Abstract
Mitochondrial morphology and dynamics maintain mitochondrial integrity by regulating its size, shape, distribution, and connectivity, thereby modulating various cellular processes. Several studies have established a functional link between mitochondrial dynamics, mitophagy, and cell death, but further investigation is needed to identify specific proteins involved in mitochondrial dynamics. Any alteration in the integrity of mitochondria has severe ramifications that include disorders like cancer and neurodegeneration. In this study, we used budding yeast as a model organism and found that Pil1, the major component of the eisosome complex, also localizes to the periphery of mitochondria. Interestingly, the absence of Pil1 causes the branched tubular morphology of mitochondria to be abnormally fused or aggregated, whereas its overexpression leads to mitochondrial fragmentation. Most importantly, pil1Δ cells are defective in mitophagy and bulk autophagy, resulting in elevated levels of reactive oxygen species and protein aggregates. In addition, we show that pil1Δ cells are more prone to cell death. Yeast two-hybrid analysis and co-immunoprecipitations show the interaction of Pil1 with two major proteins in mitochondrial fission, Fis1 and Dnm1. Additionally, our data suggest that the role of Pil1 in maintaining mitochondrial shape is dependent on Fis1 and Dnm1, but it functions independently in mitophagy and cell death pathways. Together, our data suggest that Pil1, an eisosome protein, is a novel regulator of mitochondrial morphology, mitophagy, and cell death.
Keywords: Saccharomyces cerevisiae, mitochondria, mitophagy, autophagy, protein aggregation, reactive oxygen species (ROS), cell death
Abbreviations: BAR, Bin/Amphiphysin/Rvs; ROS, reactive oxygen species; SC, synthetic complete medium; SCD, synthetic complete dextrose medium; SD-N, nitrogen starvation medium with glucose; SMD, synthetic minimal dextrose medium; SML, synthetic minimal medium with lactate
Organelles have evolved a variety of stress response processes to maintain their proteostasis and cellular homeostasis. Mitochondrion, one of the essential organelles that carry out several important cellular processes, ensures its quality control through various pathways at the molecular, organellar, and cellular levels (1). Mitochondrial fission and fusion, generally referred to as mitochondrial dynamics, maintain mitochondrial integrity by regulating its size, shape, distribution, and connectivity. Mitochondrial dynamics regulate mitochondrial quality control, metabolism, apoptosis, mitophagy, and other essential processes. Fusion of mitochondria is required to mitigate the damage and nonfunctionality by mixing components and protecting them from autophagic degradation during starvation (2, 3). Fission helps produce new mitochondria and ensures quality control by removing damaged or unwanted mitochondria through mitophagy (4). A coordinated balance of fission and fusion is critical for maintaining mitochondrial biology and therefore a plethora of important cellular processes (5, 6). In Saccharomyces cerevisiae, Fzo1, Mgm1, and Ugo1 facilitate mitochondrial fusion. Outer membrane–anchored proteins, Fzo1p and Ugo1p, carry out the outer membrane fusion of adjacent mitochondria and inner membrane–anchored Mgm1 forms transcomplexes to tether the apposing inner membranes together (7, 8, 9). Dnm1, a dynamin-related GTPase, is the major protein in mitochondrial fission (10, 11). It is predominantly present in the cytosol and recruited to mitochondria via Fis1 (12). Dnm1 assembles into oligomers which form rings and spirals at the outer membrane of mitochondria. Recruitment of Dnm1 to the mitochondrial surface is mediated through two adaptor proteins, Mdv1 and Caf4 (13, 14). These four proteins together constitute the core proteins of the mitochondrial fission machinery. However, there is a possibility that other unidentified factors still exist that take part in mitochondrial dynamics (15). Several studies have shown that any dysregulation in mitochondrial dynamics leads to neuronal disorders like Alzheimer’s, Parkinson’s, and Huntington’s (16, 17, 18). When the damage is beyond repair, mitochondria undergo mitophagy. Mitophagy is a selective autophagy where autophagosomes engulf the entire mitochondria and deliver them to the vacuole for their degradation (19, 20, 21). Mitophagy is required to eliminate the bad mitochondria from cells. Any kind of aberration in mitochondrial dynamics or mitophagy is harmful to cells. Mitochondrial dysfunction has also been linked with protein aggregation and reactive oxygen species (ROS) generation in cells which in turn leads to cell death.
Cellular organelles communicate with each other in order to cope with stress. Since the plasma membrane is positioned at the frontline to combat the external stress stimuli, it requires high degree of organization as it carries out a diverse array of functions and forms the protective barrier around the cell. The fungal plasma membrane is organized in lateral domains with specialized functions like cell wall synthesis, environmental sensing, nutrient uptake, secretion, and endocytosis (22, 23, 24). High-resolution electron microscopy of freeze-etched S. cerevisiae has shown that these domains form furrow-like invaginations in the plasma membrane (25, 26, 27). Eisosomes are filament-like complexes of peripheral membrane proteins located on the cytoplasmic side (28). They are known to be found only in a few fungal species, microalgae, and lichens (29). Pil1 and its paralog Lsp1 are the major components of eisosomes. Both these proteins contain BAR (Bin/Amphiphysin/Rvs) domains and PIP2-binding PH domains. They have been shown to bind membranes and facilitate the membrane curvature and formation of furrows (30, 31). Eisosomes regulate the lipid homeostasis at the plasma membrane and also the recruitment of proteins for subsequently protecting them from endocytosis (32). Another possible function of eisosome is to maintain membrane reservoir for plasma membrane expansion in response to various stresses (33). Recent developments in this area of research has piqued interest particularly due to association of eisosomes with RNP granules, stress granules, P-bodies, and their role in protection of nutrient transporters in response to starvation stress (34, 35, 36, 37). Though eisosomes were believed to be static structures, a recent study has shown the exchange of Pil1 molecules at the tip of eisosomes, indicating their dynamic nature (38). Also, Pil1 is reversibly phosphorylated at multiple sites which raises the possibility that this protein can be dynamic (39, 40, 41). In studies aimed to characterize the novel role of yeast mitochondrial phosphoproteome, Pil1 has been detected as one of the proteins associated with mitochondria in its phosphorylated form (42, 43). This evidence provides a possible link between mitochondria and eisosomes.
