refreshed every other day and shielded from light. After 6 d, BrdU water was replaced with regular untreated water for the duration of the experiment.
Cell Isolation.
Before euthanasia, mice were given 3 μg anti-CD8α PerCP/Cy5.5 (Tonbo Biosciences, 53–6.7) by retro-orbital injection (45, 46). After 5 min, animals were bled by cheek bleed into heparin and killed for tissue harvest. Lymphoid organs, inguinal LNs and the spleen, were collected into harvest media (either RPMI 1640 [Corning, 10–040-CV] supplemented with 5% fetal bovine serum [FBS; Atlas Biologicals, FS-0050-AD; heat inactivated before use] or 1x Hanks’ balanced salt solution [HBSS; Corning, 20–021-CV] supplemented with 2.38 g/L Hepes [4-(2-hydroxylethyl)-1-piperazineethanesulfonic acid; Thermo Fisher Scientific, BP310-1], 2.1 g/L sodium bicarbonate [Thermo Fisher Scientific, BP 328–1], and 5% FBS) and passed through a 70-μm cell strainer. For IEL, the small intestine (SI) was excised, doused in IEL media (1x HBSS supplemented with Hepes, sodium bicarbonate, and 2% FBS), divested of Peyer’s patches, and cleansed of fecal contents. After bisection to open the lumen, the sample was vortexed and left on ice in 20 mL IEL medium. IEL medium was then decanted and replaced with 30 mL of fresh IEL medium, which was then decanted. The SI was then transferred to 50 mL Erlenmeyer flasks with stir bars and 30 mL of IEL dithioerythritol (DTE) media supplemented with additional FBS to 5% and 154 mg/L dithioerythritol (EMD Millipore, 233152). After 30 min of stirring at 37 °C on a Variomag Poly 15 (Thermo Fisher Scientific, 50094595), the supernatant was decanted through a 70-μm filter. Twenty milliliters of IEL DTE media was added, and the sample was vortexed. The supernatant was filtered to combine with the previous fraction. For liver, the liver was excised, taking care to avoid the gallbladder, and placed in 5 mL harvest media on ice. It was then mechanically disrupted using a GentleMACS C tube (130-093-237) on a GentleMACS Dissociator (Miltenyi Biotec, 130–093-235) (m_spleen_01.01 twice) and filtered. For SG, kidney, FRT, SGs (submandibular, sublingual, and part of the parotid) were excised, and cervical LNs were removed if present. The capsule was removed from the kidneys during isolation. The FRT was collected, including the ovaries, cutting as close to the hindquarters as possible, and was then bisected open. All tissues were placed in harvest media on ice. Tissues were then finely minced using scissors and transferred to Erlenmeyer flasks with stir bars. Thirty milliliters of collagenase solution (RPMI 1640 supplemented with 1 mM MgCl2 [Thermo Fisher Scientific, AM9530G], 1 mM CaCl2 [Fluka Analytical, 21114–1L], 111.6 mg/L Hepes, 292 mg/L L-glutamine [Alfa Aesar, A14201], and 5% FBS) containing 0.364 mg/mL collagenase I (SG, kidney) (Worthington Biochemical, LS004197) or 0.5 mg/mL collagenase IV (FRT) (Thermo Fisher Scientific, 17104019) was added. Tissues were then incubated at 37 °C with stirring for 45 to 50 min (SG, kidney) or 60 to 70 min (FRT). The supernatant was then filtered, and the remaining volume/tissue was transferred to a GentleMACS C tube for mechanical disruption as above for liver. GentleMACS contents were then filtered and combined with the previous fraction. All NLTs were then centrifuged and resuspended in 5 mL of room temperature (RT) 44% Percoll (diluted with RPMI 1640), which was then underlaid with 3 mL of RT 67% Percoll (diluted with PBS). Percoll solution was purchased from GE Healthcare (17-0891-09), and 10x PBS (BioLegend, 926201) was added before use. Samples were spun for 20 min at 800 g at RT, with acceleration and deceleration set to minimum values. The interface was then collected, diluted with harvest media, and used for analysis. Red blood cells in blood and spleen samples were disrupted with ACK lysis buffer (150 mM ammonium chloride, 1 mM potassium bicarbonate, and 0.1 mM EDTA [ethylenediaminetetraacetic acid] in water) before staining.
