Abstract
COVID-19 is an infectious respiratory disease caused by SARS-CoV-2. Pentraxin 3 (PTX3) is involved in the activation and regulation of the complement system, demonstrating an important role in the pathogenesis of COVID-19. The aim was to evaluate the association of single nucleotide polymorphisms in PTX3 and its plasma levels with the severity of COVID-19. This is a retrospective cohort study, carried out between August 2020 and July 2021, including patients with confirmed COVID-19 hospitalized in 2 hospitals in the Northeast Region of Brazil. Polymorphisms in PTX3 (rs1840680 and rs2305619) were determined by real-time PCR. PTX3 plasma levels were measured by ELISA. Serum levels of interleukin (IL)-6, IL-8, and IL-10 were determined by flow cytometry. A multivariate logistic regression model was used to identify parameters independently associated with COVID-19 severity. P values < 0.05 were considered significant. The study included 496 patients, classified as moderate (n = 267) and severe (n = 229) cases. The PTX3 AA genotype (rs1840680) was independently associated with protection against severe COVID-19 (P = 0.037; odds ratio = 0.555). PTX3 plasma levels were significantly associated with COVID-19 severity and mortality (P < 0.05). PTX3 levels were significantly correlated with IL-6, IL-8, IL-10, C-reactive protein, total leukocytes, neutrophil-to-lymphocyte ratio, urea, creatinine, ferritin, length of hospital stay, and higher respiratory rate (P < 0.05). Our results revealed a protective effect of the PTX3 AA genotype (rs1840680) on the development of severe forms of COVID-19. Additionally, PTX3 plasma levels were associated with the severity of COVID-19. The results of this study provide evidence of an important role of PTX3 in the immunopathology of COVID-19.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10238-022-00926-w.
Keywords: Coronavirus, COVID-19, Immunity, SARS-CoV-2, PTX3, SNP
Introduction
COVID-19 is an infectious respiratory disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the first case of which was reported in December 2019, in Wuhan, China [1]. More than 470 million cases have been reported worldwide [2] as of March 2022, and, in spite of vaccination, records are still increasing daily. Although the majority of cases present mild symptoms, a smaller part of the population progresses to acute respiratory disease and shows hypoxia, resulting in the need for hospitalization and possibly developing complications such as multiple organ failure or death [3].
In the severe form of COVID-19, after SARS-CoV-2 infection, dysregulation of the immune response and exacerbated release of cytokines occur, due to increased activation of innate immunity, decreased production, and high cytotoxicity of T cells [4, 5]. These factors result in hyperinflammatory responses that directly affect lung tissue and blood vessels [6, 7]. There may also be a genetic predisposition associated with dysregulation of the complement system, since coronaviruses can activate several complement pathways and consequently result in maladjusted neutrophilia, endothelial damage, and hypercoagulability [8]. These factors may contribute to the pathogenesis of the severe form of infection.
Pentraxins are proteins involved in complement activation and amplification via communication with complement initiation pattern recognition molecules [9]. They include pentraxin 3 (PTX3), which is a member of the pentraxin family of proteins that can be synthesized by macrophages, monocytes, leukocytes, dendritic cells, adipocytes, endothelial cells, and smooth muscle cells [10].
PTX3 is capable of recognizing some types of viruses, such as cytomegalovirus [11], influenza [12], and members of the coronavirus family, including SARS-CoV-2 [13, 14]. PTX3 plasma concentrations have been shown to be increased in patients with COVID-19. They have emerged as a strong independent predictor of mortality, and the marker stood out when compared to conventional inflammation markers in hospitalized patients with COVID-19 [15]. Polymorphisms in the PTX3 gene, especially the single nucleotide polymorphisms (SNPs) rs2305619 and rs1840680 [16], have been described in the literature regarding susceptibility to pulmonary tuberculosis [17], pulmonary aspergillosis [18], hepatocellular carcinoma [19], macrophage activation syndrome in COVID-19 [20], and other clinical conditions. However, to date, no studies have been published correlating the polymorphisms of this gene with the severity of COVID-19.
Therefore, the identification of genetic factors associated with COVID-19 severity is of fundamental importance to clarifying the pathogenicity mechanisms of the disease. In light of this and given the scarcity of studies evaluating the role of PTX3 in COVID-19, studies that investigate polymorphisms in PTX3, in conjunction with PTX3 plasma levels, are noteworthy targets for determining their relationship with the pathogenesis of COVID-19.