In our quest for finding out the role of Pil1 in connection with mitochondria, we investigated the localization of Pil1 to mitochondria and found that Pil1 also localizes to mitochondrial outer membrane. In the absence of Pil1, mitochondria are majorly seen in abnormally fused, and some fractions were found to be aggregated, whereas its overexpression leads to fragmentation. We found that Pil1 also alters mitophagy. Surprisingly, absence of Pil1 not only affects mitophagy (specific autophagy) but it also shows drastic reduction in nonspecific autophagy. Additionally, we show that due to mitochondrial and autophagic abnormalities in the absence of Pil1, there is a dramatic increase in the levels of protein aggregates and ROS which renders pil1Δ cells more prone to cell death.
Results
Pil1 localizes to mitochondrial outer membrane
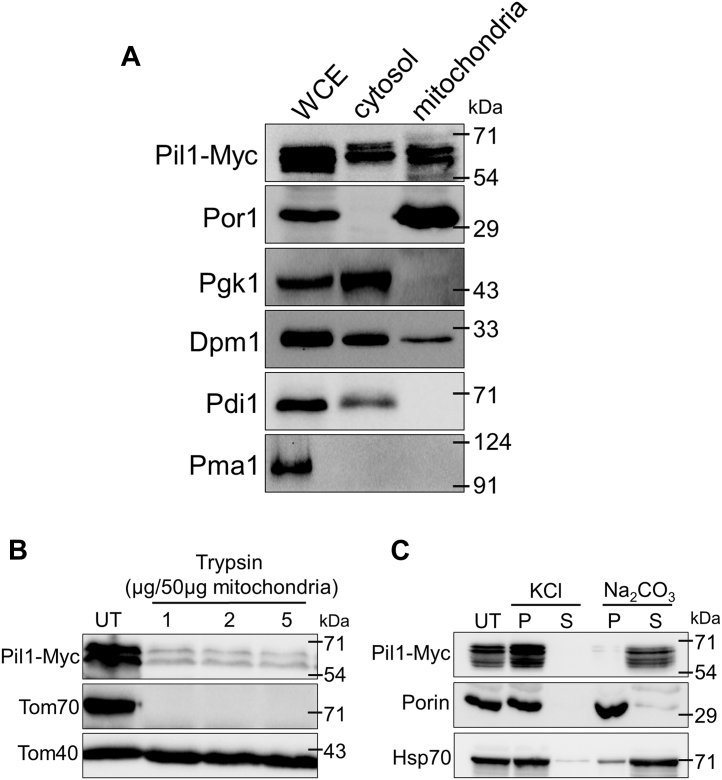
Previously, in phosphoproteome profiling of yeast mitochondria, it has been reported that Pil1 is present in mitochondria in phosphorylated state (42, 43). In order to confirm these findings, we isolated mitochondria from yeast wild-type strain where Pil1 is chromosomally tagged with 6XMyc tag at the C terminus. Whole cell extract, cytosolic fraction (postmitochondrial supernatant), and highly purified mitochondrial fraction were processed for SDS-PAGE and probed with antibodies against Myc, Porin, Pgk1, Dpm1, Pdi1, and Pma1. The immunoblotting results show that Pil1 is enriched in mitochondrial fraction as well (Fig. 1A). Absence of Pgk1 band in mitochondrial fraction shows that it is devoid of any cytosolic contamination. To further check if mitochondrial fraction is contaminated with ER, we used anti-Pdi1 antibody which is ER-lumen specific and anti-Dpm1 antibody which is ER-membrane specific. Though we did not detect Pdi1 band, we could see a small fraction of Dpm1 in highly purified mitochondrial fraction. As mitochondria are tightly associated with ER membranes, it is highly unlikely to achieve “pure” mitochondria even after sucrose gradient purification. Since eisosome complex is associated with plasma membrane, we wanted to make sure that our mitochondrial extracts do not contain any plasma membrane fractions. Absence of Pma1 band in mitochondrial fraction confirms that it does not contain plasma membrane contamination. After confirming the presence of Pil1 in mitochondria, we investigated its exact submitochondrial localization. The intact mitochondria were treated with increasing concentrations of trypsin followed by treatment with trypsin inhibitor to stop the reaction. The treated and untreated samples were probed against Myc, Tom70, and Tom40 antibodies. Significant amount of Pil1 is digested when treated with trypsin. Tom70, being a peripheral outer membrane protein gets completely digested, and Tom40, an integral outer membrane protein, remains intact (Fig. 1B). Our results clearly show that Pil1 is peripherally bound to the outer mitochondrial membrane. To further determine if Pil1 is a soluble protein or associated with the mitochondrial membrane, we carried out protein extraction with potassium chloride (KCl) and sodium carbonate (Na2CO3). When subjected to KCl, Pil1 was retrieved in the pellet fraction showing that Pil1 is tightly bound to the mitochondrial membrane. However, after Na2CO3 treatment, Pil1 was found in the supernatant fraction (Fig. 1C). This shows that Pil1, though peripherally and tightly bound to the mitochondrial outer membrane, is a soluble protein. Porin, a membrane protein and Hsp70, a soluble matrix protein were used as controls.