Flow Cytometry.
Samples and single stain controls were resuspended and washed in fluorescence-activated cell sorter (FACS) buffer (2% FBS, 2 mM EDTA in 1x PBS), followed by Fc blocking for 5 min (BD, 553142). Antibodies/viability dye for staining were then added for 20 min at 4 °C. For tetramer staining, biotinylated monomers (H-2Db KAVYNFATM [gp33/Db]; H-2Db SGVENPGGYCL [gp276/Db]; H-2Db FQPQNGQFI [NP396/Db]) were tetramerized according to the recommendations of the NIH Tetramer Core at Emory University. For gp33, PE [Phycoerythrin]/Cy7-streptavidin (Thermo Fisher Scientific, 25–4317-82) was used. For gp276 and NP396, R-PE-streptavidin (Thermo Fisher Scientific, S21388) was used. Fluorophore-conjugated streptavidin was added to 20 μg of monomer, in 10 additions of 3.18 μg, each 10 min after the other (at RT). Tetramer was then stored at 4 °C before use. Tetramer staining was done concurrently with surface antibody staining (20 min at 4 °C). Samples were then washed before fixation, washed after fixation, and stored in FACS buffer at 4 °C before intracellular staining 1 to 2 d later. Antibodies/viability dyes used fluorescein isothiocyanate (FITC) and vF450 anti-CD45.1 (BioLegend and Tonbo Biosciences, A20), FITC anti-CD45.2 (Tonbo Biosciences, 104), FITC and PE anti-CD49d (BioLegend, R1-2), PE anti-CD45.1 (Tonbo Biosciences, A20), PerCP/Cy5.5 anti-CD8α (i.v. labeling, Tonbo Biosciences, 53–6.7), PE anti-CD122 (BD Biosciences, TM-β1), Ghost Dye Red e780 (Tonbo Biosciences, 13–0865-T500), BV510 anti-CD44 (BD, IM7), BV510 anti-CD103 (BD, M290), PB [Pacific Blue] anti-CD69 (BioLegend, H1.2F3), vF450 anti-CD45.1 (Tonbo Biosciences, A20), BV605 anti-TCRβ (BD, H57-597), BV786, PE/Cy7, and BUV395 anti-CD8α (BioLegend, Tonbo Biosciences, and BD Biosciences, 53–6.7), BV786 anti-CD62L (BD Biosciences, MEL-14), rF710 anti-CD44 (Tonbo Biosciences, IM7), and BV711 anti-KLRG1 (BD, 2F1). All surface antibodies were used at a 1/200 dilution, except anti-CD69 (1/100). Viability dye was used at 1/1,000.
Intracellular Staining.
For Ki67 staining, the Foxp3/Transcription Factor Staining Buffer Kit (Tonbo Biosciences, TNB-0607-KIT) was used according to the standard protocol, with the addition of a 15-min incubation in 1x Perm buffer plus 2% Rat Serum (StemCell Technologies) before anti-Ki67 antibody staining for 45 to 60 min at RT (Thermo Fisher Scientific, 17–5698-82 [SolA15 APC, 1/200]). For BrdU staining, a modified protocol using the Cytofix/Cytoperm Fixation/Permeabilization Solution Kit (BD, 554714) was employed. Briefly, cells were washed in 1x Perm/Wash (P/W) buffer and then incubated for 10 min at 4 °C in 1x P/W buffer plus dimethyl sulfoxide (DMSO; Thermo Fisher Scientific, BP231-1) (1 part 10x P/W, 1 part DMSO, and 8 parts water). Samples were then washed in 1x P/W buffer and refixed in Cytofix/Cytoperm buffer for 5 min at RT, followed by washing in 1x P/W buffer. Cells were then digested with deoxyribonuclease (DNase) I (Sigma-Aldrich, D5025) at 37 °C for at least 50 min. DNase was stored at −80 °C as a 1 to 2 mg/mL stock in PBS and diluted to 300 to 600 μg/mL in PBS immediately before use. The concentration used for digestion was determined by titration. After digestion, cells were washed in 1x P/W buffer and stained with anti-BrdU antibody (Thermo Fisher Scientific, 17–5071-42 [APC Bu20a, 1/100]) for 50 to 60 min at RT. Cells were then washed with 1x P/W buffer and then with FACS buffer. All NLT samples were filtered, and CountBright Plus counting beads were added to BrdU flow tubes before analysis (Invitrogen, C36995). Samples were acquired using BD LSRII, LSFortessa ×20, and LSRFortessa flow cytometers and FACSDiva software.
