Abstract
Repeat-associated non-AUG (RAN) translation of expanded repeat-mutation mRNA produces toxic peptides in neurons of patients suffering from neurodegenerative diseases. Recent findings indicate that RAN translation in diverse model systems is not inhibited by cellular stressors that impair global translation through phosphorylation of the alpha subunit of eIF2, the essential eukaryotic translation initiation factor that brings the initiator tRNA to the 40S ribosome. Using in vitro, cell-based and Drosophila models, we examined the role of alternative ternary complex factors that may function in place of eIF2, including eIF2A, eIF2D, DENR and MCTS1. Among these factors, DENR knockdown had the greatest inhibitory effect on RAN translation of expanded GGGGCC and CGG repeat reporters and its reduction improved the survival of Drosophila expressing expanded GGGGCC repeats. Taken together, these data support a role for alternative initiation factors in RAN translation and suggest these may serve as novel therapeutic targets in neurodegenerative disease.
Introduction
While less efficient at initiating translation than canonical AUG start codons, ribosome profiling experiments suggest that non-AUG codons define a significant fraction of all cellular translation initiation sites (1,2). In particular, upstream open reading frames are enriched for non-AUG start codons (1–5) and proteins synthesized with non-AUG start codons often play important roles related to the cellular stress response (reviewed in ref. (6)). As such, non-AUG initiation event can be subject to different regulatory mechanisms than canonical AUG-initiated translation during the integrated stress response (ISR). The ISR reduces global cellular translation by inducing phosphorylation of the essential eukaryotic initiation factor eIF2 at serine 51 of its alpha subunit. Phosphorylation prevents eIF2 from exchanging GDP for guanosine triphosphate (GTP), which subsequently prevents eIF2 from binding and delivering the initiator methionine tRNA (Met-tRNAiMet) to the 40S ribosome. While this strategy is effective at dramatically decreasing most AUG-initiated translation, some non-AUG initiation events are spared or upregulated during the ISR (2,6,7).
One model for how specific non-AUG initiation events evade downregulation during ISR activation depends on the ability of specific non-AUG start codons to receive initiator tRNAs independent of eIF2. eIF2A, a non-essential, monomeric protein, can deliver initiator tRNAs to the pre-initiation ribosome and has been shown to act in place of eIF2 during the ISR (7–11). It represents an intriguing therapeutic target, as mice lacking eIF2A survive to adulthood with no gross abnormalities (12). Additionally, eIF2D and Density regulated re-initiation and release factor and multiple copies in T-cell lymphoma-1 are factors that normally promote ribosome recycling and translation re-initiation (13–15) but can also function in place of eIF2 to promote translation initiation and are not inhibited by ISR activation (16,17). In addition to binding and delivering the Met-tRNAiMet, each of these factors can also deliver non-methionyl tRNAs cognate to non-AUG codons used for initiation (8,11,16).
While non-AUG initiation is important in normal cellular processes, its mis-regulation has also been implicated in human disease (3,6,18). Repeat expansion mutations associated with several neurodegenerative and neuromuscular diseases undergo a process known as repeat-associated non-AUG (RAN) translation (18–22). Repeat-associated non-AUG translation initiates upstream of or within the expanded repeats and results in translation through the repetitive RNA sequence. This produces RAN peptides that contain large, repetitive amino acid sequences that are often aggregation-prone and neurotoxic in model systems.
By definition, RAN translation exclusively utilizes non-AUG start codons for synthesis of these toxic proteins. Recently, our laboratory and others showed that RAN translation of expanded GGGGCC repeats associated with C9orf72 amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD, ‘C9RAN’), as well as of expanded CGG repeats associated with fragile X-associated tremor/ataxia syndrome (FXTAS, ‘CGG RAN’), behave like some other non-AUG translation events and is increased following ISR activation, while global translation is simultaneously downregulated (23–26). Here, we investigate the ability of C9 and CGG RAN translation to utilize the eIF2 alternatives eIF2A, eIF2D and DENR/MCTS1 during translation initiation. We find evidence that DENR/MCTS1 and eIF2A support RAN translation of both CGG and GGGGCC repeats under specific conditions. Knocking down DENR, and to a lesser degree eIF2A, modestly improves repeat-mediated toxicity in Drosophila expressing GGGGCCx28 repeats. These data suggest a role for alternative ternary complex factors in RAN translation.
Results
Loss of eIF2A reduces C9 and CGG RAN translation in vitro
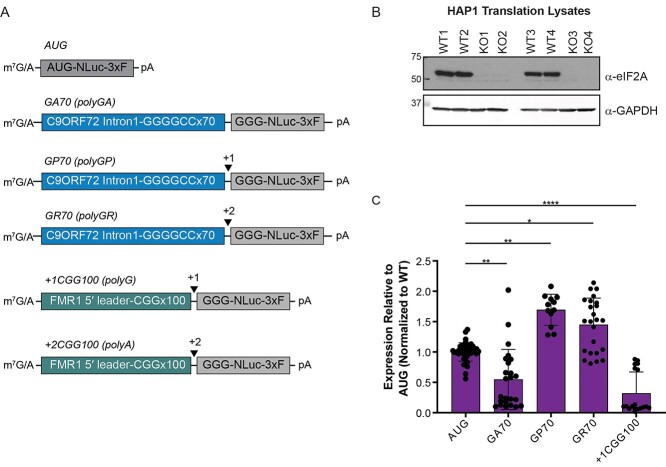
We first assessed a role of eIF2A in C9 or CGG RAN translation by expressing nanoluciferase (NLuc) reporter mRNAs containing either 70 GGGGCC repeats in the glycine–alanine (polyGA), glycine–proline (polyGP) or glycine–arginine (polyGR) reading frames, or 100 CGG repeats in the +1 poly-glycine (polyG) reading frame (23,27) in in vitro translation lysates generated from wildtype (WT) and CRISPR-mediated eIF2A knockout (KO) HAP1 cells (Fig. 1A and B, Supplementary Material, Fig. S1A) (28,29). To control for basal differences in translation activity between different lysate preparations, RAN translation expression of each reporter was normalized to the expression of an AUG-NLuc control. In doing so, we found that compared to WT lysates, in lysates lacking eIF2A expression of the polyGA C9RAN and polyG CGG RAN translation reporter mRNAs was significantly reduced in the absence of eIF2A (Fig. 1C). This is consistent with a previous report that loss of eIF2A reduces polyGA production in multiple cell lines and chick embryos (25) and promotes RAN translation from CCUG/CAGG repeats associated with myotonic dystrophy type 2 (30). Surprisingly, the opposite effect was observed for polyGP and polyGR reporter mRNAs, as their expression was significantly increased relative to AUG-NLuc in eIF2A KO versus WT lysates (Fig. 1C).
Figure 1.
RAN translation is suppressed in vitro in eIF2A KO cell lysates. (A) Schematic of nanoluciferase (NLuc) reporter mRNAs. 3xF = 3x FLAG tag, GA = glycine–alanine, GP = glycine-proline, GR = glycine-arginine, polyG = the glycine reading frame product of the CGG repeat; polyA = the alanine reading frame product of the CGG repeat. (B) Western blot of WT and eIF2A KO lysates probed with an anti-eIF2A antibody to confirm loss of eIF2A expression. GAPDH is used as a loading control. (C) Expression of indicated NLuc reporter mRNAs in eIF2A KO lysates, relative to WT lysates, after controlling for the difference in AUG-NLuc expression between paired lysates, n = 12–42. Graphs represent mean with error bars ± standard deviation. *P < 0.05, **P < 0.01, ****P < 0.0001, non-parametric analysis of variance, Kruskal–Wallis test, with Dunn’s multiple comparison test.
In HEK293 cells, siRNA-mediated knockdown (KD) of eIF2A had only modest effects on C9 and CGG RAN translation from NLuc reporter plasmids co-transfected with Firefly luciferase (FFLuc) as an internal control (Fig. 2A–C and Supplementary Material, Fig. S1C). While it modestly decreased polyGA expression relative to cells treated with a non-targeting siRNA (Fig. 2B and D), no inhibition was observed for GP and GR frames, nor for the polyG poly-alanine (polyA, +2) reading frames for the CGG repeat (Fig. 2B and C). Furthermore, no inhibition was observed when mRNA reporters like those used in the in vitro lysate experiments were directly transfected into cells with eIF2A KD (Fig. 2D). We also assessed whether eIF2A KD impaired expression of no-repeat NLuc reporters that initiate at CUG and ACG codons, as such codons were previously identified as potential initiation sites for the RAN translation of the GGGGCC and CGG repeats, respectively (23,27,31). eIF2A KD did not impair expression of these reporters (Fig. 2E).