Figure 1.
Localization of Pil1 to outer membrane of mitochondria.A, western blot of whole cell extract, cytosolic fraction (postmitochondrial fraction), and mitochondrial fraction (50 μg each) from Pil1-Myc expressing strain (YNB263). Antibodies against Myc, Porin, Pgk1, Dpm1, and Pdi1 were used for the western blot analysis. Antibodies against Por1, Pgk1, Dpm1/Pdi1, and Pma1 were used as controls for mitochondria, cytosol, ER membrane/ER lumen, and plasma membrane, respectively. B, trypsin digestion assay. Mitochondrial fraction (50 μg) isolated from Pil1-Myc strain was subjected to trypsin treatment and western blotted using antibodies against Myc, Tom70, and Tom40. Tom70 and Tom40 are peripheral and integral mitochondrial outer membrane proteins, respectively. C, salt extractions. Mitochondria (50 μg) from Pil1-Myc cells were treated with 400 mM KCl and 200 mM Na2CO3 (pH 11.4), immunoblotted using antibodies against Myc, Porin, and Hsp70. Porin and Hsp70 are used as controls for membrane and soluble mitochondrial proteins, respectively. All experiments were performed three times. P, pellet; S, supernatant; UT, untreated; WCE, whole cell extract.
Absence and abundance of Pil1 leads to abnormal morphology of mitochondria
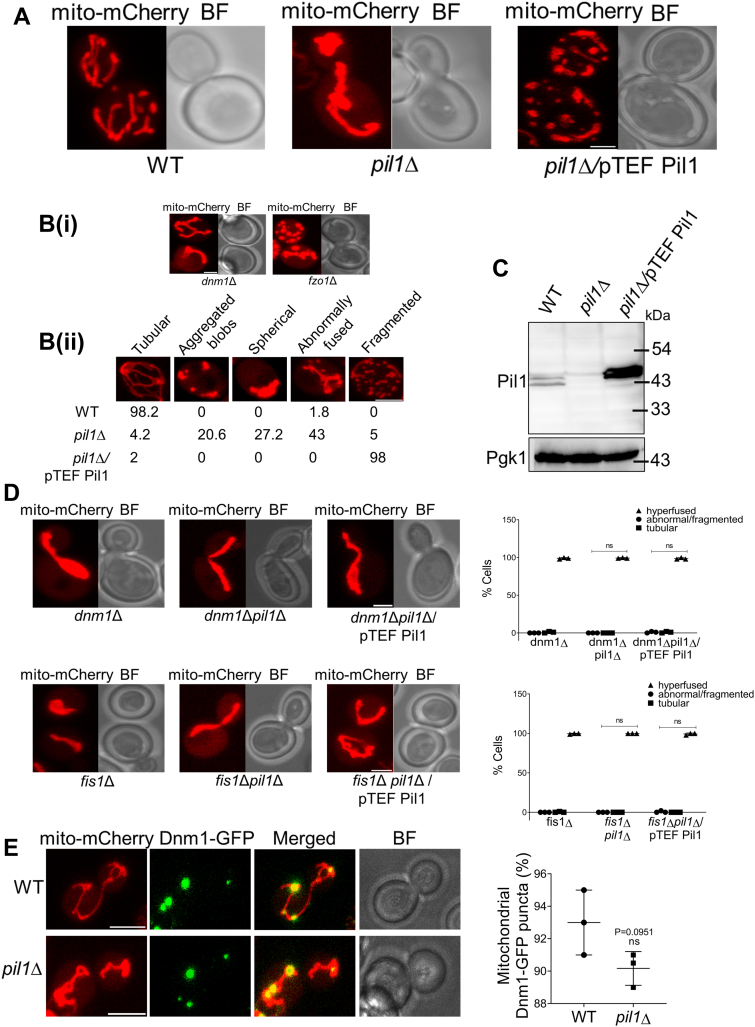
Next, we examined if Pil1 causes any kind of alteration in mitochondrial morphology by using pHS12 Mito-mCherry plasmid which contains Cox4 presequence that targets mCherry to the mitochondrial matrix. We observed the mitochondrial morphology under fluorescence microscope in WT, pil1Δ, and Pil1 overexpression (pil1Δ/pTEF Pil1) cells. The transformed cells were grown to log phase (A600=1) and mitochondrial morphology was observed under fluorescence microscope. We observed that instead of forming typical tubular structures, cells lacking Pil1 as well as those with Pil1 overexpression displayed abnormal mitochondrial morphology (Fig. 2A). Keeping the mitochondrial morphologies in dnm1Δ (hyperfused) and fzo1Δ (fragmented) as references [Fig. 2B(i)], we categorized these morphologies as abnormally fused, aggregated blobs, spherical, and fragmented. We observed that 43% of pil1Δ cells contained abnormally fused mitochondria while 27.2% cells displayed a spherical mitochondrial network. To our surprise, we also found that a fraction of pil1Δ cells (20.6%) had aggregated mitochondria [Fig. 2B(ii)]. On the other hand, when we overexpressed Pil1 by transforming pil1Δ cells with TEF-Pil1 plasmid, almost all cells (98%) contained fragmented mitochondria. The levels of Pil1 protein in WT, pil1Δ, and Pil1 overexpression cells were confirmed by immunoblotting with anti-Pil1 serum (Fig. 2C). These observations show that Pil1 indeed plays a critical role in regulating mitochondrial morphology.