Analysis was done using FlowJo v7 and v10 (Treestar). Singlet lymphocytes were gated by forward scatter area (FSC-A)/side scatter area (SSC-A) and FSC-A/FSC-width (FSC-W). Live cells were then gated according to i.v. labeling status (blood, i.v.-positive; LN, IEL, SG, FRT, i.v.-negative; spleen and kidney, i.v.-low; liver, not gated by i.v. labeling status). CD8α+/TCRβ+ cells were then divided into CD45.1+ donor P14 CD8+ T cells and CD45.1− host CD8+ T cells. Specifically for the SG in P14 memory chimeras, P14 and endogenous tetramer-specific CD8+ T cells were also gated as CD69+ to exclude possible contamination by CD69− LT–associated cells. As shown in Fig. 2 and SI Appendix, Fig. S1, tetramer-binding host cells were gated as tet+ CD44hi. As shown in SI Appendix, Fig. S3, VM and AEM cells were gated as CD44hi CD49dlo and CD44hi CD49dhi, respectively. To validate BrdU gating, in addition to comparing PBS- and IL-15c–treated samples, each experiment included a control animal not given BrdU, tissues from which were processed and stained alongside the other samples to serve as negative controls. For parabionts, samples were gated as above, with the addition of gating CD8α+ TCRβ+ T cells as CD45.1/CD45.1 (donor P14), CD45.1/CD45.2 (donor P14), and CD45.2/CD45.2 (B6 host). For pregating on CD69/CD103, CD8α+ TCRβ+ T cells were first gated on CD69 status alone (SG, kidney, liver) or by both CD69 and CD103 (IEL) and then gated on congenic markers as above. For fluorescence minus one controls for CD122 staining, cells from each sample were divided and stained with the same set of antibodies, differing only in whether the PE CD122 antibody was included.
Parabiosis.
P14 memory mice were generated as above. Sibling, cagemate females received from Charles River were randomly assigned as recipients, with several cagemates receiving CD45.1/CD45.1 P14 T cells and several other cagemates receiving CD45.1/CD45.2 P14 T cells. At a memory time point, animals were bled to determine the percent P14 T cells in the blood, and cagemates of equivalent size (to help ensure a harmonious parabiont) were paired to match P14 proportions as closely as possible to create a roughly even proportion of circulating P14 T cells from each partner after parabiosis. Seven to eight weeks after LCMV infection, animals were separated into cages of two mice several days in advance of surgery. These animals were given Dietgel Boost (ClearH2O, 72–04-5022) for 3 to 4 d before surgery, which was replaced every 2 d until at least a week after surgery. The day before surgery, animals were given ∼30 mg/kg Baytril i.p., and enriched breeder chow was placed on the cage floor for easy access.
