Abstract
pioids are commonly used for treating chronic pain. However, with continued use, they may induce tolerance and/or hyperalgesia, which limits therapeutic efficacy. The human mechanisms of opioid-induced tolerance and hyperalgesia are significantly understudied, in part, because current models cannot fully recapitulate human pathology. Here, we engineered novel human spinal microphysiological systems (MPSs) integrated with plug-and-play neural activity sensing for modeling human nociception and opioid-induced tolerance. Each spinal MPS consists of a flattened human spinal cord organoid derived from human stem cells and a 3D printed organoid holder device for plug-and-play neural activity measurement. We found that the flattened organoid design of MPSs not only reduces hypoxia and necrosis in the organoids, but also promotes their neuron maturation, neural activity, and functional development. We further demonstrated that prolonged opioid exposure resulted in neurochemical correlates of opioid tolerance and hyperalgesia, as measured by altered neural activity, and downregulation of μ-opioid receptor expression of human spinal MPSs. The MPSs are scalable, cost-effective, easy-to-use, and compatible with commonly-used well-plates, thus allowing plug-and-play measurements of neural activity. We believe the MPSs hold a promising translational potential for studying human pain etiology, screening new treatments, and validating novel therapeutics for human pain medicine.
Keywords: Microphysiological systems, Organ-on-chip, In-situ electrical sensing, Spinal cord organoids, Opioid-induced tolerance and hyperalgesia, Neural activity
Graphical abstract
Highlights
-
•
Development of novel human spinal organoid-on-a chip-systems integrating with plug-and-play in-situ neural activity measurement and bio-scaffolds
-
•
Innovative flattened organoid design for (1) avoiding hypoxia/necrosis, (2) promoting neuron maturation, activity, and functional development of organoids, and (3) enabling stable and reproducible measurement of neural activity
-
•
Fully recapitulation of human pathology (e.g., downregulation of opioid receptor expression) and electrical activity of hyperalgesia and tolerance induced by opioids
-
•
Scalable, cost-effective, easy-to-use, and compatible with commonly-used well-plates
1. Introduction
More than 20% of the general population suffers from chronic pain [1,2]. Due to the increasing frequency of chronic pain and its heavy burden on patients and society, there is increasing interest in studying pain mechanisms, treatment, and management [3]. Opioids are one of the most commonly prescribed medications for pain [4,5]. However, the administration of opioids causes undesired side effects, including opioid-induced tolerance and hyperalgesia [6,7] which limits their efficacy to relieve pain over time. The first clinical report of opioid-induced tolerance can be traced back to the 19th century [5]. More recently, human subjects and animal models have been used to study the complicated pathology of opioid tolerance and hyperalgesia [6,7]. Several human trials have been conducted to examine the existence and potential clinical impacts of opioid-induced tolerance during chronic opioid therapy [[8], [9], [10], [11]]. This approach is limited by experimental accessibility as well as moral and health concerns. Animal models have been extensively used for studying the mechanism(s) and pathology underlying opioid-induced tolerance [12]. For example, animal spinal cord slices were used to discover neuronal activity and receptor changes related to opioid-induced tolerance [[13], [14], [15]]. However, a challenge remains in translating discoveries from animal models to human treatment due to the different biology between animal models and humans [16], and concerns about the translational relevance of preclinical pain models. Thus, there is an urgent and unmet need to develop models for understanding opioid-induced tolerance in humans.
Recent advances in stem cell and organoid technologies show promise for modeling human pain and opioid-induced tolerance [17]. Human organoids are organ-like 3D in vitro cultures derived from human stem cells, recapitulating key anatomical and physiological features observed in vivo. Several pioneering studies have been reported to develop human spinal cord organoids and sensory neurons. For example, human sensory neurons [18] and sensory ganglion organiods [19] have been differentiated from induced pluripotent stem cells (iPSCs) for pain research. Human ventral spinal cord organoids have been developed to study motor neuron development and activity, and cortex-spinal-muscle assemblies were generated to study motor neuron innervation of muscles in vitro [20]. Moreover, protocols to generate spinal cord organoids with dorsal interneurons and sensory neurons have been developed for modeling spinal cord disease and neural activity [21,22]. However, limitations of current human spinal cord organoids hinder their applications for studying human neural activity and pathology of opioid-induced tolerance including the fact that (1) current human spinal cord organoids are highly heterogeneous; (2) current human spinal cord organoids suffer from hypoxia and necrosis, significantly affecting neural activity; and (3) it is hard to obtain a stable and reproducible neural electrical measurement of human spinal cord organoids due to the organoid-electrode contact issue, and the hypoxia- and necrosis-induced neuron death during the measurement.
To address the above technical barriers and to grow better organoids, microphysiological systems and organoids-on-a-chip systems have been developed. This technology permits both the regulation and monitoring of organoid development through the integration of microfluidics and various bioengineering approaches [[23], [24], [25]]. For example, efforts have been made to improve the cultural conditions of developing organoids. Pillar-based perfusion microfluidic channels have been fabricated for the on-chip formation of brain and liver organoids derived from human-induced pluripotent stem cells (iPSCs) [[26], [27], [28]]. Recently, a very innovative lung-on-a-chip design has been reported for mimicking lung function by microfabricating two microfluidic channels separated by a stretchable porous PDMS membrane sandwiched in-between the channels [[29], [30], [31]]. The lung-on-a-chip devices and membrane-based microfabricated devices have been adopted to improve the perfusion of oxygen and nutrients and generate various types of organoids, including brain, pancreas, liver, and glomerulus organoids [[32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42]]. Similarly, microfabricated devices have been made to confine the geometry of organoids for regulating organoid growth and differentiation [43]. Recently, we also employed 3D printed scaffolds to generate tubular human brain organoids [44]. Compare to conventional organoids, these engineered organoids could have reduced hypoxia/necrosis and improved physical and functional phenotypes. However, changes still remain in generating standardized organoids/3D cultures as well as characterizing their physical and functional developments. Thus, several attempts have been made to enable monitoring of development as well as the response to drug treatment. For example, long-term four-dimensional light-sheet microscopy has been established for recording the spatial lineage of developing cerebral organoids [45]. Moreover, real-time live-cell imaging has been incorporated into an automated microfluidic platform for high throughput drug screening by testing the relative apoptosis and viability of tumor organoids under different drug treatments [46]. So far, the above-mentioned pioneering works have improved the development or/and characterization of human organoids. However, there is a tremendous need to develop better models that recapitulate both human neural activity and the pathology of opioid-induced tolerance, because of technical barriers to simultaneously maintaining high-quality human spinal cord organoids and stably measuring neural activity in response to opioids.
