Significance
Many estrogen receptor–positive (ER+) breast cancers lose ER expression and become resistant to endocrine therapy and progress. Unfortunately, the molecular mechanisms underlying the loss of ER expression in breast cancer are poorly understood. This study identifies 14-3-3τ as a key driver of ER loss. We further elucidate a regulation of ERα36 by 14-3-3τ-GATA3 interaction as the mechanism of 14-3-3τ-driven ER loss in breast cancer. The in vitro tumor spheroid model established in our study mimics the evolution of ER+ to ER– human breast cancer and provides a very useful model for future investigation.
Keywords: 14-3-3τ, estrogen receptor, ERα36, GATA3, 3D tumor spheroid model
Abstract
About one-fourth of recurrent estrogen receptor–positive (ER+) breast cancers lose ER expression, leading to endocrine therapy failure. However, the mechanisms underlying ER loss remain to be fully explored. We now show that 14-3-3τ, up-regulated in ∼60% of breast cancer, drives the conversion of ER+ to ER– and epithelial-to-mesenchymal transition (EMT). We identify ERα36, an isoform of ERα66, as a downstream effector of 14-3-3τ. Overexpression of 14-3-3τ induces ERα36 in xenografts and tumor spheroids. The regulation is further supported by a positive correlation between ERα36 and 14-3-3τ expression in human breast cancers. ERα36 can antagonize ERα66 and inhibit ERα66 expression. Isoform-specific depletion of ERα36 blocks the ER conversion and EMT induced by 14-3-3τ overexpression in tumor spheroids, thus establishing ERα36 as a key mediator in 14-3-3τ-driven ER loss and EMT. ERα36 promoter is repressed by GATA3, which can be phosphorylated by AKT at consensus binding sites for 14-3-3. Upon AKT activation, 14-3-3τ binds phosphorylated GATA3 and facilitates the degradation of GATA3 causing GATA3 to lose transcriptional control over its target genes ERα66 and ERα36. We also demonstrate a role for the collaboration between 14-3-3τ and AKT in ERα36 induction and endocrine therapy resistance by three-dimensional spheroid and tamoxifen treatment models in MCF7 and T47D ER+ breast cancer cells. Thus, the 14-3-3τ-ERα36 regulation provides a previously unrecognized mechanism for ER loss and endocrine therapy failure.
Breast cancer is the most common malignant tumor and is the second leading cause of cancer-related deaths in women in the United States. Nearly 75% of breast cancers are estrogen receptor-α positive (ER+) and are commonly treated with endocrine therapies. Tamoxifen (TAM), the preferred adjuvant endocrine therapy treatment for premenopausal women (1), is a selective estrogen receptor modulator used to block ER classic signaling pathway and has been shown to improve patient survival and reduce recurrence of ER+ breast cancer (2). Unfortunately, 40% of women who receive TAM adjuvant therapy eventually develop resistance and relapse, resulting in worse prognoses and lowered overall survival (3). A meta-analysis including 4,200 patients shows 24% of ER+ breast cancers lose ERα expression when tumors relapse (4). However, the mechanisms underlying ER loss remain to be fully elucidated.
Estrogen receptor-α is a 66 kDa ligand-induced transcription factor (TF) found in many tissues of the body and promotes cell proliferation. ERα has three isoforms, all encoded by the ESR1 gene and named for their respective molecular weights: ERα66, ERα44, and ERα36. Of these isoforms, ERα66 is the most widely studied for its role in endocrine therapy treatment; however, there has been increasing interest in the ERα36 isoform for its role in the development of TAM resistance and antagonistic relationship to the full-length isoform (5).
A truncated isoform of ERα66, ERα36, is different from the canonical isoform in many ways. It has a distinct promoter located in the first intronic sequence of ESR1 and produces a messenger RNA (mRNA) transcript lacking exon 1 of the ERα66. ERα36 skips exons 7 and 8, but acquires an additional exon at the 3′ end on the coding sequence through a unique alternative splicing event (6). While ERα36 retains DNA-binding, dimerization, and partial ligand-binding domains, it lacks transactivation domains AF1 and AF2, making it act as a dominant-negative effector to ERα66 (6, 7). ERα36 primarily localizes to the cell membrane but can also be found in the cytoplasm (5). Due to different promoter usage, ERα36 is regulated differently from ERα66 (8). ERα36 is more highly expressed in many ER– breast tumors (5, 9), and its expression is repressed by ERα66 (8). Conversely, overexpression of ERα36 can down-regulate ERα66 expression (10).
ERα36 has been implicated in antiestrogen resistance in breast cancer. TAM-resistant (TAMR) cells express higher levels of ERα36 than TAM-sensitive cells, and depletion of ERα36 restored TAM sensitivity (11). Consequently, breast cancer patients with high ERα36 expression tumors benefit less from TAM treatment compared with those with low levels of ERα36 (9). High ERα36 gene expression is associated with decreased disease-free survival in breast cancer patients independent of ERα66 expression status in tumors (12). TAM resistance has also been linked to the expansion of breast cancer stem cells (13). Deng et al. found ERα36-mediated mitogenic estrogen signaling is important for maintenance of ER+ breast cancer stem cells (14). Breast cancer tumorspheres grown under TAM treatment have increased expression of ERα36 and aldehyde dehydrogenase 1 (ALDH1), a functional marker of breast cancer stem cell, as well as decreased ERα66 expression (15). While there are clear clinical and biological links between ERα36 and the development of TAM resistance, the mechanisms remain to be fully explored. The factors that lead to aberrant induction of ERα36 also have not been identified.
The 14-3-3 family includes seven isoforms (β, ε, ζ, τ, σ, η, and γ) that are ubiquitously expressed adapter proteins in eukaryotic cells (16). 14-3-3 isoforms regulate the function of hundreds of partner proteins that display a conserved phosphoserine/threonine binding motif (17, 18). Through its many binding partners, 14-3-3 facilitates a wide variety of cellular functions, including the regulation of cell cycle progression, cellular proliferation, signal transduction, and metabolite signaling (19). When 14-3-3 is dysregulated, these interactions can facilitate the development and progression of disease (20). Several 14-3-3 isoforms are up-regulated in multiple types of cancer. Specifically, 14-3-3τ is often overexpressed in breast cancer, and its overexpression is correlated with poor overall survival (21, 22). 14-3-3τ overexpression increases cancer cell invasion and metastasis by cooperating with proliferative proteins like RhoGDIα and up-regulation by tenascin-C, an extracellular matrix protein commonly found at the leading edge of growing tumors (21, 23, 24).
GATA3 is a pioneer TF that is expressed in many types of cells to regulate speciation and differentiation of many tissue types (25). GATA3 is critical in the development of the mammary gland and forms a transcriptional complex with FOXA1 and ERα66 to promote the expression of luminal cell identity genes (26). GATA3 also represses genes integral to basal-like subtypes. Dysregulation of GATA3 can lead to the loss of luminal cell integrity and progression toward more basal-like cells frequently found in breast cancer (27). However, whether GATA3 can regulate ERα36 has not been explored.
Here, we establish a correlation between high 14-3-3τ and loss of ERα66 expression and identify ERα36 as the key mediator in this process. We also determined 14-3-3τ overexpression in luminal breast cancer drives the development of earlier TAMR characteristics and a more mesenchymal-like phenotype. Additionally, we identified a mechanism for 14-3-3τ to collaborate with AKT to facilitate these changes through a binding interaction with GATA3, causing GATA3 degradation and induction of ERα36. Together, these results provide insight into 14-3-3τ’s contribution to the development of TAM resistance and progression of luminal breast cancer. We also establish a breast cancer spheroid culture recapitulating ER loss of human breast cancer, which can facilitate the discovery of new targets for therapeutic intervention.
Results
High Expression of 14-3-3τ Is Associated with Down-Regulation of ERα66 and Endocrine Therapy Failure in Breast Cancer.