On the day of surgery, sterile surgical tools and a sterile surgical site were prepared. Animals were given avertin i.p. (450 μL), and anesthesia depth was confirmed by toe pinch. If an animal was not fully anesthetized, 50 μL more of avertin was administered, and the toe pinch was repeated to confirm anesthesia depth. Opthalmic ointment was applied to prevent desiccation of the eyes. Whiskers were trimmed to avoid irritation with a dedicated set of nonsurgical scissors. Animals were shaved from behind the ear to just past the hip, rising part way up the back and onto the abdomen to expose the surgical site. Nair was then applied to remove residual hair, incubated for ∼30 s, and then removed with alcohol wipes. Betadine surgical scrub (Purdue Products/Thermo Fisher Scientific, 19–027132) was then applied and incubated for ∼60 s and then removed fully with alcohol wipes. Dedicated sterile surgical scissors were then used to cut an incision from the back of the hip to behind the cranium/ear, curving along the animal’s back, with shallow, short cuts to avoid breaking the peritoneal membrane. Sterile forceps were then used to pull up the skin and separate it from the peritoneum, releasing ∼1/2 cm of skin on each side of the incision from end to end. Animals were then carefully aligned according to the incisions, heads, shoulders, and hips, with feet touching. The dorsal skin was then gathered at the midpoint, and animals were pulled upright. Using a sterile surgical stapler (F.S.T., 12020–09) and surgical staples (BD, BBL AUTOCLIP Wound Clip System 9 mm, 427631), the skin was stapled together without folding so that the internal surfaces were in contact. Surgical staples were then applied along the length of the dorsal skin, and the animals were gently inverted so that the ventral skin could be likewise joined with surgical staples, carefully closing the ends of the incisions with both ventral and dorsal staples.
After completion of the surgery, each partner mouse received two injections of 500 μL warm PBS subcutaneously at the shoulder and the hip. Parabionts were placed in their cages with their heads slightly elevated to avoid aspiration. Paper shreds/special bedding materials were removed from the cage temporarily to avoid matting and irritation at the surgical site. Cages were placed on a heating pad, and animals were kept under observation for >1 h to confirm recovery from anesthesia. Animals were observed multiple times each day for at least a week by laboratory personnel and animal facilities staff. If parabionts showed signs of significant distress or became moribund, they were immediately killed. After healing of the surgical incision, if the mice showed signs of separating or the connecting skin was becoming too loose, additional staples were added under isoflurane anesthesia (see below) to resecure them.
After 2 wk of parabiosis, animals were transiently anesthetized using a custom isoflurane/oxygen vaporizer system. When parabionts were anesthetized, as confirmed by toe pinch, they were placed on nose cones to maintain anesthesia. Mice were bled from the cheek as above, were replaced in their cage, and were observed until recovered from anesthesia. Once anastomosis was confirmed by equilibration of donor P14s in both mice, the animals were used between ∼4 and 8 wk after parabiosis surgery.
Using the isoflurane system as above, mice were treated with BrdU, IL-15c, and anti-CD8α antibody for i.v. labeling. Each mouse received its own injection of BrdU, IL-15c, and anti-CD8α antibody at the normal dose. Rare parabiotic pairs with prolonged sickness were excluded.
Percent resident P14 T cells was calculated using the following formula, where [A] is the partner parabiont and [B] is the host parabiont, adjusting for the relative proportion of A- and B-derived P14 T cells in the blood.
Software.
As above, FACSDiva (BD) was used to acquire flow cytometry data. FlowJo v7 and v10 (Treestar) were used to analyze flow cytometry data. GraphPad Prism v8 (GraphPad Software Inc.) was used for statistical calculations and graphing data. Figures were generated in Adobe Illustrator Creative Cloud.
Statistics.
Paired or unpaired Student’s t tests were respectively used as appropriate and as specified in the figure legends. Exact P values are provided. Where no P value is provided for a relevant comparison, the result was not significant. The numbers of experiments and animals analyzed for each experiment are indicated in the figure legends. Infrequently, too few events (less than five events) were captured to accurately quantitate expression of Ki67 or BrdU on rare populations (primarily endogenous host gp33/Db tet+ cells). When this occurred, these values were excluded. Error bars represent the SEM.
