Significance
The increasing prevalence of antibiotic resistant bacterial infections represents a significant public health crisis, necessitating the development of new therapeutics that work alone or in combination to increase efficacy of standard-of-care antibiotics. We describe a new class of antibiotics, that we have termed GmPcides, that have antibacterial activity against Gram-positive bacteria including multidrug-resistant pathogens. GmPcides effectively stop the growth of dividing and kill stationary phase nondividing enterococcal cells. Additionally, sublethal doses of GmPcides have additive to synergistic effects when combined with several different classes of standard-of-care antibiotics to treat multidrug-resistant bacteria. Thus, GmPcides hold potential to be used alone and in combination with existing antibiotics to combat antibiotic resistant infections caused by Gram-positive bacteria.
Keywords: antibiotic resistance, VRE, antibiotic synergy, multidrug-resistant pathogens
Abstract
The alarming rise of multidrug-resistant Gram-positive bacteria has precipitated a healthcare crisis, necessitating the development of new antimicrobial therapies. Here we describe a new class of antibiotics based on a ring-fused 2-pyridone backbone, which are active against vancomycin-resistant enterococci (VRE), a serious threat as classified by the Centers for Disease Control and Prevention, and other multidrug-resistant Gram-positive bacteria. Ring-fused 2-pyridone antibiotics have bacteriostatic activity against actively dividing exponential phase enterococcal cells and bactericidal activity against nondividing stationary phase enterococcal cells. The molecular mechanism of drug-induced killing of stationary phase cells mimics aspects of fratricide observed in enterococcal biofilms, where both are mediated by the Atn autolysin and the GelE protease. In addition, combinations of sublethal concentrations of ring-fused 2-pyridones and standard-of-care antibiotics, such as vancomycin, were found to synergize to kill clinical strains of VRE. Furthermore, a broad range of antibiotic resistant Gram-positive pathogens, including those responsible for the increasing incidence of antibiotic resistant healthcare-associated infections, are susceptible to this new class of 2-pyridone antibiotics. Given the broad antibacterial activities of ring-fused 2-pyridone compounds against Gram-positive (GmP) bacteria we term these compounds GmPcides, which hold promise in combating the rising tide of antibiotic resistant Gram-positive pathogens.
The emergence and growing incidence of antimicrobial resistance (AMR) in common pathogens has created a global public health crisis, with 4.95 million deaths worldwide associated with AMR in 2019 (1). Some reports estimate that the number of deaths could reach 10 million per year by 2050 without the advancement of new therapies and public health interventions (2). For more than 30 years, the majority of approved antibiotics have been derived from existing chemical structures (3). Only a few new classes of clinically approved antibiotics have been discovered since the mid-1980s including: i) oxazolidinones (e.g., linezolid); ii) lipopeptides (e.g., daptomycin), both of which were discovered in 1987; and iii) the antimycobacterial diarylquinolines (e.g., bedaquiline) in 2004 (4). However, resistance to each of these three classes of antibiotics has already been reported (5–7). Overall, antibiotic resistance is reaching a tipping point, creating an urgent need to develop new therapeutics with novel mechanisms of action that work either alone or in combination with existing therapeutics as a way to successfully treat troubling AMR infections.
The Centers for Disease Control and Prevention’s 2019 AR Threats Report categorizes 18 AMR bacterial and fungal pathogens as urgent, serious, or concerning threats. Among these are vancomycin-resistant enterococci (VRE), which have been recognized as serious threats (8). The most well studied VRE primarily belong to the species Enterococcus faecalis and Enterococcus faecium and are important causes of healthcare-associated infections (HAIs), which cost up to $28.4 billion in additional healthcare expenses in the United States and are responsible for 100,000 deaths per year (9). Among the HAIs caused by enterococcal species are catheter-associated urinary tract infections (CAUTI), as well as noncatheter-associated urinary tract infections (UTI), infective endocarditis (IE), and septicemia (10, 11). The prevention and treatment of enterococcal infections in the clinical setting has been increasingly difficult due to: i) the ability of enterococci to withstand heat (12, 13), UV radiation (14), and aseptic solutions, such as chloride and alcohol preparations (15, 16); ii) their inherent resistance to antimicrobial agents, such as cephalosporins, monobactams, penicillins and aminoglycosides (17, 18); iii) their ability to form antibiotic resistant biofilms (19–21); and iv) their capacity to acquire antibiotic resistance genes, such as those that encode proteins giving resistance to vancomycin (22–24). Thus, new and innovative therapeutics to combat this serious threat are needed.
Here, we developed compounds based on a peptidomimetic bicyclic central scaffold (a dihydrothiazolo ring-fused 2-pyridone with the peptidomimetic backbone highlighted in red, Fig. 1A). Structural diversity of the 2-pyridone core is possible using synthetic methodologies (25) and the nature of the substitution pattern of the scaffold is crucial to the function of the compound. For example, compared to the early lead 2-pyridone compound, EC305, the refined compound, PS757, has an aliphatic alkyl chain connected to position 2 via a phenoxy linker with a hexyl chain, which significantly improves potency (Fig. 1). We demonstrate that PS757 has potent bacteriostatic effects against growing enterococcal cells. In addition, unlike many existing classes of antibiotics that only target actively dividing cells, PS757 is also bactericidal against stationary phase enterococcal cells. The mechanism of PS757-mediated stationary phase killing involves the enterococcal Atn autolysin and the protease GelE, which is expressed during the stationary growth phase. Further, we show that sublethal concentrations of PS757 synergize with sublethal concentrations of gentamicin and vancomycin to kill multidrug resistant enterococci, including VRE strains. PS757 also shows broad spectrum bactericidal activity against multidrug-resistant (MDR) Gram-positive pathogens, including staphylococci, methicillin-resistant Staphylococcus aureus (MRSA), and streptococcal species, suggesting that this new family of antibiotics has the potential to be broadly efficacious for treatment of Gram-positive infections, including those caused by highly antibiotic resistant pathogens.
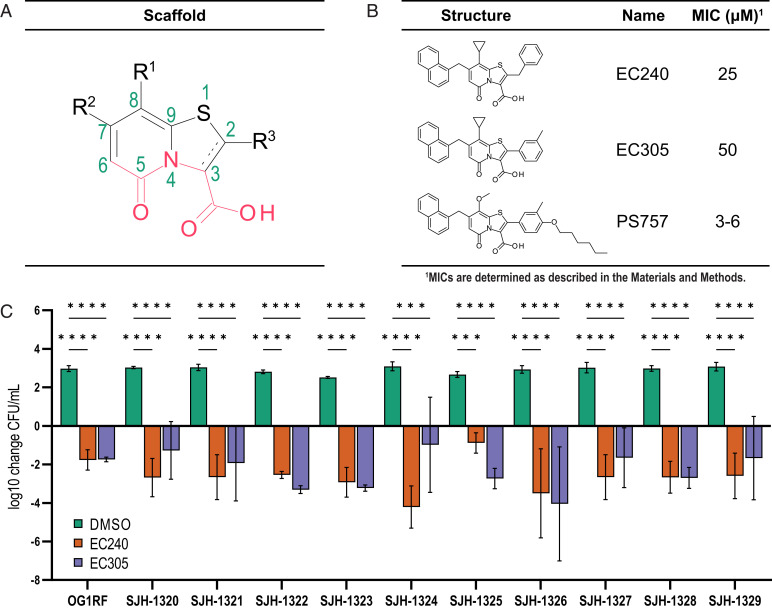
Fig. 1.
First generation 2-pyridone compounds have bactericidal activity against E. faecalis OG1RF and clinical isolates. (A) Structure of the 2-pyridone scaffold. (B) First-generation compounds EC240 and EC305, and second-generation compound PS757. MICs (see Materials and Methods) of EC240, EC305, and PS757 for E. faecalis OGR1RF are shown. (C) Net fold change (see Materials and Methods) in CFU/mL of clinical enterococcal isolates treated with DMSO (green), 100 µM EC240 (orange), and 100 µM EC305 relative to the CFU/mL of the untreated initial inoculum. Black bars represent SDs of replicates. Statistical analysis by two-way ANOVA (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Results
Ring-Fused 2-Pyridone Compounds with Antibacterial Activity Against E. faecalis OG1RF.
