Significance
Upgrading lignin, an underutilized component of biomass, is essential for sustainable biorefining. Biocatalysis has considerable potential for upgrading lignin, but our lack of knowledge of relevant enzymes and pathways has limited its application. Herein, we describe a microbial pathway that catabolizes acetovanillone, a major component of several industrial lignin streams. This pathway is unusual in that it involves phosphorylation and carboxylation before conversion to the intermediate, vanillate, which is degraded via the β-ketoadipate pathway. Importantly, the hydroxyphenylethanone catabolic pathway enables bacterial growth on softwood lignin pretreated by oxidative catalytic fractionation. Overall, these insights greatly facilitate the engineering of bacteria to biocatalytically upgrade lignin.
Keywords: bacterial catabolism, acetovanillone, hydroxyacetophenone, lignin, Rhodococcus
Abstract
Bacterial catabolic pathways have considerable potential as industrial biocatalysts for the valorization of lignin, a major component of plant-derived biomass. Here, we describe a pathway responsible for the catabolism of acetovanillone, a major component of several industrial lignin streams. Rhodococcus rhodochrous GD02 was previously isolated for growth on acetovanillone. A high-quality genome sequence of GD02 was generated. Transcriptomic analyses revealed a cluster of eight genes up-regulated during growth on acetovanillone and 4-hydroxyacetophenone, as well as a two-gene cluster up-regulated during growth on acetophenone. Bioinformatic analyses predicted that the hydroxyphenylethanone (Hpe) pathway proceeds via phosphorylation and carboxylation, before β-elimination yields vanillate from acetovanillone or 4-hydroxybenzoate from 4-hydroxyacetophenone. Consistent with this prediction, the kinase, HpeHI, phosphorylated acetovanillone and 4-hydroxyacetophenone. Furthermore, HpeCBA, a biotin-dependent enzyme, catalyzed the ATP-dependent carboxylation of 4-phospho-acetovanillone but not acetovanillone. The carboxylase’s specificity for 4-phospho-acetophenone (kcat/KM = 34 ± 2 mM−1 s−1) was approximately an order of magnitude higher than for 4-phospho-acetovanillone. HpeD catalyzed the efficient dephosphorylation of the carboxylated products. GD02 grew on a preparation of pine lignin produced by oxidative catalytic fractionation, depleting all of the acetovanillone, vanillin, and vanillate. Genomic and metagenomic searches indicated that the Hpe pathway occurs in a relatively small number of bacteria. This study facilitates the design of bacterial strains for biocatalytic applications by identifying a pathway for the degradation of acetovanillone.
Lignin is a heterogeneous aromatic biopolymer that constitutes up to 35% of lignocellulosic biomass (1, 2). Due to its abundance, lignin has considerable potential as a renewable feedstock for biobased materials and chemicals. Nevertheless, it remains underutilized due to its structural heterogeneity, which presents challenges for selective depolymerization and upgrading in biorefining (1, 3). Consequently, lignin is often burned to provide heat energy for the extraction of carbohydrates. One solution to this challenge that is gaining traction is catalytic fractionation, a process that not only separates biomass into lignin and carbohydrate fractions but also promotes conversion of the lignin into aromatic monomers and oligomers (4). The monomers can be used industrially for applications, such as polymer synthesis, or can be further converted by microbial cell factories to any of a number of useful chemicals in high atom yield (5). This biocatalytic conversion harnesses the convergent nature of bacterial aromatic catabolism whereby upper pathways transform a diversity of substrates to a small number of key intermediates, typically catechols, which are further transformed to central metabolites via lower pathways (3, 6). Biocatalytic conversion depends on elucidating and optimizing the requisite aromatic catabolic pathways.
Among catalytic fractionation strategies, oxidative catalytic fractionation (OCF) offers particular promise for softwood biomass, depolymerizing the lignin to phenolic aldehydes, acids, and ketones through cleavage of intersubunit C-C linkages (7, 8). For example, Zhu et al. (7) used OCF to convert 70% of the carbon of a softwood to useful products. Of the lignin, 29% by weight was recovered as monophenolics, mainly vanillin and acetovanillone (AV), and ∼57% by weight as small oligophenolics (<600 Da), such as methoxylated biphenyls. Using a similar approach, Abdelaziz et al. (9) oxidatively depolymerized a softwood kraft lignin to a mixture of monoaromatic compounds, including vanillate, vanillin, guaiacol, and AV (9). Such mixtures lend themselves to biocatalytic conversion, as numerous bacterial strains grow on most of these compounds, with the exception of AV. Ravi et al. (10) tested nine strains of Rhodococcus and Pseudomonas for growth on depolymerized kraft lignin containing 4-hydroxybenzoate, vanillate, vanillin, guaiacol, and AV. Of these compounds, AV was the only one not degraded by any of the tested strains.
Members of the genus Rhodococcus are of particular interest due to their ability to degrade an exceptionally wide range of aromatic compounds and their demonstrated use at industrial scale to produce thousands of tons of acrylamide annually (11, 12). Illustrative of the aforementioned convergent catabolism, rhodococci catabolize vanillate via protocatechuate using vanillate O-demethylase, encoded by vanAB; vanillin via vanillate, using vanillin dehydrogenase (vdh) (13); and guaiacol via catechol, using guaiacol O-demethylase (gcoAB) (14). Protocatechuate and catechol are converted to central metabolites via a convergent β-ketoadipate pathway (15). In contrast to these well characterized pathways, a pathway for catabolism of AV has yet to be elucidated.
We recently isolated two strains of Rhodococcus rhodochrous, GD01 and GD02, for their ability to grow on AV (16). GD02 removed the major monoaromatic compounds, including AV, present in black liquor, an industrially relevant stream generated in the kraft pulping process. GD01 and GD02 are remarkably similar to R. rhodochrous EP4, a strain that was isolated for its ability to catabolize alkylphenols and alkylguaiacols produced by the reductive catalytic fractionation of biomass (14, 17). However, EP4 did not grow on AV. Genomic analyses revealed that EP4 does not encode a predicted biotin-dependent carboxylase found in GD01 and GD02. The carboxylase, annotated as HpeCBA (formerly ApkCBA), is predicted to comprise a biotin carboxyl carrier protein (HpeC), a carboxyl transferase (HpeB), and a biotin carboxylase (HpeA) (16). The predicted carboxylase shares ∼50% amino acid sequence identity with XccBCA, a three-subunit carboxylase involved in the anaerobic catabolism of a chemical analog of AV, 4-hydroxyacetophenone (HAP), by Aromatoleum aromaticum strain EbN1 (18). The hpeC gene was up-regulated ∼100-fold during growth of GD02 on a mixture of monoaromatics that included AV (16). Collectively, these results strongly implicate HpeCBA in the catabolism of AV. However, the function of HpeCBA has yet to be validated and the AV catabolic pathway has yet to be elucidated.
Herein, we describe the catabolism of alkyl-phenyl ketones by GD02. We first assembled and annotated a high-quality genome sequence of GD02. Based on the predicted catabolic pathways, we then tested the ability of this strain to grow on a variety of aromatic compounds. To identify the relevant catabolic pathways, we analyzed the transcriptomes of GD02 growing on three alkyl-phenyl ketones: AV and HAP, both of which are hydroxyphenylethanones, and acetophenone (AP), which lacks the hydroxyl group. Bioinformatic analysis of the up-regulated genes enabled us to propose a catabolic pathway for the hydroxyphenylethanones, involving phosphorylation, carboxylation, and β-elimination, as well as a separate pathway for AP, involving a Baeyer–Villiger monooxygenase. Key steps of the former pathway were validated using purified enzymes and their hypothesized substrates. Finally, we investigated the ability of GD02 to grow on monoaromatic compounds, including AV, from OCF of lodgepole pine. The results are discussed with respect to related catabolic pathways and enzymes, as well as the engineering of biocatalysts to valorize lignin.