Supplementary Material
Acknowledgments
N.N.J. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (Grant No. DRG-2427-21). K.M.W. was supported by National Cancer Institute (NCI) (Grant No. F30 CA250321). N.V.G. was supported by NCI (Grant No. F30 CA253992). N.J.M. was supported by NCI (Grant No. K00 CA245735). H.B.D.S. was supported by National Institute of Allergy and Infectious Diseases (NIAID) (Grant No. K99/R00 AI139381). This work was funded by NIAID Grant No. R01 AI038903 to S.C.J. The authors acknowledge the S.C.J., S.E.H, D.M. and Hogquist laboratories for valuable feedback. The authors thank K.E. Block for critical reading of the manuscript. The gp33, gp276, and NP396 monomers were obtained through the NIH Tetramer Core Facility.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2209021119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix. Higher quality images are available at Figshare (https://doi.org/10.6084/m9.figshare.21334023.v1).
References
- 1.Schenkel J. M., et al. , IL-15-independent maintenance of tissue-resident and boosted effector memory CD8 T cells. J. Immunol. 196, 3920–3926 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jameson S. C., Masopust D., Understanding subset diversity in T cell memory. Immunity 48, 214–226 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Milner J. J., et al. , Delineation of a molecularly distinct terminally differentiated memory CD8 T cell population. Proc. Natl. Acad. Sci. U.S.A. 117, 25667–25678 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renkema K. R., et al. , KLRG1+ memory CD8 T cells combine properties of short-lived effectors and long-lived memory. J. Immunol. 205, 1059–1069 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wijeyesinghe S., et al. , Expansible residence decentralizes immune homeostasis. Nature 592, 457–462 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christo S. N., et al. , Discrete tissue microenvironments instruct diversity in resident memory T cell function and plasticity. Nat. Immunol. 22, 1140–1151 (2021). [DOI] [PubMed] [Google Scholar]
- 7.Zhang X., Sun S., Hwang I., Tough D. F., Sprent J., Potent and selective stimulation of memory-phenotype CD8+ T cells in vivo by IL-15. Immunity 8, 591–599 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Lodolce J. P., et al. , IL-15 receptor maintains lymphoid homeostasis by supporting lymphocyte homing and proliferation. Immunity 9, 669–676 (1998). [DOI] [PubMed] [Google Scholar]
- 9.Ku C. C., Murakami M., Sakamoto A., Kappler J., Marrack P., Control of homeostasis of CD8+ memory T cells by opposing cytokines. Science 288, 675–678 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Kennedy M. K., et al. , Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15-deficient mice. J. Exp. Med. 191, 771–780 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Becker T. C., et al. , Interleukin 15 is required for proliferative renewal of virus-specific memory CD8 T cells. J. Exp. Med. 195, 1541–1548 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldrath A. W., et al. , Cytokine requirements for acute and Basal homeostatic proliferation of naive and memory CD8+ T cells. J. Exp. Med. 195, 1515–1522 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Judge A. D., Zhang X., Fujii H., Surh C. D., Sprent J., Interleukin 15 controls both proliferation and survival of a subset of memory-phenotype CD8+ T cells. J. Exp. Med. 196, 935–946 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schluns K. S., Williams K., Ma A., Zheng X. X., Lefrançois L., Cutting edge: Requirement for IL-15 in the generation of primary and memory antigen-specific CD8 T cells. J. Immunol. 168, 4827–4831 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Rubinstein M. P., et al. , Converting IL-15 to a superagonist by binding to soluble IL-15Rα. Proc. Natl. Acad. Sci. U.S.A. 103, 9166–9171 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stoklasek T. A., Schluns K. S., Lefrançois L., Combined IL-15/IL-15Rα immunotherapy maximizes IL-15 activity in vivo. J. Immunol. 177, 6072–6080 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandau M. M., Kohlmeier J. E., Woodland D. L., Jameson S. C., IL-15 regulates both quantitative and qualitative features of the memory CD8 T cell pool. J. Immunol. 184, 35–44 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verbist K. C., Field M. B., Klonowski K. D., Cutting edge: IL-15-independent maintenance of mucosally generated memory CD8 T cells. J. Immunol. 