We have previously developed synthetically derived compounds that share a 2-pyridone scaffold (Fig. 1A) with specific substitution patterns that result in compounds that target various aspects of bacterial physiology, including an ability to block pilus biogenesis (26, 27). New synthetic methodologies were developed to more efficiently change the substitution pattern around the scaffold in all open positions, which resulted in a diverse 2-pyridone library for screening and structure activity relationship (SAR) studies. A collection of a diversely substituted 2-pyridone library was generated and screened against overnight cultures of E. faecalis OG1RF diluted 1:1,000 to an OD600 of 0.001 into fresh brain heart infusion (BHI media. We found that compounds having the 2-pyridone central fragment decorated with a large aryl group at position C7 and an aryl at C8 (linked via a methylene linker) showed inhibition of growth relative to the dimethyl sulfoxide (DMSO) vehicle control treated condition (SI Appendix, Table S1). Further, SAR revealed that by introducing an aryl or benzyl substituent at position C2, the C8 aryl could be changed to a smaller cyclopropyl substituent (chemical synthesis of these compounds is described in the Supplemental Information). This resulted in compounds with increased potency against E. faecalis OG1RF including compounds EC240 (minimal inhibitory concentration [MIC] = 25 µM) and EC305 (MIC = 50 µM) (Fig. 1B).
2-Pyridone Compounds EC240 and EC350 have Antibacterial Activity Against Clinical Isolates of E. faecalis.
The inhibitory activities of EC240 and EC305 were tested against a panel of 10 enterococcal clinical isolates (SI Appendix, Table S2 provide more information), most of which were isolated from bloodstream infections that were resistant to vancomycin. The bacterial isolates were diluted to a density of 107 CFU/mL from overnight cultures in fresh media and then treated with: i) vehicle control (DMSO); ii) EC240; or iii) EC305 at 2 to 4× the MIC determined for E. faecalis OG1RF (100 µM). The cultures were incubated for 18 h and the number of surviving CFUs was determined. Treatment with the vehicle control alone resulted in over 2.5 log increases in CFUs relative to the initial inoculum. In contrast, the CFUs for all of the EC240 and EC305 treated clinical isolates were reduced by at least 5 logs relative to the vehicle control, demonstrating that EC240 and EC305 were efficacious in killing all clinical VRE strains tested (Fig. 1C). We noted that while the MIC of EC305 was higher than that of EC240 for OG1RF, the 2-pyridones performed similarly in reducing the CFUs/mL across most clinical isolates tested (Fig. 1C) with the exception of strains SJH-1324 and SJH-1325, where SJH-1324 appeared to be more susceptible to EC240 and SJH-1325 more susceptible to EC305. These results argue strongly for further analysis investigating 2-pyridones as effective antibiotics against clinically relevant strains of VRE.
Development of More Potent EC305 Derivatives.
Taking advantage of the SAR from the first-generation 2-pyridones against E. faecalis OG1RF and VRE clinical isolates, we sought to develop derivatives with increased potency against E. faecalis. 2-pyridones equipped with a smaller group at C8 and a substituted phenyl or benzyl group at C2 displayed moderate antibacterial activity (SI Appendix, Table S1). Due to easy tractability of substituted phenyl boronic acids compared to their benzyl counterparts to be coupled at the C2 position, we chose to derivatize EC305 to improve antibacterial activity. Thus, we focused on substituting the para position of the phenyl ring at C2 in compound EC305 with lipophilic side chains. A set of analogs were synthesized that contained straight alkyl chains with one to seven carbons (SI Appendix, Table S3). In addition, the aliphatic cyclopropyl substituent in C8 was replaced with a methoxy group to potentially improve biological and physico-chemical properties. Strikingly, we observed that among the C8 methoxy- and C2 alkyl chain analogs, PS757 had substantially improved antibacterial potency (MIC = 3 to 6 µM) compared to EC240 and EC305 (4 to 8× increased potency), with little to no detectable toxicity in cell culture at concentrations up to 4× MIC (25 µM), and better solubility relative to the other active derivatives (Fig. 1B and SI Appendix, Fig. S1 and Table S3). Thus, PS757 was selected for further study.
PS757 has Potent Bacteriostatic Activity on Exponential Phase E. faecalis Cells.
Upon observing a significantly decreased MIC for the second-generation compound PS757 relative to the first-generation compounds EC240 and EC305, we sought to better understand the antibacterial mechanism(s) of action for PS757 against enterococcal cells. In the studies above, the activity of EC240 and EC305 against exponentially growing and stationary phase cells was not differentiated. Thus, here we investigated the effect of PS757 on both actively dividing exponential phase cultures of E. faecalis and on stationary phase cells. Overnight OG1RF cultures were diluted to an OD600 of 0.001 and grown to early exponential phase (OD600 0.2 to 0.3, ∼1 × 108 CFU/mL) and were subsequently: i) left untreated; ii) treated with vehicle alone (DMSO); or iii) treated with PS757 at 0.5 to 4× MIC in twofold increments (3 to 24 µM). Growth was monitored by measuring OD600 at 15, 30, 60, 120, and 1,080 min (18 h) posttreatment. Unlike the untreated and vehicle control conditions, which continued growth into stationary phase, treatment with PS757 resulted in potent growth arrest of exponentially dividing cells through the course of the experiment when treated at a concentration of 1× MIC or above (≥6 µM) and resulted in slowed growth at 0.5× MIC (3 µM) as measured by OD600 (Fig. 2A). Thus, PS757 is bacteriostatic against actively growing enterococci. No decrease in CFUs was observed over 20 h of treatment relative to the starting culture at similar concentrations of PS757 (SI Appendix, Fig. S2). Thus, while the growth of exponential phase cells was inhibited, they were not being killed at the 1 to 4× MIC concentrations (Fig. 2A and SI Appendix, Fig. S2).
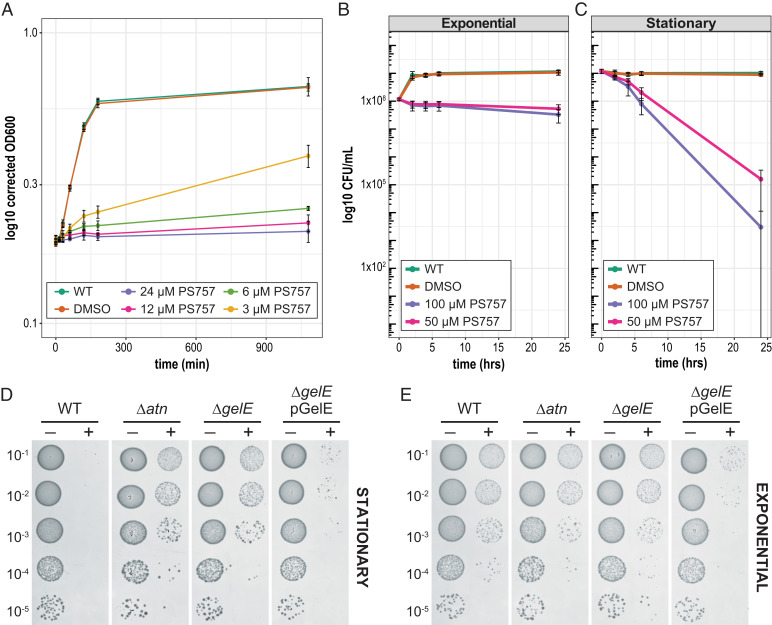
Fig. 2.
Second-generation 2-pyridone PS757 has bacteriostatic activity on exponential phase cultures and Atn autolysin-mediated bactericidal activity on nongrowing stationary phase cultures. (A) Growth of exponential phase cultures in the presence of 1/2–4× MIC (3 to 24 µM) of PS757. Exponential phase cultures (OD600= 0.2 to 0.3) of OG1RF were split and left untreated (green), treated with DMSO as a vehicle control (orange), and treated with 3 µM (yellow), 6 µM (light green), 12 µM (pink), and 24 µM (purple) PS757 at time 0. The OD600 was monitored at 15-, 30-, 60-, and 1,080-min (18 h) time points. To account for change in the OD600 due to compound addition, OD600 values were normalized by subtracting the change in OD600 at t = 0 from t = 15. Black bars represent SD of replicates. (B) Time course killing curves of exponential phase cells. Exponential phase cultures (OD600 = 0.2 to 0.3) of OG1RF were split and left untreated (green), treated with DMSO as a vehicle control (orange), and treated with 100 µM (purple) or 50 µM (pink) PS757 at time 0. The CFU/mL were enumerated at time 0, and 2 hr, 4 hr, 6 hr, and 24 hr timepoints posttreatment. (C) Time course killing curves of stationary phase cells. Stationary phase cultures (18 h) of OG1RF were split and left untreated (green), treated with DMSO as a vehicle control (orange), and treated with 100 µM (purple) or 50 µM (pink) PS757 at time 0. The CFU/mL were enumerated at time 0 and 2 hr, 4 hr, 6 hr, and 24 hr time points posttreatment. Black bars represent the SD of the replicates. (D, E) Spot titer assays of PS757-treated mutant cultures. Exponential phase (OD600 = 0.2 to 0.3) or stationary phase (18 h) cultures of indicated strains were treated with the vehicle control DMSO (−) or 100 µM PS757 (+) for 24 h. The serial dilution is indicated.
