Abstract
Objectives
We aimed to characterize and compare the viral loads (VL) of the Omicron BA.1 and BA.2 lineages and the Delta variant in nasopharyngeal samples from newly diagnosed COVID-19 patients and their kinetics over time.
Patients and methods
The kinetics of the VL were measured on the CT data from 215 SARS-CoV-2 positive patients who presented at least two positive PCRs a day apart and were screened for SARS-CoV-2 viral lineages.
Results
We observed no significant difference in median CT value during the first diagnostic test between the Delta variant and the two Omicron lineages. However, the kinetics of CT decreases for the BA.1 and BA.2 lineage were significantly lengthier in time than the kinetics for the Delta variant. The BA.2 lineage presented lower median CT value (-2 CT) (inversely proportional to the VL) than the BA.1 lineage.
Conclusions
BA.2 Omicron lineage presented higher VL than BA.1 Omicron lineage at diagnostic. Omicron BA.1 and BA.2 lineages have more prolonged replication than the Delta variant.
Keywords: SARS-CoV-2, BA.1, BA.2, Viral load, Kinetic
We have previously shown that the Delta variant presented a viral load (VL) 5 times higher than the historical variants [1]. At the end of 2021, a new variant, B.1.1.529 (BA.1 lineage), was first reported in South Africa and spread rapidly into Europe [2], [3] and throughout the world. The BA.1 Omicron lineage rapidly overtook the Delta variant and was described as 3 to 6 times more infectious than other variants [4]. In turn, however, the BA.1 Omicron lineage was rapidly overtaken by a new Omicron sublineage (BA.2). The transmission of the two Omicron lineages was more widely observed than the Delta variant in unvaccinated as well as vaccinated people and also in those previously infected by a SARS-CoV-2 variant [5]. These rapidly obtained observations suggested potential “immune escape” elicited by SARS-CoV-2 vaccines or natural infection. In order to evaluate whether high infectivity could be associated with higher VL, we measured and compared the VLs of these two Omicron lineages and the Delta SARS-CoV-2 variant.
Nasopharyngeal samples (NS) were collected at diagnosis from the Pitié-Salpêtrière hospital in Paris during the epidemic peak of each variant: from August-October 2021 for the Delta variant, from December 2021 to January 2022 for the BA.1 Omicron lineage, and from March-May 2022 for the BA.2 Omicron lineage. By that time, more than 90% of the adult French population had received a primary vaccination. A total of 215 positive SARS-CoV-2 NSs was screened to assess SARS-CoV-2 viral lineages using the previously described method [1]. The screening assessment was confirmed by full genome sequencing using the Artic V3 protocol on a GridION device (Oxford Nanopore, Oxford, UK) for samples with a Cycle threshold (CT) < 28 or by Sanger sequencing of the full Spike gene for samples with CT > 28.
To evaluate VL kinetics, we collected CT data from SARS-CoV-2-positive patients who presented at least two positive PCRs a day apart. CT values of the ORF1ab and E gene targets were obtained by the SARS-CoV-2 Cobas 6800 (Roche). GraphPad Prism8 and R software were used to perform statistical analyses (ANOVA non-parametric test with a Scheirer-Ray-Here test).
We compared the VL at diagnosis and its kinetics for these three variants over time: 71 patients for the Delta variant, 84 for the Omicron BA.1 lineage and 60 for the Omicron BA.2 lineage. The Delta, Omicron BA.1 and BA.2 lineage-positive patients presented sex ratios of 40%, 49.3% and 58.3% of females and median age of 60 years [IQR44 – 79], 55 years [IQR33 – 75] and 67 years [IQR50 – 80] respectively with no statistical difference between groups.