Figure 2.
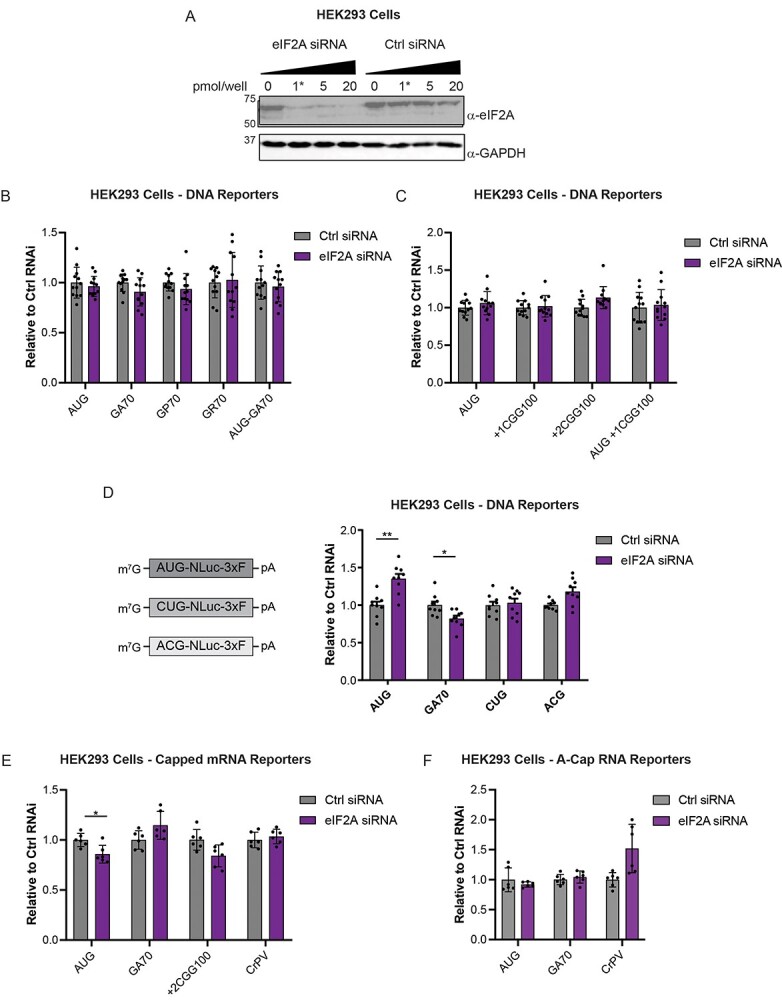
eIF2A knockdown does not alter RAN translation in transfected cells. (A) Western blot showing efficiency of eIF2A KD in HEK293 cells following transfection with increasing concentrations of eIF2A siRNA. The starred lane indicates siRNA concentration used in subsequent experiments. GAPDH was used as a loading control. (B and C) NLuc expression of indicated reporters expressed from DNA plasmids in HEK293 cells 24 h post transfection with non-targeting or eIF2A siRNAs. NLuc levels are expressed relative to levels in cells transfected with the non-targeting siRNA, n = 12. (D) Schematic of CUG and ACG-NLuc reporter mRNAs, and NLuc expression of indicated reporters expressed from DNA plasmids transfected into HEK293 cells 24 h post transfection with non-targeting or eIF2A siRNAs. NLuc levels are expressed relative to levels in cells transfected with the non-targeting siRNA, n = 9. (E) NLuc expression of indicated reporters expressed from ARCA-capped mRNAs transfected into HEK293 cells 24 h post transfection with non-targeting or eIF2A siRNAs. NLuc levels are expressed relative to levels in cells transfected with the non-targeting siRNA, n = 6. (F) NLuc expression of indicated reporters expressed from A-capped RNAs transfected into HEK293 cells 24 h post transfection with non-targeting or eIF2A siRNAs. NLuc levels are expressed relative to levels in cells transfected with the non-targeting siRNA, n = 6. All graphs represent mean with error bars ± standard deviation. *P < 0.05, **P < 0.01, Two-way ANOVA with Sidak’s multiple comparison test.
Previous studies showing a role of eIF2A in supporting C9RAN translation in the polyGA frame used bicistronic reporters with the GGGGCC repeat placed in the second cistron (25). In this sequence context, cap-independent RAN translation of the GGGGCC repeat could contribute a greater proportion of the luminescent signal than the signal generated from our functionally capped, monocistronic reporter mRNAs. To therefore test whether cap-independent translation initiation is more sensitive to eIF2A levels, we next transfected eIF2A KD HEK293 cells with A-capped reporter RNAs (23,27). Unlike the functional m7G, the A-cap cannot interact with the cap-binding factors to support cap-dependent translation initiation. As previously reported (23,27), relative to functionally capped RAN reporters, the A-capped RAN reporters supported significantly lower levels of translation (Supplementary Material, Fig. S1D). However, the remaining fraction of translation observed, which likely occurs through a cap-independent mechanism, was also not inhibited by eIF2A KD (Fig. 2F). Consequently, substantial reduction of eIF2A is not sufficient to reduce C9 or CGG RAN translation that occurs through either cap-dependent or cap-independent mechanisms in reporter transfected cells.
DENR/MCTS1 KD reduces RAN translation in HEK cells
To next determine whether either eIF2D and/or DENR/MCTS1 are involved in C9 or CGG RAN translation, we expressed RAN reporters in HEK293 cells following KD of each protein. Reduced levels of eIF2D did not inhibit C9 or CGG RAN translation in any reading frame tested (Supplementary Material, Fig. S2A–D). It did reduce expression of the ACG-initiated no-repeat reporter, but had no effect on a no-repeat CUG-initiated reporter (Supplementary Material, Fig. S2E).
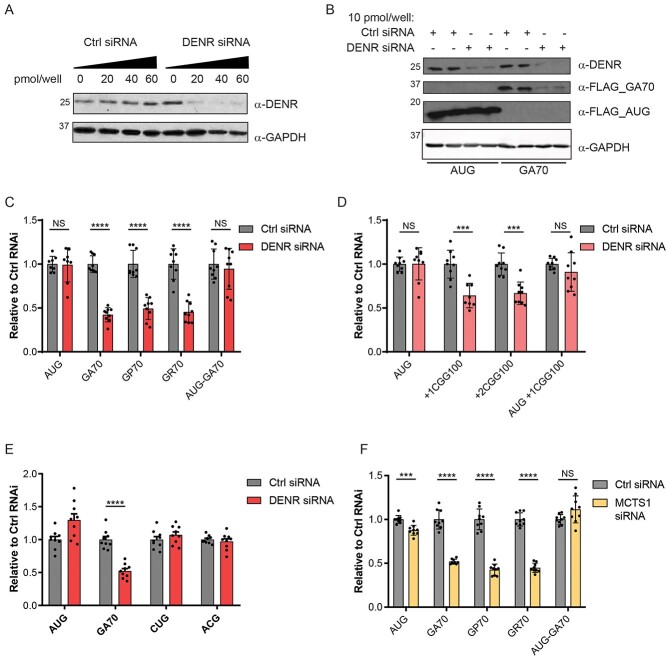
In contrast, reduction of DENR/MCTS1 levels through the use of a DENR-specific siRNA significantly reduced NLuc expression of C9 and CGG RAN reporters across all reading frames assayed, without significantly altering AUG-NLuc expression (Fig. 3A–D, Supplementary Material, Fig. S3A). The reduction of polyGA synthesis but unchanged AUG-NLuc expression following DENR KD was confirmed by western blot using an antibody against the c-terminal FLAG-tag present on each reporter (Fig. 3B).
Figure 3.
DENR/MCTS1 knockdown selectively suppresses RAN translation. (A) Western blot showing efficiency of DENR KD in HEK293 cells 48 h post transfection with increasing concentrations of DENR siRNA. GAPDH was used as a loading control. (B) Western blots showing effects of DENR KD on expression of AUG-NLuc and GA70-NLuc reporters expressed in HEK293 cells, 48 h post KD and 24 h post reporter transfection. NLuc reporters were probed with an antibody targeting their c-terminal FLAG tag. GAPDH was used as a loading control. (C–E) NLuc expression of indicated reporters expressed from DNA plasmids transfected into HEK293 cells 24 h post transfection with non-targeting or DENR targeting siRNAs. NLuc levels are expressed relative to levels in cells transfected with the non-targeting siRNA, n = 9. (F) NLuc expression of indicated reporters expressed from DNA plasmids in HEK293 cells 24 h post transfection with non-targeting or MCTS1 targeting siRNAs. NLuc levels are expressed relative to levels in cells transfected with the non-targeting siRNA, n = 9. All graphs represent mean with error bars ± standard deviation. ***P < 0.001, ****P < 0.0001, Two-way analysis of variance with Sidak’s multiple comparison test.