To test if exponential phase cells are killed at higher concentrations of PS757, exponentially growing cultures (OD600 0.2 to 0.3, ∼1 × 108 CFU/mL) were divided as previously described resulting in cultures that were: i) left untreated; ii) treated with vehicle alone (DMSO); or iii) treated with 50 to 100 µM (∼8 to 16x MIC) PS757 and CFUs were monitored at 0, 2, 4, 6, and 24 h posttreatment. Similar to the previous experiment, treatment of exponentially growing cells with 50 to 100 µM PS757 resulted in growth arrest compared to the untreated and vehicle control treated conditions. CFUs decreased <1 log even when cultures were treated with 100 µM for 24 h, suggesting there was no significant killing of exponential phase cells even with treatment of higher concentrations of PS757 (Fig. 2B).
PS757 has Bactericidal Activity on Nondividing Stationary Phase Cells.
To test the effect of PS757 on nondividing stationary phase cells, overnight OG1RF cultures were diluted to an OD600 of 0.001, grown to stationary phase (18 h), before the cultures were divided and then treated as previously described for the exponential phase cultures with 50 or 100 µM PS757. The surviving CFUs were then monitored at 0, 2, 4, 6, and 24 h post treatment. In contrast to exponential phase cultures, CFUs decreased by ∼0.5 to 1 log for the 50 µM and 100 µM PS757 treated conditions by 6 h posttreatment, whereas the CFUs of the untreated and vehicle control conditions remained unchanged (Fig. 2C). Indeed, by 24 h posttreatment, viability of treated cultures was reduced >4 logs in the presence of either 50 µM or 100 µM PS757, indicating that PS757 has bactericidal activity against stationary phase enterococcal cells (Fig. 2C).
PS757-Dependent Bactericidal Activity Partially Mimics Fratricide.
During enterococcal biofilm formation, the Atn autolysin functions in “fratricide,” a process that results in the lysis of cells and the release of extracellular DNA to form the biofilm matrix (28). During fratricide, Atn is converted from a regulator of chain length in dividing cells to an autolysin, and this conversion is dependent on the activity of proteases secreted in the stationary phase of growth (28, 29). The stationary phase GelE protease is thought to cleave Atn to release it from the cell wall in order to lyse surrounding cells (28). As PS757-mediated cell death is restricted to the stationary phase, as is the Atn and GelE-dependent process of fratricide, it was of interest to examine the contributions of Atn and GelE. An Atn mutant (Δatn) was grown to stationary phase and challenged with 50 μM or 100 μM PS757 as described above. When compared with WT, Δatn was significantly more resistant to killing by PS757 (Fig. 2D and SI Appendix, Fig. S3). Similarly, a mutant lacking GelE (ΔgelE) was also resistant to killing by PS757, with CFUs recovered at levels similar to Δatn (Fig. 2D and SI Appendix, Fig. S3). The Δatn and ΔgelE resistance to PS757 stationary phase killing did not extend to resistance to the bacteriostatic activity of PS757 for exponential phase cells, as exponential Δatn and ΔgelE cells showed the same sensitivity to PS757 as WT cells (Fig. 2E), suggesting the bactericidal mechanism of stationary phase cell death differs from the mechanism of growth inhibition in exponential phase cells. Taken together, these data indicate that because PS757-mediated stationary phase cell death is enhanced by Atn and GelE, it mimics aspects of fratricide.
In order to select for additional E. faecalis mutants with resistance to PS757, stationary phase cultures were plated on BHI-agar plates that contained 10, 20, and 40 μM concentrations of PS757-IMD (the imidazole salt of PS757), which improved the solubility of PS757 in the BHI-agar (SI Appendix, Fig. S4A). Three of the colonies that grew on the 40 μM PS757-IMD BHI-agar plates (HT061, HT062, and HT063) were selected for further analysis via whole genome sequencing. Comparison with the genome sequence from the original PS757-sensitive wild-type isolate revealed mutations in the intergenic region between the gene encoding the LmrB efflux pump, which is annotated as a lincomycin resistance protein, and a gene encoding a MerR transcriptional regulator in all three colonies (SI Appendix, Fig. S4E). Studies have shown that for the lmrB gene from Bacillus subtilis, which shares 42% identity and 62% similarity to the OG1RF LmrB at the protein level, mutations in the upstream intergenic region increases efflux activities through increased expression of lmrB (30), suggesting that the increased resistance to PS757 may be due to altered efflux in the mutant strains. Exponential phase cultures of these mutant cells could also grow in the presence of 6 to 12 μM PS757 (1 to 2× MIC) in BHI liquid media (SI Appendix, Fig. S4 B–D), suggesting that mutations in this intergenic region are also effective at overcoming the bacteriostatic effects of PS757. While the resistant isolates grew at 6 to 12 μM PS757, little to no growth was observed at concentrations greater than 12 μM in BHI liquid media (SI Appendix, Fig. S4C), despite being isolated on BHI-agar plates containing 40 μM PS757-IMD. Thus, we observed a decreased effectiveness of PS757 in solid media. Together these data suggest that altered efflux activities may also contribute to PS757 resistance in enterococcal cells.
Overcoming Vancomycin, Gentamicin, and Ciprofloxacin Resistance with Sublethal Concentrations of PS757.
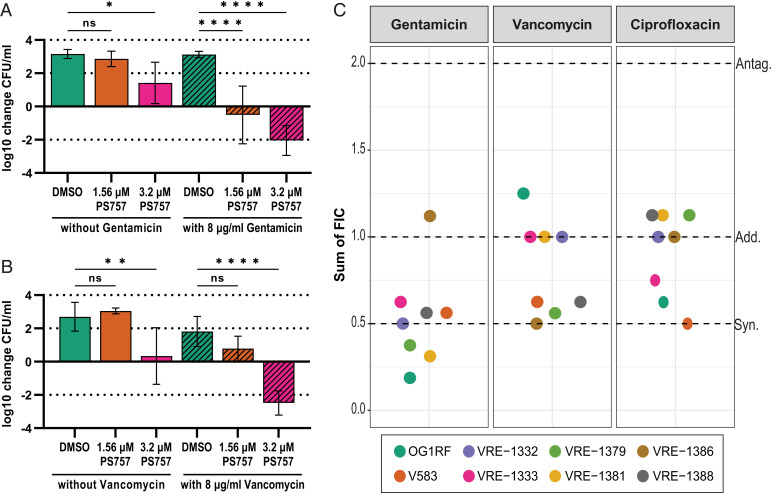
In some cases, multiple antibiotic resistant enterococci can be treated with a combination of antibiotics that act synergistically, such as aminoglycosides and β-lactam antibiotics (31). To determine if PS757 could act synergistically with standard-of-care antibiotics, we combined subinhibitory concentrations of PS757 with subinhibitory concentrations of standard-of-care antibiotics to test their efficacy against MDR enterococci. E. faecalis OG1RF, which is intrinsically resistant to gentamicin (MIC 64 µg/mL) as is characteristic of enterococci, was diluted to an OD600 of 0.001 from an overnight culture into fresh media and then treated with: i) vehicle control (DMSO) alone; ii) a subinhibitory concentration of gentamicin (8 µg/mL); iii) subinhibitory concentrations of PS757 (1.56 or 3.2 µM); or iv) a combination of subinhibitory concentrations of gentamicin and PS757 (8 µg/mL and 1.56 or 3.2 µM, respectively). Viability was then assessed by determination of CFUs following 20 h of incubation. While in vitro treatment of OG1RF with 1.56 µM PS757 or 8 µg/mL gentamicin alone had little to no effect on CFUs, combinatorial treatment with the same subinhibitory concentrations of both gentamicin and PS757 significantly reduced CFUs by ∼4 logs relative to the DMSO treated control (Fig. 3A). Similarly, treatment with 3.2 µM PS757 alone resulted in reduced growth relative to the 1.56 µM PS757 and DMSO conditions, but combinatorial treatment with both 8 µg/mL gentamicin and 3.2 µM PS757 resulted in significantly greater cell death compared to all other conditions, reducing CFUs by ∼5 logs relative to the vehicle control. Thus, PS757 can overcome the moderate intrinsic level of resistance of enterococcus to gentamicin. In addition to gentamicin resistance, the VRE strain V583 is also resistant to vancomycin (MIC >32 µg/mL). As previously described for gentamicin, we treated V583 with vancomycin (8 µg/mL) alone or subinhibitory concentrations of PS757 (1.56 µM and 3.2 µM) alone or the two in combination with one another. Similar to the gentamicin results, the combinatorial treatment of vancomycin with PS757 was significantly more effective at reducing the number of viable bacteria than any of the respective single treatment conditions (Fig. 3B).