Figure 2.
Pil1 regulates mitochondrial morphology.A, mitochondrial morphology in WT, pil1Δ, and Pil1 overexpression (pil1Δ/pTEF Pil1) was observed under fluorescence microscope. Mito-mCherry plasmid (pNB812) was used to tag mitochondria. Images are maximum projection of z-stacks. B(i), dnm1Δ and fzo1Δ were used as controls for quantification of mitochondrial morphology. B(ii), quantification of mitochondrial phenotypes in WT, pil1Δ, and pil1Δ/pTEF Pil1 cells. Mitochondrial morphology shown for each strain was collected from three independent analysis of at least 80 yeast cells. Scale bar represents 2 μm. C, Western blot showing the levels of Pil1 protein in WT, pil1Δ, and pil1Δ/pTEF Pil1 cells. WT, pil1Δ, and pil1Δ/pTEF Pil1 cells were lysed, subjected to SDS-PAGE, and immunoblotted with anti-Pil1 serum. D, Mitochondrial morphology in fis1Δ, dnm1Δ, fis1Δpil1Δ, dnm1Δpil1Δ, and fis1Δpil1Δ, dnm1Δpil1Δ strains overexpressing Pil1 was observed. Graph represents three independent trials of 100 yeast cells. Statistical analysis was done using multiple comparisons: Two-way ANOVA. ns, nonsignificant. Scale bar represents 2 μm. E, number of Dnm1 foci associated with mitochondria were analyzed in WT and pil1Δ cells transformed with mito-mCherry and Dnm1-GFP plasmids. Maximum projections of z-stacks are shown in the represented images. Data shown are the mean ± SD of three independent trials. Scale bar represents 5 μm. Statistical analysis was done using unpaired Student’s t test (p-value is indicated on the graph). BF, brightfield; ns, nonsignificant.
Next, we wanted to explore whether major mitochondrial fission factors Fis1 and Dnm1 have any role in generating mitochondrial morphology that we observed in the absence and overexpression of Pil1. We analyzed mitochondrial morphology in fis1Δpil1Δ and dnm1Δpil1Δ double mutants. Mitochondria remained hyperfused in almost all cells, as reported in fis1Δ and dnm1Δ single mutants. Additionally, overexpression of Pil1 did not rescue the fission defect in fis1Δpil1Δ and dnm1Δpil1Δ double mutants (Fig. 2D). Also, we did not see any change in distribution of Dnm1 on mitochondria in the absence of Pil1 (Fig. 2E), which shows that Pil1 does not affect the recruitment of Dnm1 to mitochondria.
Pil1 is involved in mitophagy as well as nonselective autophagy
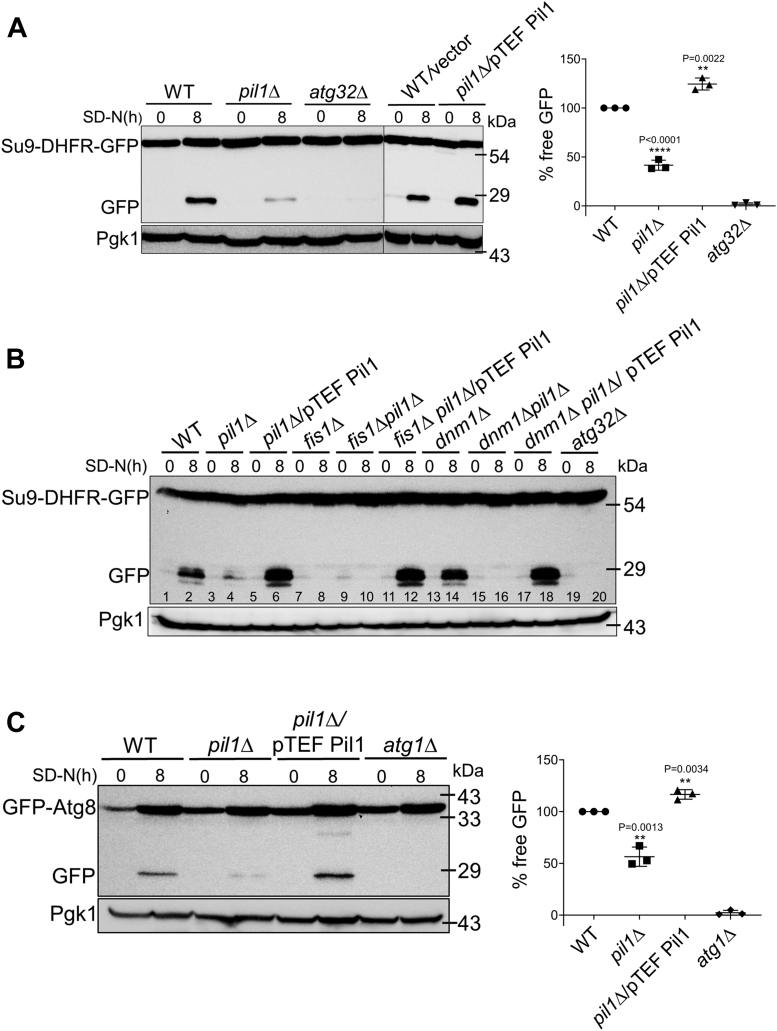
Though there is growing evidence that mitochondrial pro-fission factors play a role in mitophagy, there is still a lack of clarity behind the exact mechanism. In mammalian cells, it has been shown that mitophagy is impaired when mitochondrial fission is inhibited, which leads to accumulation of damaged mitochondria (44). Nevertheless, we decided to check whether Pil1 regulates the fission factors–mediated mitophagy or itself alters mitophagy. To measure mitophagy, we used Su9-DHFR-GFP processing assay. This plasmid has DHFR gene with Su9 presequence at the N terminal and GFP tag at the C terminal. Su9 presequence targets DHFR-GFP to mitochondria. During mitophagy, mitochondria are sequestered by autophagosomes and are targeted to the vacuole where DHFR gets degraded, and GFP moiety, being relatively resistant to proteolysis, remains intact. The amount of free GFP by immunoblotting is used as a measure to monitor mitophagy. After 8 h of nitrogen starvation to induce mitophagy, we found that free GFP release was significantly less in pil1Δ than WT and more in overexpressed Pil1 (Fig. 3A). No GFP release was seen in atg32Δ as it is essential for mitophagy (45, 46). Our data indicate that Pil1 is required for mitophagy.