Interestingly, the effect of DENR KD on GGGGCCx70 and CGGx100 reporter expression was dependent on their non-AUG initiation, as reporters with an AUG start codon inserted upstream of either repeat, to drive canonical translation initiation in the GA or + 1 CGG reading frames were not significantly inhibited (Fig. 3C and D). Furthermore, this effect was specific to repeat-associated non-AUG translation, as opposed to no-repeat non-AUG initiated translation, as DENR KD did not reduce expression of CUG-NLuc or ACG-NLuc reporters (Fig. 3E). As the CUG-NLuc and ACG-NLuc reporters are expressed at levels below that of the RAN translation reporters, this also suggests that DENR KD does not non-specifically impair all inefficient translation events (Supplementary Material, Fig. S3B).
Although DENR KD had no effect on expression of the highly stable AUG-NLuc protein, it did reduce expression of the less stable co-transfected AUG-FFLuc reporter (Supplementary Material, Fig. S3C). However, there was no significant difference in the polysome/monosome ratio in polysome profiles from control versus DENR KD cells, suggesting that DENR KD does not cause global translational inhibition (Supplementary Material, Fig. S3D).
DENR functions as a heterodimer with MCTS1 and KD of one protein leads to the depletion of the other (Supplemental Material, Fig. S3A). To verify the effect we observed on C9RAN translation using a DENR-targeting siRNA, we also knocked down this complex using a MCTS1-targeting siRNA (Supplementary Material, Fig. S3E). As expected, MCTS1 KD produced very similar results to DENR KD, having a larger inhibitory effect on C9RAN translation than on translation from either of the AUG-driven NLuc controls (Fig. 3F), but also significantly reducing expression of the FFLuc transfection control (Supplementary Material, Fig. S3F). Together, these data support a role for the DENR/MCTS1 complex in supporting C9 and CGG RAN translation.
eIF2A, eIF2D, and DENR knockdowns do not prevent increased C9 and CGG RAN translation upon ISR induction
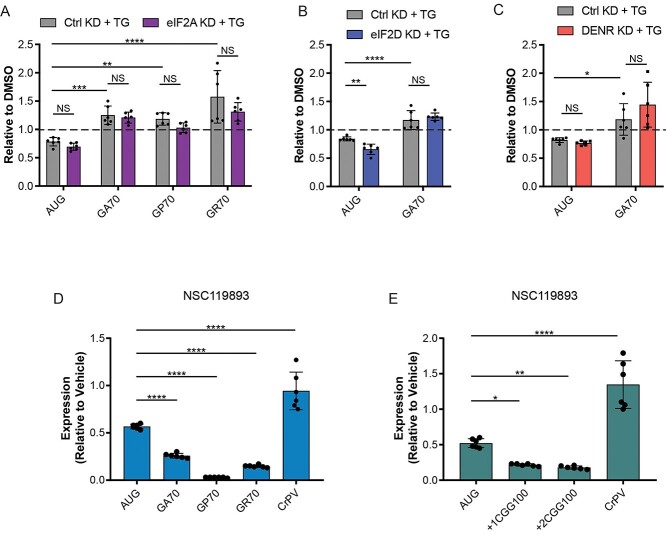
In previous studies, eIF2A was most critical for translation initiation under conditions of cellular stress, when functional eIF2 levels were limited by ISR-mediated phosphorylation of eIF2α at serine 51 (7,31,32). We thus specifically assessed the ability of eIF2A, eIF2D and DENR/MCTS1 KD to regulate C9 and CGG RAN translation following induction of endoplasmic reticulum stress. Consistent with previous findings (23–26), 2 μM thapsigargin (TG) treatment for five hours selectively increased C9RAN translation expression in HEK293 cells, while inhibiting expression of AUG-initiated control reporters (Fig. 4A–C). However, KD of eIF2A, eIF2D and DENR/MCTS1 did not prevent this increase in RAN levels (Fig. 4A–C).
Figure 4.
eIF2A, eIF2D and DENR are not required for stress-induced RAN translation. (A–C) HEK293 cells were transfected with eIF2A, eIF2D or DENR siRNAs, respectively, as well as a non-targeting control siRNA for 24 h before transfection with NLuc reporter plasmids. 19 h post reporter transfection, HEK293 cells were treated with 2 μM thapsigargin for 5 h, and then NLuc levels were measured. NLuc levels are expressed relative to vehicle (DMSO) treated cells. (D–E) NLuc expression for indicated reporter mRNAs expressed in RRL treated with 10 μM NSC119893, expressed relative to vehicle treated controls, n = 6. All graphs represent mean with error bars ± standard deviation. (A–C) Two-way analysis of variance (ANOVA) with Sidak’s multiple comparison test. (D–E) One-way ANOVA with Dunnett’s multiple comparison test, *P < 0.05, **P < 0.01, ****P < 0.0001.
These results suggest that despite limited levels of functional eIF2·guanosine triphosphate·Met-tRNAiMet ternary complex (TC), RAN translation may still utilize this factor for initiation during the ISR. Alternatively, stress-resistant RAN translation may receive an initiator tRNA through a mechanism independent of eIF2 and the additional factors assessed here. To help distinguish between these possibilities, we expressed our C9 and CGG RAN translation reporter mRNAs in a rabbit reticulocyte lysate (RRL) in the presence or absence of the small molecule NSC119893, which selectively inhibits formation of the eIF2·GTP·Met-tRNAiMet TC(33). As expected, the addition of 25 μM NSC119893 to RRL reactions greatly inhibited AUG-NLuc expression but did not affect expression of an A-capped cricket paralysis virus (CrPV)-NLuc reporter (34) that initiates translation entirely through a non-methionyl internal ribosome entry site (IRES)-mediated mechanism, in which its first tRNA is delivered by the elongation factor eEF1a (35,36) (Fig. 4D and E). Interestingly, expression of all C9 and CGG RAN translation reporters in RRL was inhibited to an even greater extent than AUG-NLuc by NSC119893 treatment (Fig. 4D and E). This suggests that in vitro RAN translation initiation of these repeats from linear mRNAs requires the canonical eIF2·GTP·Met-tRNAiMetTC.
DENR KD prolongs lifespan of Drosophila expressing GGGGCC repeat
To assess the role of these factors in an in vivo model of repeat toxicity, we used transgenic Drosophila lines conditionally expressing 28 GGGGCC repeats under the upstream activating sequence (UAS) promoter, with the repeat inserted into either the second or third chromosome (37). When expressed with a non-targeting control shRNA in the fly eye using the eye-specific GMR-GAL4 driver, the repeat causes a severe rough eye phenotype with noticeable eye shrinkage (Fig. 5A and B) (37).
Figure 5.