Fig. 3.
Sublethal concentrations of PS757 acts in combination with sublethal levels of gentamicin, vancomycin, and ciprofloxacin to kill OG1RF, V583, and VRE clinical isolates. Sublethal concentrations of PS757 combined with sublethal concentrations of gentamicin or vancomycin results in cell death. (A) Overnight cultures of OG1RF were diluted into fresh media and treated with either DMSO or sublethal concentrations of PS757 (1.56 μM or 3.2 μM) alone or in combination with a sublethal concentration of gentamicin (8 μg/mL). (B) Overnight cultures of V583 were diluted into fresh media and treated with either DMSO or sublethal concentrations of PS757 (1.56 μM or 3.2 μM) alone or in combination with a sublethal concentration of vancomycin (8 μg/mL). (A, B) Net fold change (see Materials and Methods) is presented as the average change in final CFU/mL relative to the CFU/mL of the untreated initial inoculum. Black bars represent SD of replicates. Statistical analysis by one-way ANOVA (*P < 0.05, **P < 0.01, ****P < 0.0001, ns: not significant). (C) Sums of the FIC derived from a checkerboard analysis (see Materials and Methods) of PS757 with gentamicin, vancomycin, and ciprofloxacin in clinical VRE strains. Strains OG1RF, V583, 1332, and 1333 are antibiotic resistant E. faecalis and strains 1379, 1381, 1386, and 1388 are antibiotic resistant E. faecium. FIC of ≤ 0.5 was considered synergistic (“Syn.”), >0.5 to 2.0 additive (“Add.”) and >2.0 antagonistic (“Antag.”), as indicated by the dashed lines.
Combinatorial treatment was then tested against 6 additional VRE isolates, including moderate-level (MIC = 62 to 500 µg/mL) gentamicin-resistant E. faecalis strains (1332, and 1333) and high-level (MIC = >2,000 µg/mL) gentamicin-resistant E. faecium strains (1379, 1381, 1386, and 1388) (SI Appendix, Table S2). The panel was subjected to combinatorial treatment of sublethal doses of PS757 with sublethal concentrations of either: i) gentamicin, which targets translation; ii) vancomycin, which targets cell wall synthesis; or iii) ciprofloxacin, which targets DNA synthesis. A checkerboard assay (see Materials and Methods), which is used to assess the activity of the combination of antibiotics compared to their activities alone, revealed that the fractional inhibitory concentrations (FIC) of PS757 almost always acted in either an additive or synergistic manner when combined with any of the antibiotics tested against this panel and that combined treatment lowered the MIC of the antibiotics tested up to 16-fold (Fig. 3C and SI Appendix, Table S4).
PS757 Increases Transmembrane Influx and Reduces Efflux in E. faecalis.
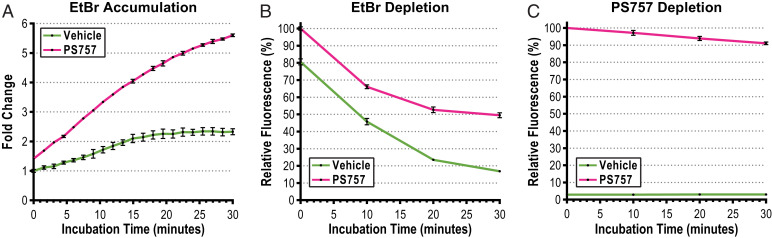
The impermeability of the membrane to many antibiotics plays an important role in enterococcal antibiotic resistance (32–35). To investigate whether PS757 can alter antibiotic influx, an ethidium bromide (EtBr) accumulation assay was performed as follows: cells from overnight cultures of E. faecalis OG1RF were resuspended in PBS and simultaneously treated with EtBr and: i) DMSO (vehicle control); or ii) PS757. EtBr becomes fluorescent when it is bound to DNA (36, 37), therefore EtBr influx was determined by monitoring the increase in fluorescence over 30 min. The cells treated with PS757 accumulated twofold to threefold more EtBr than cells treated with DMSO (Fig. 4). It was also determined that PS757 could similarly increase the influx capacity of the VRE strain V583 (SI Appendix, Fig. S5). To test whether PS757 can also alter antibiotic efflux, cells from overnight cultures were preloaded with EtBr in the absence of an energy source and in the presence of the efflux pump inhibitor reserpine (38). Cells were then washed to remove reserpine; glucose was added to reactivate efflux pumps and the decrease of fluorescence was measured over 30 min to determine EtBr efflux in the presence of: i) DMSO (vehicle control); ii) PS757; or iii) other characterized efflux inhibitors (chlorpromazine, reserpine, lansoprazole, and verapamil). We found that PS757 could inhibit efflux with an efficiency similar to chlorpromazine and more efficiently than other characterized inhibitors, including reserpine, lansoprazole or verapamil (SI Appendix, Fig. S6 A and B). None of the other characterized efflux inhibitors were capable of increasing EtBr influx similar to PS757 (SI Appendix, Fig. S6 C and D). Taken together, these data demonstrate that a sublethal concentration of PS757 can both inhibit efflux and promote influx of a small molecule across the cellular membrane, which likely contributes to its ability to synergize with standard-of-care antibiotics.
Fig. 4.
Sublethal concentrations of PS757 increase influx and decrease efflux in OG1RF. (A) EtBr accumulation in OG1RF cells treated with vehicle (DMSO, green) or PS757 (pink). EtBr accumulation results are plotted as fold-change of EtBr fluorescence over a 30-min time course. (B) Relative fluorescence of EtBr depletion of OG1RF cells pretreated with EtBr and i) vehicle (DMSO, green); or ii) PS757 (pink). (C) Relative fluorescence of PS757 depletion of OG1RF cells pretreated with EtBr and i) vehicle (DMSO, green); or ii) PS757 (pink). EtBr and PS757 depletion results are plotted as % relative EtBr fluorescence (respective to the initial fluorescence, 100% at t = 0) over a 30-min time course. Cells were pretreated with EtBr (1 µg/mL), DMSO (0.05%) and/or PS757 (5 µM). The fluorescence signals emitted from PS757 (λEx = 400 nm and λEm = 440 nm) and EtBr (λEx = 544 nm and λEm = 590 nm) were measured with SpectraMax iD3 microplate reader (Molecular Devices). All measurements were performed in triplicate and results were plotted using GraphPad Prism (version 9.3.1). Black bars represent SDs of replicates.
PS757 Associates Rapidly with E. faecalis But Is Not Depleted Efficiently.
Since PS757 can alter influx and efflux, it was of interest to determine how it interacts with cells. For this, we took advantage of its intrinsic fluorescent properties (excitation 400 nm, emission 440 nm; SI Appendix, Fig. S7). Enterococcal cells from overnight cultures were exposed to PS757 in PBS and sampled over a 30-min period to determine PS757 association with the bacteria over time. This analysis revealed that PS757 associated with cells of strains OG1RF and V583 very rapidly, with cells achieving their peak fluorescence value by 15 min postexposure (SI Appendix, Fig. S8). To simultaneously compare depletion rates of EtBr and PS757, cells from overnight cultures were preloaded with EtBr and either: i) DMSO (vehicle control) or ii) PS757 in the absence of an energy source. Cells were then washed, energized, and sampled over a 30-min period to determine depletion rates. Cells treated with the vehicle alone depleted ∼80% of the EtBr signal, while those treated with PS757 only lost ∼50% of the EtBr signal (Fig. 4B and SI Appendix, Fig. S5B). In contrast, depletion of PS757 was less efficient, with cells only losing about ∼10% of their initial PS757 fluorescent signal by 30 min (Fig. 4C and SI Appendix, Fig. S5C). Taken together, these data indicate that: i) only a brief exposure to PS757 is sufficient to inhibit EtBr efflux over a longer time scale; ii) PS757 itself rapidly associates with enterococcal cells; and iii) PS757 is not efficiently depleted from cells.
PS757 has Potent Inhibitory Activity Across a Variety of Pathogenic Gram-Positive Firmicutes.