Herein, we report novel human spinal microphysiological systems (MPSs) integrated with plug-and-play neural activity sensing to model human biology underlying opioid-induced tolerance and hyperalgesia. To avoid the hypoxia/necrosis within conventional organoids, we innovated a flattened organoid design of MPSs to enhance nutrient and oxygen perfusion. We found that the uniquely flattened organoid design not only supports neuron maturation/activity but also provides better contact between organoids and multi-electrode array (MEA) electrodes, improving the stability and reproducibility of neural activity measurement. After the integration of 3D printed organoid holder devices, the MPSs could be used for plug-and-play neural activity measurement, and are compatible with commonly-used multiwell-plates and MEA systems. As a proof-of-concept application, the human spinal MPSs were used to model neural activity and the biology of nociception and hyperalgesia induced by opioids. We demonstrated that the nociceptive stimulus induced neural activity of human spinal MPSs was rapidly suppressed by acute opioid (DAMGO or [D-Ala2,N-MePhe4, Gly-ol]-enkephalin) administration while enhanced by chronic opioid exposure. Long-term DAMGO treatment of the human spinal MPSs also reduced μ-opioid receptor levels. Thus, we believe the human spinal MPSs may hold promising potential for broad applications in basic and translational pain research.
2. Results and discussion
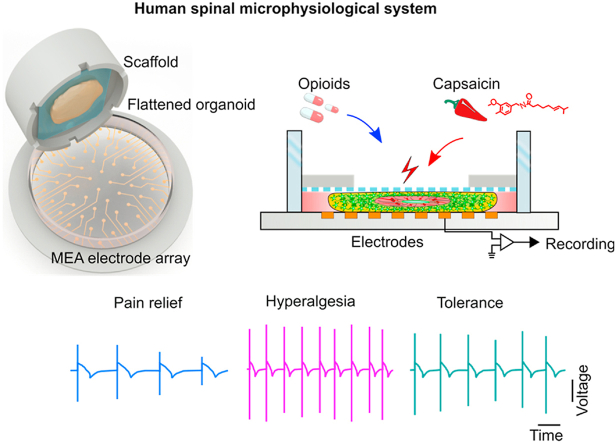
Engineering human spinal microphysiological system to model pain relief and opioid-induced hyperalgesia/tolerance. Opioids are effective analgesics, however, chronic opioid exposure is problematic for pain management for many reasons, including tolerance and hyperalgesia. To study the latter two processes, we developed novel human spinal microphysiological systems (MPSs) integrated with plug-and-play neural activity sensing. Each human spinal MPS consists of a flattened human spinal cord organoid derived from human stem cells and a 3D printed organoid holder device for plug-and-play neural activity measurement (Fig. 1a). The 3D printed organoid holder device (Fig. 1b) is made by integrating a polycarbonate membrane onto a 3D printed hollow ring (outer diameter of 15 mm, inner diameter of 11 mm, and height of 5 mm), and can be directly inserted into commonly-used cell culture well-plates or multi-electrode array (MEA) systems (Fig. S1). Since nutrients and oxygen can only penetrate about 400 μm from the surface of organoid tissue, conventional spherical organoids with a diameter up to several millimeters frequently develop central hypoxia and necrosis, detrimentally affecting neuronal function and activity. By using 3D-printed organoid holder devices, we confined the growth of human spinal cord organoids within a space (height = 500 μm) to generate flattened human spinal cord organoids. The 500 μm organoid thickness was smaller than the double side penetration depth (∼800 μm), allowing sufficient nutrients and oxygen supplement. And such thickness could also support organoid structure (VZ/SVZ or ventricular zone and subventricular zone) development [47]. We then used established human spinal MPSs to investigate their functions as well as cellular processes involved in hyperalgesia and tolerance induced by opioids (Fig. 1c).
Fig. 1.
Engineering a human spinal microphysiological system to model opioid-induced tolerance and hyperalgesia. (a) Schematics of the human spinal microphysiological system that consists of a flattened human spinal cord organoid derived from human stem cells and a 3D printed organoid holder device. (b) A representative image of the human spinal microphysiological systems compatible with a well plate-based MEA system for plug-and-play neural activity measurement. The bottom right shows a single insert. (c) Schematic for modeling anti-nociception, opioid-induced hyperalgesia, and opioid tolerance using the human spinal microphysiological systems.
