Abstract
Because society is doing significant efforts to recycle plastics, one option is to break them down into monomers with the help of specialized enzymes. Polyesters such as PLA (polylactic), PCL (polycaprolactone), PHAs (polyhydroxyalkanoates) and PET (polyethylene‐terephthalate) have been considered in more detail for these biological treatments, because they can be now produced as bio‐based polymers, and because ester bounds and esterases are very frequently found in nature. In particular since PET is the most abundant thermoplastic of the polyester family and accounts for approximately 10% of all synthetic plastics on the market, it has attracted more attention. Here we will review the patented biological recycling processes concerning the recycling of PET.
Plastic materials are composed of large polymeric molecules constructed by multiple additions of one type of a small molecule called monomer (homopolymer) or by two or more different types of monomers (heteropolymers). Most plastics in use today are obtained by using monomers derived from fossil fuels and are quite recalcitrant to biodegradation. The term non‐biodegradable applies to plastics that do not break down to a natural, environmentally safe molecule over time by biological processes. The durability of plastics is mainly based on the fact that these molecules are xenobiotics and are, therefore, a rare target for enzymes and microorganisms. Only few marketed plastics are derived from bio‐based monomers and not all of them are biodegradable and/or compostable. In fact, some bio‐based polymers can also be quite recalcitrant to biodegradation or composting as fossil‐derived plastics. The massive use of these plastics recalcitrant to biodegradation and their disposal in the environment is continuously creating serious health and socioeconomic concerns (Ali et al., 2021). Because of that society is making significant efforts to recycle these plastics and give them a new use following the principles of circular economy. In this sense, the reader is directed to a series of articles of the special section of Science “Our plastics dilemma” (Science Vol 373, issue 6550, July 2021).
One option to recycle these plastics, or at least one option to make them edible for microorganisms, is to break them down into monomers with the help of specialized enzymes. In this sense, a large number of studies can be found in the scientific literature concerning the use of enzymes capable of hydrolysing plastics that are not easily degradable, and most of them have been analysed in recent reviews (Amobonye et al., 2021; Danso et al., 2019; Filiciotto & Rothenberg, 2021; Mohanan et al., 2020; Nikolaivits et al., 2021; Prieto, 2016; Purohit et al., 2020; Sales et al., 2021; Satti & Shah, 2020; Wei & Zimmermann, 2017). Through these reviews the reader learns on the scientific state of the art in this field, however, in this article, we pay attention mainly to the knowledge that has been transferred and applied to the industrial sector, by analysing which of these findings has been patented to date. Sometimes, visiting the patent side of science and technology we can find aspects that have not been collected in the scientific journals. Moreover, since patent applications are published a few years before innovations appear on the market, this information can be generally used as a good indicator of things that would soon arrive.
To help us on this matter a recent study has been published by the European Patent Office (EPO) entitled “Patents for tomorrow's plastics: Global innovation trends in recycling, circular design and alternative sources” that registers and analyses the patenting activity worldwide related to plastic recycling and bioplastic technologies (Dossin et al., 2021). Chemical methods have led the field for decades, but EPO report looks at both biological and chemical recycling innovations. Here, we will only focus our analyses on biological recycling processes and particularly in those patented processes concerning the recycling of polyethylene terephthalate (PET) (Table 1).
TABLE 1.