Given the potent inhibitory effects we observed for E. faecalis, we analyzed the effects of treatment with PS757 against a panel of Gram-positive pathogens. Based on MICs, we found that PS757 effectively inhibits growth of all Gram-positive Firmicutes tested, including Streptococcus pyogenes, Streptococcus agalactiae, Streptococcus pneumoniae, MRSA, and the model organism Bacillus subtilis (SI Appendix, Table S5). Notably, PS757 did not affect the growth of the Gram-negative bacterium Escherichia coli, however for each of the Gram-positive pathogens tested, PS757 had more potent MIC activity across these species than in enterococcal species, suggesting that 2-pyridone derivatives can be modified to be even more efficacious in the treatment of resistant staphylococcal and streptococcal infections. We also observed that treatment of Staphylococcal aureus JE-2 cells, a plasmid-cured MRSA strain (39), with 50 μM PS757 resulted in >4 logs of cell death of both exponential and stationary phase cultures, suggesting that, unlike enterococcal cells, PS757 has bactericidal activity against both exponential and stationary phase cultures (SI Appendix, Fig. S9), making it an attractive alternative to target multidrug-resistant staphylococcal infections. The mechanism of species-specific PS757-induced cell death is the subject of future studies. Given their broad and potent bactericidal activity against Gram-positive (GmP) bacteria, GmPcides have the potential to be developed into much-needed therapies to treat MDR infections.
Discussion
Current clinically available antibiotics typically show the greatest bactericidal effects on growing bacteria (40). Indeed, for the commonly prescribed β-lactam antibiotics, a linear correlation has been shown between growth rate and bacterial cell lysis (41). While most in vitro studies examine the effects of antibiotics on exponentially growing cultures of domesticated laboratory strains in rich media, it is unlikely that this accurately recapitulates the growth of pathogenic strains of bacteria within a host, which must contend with limited nutrients and the host immune response. Here we have shown that the new class of 2-pyridone compounds, which we have named GmPcides, including the second-generation compound PS757, have bactericidal activity against nondividing stationary phase cultures of E. faecalis OG1RF and recently isolated clinical strains of Enterococcus as well as other Gram-positive pathogens. In contrast, concentrations of PS757 which reduced CFUs of stationary phase enterococcal cells by >105 instead showed potent bacteriostatic effects and little to no bactericidal activity against enterococci growing in exponential phase (Fig. 2). Thus, the bactericidal effects were predominantly observed in nondividing stationary phase enterococcal cells.
Similar to the effect of PS757 on nondividing enterococci, daptomycin also has bactericidal activity against nondividing Gram-positive S. aureus cells (42). As a lipopeptide antibiotic, daptomycin works by disrupting cell membrane integrity, causing leakage that collapses membrane potential and results in cell death (43). Daptomycin shows concentration-dependent bactericidal activity, defined as >3 log reduction in viable stationary phase S. aureus cultures (42). Gentamicin shows a slower bactericidal effect against stationary phase S. aureus relative to daptomycin over 24 h of treatment. In contrast, ciprofloxacin, nafcillin, and vancomycin did not reach the >3 log reduction in viable stationary phase cells grown under the same conditions (42). Daptomycin also has rapid bactericidal activity against exponentially growing cultures of S. aureus, with 3+ log reduction in viable cell count observed within 60 min at a concentration of 4x MIC (4 µg/mL) (42). Thus, PS757, which is very structurally distinct from daptomycin, shows similar properties of being active on both nondividing stationary phase cells and actively dividing S. aureus cells (SI Appendix, Fig. S9), but is unusual in its bacteriostatic activity toward exponential phase enterococcal cells (Fig. 2). We found that the GelE protease, which is expressed in stationary phase, and the autolysin Atn contribute to the bactericidal effect observed in stationary phase enterococcal cells. We hypothesize that treatment with PS757 results in changes to cell wall composition making it more sensitive to Atn-mediated hydrolysis, whose activity is activated by GelE during the stationary phase. It will be interesting to examine whether other physiological changes in stationary phase cultures, such as lowered pH or growth phase specific metabolites, also contribute to PS757-mediated cell death of stationary phase cells.
The bactericidal activity of compounds such as PS757 against stationary phase enterococcal cells may hold clinical promise for treatment of enterococcal infections in patients, which are unlikely to be experiencing exponential growth. Moderate level resistant (MLR) enterococcal strains show decreased permeability to aminoglycosides such as gentamicin (44). Cell wall acting antibiotics, such as a β-lactams, can increase aminoglycoside influx. Thus, combination aminoglycoside/β-lactam therapy has been used in the clinical setting for treatment of aminoglycoside resistant enterococcal infections. An additional benefit of the aminoglycoside/β-lactam combination therapy is that the synergistic effects observed between these antibiotics allow for decreased concentrations of administered aminoglycosides, which are nephrotoxic in higher sustained quantities. High-level resistance (HLR) to aminoglycosides is usually mediated by aminoglycoside modification enzymes (45). Thus, the combination therapy described above is ineffective for treatment of a substantial number of enterococcal infections that exhibit HLR to aminoglycosides. However, other combination therapies that use ampicillin with a third-generation cephalosporin have been shown to be effective in vitro and in vivo for the treatment of HLR enterococci with aminoglycosides. Further, this combination exhibits lower rates of nephrotoxicity compared to patients administered the aminoglycoside and β-lactam combination therapy. The presumed mechanism of action of the double β-lactam therapy is through the saturation of different penicillin binding proteins (PBPs).
The combination of sublethal concentrations of PS757 with the aminoglycoside gentamicin resulted in additive to synergistic effects against both MLR (OG1RF, VRE-1332, VRE-1333) and HLR (V583, VRE-1379, VRE-1381, VRE-1386, VRE-1388) enterococcal strains (Fig. 3C and SI Appendix, Table S4), raising intriguing questions about the mechanism of action for the combinatorial effects. We have shown that treatment with sublethal concentrations of PS757 results in a twofold to threefold increased accumulation of ethidium bromide compared to the vehicle control over a 30-min time-course in both an MLR strain OG1RF and a HLR strain V583 (Fig. 4 and SI Appendix, Fig S4). Thus, PS757 may work with aminoglycosides by increasing the permeability of the cell wall such that influx is increased. This suggests that the PS757-mediated increase in influx increases the intracellular concentrations of gentamicin, which would explain the observed additive to synergistic effects in the MLR strains. However, despite increased intracellular concentrations of gentamicin, HLR strains would presumably remain resistant due to the activity of the aminoglycoside modification enzymes. Thus, the cellular response to PS757 may have additional effects beyond increasing influx, such as decreased expression or inactivation of aminoglycoside modification enzymes. We demonstrate that PS757-resistant strains can be isolated with mutations in the intergenic region of the LmrB efflux pump (SI Appendix, Fig. S4), suggesting that PS757 may enter the cytoplasm and that efflux though LmrB could decrease sensitivity to intracellular PS757, supporting the hypothesis that PS757 may have intracellular targets. Future work is necessary to fully elucidate the scope of the targets and the effects of PS757 treatment.
We also observed primarily additive effects between sublethal concentrations of PS757 and the cell wall antibiotic vancomycin (Fig. 3C and SI Appendix, Table S4). Vancomycin works through binding to the peptidoglycan precursor D-alanine-D-alanine (D-Ala-D-Ala), which is involved in transpeptidation and prevents the formation of cross-bridges in peptidoglycan (46). Resistance occurs through modification of the D-Ala-D-Ala precursors to either D-alanine-D-lactate (D-Ala-D-Lac) or D-alanine-D-serine (D-Ala-D-Ser), which results in ∼1,000-fold and ∼7-fold decreased affinities with vancomycin, respectively (47, 48). Modification of the D-Ala-D-Ala precursor is achieved by a series of genes whose expression is regulated by a two-component system that responds to the presence of vancomycin or early cell wall stress induced by vancomycin. It is believed that the cross-linking reaction to incorporate the modified precursors into the cell wall also requires high-molecular weight PBPs (49). Thus, the additive effects observed with sublethal concentrations of PS757 may be due to targeting of several processes, including the interaction of PS757 with the high molecular weight PBPs required to incorporate the modified precursors (D-Ala-D-Lac or D-Ala-D-Ser), which could result in decreased peptidoglycan crosslinking and cell wall integrity (49). Another potential mechanism is PS757-mediated perturbation of the cell wall such that signaling is decreased from the two-component system required for expression of the genes that modify D-Ala-D-Ala to confer resistance to vancomycin. Alternatively, PS757 could target the cell wall precursor lipid II in a mechanism similar to Teixobactin (50). The combination effects observed for sublethal doses of PS757 and vancomycin against VRE also raise intriguing possibilities for future studies of combination therapy to treat enterococcal strains that display heteroresistance to vancomycin (51). Heteroresistance occurs when a small subpopulation of antibiotic resistant bacteria exists among a largely susceptible bacterial population (52). Recent work has shown that combination therapy against bacterial strains that display multiple heteroresistances can be used to kill these strains when monotherapy alone is insufficient (53).