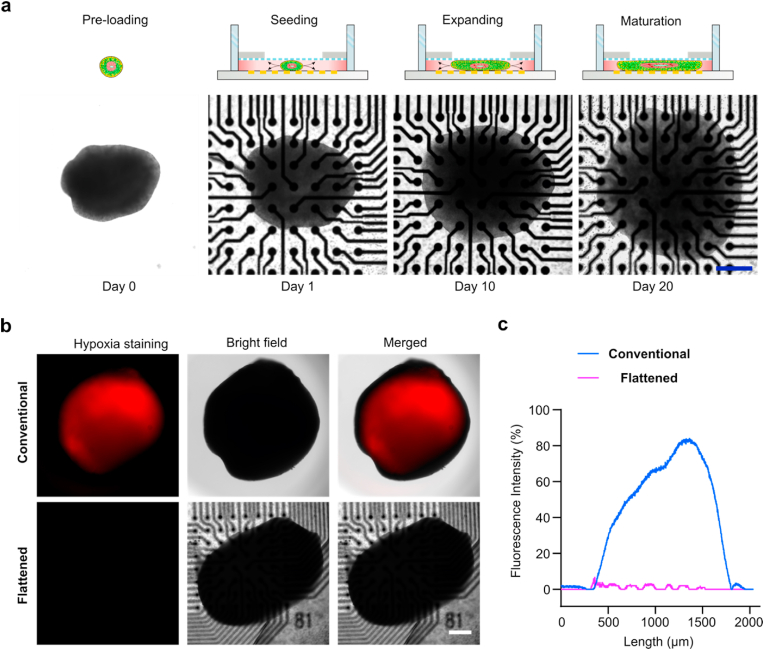
Fabrication and characterization of flattened human spinal cord organoids. To fabricate flattened human spinal cord organoids, we developed a protocol that adapted previous dorsal spinal cord organoid protocols (Fig. S2) and incorporated them into the organoid holder device. Briefly, we treated embryonic bodies (EBs) with retinoic acid and a GSK-3 inhibitor (CHIR-99021) for 4 days and then transferred them to medium containing bone morphogen protein 4 (BMP4) and retinoic acid for directing dorsal spinal cord region specification for the next 6 days. Then, EBs/organoids were transferred to the organoid holder device, and organoids were treated with N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenyl glycine t-butyl ester (DAPT) for 8 days to promote neural differentiation. Finally, the organoids were kept in a medium supplied with cyclic adenosine monophosphate (cAMP) and ascorbic acid for further neural differentiation and maturation. We tracked organoid growth into the flattened shape within the confined space (height = 500 μm) between the device membrane and the well-plate bottom (Fig. 2a). We further tested and compared hypoxia between conventional and flattened human spinal cord organoids (Fig. 2b), and quantified the hypoxia within the two types of organoids (Fig. 2c). We found a significantly reduced hypoxic core formation in the flattened organoids compared to the conventional organoids.
Fig. 2.
Fabrication and characterization of flattened human spinal cord organoids. (a) Schematics and images showing the fabrication process of the flattened human spinal cord organoids on-chip. (b) Hypoxia staining of the conventional and flattened human spinal cord organoids at 60 days. (c) Quantification of hypoxia within the conventional and flattened human spinal cord organoids at 60 days. Scale bar = 500 μm.
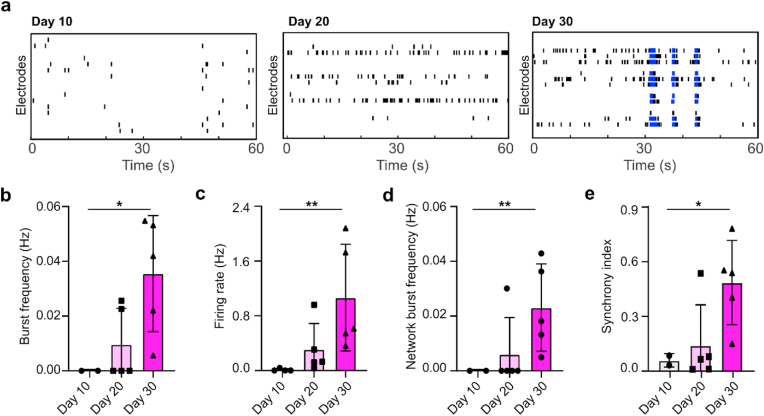
The flattened organoid design facilitates neural activity measurement and promotes neuron activity. The human spinal MPSs could be easily integrated into the MEA system for plug-and-play neural activity sensing. We showed that MPSs could be simply plugged into the wells of a 24 well MEA system (Fig. 3a), and then tested right after plugging in. The conventional organoids showed a hypoxic center with no electrical activities, while the flattened organoids showed good neuron activities across the whole organoid area. While testing the neural activity of the flattened and conventional organoids, we obtained their representative raster plots (Fig. 3b). In analyzing neural activity, we found the flattened organoids had significantly more active electrodes than the conventional organoids (Fig. 3c), indicating that the flattened organoid design provides a better organoid-electrode contact for neural activity measurement as well as reduces the necrosis and hypoxia. Moreover, we further quantified the mean firing rate of the organoids. We found that the mean firing rate of the flattened organoid (0.0658 ± 0.0234 Hz) was almost ten-fold higher than that of conventional organoids (0.0068 ± 0.0071 Hz, p = 0.00082) (Fig. 3d), implying that the human spinal MPSs have increased neural activity, perhaps due to the better perfusion afforded by the flattened organoid design. Furthermore, we characterized the neural activity of the human spinal MPSs during organoid development, with representative raster plots of human spinal MPSs obtained on days 10, 20, and 30 (Fig. 4a). We also quantified the neural activity of the human spinal MPSs including burst frequency (Fig. 4b), firing rate (Fig. 4c), synchrony index (Fig. 4d), and network burst frequency (Fig. 4e). We found all the above neural activity parameters increased during organoid development, indicating that human spinal MPSs facilitated the development and maturation of neurons.
Fig. 3.
Neural activity measurements of conventional and flattened human spinal cord organoids. (a) Schematics and representative spike activity heat map of conventional and flattened human spinal cord organoids on the MEA plates. The blue dashed line labels the brain organoids area. (b) Representative raster plots of the conventional and flattened human spinal cord organoids. (c) Active electrode number of the conventional and flattened human spinal cord organoids during the neural activity recording. (d) Quantification of the mean firing rate of the conventional and flattened human spinal cord organoids (n = 6). Scale bar = 500 μm.
Fig. 4.
Functional maturation of human spinal microphysiological systems. (a) Representative raster plots of the human spinal microphysiological system at different developmental stages. The burst frequency (b), firing rate (c), synchrony index (d), and network burst frequency (e) of human spinal microphysiological systems at different ages in culture. (n = 5) Scale bar = 500 μm.
