Dear Editor,
The Omicron variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was designated as a variant of concern about two weeks after the first infection cases were reported in Botswana and South Africa in November 2021 [1]. More than 30 of the mutations identified in Omicron are in the spike protein, including 15 in the receptor-binding domain, which affects virus transmissibility and immune escape [2, 3]. The Omicron variant has displaced the Delta variant, which previously was the most widespread variant [4]. Currently, Omicron appears to be the most infectious variant worldwide because of its high transmission rate. In Korea, genomic surveillance of SARS-CoV-2 has been conducted since the first infection was reported in January 2020 [5, 6]. On December 1, 2021, the first case of the Omicron variant was identified in a traveler returning from Nigeria. The Omicron variant rapidly outpaced the Delta variant, accounting for > 90% of cases by early February 2022. We describe how the Omicron variant became dominant in Korea and how it was classified into genetically different groups. The Institutional Review Board of the Korea Disease Control and Prevention Agency (2020-03-01-P-A) approved the study and waived the requirement for written consent.
Nasopharyngeal and oropharyngeal swabs were randomly collected from patients with SARS-CoV-2 infection. During the study period (December 1, 2021–February 3, 2022), 1.9% (8,359 cases) of the swabs collected from confirmed cases were whole-genome sequenced across the country. Sequencing libraries were prepared using the QIAseq SARS-CoV-2 Primer Panel and QIAseq FX DNA Library Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions and were sequenced on the NextSeq 2000 platform (Illumina, San Diego, CA, USA) with an average genome coverage of >1,000× for all isolates. The sequences were submitted to GISAID EpiCoV (https://www.gisaid.org/).
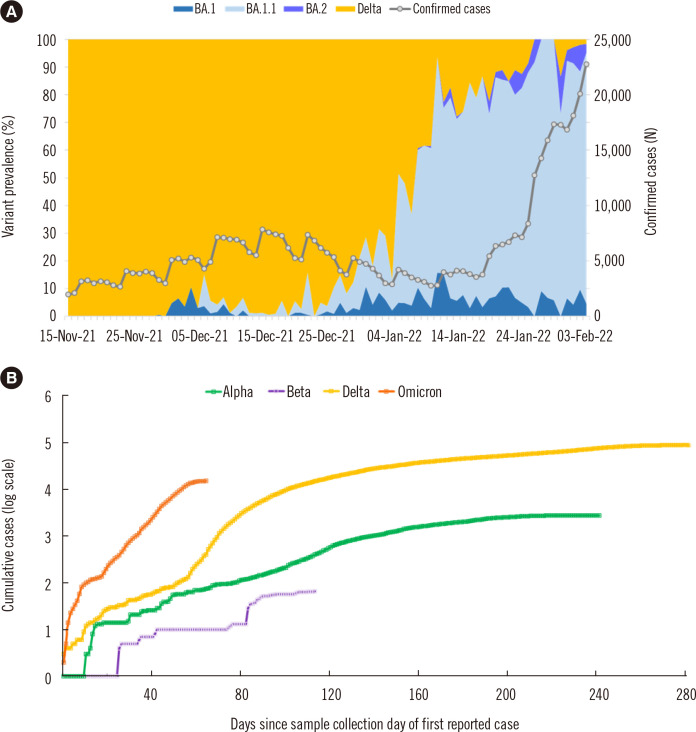
In Korea, the Delta variant increased rapidly and was identified in >90% of the samples collected in August 2021; however, in January 2022, its prevalence decreased to 36.1% because of the emergence of the Omicron variant (Fig. 1A). In February 2022, the Omicron variant accounted for 99.1% of all coronavirus disease 2019 (COVID-19) cases in Korea. With the emergence of the Omicron variant, the number of confirmed cases has increased sharply, resulting in a fifth wave of the pandemic (Fig. 1B).
Fig. 1.
Prevalence of SARS-CoV-2 variants in Korea. (A) Distribution of the Delta and Omicron variants of SARS-CoV-2 in Korea from November 15, 2021 to February 3, 2022. The number of daily reported cases is presented as a grey line. (B) Cumulative cases of four variants of concerns in Korea indexed by days. The Gamma variant has not been isolated from domestic cases in Korea.
Abbreviation: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
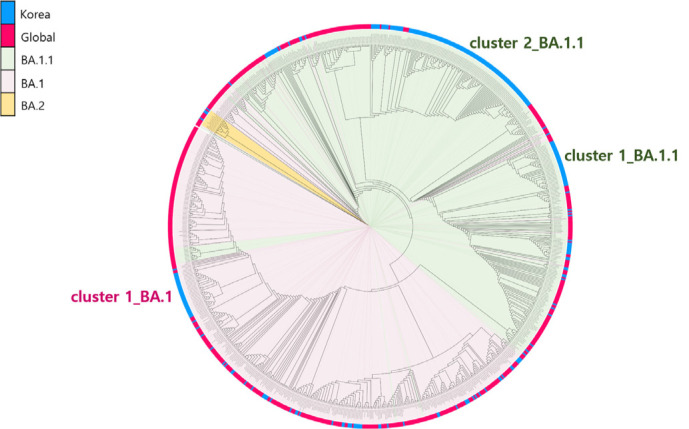
Analysis of the pangolin lineage [7] of the Omicron variant in samples from Korea identified three lineages: BA.1, BA.1.1, and BA.2. Among these, the BA.1.1 lineage was the most prevalent, accounting for 78.6% of all cases, followed by the BA.1 lineage (21.1%). A phylogenetic tree of the global and Korean Omicron variants indicated that the Omicron variant isolated in Korea grouped into three distinct clusters (Fig. 2). One of the three clusters belonged to the BA.1 lineage; the other two to the BA.1.1 lineage. The lineage of the virus isolated from the first confirmed Omicron case, a traveler returning from Nigeria, was BA.1, and it was associated with cluster 1_BA.1. Initially, the Omicron variant caused outbreaks in Incheon city. The variant then spread to other regions of Korea; however, it did not cause large-scale outbreaks (Fig. 2).