Materials and methods
Ethical considerations
This study received approval from the Ethics Committee of the Hospital das Clínicas of the Federal University of Pernambuco (HC/UFPE, acronym in Portuguese) under number CAAE: 38,196,620.0.0000.8807, and it was conducted in accordance with the provisions of the Declaration of Helsinki and the Good Clinical Practice guidelines.
Study design, location, and population
This retrospective cohort study was carried out at the Multi-User Research Laboratory (LAMUPE, acronym in Portuguese) of the University Hospital of the Universidade Federal do Vale do São Francisco (HU-UNIVASF/EBSERH, acronym in Portuguese). The study included patients over 18 years of age, with COVID-19 confirmed by molecular (RT-qPCR) or serological test, who were hospitalized between August 2020 and July 2021, at the HU-UNIVASF and the Field Hospital of the Municipality of Petrolina, which are both references for treatment of COVID-19 in the Vale do São Francisco Region, Petrolina, Pernambuco.
Cases were classified according to the criteria established by the World Health Organization, with modifications [21], as follows:
moderate cases
Hospitalized patients who did not require oxygen therapy or who required oxygen supplementation by means of a mask or nasal cannula
severe cases
Hospitalized patients who required ventilatory support by non-invasive mechanical ventilation, high-flow devices, intubation with mechanical ventilation, or mechanical ventilation with additional support (vasopressors, dialysis/hemodialysis, and extracorporeal membrane oxygenation).
Sample collection and processing
Within 24 h of hospital admission, 2 tubes of blood were collected by vacuum venipuncture in order to obtain whole blood, serum, and plasma. The samples were centrifuged at 3,500 RPM for 10 min.
Genetic study
Genomic DNA was obtained from peripheral blood samples, using a commercial extraction kit (ReliaPrep™ Blood gDNA Miniprep System, Promega, Madison, WI, USA), following manufacturer instructions. For genotyping and detection of polymorphisms in PTX3 (rs2305619 and rs1840680), the method of real-time PCR was used, with pre-designed TaqMan® assays and QuantStudio™ 5 equipment (Thermo Fisher).
Determination of PTX3 and cytokine levels
PTX3 was measured by enzyme-linked immunosorbent assay (ELISA) in the plasma of patients included in the study, using Human PTX3/Pentraxin 3 ELISA Kit PicoKine™ (Boster Biological Technology®) commercial kits, following manufacturer instructions.
Concentrations of interleukin (IL)-6, IL-8, and IL-10 were measured using cytometric bead array (CBA) kits (BD Biosciences, San Diego, CA, USA), following manufacturer instructions. CBA was performed with a BD™ FACSCalibur flow cytometer. Quantitative analysis was performed using FCAP Array™ Software.
Patients were randomly selected between the groups (moderate and severe), according to the availability of biological material for use.
Data collection
Demographic, clinical, and laboratory data were obtained by analysis of electronic medical records. For characterization of patients, data were collected regarding sex, age, comorbidities, length of stay, time from onset of symptoms to hospitalization, and outcome. Clinical signs (lower systolic and diastolic pressure, higher respiratory rate, and lower O2 saturation) were collected upon hospitalization.
Regarding laboratory tests, which were conducted within 24 h after admission, the following were included: complete blood count, activated partial thromboplastin time (APTT), D-dimer, aspartate aminotransferase (AST), alanine aminotransferase (ALT), total, direct and indirect bilirubin, creatinine, ferritin, C-reactive protein (CRP), troponin T, urea, prothrombin time, and international normalized ratio (INR). In addition, the neutrophil-to-lymphocyte ratio (NLR) and monocyte-to-lymphocyte ratio (MLR) were calculated by dividing the absolute value of neutrophils by lymphocytes for NLR and the absolute value of monocytes by lymphocytes for MLR.
Statistical analysis
The program SPSS Statistics version 22.0 (SPSS, Inc., Chicago, IL, USA) was used for data analysis. The program GraphPad Prism version 8.0 and 9.0 (GraphPad, San Diego, CA, USA) was used to construct the graphs. Categorical data were expressed as absolute frequency and percentage. Continuous variables were presented as median with interquartile range.
The Kolmogorov–Smirnov test was used to verify normal distribution of continuous variables. Comparisons between 2 groups were performed using Student’s t test and Mann–Whitney test for parametric and nonparametric variables, respectively. For categorical variables, Pearson’s chi-square test and Fisher’s exact test were used. For comparisons between more than 2 groups, ANOVA or Kruskal–Wallis tests were applied to parametrically or nonparametrically distributed data.
A multivariate logistic regression model was used, with the backward method, including possible confounding factors associated with COVID-19 severity described in previous studies [22–24]. The odds ratio (OR) was estimated with a 95% confidence interval (CI). P values < 0.05 were considered significant.