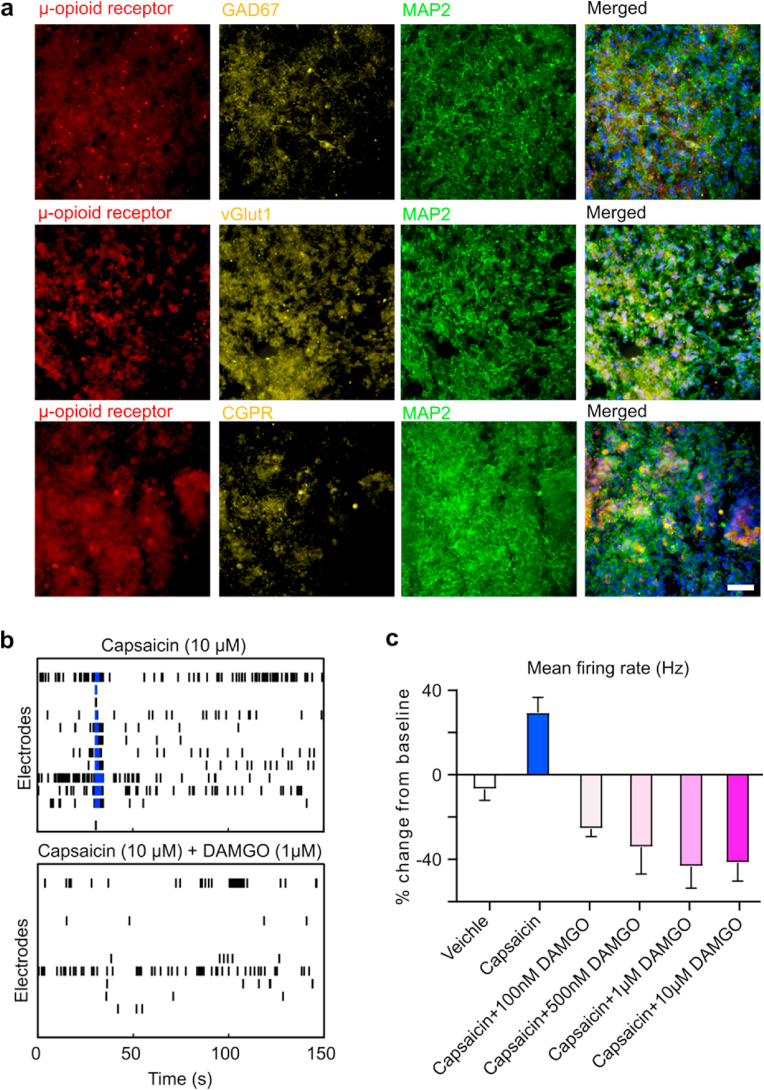
MPS response to short-term opioid administration. Opioids are widely used to relieve acute pain. To validate that the human spinal MPSs can be utilized to model the molecular mechanisms underlying pain relief during short-term opioid treatment, we first employed immunofluorescence staining to confirm the presence of neuronal populations that express μ-opioid receptors. The staining results showed that μ-opioid receptors were widely expressed in various types of neurons of human spinal MPSs, including excitatory, inhibitory, and sensory neurons (Fig. 5a), similar to the findings from in vivo animal studies [[48], [49], [50]]. To further validate the functional expression of μ-opioid receptors, we treated human spinal MPSs with increasing concentrations of the μ-opioid receptor agonist, DAMGO. DAMGO treatment inhibited organoid neural activity in a dose-dependent fashion (Fig. S3). As the presence of μ-opioid receptors in neurons of human spinal MPSs was confirmed, we then tested the response of human spinal MPSs to pain modulators and opioid agonist treatments (Fig. 5b). Capsaicin, a transient receptor potential cation channel subfamily V member 1 (TRPV1) agonist, was applied to MPSs to model nociception. Capsaicin stimulation increased the mean firing rate of organoids by 35.4%. Then we combined capsaicin and different concentrations of DAMGO to determine if opioids would decrease capsaicin-induced neuronal firing. The opioid treatment effectively reduced capsaicin responses and the mean firing rate of spinal cord organoids dropped below baseline even under a low dose of DAMGO (100 nM) (Fig. 5c, Video S1). As the opioid concentration increased, the decrease of MPS firing rate became more apparent, and the organoid firing rate was minimal when the opioid concentration was greater than 1 μM. Thus, our MPSs could model capsaicin elicited pain activity and its treatment by opioids, recapitulating the in vivo situation.
Fig. 5.
Model pain relief using human spinal microphysiological systems. (a) Immunofluorescence of human spinal microphysiological systems showing the expression of μ-opioid receptors on sensory neurons (CGRP), GABAergic (GAD67), and glutamate neurons (vGlut1). (b) Representative raster plots showing the neural activity signals of human spinal microphysiological systems treated with a noxious agent (capsaicin) only and capsaicin and opioid (DAMGO) together. (c) Percentage changes of mean firing rates after treatment with capsaicin only or by capsaicin with increasing concentrations of DAMGO (n = 6). Scale bar = 50 μm.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.bioactmat.2022.10.007.