Given the broad-spectrum activity of PS757 against Gram-positive pathogens, coupled with the unusual activities against nondividing enterococcal cells and additive to synergistic effects observed with standard-of-care antibiotics, GmPcides have the potential to be a valuable addition to our pharmacopeia. Additionally, synthetic methodologies will allow for fine-tuning of future generations of GmPcides to select for species-specific properties that are most advantageous in treatment. Further analysis of GmPcides, including the identification of their target, their species-specific mechanism of action, and the mechanisms by which they act additively to synergistically with standard-of-care antibiotics will also provide a valuable chemical tool for the identification of novel pathways that can be exploited for future drug development.
Materials and Methods
Bacterial Strains and Growth Conditions.
Unless otherwise specified, all bacterial cultures were inoculated from a single bacterial colony grown on Brain Heart Infusion (BHI) agar plates and grown overnight in BHI broth (BD and Company). Liquid cultures were grown static (enterococci and streptococci) or shaking (staphylococci, B. subtilis) at 37 °C for 18 h, as described (54–56). Other growth conditions and the concentration of the antibiotics used are described in the text or below. Number of CFUs was determined by serial dilution and quantitative plating on BHI agar plates. Bacterial strains are listed in SI Appendix, Table S2.
Determination of MIC and Minimum Bactericidal Concentration.
Broth microdilution MIC and minimum bactericidal conentratrion (MBC) assays were performed in BHI medium following the Clinical and Laboratory Standards Institute (CLSI) methods for antimicrobial susceptibility testing (57). In accordance with CLSI standards, the MIC of each antimicrobial was defined as the lowest concentration of compound that prevented bacterial growth relative to the inoculum (107 CFU), as determined by optical density (OD600) using a spectrophotometer. Validation of assays included routine testing of quality control standard cultures, including E. faecalis ATCC 29212 and E. faecalis ATCC 51299. MIC and MBC values for experimental compounds were rejected if MICs and MBCs of quality control strains were outside of CLSI determined quality control ranges when tested against a standardized panel of antimicrobials (57).
Susceptibility of clinical isolates.
Frozen stocks of clinical VRE bloodstream isolates (SI Appendix, Table S2) were used to inoculate 10 mL of BHI in 15 mL conical tubes. Caps were tightened and cultures were grown overnight at 37 °C. Overnight cultures were normalized to OD600 = 1 and diluted 1:1,000 in fresh BHI, constituting the initial inoculum. Ten-fold serial dilutions in PBS (pH 7.4) were plated to calculate the CFUs of the initial inoculum (∼1 × 106 CFUs/mL). For each diluted culture, 990 µL was aliquoted into 1.5 mL microcentrifuge tubes containing 10 µL of 100% DMSO, 10 mM EC240, or 10 mM EC305 (final concentration of 100 µM) and mixed gently by pipetting. Sealed tubes were then placed at 37 °C for 18 h. Tenfold serial dilutions of each culture in PBS (pH 7.4) were subsequently plated on BHI agar plates and resulting final CFUs were enumerated. The experiment was repeated in triplicate on three separate days. The plotted net change values were calculated as the average of the log10 (final CFU/mL) – log10 (inoculum CFU/mL) values for each replicate.
Growth Curves of Exponential Phase Cultures.
Overnight cultures of OG1RF were inoculated into 10 mL of BHI in 15 mL conical tubes from frozen stocks with the caps tightened and grown static at 37 °C overnight. Resulting cultures were normalized to OD600 = 1 and then diluted 1:1,000 into two 15 mL conical tubes with 10 mL of fresh BHI and grown static with the caps tightened at 37 °C to an OD600 = 0.2 to 0.3. The cultures were pooled and the initial OD600 was measured (t = 0). The culture was then split and 1.98 mL of culture was added to the wells of a 12-well tissue culture test plate (TPP, 92012). A 2.4 mM solution of PS757 was prepared in DMSO and serially diluted twofold into 100% DMSO to create a range of 100× solutions (1.2 mM, 0.6 mM, 0.3 mM). Twenty microliters of the DMSO vehicle control or 100× solutions were added into the culture containing wells. The top and bottom rows of wells contained 2 mL BHI with no compound or bacteria to serve as blanks. Cultures were incubated at 37 °C and at specified time intervals cultures were mixed by pipetting and OD600 was measured with the BioTek Synergy H1 Microplate Reader. The OD600 values were normalized between the t = 15 and t = 0 timepoints to account for the PS757-dependent increase in optical density (OD600t15 – OD600t0), as PS757 gives an OD600 signal at high concentrations, and this correction value was subtracted from all subsequent readings. The average of three independent replicates performed on separate days was plotted and the SD of the replicates are indicated.
Killing Curves of Exponential and Stationary Phase Cultures.
Overnight cultures of specified enterococcal strains were inoculated into 10 mL of BHI in 15 mL conical tubes with the caps tightened from frozen stocks and grown overnight. Cultures were then normalized to OD600 = 1 and diluted 1:1,000 into 10 mL of fresh BHI in 15 mL conical tubes with the caps tightened. For stationary phase culture experiments, diluted cultures were then grown for 18 h overnight at 37 °C, multiple 18-h cultures of the same strain were pooled, and tenfold serial dilutions in PBS (pH 7.4) were performed to calculate CFUs of the initial inoculum. For exponential phase culture experiments, diluted cultures were grown at 37 °C to an OD600 = 0.2 to 0.3 (∼4.5 h), pooled, and tenfold serial dilutions in PBS (pH 7.4) were performed to calculate CFUs of the initial inoculum. For both stationary and exponential phase experiments, 990 µL of the initial inoculum was aliquoted into 1.5 mL microcentrifuge tubes containing 10 µL of 100% DMSO, 10 mM PS757, or 5 mM PS757 (final concentration of 100 µM and 50 µM, respectively), mixed gently by pipetting, and placed at 37 °C. For time course experiments, at each timepoint (2, 4, 6 and 24 h) the cultures were gently inverted four times and 50 µL of culture was taken to perform ten-fold serial dilutions in PBS (pH 7.4). The serial dilutions were subsequently plated and CFUs were enumerated at dilutions that yielded 10 to 80 colonies. For the mutant killing curve experiments, the CFUs were enumerated at 20 h posttreatment. Each experiment was repeated in triplicate on 3 separate days and included at least two technical replicates. Where the results are plotted as net fold changes, the values were calculated as the average of the log10 (final CFU/mL) – log10 (inoculum CFU/mL) values for each replicate, where the results are not reported as net fold-changes they are average of the replicates with the SD indicated.
Spot Titer Assays.
Exponential and stationary phase cultures were prepared as described in the previous section. Overnight cultures of the complementation strain OG1RF ΔgelEpgelE were grown as previously described (58). Briefly, strains were streaked from frozen stocks onto BHI agar plates with kanamycin (500 μg/mL) and resulting colonies were restreaked twice onto BHI agar plates with kanamycin (500 μg/mL). Colonies were then inoculated into liquid BHI containing kanamycin (500 μg/mL) and chloramphenicol (25 μg/mL) and grown overnight at 37 °C. After 24 h of treatment with 100 μM PS757 or vehicle control (DMSO), tenfold serial dilutions in PBS (pH 7.4) were prepared and 5 µL of culture was spotted onto BHI agar plates. The plates were incubated overnight at 37 °C and subsequently imaged on the ChemiDoc Imaging System using the Bio-Rad Image Touch Software (version 3.0.1.14).
EtBr Depletion and Accumulation Assays.
Bacterial cultures were grown overnight in 12 mL of BHI in 50 mL conical tubes with loosely closed caps at 37 °C statically. For the EtBr depletion assay, cultures were pelleted, washed twice with PBS (pH 7.4) and normalized to OD600 = 1 in 12 mL of PBS (pH 7.4) with 0.5 µg/mL EtBr and 20 µg/mL reserpine followed by 1-h static incubation at 37 °C. The cells were then harvested by centrifugation and resuspended with 12 mL of PBS (pH 7.4). Cell suspensions (100 µL) were aliquoted to flat-bottomed black 96-well plates (Grenier, M4936) containing 100 µL of PBS (pH 7.4) with 0.4% glucose and/or 10 µM of assay compounds. For EtBr accumulation assay, the PBS-washed cells were normalized to OD600 = 1 in 12 mL of PBS (pH 7.4) and 100 µL of cell suspensions were aliquoted to flat-bottomed black 96-well plates (Grenier, M4936) containing 100 µL of PBS (pH 7.4) with 1.0 µg/mL EtBr, 0.4% glucose and/or 10 µM of assay compounds. The fluorescence signal emitted from EtBr (λEx = 544 nm and λEm = 590 nm) was measured every 1.5 min for 30 min by shaking 10 seconds without lid in SpectraMax Id3 microplate reader (Molecular Devices) that was preheated to 37 °C. All measurements were performed in triplicates. The fluorescence signals from EtBr were normalized with the average EtBr signal of vehicle control samples at zero minute. The normalized data were then plotted using GraphPad Prism (version 9.3.1).