Non‐exhaustive list of patents related to PET enzymatic degradation
| System | Patent | Priority date | Proprietary | Main claim |
|---|---|---|---|---|
| Cutinase | WO 2000/034450 A1 | Dec 4 1998 | Novo Nordisk | H. insolensis enzyme mutants with higher thermostability produced in Aspergillus oryzae. |
| CN101792729A | Dec 18 2009 | Jiangnan University | T. fusca enzyme produced in E. coli. | |
| WO 2012/075662 A1 | Dec 8 2010 | Jiangnan University | T. fusca enzyme produced in E. coli. | |
| WO 2012/099018 A1 | Jan 19 2011 | Amano Enzyme & Osaka Universtity | T. fusca enzyme produced in E. coli. | |
| WO 2014/079844 A1 | Nov 20 2012 | Carbios | T. cellulosilytica enzyme produced in E. coli. | |
| WO 2014/097321 Al | Dec 17 2012 | Veeranki, Venkata D. & Hedge, Krishnamoorthy | Cut1 and Cut2 cutinases from T. fusca produced in E. coli. | |
| WO 2015/067619 A2 | Nov 5 2013 | Carbios | Use of enzymes to degrade polyesters. | |
| JP2015119670A | Dec 24 2013 | Kyoto Institute of Technology | Thermostable Saccharomonospora viridis enzyme produced in E. coli. | |
| WO 2015/173265 A1 | May 16 2014 | Carbios | T. cellulosilytica enzyme produced in E. coli | |
| WO 2015/097104A1 | Dec 19 2014 | Carbios | Use of enzymes to degrade polyesters. | |
| US 2018/0142097 A1 | Jun 12 2015 | Carbios | Plastic composites with enzymes. | |
| WO 2017/198786 A1 | May 19 2016 | Carbios | Amorphization of PET. | |
| WO 2017/204615 A2 | May 26 2016 | Universidad Nacional Autónoma de Méexico | Aspergillus nidulans enzyme produced in yeasts and E. coli. | |
| WO 2018/011281 A1 | Jul 12 2016 | Carbios | Thermobifida sp. enzyme mutants. | |
| WO 2018/011284 A1 | Jul 12 2016 | Carbios | Thermobifida sp. enzyme mutants. | |
| EP 3517608 A1 | Jan 30 2018 | Carbios | Thermobifida sp. enzyme mutants. | |
| WO 2020/169633 A1 | Feb 21 2019 | Evoxx Technologies & Henkel AG&Co | Thermobifida sp. enzyme mutants. | |
| WO 2021/181205 A2 | Mar 9 2020 | Daniel E. Rodriguez | T. fusca enzyme mutants with higher activity and thermostability. | |
| CN112159478A | Sep 28 2020 | Jiangnan University | Fusion protein of cutinase and a carbohydrate‐binding module produced in E. coli and B. subtilis. | |
| PETase | WO 2017/198463 A1 | May 17 2016 | Henkel AG&Co | Combination of enzyme plus a surfactant. |
| CN107794252A | Oct 18 2016 | University of Electronic Science and Technology of China | Wild‐type enzyme produced in E. coli. | |
| CN107674866A | Oct 18 2016 | University of Electronic Science and Technology of China | Mutant enzyme expressed in E. coli. | |
| WO 2018/168679 A1 | Mar 14 2017 | Keio University | Combination of mutant enzymes and a surfactant. | |
| WO 2019/168811 A1 | Feb 28 2018 | Alliance for Sustainable Energy & University of Portsmouth | Mutant enzyme expressed in E. coli. | |
| CN106367408A | May 11 2018 | Tianjin University | Mutant enzyme expressed in E. coli. | |
| CN108588052A | May 11 2018 | Tianjin University | Mutant enzyme expressed in E. coli. | |
| CN108467857A | Mar 14 2018 | Sichuan University | Mutant enzyme expressed in E. coli. | |
| US 2020/0048621 A1 | Aug 8 2018 | Kyungpook National University Industry‐Academic Cooperation Foundation | Preparation of crystals of a mutant enzyme. | |
| CN110241097A | May 24 2019 | Shandong University | Mutant enzyme produced in E. coli | |
| CN111057693A | Dec 31 2019 | Tianjin Institute of Industrial Biotechnology of CAS | Mutant enzyme produced in E. coli. | |
| CN112852800A | Jan 9 2020 | Tianjin Institute of Industrial Biotechnology of CAS | Enzyme with new signal peptide secreted in E. coli. | |