RESULTS
Genome Sequence.
To better understand how GD02 catabolizes AV and related compounds, we first generated an improved genome assembly for the strain. Previously existing Illumina short-read data (16) plus new Oxford Nanopore long-read data were assembled with Unicycler as a short-read–first hybrid assembly pipeline. Overall, the hybrid assembly improved the quality of the genome sequence and increased completeness from 99.2 to 99.8%. More significantly, this closed the assembly previously comprised of 24 scaffolds, revealing three genomic elements (SI Appendix, Fig. S1): a circular 5,587,176-bp chromosome, a 721,409-bp linear plasmid (pRGD1), and a 165,007-bp circular plasmid (pRGD2). The new assembly is 6.47 Mb and contains 5,932 predicted genes, 147 more genes than the previous assembly.
Analysis of the annotated genome of GD02 revealed numerous genes predicted to encode aromatic catabolism (Table 1 and SI Appendix, Table S1), which are mainly concentrated in one region of the genome (SI Appendix, Fig. S1) and which were mostly previously reported (16). The genomes of GD01 and GD02 are nearly identical (average nucleotide identity, 99.1%). However, GD01 lacks pRGD2, which contains a second set of genes encoding vanillate-O-demethylase, vanA2B2. The encoded proteins VanA and VanA2 share 97.8% amino acid sequence identity, while VanB and VanB2 share 68.9% identity. Proximal to vanA2B2 on pRGD2 are genes predicted to encode mycothiol-dependent oxidation of formaldehyde, produced in the O-demethylation of vanillate.
Table 1.
Annotated hydroxyphenylethanone and AP catabolic genes of GD02
| Catabolism type and genes | Locus* | Gene product | Closest homolog† | %ID‡ |
|---|---|---|---|---|
| Hydroxyphenylethanone catabolism | ||||
| hpeH | 02990 | Hydroxyphenylethanone kinase, α subunit | creH, Corynebacterium glutamicum, WP_011013724.1 | 46 |
| hpeI | 02985 | Hydroxyphenylethanone kinase, β subunit | creI, C. glutamicum, WP_011013725.1 | 40 |
| hpeC | 02980 | Phosphophenylethanone carboxylase, biotin carboxyl carrier protein | xccB, Aromatoleum aromaticum EbN1, WP_011236025.1 | 51 |
| hpeB | 02975 | Phosphophenylethanone carboxylase, carboxyltransferase | xccC, A. aromaticum EbN1, WP_011236024.1 | 51 |
| hpeA | 02970 | Phosphophenylethanone carboxylase, biotin carboxylase | xccA, A. aromaticum EbN1, WP_011236023.1 | 45 |
| hpeD | 02965 | Phosphophenyl-β-ketopropionate phosphatase | nagD, Escherichia coli K12, WP_000153129.1 | 24 |
| hpeE | 02960 | Hydroxyphenyl-β-ketopropionyl-coA synthetase | acsA2, Sinorhizobium meliloti, WP_010968769.1 | 42 |
| hpeF | 02955 | Hydroxyphenyl-β-ketopropionyl-coA hydrolase | couO, Rhodococcus jostii RHA1, WP_011597356.1 | 95 |
| AP catabolism | ||||
| acpB | 09510 | Phenyl acetate esterase | nlhH, Mycobacterium tuberculosis ATCC 25618, WP_003407276.1 | 41 |
| acpA | 09515 | Acetophenone monooxygenase | pamO, Thermobifida fusca YX, WP_011291921.1 | 55 |
ID, identity.
*Complete GD02 loci include the following prefix, LCH94_XXXXX.
†Closest homolog of experimentally verified function (gene name, strain, encoded protein identifier).
‡Percent amino acid sequence identity calculated over the entire length of the proteins encoded by the genes.
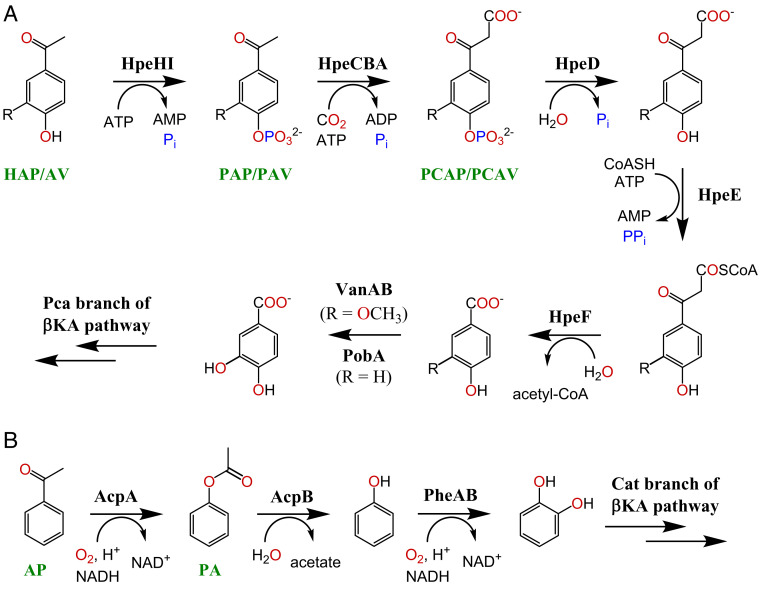
We found that the hpeCBA genes (formerly apkCBA), encoding the carboxylase previously associated with AV degradation (16), were part of a cluster containing eight catabolic genes in the same orientation (SI Appendix, Fig. S2). Based on the following annotations, we hypothesized that this cluster encodes a pathway responsible for catabolism of hydroxyphenylethanones, including AV and HAP (Fig. 1A). The first two genes in this cluster were predicted to encode a kinase, HpeHI, whose closest characterized homolog, CreHI from Corynebacterium glutamicum (Table 1), initiates degradation of 4-cresol by phosphorylation (19). HpeHI and CreHI are homologs of phenylphosphate synthase, which initiates anaerobic phenol catabolism in Thauera aromatica (20), Geobacter metallireducens (21), and Desulfatiglans anilini (22). Phylogenetic analyses revealed that the kinases predicted to catalyze the phosphorylation of hydroxyphenylethanones, phenol, and 4-alkylphenols form distinct respective clades (SI Appendix, Figs. S3 and S4). In addition, the two components share congruent phylogenies. Based on these observations, we hypothesized that HpeHI similarly initiates AV degradation.
Fig. 1.
Proposed pathways for degradation of hydroxyphenylethanones (A) and AP (B). In the catabolism of HAP (R = H), HAP is phosphorylated to PAP, then carboxylated to PCAP. In the catabolism of AV (R = OCH3), AV is phosphorylated to PAV, then carboxylated to PCAV. Pca, protocatechuate; βKA, β-keto adipate; Cat, catechol.