186, 6667–6671 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mackay L. K., et al. , The developmental pathway for CD103+CD8+ tissue-resident memory T cells of skin. Nat. Immunol. 14, 1294–1301 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Adachi T., et al. , Hair follicle-derived IL-7 and IL-15 mediate skin-resident memory T cell homeostasis and lymphoma. Nat. Med. 21, 1272–1279 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mackay L. K., et al. , T-box transcription factors combine with the cytokines TGF-β and IL-15 to control tissue-resident memory T cell fate. Immunity 43, 1101–1111 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Intlekofer A. M., et al. , Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 6, 1236–1244 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Masopust D., Vezys V., Wherry E. J., Barber D. L., Ahmed R., Cutting edge: Gut microenvironment promotes differentiation of a unique memory CD8 T cell population. J. Immunol. 176, 2079–2083 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Casey K. A., et al. , Antigen-independent differentiation and maintenance of effector-like resident memory T cells in tissues. J. Immunol. 188, 4866–4875 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doedens A. L., et al. , Molecular programming of tumor-infiltrating CD8+ T cells and IL15 resistance. Cancer Immunol. Res. 4, 799–811 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma L. J., Acero L. F., Zal T., Schluns K. S., Trans-presentation of IL-15 by intestinal epithelial cells drives development of CD8αα IELs. J. Immunol. 183, 1044–1054 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tough D. F., Borrow P., Sprent J., Induction of bystander T cell proliferation by viruses and type I interferon in vivo. Science 272, 1947–1950 (1996). [DOI] [PubMed] [Google Scholar]
- 28.Boyman O., Kovar M., Rubinstein M. P., Surh C. D., Sprent J., Selective stimulation of T cell subsets with antibody-cytokine immune complexes. Science 311, 1924–1927 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Boyman O., Ramsey C., Kim D. M., Sprent J., Surh C. D., IL-7/anti-IL-7 mAb complexes restore T cell development and induce homeostatic T cell expansion without lymphopenia. J. Immunol. 180, 7265–7275 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Lodolce J. P., Burkett P. R., Boone D. L., Chien M., Ma A., T cell-independent interleukin 15Rα signals are required for bystander proliferation. J. Exp. Med. 194, 1187–1194 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dubois S., Mariner J., Waldmann T. A., Tagaya Y., IL-15Rα recycles and presents IL-15 In trans to neighboring cells. Immunity 17, 537–547 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Burkett P. R., et al. , IL-15Rα expression on CD8+ T cells is dispensable for T cell memory. Proc. Natl. Acad. Sci. U.S.A. 100, 4724–4729 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burkett P. R., et al. , Coordinate expression and trans presentation of interleukin (IL)-15Rα and IL-15 supports natural killer cell and memory CD8+ T cell homeostasis. J. Exp. Med. 200, 825–834 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandau M. M., Schluns K. S., Lefrancois L., Jameson S. C., Cutting edge: Transpresentation of IL-15 by bone marrow-derived cells necessitates expression of IL-15 and IL-15Rα by the same cells. J. Immunol. 173, 6537–6541 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Schluns K. S., Klonowski K. D., Lefrançois L., Transregulation of memory CD8 T-cell proliferation by IL-15Rα+ bone marrow-derived cells. Blood 103, 988–994 (2004). [DOI] [PubMed] [Google Scholar]
- 36.Anthony S. M., Howard M. E., Hailemichael Y., Overwijk W. W., Schluns K. S., Soluble interleukin-15 complexes are generated in vivo by type I interferon dependent and independent pathways. PLoS One 10, e0120274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levin A. M., et al. , Exploiting a natural conformational switch to engineer an interleukin-2 ‘superkine’. Nature 484, 529–533 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silva D. A., et al. , De novo design of potent and selective mimics of IL-2 and IL-15. Nature 565, 186–191 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spangler J. B., Moraga I., Mendoza J. L., Garcia K. C., Insights into cytokine-receptor interactions from cytokine engineering. Annu. Rev. Immunol. 33, 139–167 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waldmann T. A., Miljkovic M. D., Conlon K. C., Interleukin-15 (dys)regulation of lymphoid homeostasis: Implications for therapy of autoimmunity and cancer. J. Exp. Med. 217, e20191062 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller J. S., et al. , Safety and virologic impact of the IL-15 superagonist N-803 in people living with HIV: A phase 1 trial. Nat. Med. 28, 392–400 (2022). [DOI] [PubMed] [Google Scholar]