| WO 2021/145822 A1 | Jan 16 2020 | Agency for Science, Technology & Research | Cyclic PETase with higher thermostability. | |
| KR102298651B1 | May 20 2020 | Kyungpook National University Industry‐Academic Cooperation Foundation | Mutant enzyme with higher thermostability. | |
| WO 2021/148713 A1 | Jan 24 2021 | VTT Technical Research Centre of Finland | Degradation of PEF. | |
| CN113774041A | Aug 6 2021 | University of Tianjin | IsPETase mutant enzyme. | |
| MHETase | KR102261596B1 | Jan 6 2020 | Kyungpook National University Industry‐Academic Cooperation Foundation | MHETase mutant with exo‐PETase activity. |
| PETase & MHETase | WO 2015/025861 A1 | Aug 21 2013 | Keio University | Enzymes from I. sakaeinsis. |
| WO 2020/055369 A3 | Sep 11 2018 | Istanbul Medipol University | Enzymes produced in E. coli and Bacillus subtilis | |
| KR102176506B1 | Dec 4 2018 | Kyungpook National University Industry‐Academic Cooperation Foundation | Enzymes produced in E. coli. | |
| CN113549244A | Jul 16 2021 | Dalian Ocean University | Enzymes produced in E. coli. | |
| Whole organisms | ||||
| E. coli | CN106636158A | Oct 20 2016 | Tianjin University | Expression of PETase in cell surface. |
| Pichia pastoris | CN106497963A | Oct 20 2016 | Tianjin University | Expression of PETase in cell surface. |
| P. pastoris | CN106497964B | Oct 20 2016 | Tianjin University | Expression of PETase in cell surface. |
| P. putida | WO 2019/222396 A1 | May 15 2018 | Alliance for Sustainable Energy & UT‐Batelle | Strain able to metabolize PET. |
| E. coli | CN109402037A | Nov 7 2018 | Shenlun Biotech Shenzhen Co ltd | PETase and MHETase are expressed together. |
| B. subtilis + (P. putida & Rhodococcus sp.) | CN109929789A | Mar 26 2019 | Tianjin University | PETase and MHETase. are expressed together in B. subtilis and mixed with P. putida and Rhodococcus sp. to degrade PET. |
| Clostridium thermocellum | CN111100835A | Jan 7 2020 | Qingdao Institute of Bioenergy and Bioprocess Technology of CAS | PETase and MHETase are expressed together. |
| Mechanoenzymatic | WO 2021/081633 A1 | Oct 28 2019 | The Royal Institution for the Advancement of Learning/McGill University | Combination of enzymatic and mechanical treatments. |
Within the large universe of plastic polymers, polyesters such as PLA (polylactic), PCL (polycaprolactone), PHAs (polyhydroxyalkanoates) and PET have been generally considered in more detail for biological treatments, probably because they can be now produced as bio‐based polymers, and because ester bounds and esterases are very frequently found in nature. In particular, since PET is the most abundant thermoplastic of the polyester family and accounts for approximately 10% of all synthetic plastics on the market (Ali et al., 2021), it has attracted more attention. Although lipases, carboxylesterases and polyester hydrolases can hydrolyse PET, the best hydrolytic yields have been found using cutinases or a mixture of two enzymes, i.e., a PET hydrolase (PETase) (very similar to cutinases) and a mono‐(2‐hydroxyethyl) terephthalate (MHET) hydrolase (MHETase) (Ahmaditabatabaei et al., 2021; Carniel et al., 2021; Carr et al., 2020; Damayanti & Wu, 2021; Maity et al., 2021; Maurya et al., 2020; Salvador et al., 2019; Samak et al., 2020; Tiso et al., 2021; Tournier et al., 2020; Urbanek et al., 2021; Zimmermann, 2020). An additional enzyme (BHETase) will be required to hydrolyse a trace by product bis‐(2‐hydroxyethyl) terephthalate (BHET) (Figure 1).
FIGURE 1.