The Spearman correlation test was performed to correlate plasma levels of PTX3 with other continuous variables. Significant results (P < 0.05) were presented in the form of correlation graphs and a heat map, where the magnitude of the correlation coefficients is shown in a color scale.
Results
Clinical and laboratory characterization
This study included a total of 496 patients with COVID-19 admitted to 2 reference hospitals in the Northeast Region of Brazil. These patients were grouped into moderate (n = 267) and severe (n = 229) cases, according to the classification established by the World Health Organization in 2020 (with modifications) [21].
Patient characteristics are described in Table 1. A higher prevalence of male sex was observed in both groups, with mean age of 51 years in the moderate group and 54 years in the severe group (P > 0.05). Previous comorbidities, such as diabetes mellitus, systemic arterial hypertension, chronic kidney disease, and obesity, were more prevalent in the severe group (P < 0.05). The severe group showed longer hospital stay (11 days) when compared to the moderate group (4 days) (P < 0.0001). The mortality rate in the severe group was 30.6%. In contrast, there were no deaths in the moderate group (P < 0.0001).
Table 1.
Characterization of hospitalized patients with COVID-19 according to disease severity
| Moderate cases (n = 267) | Severe cases (n = 229) | p value | |
|---|---|---|---|
| Sex | 0.52 | ||
| Male (n, %) | 156 (58.4) | 141 (61.6) | |
| Female (n, %) | 111 (41.6) | 88 (38.4) | |
| Age (years), median (IQR) | 51 (41–63) | 54 (44–65) | 0.109 |
| Comorbidities | |||
| Systemic arterial hypertension (n, %) | 113 (42.3) | 118 (51.5) | 0.047 |
| Diabetes mellitus (n, %) | 60 (22.5) | 78 (34.1) | 0.005 |
| Obesity (n, %) | 53 (19.9) | 64 (27.9) | 0.044 |
| History of tobacco use (n, %) | 58 (21.7) | 56 (24.5) | 0.521 |
| Chronic kidney disease (n, %) | 3 (1.1) | 11 (4.8) | 0.026 |
| COPD (n, %) | 9 (3.4) | 8 (3.5) | 1 |
| Asthma (n, %) | 8 (3.0) | 6 (2.6) | 1 |
| Length of hospital stay, median (IQR) | 4 (3–6) | 11 (6–20) | < 0.0001 |
| Time from symptom onset to hospitalization, median (IQR) | 9 (6–11) | 9 (6–11) | 0.987 |
| Outcome | < 0.0001 | ||
| Recovery (n, %) | 267 (100.0) | 159 (69.4) | |
| Death (n, %) | – | 70 (30.6) | |
COPD: chronic obstructive pulmonary disease; IQR: interquartile range. Significant values (p < 0.05) are highlighted in bold
Laboratory data at the time of admission are summarized in Supplementary Table 1. Serum levels of IL-6, IL-8, and IL-10 were higher in the severe group (P < 0.05) (Supplementary Table 1).
Polymorphisms in PTX3 and COVID-19 severity
The distribution of allelic and genotypic frequencies of the PTX3 polymorphisms rs1840680 and rs2305619 according to COVID-19 severity are shown in Table 2. The polymorphisms studied were in accordance with Hardy–Weinberg equilibrium. A higher frequency of the AA genotype (rs1840680) was observed in the moderate group when compared to the severe group (16.10% versus 10.0%; P = 0.047). No significant association was observed between the polymorphism rs2305619 and COVID-19 severity (Table 2).
Table 2.
Allele and genotype distribution of the rs1840680 and rs2305619 polymorphisms in PTX3 according to COVID-19 severity
| Genotype/allele | Moderate cases (n = 267) | Severe cases (n = 229) | p value | |
|---|---|---|---|---|
| rs1840680 | GG | 103 (38.6) | 91 (39.70) | AA versus AG + GG |
| AG | 121 (45.30) | 115 (50.20) | 0.047 | |
| AA | 43 (16.10) | 23 (10.00) | ||
| G | 327 (61.24) | 297 (64.85) | 0.24 | |
| A | 207 (38.76) | 161 (35.15) | ||
| rs2305619 | GG | 69 (25.80) | 65 (28.40) | AA versus AG + GG |
| AG | 144 (53.90) | 122 (53.30) | 0.596 | |
| AA | 54 (20.20) | 42 (18.30) | ||
| G | 282 (52.81) | 252 (55.02) | 0.485 | |
| A | 252 (47.19) | 206 (44.98) |
Values are presented as n (%). Significant values (p < 0.05) are highlighted in bold
In order to verify whether the PTX3 polymorphism rs1840680 is independently associated with COVID-19 severity, analysis was performed using the multivariate logistic regression method (Table 3). The following possible confounding factors associated with COVID-19 severity established in previous studies were included in the model: age, sex, diabetes mellitus, systemic arterial hypertension, chronic kidney disease, neoplasia, and obesity. After excluding the variables using the backward regression method, the following variables remained in the model: diabetes mellitus (P = 0.040; OR = 1.54; 95% CI 1.02 to 2.33), chronic kidney disease (P = 0.046; OR = 3.83; 95% CI 1.02 to 14.33), and rs1840680 (AA) (P = 0.037; OR = 0.555; 95% CI 0.32 to 0.96).