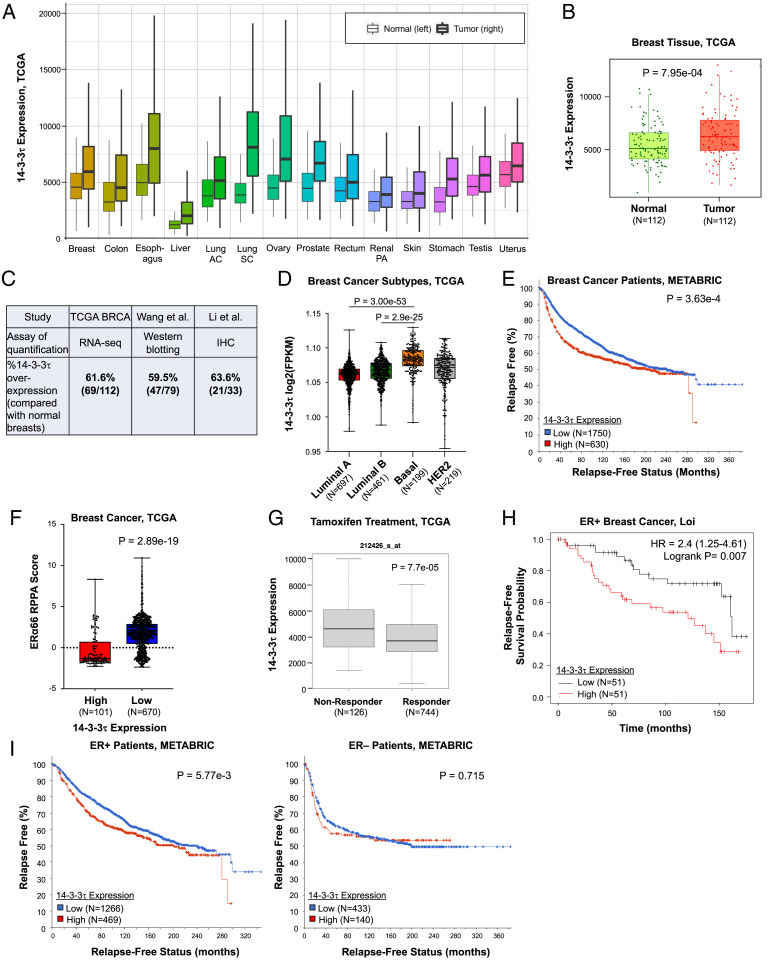
Encoded by gene YWHAQ, 14-3-3τ is up-regulated across many cancer types, including breast cancer (Fig. 1A). To examine the clinical impact of 14-3-3τ in breast cancer, we investigated datasets from The Cancer Genome Atlas (TCGA) and the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC). We noted the significant increase of 14-3-3τ expression in breast cancer compared with paired, normal breast tissues (Fig. 1B), which is consistent with the results of 14-3-3τ protein examination from two smaller cohorts of breast cancer patients (21, 22). Overall, about 60% of breast tumors express higher levels of 14-3-3τ than matched, adjacent normal tissues, regardless the methods of quantification (Fig. 1C). Next, we investigated 14-3-3τ expression across breast cancer subtypes. We found 14-3-3τ expression significantly increased in more aggressive subtypes, such as luminal B, human epidermal growth factor receptor 2–positive, and basal-like subtypes (Fig. 1D). Patients with high 14-3-3τ expression in breast cancer also have significantly reduced, relapse-free survival (Fig. 1E) and overall survival (SI Appendix, Fig. S1A) in the METABRIC cohort.
Fig. 1.
14-3-3τ is up-regulated in breast cancer, and its up-regulation is significantly associated with decreased ERα66 expression, tamoxifen resistance, and relapse-free survival. (A) 14-3-3τ expression is significantly elevated in 14 cancers from TCGA database compared with normal tissue expression. Tumor samples are denoted with bolded lines. Significance calculated by Mann-Whitney U test. AC, adenocarcinoma; SC, squamous cell carcinoma; PA, papillary cell carcinoma. (B) 14-3-3τ expression is significantly (P = 7.95e-4) increased in breast cancer compared with their matched, normal breast tissues in the TCGA BRCA dataset. (C) 14-3-3τ is overexpressed in about 60% of breast tumors when compared with their matched, adjacent normal breast tissues in three cohorts: TCGA, Wang et al. (22) and Li et al. (21). 14-3-3τ overexpression in Wang et al. is defined by ≥ 1.5X of their matched normal breast tissues on Western blot analysis. The cutoff for TCGA and Li et al. is the 14-3-3τ expression level in the respective matched normal breast tissues. IHC, immunohistochemistry. (D) 14-3-3τ expression significantly increases from luminal A and luminal B to basal-like breast cancer subtype (P = 3.00e-53 and P = 2.9e-25, respectively). (E) METABRIC relapse-free survival analysis showing breast cancer patients with high 14-3-3τ expression (red) are significantly (P = 3.63e-4) more likely to relapse than patients with low expression (blue). Patients were split by upper tertile of 14-3-3τ expression in their breast cancers. Similar results were obtained when patients were split by upper quartile or median. (F) High 14-3-3τ expression is significantly (P = 2.89e-19) associated with low ERα66 protein expression in breast cancer RPPA analysis of TCGA BRCA samples. Cutoff was defined by 14-3-3τ expression Z score >1. Similar results were obtained when using other cut-offs. (G) ER+ patients with high 14-3-3τ expression are significantly (P = 7.7e-05) less likely to respond to TAM treatment (TCGA). (H) ER+ patients treated with TAM who have high 14-3-3τ expression (red) are significantly (P = 0.007) more likely to relapse than patients with low 14-3-3τ expression (black) (Loi cohort (28)). (I) High 14-3-3τ expression (red) only significantly impacts relapse-free survival in ER+ patients compared with ER– patients breast cancer (METABRIC). Patients were split by upper tertile of 14-3-3τ expression in their breast cancers. HER2, human epidermal growth factor receptor 2; HR, hazard ratio; RNA-seq, RNA sequencing.
We next queried TCGA database to identify clinically relevant factors associated with high 14-3-3τ expression. We found high 14-3-3τ expression is very significantly correlated to low ERα66 expression in breast cancer patients (Fig. 1F). We then analyzed ER+ breast tumor response to TAM in the context of high 14-3-3τ in TCGA. The nonresponders (n = 126) had significantly higher 14-3-3τ expression than the treatment responders (n = 744) (Fig. 1G). The association between high 14-3-3τ expression and endocrine therapy failure in ER+ breast cancer can be observed in two additional cohorts, Loi cohort (28) (Fig. 1H) and METABRIC cohort (29) (Fig. 1 I, Left). The association of 14-3-3τ overexpression with poor response to therapy is only seen in endocrine therapy, but not in chemotherapy in both the TCGA dataset and METABRIC dataset (SI Appendix, Fig. S1 B and C), suggesting that the effect of 14-3-3τ overexpression is predictive of endocrine failure. We also examined ER– patients in the METABRIC cohort. Contrary to ER+ breast cancer, there is no significant difference in the relapse-free survival of ER– breast cancer patients with high 14-3-3τ expression (Fig. 1 I, Right). The association of high 14-3-3τ expression with shorter relapse-free survival, particularly in ER+ breast cancer, was further supported by a meta-analysis (SI Appendix, Fig. S2) comprising 3,554 breast cancer patients (30). Collectively, these analyses identify an association between high expression of 14-3-3τ and down-regulation of ERα66, as well endocrine therapy failure in breast cancer.
Overexpression of 14-3-3τ in MCF7 Xenografts Induces ERα66 Loss and Epithelial-to-Mesenchymal Transition (EMT) Characteristics.
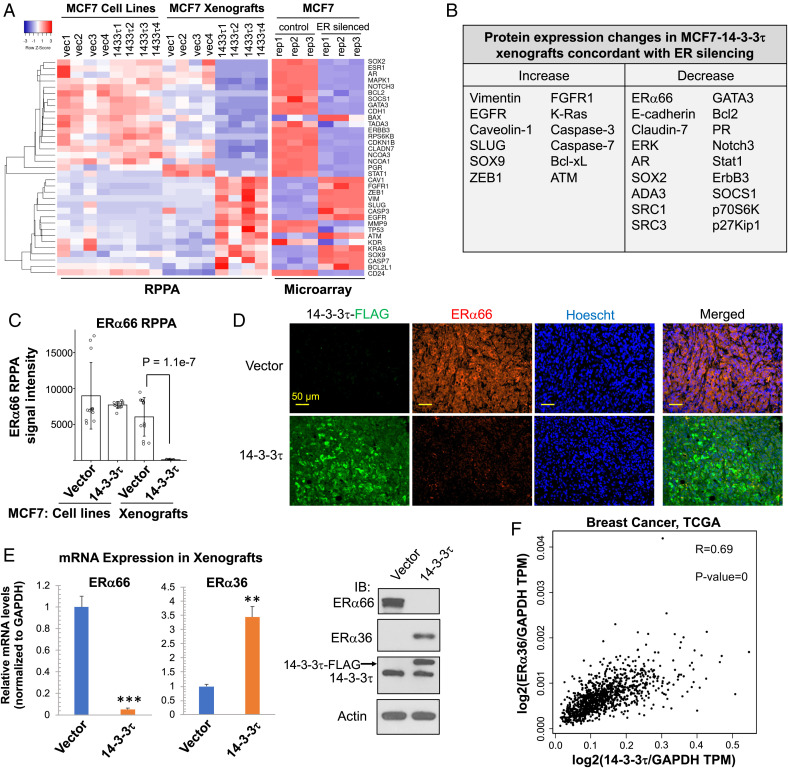
We previously established stable overexpression of 14-3-3τ-FLAG in luminal breast cancer cell line MCF7 and showed that 14-3-3τ promoted MCF7 xenograft progression and metastasis (23). To systemically identify the pathways affected by 14-3-3τ in breast cancer, we performed reverse phase protein array (RPPA) in control and 14-3-3τ-overexpressing MCF7 cells (referred to as MCF7-14-3-3τ hereafter) and primary xenografts. We found no significant changes between MCF7-vector and MCF7-14-3-3τ cell lines but found that all MCF7-14-3-3τ xenografts lost ERα66 expression compared with control xenografts. 14-3-3τ-overexpression also up-regulated EMT marker proteins, such as SOX9, SLUG, caveolin-1, and vimentin, and down-regulated negative regulators of EMT, such as E-cadherin, claudin-7, and GATA3 in MCF7 xenografts (Fig. 2 A, Left). These EMT features were not significantly changed by 14-3-3τ overexpression in two-dimensional (2D) cultured MCF7 or T47D cell lines (SI Appendix, Fig. S3A). In contrast, all 14-3-3τ-overexpressing MCF7 xenografts lost E-cadherin expression (SI Appendix, Fig. S3B). We performed gene set analyses using multiple software programs and found that most of these proteins are regulated by ERα66. We also analyzed an Affymetrix microarray database GSE27473 performed in ERα-depleted MCF7 cells (31), which showed that depletion of ERα in MCF7 cells altered the expression of a number of proteins in a manner highly similar to that observed in MCF7-14-3-3τ xenografts (Fig. 2 A, Right) and also induced EMT-like phenotypes. The protein expression changes in MCF7-14-3-3τ xenografts that are concordant with changes induced by ERα66 silencing are summarized in Fig. 2B. These data suggest that down-regulation of ERα66 (Fig. 2C) is the key event in MCF7-14-3-3τ xenografts.