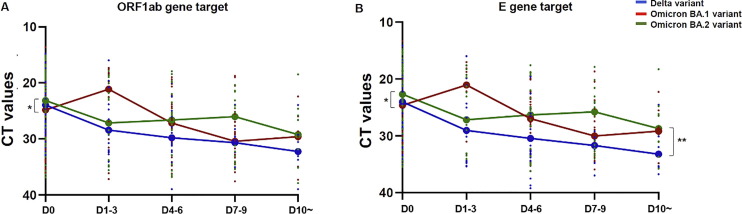
Similar median CT value was observed at the time of first diagnostic test for the Omicron BA.1 lineage and Delta variants: ORF1ab gene target, median CT values 24.84 [IQR20.28 – 30.97] and 23.97 [IQR19.60 – 28.19] respectively (Fig. 1 A); E gene targets, 24.64 [IQR20.09 – 30.62] and 24.03 [IQR19.75 – 28.27]), respectively (Fig. 1B). An increase of relative VL (corresponding to decreased CT values) was observed only for the Omicron BA.1 lineage at days 1–3 (ORF1ab gene target median CT value 21.13 [IQR18.73 – 31.19] and E gene target 21.07 [IQR18.89 – 30.55]) compared to the Delta and the BA.2 lineages (ORF1ab gene target 28.45 [IQR20.24 – 32.15] and 27.16 [IQR20.08 – 33.78 and E gene target 29.06 [IQR20.50 – 34.43] and 27.20 [IQR19.70 – 33.10], respectively) (Fig. 1A-B).
Fig. 1.
Omicron and Delta variant’s viral load kinetics. The graphs represent the kinetics of the VL, expressed as CT, (A) of the ORF1ab gene target and (B) of the E gene for the Delta variant (in blue) for the Omicron BA.1 lineage (in red) and for the Omicron BA.2 lineage (in green). ANOVA non-parametric test shows a significant effect of the variant type on the CT decrease over time for the E gene target. *p < 0,05, **p < 0,005.
The Omicron BA.2 lineage presented higher VL (inversely proportional to the CT values) for the ORF1ab gene target (median CT value 23.17 [IQR18.77 – 25.84]) and the E gene target (22.70 [IQR18.53 – 25.58]) than the BA.1 lineage at diagnosis (ORF1ab gene p = 0.038 and E gene p = 0.042) but was not significantly different from the VL of the Delta variant. However, similar median CT value was achieved at day 10 between the two Omicron sub-lineages (BA.1: 29.61 [IQR26 – 34.95], BA.2: 29.25 [IQR24.94 – 34.34] for ORF1ab gene target and BA.1: 29.18 [IQR25.53 – 33.71] and BA.2 28.75 [IQR25.15 – 33.05] for E gene target) (Fig. 1A-B). Moreover, we observed a significant influence of the variant on CT decay over time for the E gene target (p < 0.01).
Our results show no significant difference between the relative viral loads of the BA.1 and BA.2 Omicron lineages and the Delta variants at time of diagnosis. This result suggested that the more pronounced infectivity of Omicron BA.1 lineage was not correlated with higher VL values, a finding consistent with previous studies [6]. Our results also showed that VL of the BA.2 lineage was higher than that of the BA.1 at diagnosis, which is likewise coherent with the literature [7]. We also observed that the CT values of the Delta variant decreased more rapidly than those of the BA.1 and BA.2 Omicron lineage over time. Of note, we found that the median CT value of the Omicron BA.1 lineage increased from one to three days after the first diagnostic test. These results suggest that unlike the Delta and the BA.2 lineage, the VL peak for the BA.1 Omicron lineage does not occur, at symptom onset, but rather a few days later. However, this “delayed” VL peak may have resulted from earlier initial diagnostic testing for the BA.1 omicron lineage group. It also bears mentioning that show a longer phase of replication for the BA.1 and BA.2 Omicron lineages in comparison with the Delta variant. More specifically, we observed that contrary to the Delta variant, until day 10 the CT values of the BA.1 and BA.2 Omicron lineages remained below 33, which has been defined as the threshold value for moderate viral shedding [8], and is consistent with the literature [9]. While these differences may be associated with the immune status (vaccine booster dose or not, serological response) of our cohort, they may also be due to the intrinsic properties of Spike proteins of the BA.1 and BA.2 Omicron lineages, or to the immune escape of the Omicron lineages with regard to neutralizing antibodies. In fact, the Omicron BA.1 and BA.2 lineages harbored several mutations in the RBD region, the primary target of plasma-neutralizing activity, which in comparison to other SARS-CoV-2 variants leads to immune escape from almost all of the natural and therapeutic antibodies [10]. Moreover, it has been shown that the BA.2 Omicron lineage is 4.5 more contagious than the Delta variant and 1.5 more contagious than the BA.1 Omicron lineage, and it seems to be 30% more capable of escaping the immune system than the BA.1 Omicron lineage [11], a factor that could help to explain its predominance. One of the limitations of our study consists in imprecise information about the date of symptom onset. Furthermore, the patient cohorts were selected in different proportions according to Delta and Omicron lineage. Moreover, it has been observed in several studies that while viral load may be an independent predictor of disease severity, it is more closely associated with transmissibility [12], [13].