- 42.Propper D. J., Balkwill F. R., Harnessing cytokines and chemokines for cancer therapy. Nat. Rev. Clin. Oncol. 19, 237–253 (2022). [DOI] [PubMed] [Google Scholar]
- 43.Beura L. K., et al. , Intravital mucosal imaging of CD8+ resident memory T cells shows tissue-autonomous recall responses that amplify secondary memory. Nat. Immunol. 19, 173–182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park S. L., et al. , Local proliferation maintains a stable pool of tissue-resident memory T cells after antiviral recall responses. Nat. Immunol. 19, 183–191 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Skon C. N., et al. , Transcriptional downregulation of S1pr1 is required for the establishment of resident memory CD8+ T cells. Nat. Immunol. 14, 1285–1293 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anderson K. G., et al. , Intravascular staining for discrimination of vascular and tissue leukocytes. Nat. Protoc. 9, 209–222 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murali-Krishna K., et al. , Counting antigen-specific CD8 T cells: A reevaluation of bystander activation during viral infection. Immunity 8, 177–187 (1998). [DOI] [PubMed] [Google Scholar]
- 48.Kaech S. M., Ahmed R., Memory CD8+ T cell differentiation: Initial antigen encounter triggers a developmental program in naïve cells. Nat. Immunol. 2, 415–422 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Masopust D., Ha S. J., Vezys V., Ahmed R., Stimulation history dictates memory CD8 T cell phenotype: Implications for prime-boost vaccination. J. Immunol. 177, 831–839 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Jameson S. C., Lee Y. J., Hogquist K. A., Innate memory T cells. Adv. Immunol. 126, 173–213 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kedl R. M., White J. T., Foreign antigen-independent memory-phenotype CD4+ T cells: A new player in innate immunity? Nat. Rev. Immunol. 18, 1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haluszczak C., et al. , The antigen-specific CD8+ T cell repertoire in unimmunized mice includes memory phenotype cells bearing markers of homeostatic expansion. J. Exp. Med. 206, 435–448 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beura L. K., et al. , T cells in nonlymphoid tissues give rise to lymph-node-resident memory T cells. Immunity 48, 327–338.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steinert E. M., et al. , Quantifying memory CD8 T cells reveals regionalization of immunosurveillance. Cell 161, 737–749 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walsh D. A., et al. , The functional requirement for CD69 in establishment of resident memory CD8+ T cells varies with tissue location. J. Immunol. 203, 946–955 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.White J. T., et al. , Virtual memory T cells develop and mediate bystander protective immunity in an IL-15-dependent manner. Nat. Commun. 7, 11291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rowley J., Monie A., Hung C.-F., Wu T.-C., Expression of IL-15RA or an IL-15/IL-15RA fusion on CD8+ T cells modifies adoptively transferred T-cell function in cis. Eur. J. Immunol. 39, 491–506 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sowell R. T., et al. , IL-15 complexes induce migration of resting memory CD8 T cells into mucosal tissues. J. Immunol. 199, 2536–2546 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Verbist K. C., Cole C. J., Field M. B., Klonowski K. D., A role for IL-15 in the migration of effector CD8 T cells to the lung airways following influenza infection. J. Immunol. 186, 174–182 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Epardaud M., et al. , Interleukin-15/interleukin-15Rα complexes promote destruction of established tumors by reviving tumor-resident CD8+ T cells. Cancer Res. 68, 2972–2983 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Burrack K. S., et al. , Interleukin-15 complex treatment protects mice from cerebral malaria by inducing interleukin-10-producing natural killer cells. Immunity 48, 760–772.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix. Higher quality images are available at Figshare (https://doi.org/10.6084/m9.figshare.21334023.v1).