Fig. 2.
14-3-3τ overexpression down-regulates ERα66, but up-regulates ERα36 and induces EMT changes in MCF7 xenografts. (A, Left) Heatmap of RPPA analyses of MCF7-vector and MCF7-14-3-3τ cell lines (n = 4 biological replicates per group) and xenografts (n = 4 xenografts per group). Right: Heatmap of microarray analysis of control MCF7 and ERα66-silenced MCF7 cells (n = 3 biological replicates per cell line). The data were extracted from GSE27473. (B) Summary of RPPA protein changes exclusively in 14-3-3τ xenografts relevant to breast cancer progression and EMT and concordant to changes induced by ERα66 silencing. (C) RPPA data of ERα66 protein expression. Data are represented as means ± SD. N = 12 per group (three RPPA technical replicates per biological sample, four biological replicates per group). (D) Representative immunohistochemistry images of vector and 14-3-3τ xenograft samples illustrating ERα66 loss in 14-3-3τ tumors. Imaged at 20X. (Scale bar, 50 μm.) (E) Representative MCF7 vector and 14-3-3τ xenograft mRNA and lysate analyzed by RT-qPCR and Western blot demonstrating significant loss of ERα66 and induction of ERα36. Data are represented as means ± SEM, n = 3 xenografts from each group; **P < 0.01; ***P < 0.001 (two-tailed t test). IB, immunoblotting. The individual data from each xenograft are presented in SI Appendix, Fig. S4. (F) A strong, positive correlation (R = 0.69, Pearson) exists between 14-3-3τ and ERα36 gene expression in breast cancer TCGA dataset (n = 1085). Gene expression is normalized to GAPDH. TPM, transcripts per million.
To validate these RPPA results, we confirmed the loss of ERα66 in MCF7-14-3-3τ xenografts with immunohistochemistry, staining for ERα66 and 14-3-3τ-FLAG (Fig. 2D). Additionally, we performed Western blot analysis and RT-qPCR to confirm ERα66 protein was completely lost in all MCF7-14-3-3τ xenografts (Fig. 2E and SI Appendix, Fig. S4A). Interestingly, we also discovered induction of ERα36 expression exclusively in MCF7-14-3-3τ xenografts (Fig. 2E and SI Appendix, Fig. S4B). We examined a TCGA breast cancer dataset and found a strong, positive correlation (R = 0.69) between 14-3-3τ and ERα36 gene expression (Fig. 2F). Together, these data demonstrate that 14-3-3τ overexpression is able to drive ERα loss and EMT induction in luminal breast cancer in vivo and suggest that 14-3-3τ promotes ERα36 expression.
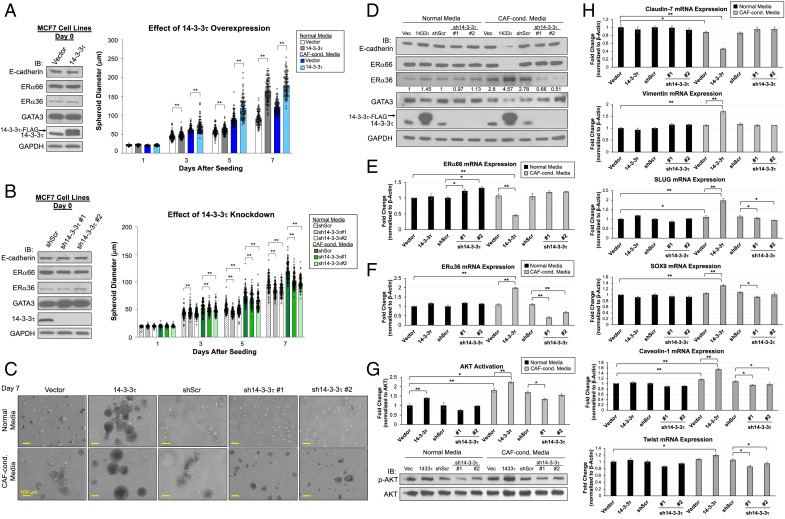
14-3-3τ-Driven ERα66 Loss and EMT Induction Can Be Recapitulated in Three-Dimensional (3D) Spheroid Model.
As ERα expression was not spontaneously diminished by 14-3-3τ overexpression in 2D cultured MCF7 cells, we sought to recapitulate the tumor microenvironment (TME) of xenografts by recreating an in vitro TME that facilitates 3D growth and mimics physiological signaling pathways. To address these requirements, we used Matrigel to simulate the extracellular structure of the TME and supplemented the spheroid culture media (normal media [NM]) with cancer-associated fibroblasts (CAF)-conditioned media (CCM) (SI Appendix, Fig. S5A). We established two MCF7-14-3-3τ knockdown (KD) lines using short hairpin RNA (shRNA), sh14-3–3τ#1 and #2, as well as a scrambled control shRNA (shScr) as previously described (23). On day 0, we plated MCF7 vector, 14-3-3τ overexpression, shScr, and sh14-3–3τ#1 and #2 cell lines onto six-well plates coated with lactose dehydrogenase-elevating virus (LDEV)-free Matrigel Matrix and grew in NM or CCM for 8 d (SI Appendix, Fig. S5B). Spheroid diameters were measured every other day, and diameter significance was calculated through a Student t test within each media group to assess 14-3-3τ impact on spheroid growth. Average diameter measurements by day are shown in SI Appendix, Fig. S5C. Compared with NM, CCM facilitated spheroid growth. 14-3-3τ overexpression further promoted the growth of MCF7 spheroids, as spheroid diameter increased by 1.5-fold (Fig. 3 A, Right and C), whereas 14-3-3τ depletion inhibited growth (Fig. 3 B, Right and C). Moreover, 14-3-3τ overexpression was able to induce ERα36 and greatly repress ERα66 expression in CAF-conditioned spheroid culture at both protein (Fig. 3D) and mRNA levels (Fig. 3 E and F). We also observed the induction of EMT features exclusively in MCF7-14-3-3τ spheroids grown in CCM (referred to as 14-3-3τ-CCM spheroids hereafter). These spheroids demonstrated the reduction of epithelial markers E-cadherin, GATA3, and claudin-7 (Fig. 3 D and H) and induction of mesenchymal markers vimentin, Twist, SOX9, SLUG, and caveolin-1 (Fig. 3H). Importantly, these changes were not seen in spheroid culture without CCM or in cell lines at time of seeding on day 0 (Fig. 3 A and B, Left). Conversely, depletion of 14-3-3τ reduced the ERα36 mRNA levels in CAF-conditioned spheroid culture (Fig. 3F). We also found CCM enhanced AKT activation, which was further promoted by 14-3-3τ overexpression, but inhibited by 14-3-3τ depletion (Fig. 3G). This is consistent with a role for 14-3-3τ in activation of the Jak/PI3K/AKT pathway (32). Overall, 14-3-3τ-CCM spheroids display characteristics that are consistent with our xenograft data and allow us to study the mechanisms involved in 14-3-3τ-driven ERα66 loss and EMT induction, specifically the relationship between 14-3-3τ and ERα36.
Fig. 3.
Spheroid model of 14-3-3τ-driven ERα66 loss and EMT induction using CAF-conditioned media. (A, Left) Western blot analyses of MCF7 cell lysates seeded in 3D model on day 0. IB, immunoblotting. Right: Spheroid growth curves of MCF7 vector and 14-3-3τ grown for 8 d in normal media (NM) or CAF-conditioned media (CCM). Graph represents average spheroid diameter (n = 103) measured every other day. All cell lines grew significantly larger in CCM compared with NM with 14-3-3τ-CCM growing the largest. *P < 0.05, **P < 0.01. (B, Left) Western blot analyses of MCF7 cell lysates seeded in 3D model on day 0. Right: Spheroid growth curves of MCF7 shScr and 14-3-3τKDs grown for 8 d in NM or CCM. Graph represents average spheroid diameter (n = 103) measured every other day. All cell lines grew significantly larger in CCM compared with NM, with KD lines growing the smallest. *P < 0.05, **P < 0.01. (C) Representative images of spheroids from all cell lines on day 7 taken at 10X magnification. (Scale bar, 100 μm.) (D) Western blot analyses of spheroids grown in NM or CCM harvested at day 8. 14-3-3τ-CCM cells had reduction of ERα66, GATA3, and E-cadherin and an induction of ERα36. The relative intensities of ERα36 were quantified (NIH ImageJ software (33)) and normalized to NM control line. (E) ERα66 mRNA expression was significantly decreased upon 14-3-3τ overexpression when grown in CCM, while sh14-3–3τ increased its expression. (F) ERα36 expression was significantly increased in 14-3-3τ-CCM spheroids while sh14-3–3τ-CCM spheroids significantly hindered its expression. (G) AKT activity was significantly increased in all spheroids grown in CCM compared with NM with highest activity in 14-3-3τ-CCM spheroids. Spheroids with 14-3-3τ depletion grown in CCM had reduced AKT activation compared with shScr-CCM. AKT activation was determined by p-AKT protein quantification (NIH ImageJ software) normalized to AKT and relative to respective control line. (H) Genes that demonstrate EMT features are most significantly changed in 14-3-3τ-CCM spheroids, including significant down-regulation in epithelial marker claudin-7 and induction of mesenchymal markers (vimentin, SLUG, SOX9, caveolin-1, and Twist). (E–H) Data shown are mean ± SD, (n = 3 biological replicates); *P < 0.05, **P < 0.01 (two-tailed t test).