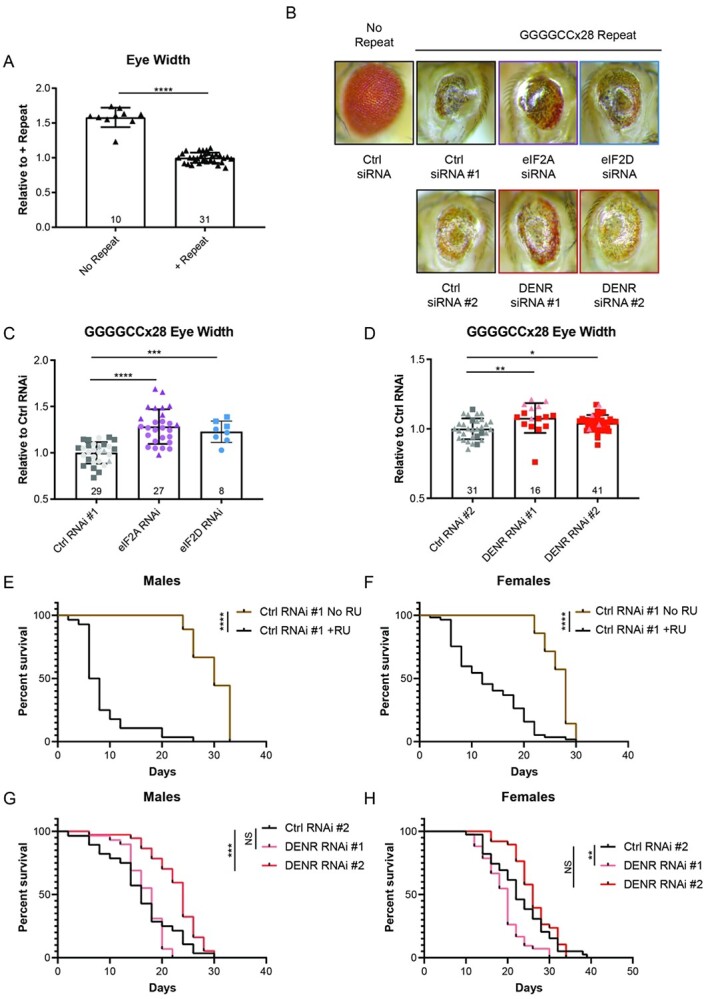
Knockdown of DENR suppresses GGGGCC associated toxicity in Drosophila. (A) Quantification of eye width in flies expressing control (ctrl) shRNA #1 in the absence or presence of the GGGGCCx28 repeat, under the GMR-Gal4 driver. Experimental numbers are indicated within bars and each fly is represented by a single data point. (B) Representative images of fly eyes expressing control or CG7414 (‘eIF2A’), eIF2D, or DENR shRNAs in the presence of the GGGGCCx28 repeat. (C–D) Comparison of eye width in flies expressing indicated shRNAs in the presence of the GGGGCCx28 repeat. Experimental numbers are indicated within bars, and each fly is represented by a single data point. Shape of data points represents the experimental trials that data points were collected from. (E–F) Survival curve of control male and female Tub5-GS-Gal4 flies, respectively. Survival of flies with induced expression of the GGGGCCx28 repeat and control shRNA through RU486 treatment (+RU, male n = 28, female n = 57), is compared to survival of flies without induced expression (No RU, male n = 9, female n = 14). (G–H) Survival curve of RU486-treated GGGGCCx28 male and female Tub5-GS-Gal4 flies, respectively, expressing ctrl (male n = 28, female n = 39) or DENR targeting shRNA #1 (male n = 29, female n = 42) and shRNA #2 (male n = 37, female n = 38). Bar graphs represent mean with error bars ± standard deviation, * P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by one-way ANOVA with Dunnett’s multiple comparison test. For survival curves, **P < 0.01, ***P < 0.001, ****P < 0.0001, Mantel-Cox test.
We next co-expressed the GGGGCC repeat with shRNAs targeting the predicted Drosophila homolog of eIF2A (CG7414), eIF2D and DENR using the GMR-GAL4 driver, and imaged the fly eyes 1–3 days post eclosion (Fig. 5B). Using ImageJ, we measured the width of repeat-expressing fly eyes in the presence of the control versus targeting shRNAs. RNAi against eIF2A, eIF2D and DENR all modestly improved the rough of phenotype in flies maintained at 29°C (Fig. 5C and D).
We next expressed the GGGGCCx28 repeat along with a non-targeting control shRNA ubiquitously throughout the fly using the RU-486 inducible tubulin driver, Tub5-GS GAL4. This results in a substantial decrease in the lifespan of both male and female flies (Fig. 5E and F). We then tracked the survival of the GGGGCCx28 flies ubiquitously expressing shRNAs against eIF2A (CG7414), eIF2D, DENR and MCTS1 compared to control shRNAs (Average KD efficiencies in Table 3). While shRNAs against eIF2A, eIF2D and MCTS1 did not confer any survival benefit, we found that one shRNA targeting DENR significantly prolonged male fly survival in the presence of the repeat, although its benefit was not observed in female flies (Fig. 5G and H and Supplementary Material, Fig. S4A–F). Together, these data suggest that modulating levels of non-canonical initiation factors can reduce toxicity caused by GGGGCC repeat expression in model organisms.
Table 3.
Primary antibody information
| Antibody target | Product information | Species | Probing conditions |
|---|---|---|---|
| eIF2A | Protein Tech, 11 233-1-AP | Rabbit | 1:8000 – 1 h at room temperature |
| eIF2A | Abcam; ab169528 | Rabbit | 1:1000 – overnight at 4°C |
| eIF2D | Protein Tech, 12 840–1-AP | Rabbit | 1:1000 – overnight at 4°C |
| DENR | Sigma, Clone 1H3, WH0008562M1 | Mouse | 1:1000 – overnight at 4°C |
| MCTS1 | Sigma, SAB2701331 (discontinued) | Rabbit | 1:500 – overnight at 4°C |
| FLAG | Sigma, M2, F1804 | Mouse | 1:1000 – overnight at 4°C |
| GAPDH | SCBT, sc-32 233 | Mouse | 1:1000–1 h at room temperature |
To next determine whether DENR KD reduces RAN translation in Drosophila, we used a well-characterized fly model of CGG RAN translation, which uses the GMR-GAL4 driver to express within the fly eye a UAS-CGG90 insert with enhanced green fluorescent protein tagging the polyG RAN product (20,38,39). We crossed this fly to those expressing either DENR shRNA #1 or control shRNA #2, and by fluorescence microscopy found that DENR KD significantly reduced total EGFP levels and polyG aggregates in the fly eye (Supplementary Material, Fig. S4G). This suggests that DENR KD reduces RAN translation in vivo within the Drosophila eye.
Discussion
Characterizing ways in which RAN translation differs mechanistically from canonical translation has great potential for uncovering new strategies for therapeutic targeting. Previous work from our group and others revealed the ability of C9 and CGG RAN translation to continue during ISR activation, a condition that impairs canonical translation (23–26). As the ISR inhibits functional eIF2 TC formation, we hypothesized that RAN translation uses factors that can function in place of eIF2. We thus investigated the role of eIF2A, eIF2D and DENR/MCTS1 in supporting C9 and CGG RAN translation. While our results indicate that these factors are not responsible for promoting increased RAN translation during the ISR, they do support a role for non-canonical initiation factors, particularly the DENR/MCTS1 complex, in RAN translation initiation.
Despite being first identified nearly 50 years ago (8,9), eIF2A is still a poorly understood translation factor. Recent work established it as a modifier of both C9RAN and RAN translation of CCUG and CAGG repeats causative of myotonic dystrophy type 2 (25,30). Here we found that in in vitro lysates, deletion of eIF2A reduces RAN translation relative AUG-initiated translation for the polyGA C9RAN product and the polyG CGG product but increases RAN translation of the C9RAN polyGP and polyGR products. Intriguingly, while translation initiation of both the polyGA and polyG products predominantly occurs at near-AUG codons (CUG and ACG, respectively), positioned above the expanded repeats (23,27,40), the initiation sites of the polyGP and polyGR products are less well-defined and may be located within the repeat itself (23,24,27,40,41). This suggests that eIF2A’s role in RAN translation may be associated with the specific non-AUG codon used.
However, we did not see a robust or consistent inhibition following eIF2A KD in HEK293 cells. One possible explanation for the lack of effect in a cell-based system is that the interplay between eIF2A’s role in regulating translation during cellular stress (7,31,32), the cellular stress resulting from transient transfections, and the increase in RAN translation during cellular stress conditions (23–26), could obscure the role eIF2A plays in regulating RAN translation under the non-stressed conditions in in vitro translation lysates. In the context of previously published work (25,30), this suggests that eIF2A function may be highly context-dependent, with the factors that regulate its activity still unknown. This is consistent with conflicting reports on whether eIF2A is involved in IRES-mediated translation of the hepatitis C virus mRNA (28,31,42).
Similar principles may explain our findings related to eIF2D. While we did not observe a loss of RAN translation with eIF2D KD, we did observe increased polyGR expression compared to controls. It should be noted, however, that our constructs gave different results than a recent study demonstrating a role for eIF2D in C9RAN translation. These studies were most notable for a protective effect of eIF2D in a Caenorhabditis elegans model, which appeared to be largely dependent on polyGA production (43) and CUG-mediated initiation. We similarly observed a modest improvement in the rough eye phenotype with eIF2D KD in Drosophila, although survival in the setting of post-eclosion ubiquitous expression was not significantly impacted (Fig. 5B and C, Supplementary Material, Fig. S4C and D).