Fig. 2.
Phylogenetic analysis of SARS-CoV-2 sequences. In total, 858 sequences, including 286 sequences from isolates from Korea, were used to construct the tree. All sequences were aligned to the Wuhan-Hu-1 reference sequence using Geneious Prime software (https://www.geneious.com/) and then manually trimmed to equal lengths. A maximum-likelihood phylogenetic tree was reconstructed using FastTree under the GRT+γ nucleotide substitution model, and the phylogenetic tree was visualized using iTOL (https://itol.embl.de/).
Abbreviation: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
The BA.1.1 lineage isolates from Korean samples were grouped into two genetic clusters based on whether or not they harbored the open reading frame (ORF)1a E775G mutation (Fig. 2). Cluster 1_BA.1.1, which does not have this mutation, is related to a previously reported outbreak [8]. This outbreak resulted from a single case from Iran and started with household transmission to a kindergarten in the local community. Cluster 1_BA.1.1 subgroup viruses were concentrated in the Honam region and accounted for 15.2% of all sequences. Cluster 2_BA.1.1 was the largest of the three clusters (56.4% of all sequences). The metropolitan area was a major source of viruses in this cluster, which then spread throughout the country. Mutational analysis indicated that the most common non-synonymous mutations were found in the ORF9b protein (P10S), regardless of the lineage.
We described the displacement of the Delta variant by the Omicron variant in Korea. The rate of transmission of the Omicron variant is higher than those of previous variants in Korea, driving the fifth wave of the COVID-19 pandemic in the country. It has been suggested that immune evasion contributes to the rapid spread of the Omicron variant [9, 10]. However, more research is needed to better understand the link between immune system evasion and the transmissibility of SARS-CoV-2 variants. Considering the high transmission rates of emerging variants worldwide, monitoring of new SARS-CoV-2 variants must be conducted rapidly and regularly.
ACKNOWLEDGEMENTS
We acknowledge both the originating and submitting laboratories who have deposited and shared genome data from the GISAID EpiCoV database (https://www.gisaid.org/).
Footnotes
AUTHOR CONTRIBUTIONS
Park AK, Kim I-H, Lee CY, Kim J-A, Lee H, Kim HM, Lee N-J, Woo S, and Lee J contributed to sample preparation and carried out the experiments. Kim I-H, Yoo C-K, and Kim E-J conceived and planned the experiments. Park AK, Rhee JE, Kim I-H, and Kim E-J interpreted the results. Park AK took the lead in writing the manuscript. All authors provided critical feedback that helped shape the research, analyses, and final manuscript.
CONFLICTS OF INTEREST
None declared.
RESEARCH FUNDING
This study was supported by a grant from the Korea Disease Control and Prevention Agency (grant number 4800-4837-301).
REFERENCES
- 1.WHO, author. Classification of Omicron (B.1.1.529): SARS-CoV-2 variant of concern. [Updated on Nov 2021]. https://www.who.int/news/item/26-11-2021-classification-of-omicron-(B.1.1.529)-sars-cov-2-variant-of-concern.
- 2.Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19:409–24. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alenquer M, Ferreira F, Lousa D, Valério M, Medina-Lopes M, Bergman ML, et al. Signatures in SARS-CoV-2 spike protein conferring escape to neutralizing antibodies. PLoS Pathog. 2021;17:e1009772. doi: 10.1371/journal.ppat.1009772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shu Y, McCauley J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park AK, Kim IH, Kim J, Kim JM, Kim HM, Lee CY, et al. Genomic surveillance of SARS-CoV-2: distribution of clades in the Republic of Korea in 2020. Osong Public Health Res Perspect. 2021;12:37–43. doi: 10.24171/j.phrp.2021.12.1.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park AK, Kim IH, Man Kim H, Lee H, Lee NJ, Kim JA, et al. SARS-CoV-2 B.1.619 and B.1.620 lineages, South Korea, 2021. Emerg Infect Dis. 2022;28:415–9. doi: 10.3201/eid2802.211653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rambaut A, Holmes EC, Hill V, O'Toole Á, McCrone JT, Ruis C, et al. A dynamic nomenclature proposal for SARS-CoV-2 to assist genomic epidemiology. Nat Microbiol. 2020;5:1403–7. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim EY, Choe YJ, Park H, Jeong H, Chung JH, Yu J, et al. Community transmission of SARS-CoV-2 Omicron variant, South Korea, 2021. Emerg Infect Dis. 2022;28:898–900. doi: 10.3201/eid2804.220006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu L, Iketani S, Guo Y, Chan JF, Wang M, Liu L, et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022;602:676–81. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- 10.Cao Y, Wang J, Jian F, Xiao T, Song W, Yisimayi A, et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602:657–63. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]