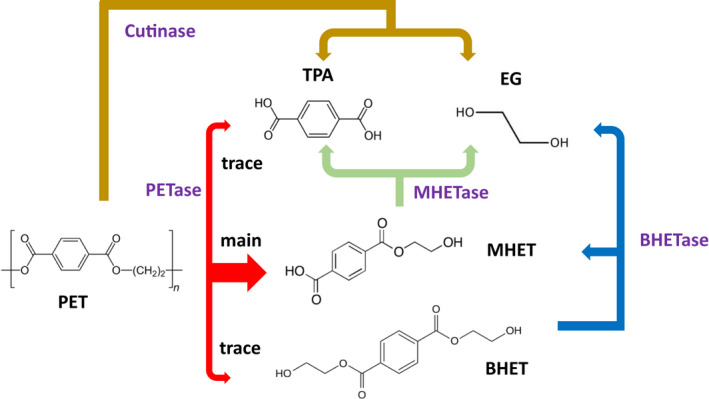
Enzymatic hydrolysis of polyethylene terephthalate (PET). Terephthalic acid (TPA). Ethylene glycol (EG). Bis‐(2‐hydroxyethyl) terephthalate (BHET). Mono‐(2‐hydroxyethyl) terephthalate (MHET)
One of the companies that has been more active in the development of applications of PET degrading enzymes is Carbios (France). In fact, one of the first patents on PET degrading enzymes was filled by this company using a cutinase isolated from the actinomycete Thermobifida sp. (Maille, 2015). Carbios also protected other industrial development related to PET degradation using this cutinase with a previous amorphizing of the plastic product prior to depolymerization (Desrousseaux et al., 2017). Carbios also protected the use of a cutinase from Thermobifida cellulosilytica expressed in Escherichia coli (Boisart & Maille, 2014; Herrero Acero et al., 2015) to break down the crystalline structure of PET and generate high methane potential intermediates (Boisart et al., 2016). However, the treatment of PET with cutinase leads to only 10% hydrolysis after 7 days at 50°C. In addition, in this patent Carbios protected the use of many other hydrolytic enzymes and microorganisms to degrade a large number of plastics (e.g., PLA, polyamides, polyolefines) (Boisart et al., 2016). At the time, it was clear that the hydrolytic yield was not sufficient for industrial recycling, so Carbios worked to develop a new highly efficient recombinant enzyme that, upon publication in Nature, became a breakthrough in the field (Tournier et al., 2020). The new PET hydrolase (e.g., the variant ICCG, F243I/D238C/S283C/Y127G) achieves, over 10 h, a minimum of 90% PET depolymerization into monomers, showing a productivity of 16.7 g of terephthalate/L h (200 g/kg of PET suspension), with an enzyme concentration of 3 mg/g of PET. The new enzyme was developed using a cutinase described by Sulaiman et al. (2012) and the results were translated first to three patents (Topham et al., 2019; Tournier et al., 2018; Zimmermann et al., 2019) opening a new window for its industrial utilization. It is also worth to mention that in a different patent Carbios claims to improve the biodegradability of polyester plastics (e.g., PLA, PCL, PBAT, PHAs and PBS) under environmental conditions by creating plastic composites that integrate one hydrolytic enzyme as well as an anti‐acid filler such as calcium carbonate (Guemard et al., 2018). Interestingly, these findings that protected a new friendlier environmental alternative for plastics have not been published in the scientific literature yet, demonstrating that patents can provide complementary information.
In this sense, it is worth to mention that the first cutinases used to degrade PET were discovered many years ago in Fusarium solani pisi (Vertommen et al., 2005) and in Thermobifida fusca DSM43793 (former name: Thermomonospora fusca) (Müller et al., 2005). Remarkably, the cutinase from F. solani pisi had been patented by Plant Genetic Systems (Belgium) for other uses not related to PET degradation (De Geus, 1990). Thermostable variants of a similar enzyme isolated from Humicola insolensis were patented by Novo Nordisk and used for PET degradation (Abo et al., 2000). However, although the cutinase from T. fusca was discovered in 1999 by Fett et al. (1999), it was not protected at that time. Nevertheless, a recombinant form of cutinase from T. fusca produced in E. coli was protected by two different patents, one from the University of Osaka and Amano Enzyme Inc. (Japan) (Shigenori et al., 2012) and the other from the University of Jiangnan (China), although curiously the inventors did not claim for any specific use (Chen et al., 2012).
Since these cutinases were not very efficient, new efforts were directed to find new enzymes and thus, Sulaiman et al. (2012) isolated a more efficient PET degrading enzyme from a metagenome library that showed 57%–60% amino acid similarity to Thermomonospora curvata lipase and T. fusca cutinase. Surprisingly, this metagenomic enzyme was not patented when discovered, however, it was further modified and patented by Carbios, as mentioned above (Topham et al., 2019; Tournier et al., 2018; Zimmermann et al., 2019). Other enzymes that degrade PET and have been cloned and patented are a cutinase from Aspergillus nidulans (Farres et al., 2017) and a thermostable cutinase from Saccharomonospora viridis (Kawai et al., 2015).