The next three genes encode the three-component, biotin-dependent carboxylase, HpeCBA. Because its homolog, XccBCA, is involved in the catabolism of HAP (18), we hypothesized that the Hpe pathway might degrade HAP in addition to AV. The closest characterized homolog of the product of the fifth gene, HpeD, is NagD from Escherichia coli K12, which catalyzes the dephosphorylation of a broad range of C5 and C6 sugars (23). Thus, we hypothesized that HpeD dephosphorylates the product of the HpeCBA reaction. The final two genes in the cluster, hpeE and hpeF, are predicted to encode an adenosine monophosphate (AMP)–forming acyl-coenzyme A (CoA) synthetase and a β-ketoacyl-CoA hydrolase, respectively. We propose that these catalyze CoA thioesterification of the carboxylated products of HpeD and the subsequent β-elimination of acetyl-CoA. Homologs of HpeE and HpeF, CouL and CouO, respectively, catalyze identical or analogous reactions in the degradation of coumarate and ferulate by various rhodococci (24). The proposed Hpe pathway produces vanillate from AV and 4-hydroxybenzoate from HAP. Vanillate is catabolized by GD02, presumably via O-demethylation to protocatechuate by VanAB (16). We identified a gene encoding a predicted 4-hydroxybenzoate 3-monooxygenase, PobA, which hydroxylates 4-hydroxybenzoate to protocatechuate (SI Appendix, Table S1).
The hpe gene cluster ends with a gene encoding a transposase and a duplicated fragment of hpeF (SI Appendix, Fig. S2). There is also a transposase gene immediately upstream of the cluster as well as integrase and recombinase genes downstream. Blast searches identified complete, syntenous hpe catabolic gene clusters in only four additional genomes, those of R. rhodochrous J4, Rhodococcus sp. JVH1, Rhodococcus sp. (metagenome assembled genome), and Actinomadura macra NBRC 14102. These genomes all contain both pobA and vanAB genes, so they might use both AV and HAP. Diverse bacteria have gene clusters containing various combinations of hpe gene homologs (SI Appendix, Fig. S5).
Growth Experiments.
We next tested the ability of GD02 to grow on a range of aromatic substrates. GD02 was previously reported to grow on AV, guaiacol, vanillate, and vanillin (16). We confirmed growth on these substrates and identified eight additional aromatic compounds supporting growth: AP, benzoate, HAP, 2-hydroxyacetophenone, 4-hydroxybenzoate, 4-methylguaiacol, 4-methylphenol, and phenol (SI Appendix, Table S2). Growth on guaiacol and 4-methylguaiacol is consistent with the presence of gcoAB in the GD02 genome. GD02 did not grow on 4-propylguaiacol or 4-ethylphenol, which is consistent with the absence of agcAB and aphAB, respectively (14, 17). GD02 did not grow on syringate, syringaldehyde, or 4-alkyl syringols, which is consistent with the absence of genes predicted to encode the degradation of these compounds. GD02 also did not grow on two biaryl compounds that are model lignin degradation intermediates: 2,2'-dihydroxy-3,3′-dimethoxy-5,5′-dicarboxybiphenyl and guaiacylglycerol-β-guaiacyl ether. In accordance with their highly similar genomes, GD01 had the same aromatic substrate profile as GD02. However, GD02 grew on higher concentrations of AV and vanillin than did GD01: 5 mM versus 1 mM for both compounds.
Transcriptomic Analysis.
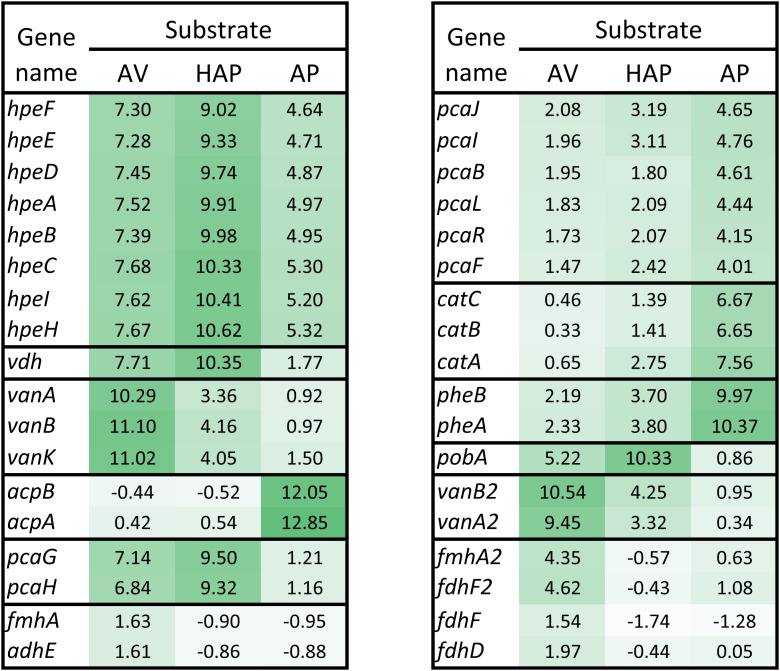
To gain further insight into the basis of the ability of GD02 to grow on AV, we performed transcriptomic experiments with cells grown on AV, HAP, AP, and as a control, citrate. The hpe genes were among those most highly up-regulated on both AV (>150-fold) and HAP (>500-fold; Fig. 2). The hpe gene cluster appears to be expressed as an operon, based on similar up-regulation of the genes, cotranscription, and four-nucleotide overlaps of seven of the eight predicted genes (SI Appendix, Fig. S2). Two genes upstream of the hpe cluster (LCH94_03000 and LCH94_03005) encode putative transcriptional regulators. Although these two genes were not as highly up-regulated as the hpe genes, their patterns of expression on the various substrates were congruent with those of the hpe genes, suggesting they may be involved in regulating the putative operon. Consistent with vanillate as a catabolic intermediate of AV but not HAP, the vanAB and vanA2B2 genes were highly up-regulated (>670-fold) on only AV. Interestingly, the expression pattern of vdh, encoding vanillin dehydrogenase, resembled that of the hpe genes, not vanAB, being most highly up-regulated on HAP. Consistent with 4-hydroxybenzoate as a catabolic intermediate of HAP but not AV, the pobA gene was highly up-regulated (>1,200-fold) on only HAP. The pcaGH genes were highly up-regulated (>110-fold) on both AV and HAP, consistent with protocatechuate as a catabolic intermediate of both substrates. However, the remaining pca genes, located in a separate gene cluster (SI Appendix, Fig. S1 and Table S1), were much less up-regulated. Read counts for the pcaGH and pcaJIBLRF genes were uniformly low on the control substrate, citrate (range, 65 to 213 average counts per gene), indicating that the latter genes were not constitutively expressed at a high level. Genes encoding mycothiol-dependent formaldehyde oxidation were also significantly up-regulated on AV.
Fig. 2.
Key genes up-regulated during growth on AV, HAP, and AP. Heat map shows log2-fold increase in gene expression during growth on the aromatic substrates versus on citrate. Horizontal lines separate clusters of genes. Genes are fully identified in Table 1 and SI Appendix, Table S1.