Unlike eIF2A KD or eIF2D KD, KD of the DENR/MCTS1 complex, through use of either DENR or MCTS1 targeting siRNAs, significantly reduced RAN translation across all three reading frames of GGGGCCx70 reporters and in both the +1 and + 2 reading frames of CGGx100 reporters. The greater dependence of RAN translation in HEK293 cells on DENR/MCTS1 compared to eIF2D or eIF2A is likely not simply a result of DENR/MCTS1 being more abundant in this system, as data from the Human Atlas Project suggests these factors are expressed at similar levels (44). The inhibitory effect of DENR/MCTS1 KD was specific to RAN translation, as it had little effect on near-AUG reporters lacking expanded repeats or on AUG-initiated repeat-containing reporters. The dependence on both non-AUG initiation and the expanded repeat is intriguing. While the DENR/MCTS1 complex can support translation initiation through delivery of initiator tRNAs to the ribosome (16), its main described functions to date are in ribosome recycling and re-initiation (13–15). As translation through GC-rich repeats may trigger ribosome stalling where choices must be made between re-initiation and ribosomal recycling pathways, RAN translation may uniquely require the combination of non-canonical initiation and assistance with translational elongation.
Unfortunately, unlike eIF2A, the DENR/MCTS1 complex is essential to life (15). While the conditions of our KD do not impair global translation as measured by polysome profiling and our AUG-NLuc reporters, they do reduce FFLuc reporter expression, which is less stable and may be an indication of reduced cellular fitness. Future work aimed at better understanding the mechanism by which DENR/MCTS1 supports translation may reveal ways to specifically target RAN translation without interfering with DENR/MCTS1’s essential functions.
Our work in Drosophila demonstrated partial rescue associated with KD of either eIF2A or DENR expression, with the effects of DENR being more robust. While these findings are promising and provide evidence for DENR KD-mediated effects on repeat associated toxicity across multiple assays, these models lack the normal genomic sequence context for the C9orf72 repeat (37), which places limitations on its transferability into more complex organisms or human patients. Future studies will be needed to understand the strength of its selective effects in patient-derived cells with full-length repeats in their endogenous sequence context.
In sum, we have studied the role of three alternative TCs for their ability to selectively modulate RAN translation at two different repeat elements and in a Drosophila model of C9 ALS/FTD and FXTAS. These studies suggest that the DENR/MCTS1 complex in particular has roles in RAN translation and is worthy of further study in human neuronal model systems.
Materials and Methods
RNA synthesis
RNAs were in vitro transcribed from PspOMI-linearized pcDNA3.1(+) reporter plasmids using HiScribe T7 High Yield RNA Synthesis Kit (NEB) with 3'-O-Me-m7GpppG anti-reverse cap analog (ARCA) or ApppG cap (NEB) added at 8:1 to GTP for a capping efficiency of ~90%, as previously described (23,27). After RNA synthesis, DNA templates were removed with RNase-free DNaseI (NEB), and RNAs were poly-adenylated with Escherichia coli poly-A polymerase (NEB) as previously described (23,27). mRNAs were then cleaned and concentrated with RNA Clean and Concentrator-25 Kit from Zymo Research and run on a denaturing formaldehyde RNA gel to verify mRNA size and integrity. Previously unpublished sequences for the polyGP and polyGR reporters used in Figure 1 and Supplementary Material, Figure S1 are in Table 1. All other reporter sequences have been previously published (23,27).
Table 1.
Reporter sequences
| Reporter name | Sequence from T7 to PspOMI cut site |
|---|---|
| polyGP | GGTGTGTGTTTTTGTTTTTCCCACCCTCTCTCCCCACTACTTGCTCTCACAGTACTCGCTGAGGGTGAACAAGAAAAGACCTGATAAAGATTAACCAGAAGAAAACAAGGAGGGAAACAACCGCAGCCTGTAGCAAGCTCTGGAACTCAGGAGTCGCGCGCTAgCGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCAGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGTTGGGGTCTTCACACTCGAAGATTTCGTTGGGGACTGGCGACAGACAGCCGGCTACAACCTGGACCAAGTCCTTGAACAGGGAGGTGTGTCCAGTTTGTTTCAGAATCTCGGGGTGTCCGTAACTCCGATCCAAAGGATTGTCCTGAGCGGTGAAAATGGGCTGAAGATCGACATCCATGTCATCATCCCGTATGAAGGTCTGAGCGGCGACCAAATGGGCCAGATCGAAAAAATTTTTAAGGTGGTGTACCCTGTGGATGATCATCACTTTAAGGTGATCCTGCACTATGGCACACTGGTAATCGACGGGGTTACGCCGAACATGATCGACTATTTCGGACGGCCCTATGAAGGCATCGCCGTGTTCGACGGCAAAAAGATCACTGTAACAGGGACCCTGTGGAACGGCAACAAAATTATCGACGAGCGCCTGATCAACCCCGACGGCTCCCTGCTGTTCCGAGTAACCATCAACGGAGTGACCGGCTGGCGGCTGTGCGAACGCATTCTGGCGGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAGtaaGGCCGCGACTCGAGAG |
| polyGR | GGTGTGTGTTTTTGTTTTTCCCACCCTCTCTCCCCACTACTTGCTCTCACAGTACTCGCTGAGGGTGAACAAGAAAAGACCTGATAAAGATTAACCAGAAGAAAACAAGGAGGGAAACAACCGCAGCCTGTAGCAAGCTCTGGAACTCAGGAGTCGCGCGCTAgCGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCAGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGGGCCGGTTTGGGGTCTTCACACTCGAAGATTTCGTTGGGGACTGGCGACAGACAGCCGGCTACAACCTGGACCAAGTCCTTGAACAGGGAGGTGTGTCCAGTTTGTTTCAGAATCTCGGGGTGTCCGTAACTCCGATCCAAAGGATTGTCCTGAGCGGTGAAAATGGGCTGAAGATCGACATCCATGTCATCATCCCGTATGAAGGTCTGAGCGGCGACCAAATGGGCCAGATCGAAAAAATTTTTAAGGTGGTGTACCCTGTGGATGATCATCACTTTAAGGTGATCCTGCACTATGGCACACTGGTAATCGACGGGGTTACGCCGAACATGATCGACTATTTCGGACGGCCCTATGAAGGCATCGCCGTGTTCGACGGCAAAAAGATCACTGTAACAGGGACCCTGTGGAACGGCAACAAAATTATCGACGAGCGCCTGATCAACCCCGACGGCTCCCTGCTGTTCCGAGTAACCATCAACGGAGTGACCGGCTGGCGGCTGTGCGAACGCATTCTGGCGGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGATTACAAGGATGACGATGACAAGtaaGGCCGCGACTCGAGAG |
Rabbit reticulocyte lysate in vitro translation
mRNAs were in vitro translated with Flexi RRL System (Promega), as previously described (23,27). Around 10 μL reactions for luminescence assays were programmed with 3 nM mRNA and contained 30% RRL, 10 μM amino acid mix minus methionine, 10 μM amino acid mix minus leucine, 0.5 mM MgOAc, 100 mM KCl and 0.8 U/μL Murine RNAse Inhibitor (NEB). Around 25 μM NSC119893 (the Chemical Repository at the National Cancer Institute’s Developmental Therapeutics Program) or equal volume dimethyl sulfoxide was added to the RRL reaction mix to constitute 1% of final reaction volume (0.1 μL/reaction). Reactions were incubated at 30°C for 30 min before termination by incubation at 4°C. Samples were then diluted 1:7 in Glo Lysis Buffer (Promega) and incubated 1:1 for 5 min in the dark in opaque 96-well plates with NanoGlo Substrate freshly diluted 1:50 in NanoGlo Buffer (Promega). Luminescence was measured on a GloMax 96 Microplate Luminometer.
Cell line information and maintenance
HEK293 cells were purchased from American Type Culture Collection (ATCC CRL-1573). They were maintained within 37°C incubators at 5% CO2 in DMEM + high glucose (GE Healthcare Bio-Science, SH30022FS) supplemented with 9.09% fetal bovine serum (50 mL added to 500 mL DMEM; Bio-Techne, S11150).
eIF2A KO (Cat # HZGHC002650c001) and isogenic control HAP1 cells were purchased from Horizon Discovery (Cambridge, UK) (28,29). They were maintained within 37°C incubators at 5% CO2 in IMDM (Invitrogen, 124400–61) supplemented with 9.09% fetal bovine serum (50 mL added to 500 mL IMDM; Bio-Techne, S11150).