More recently, the interest in PET hydrolases was greatly boosted by the work of Yoshida et al. (2016) who reported that Ideonella sakaiensis 201‐F6 was capable of growing using low‐crystallinity PET, thanks to two enzymes that hydrolysed PET and MHET. This finding was patented by the University of Keio (Miyamoto et al., 2015, 2018). Since then, several patents have been filed using these enzymes. For instance, the group of G. Beckham (USA) has developed an intense activity on these enzymes (Austin et al., 2018; Erickson et al., 2021; Knott et al., 2020; Werner et al., 2021). Interestingly, chimeric enzymes fusing the PETase and MHETase proteins with different linkers have been constructed to improve the PET degradation yield (Knott et al., 2020). This group has filled two patents, one shared by the Alliance for Sustainable Energy (USA) and the University of Portsmouth (UK) claiming for a PETase mutant with increasing PET degrading efficiency (Beckham, Jayakody, et al., 2019) and the other shared with UT‐Batelle (USA) claiming for a recombinant Pseudomonas putida that expresses PET and MHET enzymes (Beckham, Johnson, et al., 2019). Remarkably, the Agency for Science, Technology and Research (Singapore) has constructed a cyclic enzyme using the SpyTag/SpyCatcher system to increase its thermostability (Ghadessy & Sana, 2021). In addition, other patents have been filled concerning the use of these recombinant enzymes (Demircan & Keskin, 2020; Jin, 2020). Despite the intense work done in recent years with these enzymes, none of these patents have hit the market yet.
CONCLUSIONS
PET is the most abundant polyester plastic, with almost 70 Mt manufactured annually worldwide for use in textiles and packaging, and thus, the possibility of its continuous recycling is currently considered as a critical challenge. In this review, we have learned that PET biological recycling is getting closer and closer to the market, thanks to improvements in the genetically modified cutinases and PETases/MHETases. In fact, biological PET recycling has been claimed to very close to a happy ending, since a consortium of Carbios, L'Oréal, Nestlé Waters, PepsiCo and Suntory Beverage and Food Europe has announced last July the successful production of the world's first food‐grade PET plastic bottles produced entirely from enzymatically recycled plastic. Bottles have been manufactured based on Carbios' PET enzymatic recycling technology (C‐ZYME® technology) and according to the company the process can be achieved at high speed, i.e., breaking down 97% of PET in just 16 h. Nevertheless, a techno‐economic analysis by Singh et al. (2021) on the enzymatic recycling of PET demonstrates that current processes still consume large amounts of energy and generate some undesirable waste which will require additional efforts to address these and other important limitations. In addition, more research is still required to improve the development of the enzymes.
The enzymatic recycling of PET paves the way for applying this technology not only to other polyesters, but also to all fossil oil derived plastics. If we were able to combine the production of conventional plastics using bio‐based monomers with their enzymatic recycling, we will be closer to envisioning a more sustainable future for plastics. Furthermore, if we were able to substitute these conventional plastics, even if recyclable, by a new generation of plastics that can be easily biodegradable when accidently released to the environment, we will get a cleaner and safer planet.
CONFLICT OF INTEREST
None declared.
ACKNOWLEDGEMENT
I thank M. A. Prieto for the critical reading of the manuscript and her advise.
REFERENCES
- Abo, M. , Fukuyama, S. , Svendsen, A. & Matsui, T. (2000) Cutinase variants . Patent WO 2000/034450 A1.