The transcriptomic experiment indicated a distinct pathway for catabolism of AP (Fig. 1B). The most highly up-regulated genes on AP (>4,000-fold) were acpAB (Fig. 2). The acpA gene encodes a Baeyer–Villiger monooxygenase whose closest functionally characterized homolog is phenylacetone monooxygenase (Table 1). The acpB gene encodes an esterase. This suggested that AcpA oxygenates AP to form phenyl acetate (PA) and that AcpB hydrolyses PA to phenol plus acetate. Consistent with this hypothesis, GD02 grew on phenol, and the fourth and fifth most highly up-regulated genes (950-fold) on AP were pheAB, encoding a phenol monooxygenase that forms catechol. The genes encoding the Cat branch of the β-ketoadipate pathway, catABC, were also among the most up-regulated genes on AP (>100-fold), as were the pca genes encoding the subsequent steps of the β-ketoadipate pathway (>16-fold). Notably, none of the acp, phe, or cat genes was up-regulated on AV or HAP. Unexpectedly, the hpe gene cluster was up-regulated on AP. Blast searches revealed probable orthologs of the acpAB genes in diverse Actinobacteria, including six Rhodococcus spp. Homologs of the acpAB genes were also identified in diverse Proteobacteria.
Hpe Pathway Validation.
To validate the Hpe pathway, we characterized the pathway’s predicted kinase, carboxylase, and phosphatase. The genes predicted to encode the kinase, hpeH and hpeI, and phosphatase, hpeD, were individually cloned and expressed in E. coli. HpeH, HpeI, and HpeD were purified as His-tagged proteins and the molecular masses observed in sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) matched the expected masses of 71.2 kDa, 40.6 kDa, and 30.2 kDa, respectively (SI Appendix, Fig. S6). The three genes predicted to encode the carboxylase, hpeCBA, were expressed together in Rhodococcus jostii RHA1. HpeCBA was purified to greater than 95% apparent homogeneity using ammonium sulfate precipitation followed by anion exchange chromatography. In this preparation, the molecular masses of HpeA and HpeB as estimated by SDS-PAGE matched their calculated molecular masses of 57.4 kDa and 49.3 kDa, respectively. In contrast, the molecular mass of HpeC was estimated to be ∼28 kDa, which is considerably higher than the molecular mass of 17.4 kDa calculated from its amino acid sequence. However, a His-tagged version of HpeC, which also ran on SDS-PAGE as a band higher than 25 kDa, had a molecular mass of 19,471.4 Da according to mass spectrometry, matching its predicted molecular mass. This confirmed that the smallest band present in the HpeCBA complex corresponded to the HpeC subunit. A streptavidin-shift assay indicated that HpeC contained ∼0.7 equivalents of biotin after normalizing using the intensity of the HpeAB band (SI Appendix, Fig. S7). Attempts to produce HpeCBA in E. coli were unsuccessful. Moreover, omitting dithiothreitol and glycerol from the purification buffers led to preparations of lower specific activity.
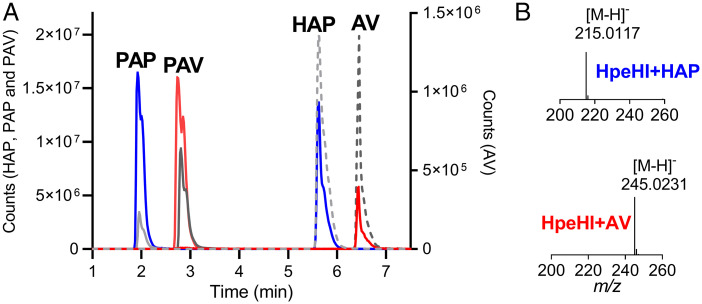
We used liquid chromatography–mass spectrometry (LC–MS) to show that HpeHI catalyzed the phosphorylation of AV and HAP to 4-phospho-acetovanillone (PAV) and 4-phospho-acetophenone (PAP), respectively. The retention times (tRs) and mass spectra of the reaction products corresponded to those of authentic standards (Fig. 3). Both protein components were required for phosphorylation. We next used a spectrophotometric assay to compare the relative ability of HpeHI to turn over the two compounds. This assay was based on the strong absorption of the phenolate ions of HAP and AV at pH 8 (SI Appendix, Fig. S8). In assays containing 1 μM of each kinase component, the enzyme turned over AV and HAP at rates (± SD) of 4.1 ± 0.4 min−1 and 3.4 ± 0.7 min−1, respectively.
Fig. 3.
Phosphorylation of AV and HAP by HpeHI. (A) Extracted ion chromatograms for substrates and products of the HpeHI reaction are labeled, corresponding to both the retention time and m/z of standards. Reaction mixtures of HpeHI with HAP (blue) or AV (red) or no enzyme controls for HAP (dashed, light gray) or AV (dashed, dark gray) are shown. Synthesized standards for the expected products PAP (solid, light gray) and PAV (solid, dark gray) are also shown. (B) Mass spectra of PAP and PAV produced in the reaction (Right) match the expected m/z with <5 ppm error.
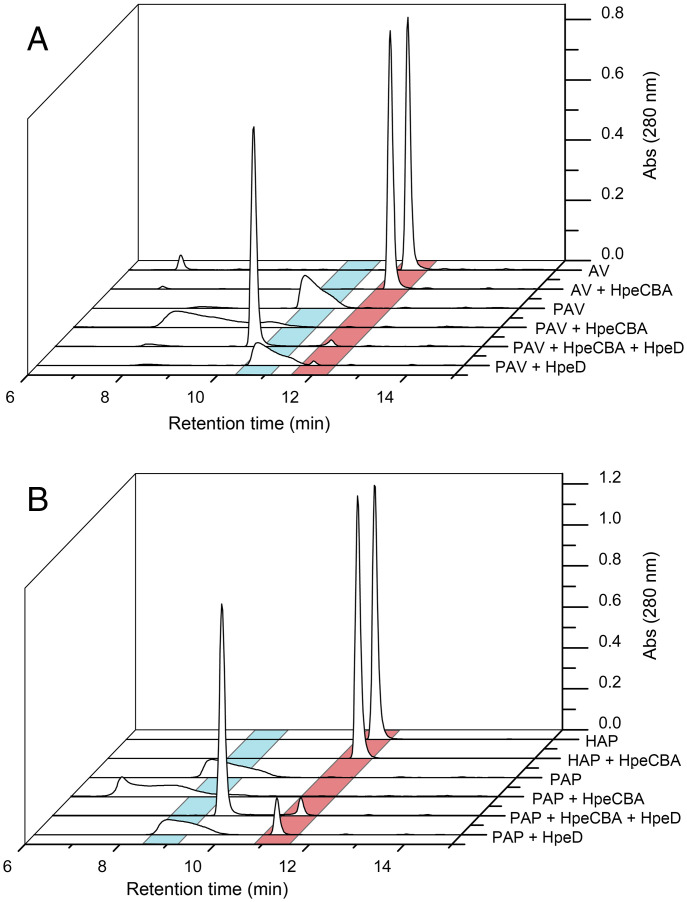
We investigated the activities of HpeCBA in vitro using a variety of phosphorylated and unphosphorylated substrates. To this end, we synthesized PAV and PAP. High-performance liquid chromatography (HPLC) analyses indicated that reaction mixtures containing HpeCBA and PAV, depleted PAV (tR = 10.3 min) and produced a compound with a tR of ∼8.3 min (Fig. 4A). Both peaks were relatively broad. Control reactions confirmed that conversion depended on the presence of the enzyme, adenosine triphosphate (ATP), Mg2+, and HCO3−, as did the carboxylase activity in a coupled reaction (see below). LC–MS analysis demonstrated that the product had a mass to charge (m/z) value of 289.0018, corresponding to that of the predicted carboxylated product, 4-phospho-carboxyacetovanillone (PCAV; SI Appendix, Fig. S9A). Similarly, HpeCBA catalyzed the transformation of PAP to a compound with a tR of 11.9 min (Fig. 4B) and m/z value of 259.0017, corresponding to that of the predicted carboxylated product, 4-phospho-carboxyacetophenone (PCAP; SI Appendix, Fig. S10A). Finally, under these reaction conditions, HpeCBA did not detectably transform either AV or HAP, indicating that phosphorylation is required for carboxylation of these compounds.