The following is the supplementary data related to this article:
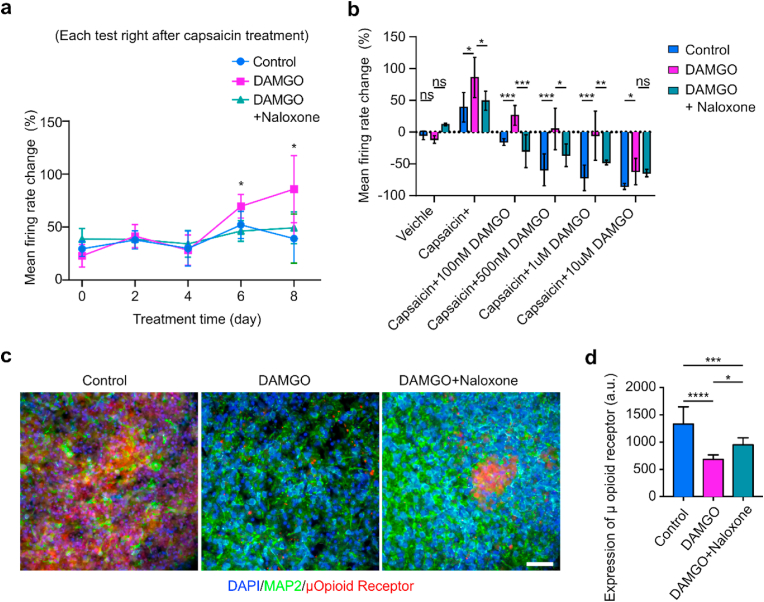
Chronic opioids induced hyperalgesia and tolerance. Extended opioid usage can induce a paradoxically increased sensitivity to pain (or opioid-induced hyperalgesia) as well as tolerance to opioid-mediated pain relief. Both are major limitations when using opioids to treat chronic pain. To validate MPSs as a system for recapitulating opioid-induced hyperalgesia and tolerance observed following prolonged opioid administration, we continuously treated the human spinal MPSs with DAMGO (500 nM) for 8 days and tested their responses to pain modulators every other day (Fig. 6a, Video S2). In the first 4 days, capsaicin increased the mean firing rate to an equivalent extent in the DAMGO-treated and the control groups. However, chronic DAMGO-treated organoids showed a significantly heightened firing rate increase on day 6 (70% ± 11% versus 51% ± 13%) and day 8 (87% ± 32% versus 39% ± 23%) of treatment. The results indicated a heightened sensitivity to capsaicin in human spinal MPSs during prolonged opioid administration. Thus, MSPs may be a good system for modeling opioid-induced tolerance. We also tested whether organoids treated together with DAMGO and naloxone, a μ-opioid receptor antagonist, could reduce neuronal activity in the model of opioid-induced hyperalgesia. The DAMGO + naloxone group showed no significant difference to control group in capsaicin-induced mean firing change over eight days, indicating a requirement of mu-opioid receptors in this model of hyperalgesia. We further measured the responses of human spinal MPS to capsaicin and DAMGO (with different doses) to test opioid tolerance after extended opioid administration (Fig. 6b). A low dose of DAMGO (100 nM) was able to reduce the firing rate below the baseline and effectively relieve the neuronal firing induced by capsaicin in the control and DAMGO + Naloxone group, while organoids chronically treated with DAMGO needed higher concentrations of DAMGO to return neuronal activity to baseline. The results from the human spinal MPSs are similar to those findings on spinal cord slices from animals with both opioid-induced hyperalgesia and tolerance. We further validated human spinal MPSs as able to recapitulate the key receptor and pathological changes with prolonged opioid administration via immunofluorescence staining of the three groups (Fig. 6c). We found that μ-opioid receptor expression was downregulated after prolonged DAMGO treatment.
Fig. 6.
Modeling opioid-induced hyperalgesia and μ-opioid receptor down-regulation using human spinal microphysiological systems. (a) Percentage change of mean firing rates for the three groups including vehicle treatment only (Control), opioid treatment only (500 nM DAMGO), and the combination of opioid and naloxone treatment (500 nM DAMGO + 1 μM naloxone) over 8 days. (b) After the 8-day treatment, the percentage changes of mean firing rates from baseline by treating with a noxious agent (capsaicin), and capsaicin together with different doses of DAMGO. (c) Immunofluorescence showing the expression of μ-opioid receptors within human spinal microphysiological systems for the three groups after the 8-day treatment. (d) Quantification of μ-opioid receptor expression within the three groups as measured by fluorescent intensity (n = 6). Scale bar = 50 μm.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.bioactmat.2022.10.007.
The following is the supplementary data related to this article:
3. Conclusion
In summary, we report novel human spinal microphysiological systems (MPSs) integrated with neural activity sensing for modeling human opioid-induced analgesia, tolerance and hyperalgesia. The MPSs have several unique advantages: (1) the innovative flattened organoid design of MPSs enhances nutrients and oxygen entry to avoid typical organoid hypoxia/necrosis and to support neuron maturation/activity; (2) the flattened organoid design of MPSs also provides better contact between the organoid and the electrodes of a multi-electrodes array (MEA), improving the stability and reproducibility of neural activity measurement; (3) the MPSs within 3D printed devices are compatible with commonly-used well-plates or MEA systems, highlighting their potential for broad applications in basic and translational pain research. Using the engineered system, we not only demonstrated that acute opioid exposure could rapidly reduce neural activity evoked by capsaicin, but also showed that prolonged opioid exposure downregulates μ-opioid-receptor levels and produces neurophysiological and neurochemical features associated with both tolerance and opioid-induced hyperalgesia. One limitation of our current study is that the neurons developed within our in vitro MPS system could differ from in vivo human spinal cord and sensory neurons. Future efforts are underway to compare the phenotypes of the neurons and glial cells within our MPS system and the counterparts in human to understand the gap and further fine tuning our differentiation protocol. We believe the human spinal MPSs could be broadly applied to studies of pain etiology and its treatment such as assessments of biased agonism, receptor internalization, etc. Successful implementation of MPS system will facilitate drug discovery efforts aimed at identifying and validating new pain therapeutics while enhancing prospects for successful clinical translation.
4. Experimental section
Fabrication of 3D printed organoid holder devices. The organoid holder devices were designed employing AutoCAD software and fabricated using our well-developed 3D printing protocol [51,52] through a stereolithography 3D printer (Form 3B, Formlabs) and FormLabs Clear Resin V4. The detailed device design is shown in Fig. S2a.
Human stem cell culture. We obtained human embryonic stem cells (WA09) from WiCell institute and followed the guidelines of both WiCell Institute and Indiana University when handling these cells. Matrigel (Corning) coated 6 well plates were used to culture WA09 cells with mTESR plus medium (Stemcell Technologies) in a humidified incubator at 37° Celsius and 5% CO2. The medium was changed every other day. ReLeSR (Stemcell Technologies) was used to passage WA09 cells weekly.