- Ahmaditabatabaei, S. , Kyazze, G. , Iqbal., H.M.N. & Keshavarz, T. (2021) Fungal enzymes as catalytic tools for polyethylene terephthalate (PET) degradation. Journal of Fungi, 7, 931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, S.S. , Elsamahy, T. , Al‐Tohamy, R. , Zhu, D. , Mahmoud, Y.A. , Koutra, E. et al. (2021) Plastic wastes biodegradation: mechanisms, challenges and future prospects. Science of the Total Environment, 780, 146590. [DOI] [PubMed] [Google Scholar]
- Amobonye, A. , Bhagwat, P. , Singh, S. & Pillai, S. (2021) Plastic biodegradation: frontline microbes and their enzymes. Science of the Total Environment, 759, 143536. [DOI] [PubMed] [Google Scholar]
- Austin, H.P. , Allen, M.D. , Donohoe, B.S. , Rorrer, N.A. , Kearns, F.L. , Silveira, R.L. et al. (2018) Characterization and engineering of a plastic‐degrading aromatic polyesterase. Proceedings of the National Academy of Sciences of the United States of America, 115, 4350–4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham, G.T. , Jayakody, L.N. , Guss, A.M. , Mand, T.D. , Johnson, C.W. & Pardo Mendoza, I. (2019) Engineered microorganisms for the deconstruction of polymers . Patent WO 2019a/222396 A1.
- Beckham, G.T. , Johnson, C.W. , Donohoe, B.S. , Rorrer, N. , McGeehan, J.E. , Austin, H.P. & Allen, M.D. (2019) Enzymes for polymer degradation . Patent WO 2019b/168811 A1.
- Boisart, C. & Maille, E. (2014) Method for recycling plastic products . Patent WO 2014/079844 A1.
- Boisart, C. , Maille, E. & Guillamot, F. (2016) A method for degrading a plastic . Patent US 2016/0280881 A1.
- Carniel, A. , Waldow, V.A. & Castro, A.M. (2021) A comprehensive and critical review on key elements to implement enzymatic PET depolymerization for recycling purposes. Biotechnology Advances, 52, 107811. [DOI] [PubMed] [Google Scholar]
- Carr, C.M. , Clarke, D.J. & Dobson, A.D.W. (2020) Microbial polyethylene terephthalate hydrolases: current and future perspectives. Frontiers in Microbiology, 11, 571265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. , Wu, J. , Wu, D. & Wang, L. (2012) Cutinase‐producing genetically engineered microorganisms and use thereof . Patent WO 2012/075662 A1.
- Damayanti, D. & Wu, H.S. (2021) Strategic possibility routes of recycled PET. Polymers (Basel), 213, 1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danso, D. , Chow, J. & Streit, W.R. (2019) Plastics: environmental and biotechnological perspectives on microbial degradation. Applied and Environmental Microbiology, 85, e01095–e01019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Geus, P. (1990) Cutinase . Patent WO 90/09446.
- Demircan, T. & Keskin, E. (2020) Method for preparing recombinant PETase and METHase enzymes for use in decomposition of plastics . Patent WO 2020/055369.
- Desrousseaux, M.‐L. , Teixer, H. , Duquesne, S. , Marty, A. , Aloui Dalibey, M. & Chateau, M. (2017) A process for degrading plastic products . Patent WO 2017/198786 A1.
- Dossin, M. , Grilli, M. , Marsitzky, D. , Meiser, W. , Ménière, Y. , Philpott, J. et al. (2021) Patents for tomorrow's plastics: global innovation trends in recycling, circular design and alternative sources. Munich: European Patent Office. [Google Scholar]
- Erickson, E. , Shakespeare, T.J. , Bratti, F. , Buss, B.L. , Graham, R. , Hawkins, M.A. et al. (2021) Comparative performance of PETase as a function of reaction conditions, substrate properties, and product accumulation. ChemSusChem, 15, e202101932. [DOI] [PubMed] [Google Scholar]
- Farres, A.M. , Peña, C. , Hernández, E.E. , Morales, S.L. , Sánchez, M. & Solis, I. (2017) Recombinant cutinases from aspergillus nidulans for the biodegradation of polyesters . Patent WO 2017/204615 A2.
- Fett, W.F. , Wijey, C. , Moreau, R.A. & Osman, S.F. (1999) Production of cutinase by Thermomonospora fusca ATCC 27730. Journal of Applied Microbiology, 86, 561–568. [Google Scholar]
- Filiciotto, L. & Rothenberg, G. (2021) Biodegradable plastics: standards, policies, and impacts. ChemSusChem, 14, 56–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadessy, F.J. & Sana, B. (2021) Thermostable PETase enzyme . Patent WO 2021/145822 A1.