Table 3.
Multivariate logistic regression to determine risk factors independently associated with COVID-19 severity
| Multivariate analysis | ||
|---|---|---|
| OR (95% CI) | p value | |
| Diabetes mellitus | 1.54 (1.02–2.33) | 0.040 |
| Chronic kidney disease | 3.83 (1.02–14.33) | 0.046 |
| rs1840680 (AA) | 0.555 (0.32–0.96) | 0.037 |
CI: confidence interval; OR: odds ratio
When we analyzed the association of PTX3 polymorphisms with clinical characteristics presented during hospitalization and outcome, it was observed that individuals with the AA genotype (rs1840680) had a shorter hospital stay when compared to those with the AG or GG genotypes (P < 0.004) (Table 4).
Table 4.
Genotype distribution of rs1840680 and rs2305619 in PTX3 according to clinical characteristics and outcome
| Variable | rs1840680 (PTX3) | rs2305619 (PTX3) | ||||
|---|---|---|---|---|---|---|
| AA (n = 66) | AG + GG (n = 430) | p value | AA (n = 96) | AG + GG (n = 400) | p value | |
| Lower systolic pressure (mmHg), median (IQR) | 101.5 (94.7–110.0) | 100 (91.25–110.00) | 0.326 | 100 (94–110) | 100 (91–110) | 0.634 |
| Lower diastolic pressure (mmHg), median (IQR) | 61.5 (59.2–72.7) | 60 (58–70) | 0.370 | 60.00 (56.25–76.00) | 60 (58–70) | 0.636 |
| Higher respiratory rate (RPM), median (IQR) | 25 (22–31) | 27 (23–32) | 0.133 | 26 (23–32) | 26 (23–32) | 0.802 |
| Lower O2 saturation (%), median (IQR) | 90.5 (88.7–93.0) | 90 (88–93) | 0.738 | 90.00 (87.25–93.00) | 90.5 (88.0–93.0) | 0.713 |
| Length of hospital stay (days), median (IQR) | 3.50 (2–7) | 6 (3–12) | 0.004 | 5 (3–10.75) | 5 (3–12) | 0.298 |
| Death (n, %) | 7 (10.6) | 63 (14.7) | 0.452 | 14 (14.6) | 56 (14.0) | 0.871 |
Significant values (p < 0.05) are highlighted in bold. IQR: interquartile range; mmHg, millimeters of mercury; O2, oxygen; RPM: respirations per minute
Although the AA genotype of rs1840680 had lower plasma levels of PTX3 when compared to the other genotypes, this association was not statistically significant (Supplementary Fig. 1).
Plasma levels of PTX3
For measurement of PTX3, 169 patients were included, 86 from the severe group and 83 from the moderate group. Figure 1A displays the distribution of protein plasma levels according to COVID-19 severity classification. The severe group had higher PTX3 levels (median: 987.0 pg/mL) when compared to the moderate group (median: 570.5 pg/ml) (P = 0.0004). Regarding the outcome, PTX3 levels at admission were observed to be 3.3 times higher in patients who died than in those who survived (2,233 pg/mL [n = 25] versus 663.2 pg/mL [n = 144], P < 0.0001) (Fig. 1B).
Fig. 1.