Fabrication of flattened human spinal cord organoids. Similarly, to our early protocols [53,54], embryonic bodies (EBs) were fabricated by aggregation of ∼9000 WA09 cells each well using a 96-well U bottom microplate (Corning). EBs were cultured in 100 μL EB formation medium (Stemcell Technologies) supplemented with 10 μM Y-27632 (SelleckChem). After one day of EB formation, the EBs were transferred to the spinal cord medium I (ScM I) supplied with 10 nM retinoic acid (Sigma Aldrich) and 3 μM CHIR-99021 (Stemcell Technologies). After 4-day culture in SCM I, the spheroids were then transferred to spinal cord medium II (ScM II) with 10 nM retinoic acid and 5 ng/mL recombinant human BMP4 (Peprotech) for the next 6 days. On day 10, we embedded the spinal cord organoids into Matrigel (corning). The organoids were switched to spinal cord medium III (ScM III), supplemented with 10 μM DAPT, and cultured for 8 more days. During this period, spinal cord organoids were transferred to our microfluidic devices integrated with 24 well ultra-low attachment plates (Corning) or 24 well MEA systems for the generation of flattened organoids. For traditional spherical organoids, the organoids were cultured in 6 well ultra-low attachment plates (Corning) shaking at 60RPM with an orbital shaker (OrbiShaker JR, Benchmark Scientific). On Day 18, the organoids were finally transferred to spinal cord medium IV (ScM IV), with 1 μM cAMP (Sigma Aldrich) and 20 μg/mL ascorbic acid (Sigma Aldrich) for subsequent continuous culture. Medium change was performed every other day during this process except when specified. Detailed medium composition is listed in Supplementary Table S1.
Neural activity measurement of human spinal cord organoids. On Day 10, the human spinal cord organoids were transferred into devices. After having settled for 10 days, human spinal cord organoids were attached to wells on the MEA plates and prepared for electrical measurements. Recording of human spinal cord organoids’ electrical activity was performed by using the Axion Maestro Edge (Axion inc). The spinal cord organoids were kept at 37 °C maintained by an internal heater and infused with 5% CO2. To determine the mean firing rate of a spinal cord organoid, we repeated 3-min MEA recordings 5 times after the treatment administration. The final mean firing rate of the organoid was calculated as the average mean firing rate of the 3 most consistent recordings among the 5 recordings. To reduce the variation brought by adding substance, the medium was also refreshed before measuring the mean firing rate baseline. The substance was first diluted into SCM IV at 2x of the desired concentration, then half of the medium was replaced with the 2x concentrated solution to reach the desired concentration.
Cryo-section of organoids. After treating with different drugs as mentioned, the culture samples were washed three times in 1x Phosphate-Buffered Saline (PBS, Gibco) and then fixed in 4% paraformaldehyde in 1x PBS (Thermo Scientific) overnight at 4 °C. The samples were rinsed three times with 1x PBS and then transferred into a 30% sucrose (w/v) solution overnight at 4 °C for cryoprotection. The organoids were then incubated with 7.5% gelatin (w/v) and 10% sucrose (w/v) in PBS solution at 37 °C for 1 h. Finally, the samples were snap-frozen onto a cryomold (Sakura Finetek) in a dry ice/ethanol slurry. The frozen samples were then sectioned at 30 μm thickness on a cryostat (Leica).
Immunofluorescence staining. We characterized the human spinal cord organoids using protocols previously developed in the lab [[55], [56], [57], [58]]. The cryosectioned samples plated onto glass slides were washed three times with 1x PBS. Then, for antigen retrieval, the slides were treated with 2 N hydrochloric acid (HCl) for 15 min. After HCl treatment, the samples were further washed twice using 1XPBS and treated with a blocking solution (0.3% Triton-X100, 5% normal goat serum in 1XPBS) for 1 h. Following blocking, the samples were immersed in a blocking solution containing primary antibodies at 4 °C overnight. We then washed sections 3 times with 1x PBS, and incubated the samples with a blocking solution containing secondary antibodies for 1 h at room temperature. Finally, the samples were washed with 1x PBS another 3 times and coverslipped with gold anti-fade mounting medium (Invitrogen). Detailed antibody information and dilution factors can be found in Supplementary Table S2.
qPCR analysis. Mature organoids were lysed for RNA extraction using RNeasy Mini Kit (Qiagen). RNA was first reversed and transcribed into complementary DNA (cDNA) using a cDNA synthesis kit (Quantabio). The cDNA was then analyzed by quantitative PCR using the SYBR green kit (Applied Biosystems). Gene expression fold change was analyzed by normalizing against the housekeeping gene GAPDH.
Statistical analysis. Statistical analysis was carried out using GraphPad Prism 8. Two sample groups were compared using the paired Students' t-test. Statistical significance was denoted as: *p < 0.05, **p < 0.01, ***p < 0.005. ****p < 0.001.
Ethics approval and consent to participate
Manuscript reporting studies don't involve any human participants, human data, or human tissue.
CRediT authorship contribution statement
Hongwei Cai: Methodology, Formal analysis, Conceptualization, Data curation, Investigation, Writing - original draft. Zheng Ao: Methodology, Conceptualization, Investigation, Validation, Writing - review & editing. Chunhui Tian: Methodology, Visualisation. Zhuhao Wu: Methodology, Visualisation. Connor Kaurich: Methodology, Writing - review & editing. Zi Chen: Funding acquisition, Data curation, Writing - review & editing. Mingxia Gu: Formal analysis, Investigation, Validation, Writing - review & editing. Andrea G. Hohmann: Formal analysis, Funding acquisition, Writing - review & editing. Ken Mackie: Formal analysis, Funding acquisition, Writing - review & editing. Feng Guo: Formal analysis, Conceptualization, Funding acquisition, Project administration, Supervision, Writing - review & editing.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgment
The project was supported by the departmental start-up funds of Indiana University Bloomington, and in part by NSF grants (CCF-1909509, and CMMI-2025434) and NIH awards (DP2AI160242, DA056242, and DA047858).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2022.10.007.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Von Korff M., et al. United States national pain strategy for population research: concepts, definitions, and pilot data. J. Pain. 2016;17:1068–1080. doi: 10.1016/j.jpain.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 2.Mills S.E.E., Nicolson K.P., Smith B.H. Chronic pain: a review of its epidemiology and associated factors in population-based studies. Br. J. Anaesth. 2019;123:e273–e283. doi: 10.1016/j.bja.2019.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dahlhamer J., et al. Prevalence of chronic pain and high-impact chronic pain among adults—United States, 2016. MMWR (Morb. Mortal. Wkly. Rep.) 2018;67:1001. doi: 10.15585/mmwr.mm6736a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trescot A.M., et al. Opioids in the management of chronic non-cancer pain: an update of American society of the interventional pain physicians'(ASIPP) guidelines. Pain Physician. 2008;11:S5–S62. [PubMed] [Google Scholar]
- 5.Marion Lee M., Sanford Silverman M., Hans Hansen M., Vikram Patel M., Laxmaiah Manchikanti M. A comprehensive review of opioid-induced hyperalgesia. Pain Physician. 2011;14:145–161. [PubMed] [Google Scholar]
- 6.Chu L.F., Angst M.S., Clark D. Opioid-induced hyperalgesia in humans: molecular mechanisms and clinical considerations. Clin. J. Pain. 2008;24:479–496. doi: 10.1097/AJP.0b013e31816b2f43. [DOI] [PubMed] [Google Scholar]
- 7.Silverman S.M. Opioid induced hyperalgesia: clinical implications for the pain practitioner. Pain Physician. 2009;12:679–684. [PubMed] [Google Scholar]
- 8.Li X., Angst M.S., Clark J.D. Opioid-induced hyperalgesia and incisional pain. Anesth. Analg. 2001;93:204–209. doi: 10.1097/00000539-200107000-00040. [DOI] [PubMed] [Google Scholar]
- 9.Doverty M., et al. Hyperalgesic responses in methadone maintenance patients. Pain. 2001;90:91–96. doi: 10.1016/s0304-3959(00)00391-2. [DOI] [PubMed] [Google Scholar]
- 10.Guignard B., et al. Acute opioid tolerance: intraoperative remifentanil increases postoperative pain and morphine requirement. The Journal of the American Society of Anesthesiologists. 2000;93:409–417. doi: 10.1097/00000542-200008000-00019. [DOI] [PubMed] [Google Scholar]
- 11.Koppert W., et al. Differential modulation of remifentanil-induced analgesia and postinfusion hyperalgesia by S-ketamine and clonidine in humans. The Journal of the American Society of Anesthesiologists. 2003;99:152–159. doi: 10.1097/00000542-200307000-00025. [DOI] [PubMed] [Google Scholar]
- 12.Angst M.S., Clark J.D. Opioid-induced hyperalgesia: a qualitative systematic review. The Journal of the American Society of Anesthesiologists. 2006;104:570–587. doi: 10.1097/00000542-200603000-00025. [DOI] [PubMed] [Google Scholar]
- 13.Ossipov M.H., Lai J., King T., Vanderah T.W., Porreca F. Underlying mechanisms of pronociceptive consequences of prolonged morphine exposure. Peptide Science: Original Research on Biomolecules. 2005;80:319–324. doi: 10.1002/bip.20254. [DOI] [PubMed] [Google Scholar]
- 14.Vanderah T.W., et al. Dynorphin promotes abnormal pain and spinal opioid antinociceptive tolerance. J. Neurosci. 2000;20:7074–7079. doi: 10.1523/JNEUROSCI.20-18-07074.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X., Clark J.D. Hyperalgesia during opioid abstinence: mediation by glutamate and substance p. Anesth. Analg. 2002;95:979–984. doi: 10.1097/00000539-200210000-00035. [DOI] [PubMed] [Google Scholar]
- 16.Vuong C., Van Uum S.H.M., O'Dell L.E., Lutfy K., Friedman T.C. The effects of opioids and opioid analogs on animal and human endocrine systems. Endocr. Rev. 2010;31:98–132. doi: 10.1210/er.2009-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ao Z., et al. Human spinal organoid-on-a-chip to model nociceptive circuitry for pain therapeutics discovery. Anal. Chem. 2022;94:1365–1372. doi: 10.1021/acs.analchem.1c04641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mis M.A., et al. Resilience to pain: a peripheral component identified using induced pluripotent stem cells and dynamic clamp. J. Neurosci. 2019;39:382–392. doi: 10.1523/JNEUROSCI.2433-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao D., et al. Generation of self-organized sensory ganglion organoids and retinal ganglion cells from fibroblasts. Sci. Adv. 2020;6:eaaz5858. doi: 10.1126/sciadv.aaz5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andersen J., et al. Generation of functional human 3D cortico-motor assembloids. Cell. 2020;183:1913–1929. doi: 10.1016/j.cell.2020.11.017. e1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogura T., Sakaguchi H., Miyamoto S., Takahashi J. Three-dimensional induction of dorsal, intermediate and ventral spinal cord tissues from human pluripotent stem cells. Development. 2018;145:dev162214. doi: 10.1242/dev.162214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duval N., et al. in Development (Cambridge, England) 2019;146 doi: 10.1242/dev.175430. [DOI] [PubMed] [Google Scholar]
- 23.Huh D., Torisawa Y.-s., Hamilton G.A., Kim H.J., Ingber D.E. Microengineered physiological biomimicry: organs-on-chips. Lab Chip. 2012;12:2156–2164. doi: 10.1039/c2lc40089h. [DOI] [PubMed] [Google Scholar]
- 24.Ma C., Kuzma M.L., Bai X., Yang J. Biomaterial‐based metabolic regulation in regenerative engineering. Adv. Sci. 2019;6 doi: 10.1002/advs.201900819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim G.B., et al. The critical chemical and mechanical regulation of folic acid on neural engineering. Biomaterials. 2018;178:504–516. doi: 10.1016/j.biomaterials.2018.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu Y., et al. A hollow fiber system for simple generation of human brain organoids. Integrative Biology. 2017;9:774–781. doi: 10.1039/c7ib00080d. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y., Wang L., Zhu Y., Qin J. Human brain organoid-on-a-chip to model prenatal nicotine exposure. Lab Chip. 2018;18:851–860. doi: 10.1039/c7lc01084b. [DOI] [PubMed] [Google Scholar]
- 28.Zhu Y., et al. In situ generation of human brain organoids on a micropillar array. Lab Chip. 2017;17:2941–2950. doi: 10.1039/c7lc00682a. [DOI] [PubMed] [Google Scholar]