- Guemard, E. , Chateau, M. & Marty, A. (2018) Biodegradable polyester composition and uses thereof . Patent US 2018/0142097 A1.
- Herrero Acero, E. , Guebitz, G. & Ribitsch, C. (2015) Method for recycling plastic products . Patent WO 2015/097104 A1.
- Jin, K.K. (2020) Mhetase mutant having exo‐hydrolytic activity for polyethylene terephthalate polymer having terminus . Patent WO 2021/141280 A1.
- Kawai, F. , Tanokura, M. , Miyagawa, T. & Mizushima, H. (2015) Novel cutinase, gene encoding the cutinase, and method for decomposing polyester‐ or esther‐compound by using the cutinase . Patent JP2015119670A.
- Knott, B.C. , Erickson, E. , Allen, M.D. , Gado, J.E. , Graham, R. , Kearns, F.L. et al. (2020) Characterization and engineering of a two‐enzyme system for plastics depolymerization. Proceedings of the National Academy of Sciences of the United States of America, 117, 25476–25485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maille, E. (2015) Process of recycling mixed PET plastic articles . Patent WO 2015/173265 A1.
- Maity, W. , Maity, S. , Bera, S. & Roy, A. (2021) Emerging roles of PETase and MHETase in the biodegradation of plastic wastes. Applied Biochemistry and Biotechnology, 193, 2699–2716. [DOI] [PubMed] [Google Scholar]
- Maurya, A. , Bhattacharya, A. & Khare, S.K. (2020) Enzymatic remediation of polyethylene terephthalate (PET)‐based polymers for effective Management of Plastic Wastes: an overview. Frontiers in Bioengineering and Biotechnology, 8, 602325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, K. , Furukawa, M. , Kawakami, N. & Oda, K. (2018) Method for improving activity of PET‐degrading enzyme using additive . Patent WO 2018/168679 A1.
- Miyamoto, K. , Yhosida, S. , Oda, K. , Kimura, Y. & Kazumi, H. (2015) Aromatic polyester decomposition enzyme and method for decomposing aromatic polyesters using said enzyme . Patent WO 2015/025861 A1.
- Mohanan, N. , Montazer, Z. , Sharma, P.K. & Levin, D.B. (2020) Microbial and enzymatic degradation of synthetic plastics. Frontiers in Microbiology, 11, 580709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, R.J. , Schrader, H. , Profe, J. , Dresler, K. & Deckwer, W.D. (2005) Enzymatic degradation of poly(ethylene terephthalate): rapid hydrolysis using a hydrolase from Thermobifida fusca . Macromolecular Rapid Communications, 26, 1400–1405. [Google Scholar]
- Nikolaivits, E. , Pantelic, B. , Azeem, M. , Taxeidis, G. , Babu, R. , Topakas, E. et al. (2021) Progressing plastics circularity: a review of mechano‐biocatalytic approaches for waste plastic (re)valorization. Frontiers in Bioengineering and Biotechnology, 9, 696040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto, A. (2016) To be, or not to be biodegradable that is the question for the bio‐based plastics. Microbial Biotechnology, 9, 652–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purohit, J. , Chattopadhyay, A. & Teli, B. (2020) Metagenomic exploration of plastic degrading microbes for biotechnological application. Current Genomics, 21, 253–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sales, J.C.S. , Santos, A.G. , de Castro, A.M. & Coelho, M.A.Z. (2021) A critical view on the technology readiness level (TRL) of microbial plastics biodegradation. World Journal of Microbiology and Biotechnology, 37, 116. [DOI] [PubMed] [Google Scholar]
- Salvador, M. , Abdulmutalib, U. , Gonzalez, J. , Kim, J. , Smith, A.A. , Faulon, J.L. et al. (2019) Microbial genes for a circular and sustainable bio‐PET economy. Genes (Basel), 10, 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samak, N.A. , Jia, Y. , Sharshar, M.M. , Mu, T. , Yang, M. , Peh, S. et al. (2020) Recent advances in biocatalysts engineering for polyethylene terephthalate plastic waste green recycling. Environment International, 145, 106144. [DOI] [PubMed] [Google Scholar]
- Satti, S.M. & Shah, A.A. (2020) Polyester‐based biodegradable plastics: an approach towards sustainable development. Letters in Applied Microbiology, 70, 413–430. [DOI] [PubMed] [Google Scholar]
- Shigenori, K. , Eiko, K. , Yuichi K., Kazufumi, T. & Satoshi, K. (2012) Novel esterase derived from foliage compost . Patent WO 2012/099018A1.