Figure 3.
Pil1 regulates mitophagy and bulk autophagy.A and B, mitophagy assay. Indicated strains transformed with Su9-DHFR-GFP plasmid were grown in SML media (SC with lactate lacking leucine and uracil) to proliferate mitochondria. Cells were then shifted to SD-N media for 8 h. Samples were collected before and after nitrogen starvation. Protein extracts were immunoblotted with anti-GFP antibody. Positions of full length Su9-DHFR-GFP and free GFP are indicated. Anti-Pgk1 serum was used as loading control. Quantification of mitophagy after 8 h of nitrogen starvation is represented as a percentage of the ratio of free GFP to total GFP signal. Data are presented as mean ± SD of three independent assays. C, autophagy assay. Indicated strains carrying pRS423 GFP-Atg8 plasmid were grown in SMD media (SCD lacking histidine and leucine) and then shifted to SD-N media for 8 h. Cells were collected before and after nitrogen starvation. Immunoblotting was done with anti-GFP, and positions of full length GFP-Atg8 and free GFP are indicated. Anti-Pgk1 was used as a loading control. Quantification of autophagy is the percentage of free GFP/total GFP signal. Data are presented as mean ± SD of three independent experiments. Statistical analysis was done using unpaired Student’s t test (p-value is indicated on the graph) ∗∗∗∗p < 0.0001, ∗∗p < 0.01 versus WT. SC, synthetic complete medium; SCD, synthetic complete dextrose medium; SD-N, nitrogen starvation medium with glucose; SMD, synthetic minimal dextrose medium; SML, synthetic minimal medium with lactate.
We sought to find out if the function of Pil1 in mitophagy is dependent on Fis1 and Dnm1. For this, we monitored mitophagy levels in fis1Δ, dnm1Δ, fis1Δpil1Δ, and dnm1Δpil1Δ strains. To our surprise, we found that fis1Δ and fis1Δpil1Δ cells were defective in mitophagy (Fig. 3B, lanes 7, 8 & 9, 10). On the other hand, dnm1Δ cells showed almost normal mitophagy levels (lanes 13, 14), which were completely inhibited upon Pil1 depletion, i.e., in dnm1Δpil1Δ cells (lanes 15, 16). To get further clarification, we checked mitophagy after overexpressing Pil1 in fis1Δpil1Δ and dnm1Δpil1Δ cells, and found that it significantly increased (lanes 11, 12 & 17, 18). Hence, overexpression of Pil1 can complement the mitophagy defect in fis1Δ cells. Altogether, our data suggest that the role of Pil1 in mitophagy is independent of Fis1 and Dnm1.
Considering that Pil1 has a role in mitophagy, we next wanted to check if role of Pil1 is limited to mitophagy or it regulates nonselective (bulk) autophagy as well. We utilized the GFP-Atg8 processing assay to monitor autophagy. Atg8 is considered as autophagosome marker and is known to present on both the sides of the phagophore, the precursor to the autophagosome. After completion of autophagosome formation, Atg8 present on the surface is removed, whereas Atg8 present on the inner side is carried to the vacuole and degraded after lysis of the autophagic body. Hence, monitoring of free GFP processed from GFP-Atg8 fusion directly measures the level of autophagy (47). After 8 h of autophagy induction in nitrogen starvation medium, we could see that free GFP band was much less intense in pil1Δ. In contrast, it was more intense in Pil1 overexpression cells when compared to WT (Fig. 3C). atg1Δ is used as a control. These results suggest that Pil1 is also required for bulk autophagy. However, the mechanism behind its role in the processes of selective and nonselective autophagy needs to be studied in more detail.