HAP1 cell lysate in vitro translation
Lysates were prepared using a previously developed protocol within our laboratory (23). HAP1 cells were trypsinized, centrifuged at 200xg for 5 min and the cell pellet washed once with 1X PBS. Cell pellets were then weighed and resuspended in RNAse-free hypotonic buffer containing 10 mM HEPES-KOH (pH 7.6), 10 mM potassium acetate, 0.5 mM magnesium acetate, 5 mM DTT and EDTA-free protease inhibitor cocktail (Roche), 250 μL buffer added per 200 mg cells (23). Resuspended cells were incubated on ice for 20 min, passed 10X through a 27G syringe, incubated for another 20 min on ice and centrifuged at 10 000xg for 10 min at 4°C to pellet cell debris. The supernatant was recovered and total protein quantified with a BCA assay. Lysates were diluted to 8 μg/μl protein in the hypotonic lysis buffer and stored in single use aliquots at -80°C.
For in vitro translation reactions, 8 μg lysate was supplemented to final concentrations of 20 mM HEPES-KOH (pH 7.6), 44 mM potassium acetate, 2.2 mM magnesium acetate, 2 mM DTT, 20 mM creatine phosphate (Roche), 0.1 μg/μl creatine kinase (Roche), 0.1 mM spermidine and on average 0.1 mM of each amino acid (23). In vitro transcribed reporter mRNAs were added to 4 nM. Translation assays and luminescence measurements were then performed as with RRL reactions.
Cell transfection, drug treatments and analysis
For luminescence assays, HEK293 cells were seeded in 96-well plates and transfected 24 h later at 40–50% confluency with siRNAs using Lipofectamine RNAiMax. The following amount of stealth siRNA (Thermo) was added per well, to achieve maximum KD efficiency: 0.1 pmol eIF2A siRNA, 1 pmol DENR siRNA, 2 pmol MCTS1 siRNA and 4 pmol eIF2D siRNA. The control non-targeting siRNA was added at same concentration as the targeting siRNA was compared to. siRNA sequences are listed in Table 2.
Table 2.
siRNA product information
| siRNA target | Product information | Sequence sense/antisense |
|---|---|---|
| eIF2A | Thermo stealth siRNA – HSS130478 | ACGAAACACUGUCUCUCAGUCAAUU/ AAUUGACUGAGAGACAGUGUUUCGU |
| eIF2D | Thermo stealth siRNA – HSS103085 | GGACAGGAGAAAGCUUCGAGCUGAU/ AUCAGCUCGAAGCUUUCUCCUGUCC |
| DENR | Thermo stealth siRNA – HSS112532 | ACCAACAGAGUACUGUGAAUAUAUG/ CAUAUAUUCACAGUACUCUGUUGGU |
| MCTS1 | Thermo stealth siRNA – HSS178943 | CAGGUUGAUAAAGGAGCCAUCAAAU/AUUUGAUGGCUCCUUUAUCAACCUG |
| EGFP (non-targeting control) | Thermo stealth siRNA | CACAUGAAGCAGCACGACUUCUUCA/UGAAGAAGUCGUGCUGCUUCAUGUG |
Reporters were transfected into cells ~24 h post siRNA transfections, when cells were 80–90% confluent. DNA transfections were performed in triplicate with FuGene HD at a 3:1 ratio to DNA, with 50 ng NLuc reporter DNA and 50 ng pGL4.13 FLuc reporter was added per well. RNA transfections were performed with TransIT-mRNA Transfection Kit from Mirus Bio, as per manufacturer’s recommended protocol, with 90 ng reporter mRNA and 200 ng pGL4.13 FLuc control DNA added to each well in triplicate.
For TG experiments, 2 μM TG or equal volume DMSO was added to HEK293 cells 19 h post transfection for 5 h.
Cells were lysed 24 h post transfection with 60 μL Glo Lysis buffer for 5 min at room temperature. Around 25 μL of lysate was mixed with NanoGlo substrate prepared as for RRL reactions, and 25 μL of ONE-Glo Luciferase Assay System (Promega), for 5 min in the dark, in opaque 96-well plates. Luminescence measurements were obtained as with RRL reactions.
For western blots, HEK293 cells were seeded in 12-well plates and transfected 24 h later at 40–50% confluency, with siRNAs using RNAiMax. Where indicated, 24 h post transfections, at 80–90% confluency cells, were transfected with 500 ng NLuc reporter DNAs and 4:1 FuGene HD. Around 48 h post siRNA transfection, cells were lysed in 300 μL RIPA buffer with protease inhibitor for 30 min at 4°C. Lysates were homogenized by passing through a 28G syringe, mixed with 6X sample buffer and stored at –20°C.
Western blots
All samples for western blot were run on 12% sodium dodecyl sulfate-polyacrylamide gels at 150 V for ~90 min. For western blot analysis of HAP1 lysates, 60 μg protein was loaded per well. For HEK293 western blots, 30 μL lysate for each sample was loaded. Gels were transferred to PVDF membranes either overnight at 30 V and 4°C, or for 2.5 h at 320 mAmps and 4°C. Membranes were blocked with 5% non-fat dairy milk, and all antibodies diluted in 5% non-fat dairy milk. Primary antibodies information and probing conditions are listed in Table 3. For eIF2A, eIF2D, DENR, MCTS1 and FLAG western blots, horseradish peroxidase secondary antibodies were applied at 1:10000, with 1 h incubations at room temperature. Bands were then visualized on film. For GAPDH loading controls, LiCor IRDye secondary antibodies were applied at 1:10 000, with 1 h incubations at room temperature and bands visualized with LiCor Odyssey CLx Imaging Systems.
Polysome profiling
24 h post indicated KD, HEK293 cells were treated with 100 μg/mL cycloheximide (CHX) for 5 min at 37°C. They were then harvested as previously described (37). Briefly, they were transferred to ice and washed with 5.0 mL ice-cold PBS containing 100 μg/mL CHX, collected by scraping in cold PBS + CHX and pelleted at 234xg and 4°C for 5 min. PBS was aspirated and pellets re-suspended in polysome-profiling lysis buffer (20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 15 mM MgCl2, 8% (vol/vol) glycerol, 20 U/ml SUPERase, 80 U/ml murine RNase inhibitor, 0.1 mg/ml heparin, 100 μg/ml CHX, 1 mM DTT, 1× EDTA-free protease inhibitor cocktail, 20 U/ml Turbo DNase, 1% Triton X-100) (37). Lysates were passed through a 20G needle 10x and incubated on ice for 5 min. Cellular debris was pelleted at 14 000xg and 4°C for 5 min, and supernatant transferred to a fresh tube. Total lysate RNA was estimated by NanoDrop. Lysates were flash-frozen in liquid nitrogen and stored at –80°C until fractionation.
Sucrose gradients were prepared by successively freezing equal volumes of 50, 36.7, 23.3 and 10% sucrose (wt/vol) in 12 ml Seton tubes. Sucrose-gradient buffer consisted of 20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 15 mM MgCl2, 10 U/ml SUPERase, 20 U/ml murine RNase inhibitor, 100 μg/ml CHX and 1 mM DTT (37). Prior to use, gradients were allowed to thaw and linearize overnight at 4°C. For fractionation, approximately 250 μg total RNA was applied to the top of the sucrose gradient. Gradients were spun at 151 263xg and 4°C for 3 h using a Beckman Coulter Optima L-90 K ultracentrifuge and SW 41 Ti swinging-bucket rotor. Gradients were fractionated with Brandel’s Gradient Fractionation System, measuring absorbance at 254 nm. The detector was baselined with 60% sucrose chase solution and its sensitivity set to 1.0. For fractionation, 60% sucrose was pumped at a rate of 1.5 mL/min. Brandel’s PeakChart software was used to collect profile data.
Drosophila work
The GGGGCC x 28 repeat, along with 30 nt on both ends in the first intron of the gene C9ORF72, were PCR cloned from the genomic DNA from fibroblasts of an ALS patient of Central Biorepository of University of Michigan and placed to the 5′ upstream in the +1 reading frame (GP) relative to the GFP gene in the vector PGFPN1 (Clonetech). The repeat and GFP were then subcloned into the NotI site of vector pUAST and the sequence was verified for the repeat length and relative reading frame. This vector was used to generate transgenic flies by standard p-element insertion (Best Gene, CA) (35).
Information on the GAL4 and shRNA lines interrogated is included in Table 4. For eye shrinkage experiments, male flies containing the UAS-GGGGCCx28 and a UAS-shRNA transgene were crossed to GMR-GAL4 virgin females at 29°C. One eye of each resulting progeny was imaged 0–2 days post eclosion using Leica M125 stereomicroscope and a Leica DFC425 digital camera. Eye widths were then measured with ImageJ and normalized to values obtained to flies expressing control shRNAs.
Table 4.