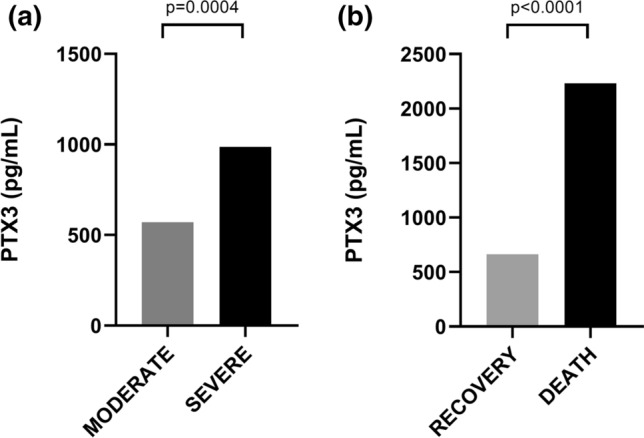
Median plasma levels of PTX3 (pg/mL) in individuals hospitalized with COVID-19. A comparison between moderate (n = 83) and severe (n = 86) forms. B comparison between recovery (n = 144) and death (n = 25) groups. Statistical significance was determined using Mann–Whitney U-test. Significance was considered when p < 0.05
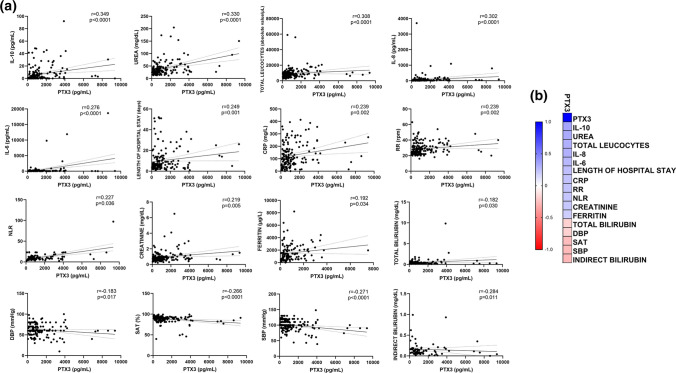
In order to analyze the relationship between PTX3 plasma levels and patients’ clinical and laboratory variables, we conducted a correlation study. Figure 2 displays the correlation graphs that demonstrated statistically significant values. A positive correlation was observed for PTX3 levels with IL-6, IL-8, IL-10, CRP, total leukocytes, NLR, urea, creatinine, ferritin, length of hospital stay, and higher respiratory rate. Total bilirubin, lower diastolic pressure, lower oxygen saturation, lower systolic blood pressure, and indirect bilirubin were inversely correlated with PTX3 (P < 0.05 for all analyses) (Fig. 2).
Fig. 2.
Relationship between plasma levels of PTX3 (pg/mL) and the continuous variables included in the study. A A linear regression line shows the relationship between plasma levels of PTX3 and continuous variables. The spline chart demonstrates the mean with 95% confidence intervals. B Heat map representing the degree of correlation between PTX-3 levels and the variables analyzed. The degree of correlation was assessed using Spearman’s rank correlation coefficient test. Significance was considered when p < 0.05. CRP: C-reactive protein; DBP: diastolic blood pressure; IL: interleukin; mmHg, millimeters of mercury; NLR: neutrophil-to-lymphocyte ratio; RPM: respirations per minute; RR: respiratory rate; SAT: lower oxygen saturation; SBP: systolic blood pressure
Discussion
Innate immunity plays an important role in the clinical course of COVID-19, and the complement system is an important component involved in the immunopathological mechanism of the disease. The factors involved in COVID-19 severity have not yet been fully clarified, but they seem to be related to viral, environmental, and genetic factors of the host.
We have demonstrated, for the first time, that the AA genotype in PTX3 (rs1840680) was associated with protection against the development of severe COVID-19 in hospitalized patients, regardless of other possible confounding factors. Furthermore, patients with the AA genotype spent less time in the hospital when compared to those with the GA + GG genotypes. Several previous studies have reported an association of the rs1840680 polymorphism in the context of lung infectious diseases [16–18]. Additionally, Kerget et al. (2021), evaluating a small cohort of 94 patients with COVID-19, observed a higher frequency of the AA genotype in those who developed macrophage activation syndrome [20].
In this study, although individuals carrying the AA genotype showed lower levels of PTX-3, the difference was not statistically significant. The rs1840680 polymorphism is located in an intronic region in the PTX3 gene. Despite its location, previous studies have shown its influence on PTX3 levels, possibly explained by a linkage disequilibrium with another variant located in a regulatory region [25]. However, the studies show discordant results. Barbati et al. [25] and Carmo et al. [19], observed that AA carriers for rs1840680 (vs. AG and GG genotypes) had higher PTX3 plasma levels in healthy individuals and in patients with chronic hepatitis C, respectively. In contrast, Sheneef et al. [26], analyzing Egyptian patients with pulmonary tuberculosis, observed lower PTX-3 levels in patients carrying the AA genotype [26]. A previous study with Turkish patients hospitalized due to COVID-19 found a significant association between the AA genotype of rs1840680 and decreased levels of PTX3 [20]. Furthermore, a study investigating pulmonary aspergillosis in patients with chronic obstructive pulmonary disease also showed a relationship between the AA genotype of rs1840680 and decreased plasma levels of PTX3 [18]. Given that PTX3 is an acute-phase protein, it is possible that its supply is depleted depending on the phase of the disease, resulting in a smaller difference between the genotypes, which may explain the differences found between the studies. In addition, it is possible that polymorphisms in PTX3 have a greater influence on its levels in other tissues than in plasma, as observed previously in samples of bronchoalveolar-lavage fluid from patients with invasive aspergillosis [27].