Pil1 interacts with Fis1 and Dnm1
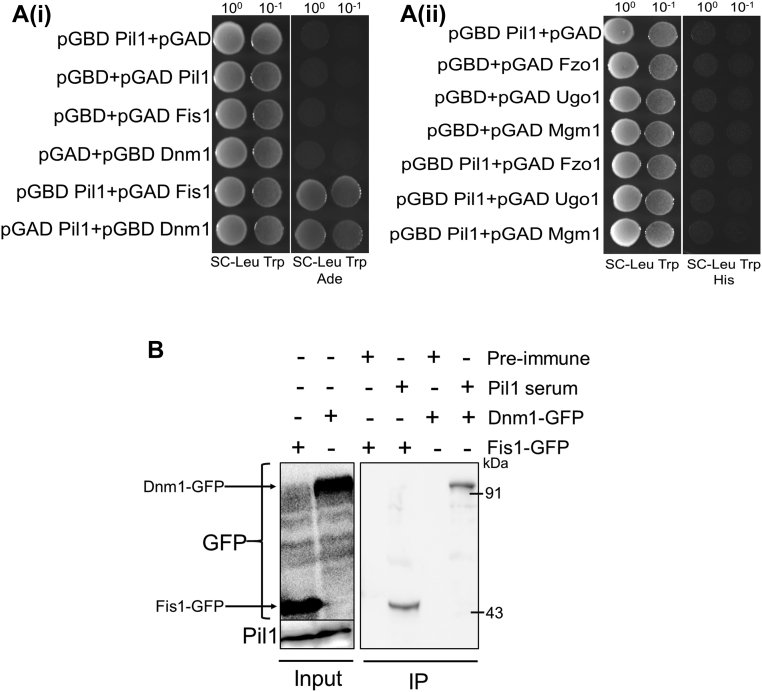
Role of Fis1 and Dnm1 in mitochondrial fission is very well established. but their role in mitophagy is unclear. According to our data, Fis1 is required for mitophagy, but not Dnm1. However, role of Pil1 in mitophagy is not dependent on these proteins. But there is a possibility that these mitochondrial fission proteins regulate the process of mitophagy indirectly through involvement of other factors. Intrigued by the change in mitochondrial morphology induced by Pil1 and its role in mitophagy, we extended our analysis to find out if Pil1 interacts with Fis1 and Dnm1. We performed yeast two-hybrid assay and found that Pil1 strongly interacts with both the proteins [Fig. 4A(i)]. Since in yeast two-hybrid analysis, there is a possibility of nonspecific interactions with proteins having hydrophobic domains, we also checked interaction of Pil1 with mitochondrial fusion proteins Fzo1, Ugo1, and Mgm1 but did not find even weak interaction [Fig. 4A(ii)]. To confirm the interaction of Pil1 with Fis1 and Dnm1, we expressed Fis1 and Dnm1 in pUG35-GFP vector under MET25 promoter and immunoprecipitated with Pil1 antibody. We find that Pil1 specifically pulled down Fis1-GFP and Dnm1-GFP (Fig. 4B). Based on these data, we conclude that Pil1 physically interacts with major components of mitochondrial fission machinery, Fis1 and Dnm1.
Figure 4.
Pil1 interacts with mitochondrial fission protein Fis1 and Dnm1.A(i), yeast two-hybrid analysis between Fis1 and Pil1 and between Dnm1 and Pil1. A(ii), yeast two-hybrid analysis of Pil1 with Fzo1, Ugo1, and Mgm1. The PJ69-4A strain was transformed with pGAD-C1 and pGBD-C1 plasmids, which can express the indicated proteins. Cells were grown on +Ade/–Ade plates for (i) and +His/–His for (ii) at 30 °C for 2 to 3 days. B, co-immunoprecipitation assays to test the interaction of Pil1 with Fis1-GFP and Dnm1-GFP. fis1Δ cells expressing Fis1-GFP and dnm1Δ cells expressing Dnm1-GFP were grown in SMD (SCD lacking uracil and methionine) media until log phase. Anti-Pil1 serum was used to precipitate Fis1-GFP and Dnm1-GFP in respective samples. Preimmune serum was used as negative control. Western blotting was done using anti-Pil1 and anti-GFP antibodies. Both the experiments were performed twice. SCD, synthetic complete dextrose medium; SMD, synthetic minimal medium.