- Singh, A. , Rorrer, N.A. , Nicholson, S.R. , Erickson, E. , DesVeaux, J.S. , Avelino, A.F.T. et al. (2021) Techno‐economic, life‐cycle, and socioeconomic impact analysis of enzymatic recycling of poly(ethylene terephthalate). Joule, 5, 2479–2503. [Google Scholar]
- Sulaiman, S. , Yamato, S. , Kanaya, E. , Kim, J.J. , Koga, Y. , Takano, K. et al. (2012) Isolation of a novel cutinase homolog with polyethylene terephthalate degrading activity from leaf‐branch compost by using a metagenomic approach. Applied and Environmental Microbiology, 78, 1556–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiso, T. , Narancic, T. , Wei, R. , Pollet, E. , Beagan, N. , Schröder, K. et al. (2021) Towards bio‐upcycling of polyethylene terephthalate. Metabolic Engineering, 66, 167–178. [DOI] [PubMed] [Google Scholar]
- Topham, C. , Texier, H. , Tournier, V. , Desrousseaux, M.L. , Duquesne, S. , Andre, I. et al. (2019) Esterases and uses thereof . Patent US 2019/0233803 A1.
- Tournier, V. , Texier, H. , Desrousseaux, M.‐L. , Tophan, C. , Andre, I , Barbe, S. et al. (2018) Novel esterases and uses thereof . Patent WO 2018/011281 A1.
- Tournier, V. , Topham, C.M. , Gilles, A. , David, B. , Folgoas, C. , Moya‐Leclair, E. et al. (2020) An engineered PET depolymerase to break down and recycle plastic bottles. Nature, 580, 216–219. [DOI] [PubMed] [Google Scholar]
- Urbanek, A.K. , Kosiorowska, K.E. & Mirończuk, A.M. (2021) Current knowledge on polyethylene terephthalate degradation by genetically modified microorganisms. Frontiers in Bioengineering and Biotechnology, 9, 771133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertommen, M.A.M.E. , Nierstrasz, V.A. , van der Veer, M. & Warmoeskerken, M.M.C.G. (2005) Enzymatic surface modification of poly(ethylene terephthalate). Journal of Biotechnology, 120, 376–386. [DOI] [PubMed] [Google Scholar]
- Wei, R. & Zimmermann, W. (2017) Microbial enzymes for the recycling of recalcitrant petroleum‐based plastics: how far are we? Microbial Biotechnology, 10, 1308–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, A.Z. , Clare, R. , Mand, T.D. , Pardo, I. , Ramirez, K.J. , Haugen, S.J. et al. (2021) Tandem chemical deconstruction and biological upcycling of poly(ethylene terephthalate) to β‐ketoadipic acid by pseudomonas putida KT2440. Metabolic Engineering, 67, 250–261. [DOI] [PubMed] [Google Scholar]
- Yoshida, S. , Hiraga, K. , Takehana, T. , Taniguchi, I. , Yamaji, H. , Maeda, Y. et al. (2016) A bacterium that degrades and assimilates poly (ethylene terephthalate). Science, 351, 1196–1199. [DOI] [PubMed] [Google Scholar]
- Zimmermann, W. (2020) Biocatalytic recycling of polyethylene terephthalate plastic. Philosophical Transactions. Series A, Mathematical, Physical, and Engineering Sciences, 378, 20190273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, W. , Wey, R. , Hille, P. , Oeser, T. & Schmidt, J. (2019) New polypeptides having a polyester degrading activity and uses thereof . Patent EP 3517608 A1.