Drosophila line information
| Name | Gene target | Vendor | Stock # | Average target KD percentage |
|---|---|---|---|---|
| GMR-GAL4 | NA | BDSC | 8605 | NA |
| Tub5-GS-GAL4 | NA | Internal | NA | NA |
| Ctrl shRNA #1 | Luciferase | BDSC | 31 603 | NA |
| Ctrl shRNA #2 | LexA | BDSC | 67 947 | NA |
| eIF2A shRNA | CG7414 | BDSC | 50 649 | M: 22%, F: 44% |
| eIF2D shRNA | eIF2D | BDSC | 33 995 | M: 70%, F: 88% |
| DENR shRNA #1 | DENR | VDRC | 101 746 | M: 54%, F: 37% |
| DENR shRNA #2 | DENR | VDRC | 49 895 | M: 70%, F: 40% |
| MCTS1 shRNA | MCTS1 | BDSC | 55 920 | M: 79%, F: 94% |
For survival experiments, male flies containing the UAS-GGGGCCx28 and a UAS-shRNA transgene were crossed with Tub5-GAL4 GeneSwitch (GS) virgin females at 25°C. Zero to two days post eclosion, resulting progeny were placed on SY10 food containing 200 μM RU486. Male and female progeny were housed separately, with no more than 28 flies per tube. RU486 food was changed every 2 days, with number of dead flies counted during each flip.
Eye inclusion experiments were performed as described previously (39). Male flies containing the UAS-shRNA transgene against DENR #1 or a non-targeting control (Ctrl #2) were crossed to GMR-GAL4 virgin females containing the UAS-FMR1(CGGx90)-EGFP transgene at 25°C. Zero to two days post eclosion, eyes of resulting progeny were imaged using a Leica M125 stereomicroscope and a Leica DFC425 digital camera with GFP filter. All images were taken at the same exposure. GFP images were converted to greyscale and total intensity was measured using ImageJ. For real-time quantitative PCR, total RNA was isolated from 19 to 26 flies per sex per genotype with TRIzol reagent (Invitrogen). Ten micrograms of isolated RNA was treated with TURBO DNase (Ambion) to remove contaminating DNA and was clean and concentrated with RNA Clean and Concentrator-25 Kit (Zymo Research). Around 1 μg of cleaned RNA was used to generate cDNA with iScript reverse transcriptase per the manufacturer’s protocol (Bio-Rad). 20 μL quantitative real-time PCR reactions with 100 ng of cDNA input were carried out using TaqMan Fast Advance Master Mix (Applied Biosystems, 4 444 557) on an Applied Biosystems Quant Studio 3 machine for 40 cycles using fast cycling parameters (95°C for 20 s, 95°C for 1 s, 60°C for 20 s). All runs included a standard dilution curve representing 2x to 0.02x of the RNA concentration utilized for all primer sets to ensure linearity. Equivalent efficiency of individual primer sets was confirmed prior to data analysis. All samples were run in triplicate. The level of mRNA of interest was normalized to RPL32 mRNA and expressed as the change in gene expression to control lines.
Supplementary Material
Acknowledgements
We thank all members of the Todd Lab for their input and assistance on this project. In particular, we thank Udit Sheth, Kristina Zheng, Katyanne Calleja and Sofia Bennetts for assistance maintaining fly lines used in these studies. We also thank the Pletcher Lab at the University of Michigan for making our fly food and Dr Michael Kearse of the Ohio State University for kindly sharing the CrPV-NLuc reporter used in these studies and for providing guidance on studies involving NSC119893.
Conflict of Interest statement. The authors have no financial conflicts of interest associated with the work presented in this manuscript.
Contributor Information
Katelyn M Green, Department of Neurology, University of Michigan, Ann Arbor, MI, USA; Cellular and Molecular Biology Graduate Program, University of Michigan, Ann Arbor, MI, USA.
Shannon L Miller, Department of Neurology, University of Michigan, Ann Arbor, MI, USA; Cellular and Molecular Biology Graduate Program, University of Michigan, Ann Arbor, MI, USA.
Indranil Malik, Department of Neurology, University of Michigan, Ann Arbor, MI, USA.
Peter K Todd, Department of Neurology, University of Michigan, Ann Arbor, MI, USA; Cellular and Molecular Biology Graduate Program, University of Michigan, Ann Arbor, MI, USA; VA Ann Arbor Healthcare System, Ann Arbor, MI, USA.
Funding
National Institute of Health (grants P50HD104463, R01NS099280 and R01NS086810 to P.K.T., and F31NS100302 to K.M.G.). P.K.T. was also supported by Veterans Affairs grant BLRD BX004842, and by private philanthropic support. K.M.G. and S.L.M. were additionally supported by the Cellular and Molecular Biology Graduate Program at the University of Michigan (T32GM007315). I.M. was supported by an Alzheimer’s Association Research Fellowship.
References
- 1. Ingolia, N.T., Lareau, L.F. and Weissman, J.S. (2011) Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell, 147, 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ingolia, N.T., Ghaemmaghami, S., Newman, J.R. and Weissman, J.S. (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science, 324, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sendoel, A., Dunn, J.G., Rodriguez, E.H., Naik, S., Gomez, N.C., Hurwitz, B., Levorse, J., Dill, B.D., Schramek, D., Molina, H. et al. (2017) Translation from unconventional 5′ start sites drives tumour initiation. Nature, 541, 494–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brar, G.A., Yassour, M., Friedman, N., Regev, A., Ingolia, N.T. and Weissman, J.S. (2012) High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science, 335, 552–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rodriguez, C.M., Chun, S.Y., Mills, R.E. and Todd, P.K. (2019) Translation of upstream open reading frames in a model of neuronal differentiation. BMC Genomics, 20, 391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kearse, M.G. and Wilusz, J.E. (2017) Non-AUG translation: a new start for protein synthesis in eukaryotes. Genes Dev., 31, 1717–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Starck, S.R., Tsai, J.C., Chen, K., Shodiya, M., Wang, L., Yahiro, K., Martins-Green, M., Shastri, N. and Walter, P. (2016) Translation from the 5′ untranslated region shapes the integrated stress response. Science, 351, aad3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Merrick, W.C. and Anderson, W.F. (1975) Purification and characterization of homogeneous protein synthesis initiation factor M1 from rabbit reticulocytes. J. Biol. Chem., 250, 1197–1206. [PubMed] [Google Scholar]
- 9. Adams, S.L., Safer, B., Anderson, W.F. and Merrick, W.C. (1975) Eukaryotic initiation complex formation. Evidence for two distinct pathways. J. Biol. Chem., 250, 9083–9089. [PubMed] [Google Scholar]
- 10. Zoll, W.L., Horton, L.E., Komar, A.A., Hensold, J.O. and Merrick, W.C. (2002) Characterization of mammalian eIF2A and identification of the yeast homolog. J. Biol. Chem., 277, 37079–37087. [DOI] [PubMed] [Google Scholar]
- 11. Starck, S.R., Jiang, V., Pavon-Eternod, M., Prasad, S., McCarthy, B., Pan, T. and Shastri, N. (2012) Leucine-tRNA initiates at CUG start codons for protein synthesis and presentation by MHC class I. Science, 336, 1719–1723. [DOI] [PubMed] [Google Scholar]
- 12. Golovko, A., Kojukhov, A., Guan, B.J., Morpurgo, B., Merrick, W.C., Mazumder, B., Hatzoglou, M. and Komar, A.A. (2016) The eIF2A knockout mouse. Cell Cycle, 15, 3115–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bohlen, J., Harbrecht, L., Blanco, S., Clemm von Hohenberg, K., Fenzl, K., Kramer, G., Bukau, B. and Teleman, A.A. (2020) DENR promotes translation reinitiation via ribosome recycling to drive expression of oncogenes including ATF4. Nat. Commun., 11, 4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ahmed, Y.L., Schleich, S., Bohlen, J., Mandel, N., Simon, B., Sinning, I. and Teleman, A.A. (2018) DENR-MCTS1 heterodimerization and tRNA recruitment are required for translation reinitiation. PLoS Biol., 16, e2005160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schleich, S., Strassburger, K., Janiesch, P.C., Koledachkina, T., Miller, K.K., Haneke, K., Cheng, Y.S., Kuechler, K., Stoecklin, G., Duncan, K.E. et al. (2014) DENR-MCT-1 promotes translation re-initiation downstream of uORFs to control tissue growth. Nature, 512, 208–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Skabkin, M.A., Skabkina, O.V., Dhote, V., Komar, A.A., Hellen, C.U. and Pestova, T.V. (2010) Activities of ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev., 