Fig. 4.
Transformations catalyzed by HpeCBA and HpeD. (A) AV or PAV were incubated with either HpeCBA or HpeCBA + HpeD as indicated. The product of PAV plus HpeCBA was PCAV, identified by mass spectrometry (SI Appendix, Fig. S9A). The product of PAV plus HpeCBA plus HpeD was 4-hydroxy-3-methoxyphenyl-β-ketopropionate, identified by mass spectrometry (SI Appendix, Fig. S9B). (B) HAP or PAP were incubated with either HpeCBA or HpeCBA plus HpeD, as indicated. The product of PAP plus HpeCBA was PCAP, identified by mass spectrometry (SI Appendix, Fig. S10A). The product of PAV plus HpeCBA plus HpeD was 4-hydroxyphenyl-β-ketopropionate, identified by mass spectrometry (SI Appendix, Fig. S10B). (A and B) Reactions were incubated for 20 min at 30 °C using 250 µM substrate in 20 mM (3-(N-morpholino)propanesulfonic acid (MOPS; I = 0.1 M), pH 7.5, containing 0.5 mM ATP, 4 mM MgCl2, and 40 mM NaHCO3. HPLC traces were recorded at 280 nm. Abs, absorbance.
To investigate the activity of HpeD, we added the enzyme to the above-described carboxylation reactions. When PAV was incubated in the presence of HpeD and HpeCBA, an HPLC peak corresponding to PCAV was not detected (Fig. 4A). Instead, we observed a compound with a tR of 10.1 min and an m/z value of 211.0613, corresponding to that of 4-hydroxy-3-methoxyphenyl-β-ketopropionate (SI Appendix, Fig. S9B). In addition, a small peak corresponding to AV was observed. Incubation of PAV with HpeD alone yielded similarly small amounts of AV. Overall, these results indicate that HpeD is a phosphohydrolase that catalyzes the conversion of PCAV to 4-hydroxy-3-methoxyphenyl-β-ketopropionate. The results further indicate that the enzyme can catalyze the hydrolysis of PAV but that its specific activity for PCAV is much higher. Finally, similar results were obtained using PAP (Fig. 4B, SI Appendix, Fig. S10B). That is, HpeD catalyzed the hydrolysis of PCAP to 4-hydroxyphenyl-β-ketopropionate and, at a much lower rate, PAP to HAP. Overall, these results consistently support the proposed Hpe pathway (Fig. 1A).
Kinetic Properties of HpeCBA Carboxylase.
To gain insight into the overall energetics and preferred substrate of the Hpe pathway, we further characterized the novel HpeCBA carboxylase with respect to its utilization of ATP and substrate specificity. HPLC analysis of HpeCBA reactions performed using PAV and ATP revealed that adenosine diphosphate (ADP) was exclusively produced: no AMP was detected (SI Appendix, Fig. S11A). To determine the stoichiometry of ADP production, we used a continuous spectrophotometric assay in which ADP production was coupled to NADH oxidation using pyruvate kinase and lactate dehydrogenase (25). In assays containing limiting amounts of PAV, 0.93 ± 0.05 mol NADH was oxidized per mole of PAV consumed (SI Appendix, Fig. S11B). This indicates that HpeCBA catalyzes the hydrolysis of 1 mol of ATP to ADP per mole of PAV carboxylated.
The substrate specificity of HpeCBA was evaluated using the aforementioned spectrophotometric assay. Under the described conditions, the rate of NADH oxidation was directly proportional to the concentration of HpeCBA. The reaction was dependent on the presence of HCO3−, ATP, and Mg2+, and the pH optimum was 7.5 (SI Appendix, Fig. S12). This pH was used for additional steady-state kinetic studies. HpeCBA obeyed Michaelis–Menten kinetics in transforming both PAV and PAP (SI Appendix, Fig. S13). The enzyme’s specificity (kcat/KM) for PAP was an order of magnitude higher than for PAV (Table 2), indicating a strong preference for the former substrate. Nevertheless, the kcat of the carboxylase for the two substrates is remarkably similar.
Table 2.
Steady-state kinetic parameters of HpeCBA*
| Substrate | kcat, s-1† | KM, μM | kcat/KM, mM−1 s−1 |
|---|---|---|---|
| PAV | 2.25 (0.06) | 560 (30) | 3.9 (0.1) |
| PAP | 2.40 (0.07) | 69 (6) | 34 (2) |
*Experiments were performed in 20 mM MOPS (I = 0.1 M), pH 7.5, at 25 °C.
†kcat values are based on protein concentrations and may be ∼25% higher based on the biotin content of HpeC (SI Appendix, Fig. S7). SD of triplicates is shown in parentheses.
Finally, we tested whether the phosphatase facilitates the activity of the carboxylase, for example by generating a phenolate anion that serves as the substrate of the carboxylase. When added at a 1:1 molar ratio with respect to HpeCBA, HpeD did not detectably affect the specific activity of the carboxylase, 1.42 ± 0.05 U/mg with HpeD versus 1.42 ± 0.04 U/mg without HpeD (SI Appendix, Fig. S14). This indicates that HpeD does not generate a substrate for HpeCBA or interact with the carboxylase to accelerate the reaction. This result is consistent with the pH profile of HpeCBA (SI Appendix, Fig. S12B), whose specific activity declined above pH 7.5 despite the fact that the concentration of phenolate anion increases above this pH value. It is further noted that under these reaction conditions, no HpeD-catalyzed dephosphorylation of PAP was detected (SI Appendix, Fig. S14), indicating that the phosphatase has a much stronger substrate preference for the carboxylated product. Interestingly, the carboxylase progress curves remained linear for longer in the presence of HpeD, indicating that HpeCBA might be subject to product inhibition.
Growth of GD02 on Pine OCF Extracts.
To investigate the potential of GD02 to transform an industrially relevant lignin stream, the strain was tested for growth on an extract of softwood (namely, lodgepole pine) biomass treated by OCF. The slurry resulting from OCF was acidified and extracted with ethyl acetate. Extraction yielded up to 15.0 ± 0.2 mM monoaromatic compounds, predominantly vanillin, AV, and vanillic acid present in a ∼5:1:1 molar ratio (SI Appendix, Table S3), which represented 75% of the monoaromatic compounds present in the slurry and indicated 33.9% conversion of lignin into monoaromatic compounds. The recovery of monoaromatics from OCF by ethyl acetate extraction differed based upon conditions (solvent ratios) and process (repeated extraction of liquor). OCF extracts additionally contained small organic acids, including fumaric, 2-hydroxybutyric, glycolic, and lactic acids. For growth studies, the extract was evaporated, and the residue was suspended in dimethyl sulfoxide, which does not support growth of GD02 (SI Appendix, Table S2). The dimethyl sulfoxide solution was added to liquid medium to yield a final concentration of up to 3 mM total aromatic compounds.