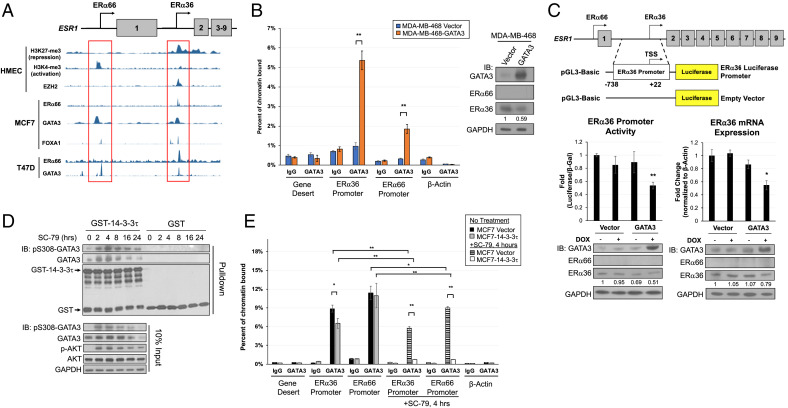
AKT Activation Induces the Binding of 14-3-3τ to GATA3, Leading to the Dissociation of GATA3 from the ERα36 Promoter.
Both our xenograft model and 3D spheroid model demonstrate a role for 14-3-3τ in the regulation of ERα36 expression. We next sought to elucidate the specific mechanism. In human mammary epithelial cells, the ERα66 promoter is marked by active histone markers, such as H3K4me3. On the contrary, the ERα36 promoter is in a repressive state as marked by H3K27me3 and is bound by H3K27 methylase EZH2 (Fig. 4A). To identify potential regulatory elements, we began by examining MCF7 and T47D cell lines in the Encyclopedia of DNA Elements (ENCODE) database looking for TFs that bind either or both ERα66 and ERα36 promoter regions. Indeed, we found several TFs that bind these regions, including ERα66, FOXA1, and GATA3 (Fig. 4A). It is well documented that ERα66/FOXA1/GATA3 form a transcriptional complex that promotes luminal cell identity genes (34, 35). Because ERα36 is characteristic of more basal-like breast cancer cells and antagonistic to ERα66, we hypothesized one of these TFs could bind ERα36 region to repress its transcription in luminal cell lines MCF7 and T47D. Because we observed significantly reduced GATA3 expression in both 14-3-3τ xenograft and CCM spheroids and it had the most robust, predicted peaks at both promoter sites in ENCODE, we focused on this TF.
Fig. 4.
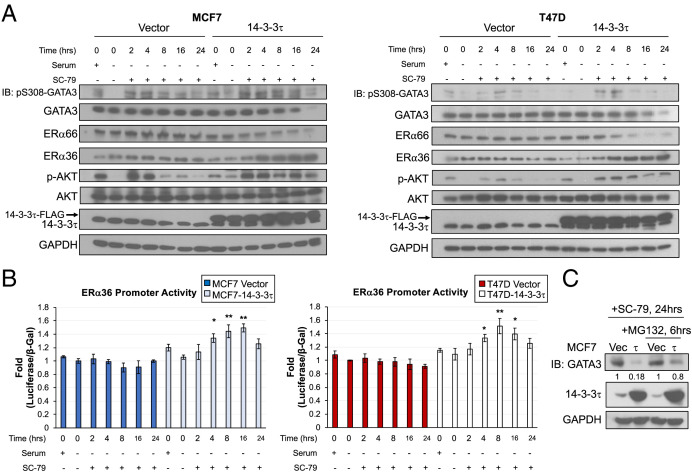
14-3-3τ binds GATA3 after AKT activation, and this interaction dissociates GATA3 from the ERα36 promoter. (A) ENCODE data show ERα66 and ERα36 promoter regions exhibit corresponding histone marks for activation or repression, respectively, in human mammary epithelial cells (HMEC). In MCF7 and T47D cell lines, several transcription factors bind to ERα66 and ERα36 promoter regions. (B) ChIP assay demonstrates the binding of GATA3 to both ERα36 and ERα66 promoter regions in MDA-MB-468 cells overexpressing DOX-inducible GATA3 following 1 μM DOX treatment for 48 h. Western blot confirms DOX-inducible GATA3 overexpression and reduction of ERα36 protein. ERα36 protein expression was quantified (NIH ImageJ software), normalized to GAPDH and relative to vector. Data represent mean ± SD, (n = 3 biological replicates); *P < 0.05, **P < 0.01 (two-tailed t test). IB, immunoblotting. (C) Schematic of luciferase reporter construct for activity at the ERα36 promoter region. GATA3 induction in MDA-MB-468 reduces ERα36 promoter activity, transcript, and protein when treated with 1 μM of DOX for 48 h. ERα36 protein expression was quantified (NIH ImageJ software), normalized to GAPDH and relative to vector/no DOX. Data represent mean ± SD, (n = 3 biological replicates); *P < 0.05, **P < 0.01 (two-tailed t test). (D) AKT phosphorylates GATA3 at S308 and creates a 14-3-3τ-binding motif. 14-3-3τ binds GATA3 after activation of AKT with 10 μM SC-79; the interaction of 14-3-3τ with GATA3 peaks at 4 h after treatment. (E) ChIP assay demonstrates significant binding of GATA3 to both ERα36 and ERα66 promoter regions in MCF7 vector and 14-3-3τ cells. GATA3 binding to both promoters is significantly eliminated in MCF7-14-3-3τ cells treated with SC-79 at 4 h. Data shown are mean ± SD of a representative experiment (n = 6 technical replicates); *P < 0.05, **P < 0.01 (two-tailed t test).
To test whether GATA3 binds the ERα36 promoter, we established stable MDA-MB-468 cell lines, which express exogenous GATA3 in a doxycycline-inducible manner and performed chromatin immunoprecipitation (ChIP) assay. The MDA-MB-468 triple-negative breast cancer (TNBC) cell line expresses low levels of GATA3 and high levels of ERα36 compared with luminal cell lines, making it an ideal system to study the relationship between GATA3 and ERα36 (36, 37). Indeed, GATA3 bound both ERα36 and ERα66 promoters (Fig. 4B). Correspondingly, ERα36 protein levels were reduced when GATA3 was overexpressed (Fig. 4 B, Right). We did not see an increase in ERα66 protein levels by GATA3 overexpression. This is likely due to DNA methylation at this locus, a common characteristic in TNBC (38, 39). Indeed, the promoter of ESR1 gene in MDA-MB-468 is hypermethylated according to the Cancer Cell Line Encyclopedia (CCLE) database (40).
We next performed a luciferase reporter assay to measure the activity of the ERα36 promoter (–738bp to +22bp of the transcription start site (TSS)) (Fig. 4 C, Upper Top). We found GATA3 overexpression significantly decreased the transcriptional activity and mRNA expression of ERα36 (Fig. 4C). Together, these data support our hypothesis that GATA3 binds to the ERα36 promoter and represses its activity.
Next, we determined whether 14-3-3τ could interact with GATA3. 14-3-3τ binds its partners through highly conserved phosphoserine binding motifs (17, 18). Hosokawa et al. recently discovered that AKT-mediated GATA3 phosphorylation resulted in the loss of GATA3-controlled transcriptional repression (41). Since this Ser-308 phosphorylation creates a 14-3-3τ consensus binding site, we next examined this potential interaction. We first starved MCF7-14-3-3τ cells in serum-free condition for 24 h and then treated them with 10 μM SC-79, an AKT activator (42), for various times. We next performed glutathione S-transferase (GST)-pulldown assay by incubating the cell lysates with GST or GST-14-3-3τ (Fig. 4D). Indeed, 14-3-3τ was able to bind and pull down S308-phosphorylated GATA3 with peak interaction at 4 h after treatment.
To further explore the transcriptional consequences of 14-3-3τ-GATA3 interaction, we performed a ChIP assay to determine the GATA3 binding to the ERα36 or ERα66 promoter regions in control or 14-3-3τ-overexpressing MCF7 cells with or without SC-79 treatment for 4 h. Indeed, 14-3-3τ overexpression decreased GATA3 binding to the ERα36 promoter under normal growth conditions (Fig. 4E). The effect was further aggravated upon treatment with SC-79. In fact, the binding of GATA3 to both ERα36 and ERα66 promoters was almost completely inhibited in SC-79-treated MCF7-14-3-3τ cells. Together, these data demonstrate that 14-3-3τ overexpression inhibits ERα promoter occupancy by GATA3, particularly upon AKT activation.
14-3-3τ Facilitates GATA3 Degradation Resulting in ERα36 Transcriptional Activation.
We next examined how the interaction between 14-3-3τ and GATA3 impacts ERα and ERα36 expression. After serum starvation for 24 h, vector and 14-3-3τ-overexpressing MCF7 and T47D cell lines were treated with SC-79 for various times, followed by Western blot analysis. We found that the levels of GATA3 and ERα66 were not altered in vector control cells, but were gradually decreased in 14-3-3τ-overexpressing cells (Fig. 5A). Conversely, the levels of ERα36 were gradually increased following treatment in 14-3-3τ-overexpressing cells, but not in vector control cells. To further explore ERα36 transcriptional activity after AKT activation, the SC-79-treated vector and 14-3-3τ-overexpressing MCF7 and T47D cells were subjected to the ERα36 promoter-driven luciferase reporter assay. While the ERα36 promoter activity was not altered by SC-79 treatment in the vector cells, it was significantly increased in 14-3-3τ-overexpressing cell lines after 4-, 8-, and 16-h treatment (Fig. 5B). To understand how GATA3 expression was reduced by SC-79 treatment in 14-3-3τ-overexpressing cells, we treated these cells with MG132, a proteasome inhibitor, for 6 h before harvesting (Fig. 5C). We found GATA3 protein levels were partially recovered by MG132 treatment, indicating that SC-79 promotes GATA3 degradation through the proteasomal pathway. We also observed GATA3 mRNA decreased at 24 h in MCF7-14-3-3τ cells treated with SC-79 (SI Appendix, Fig. S6), which could be due to the fact that GATA3 positively autoregulates its own expression (43).