Our genetic study results are corroborated by analysis of PTX3 plasma levels, where we observed that patients with severe disease had increased PTX3 levels when compared to moderate cases. In addition, at admission, the PTX3 levels of patients who died were 3.3 times higher than the levels of patients who survived. PTX3 levels were also significantly correlated with laboratory parameters of hematologic, renal, and hepatic function, as well as clinical parameters such as blood pressure, O2 saturation, and length of hospital stay.
Previous studies have demonstrated that PTX3 plasma level is a good predictor of mortality in patients with COVID-19 [15, 28, 29]. Corroborating our findings, in 2021, Brunetta et al. [15] observed that patients with COVID-19 admitted to the intensive care unit and patients who did not survive had significantly higher PTX3 levels than patients admitted to wards or patients who survived. Furthermore, they observed that PTX3 levels were significantly correlated with laboratory parameters such as CRP, procalcitonin, IL-6, ferritin, D-dimer, lactate dehydrogenase, troponin-I, lymphocyte count, and platelet count. The authors concluded that PTX3 is an important predictor of mortality, showing better results than other inflammatory markers, such as IL-6 and CRP [15]. In 2021, Gutmann et al. [28] demonstrated that PTX3 was an important predictor of 28-day intensive care unit mortality, and Hansen et al., in 2022 [30], demonstrated that PTX3 appears to be a useful clinical biomarker for predicting respiratory failure and risk of death at 30 days in patients with COVID-19 treated with and without remdesivir and dexamethasone [30].
Our findings indicate a possible role of PTX3 in the immunopathology of severe forms of COVID-19, possibly associated with increased activation of the complement system and inflammatory cytokines. In line with this, our findings have demonstrated a positive correlation of PTX3 levels with IL-6, IL-8, and IL-10 levels, as well as with an increase in leukocyte count and NLR. Previous studies also demonstrated that increased concentrations of IL-6 and IL-8 were correlated with worse prognosis, hospitalization, transfer to the intensive care unit, mechanical ventilation, and death [31–33].
Pentraxins are involved in the activation and amplification of the complement system via communication with complement initiation pattern recognition molecules, such as C1q and ficolins [9]. This activation contributes to increased levels of C3a and C5a, which can contribute to the release of pro-inflammatory cytokines with consequent impairment in fibrinolysis and increased procoagulant activity, which contribute to microvascular thrombosis and endothelial damage [34]. In COVID-19, the accumulation of C5a in circulation and the deposition of the membrane attack complex in pulmonary tissue contribute to the pathogenesis of the disease [8, 35–38]. Furthermore, myeloid cells from respiratory samples of patients with severe COVID-19 exhibit transcriptome signatures for the Fcγ receptor and complement activation [35], emphasizing the importance of these mechanisms in the pathogenesis of COVID-19.
Accordingly, there is a possibility that high levels of PTX3 in COVID-19 may be a reflection of a failure in the immune system regarding inhibition of factors that act against uncontrolled inflammation [15]. Thus, individuals who are genetically predisposed to produce lower amounts of PTX3, as shown by the rs1840680 AA genotype in the present study, would be more protected from developing severe forms of the disease.
The limitations to this study include lack of information on viral aspects, such as viral load and variants. Although there is not sufficient evidence to demonstrate the association between variants of concern and COVID-19 severity, the recruitment period for the majority of patients included in this study coincides with the circulation of the gamma variant in Brazil [39]. Furthermore, this study was carried out in 2 reference centers in the same municipality, making further studies necessary to confirm our genetic findings in other populations.
Conclusion
Our findings have demonstrated, for the first time, a protective effect of the PTX3 AA genotype (rs1840680) on the development of severe forms of COVID-19. Additionally, PTX3 plasma levels were associated with the severity of COVID-19. These results reinforce previous findings that PTX3 plays an important role in the immunopathology of COVID-19 and is able to predict the risk of developing severe forms of the disease.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank the Multi-User Research Laboratory of HU-UNIVASF (EBSERH) and the Secretary of Health of the State of Pernambuco for the support provided.