UV-Vis Spectra.
The absorbance of PS757 was measured in T90+ UV/VIS spectrophotometer. The compound was dissolved in DMSO at 10 mM concentration. The baseline was corrected against blank DMSO control. The spectral scan was recorded in the range of 280 to 520 nm with a scanning speed of 100 nm/min and plotted using the Spectragryph software (59). The absorption maxima (lmax) was determined from the maximum absorbance value against the wavelength.
General Fluorescence Measurements.
The fluorescence emission of PS757 was measured using a JASCO FP-6500 spectrofluorometer. Ten micrometers of PS757 in DMSO was excited at 400 nm and the emission spectra was recorded in the range of 410 to 650 nm at 25 °C. The baseline was corrected with the blank DMSO control. The emission maxima (lmax) was determined from the maximum emission value against the wavelength.
PS757 Accumulation and Depletion Assays.
Bacteria cultures were grown overnight in 12 mL of BHI in 50 mL conical tubes with loosely closed caps at 37 °C statically. For the PS757 accumulation assay, cultures were pelleted, washed twice with PBS (pH 7.4) and normalized to OD600 = 1 in 6 mL of PBS (pH 7.4) with 0.4% glucose. Then the cells were incubated with either i) 0.01% DMSO (vehicle); or ii) 1 µM PS757 for 30-min under static incubation at 37 °C. At 0, 15 and 30 min, 1 mL of these cell suspensions were harvested by centrifugation for 3 min, resuspended with 1 mL of PBS (pH 7.4) and 200 µL of cell suspensions were aliquoted to flat-bottomed black 96-well plates with clear bottom (Grenier, M0562). For the PS757-EtBr depletion assay, the PBS-washed cells were normalized to OD600 = 1 in 5 mL of PBS (pH 7.4) with 0.5 µg/mL EtBr followed by 30-min static incubation at 37 °C. As follows, DMSO (vehicle) or PS757 was added to 0.05% or 5 µM final concentration to the EtBr preloaded cells and the samples were incubated at room temperature for 3 to 4 min. The cells were then harvested by centrifugation and resuspended with 5 mL of PBS (pH 7.4) with 0.4% glucose followed by static incubation at 37 °C for 30 min. At 0, 15 and 30 min, 1 mL of these cell suspensions were harvested by centrifugation and resuspended with 1 mL of PBS (pH 7.4). As follows, 200 µL of cell suspensions were aliquoted to flat-bottomed black 96-well plates with clear bottom (Grenier, M0562). The fluorescence signals emitted from PS757 (λEx = 400 nm and λEm = 440 nm) and EtBr (λEx = 544 nm and λEm = 590 nm) as well as OD600 were measured without lid in SpectraMax iD3 microplate reader (Molecular Devices) at each time point. All measurements were performed in triplicates. The fluorescence signals from PS757 and EtBr were first normalized with OD600 measurements, and then normalized with the average of vehicle control at zero minute. The normalized data were then plotted using GraphPad Prism (version 9.3.1).
Combination Treatment with Sublethal Concentrations of PS757 and Standard of Care Antibiotics.
Colonies were inoculated into 10 mL of BHI in a 15 mL conical tube with the caps tightly secured for overnight growth at 37 °C. The cultures were then normalized to an OD600 = 1.0 and diluted 1:1,000 in fresh BHI. This initial inoculum was then serially diluted to determine the initial CFUs/mL Gentamicin, vancomycin, and PS757 solutions were prepared at 40x concentrations and 5 µL of each drug was aliquoted into a 13 × 100mm mL glass tube. For the vehicle control conditions, 10 µL of 10% DMSO was added instead. The initial inoculum was then added to each tube at a volume of 190 µL, gently mixed with the antibiotic, and the culture was then grown statically at 37 °C for 18 to 20 h. Following the treatment period, the cultures were serially diluted and plated for CFUs on BHI agar plates. Each condition is represented by at least 4 replicates. The net fold change values were calculated as the average of the log10 (final CFU/mL) – log10 (inoculum CFU/mL) values for each replicate.
Assessment of Antimicrobial Synergy.
The checkerboard method was employed to evaluate whether treatment with two different antimicrobials in an in vitro assay had a synergistic effect by testing a cross-section of interceding antimicrobial concentration gradients following a 1:2 dilution scheme across a 96-well assay plate (60). The interaction of two antimicrobial compounds was determined to be antagonistic, synergistic, or indifferent by calculating the sum of fractional inhibition combinations (FIC). FIC values for each concentration of a compound were calculated as the ratio of the MIC of a compound in combination versus the MIC of the compound alone. An interaction was considered antagonistic when the FIC is ≥2, indifferent when FIC is between 0.5 and 2.0, and synergistic when FIC is ≤0.5. Controls included single compound gradients and untreated cultures.
Supplementary Material
Acknowledgments
This work was supported by the NIH R01AI134847-01A1 (F.A.), RO1DK51406 (S.J.H and M.G.C), and 1U19AI157797-01 (S.J.H, F.A., and M.G.C) and R01DK128805 (A.L.F.-M.). It was also supported by the Swedish Research Council 2018-04589 and 2021-05040J (F.A.), the Kempe Foundation SMK-1755 (F.A.), and the Erling-Persson Foundation (J.J., H.T., M.B., F.A., P.S., S.S., A.E.G.L.). Parts of this project have been supported under the framework of the JPIAMR–Joint Programming Initiative on Anti-microbial Resistance 2018-00969 (F.A.). T.M.N. was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number T32AI007172.
Footnotes
J.J., F.A., A.L.F.M., M.G.C., and S.J.H. have ownership interest in QureTech Bio AB, which licenses EC240, EC305, PS757, FN075, EC260, EC312, PS579, PS581, PS583, PS623, PS624, PS625, PS627, PS631, PS749, and PS756, and may benefit if the company is successful in marketing GmPcides. S.J.H, F.A., and J.J. serve on the Board of Directors for QureTech Bio AB. The remaining authors declare no competing interests.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2210912119/-/DCSupplemental.
Data, Materials, and Software Availability
DNA sequencing data have been deposited in SRA [PRJNA876221 (61)].
All study data are included in the article and/or supporting information.
References
- 1.Antimicrobial Resistance Collaborators, Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 399, 629–655 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Neill J., Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Review on Antimicrobial Resistance. Wellcome Trust and HM Government. https://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf, Accessed 23 May 2022).