Fig. 5.
14-3-3τ facilitates GATA3 degradation resulting in ERα36 transcriptional activation. (A) Activation of AKT with 10 μM SC-79 treatment decreased the levels of GATA3 and ERα66, but increased ERα36 levels in 14-3-3τ overexpressing MCF7 and T47D cells; however, this effect was not observed in vector control cells. IB, immunoblotting. (B) The transcriptional activity of ERα36 was enhanced by SC-79 treatment in 14-3-3τ-overexpressing MCF7 and T47D cells, but not in vector control cells. Data represent mean ± SD, (n = 3 biological replicates); *P < 0.05, **P < 0.01 (two-tailed t test). (C) SC-79 treatment for 24 h decreased the levels of GATA3 in MCF7-14-3-3τ cells; however, this effect was partially rescued by treatment with 10 μM MG132 proteasome inhibitor. GATA3 was quantified (NIH ImageJ software), normalized to GAPDH, and the fold change was calculated relative to that in vector control cells.
Collectively, these experiments demonstrate that 14-3-3τ binds AKT-phosphorylated GATA3. This interaction leads to the inhibition of GATA3 recruitment to ERα promoters causing down-regulation of ERα66, and a corresponding increase in ERα36 expression. The inhibition of GATA3 activity by 14-3-3τ is at least in part due to GATA3 protein degradation.
14-3-3τ Facilitates the Development of TAM-Resistant Characteristics.
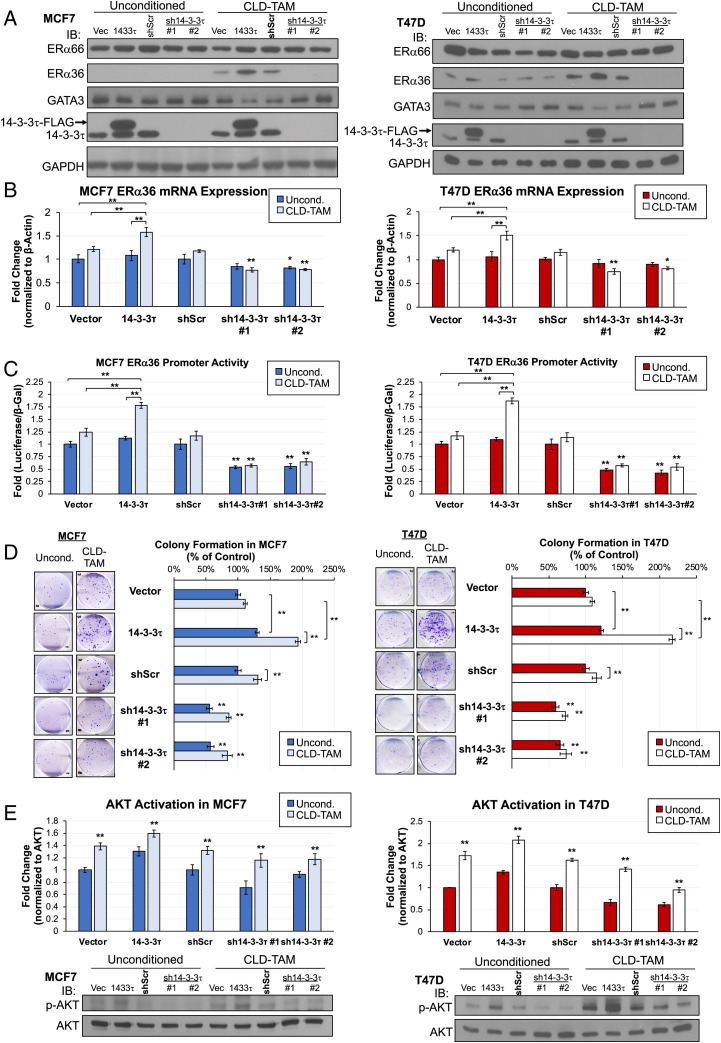
Previously, we showed that transient overexpression of 14-3-3τ in MCF7 to a similar level of some primary breast tumors inhibited the response to TAM (22). On the contrary, depletion of 14-3-3τ enhanced TAM response (22). Those data demonstrate a role for 14-3-3τ in modulating the TAM response. Since chronic treatment of TAM (over 6 mo) can induce ERα36 and down-regulate ERα, and result in TAMR ER+ breast cancer cells (10), we investigated a role for 14-3-3τ in the induction of ERα36 during this process. Previous methodology to generate TAMR cells required treating ER+ cell lines with slowly increasing molarities of TAM for 6–12 mo (44). To determine the contribution of 14-3-3τ during TAMR development, we modified this methodology by treating vector control, 14-3-3τ-overexpressing or -depleted MCF7 or T47D cells with 1 μM TAM for a shortened period of 80 d. We name this treatment “continuous low-dose” (CLD)-TAM treatment.
We found that CLD-TAM modestly induced ERα36 expression, which was augmented by 14-3-3τ overexpression in both MCF7 and T47D cell lines (Fig. 6A). We also observed a marked decrease of GATA3 expression in these cell lines. On the contrary, depletion of 14-3-3τ inhibited the induction of ERα36 by CLD-TAM. Down-regulation of GATA3 expression by CLD-TAM was also rescued by 14-3-3τ depletion. ERα expression remained unchanged in these cell lines. This is not surprising, as ERα loss is a hallmark of TAM resistance, but that loss requires longer TAM treatment as demonstrated by other groups (45, 46). Additionally, GATA3 is still expressed in these cells and is still able to promote ERα66 expression. We predict longer treatment of these cells will eventually create the canonical TAMR phenotype hallmarked by decreased ERα66.
Fig. 6.
Continuous low-dose tamoxifen (CLD-TAM) treatment of luminal breast cancer cells with 14-3-3τ overexpression reduces GATA3 expression while inducing ERα36 expression and significantly increasing resistance to TAM. (A) CLD-TAM treatment reduced GATA3 expression with a corresponding ERα36 protein induction while did not affect ERα66 levels in 14-3-3τ overexpressing MCF7 or T47D cells. IB, immunoblotting. (B) ERα36 mRNA expression was significantly increased in CLD-TAM-treated MCF7 or T47D cells with 14-3-3τ overexpression, while 14-3-3τ knockdown cells showed significant reduction of ERα36 transcript in both treatment groups compared with scrambled control. Data represent mean ± SD, (n = 4 biological replicates); *P < 0.05, **P < 0.01 (two-tailed t test). (C) 14-3-3τ promoted the induction of ERα36 promoter activity by CLD-TAM, while 14-3-3τ knockdown significantly inhibited the activity. Data represent mean ± SD, (n = 4 biological replicates); *P < 0.05, **P < 0.01 (two-tailed t test). (D) All CLD-TAM-conditioned cells showed better cell viability than unconditioned cells when treated with 5 μM TAM for 14 d. CLD-TAM-treated 14-3-3τ cells had the most resistance to TAM treatment while 14-3-3τ knockdown cells remained the most sensitive. (E) 14-3-3τ modulated AKT activation after CLD-TAM treatment. D and E: Data represent mean ± SD, (n = 3 biological replicates); *P < 0.05, **P < 0.01 (two-tailed t test).
We next explored ERα36 mRNA and transcriptional activity in CLD-TAM-treated cells. Consistent with protein analysis, 14-3-3τ overexpression potentiated ERα36 mRNA induction by CLD-TAM; on the contrary, depletion of 14-3-3τ inhibited ERα36 transcript expression (Fig. 6B). Next, we measured ERα36 transcriptional activity using ERα36 promoter-driven luciferase reporter assay (Fig. 6C). Consistently, CLD-TAM-treated 14-3-3τ cells showed the highest increase of ERα36 promoter activity; in contrast, this activity was significantly decreased in 14-3-3τ KD cells compared with shScr control treatment group.
We then investigated the impact of CLD-TAM conditioning on TAM response using the colony formation assay (Fig. 6D). When treated with 5 μM TAM, CLD-TAM-conditioned 14-3-3τ cells showed the most significant colony formation, whereas sh14-3–3τ#1 and #2 cells remained the most sensitive to treatment despite prior TAM exposure. Finally, we confirmed significantly increased AKT activation across all CLD-TAM cell lines compared with unconditioned lines, which was enhanced by 14-3-3τ overexpression and reduced by 14-3-3τ depletion (Fig. 6E). Together, we conclude that 14-3-3τ overexpression facilitates earlier TAM resistance as seen by a significant induction of ERα36 expression and greater resistance to additional TAM treatment.
KD of ERα36 in MCF7-14-3-3τ 3D Spheroids Rescues ERα Expression and Abrogates EMT.