Absence of Pil1 leads to elevated ROS levels and protein aggregation
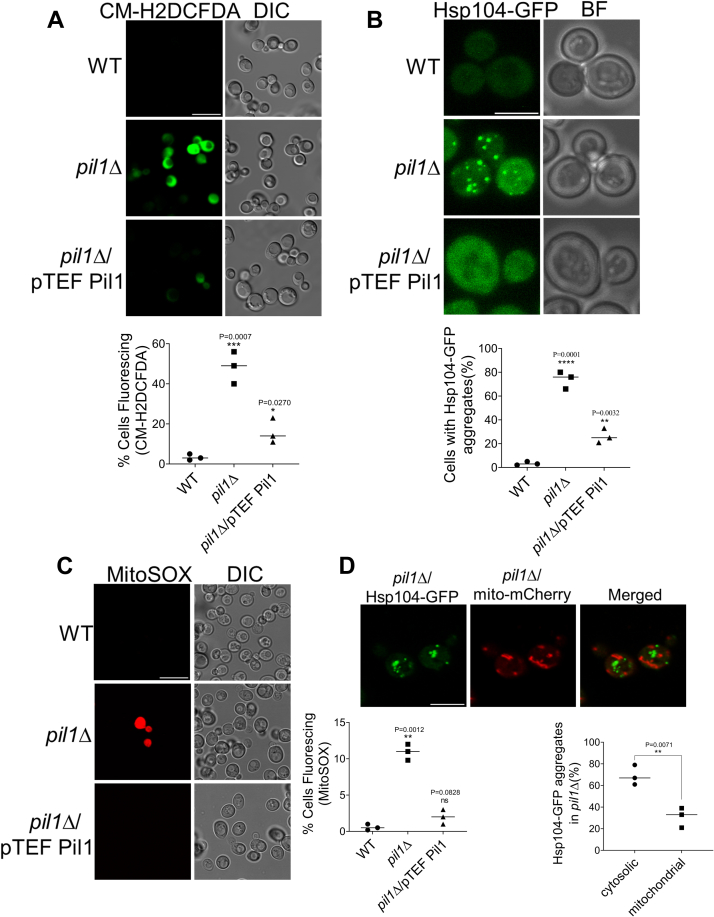
Because altered mitochondrial network and autophagy dysregulation are linked with abnormal ROS levels and protein aggregation in the cell (48, 49, 50), we next investigated if Pil1 regulates ROS levels and protein aggregation. To measure ROS levels, we used CM-H2DCFDA, a fluorescent ROS indicator. We found that ROS levels were significantly high in pil1Δ cells compared to WT (Fig. 5A). In order to study protein aggregation, we used Hsp104, endogenously tagged at the C terminus with GFP. Hsp104 is a hexameric AAA+ ATPase based chaperone in yeast, a component of disaggregase machinery which is known to bind with protein aggregates and cause their dissolution (51, 52, 53, 54, 55). Brightness intensity, size, and number of Hsp104-GFP foci increase under the abundance of protein aggregates. Based on the images obtained using confocal microscopy, we quantified the number of Hsp104-GFP aggregates. Surprisingly, we found that Hsp104-GFP foci are much more abundant in pil1Δ, whereas it was negligible in WT cells (Fig. 5B). However, cells with Pil1 overexpression also showed moderate elevation in Hsp-104 aggregates and ROS levels compared to WT, which could be attributed to the fragmented mitochondrial network upon Pil1 overexpression (56, 57).
Figure 5.
Absence of Pil1 leads to elevated ROS and protein aggregates.A, measurement of cellular ROS. Indicated strains were grown to log phase in SMD media (SCD lacking uracil) and 1 A cells were treated with 10 mM CM-H2DCFDA for 30 min at 30 °C. Imaging by fluorescence microscopy was performed as described in Experimental procedures. Quantification represents the percentage of DCF-positive fluorescing cells, where a minimum of 100 cells for each strain were counted. Scale bar represents 10 μm. B, analysis of Hsp104-GFP aggregates. Indicated strains endogenously expressing HSP104-GFP were grown till log phase, and imaging was performed by fluorescence microscopy as described in Experimental procedures. Represented images are maximum projections of z-stacks. Cells containing at least two Hsp104-GFP foci were counted in each strain. Minimum of 100 cells in three independent trials were counted, and data are presented as mean ± SD. Scale bar represents 5 μm. C, measurement of mitochondrial ROS (mtROS). Indicated strains were grown to log phase in SMD media (lacking uracil) and treated with 5 μM MitoSOX Red for 10 min at 30 °C before imaging by fluorescence microscopy. Quantification represents the percentage of MitoSOX-positive fluorescing cells, where a minimum of 100 cells for each strain were counted. Data are presented as mean ± SD of three independent experiments. Scale bar represents 10 μm. D, analysis of mitochondrial-associated Hsp104-GFP aggregates in pil1Δ. HSP104-GFP pil1Δ strain transformed with mito-mCherry and grown till log phase in SMD media. Imaging was performed by fluorescence microscopy, and represented images are flattened z-stacks. Scale bar represents 5 μm. Ratio of Hsp104-GFP foci localized with cytosol or mitochondria to total Hsp104-GFP foci was calculated as percentage. Data are presented as mean ± SD of three independent experiments. Statistical analysis was done using unpaired Student’s t test (p-value is indicated on the graph) ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, and ∗p < 0.05 versus WT. BF, brightfield; DIC, differential interference contrast; ROS, reactive oxygen species; SCD, synthetic complete dextrose media; SMD, synthetic minimal media.