- 3.Miethke M., et al. , Towards the sustainable discovery and development of new antibiotics. Nat. Rev. Chem. 5, 726–749 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hutchings M. I., Truman A. W., Wilkinson B., Antibiotics: Past, present and future. Curr. Opin. Microbiol. 51, 72–80 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Tsiodras S., et al. , Linezolid resistance in a clinical isolate of Staphylococcus aureus. Lancet 358, 207–208 (2001). [DOI] [PubMed] [Google Scholar]
- 6.J. S. Lewis, 2nd, et al. , Emergence of daptomycin resistance in Enterococcus faecium during daptomycin therapy. Antimicrob. Agents Chemother. 49, 1664–1665 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Somoskovi A., Bruderer V., Hömke R., Bloemberg G. V., Böttger E. C., A mutation associated with clofazimine and bedaquiline cross-resistance in MDR-TB following bedaquiline treatment. Eur. Respir. J. 45, 554–557 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention (U.S.), Antibiotic resistance threats in the United States, 2019 (National Center for Emerging Zoonotic and Infectious Diseases, U.S., 2019), 10.15620/cdc:82532. [DOI] [Google Scholar]
- 9.Roberts R. R., et al. , Costs attributable to healthcare-acquired infection in hospitalized adults and a comparison of economic methods. Med. Care 48, 1026–1035 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Fiore E., Van Tyne D., Gilmore M. S., Pathogenicity of Enterococci. Microbiol. Spectr. 7, 7.4.9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilmore M. S., Clewell D. B., Ike Y., Shankar N., Eds., Enterococci: From Commensals to Leading Causes of Drug Resistant Infection (Massachusetts Eye and Ear Infirmary, 2014). [PubMed] [Google Scholar]
- 12.Bradley C. R., Fraise A. P., Heat and chemical resistance of Enterococci. J. Hosp. Infect. 34, 191–196 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Panagea S., Chadwick P. R., Heat tolerance of vancomycin resistant Enterococcus faecium. J. Clin. Pathol. 49, 687–689 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maraccini P. A., Ferguson D. M., Boehm A. B., Diurnal variation in Enterococcus species composition in polluted ocean water and a potential role for the Enterococcal carotenoid in protection against photoinactivation. Appl. Environ. Microbiol. 78, 305–310 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pidot S. J., et al. , Increasing tolerance of hospital Enterococcus faecium to handwash alcohols. Sci. Transl. Med. 10, eaar6115 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Alotaibi S. M. I., et al. , Susceptibility of vancomycin-resistant and -sensitive Enterococcus faecium obtained from Danish hospitals to benzalkonium chloride, chlorhexidine and hydrogen peroxide biocides. J. Med. Microbiol. 66, 1744–1751 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Rice L. B., et al. , Role of class A penicillin-binding proteins in the expression of beta-lactam resistance in Enterococcus faecium. J. Bacteriol. 191, 3649–3656 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arbeloa A., et al. , Role of class A penicillin-binding proteins in PBP5-mediated beta-lactam resistance in Enterococcus faecalis. J. Bacteriol. 186, 1221–1228 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otto M., Bacterial evasion of antimicrobial peptides by biofilm formation. Curr. Top. Microbiol. Immunol. 306, 251–258 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Toledo-Arana A., et al. , The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl. Environ. Microbiol. 67, 4538–4545 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCormick J. K., Tripp T. J., Dunny G. M., Schlievert P. M., Formation of vegetations during infective endocarditis excludes binding of bacterial-specific host antibodies to Enterococcus faecalis. J. Infect. Dis. 185, 994–997 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Van Tyne D., Gilmore M. S., Friend turned foe: Evolution of enterococcal virulence and antibiotic resistance. Annu. Rev. Microbiol. 68, 337–356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller W. R., Munita J. M., Arias C. A., Mechanisms of antibiotic resistance in enterococci. Expert Rev. Anti Infect. Ther. 12, 1221–1236 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leclercq R., Derlot E., Duval J., Courvalin P., Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N. Engl. J. Med. 319, 157–161 (1988). [DOI] [PubMed] [Google Scholar]
- 25.Chorell E., Das P., Almqvist F., Diverse functionalization of thiazolo ring-fused 2-pyridones. J. Org. Chem. 72, 4917–4924 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Chorell E., et al. , Design and synthesis of C-2 substituted thiazolo and dihydrothiazolo ring-fused 2-pyridones: Pilicides with increased antivirulence activity. J. Med. Chem. 53, 5690–5695 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aberg V., Almqvist F., Pilicides-small molecules targeting bacterial virulence. Org. Biomol. Chem. 5, 1827–1834 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Thomas V. C., et al. , A fratricidal mechanism is responsible for eDNA release and contributes to biofilm development of Enterococcus faecalis. Mol. Microbiol. 72, 1022–1036 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mesnage S., Chau F., Dubost L., Arthur M., Role of N-acetylglucosaminidase and N-acetylmuramidase activities in Enterococcus faecalis peptidoglycan metabolism. J. Biol. Chem. 283, 19845–19853 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Kumano M., et al. , Lincomycin resistance mutations in two regions immediately downstream of the -10 region of lmr promoter cause overexpression of a putative multidrug efflux pump in Bacillus subtilis mutants. Antimicrob. Agents Chemother. 47, 432–435 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arias C. A., Contreras G. A., Murray B. E., Management of multidrug-resistant enterococcal infections. Clin. Microbiol. Infect. 16, 555–562 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X.-Z., Nikaido H., Efflux-mediated drug resistance in bacteria. Drugs 64, 159–204 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Higgins C. F., Multiple molecular mechanisms for multidrug resistance transporters. Nature 446, 749–757 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Poole K., Efflux-mediated antimicrobial resistance. J. Antimicrob. Chemother. 56, 20–51 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Alekshun M. N., Levy S. B., Molecular mechanisms of antibacterial multidrug resistance. Cell 128, 1037–1050 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Viveiros M., et al. , Demonstration of intrinsic efflux activity of Escherichia coli K-12 AG100 by an automated ethidium bromide method. Int. J. Antimicrob. Agents 31, 458–462 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Paixão L., et al. , Fluorometric determination of ethidium bromide efflux kinetics in Escherichia coli. J. Biol. Eng. 3, 18 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang X., et al. , Role of efflux pumps in the in vitro development of ciprofloxacin resistance in Listeria monocytogenes. Front. Microbiol. 9, 2350 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fey P. D., et al. , A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio 4, e00537-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eng R. H., Padberg F. T., Smith S. M., Tan E. N., Cherubin C. E., Bactericidal effects of antibiotics on slowly growing and nongrowing bacteria. Antimicrob. Agents Chemother. 35, 1824–1828 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee A. J., et al. , Robust, linear correlations between growth rates and β-lactam-mediated lysis rates. Proc. Natl. Acad. Sci. U.S.A. 115, 4069–4074 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mascio C. T. M., Alder J. D., Silverman J. A., Bactericidal action of daptomycin against stationary-phase and nondividing Staphylococcus aureus cells. Antimicrob. Agents Chemother. 51, 4255–4260 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silverman J. A., Perlmutter N. G., Shapiro H. M., Correlation of daptomycin bactericidal activity and membrane depolarization in Staphylococcus aureus. Antimicrob. Agents Chemother. 47, 2538–2544 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cetinkaya Y., Falk P., Mayhall C. G., Vancomycin-resistant enterococci. Clin. Microbiol. Rev. 13, 686–707 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shete V., Grover N., Kumar M., Analysis of aminoglycoside modifying enzyme genes responsible for high-level aminoglycoside resistance among enterococcal isolates. J. Pathogens 2017, 3256952 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perkins H. R., Vancomycin and related antibiotics. Pharmacol. Ther. 16, 181–197 (1982). [DOI] [PubMed] [Google Scholar]
- 47.Bugg T. D., et al. , Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: Biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry 30, 10408–10415 (1991). [DOI] [PubMed] [Google Scholar]
- 48.Arthur M., Reynolds P., Courvalin P., Glycopeptide resistance in enterococci. Trends Microbiol. 4, 401–407 (1996). [DOI] [PubMed] [Google Scholar]
- 49.al-Obeid S., Billot-Klein D., van Heijenoort J., Collatz E., Gutmann L., Replacement of the essential penicillin-binding protein 5 by high-molecular mass PBPs may explain vancomycin-β-lactam synergy in low-level vancomycin-resistant Enterococcus faecium D366. FEMS Microbiol. Lett. 70, 79–84 (1992). [DOI] [PubMed] [Google Scholar]
- 50.Ling L. L., et al. , A new antibiotic kills pathogens without detectable resistance. Nature 517, 455–459 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou Y., et al. , Vancomycin heteroresistance in vanM-type Enterococcus faecium. Microb. Drug Resist. 26, 776–782 (2020). [DOI] [PubMed] [Google Scholar]
- 52.El-Halfawy O. M., Valvano M. A., Antimicrobial heteroresistance: An emerging field in need of clarity. Clin. Microbiol. Rev. 28, 191–207 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Band V. I., et al. , Antibiotic combinations that exploit heteroresistance to multiple drugs effectively control infection. Nat. Microbiol. 4, 1627–1635 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walker J. N., et al. , Catheterization alters bladder ecology to potentiate Staphylococcus aureus infection of the urinary tract. Proc. Natl. Acad. Sci. U.S.A. 114, E8721–E8730 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guiton P. S., Hung C. S., Hancock L. E., Caparon M. G., Hultgren S. J., Enterococcal biofilm formation and virulence in an optimized murine model of foreign body-associated urinary tract infections. Infect. Immun. 78, 4166–4175 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Port G. C., Cusumano Z. T., Tumminello P. R., Caparon M. G., SpxA1 and SpxA2 act coordinately to fine-tune stress responses and virulence in Streptococcus pyogenes. MBio 8, e00288-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.CLSI Performance standards for antimicrobial susceptibility testing. 30th ed. CLSI Supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute (2020).
- 58.Xu W., et al. , Host and bacterial proteases influence biofilm formation and virulence in a murine model of enterococcal catheter-associated urinary tract infection. NPJ Biofilms Microbiomes 3, 28 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Menges F., Spectragryph—Optical spectroscopy software. Spectroscopy (Springf.) (2022). [Google Scholar]
- 60.Orhan G., Bayram A., Zer Y., Balci I., Synergy tests by E test and checkerboard methods of antimicrobial combinations against Brucella melitensis. J. Clin. Microbiol. 43, 140–143 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tükenmez H., Sequencing of PS757-tolerant Enterococcus faecalis OG1RF mutants. BioProject: SRA. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA876221. Deposited 2 September 2022. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequencing data have been deposited in SRA [PRJNA876221 (61)].
All study data are included in the article and/or supporting information.