Author contributions
RFC conceptualized and designed the study with support from TAF. TAF and MVSS were involved in data collection, sample processing, and experimental trials, with support from VCP, MKAC, and VRAP. TAF and RFC conducted the statistical analysis. TAF wrote the initial draft, with support from RFC and ACA. All authors critically reviewed the manuscript and approved the final version.
Funding
This work was supported by the Foundation for the Support of Science and Technology of the State of Pernambuco (FACEPE, acronym in Portuguese) (grant numbers APQ-0422–2.02/19 and IBPG-0010–2.02/20) and by the Brazilian Coordination for the Improvement of Higher Education Personnel (CAPES, acronym in Portuguese), Finance Code 001.
Declarations
Conflict of interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Consent to Participate
Informed consent was obtained from all individual participants included in the study.
Consent for Publication
Not applicable.
Ethics Approval
This study received approval from the Ethics Committee of the Hospital das Clínicas of the Federal University of Pernambuco (HC/UFPE, acronym in Portuguese) under number CAAE: 38196620.0.0000.8807.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zhu N, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Worldometer. COVID Live Update: 475,797,234 Cases and 6,126,926 Deaths from the Coronavirus - Worldometer. 2022. https://www.worldometers.info/coronavirus/. Accessed 23 March 2022.
- 3.Yong Hu, et al. Prevalence and severity of corona virus disease 2019 (COVID-19): A systematic review and meta-analysis. J Clin Virol. 2020;127:104371. doi: 10.1016/j.jcv.2020.104371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim Kwang Dong, et al. Adaptive immune cells temper initial innate responses. Nat Med. 2007;13(10):1248–1252. doi: 10.1038/nm1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Z, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420–422. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cecconi M, et al. Ten things we learned about COVID-19. Intensive Care Med. 2020;46:1590–1593. doi: 10.1007/s00134-020-06140-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, et al. Cytokine storm and leukocyte changes in mild versus severe SARS-CoV-2 infection: review of 3939 COVID-19 patients in China and emerging pathogenesis and therapy concepts. J Leukoc Biol. 2020;108:17–41. doi: 10.1002/JLB.3COVR0520-272R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Java A, et al. The complement system in COVID-19: friend and foe? JCI insight. 2020;5:e140711. doi: 10.1172/jci.insight.140711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma YJ, Garred P. Pentraxins in complement activation and regulation. Front Immunol. 2018;9:1–8. doi: 10.3389/fimmu.2018.03046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Porte Rémi, et al. The long pentraxin PTX3 as a humoral innate immunity functional player and biomarker of infections and sepsis. Frontiers Immunol. 2019 doi: 10.3389/fimmu.2019.00794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bozza S, et al. Pentraxin 3 protects from MCMV infection and reactivation through TLR sensing pathways leading to IRF3 activation. Blood. 2006;108:3387–3396. doi: 10.1182/blood-2006-03-009266. [DOI] [PubMed] [Google Scholar]
- 12.Reading PC, et al. Antiviral activity of the long chain pentraxin PTX3 against influenza viruses. J. Immunol. 2008;180:3391–3398. doi: 10.4049/jimmunol.180.5.3391. [DOI] [PubMed] [Google Scholar]
- 13.Han B, et al. Protective effects of long pentraxin PTX3 on lung injury in a severe acute respiratory syndrome model in mice. Lab Invest. 2012;92:1285–1296. doi: 10.1038/labinvest.2012.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stravalaci M, et al. Recognition and inhibition of SARS-CoV-2 by humoral innate immunity pattern recognition molecules. Nat Immunol. 2022;23:275–286. doi: 10.1038/s41590-021-01114-w. [DOI] [PubMed] [Google Scholar]
- 15.Brunetta E, et al. Macrophage expression and prognostic significance of the long pentraxin PTX3 in COVID-19. Nat immunol. 2021;22:19–24. doi: 10.1038/s41590-020-00832-x. [DOI] [PubMed] [Google Scholar]
- 16.Chiarini M, et al. PTX3 genetic variations affect the risk of Pseudomonas aeruginosa airway colonization in cystic fibrosis patients. Genes Immun. 2010;11:665–670. doi: 10.1038/gene.2010.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olesen R, et al. DC-SIGN (CD209), pentraxin 3 and vitamin D receptor gene variants associate with pulmonary tuberculosis risk in West Africans. Genes Immun. 2007;8:456–467. doi: 10.1038/sj.gene.6364410. [DOI] [PubMed] [Google Scholar]
- 18.He Q, et al. Pentraxin 3 gene polymorphisms and pulmonary aspergillosis in chronic obstructive pulmonary disease patients. Clin Infect Dis. 2018;66:261–267. doi: 10.1093/cid/cix749. [DOI] [PubMed] [Google Scholar]
- 19.Carmo RF, et al. Genetic variation in PTX 3 and plasma levels associated with hepatocellular carcinoma in patients with HCV. J Viral Hepat. 2016;23:116–122. doi: 10.1111/jvh.12472. [DOI] [PubMed] [Google Scholar]
- 20.Kerget F, et al. Evaluation of the relationship between pentraxin 3 (PTX3) rs2305619 (281A/G) and rs1840680 (1449A/G) polymorphisms and the clinical course of COVID-19. J. Med. Virol. 2021;93:6653–6659. doi: 10.1002/jmv.27238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.World Health Organization. COVID-19 Therapeutic Trial Synopsis. 2020. https://www.who.int/publications/i/item/covid-19-therapeutic-trial-synopsis. Acessed 17 Dec 2021.