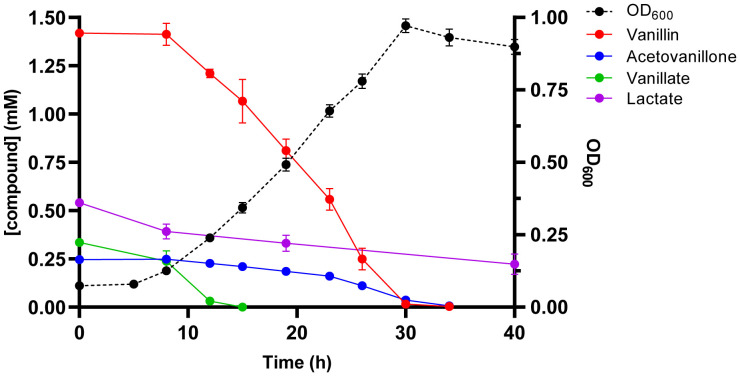
GD02 grew on liquid minimal medium supplemented with OCF extract up to a concentration of 2 mM total aromatic compounds. The doubling time of the bacterium was ∼9 h (Fig. 5), which was faster than on 2 mM vanillin (∼15 h) but slower than on 2 mM vanillate (∼3.3 h). HPLC analysis revealed that the three major monoaromatic compounds were efficiently removed, with vanillate depleted well before vanillin and AV. Among the small organic acids, GD02 removed 59% of the lactate but did not detectably deplete the others. GD02 did not grow on higher concentrations of OCF extract (SI Appendix, Fig. S15), and further experiments revealed that extract at a concentration of 3 mM total aromatics inhibited growth of GD02 on rich medium.
Fig. 5.
Growth of GD02 on lodgepole pine OCF extracts. GD02 was grown at 30 °C in M9+minerals containing OCF extracts at a concentration of 2 mM total aromatic compounds. Depletion of aromatic compounds from culture supernatants was measured using HPLC. Data points represent the average of triplicate experiments and the error bars represent the SD.
Discussion
This study provides a consistent body of evidence supporting the proposed pathway for the catabolism of hydroxyphenylethanones (Fig. 1A). Transcriptomic analysis revealed that the hpe genes plus additional genes predicted to encode AV and HAP catabolism were among the most highly up-regulated genes during growth on these substrates (Fig. 2). The expected intermediate of AV catabolism, vanillate, supported growth of GD02, as did the expected intermediate of HAP catabolism, 4-hydroxybenzoate (SI Appendix, Table S2). The kinase (HpeHI), carboxylase (HpeCBA), and phosphatase (HpeD) were all shown to have their predicted activities on AV, HAP, and their respective intermediates (Figs. 3 and 4). Importantly, HpeCBA carboxylated only the phosphorylated intermediates, PAV and PAP, and not AV or HAP. Thus, the evidence that AV and HAP are catabolized via the Hpe pathway as proposed is strong. It is possible that AcpA might have some ability to transform AV or HAP. However, this appears unlikely, as R. jostii RHA1 also has acpAB and is able to grow on AP, but it is not able to grow on AV or HAP.
HpeHI appears to belong to a group of kinases involved in aromatic catabolism (SI Appendix, Figs. S3 and S4). The rationale for the energetically costly phosphorylation reaction in the Hpe pathway is unclear. One possibility is that phosphorylation transforms the hydrophobic substrate, maintaining a concentration gradient favorable for its uptake via passive diffusion while simultaneously preventing its loss from the cell via passive diffusion. Additional or alternative possibilities are that phosphorylation detoxifies the substrate or phosphorylation permits the use of a carboxylase that consumes one ATP, rather than two, per reaction.
An unusual feature of the Hpe pathway is carboxylation of the acetyl substituent of AV and HAP. Carboxylation is a common strategy in the assimilation of compounds that lack a functionalizable terminal carbon group (26). Also, several anaerobic aromatic degradation pathways utilize carboxylation. Following the phosphorylation of phenol by T. aromatica, 4-phenylphosphate is subsequently carboxylated by an enzyme not homologous to HpeCBA, with concomitant release of the phosphate group, forming 4-hydroxybenzoate (27). A. aromaticum EbN1 uses an ATP-dependent carboxylase not homologous to HpeCBA for anaerobic degradation of ethylbenzene and AP (28). As noted in the introductory text, XccBCA, a homolog of HpeCBA, is used by EbN1 for anaerobic degradation of 1-ethylphenol (18). Interestingly, the genes encoding XccBCA are clustered with homologs of hpeHI (SI Appendix, Fig. S5). Thus, while the substrate of XccBCA was not previously proposed to be phosphorylated, this may be the case. We are not aware of any aerobic aromatic degradation pathways, other than the Hpe pathway, that involve carboxylation. Notably, the catabolic steps encoded by the hpe genes do not require O2, which means the pathway may have evolved in an anaerobic organism.
GD02 appears to have acquired the Hpe pathway via horizontal gene transfer. This is consistent with the absence of the hpe genes in the genomes of most Rhodococcus strains. The presence of transposase and integrase genes upstream and downstream of the hpe gene cluster (SI Appendix, Fig. S2) is also consistent with horizontal transfer. Since the hpe genes are contiguous and cotranscribed, it appears that the complete gene cluster transferred. Since HAP is the preferred substrate for HpeCBA, it may be that the ability of GD02 to grow on AV is a fortuitous consequence of the cooccurrence of the Hpe pathway and the vanillate pathway.
It is surprising that the hpe genes are substantially up-regulated on AP (Fig. 2). Since the Hpe pathway requires a hydroxyl group on the aromatic ring, it cannot transform AP. Thus, it seems likely that AP coincidentally functions as an effector for transcriptional regulation of the hpe genes. Interestingly, on the substrates tested, the expression pattern of vdh, encoding vanillin dehydrogenase, closely resembled that of the hpe genes, despite the fact that Vdh is not required for catabolism of AV or HAP. Possibly, the hpe and vdh genes are coregulated as an adaptation to environments where vanillin cooccurs with hydroxyphenylethanones.
We have identified genes and proteins likely responsible for a pathway for AP catabolism, which was first proposed in 1975 (29). The previous study reported enzymes in cell-free extracts that oxidized AP to PA and hydrolyzed PA to phenol plus acetate. Enzymological studies (30) reported transformation of AP and some chemical analogs by Baeyer–Villiger monooxygenases but did not link this transformation to growth on AP. It would be informative to express and characterize acetophenone monooxygenase to confirm the Acp pathway and determine the substrate range of the enzyme. The initial steps catalyzed by AcpA and AcpB are analogous to those proposed for catabolism of HAP by Pseudomonas fluorescens ACB (31, 32).
The transcriptomic results indicate that AV, HAP, and AP are all funneled into the branched β-ketoadipate pathway, with AV and HAP entering via protocatechuate and AP entering via catechol. Accordingly, the pcaGH genes, encoding protocatechuate 3,4-dioxygenase, were highly up-regulated on both AV and HAP but not on AP (Fig. 2). Surprisingly, the pca genes encoding the remainder of the β-ketoadipate pathway, located in a separate cluster, were much less up-regulated on AV and HAP. It appears that the latter genes are less highly expressed than the former on AV and HAP, since they also did not have a higher constitutive level of expression. In most Rhodococcus strains, other than GD02, the pca genes occur in a single cluster (12).