24, 1787–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vasudevan, D., Neuman, S.D., Yang, A., Lough, L., Brown, B., Bashirullah, A., Cardozo, T. and Ryoo, H.D. (2020) Translational induction of ATF4 during integrated stress response requires noncanonical initiation factors eIF2D and DENR. Nat. Commun., 11, 4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zu, T., Gibbens, B., Doty, N.S., Gomes-Pereira, M., Huguet, A., Stone, M.D., Margolis, J., Peterson, M., Markowski, T.W., Ingram, M.A. et al. (2011) Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl. Acad. Sci. U. S. A., 108, 260–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bañez-Coronel, M., Ayhan, F., Tarabochia, A.D., Zu, T., Perez, B.A., Tusi, S.K., Pletnikova, O., Borchelt, D.R., Ross, C.A., Margolis, R.L. et al. (2015) RAN translation in Huntington disease. Neuron, 88, 667–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Todd, P.K., Oh, S.Y., Krans, A., He, F., Sellier, C., Frazer, M., Renoux, A.J., Chen, K.C., Scaglione, K.M., Basrur, V. et al. (2013) CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron, 78, 440–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mori, K., Weng, S.M., Arzberger, T., May, S., Rentzsch, K., Kremmer, E., Schmid, B., Kretzschmar, H.A., Cruts, M., Van Broeckhoven, C. et al. (2013) The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science, 339, 1335–1338. [DOI] [PubMed] [Google Scholar]
- 22. Ash, P.E., Bieniek, K.F., Gendron, T.F., Caulfield, T., Lin, W.L., Dejesus-Hernandez, M., van Blitterswijk, M.M., Jansen-West, K., Paul, J.W., Rademakers, R. et al. (2013) Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron, 77, 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Green, K.M., Glineburg, M.R., Kearse, M.G., Flores, B.N., Linsalata, A.E., Fedak, S.J., Goldstrohm, A.C., Barmada, S.J. and Todd, P.K. (2017) RAN translation at C9orf72 -associated repeat expansions is selectively enhanced by the integrated stress response. Nat. Commun., 8, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cheng, W., Wang, S., Mestre, A.A., Fu, C., Makarem, A., Xian, F., Hayes, L.R., Lopez-Gonzalez, R., Drenner, K., Jiang, J. et al. (2018) C9ORF72 GGGGCC repeat-associated non-AUG translation is upregulated by stress through eIF2α phosphorylation. Nat. Commun., 9, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sonobe, Y., Ghadge, G., Masaki, K., Sendoel, A., Fuchs, E. and Roos, R.P. (2018) Translation of dipeptide repeat proteins from the C9ORF72 expanded repeat is associated with cellular stress. Neurobiol. Dis., 116, 155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Westergard, T., McAvoy, K., Russell, K., Wen, X., Pang, Y., Morris, B., Pasinelli, P., Trotti, D. and Haeusler, A. (2019) Repeat-associated non-AUG translation in C9orf72 -ALS/FTD is driven by neuronal excitation and stress. EMBO Mol. Med., 11, e9423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kearse, M.G., Green, K.M., Krans, A., Rodriguez, C.M., Linsalata, A.E., Goldstrohm, A.C. and Todd, P.K. (2016) CGG repeat-associated non-AUG translation utilizes a cap-dependent scanning mechanism of initiation to produce toxic proteins. Mol. Cell, 62, 314–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. González-Almela, E., Williams, H., Sanz, M.A. and Carrasco, L. (2018) The initiation factors eIF2, eIF2A, eIF2D, eIF4A, and eIF4G are not involved in translation driven by hepatitis C virus IRES in human cells. Front. Microbiol., 9, 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sanz, M.A., González Almela, E. and Carrasco, L. (2017) Translation of Sindbis subgenomic mRNA is independent of eIF2, eIF2A and eIF2D. Sci. Rep., 7, 43876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tusi, S.K., Nguyen, L., Thangaraju, K., Li, J., Cleary, J.D., Zu, T. and Ranum, L.P.W. (2021) The alternative initiation factor eIF2A plays key role in RAN translation of myotonic dystrophy type 2 CCUG•CAGG repeats. Hum. Mol. Genet., 30, 1020–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim, J.H., Park, S.M., Park, J.H., Keum, S.J. and Jang, S.K. (2011) eIF2A mediates translation of hepatitis C viral mRNA under stress conditions. EMBO J., 30, 2454–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ventoso, I., Sanz, M.A., Molina, S., Berlanga, J.J., Carrasco, L. and Esteban, M. (2006) Translational resistance of late alphavirus mRNA to eIF2alpha phosphorylation: a strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev., 20, 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Robert, F., Kapp, L.D., Khan, S.N., Acker, M.G., Kolitz, S., Kazemi, S., Kaufman, R.J., Merrick, W.C., Koromilas, A.E., Lorsch, J.R. et al. (2006) Initiation of protein synthesis by hepatitis C virus is refractory to reduced eIF2.GTP.Met-tRNA(i)(Met) ternary complex availability. Mol. Biol. Cell, 17, 4632–4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kearse, M.G., Goldman, D.H., Choi, J., Nwaezeapu, C., Liang, D., Green, K.M., Goldstrohm, A.C., Todd, P.K., Green, R. and Wilusz, J.E. (2019) Ribosome queuing enables non-AUG translation to be resistant to multiple protein synthesis inhibitors. Genes Dev., 33, 871–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kou 1, Y.-H., Chou, S.-M., Wang, Y.-M., Chang, Y.-T., Huang, S.-Y., Jung, M.-Y., Huang, Y.-H., Chen, M.-R., Chang, M.-F. and Chang, S.C. (2006) Hepatitis C virus NS4A inhibits cap-dependent and the viral IRES-mediated translation through interacting with eukaryotic elongation factor 1A. J. Biomed. Sci., 13(6), 861–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jan, E. and 1, Terri Goss Kinzy, Peter Sarnow. (2003) Divergent tRNA-like element supports initiation, elongation, and termination of protein biosynthesis. Proc. Natl. Acad. Sci. U. S. A., 100(26), 15410–15415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. He, F., Flores, B.N., Krans, A., Frazer, M., Natla, S., Niraula, S., Adefioye, O., Barmada, S.J. and Todd, P.K. (2020) The carboxyl termini of RAN translated GGGGCC nucleotide repeat expansions modulate toxicity in models of ALS/FTD. Acta Neuropathol. Commun., 8, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Linsalata, A.E., He, F., Malik, A.M., Glineburg, M.R., Green, K.M., Natla, S., Flores, B.N., Krans, A., Archbold, H.C., Fedak, S.J. et al. (2019) DDX3X and specific initiation factors modulate FMR1 repeat-associated non-AUG-initiated translation. EMBO Rep., 20, e47498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Malik, I., Tseng, Y.J., Wright, S.E., Zheng, K., Ramaiyer, P., Green, K.M. and Todd, P.K. (2021) SRSF protein kinase 1 modulates RAN translation and suppresses CGG repeat toxicity. EMBO Mol. Med., 13, e14163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tabet, R., Schaeffer, L., Freyermuth, F., Jambeau, M., Workman, M., Lee, C.Z., Lin, C.C., Jiang, J., Jansen-West, K., Abou-Hamdan, H. et al. (2018) CUG initiation and frameshifting enable production of dipeptide repeat proteins from ALS/FTD C9ORF72 transcripts. Nat. Commun., 9, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang, S., Latallo, M.J., Zhang, Z., Huang, B., Bobrovnikov, D.G., Dong, D., Livingston, N.M., Tjoeng, W., Hayes, L.R., Rothstein, J.D. et al. (2018) Nuclear export and translation of circular repeat-containing intronic RNA in C9ORF72 -ALS/FTD. Nat. Commun., 12(1), 4908, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jaafar, Z.A., Oguro, A., Nakamura, Y. and Kieft, J.S. (2016) Translation initiation by the hepatitis C virus IRES requires eIF1A and ribosomal complex remodeling. elife, 5, e21198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sonobe, Y., Aburas, J., Krishnan, G., Fleming, A.C., Ghadge, G., Islam, P., Warren, E.C., Gu, Y., Kankel, M.W., Brown, A.E.X. et al. (2021) A C. elegans model of C9orf72 -associated ALS/FTD uncovers a conserved role for eIF2D in RAN translation. Nat. Commun., 12, 6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Uhlén, M., Fagerberg, L., Hallström, B.M., Lindskog, C., Oksvold, P., Mardinoglu, A., Sivertsson, A., Kampf, C., Sjöstedt, E., Asplund, A. et al. Proteomics. Tissue-based map of the human proteome. Science, 347, 1260419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.