- 22.Dorjee K, et al. Prevalence and predictors of death and severe disease in patients hospitalized due to COVID-19: a comprehensive systematic review and meta-analysis of 77 studies and 38,000 patients. PloS one. 2020;15:e0243191. doi: 10.1371/journal.pone.0243191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, et al. Epidemiology of COVID-19: a systematic review and meta-analysis of clinical characteristics, risk factors, and outcomes. J Med Virol. 2021;93:1449–1458. doi: 10.1002/jmv.26424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu K, et al. Application of ordinal logistic regression analysis to identify the determinants of illness severity of COVID-19 in China. Epidemiol. Infect. 2020;148:E146. doi: 10.1017/S0950268820001533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barbati E, et al. Influence of pentraxin 3 (PTX3) genetic variants on myocardial infarction risk and PTX3 plasma levels. PloS one. 2012;7:e53030. doi: 10.1371/journal.pone.0053030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheneef A, et al. Pentraxin 3 genetic variants and the risk of active pulmonary tuberculosis. Egypt J Immunol. 2017;24:21–27. [PubMed] [Google Scholar]
- 27.Cunha C, et al. Genetic PTX3 deficiency and aspergillosis in stem-cell transplantation. N Engl J Med. 2014;370(5):421–432. doi: 10.1056/NEJMoa1211161. [DOI] [PubMed] [Google Scholar]
- 28.Gutmann C, et al. SARS-CoV-2 RNAemia and proteomic trajectories inform prognostication in COVID-19 patients admitted to intensive care. Nat Commun. 2021;12:1–17. doi: 10.1038/s41467-021-23494-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hansen CB, et al. Complement related pattern recognition molecules as markers of short-term mortality in intensive care patients. Infection. 2020;80:378–387. doi: 10.1016/j.jinf.2020.01.010. [DOI] [PubMed] [Google Scholar]
- 30.Hansen CB, et al. Prediction of Respiratory Failure and Mortality in COVID-19 Patients Using Long Pentraxin PTX3. J Innate Immun. 2022 doi: 10.1159/000521612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buszko M, et al. Lessons learned: new insights on the role of cytokines in COVID-19. Nat. Immunol. 2021;22:404–411. doi: 10.1038/s41590-021-00901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Del Valle DM, et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson PC, et al. Accumulating evidence suggests anti-TNF therapy needs to be given trial priority in COVID-19 treatment. Lancet Rheumatol. 2020;2:e653–e655. doi: 10.1016/S2665-9913(20)30309-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chauhan AJ, Wiffen LJ, Brown TP. COVID-19: a collision of complement, coagulation and inflammatory pathways. J. Thromb. Haemost. 2020;18:2110–2117. doi: 10.1111/jth.14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim DM, et al. Enhanced eosinophil-mediated inflammation associated with antibody and complement-dependent pneumonic insults in critical COVID-19. Cell Rep. 2021;37:109798. doi: 10.1016/j.celrep.2021.109798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma Lina, et al. Increased complement activation is a distinctive feature of severe SARS-CoV-2 infection. Sci Immunol. 2021 doi: 10.1126/sciimmunol.abh2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Magro C, et al. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: a report of five cases. Transl Res. 2020;220:1–13. doi: 10.1016/j.trsl.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen B, et al. Proteomic and metabolomic characterization of COVID-19 patient sera. Cell. 2020;182:59–72. doi: 10.1016/j.cell.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sabino EC, et al. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet. 2021;397:452–455. doi: 10.1016/S0140-6736(21)00183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.