- 29.Jalili-Firoozinezhad S., et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nature biomedical engineering. 2019;3:520–531. doi: 10.1038/s41551-019-0397-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mammoto A., et al. A mechanosensitive transcriptional mechanism that controls angiogenesis. Nature. 2009;457:1103–1108. doi: 10.1038/nature07765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benam K.H., et al. Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat. Methods. 2016;13:151–157. doi: 10.1038/nmeth.3697. [DOI] [PubMed] [Google Scholar]
- 32.Esfahani S.N., et al. Microengineered human amniotic ectoderm tissue array for high-content developmental phenotyping. Biomaterials. 2019;216 doi: 10.1016/j.biomaterials.2019.119244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamei K.-i., et al. Integrated heart/cancer on a chip to reproduce the side effects of anti-cancer drugs in vitro. RSC Adv. 2017;7:36777–36786. [Google Scholar]
- 34.Kitsara M., Kontziampasis D., Agbulut O., Chen Y. Heart on a chip: micro-nanofabrication and microfluidics steering the future of cardiac tissue engineering. Microelectron. Eng. 2019;203:44–62. [Google Scholar]
- 35.Lee K.-H., Lee J., Lee S.-H. 3D liver models on a microplatform: well-defined culture, engineering of liver tissue and liver-on-a-chip. Lab Chip. 2015;15:3822–3837. doi: 10.1039/c5lc00611b. [DOI] [PubMed] [Google Scholar]
- 36.Neal J.T., et al. Organoid modeling of the tumor immune microenvironment. Cell. 2018;175:1972–1988. doi: 10.1016/j.cell.2018.11.021. e1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shin T.H., Kim M., Sung C.O., Jang S.J., Jeong G.S. A one-stop microfluidic-based lung cancer organoid culture platform for testing drug sensitivity. Lab Chip. 2019;19:2854–2865. doi: 10.1039/c9lc00496c. [DOI] [PubMed] [Google Scholar]
- 38.Taghibakhshi A., Barisam M., Saidi M.S., Kashaninejad N., Nguyen N.-T. Three-dimensional modeling of avascular tumor growth in both static and dynamic culture platforms. Micromachines. 2019;10:580. doi: 10.3390/mi10090580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tao T., et al. Engineering human islet organoids from iPSCs using an organ-on-chip platform. Lab Chip. 2019;19:948–958. doi: 10.1039/c8lc01298a. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y., et al. In situ differentiation and generation of functional liver organoids from human iPSCs in a 3D perfusable chip system. Lab Chip. 2018;18:3606–3616. doi: 10.1039/c8lc00869h. [DOI] [PubMed] [Google Scholar]
- 41.Zheng Y., et al. Controlled modelling of human epiblast and amnion development using stem cells. Nature. 2019;573:421–425. doi: 10.1038/s41586-019-1535-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karzbrun E., Kshirsagar A., Cohen S.R., Hanna J.H., Reiner O. Human brain organoids on a chip reveal the physics of folding. Nat. Phys. 2018;14:515–522. doi: 10.1038/s41567-018-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rothenbucher T.S.P., et al. Next generation human brain models: engineered flat brain organoids featuring gyrification. Biofabrication. 2021;13 doi: 10.1088/1758-5090/abc95e. [DOI] [PubMed] [Google Scholar]
- 44.Ao Z., Cai H., Wu Z., Song S., Karahan H., Kim B., Lu H., Kim J., Mackie K., Guo F., et al. Tubular human brain organoids to model microglia-mediated neuroinflammation. Lab Chip. 2021;21:2751–2762. doi: 10.1039/d1lc00030f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He Z., et al. Lineage recording in human cerebral organoids. Nat. Methods. 2022;19:90–99. doi: 10.1038/s41592-021-01344-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuster B., et al. Automated microfluidic platform for dynamic and combinatorial drug screening of tumor organoids. Nat. Commun. 2020;11:5271. doi: 10.1038/s41467-020-19058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qian X., et al. Sliced human cortical organoids for modeling distinct cortical layer formation. Cell Stem Cell. 2020;26:766–781. doi: 10.1016/j.stem.2020.02.002. e769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun J., Chen S.-R., Chen H., Pan H.-L. μ-Opioid receptors in primary sensory neurons are essential for opioid analgesic effect on acute and inflammatory pain and opioid-induced hyperalgesia. J. Physiol. 2019;597:1661–1675. doi: 10.1113/JP277428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hohmann A.G., Herkenham M. Regulation of cannabinoid and mu opioid receptors in rat lumbar spinal cord following neonatal capsaicin treatment. Neurosci. Lett. 1998;252:13–16. doi: 10.1016/s0304-3940(98)00534-5. [DOI] [PubMed] [Google Scholar]
- 50.Hohmann A.G., Briley E.M., Herkenham M. Pre- and postsynaptic distribution of cannabinoid and mu opioid receptors in rat spinal cord. Brain Res. 1999;822:17–25. doi: 10.1016/s0006-8993(98)01321-3. [DOI] [PubMed] [Google Scholar]
- 51.Ao Z., et al. Rapid profiling of tumor-immune interaction using acoustically assembled patient-derived cell clusters. Adv. Sci. 2022;9 doi: 10.1002/advs.202201478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cai H., et al. Acoustofluidic assembly of 3D neurospheroids to model alzheimer's disease. Analyst. 2020;145:6243–6253. doi: 10.1039/d0an01373k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ao Z., et al. Understanding immune-driven brain aging by human brain organoid microphysiological analysis platform. Adv. Sci. 2022 doi: 10.1002/advs.202200475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ao Z., et al. One-Stop microfluidic assembly of human brain organoids to model prenatal cannabis exposure. Anal. Chem. 2020;92:4630–4638. doi: 10.1021/acs.analchem.0c00205. [DOI] [PubMed] [Google Scholar]
- 55.Cai H., et al. Intelligent acoustofluidics enabled mini-bioreactors for human brain organoids. Lab Chip. 2021;21:2194–2205. doi: 10.1039/d1lc00145k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ao Z., et al. Controllable fusion of human brain organoids using acoustofluidics. Lab Chip. 2021;21:688–699. doi: 10.1039/d0lc01141j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ao Z., et al. Human spinal organoid-on-a-chip to model nociceptive circuitry for pain therapeutics discovery. Anal. Chem. 2022;94:1365–1372. doi: 10.1021/acs.analchem.1c04641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cai H., et al. Trapping cell spheroids and organoids using digital acoustofluidics. Biofabrication. 2020;12 doi: 10.1088/1758-5090/ab9582. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.