Strain GD02 has metabolic capacities with strong potential for use in biocatalytic valorization of lignin. The Hpe pathway converges with the vanillin pathway, facilitating metabolism of vanillin, vanillate, and AV, the three main monoaromatic products of OCF. Accordingly, GD02 completely degraded these three components of pine OCF (Fig. 5). In a previous study, we showed that GD02 removes these three compounds from black liquor (16). These convergent pathways comprise an attractive metabolic module potentially useful for developing biocatalysts to valorize lignin treated by OCF or to valorize black liquor. Additionally, the plasmid pRGD2 encodes two processes that may protect GD02 from toxicity associated with growth on vanillin, vanillate, and AV. This protective effect is suggested by the ability of GD02 to grow on much higher concentrations of AV or vanillin than does GD01, which lacks the plasmid. The extra copy of vanAB genes on pRGD2 may reduce intracellular accumulation of toxic vanillin and potentially toxic AV, or Hpe pathway intermediates. Additionally, oxidation of formaldehyde encoded on pRGD2 may reduce toxicity of this product from O-demethylation of vanillate, occurring during degradation of vanillin, vanillate, and AV. This detoxication was previously proposed to facilitate growth of R. jostii RHA1 on p-hydroxycinnamates (24).
Higuchi et al. (33) recently described a pathway used by Sphingobium sp. SYK-6 to catabolize AV and acetosyringone. The SYK-6 pathway is encoded by the acvABCDEF genes, which are homologous and syntenous to hpeHICBAD (SI Appendix, Fig. S5). While we did not test it, growth of GD02 on acetosyringone is unlikely given the inability of GD02 to grow on syringate, an intermediate in acetosyringone degradation by SYK-6. Importantly, Higuchi et al. demonstrated that the acv genes are essential for growth of SYK-6 on AV, and that they conferred the ability to grow on AV when heterologously expressed in Pseudomonas sp. NGC7. Interestingly, SYK-6 lacks homologs of HpeE and HpeF and appears to use the functionally analogous, but non-homologous, enzymes, VceA and VceB, which were previously shown to transform hydroxyphenyl-β-ketopropionates to hydroxybenzoates (34).
Materials and Methods
Further details of all materials and methods are provided in the SI Appendix.
Chemicals and Reagents.
All reagents were purchased from Sigma-Aldrich Co. and were of analytical grade unless otherwise noted. PAP and PAV were synthesized by a published method (35) with modifications, as described in the SI Appendix. Enzymes for cloning were purchased from New England Biolabs. Oligonucleotides were ordered from Integrated DNA Technologies.
Genome Sequencing, Assembly, and Annotation.
Genomic DNA for long-read sequencing was extracted from GD02 using the Genomic-tip 500/G kit (Qiagen) according to the manufacturer’s protocol, with modifications. Oxford Nanopore sequencing was performed by the Microbial Genome Sequencing Center. Long and short reads were assembled with Unicycler as a short-read first-assembly pipeline (36). The genome was annotated with the NCBI Prokaryotic Genome Annotation Pipeline.
Growth Experiments.
R. rhodochrous GD01 and GD02 were grown on M9 medium (37) amended with an HCl-solubilized mineral solution (M9+minerals) described previously (38) plus organic growth substrates. The optical density at a wavelength of 600 nm (OD600) was monitored spectrophotometrically approximately every 24 h. Protein was quantified using the Micro BCA Protein Assay (Thermo Fisher Scientific Inc.) and a VersaMax microplate reader (Molecular Devices). Colony-forming units were determined on Luria–Bertani plates. Substrate depletion was determined by gas chromatography–mass spectrometry (GC-MS).
Transcriptomic Experiment.
GD02 was grown on M9+minerals plus AV, HAP, AP, or citrate, and cells were harvested during midlog phase. RNA was extracted using TRIzol (Thermo-Fisher) and the Qiagen RNeasy mini kit (catalog 74104). Complementary DNA was generated with Turbo DNase (Thermo-Fisher). RNA sequencing was performed by the Sequencing and Bioinformatics Consortium at The University of British Columbia.
Protein Production and Enzyme Assays.
HpeH, HpeI, and HpeD were produced by heterologous expression of hpeH, hpeI, and hpeD in E. coli BL-21 λ (DE3). HpeC and HpeCBA were produced by heterologous expression of hpeC and hpeCBA in R. jostii RHA1. HpeH, HpeI, HpeC, and HpeD were produced with N-terminal His tags and were purified using immobilized metal-affinity chromatography. The HpeCBA complex was precipitated with ammonium sulfate and purified using anion-exchange chromatography. Substrate removal and product formation by enzymes were determined by HPLC–mass spectrometry. The activity of HpeHI was evaluated spectrophotometrically based on the difference in absorbance between the substrates and phosphorylated products. Steady-state kinetic assays of HpeCBA were performed spectrophotometrically using a coupled assay and monitoring NADH oxidation.
OCF Extract Transformation Experiments.
Lodgepole pine, Pinus contorta, was subjected to OCF according to the method of Zhu et al. (7). Low-molecular-weight compounds were extracted with ethyl acetate, the solvent was evaporated under N2, and the extracted compounds were dissolved in dimethyl sulfoxide. GD02 was grown on M9+minerals supplemented with OCF extract. Growth was monitored at OD600, and removal of compounds in the extract was measured by GC–MS and HPLC.
Note added in proof.
A paper by Higuchi et al. (33) describing catabolism of AV and acetosyringone by Sphingobium sp. SYK-6 was published while the current manuscript was in review.
Supplementary Material
Acknowledgments
We thank Gordon Stewart for assisting with chemical analyses, Logan Robeck for assisting with protein production, Dr. Shawn Mansfield for assisting with extraction of OCF, and Dr. Steven Withers for chemical synthesis. This study was supported by a research contract from Genome BC (GEN005) and the BC BioProducts Alliance. L.D.E. is the recipient of a Canada Research Chair. G.N.D. is the recipient of a University of British Columbia 4-Year Fellowship. N.A.L. was the recipient of a Natural Sciences and Engineering Research Council of Canada Undergraduate Student Research Award.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2213450119/-/DCSupplemental.
Data, Materials, and Software Availability
DNA and RNA data have been deposited with the National Center for Biotechnology Information, accession numbers JAHRXG000000000 and PRJNA742134, respectively (39, 40). All study data are included in the article and/or supporting information.