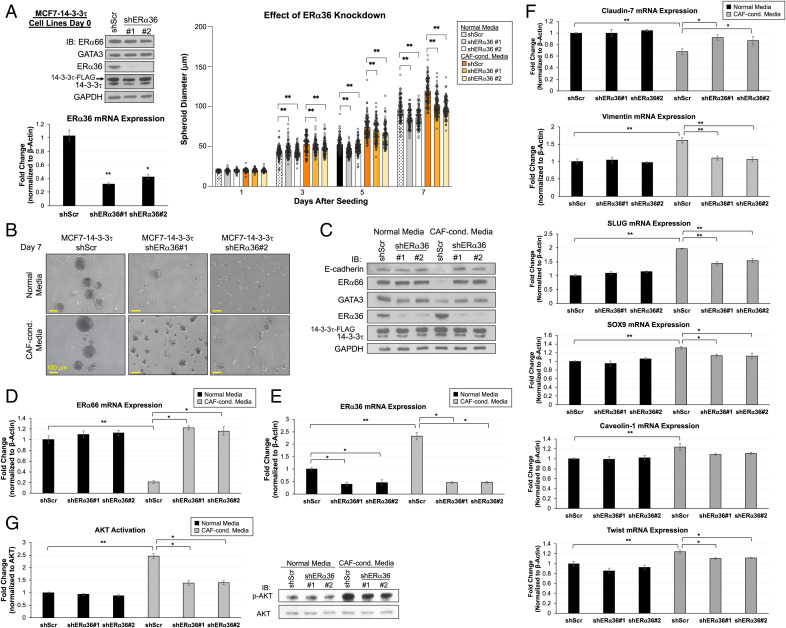
To further establish a role for ERα36 in 14-3-3τ-induced loss of ERα, we repeated a 3D model experiment using MCF-14-3-3τ cells stably expressing one of two ERα36-specific shRNA constructs. ERα36 KD was confirmed by Western blot analysis and qPCR in MCF7-14-3-3τ on day 0 (Fig. 7 A, Left). These two shERα36 constructs specifically depleted the expression of ERα36 without affecting ERα expression in 2D cell culture and in NM (Fig. 7 A, Left and C). The effect of 14-3-3τ on promoting spheroid growth was attenuated by depletion of ERα36 (Fig. 7 A and B and SI Appendix, Fig. S7).
Fig. 7.
Knockdown of ERα36 in MCF7-14-3-3τ grown in 3D model rescues ERα66 expression and abrogates EMT. (A, Right) Spheroid growth curves of MCF7-14-3-3τ stably expressing shScr or shERα36#1 or #2 in normal media (NM) or CAF-conditioned media (CCM). Graph represents average spheroid diameter (n = 103) measured every other day for 7 d. Left : Western blot analyses and ERα36 mRNA expression in MCF7-14-3-3τ stably expressing shScr or shERα36 at time of seeding. *P < 0.05, **P < 0.01. IB, immunoblotting. (B) Representative microscope images of spheroids from all cell lines on day 7 taken at 10X magnification. (Scale bar, 100 μm.) (C) Western blot analyses of spheroids grown in NM or CCM harvested at day 8. MCF7-14-3-3τ-shScr showed reduction of ERα66, GATA3, and E-cadherin expressions and an induction of ERα36 expression in CCM compared with in NM spheroids. Both shERα36 spheroids grown in CCM showed comparable expression of these proteins as those grown in NM. (D) ERα66 mRNA expression was significantly reduced in MCF7-14-3-3τ shScr spheroids grown in CCM but not in any other spheroids. (E) ERα36 mRNA expression was significantly increased in MCF7-14-3-3τ shScr spheroids grown in CCM. (F) Genes that demonstrate EMT features are most significantly changed in MCF7-14-3-3τ shScr spheroids grown in CCM including down-regulation in epithelial marker claudin-7 and induction of mesenchymal markers (vimentin, SLUG, SOX9, caveolin-1, and Twist). Spheroids with depleted ERα36 grown in CCM demonstrated significant induction of claudin-7 and reduction of mesenchymal markers compared with MCF7-14-3-3τ shScr-CCM spheroids. (G) AKT activation was greater in all spheroids grown in CCM compared with NM, with the largest activity in MCF7-14-3-3τ shScr-CCM spheroids. The activation of AKT was significantly reduced by ERα36 depletion only in spheroids grown in CCM. AKT activation was determined by p-AKT protein quantification (NIH ImageJ software) normalized to AKT and relative to shScr/NM control. D–G: Data represent mean ± SD, (n = 3 biological replicates); *P < 0.05, **P < 0.01 (two-tailed t test).
We next harvested the spheroids on day 8 and performed both protein and mRNA analyses. The expression of ERα66 and GATA3 in all of the spheroids grown in NM was not significantly altered (Fig. 7C). In contrast, there was a significant loss of ERα66 protein and transcript along with a clear induction of ERα36 protein and transcript in MCF7-14-3-3τ-shScr spheroids grown in CCM (Fig. 7 C–E). We again observed the development of EMT characteristics in MCF7-14-3-3τ-shScr spheroids grown in CCM. These spheroids demonstrated the greatest reduction of epithelial markers E-cadherin, GATA3, and claudin-7 and the largest induction of mesenchymal markers vimentin, SLUG, SOX9, caveolin-1, and Twist (Fig. 7 C and F). The depletion of ERα36 in CCM spheroids mitigated these changes. Importantly, KD of ERα36 in MCF7-14-3-3τ spheroids grown in CCM almost completely rescued ERα66, GATA3, and E-cadherin expressions (Fig. 7 C and D) and reduced the increase of mesenchymal markers compared with MCF7-14-3-3τ-shScr-CCM (Fig. 7F). These findings demonstrate a role of ERα36 induction for the transition to more mesenchymal-like cells in 14-3-3τ-overexpressing breast cancer cells.
We also investigated AKT activation in the context of ERα36 KD. Consistent with our previous findings, we found AKT activity was increased in all spheroids grown in CCM compared with those grown in NM (Fig. 7G). We found 14-3-3τ-shERα36#1 and #2 CCM spheroids had significantly lower AKT activation compared with 14-3-3τ-shScr-CCM spheroids. This is consistent with the literature in that ERα36 contributes to the propagation of AKT activation (47–49) and, in this case, depletion of ERα36 mitigates the effect of 14-3-3τ on AKT activation. Together, these results demonstrate that silencing of ERα36 in the context of 14-3-3τ overexpression is sufficient to rescue ER+ phenotype and reduce the emergence of EMT characteristics.
Discussion
The evolution of breast cancer from ER+ to ER– in patients not only takes a long time, but likely requires many steps and involves a sequence of different molecular events. Here, we develop xenograft and 3D breast cancer spheroid models to recapitulate the process of ER loss in breast cancer. Since our data demonstrate that up-regulation of 14-3-3τ alone in ER+ breast cancer cells can convert ER+ to ER– tumors in vivo, we argue that the 14-3-3τ/GATA3/ERα36 pathway uncovered in this study plays a pivotal, if not the first, step in this evolution.
Through the combined approaches of RPPA and microarray bioinformatics, we discovered the switch of ERα expression (induction of ERα36 and down-regulation of ERα66) as the key event caused by 14-3-3τ overexpression in vivo, which is found in about 60% of human breast cancer. ERα36 has recently garnered interest for its role in antiestrogen resistance and corresponding poor patient outcomes (8, 9, 12). Despite its clinical relevance, ERα36’s exact regulation and mechanisms of action remain mostly undiscovered. Here, we elucidate a mechanism driven by 14-3-3τ to drive loss of ERα66 and induce ERα36 to generate a more aggressive, less TAM-sensitive breast cancer.
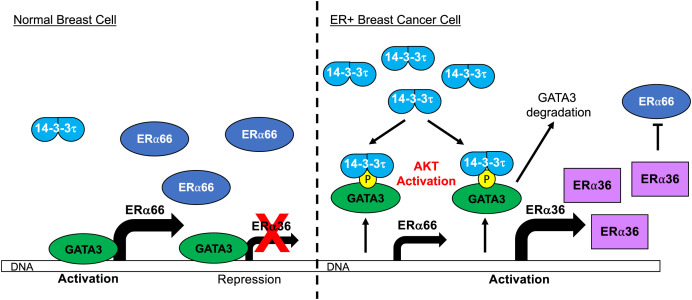
Based on our data, we propose the following mechanism for 14-3-3τ-driven ERα66 loss. Under normal breast cell conditions, we demonstrate GATA3 binds both the ERα66 and ERα36 promoter regions. Other groups have already demonstrated that GATA3 is a key TF for luminal cell identity genes and therefore promotes ERα66 expression. Our data add to this understanding, as we found GATA3 also binds the ERα36 promoter but represses transcription of this antagonistic isoform, thus further maintaining luminal cell characteristics (Fig. 8, Left). The differential activity of GATA3 on the ERα66 and ERα36 promoters is likely due to the different epigenetic modifications on these two promoters (Fig. 4A). GATA3 can function as either a transcriptional activator or repressor, depending on the epigenetic status of the promoters and whether GATA3 works in concert with coactivators or repressors (50). Transcriptional coactivator p300 is recruited along with GATA3 to the ERα66 promoter for ERα66 activation (43). On the contrary, the ERα36 promoter is bound by H3K27 methylase EZH2 and is in repressive state. The repression of ERα36 by GATA3 is consistent with previous reports that demonstrated GATA3 represses genes associated with basal-like pathogenesis in the mammary gland (51, 52). Thus, GATA3 acts as a transcriptional activator for ERα66 to support luminal cell identity, but as a transcriptional repressor for ERα36 to inhibit basal-like characteristics.
Fig. 8.