In conclusion, our study reinforces the hypothesis that the higher infectivity of the Omicron BA.1 and BA.2 lineages was primarily associated not with high VL, but rather with prolonged replication and capacity for escape from the immune system. In addition, our results suggest that the period of infectivity of these lineages may be longer than was the case with previous SARS-COV-2 variants. This finding could have appreciable implications in diagnosis and isolation strategy.
Authors’ contributions
ET, CS, AGM, VC contributed to the design of the study and the manuscript. SM, BA, TC, ET, SA, VP, AF participated in the data collection. ET, CS carried out analyses of data. All the authors have reviewed and approved the submitted manuscript.
Funding
ET was supported by the ANRS-MIE (Agence Nationale de Recherches sur le SIDA et les hépatites virales-Maladies Infectieuses Emergentes) (AC43, Medical Virology).
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- 1.Teyssou E., Delagrèverie H., Visseaux B., Lambert-Niclot S., Brichler S., Ferre V., et al. The Delta SARS-CoV-2 variant has a higher viral load than the Beta and the historical variants in nasopharyngeal samples from newly diagnosed COVID-19 patients. J Infect. 2021;83(4):e1–e3. doi: 10.1016/j.jinf.2021.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern; n.d. https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern (accessed February 4, 2022).
- 3.Weekly epidemiological update: Omicron variant of concern (VOC) – week 50 (data as of 19 December 2021). Eur Cent Dis Prev Control 2021. https://www.ecdc.europa.eu/en/news-events/weekly-epidemiological-update-omicron-variant-concern-voc-week-50-data-19-december-2021 (accessed January 28, 2022).
- 4.Callaway E., Ledford H. How bad is Omicron? What scientists know so far. Nature. 2021;600:197–199. doi: 10.1038/d41586-021-03614-z. [DOI] [PubMed] [Google Scholar]
- 5.Lyngse F.P., Kirkeby C.T., Denwood M., Christiansen L.E., Mølbak K., Møller C.H., et al. Transmission of SARS-CoV-2 Omicron VOC subvariants BA.1 and BA.2: Evidence from Danish Households. Infect Dis (except HIV/AIDS) 2022 doi: 10.1101/2022.01.28.22270044. [DOI] [Google Scholar]
- 6.Puhach O, Adea K, Hulo N, Sattonnet P, Genecand C, Iten A, et al. Infectious viral load in unvaccinated and vaccinated patients infected with SARS-CoV-2 WT, Delta and Omicron 2022:2022.01.10.22269010. 10.1101/2022.01.10.22269010. [DOI] [PubMed]
- 7.Hirotsu Y., Maejima M., Shibusawa M., Natori Y., Nagakubo Y., Hosaka K., et al. to March 2022. J Infect. 2022;85(2):174–211. doi: 10.1016/j.jinf.2022.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avis-SFM-valeur-Ct-excrétion-virale-_-Version-def-14012021_V4.pdf; n.d.
- 9.Marking U., Havervall S., Greilert Norin N., Bladh O., Christ W., Gordon M., et al. Correlates of protection, viral load trajectories and symptoms in BA.1, BA.1.1 and BA.2 breakthrough infections in triple vaccinated healthcare workers. Infect Dis (except HIV/AIDS) 2022 doi: 10.1101/2022.04.02.22273333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mannar D., Saville J.W., Zhu X., Srivastava S.S., Berezuk A.M., Tuttle K.S., et al. SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein-ACE2 complex. Science. 2022;375(6582):760–764. doi: 10.1126/science.abn7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, Wei G-W. Omicron BA.2 (B.1.1.529.2): high potential to becoming the next dominating variant. ArXiv 2022:arXiv:2202.05031v1.
- 12.Shenoy S. SARS-CoV-2 (COVID-19), viral load and clinical outcomes; lessons learned one year into the pandemic: A systematic review. World J Crit Care Med. 2021;10:132–150. doi: 10.5492/wjccm.v10.i4.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dadras O., Afsahi A.M., Pashaei Z., Mojdeganlou H., Karimi A., Habibi P., et al. The relationship between COVID-19 viral load and disease severity: A systematic review. Immun Inflamm Dis. 2022;10(3):e580. doi: 10.1002/iid3.580. [DOI] [PMC free article] [PubMed] [Google Scholar]