References
- 1.Ragauskas A. J., et al. , Lignin valorization: Improving lignin processing in the biorefinery. Science (1979) 344, 1246843 (2014). [DOI] [PubMed] [Google Scholar]
- 2.del Río J. C., et al. , Lignin monomers from beyond the canonical monolignol biosynthetic pathway: Another brick in the wall. ACS Sustain. Chem.& Eng. 8, 4997–5012 (2020). [Google Scholar]
- 3.Becker J., Wittmann C., A field of dreams: Lignin valorization into chemicals, materials, fuels, and health-care products. Biotechnol. Adv. 37, 107360 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Abu-Omar M. M., et al. , Guidelines for performing lignin-first biorefining. Energy Environ. Sci. 14, 262–292 (2021). [Google Scholar]
- 5.Johnson C. W., et al. , Innovative chemicals and materials from bacterial aromatic catabolic pathways. Joule 3, 1523–1537 (2019). [Google Scholar]
- 6.Eltis L. D., Singh R., “Biological funneling as a means of transforming lignin-derived aromatic compounds into value-added chemicals” in Lignin Valorization, G. T. Beckham, Ed. (The Royal Society of Chemistry, 2018), pp. 290–313. [Google Scholar]
- 7.Zhu Y., et al. , Complementing vanillin and cellulose production by oxidation of lignocellulose with stirring control. ACS Sustain. Chem. Eng. 8, 2361–2374 (2020). [Google Scholar]
- 8.Luo H., et al. , Oxidative catalytic fractionation of lignocellulosic biomass under non-alkaline conditions. J. Am. Chem. Soc. 143, 15462–15470 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abdelaziz O. Y., et al. , Oxidative depolymerization of kraft lignin for microbial conversion. ACS Sustain. Chem. Eng. 7, 11640–11652 (2019). [Google Scholar]
- 10.Ravi K., et al. , Bacterial conversion of depolymerized Kraft lignin. Biotechnol. Biofuels 12, 56 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes J., Armitage Y. C., Symes K. C., Application of whole cell rhodococcal biocatalysts in acrylic polymer manufacture. Antonie van Leeuwenhoek 74, 107–118 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Yam K. C., van der Geize R., Eltis L. D., “Catabolism of aromatic compounds and steroids by rhodococcus” in H. M. Alvarez,Biology of Rhodococcus, H. M. Alvarez, Ed. (Springer, Berlin, Heidelberg, 2010), pp. 133–169. [Google Scholar]
- 13.Chen H.-P., et al. , Vanillin catabolism in Rhodococcus jostii RHA1. Appl. Environ. Microbiol. 78, 586–588 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fetherolf M. M., et al. , Characterization of alkylguaiacol-degrading cytochromes P450 for the biocatalytic valorization of lignin. Proc. Natl. Acad. Sci. U.S.A. 117, 25771–25778 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patrauchan M. A., et al. , Catabolism of benzoate and phthalate in Rhodococcus sp. strain RHA1: Redundancies and convergence. J. Bacteriol. 187, 4050–4063 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Navas L. E., et al. , Bacterial transformation of aromatic monomers in softwood black liquor. Front. Microbiol. 12, 735000 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levy-Booth D. J., et al. , Catabolism of alkylphenols in Rhodococcus via a meta-cleavage pathway associated with genomic islands. Front. Microbiol. 10, 1862 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wöhlbrand L., Wilkes H., Halder T., Rabus R., Anaerobic degradation of p-ethylphenol by “Aromatoleum aromaticum” strain EbN1: Pathway, regulation, and involved proteins. J. Bacteriol. 190, 5699–5709 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du L., et al. , Characterization of a unique pathway for 4-Cresol catabolism initiated by phosphorylation in Corynebacterium glutamicum. J. Biol. Chem. 291, 6583–6594 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmeling S., et al. , Phenylphosphate synthase: A new phosphotransferase catalyzing the first step in anaerobic phenol metabolism in Thauera aromatica. J. Bacteriol. 186, 8044–8057 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schleinitz K. M., et al. , Phenol degradation in the strictly anaerobic iron-reducing bacterium Geobacter metallireducens GS-15. Appl. Environ. Microbiol. 75, 3912–3919 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie X., Müller N., Enzymes involved in the anaerobic degradation of phenol by the sulfate-reducing bacterium Desulfatiglans anilini. BMC Microbiol. 18, 93 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plumbridge J. A., Sequence of the nagBACD operon in Escherichia coli K12 and pattern of transcription within the nag regulon. Mol. Microbiol. 3, 505–515 (1989). [DOI] [PubMed] [Google Scholar]
- 24.Otani H., Lee Y.-E., Casabon I., Eltis L. D., Characterization of p-hydroxycinnamate catabolism in a soil Actinobacterium. J. Bacteriol. 196, 4293–4303 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venkatraman J., et al. , Screening, identification, and characterization of mechanistically diverse inhibitors of the Mycobacterium tuberculosis enzyme, pantothenate kinase (CoaA). J. Biomol. Screen. 17, 293–302 (2012). [DOI] [PubMed] [Google Scholar]
- 26.Erb T. J., Carboxylases in natural and synthetic microbial pathways. Appl. Environ. Microbiol. 77, 8466–8477 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schühle K., Fuchs G., Phenylphosphate carboxylase: A new C-C lyase involved in anaerobic phenol metabolism in Thauera aromatica. J. Bacteriol. 186, 4556–4567 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rabus R., Kube M., Beck A., Widdel F., Reinhardt R., Genes involved in the anaerobic degradation of ethylbenzene in a denitrifying bacterium, strain EbN1. Arch. Microbiol. 178, 506–516 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Cripps R. E., The microbial metabolism of acetophenone. Metabolism of acetophenone and some chloroacetophenones by an Arthrobacter species. Biochem. J. 152, 233–241 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamerbeek N. M., Janssen D. B., van Berkel W. J. H., Fraaije M. W., Baeyer–Villiger monooxygenases, an emerging family of flavin-dependent biocatalysts. Adv. Synth. Catal. 345, 667–678 (2003). [Google Scholar]
- 31.Moonen M. J. H., et al. , Elucidation of the 4-hydroxyacetophenone catabolic pathway in Pseudomonas fluorescens ACB. J. Bacteriol. 190, 5190–5198 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamerbeek N. M., et al. , 4-Hydroxyacetophenone monooxygenase from Pseudomonas fluorescens ACB. A novel flavoprotein catalyzing Baeyer-Villiger oxidation of aromatic compounds. Eur. J. Biochem. 268, 2547–2557 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Higuchi Y., et al. , The catabolic system of acetovanillone and acetosyringone in Sphingobium sp. strain SYK-6 useful for upgrading aromatic compounds obtained through chemical lignin depolymerization. Appl. Environ. Microbiol. 88, e0072422 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Higuchi Y., et al. , Discovery of novel enzyme genes involved in the conversion of an arylglycerol-β-aryl ether metabolite and their use in generating a metabolic pathway for lignin valorization. Metab. Eng. 55, 258–267 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Silverberg L. J., Dillon J. L., Vemishetti P., A simple, rapid and efficient protocol for the selective phosphorylation of phenols with dibenzyl phosphite. Tetrahedron Lett. 37, 771–774 (1996). [Google Scholar]
- 36.Wick R. R., Judd L. M., Gorrie C. L., Holt K. E., Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput. Biol. 13, e1005595 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ausubel F. M., Short Protocols in Molecular Biology: A Compendium of Methods from Current Protocols in Molecular Biology (Wiley, ed. 5, 2002). [Google Scholar]
- 38.Vaillancourt F. H., Han S., Fortin P. D., Bolin J. T., Eltis L. D., Molecular basis for the stabilization and inhibition of 2, 3-dihydroxybiphenyl 1,2-dioxygenase by t-butanol. J. Biol. Chem. 273, 34887–34895 (1998). [DOI] [PubMed] [Google Scholar]
- 39.Dexter G. N., et al. , Rhodococcus rhodochrous Genome sequencing and assembly. BioProject. https://www.ncbi.nlm.nih.gov/bioproject?term=JAHRXG000000000. Deposited 26 October 2021.
- 40.Dexter G. N., et al. , Rhodococcus rhodochrous Genome sequencing and assembly. BioProject. https://www.ncbi.nlm.nih.gov/bioproject?term=PRJNA742134. Deposited 26 October 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA and RNA data have been deposited with the National Center for Biotechnology Information, accession numbers JAHRXG000000000 and PRJNA742134, respectively (39, 40). All study data are included in the article and/or supporting information.