Schematic model of the molecular mechanism controlling ERα36 transcription in normal versus luminal breast cancer cells. Under normal cell conditions, GATA3 binds both ERα66 and ERα36 promoters to promote ERα66 transcription and to repress ERα36 transcription. During the evolution of breast cancer, some ER+ breast cancer may acquire both high 14-3-3τ expression and aberrant AKT activation, either due to intrinsic factors or interactions with TME. These events facilitate GATA3 phosphorylation leading to 14-3-3τ-GATA3 interaction which causes GATA3 to lose transcriptional control of ERα66 and ERα36. ERα36 is now transcribed and antagonizes ERα66 expression leading to down-regulation of ERα66 and more basal-like cell characteristics.
In ER+ breast cancer cells with high 14-3-3τ expression (Fig. 8, Right), we demonstrate aberrant AKT activation, due to stimulation from TME or adaptive response to chronic TAM treatment, leads to GATA3 phosphorylation and creates a consensus 14-3-3 binding motif. 14-3-3τ-GATA3 interaction facilitates the dissociation (and subsequent degradation) of GATA3 from both ER promoter regions such that ERα66 is no longer promoted and ERα36 is no longer repressed. ERα36 can then further antagonize ERα66 expression. The reduction of both ERα66 and GATA3, both epithelial markers, paired with the induction of ERα36 create a more basal-like breast cancer cell that is less sensitive to endocrine therapy.
We also elucidate the regulation of ERα36 by GATA3. GATA3 has been shown to promote luminal cell identity by participating in a positive and reciprocal regulatory loop with ERα66 (43, 53). Based on the predicted GATA3 binding in the ENCODE database in Fig. 4A, we hypothesized GATA3 binds both ERα66 and ERα36 promoter regions to promote the former, a luminal protein, and repress the latter, a basal-like protein. We confirmed GATA3 binds these promoters through ChIP assays (Fig. 4 B and E) and confirmed its dual regulation through luciferase reporter assays (Figs. 4C and 5B) that demonstrated GATA3 not only represses ERα36 transcription, but also loses that repressive control when bound by 14-3-3τ. We see the downstream consequences of this as ERα36 transcript and protein levels increase. This 14-3-3τ-GATA3 interaction and its resulting transcriptional and protein changes are dependent on AKT phosphorylation of GATA3 on S308, an event that not only produces a consensus binding motif for 14-3-3τ but has previously been shown to change GATA3 transcriptional regulation (41).
The aberrant AKT activation that facilitates our proposed mechanism is provided by abnormal growth stimuli from supporting stromal cells in the xenograft TME, CAF-secreted factors in the 3D model, and characteristic of TAMR cells. CAF secretions, such as TGF-β, SDF-1, IL-6/11, and Wnt, have also been shown to influence the metabolism, motility, and characteristics of cancer cells, including the up-regulation of PI3K/AKT pathway (54). CAF-induced AKT activation is supported by the significantly increased levels of p-AKT in spheroids grown in CCM compared with NM (Figs. 3G and 7G). TAMR cells have characteristically high AKT activation (55), which we confirmed in cell lines treated with CLD-TAM, especially in CLD-TAM 14-3-3τ cells, compared with unconditioned cell lines (Fig. 6E). While we do not observe the full TAMR phenotype in our study due to the relatively short time period of TAM exposure, our proposed mechanism contributes to the understanding and clinical relevance of increased ERα36 expression in the development of TAMR tumors.
In addition to these external stimuli leading to abnormal AKT activity, the 14-3-3 family is known to control and propagate AKT signaling in many cancers (32, 56–58). Across our experiments, we consistently observe 14-3-3τ overexpression has the greatest AKT activity, while 14-3-3τKDs have lowered activity. The combination of aberrant AKT activation from extracellular sources and elevated 14-3-3τ are sufficient to allow increased GATA3 phosphorylation and interaction with 14-3-3τ causing GATA3 to lose transcriptional control of ERα66 and ERα36. A large proteomic and transcriptome profiling study found AKT pathway activation is associated with lowered ERα66 expression in ER+ breast cancers (59), further supporting our proposed mechanism. Additionally, many other groups have demonstrated ERα36 propagates AKT activation, thus bolstering AKT activation (47–49). This activation causes the further phosphorylation of GATA3 and its transcriptional consequences in the context of high 14-3-3τ expression. It is possible that other factors secreted by CAF in TME also contribute to permanent establishment of EMT-like, ER– breast cancer.
In sum, our study identifies a role for 14-3-3τ in driving ERα66 loss in breast cancer and elucidates a pathway of 14-3-3τ/GATA3/ERα36 in conjunction with AKT activation to promote ERα66 loss in ER+ breast cancer. The 3D model we developed here also allows for the further investigation of other contributing mechanisms to this phenotype. Additionally, we can also use this 3D model to identify targets or pathways that are amenable for therapeutic intervention to prevent the switch of ERα expression and the development of endocrine therapy resistance.
Materials and Methods
Spheroid Culture.
Spheroid medium was prepared by supplementing 500 mL Ham’s F-12 Nutrient Mixture (Millipore Sigma) with 4.5 g/L L-glutamine, 2.5 mg insulin, 500 μg hydrocortisone, 500 ng epidermal growth factor, and 50 mL bovine calf serum. CAF-CCM was collected daily. Six-well plates were coated evenly with 300 μL of Matrigel Matrix-LDEV free (Corning) per well and seeded with 5 × 104 cells per well. The medium was changed daily with either 100% spheroid media as described above or 1:1 spheroid media to CAF-CCM. Spheroid growth was monitored using light microscopy. At 10X magnification, the diameters of more than 100 spheroids were measured and averages reported on days 1, 3, 5, and 7 after seeding.
ChIP.
Cells were fixed, harvested, and nuclei were isolated as previously described (60). Nuclei were suspended in shearing buffer (0.1 mM ethylenediaminetetraacetic acid (EDTA), pH 8.0, 0.1 mM ethylene glycol tetraacetic acid (EGTA), 10 mM Tris⋅HCl, pH 6.8, 100 mM NaCl, 0.1% sodium deoxycholate, 0.5% N-lauroylsarcosine, protease inhibitors) and sonicated until an average fragment size of 800 bp was reached. Chromatin concentrations were calculated based on processed aliquots, and equal chromatin amounts were used in each subsequent immunoprecipitation. All ChIP, washes, and downstream processing were performed as described previously (60). Immunoprecipitation samples were incubated with 4 μg GATA3 antibody or 4 μg immunoglobulin G antibody control and precipitated with protein G magnetic dynabeads (Invitrogen). Samples were analyzed via qPCR with ChIP primer sequences as indicated in SI Appendix, Table 3. Primer sequence for Gene Desert was designed by Active Motif.
RPPA Analysis.
The RPPA was performed and analyzed by Antibody-Based Proteomics Core Facility at Baylor College of Medicine as previously described (61). Samples were probed with 183 antibodies.
Cell culture, immunofluorescence staining, mesenchymal stem cells, plasmid construction, virus production and stable cell line generation, TAM treatment, luciferase reporter assay, Western blotting, GST-pulldown assay, proteasome degradation assay, antibodies, RNA extraction and RT-qPCR, colony formation assay, and statistical analysis are described in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
The authors thank Dr. Shiuh-Rong Ho for technical support in cloning and other members in Lin Laboratory for discussion and advice, Drs. Xiang Zhang and Hai Wang for mesenchymal stem cells and methodology on differentiation to cancer-associated fibroblasts, Dr. Xiaoyong Fu for plasmids, Antibody-Based Proteomics Core at Baylor College of Medicine (Dr. Shixia Huang, Cancer Prevention and Research Institute of Texas Core Facility Award RP120092) for reverse phase protein array (RPPA) experiments, and Drs. Kimal Rajapakshe and Cristian Coarfa for RPPA data processing and analyses. We also thank Fannie Lin for valuable discussion and comments. This work was supported by NIH Grants R01CA203824, R01CA100857, and R21CA198041, and Department of Defense Grants W81XWH-18-1-0329 and W81XWH-19-1-0369. L.A.W.G. was supported by NIH T32GM136560, and Y.X. was supported by NIH T32CA174647.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2209211119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and/or supporting information.
References
- 1.Burstein H. J., et al. , Adjuvant endocrine therapy for women with hormone receptor-positive breast cancer: ASCO clinical practice guideline focused update. J. Clin. Oncol. 37, 423–438 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Davies C., et al. ; Adjuvant Tamoxifen: Longer Against Shorter (ATLAS) Collaborative Group, Long-term effects of continuing adjuvant tamoxifen to 10 years versus stopping at 5 years after diagnosis of oestrogen receptor-positive breast cancer: ATLAS, a randomised trial. Lancet 381, 805–816 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rondón-Lagos M., Villegas V. E., Rangel N., Sánchez M. C., Zaphiropoulos P. G., Tamoxifen resistance: Emerging molecular targets. Int. J. Mol. Sci. 17, E1357 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aurilio G., et al. , A meta-analysis of oestrogen receptor, progesterone receptor and human epidermal growth factor receptor 2 discordance between primary breast cancer and metastases. Eur. J. Cancer 50, 277–289 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Wang Z. Y., Yin L., Estrogen receptor alpha-36 (ER-α36): A new player in human breast cancer. Mol. Cell. Endocrinol. 418, 193–206 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Wang Z., et al. , Identification, cloning, and expression of human estrogen receptor-alpha36, a novel variant of human estrogen receptor-alpha66. Biochem. Biophys. Res. Commun. 336, 1023–1027 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Chaudhri R. A., Hadadi A., Lobachev K. S., Schwartz Z., Boyan B. D., Estrogen receptor-alpha 36 mediates the anti-apoptotic effect of estradiol in triple negative breast cancer cells via a membrane-associated mechanism. Biochim. Biophys. Acta 1843, 2796–2806 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zou Y., Ding L., Coleman M., Wang Z., Estrogen receptor-alpha (ER-alpha) suppresses expression of its variant ER-alpha 36. FEBS Lett. 583, 1368–1374 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi L., et al. , Expression of ER-alpha36, a novel variant of estrogen receptor alpha, and resistance to tamoxifen treatment in breast cancer. J. Clin. Oncol. 27, 3423–3429 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li G., et al. , Estrogen receptor-α36 is involved in development of acquired tamoxifen resistance via regulating the growth status switch in breast cancer cells. Mol. Oncol. 7, 611–624 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X., Wang Z. Y., Estrogen receptor-α variant, ER-α36, is involved in tamoxifen resistance and estrogen hypersensitivity. Endocrinology 154, 1990–1998 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Nagel A., et al. , Clinical and biological significance of ESR1 gene alteration and estrogen receptors isoforms expression in breast cancer patients. Int. J. Mol. Sci. 20, E1881 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ojo D., et al. , Factors promoting tamoxifen resistance in breast cancer via stimulating breast cancer stem cell expansion. Curr. Med. Chem. 22, 2360–2374 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Deng H., et al. , ER-α36-mediated rapid estrogen signaling positively regulates ER-positive breast cancer stem/progenitor cells. PLoS One 9, e88034 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng H., et al. , ER-alpha variant ER-alpha36 mediates antiestrogen resistance in ER-positive breast cancer stem/progenitor cells. J. Steroid Biochem. Mol. Biol. 144, 417–426 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Yang X., et al. , Structural basis for protein-protein interactions in the 14-3-3 protein family. Proc. Natl. Acad. Sci. U.S.A. 103, 17237–17242 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muslin A. J., Tanner J. W., Allen P. M., Shaw A. S., Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84, 889–897 (1996). [DOI] [PubMed] [Google Scholar]
- 18.Yaffe M. B., et al. , The structural basis for 14-3-3:phosphopeptide binding specificity. Cell 91, 961–971 (1997). [DOI] [PubMed] [Google Scholar]
- 19.Aitken A., 14-3-3 proteins: A historic overview. Semin. Cancer Biol. 16, 162–172 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Hermeking H., Benzinger A., 14-3-3 proteins in cell cycle regulation. Semin. Cancer Biol. 16, 183–192 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Li N., et al. , Overexpression of 14-3-3θ promotes tumor metastasis and indicates poor prognosis in breast carcinoma. Oncotarget 5, 249–257 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang B., et al. , 14-3-3Tau regulates ubiquitin-independent proteasomal degradation of p21, a novel mechanism of p21 downregulation in breast cancer. Mol. Cell. Biol. 30, 1508–1527 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao Y., et al. , 14-3-3τ promotes breast cancer invasion and metastasis by inhibiting RhoGDIα. Mol. Cell. Biol. 34, 2635–2649 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin D., Brown-Luedi M., Chiquet-Ehrismann R., Tenascin-C signaling through induction of 14-3-3 tau. J. Cell Biol. 160, 171–175 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chou J., Provot S., Werb Z., GATA3 in development and cancer differentiation: Cells GATA have it! J. Cell. Physiol. 222, 42–49 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kouros-Mehr H., Kim J. W., Bechis S. K., Werb Z., GATA-3 and the regulation of the mammary luminal cell fate. Curr. Opin. Cell Biol. 20, 164–170 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coradini D., Boracchi P., Oriana S., Biganzoli E., Ambrogi F., Differential expression of genes involved in the epigenetic regulation of cell identity in normal human mammary cell commitment and differentiation. Chin. J. Cancer 33, 501–510 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loi S., et al. , Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J. Clin. Oncol. 25, 1239–1246 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Curtis C., et al. ; METABRIC Group, The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Györffy B., et al. , An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 123, 725–731 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Al Saleh S., Al Mulla F., Luqmani Y. A., Estrogen receptor silencing induces epithelial to mesenchymal transition in human breast cancer cells. PLoS One 6, e20610 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Y., et al. , 1,3-Dicaffeoylquinic acid targeting 14-3-3 tau suppresses human breast cancer cell proliferation and metastasis through IL6/JAK2/PI3K pathway. Biochem. Pharmacol. 172, 113752 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akbari V., Kallhor M., Akbari M. T., Transcriptome mining of non-BRCA1/A2 and BRCA1/A2 familial breast cancer. J. Cell. Biochem. 120, 575–583 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Kong S. L., Li G., Loh S. L., Sung W. K., Liu E. T., Cellular reprogramming by the conjoint action of ERα, FOXA1, and GATA3 to a ligand-inducible growth state. Mol. Syst. Biol. 7, 526 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu B., et al. , Huaier polysaccharide inhibits the stem-like characteristics of ERα-36high triple negative breast cancer cells via inactivation of the ERα-36 signaling pathway. Int. J. Biol. Sci. 15, 1358–1367 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X., et al. , Estrogen receptor-α36 is involved in icaritin induced growth inhibition of triple-negative breast cancer cells. J. Steroid Biochem. Mol. Biol. 171, 318–327 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Prabhu J. S., et al. , The epigenetic silencing of the estrogen receptor (ER) by hypermethylation of the ESR1 promoter is seen predominantly in triple-negative breast cancers in Indian women. Tumour Biol. 33, 315–323 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Quintas-Granados L. I., et al. , The high methylation level of a novel 151-bp CpG island in the ESR1 gene promoter is associated with a poor breast cancer prognosis. Cancer Cell Int. 21, 649 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nusinow D. P., et al. , Quantitative proteomics of the cancer cell line encyclopedia. Cell 180, 387–402.e16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hosokawa H., et al. , Akt1-mediated Gata3 phosphorylation controls the repression of IFNγ in memory-type Th2 cells. Nat. Commun. 7, 11289 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jo H., et al. , Small molecule-induced cytosolic activation of protein kinase Akt rescues ischemia-elicited neuronal death. Proc. Natl. Acad. Sci. U.S.A. 109, 10581–10586 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eeckhoute J., et al. , Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res. 67, 6477–6483 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Fu X., et al. , FOXA1 overexpression mediates endocrine resistance by altering the ER transcriptome and IL-8 expression in ER-positive breast cancer. Proc. Natl. Acad. Sci. U.S.A. 113, E6600–E6609 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moon S. Y., et al. , Inhibition of STAT3 enhances sensitivity to tamoxifen in tamoxifen-resistant breast cancer cells. BMC Cancer 21, 931 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clarke R., Leonessa F., Welch J. N., Skaar T. C., Cellular and molecular pharmacology of antiestrogen action and resistance. Pharmacol. Rev. 53, 25–71 (2001). [PubMed] [Google Scholar]
- 47.Lin S. L., et al. , ER-alpha36, a variant of ER-alpha, promotes tamoxifen agonist action in endometrial cancer cells via the MAPK/ERK and PI3K/Akt pathways. PLoS One 5, e9013 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mansouri S., Farahmand L., Teymourzadeh A., Majidzadeh-A K., Clinical evidence on the magnitude of change in growth pathway activity in relation to tamoxifen resistance is required. Curr. Cancer Drug Targets 18, 668–676 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Zhao Y., et al. , let-7 microRNAs induce tamoxifen sensitivity by downregulation of estrogen receptor α signaling in breast cancer. Mol. Med. 17, 1233–1241 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yagi R., Zhu J., Paul W. E., An updated view on transcription factor GATA3-mediated regulation of Th1 and Th2 cell differentiation. Int. Immunol. 23, 415–420 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pei X. H., et al. , CDK inhibitor p18(INK4c) is a downstream target of GATA3 and restrains mammary luminal progenitor cell proliferation and tumorigenesis. Cancer Cell 15, 389–401 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tkocz D., et al. , BRCA1 and GATA3 corepress FOXC1 to inhibit the pathogenesis of basal-like breast cancers. Oncogene 31, 3667–3678 (2012). [DOI] [PubMed] [Google Scholar]
- 53.Porras L., Ismail H., Mader S., Positive regulation of estrogen receptor alpha in breast tumorigenesis. Cells 10, 2966 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ziani L., Chouaib S., Thiery J., Alteration of the antitumor immune response by cancer-associated fibroblasts. Front. Immunol. 9, 414 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jordan N. J., Gee J. M., Barrow D., Wakeling A. E., Nicholson R. I., Increased constitutive activity of PKB/Akt in tamoxifen resistant breast cancer MCF-7 cells. Breast Cancer Res. Treat. 87, 167–180 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Gómez-Suárez M., et al. , 14-3-3 Proteins regulate Akt Thr308 phosphorylation in intestinal epithelial cells. Cell Death Differ. 23, 1060–1072 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li J., Xu H., Wang Q., Wang S., Xiong N., 14-3-3ζ promotes gliomas cells invasion by regulating Snail through the PI3K/AKT signaling. Cancer Med. 8, 783–794 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Wu Q., et al. , Overexpression of 14-3-3σ modulates cholangiocarcinoma cell survival by PI3K/Akt signaling. BioMed Res. Int. 2020, 3740418 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59.Creighton C. J., et al. , Proteomic and transcriptomic profiling reveals a link between the PI3K pathway and lower estrogen-receptor (ER) levels and activity in ER+ breast cancer. Breast Cancer Res. 12, R40 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paik J. C., Wang B., Liu K., Lue J. K., Lin W. C., Regulation of E2F1-induced apoptosis by the nucleolar protein RRP1B. J. Biol. Chem. 285, 6348–6363 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee Y. J., et al. , CGRRF1, a growth suppressor, regulates EGFR ubiquitination in breast cancer. Breast Cancer Res. 21, 134 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.