Mitochondria are the major source of ROS production in cells. Because we found that the absence of Pil1 leads to abnormal mitochondrial morphology and reduced mitophagy, we decided to check if ROS and protein aggregates that we have observed in pil1Δ are associated with mitochondria. Using MitoSOX red dye to measure mitochondrial ROS, we found a small but statistically significant increase (10.9%) in mitochondrial ROS between WT and pil1Δ (Fig. 5C) which could be due to the defective mitochondrial morphology and mitophagy in pil1Δ. Using confocal microscopy, we analyzed the number of Hsp104-GFP aggregates associated with mitochondria in pil1Δ cells. We found that mitochondrial-associated aggregates (33%) in pil1Δ cells are much less than those of cytosolic aggregates (67%) (Fig. 5D). Our data suggest that Pil1-depleted cells have higher levels of ROS and protein aggregates, which might be a result of decreased mitophagy/autophagy.
pil1Δ cells are more prone to cell death
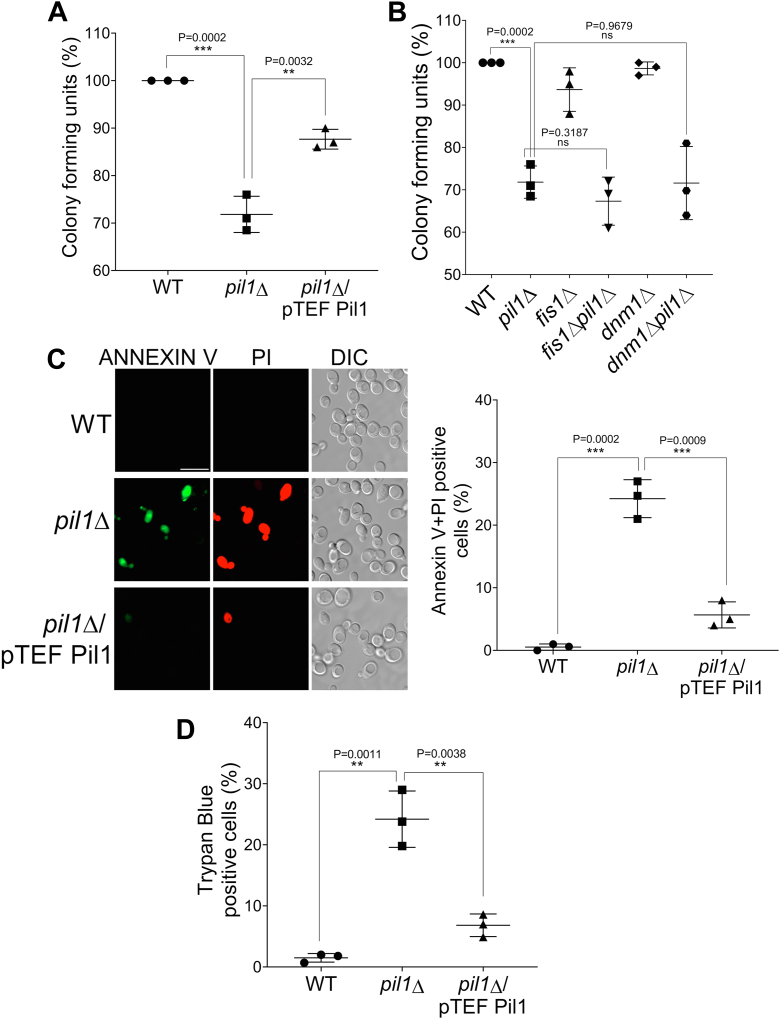
Abnormalities in mitochondria and ROS have been directly linked with cell death. We wanted to see if the higher ROS levels that we observed in pil1Δ cells make them more prone to cell death. To study viability, we applied the survival assay which is based on the ability of viable cells to form a colony (clonogenicity) on a nutrient-rich solid medium. We found a 29% reduction in colony formation in pil1Δ compared to WT which was rescued to some extent by overexpressing Pil1 (13%) (Fig. 6A). A study has well established the role of Fis1 and Dnm1 in programmed cell death induced by different kinds of death stimuli (58). However, our aim is to check if role of Pil1 in cell death under normal conditions is dependent on Dnm1 and Fis1. To this end, we applied colony survival assay to fis1Δ, dnm1Δ, fis1Δpil1Δ, and dnm1Δpil1Δ cells in the absence of any death stimuli. We found that there was a similar pattern of reduction in colony forming units in fis1Δpil1Δ (31%) and dnm1Δpil1Δ (30.2%) cells as in pil1Δ, indicating that they are more prone to cell death compared to WT (Fig. 6B).
Figure 6.
Pil1 regulates cell death.A and B, indicated strains were grown till log phase in SMD (in A) or SCD (in B), equalized to optical density at A600=0.5/ml, serially diluted, and then spotted onto agar plates. Plates were incubated at 30 °C for 2 days and colonies were counted. C, viability was assessed by annexin V/PI co-staining. annexin V+/PI+ yeast cells referred to as late apoptotic or early necrotic cells were counted and presented as mean ± SD for approximately 100 cells per sample in two biological replicates. Scale bar represents 10 μm. D, cell viability assay with trypan blue. Indicated cells were grown till log phase in SMD media, and 10 μl cells were mixed with 10 μl 0.4% trypan blue solution and number of cells trypan blue-positive cells were counted. Data are presented as mean ± SD of three independent assays. Student’s t test was used to calculate p-values (as indicated on the graph) ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, and ∗p < 0.05. DIC, differential interference contrast; SCD, synthetic complete dextrose medium; SMD, synthetic minimal medium with dextrose.
To determine the mode of cell death, we performed annexin V/PI co-staining. Externalization of phosphatidylserine on the surface of plasma membrane is one of the characteristics of cells which are in the early to mid-stage of apoptosis. Annexin V binds to the exposed phosphatidylserine on the surface of apoptotic cells, whereas PI stains necrotic cells which have lost the plasma membrane integrity. Cells that are co-stained with annexin V and PI are considered either in late apoptosis or early necrosis stage. We found that 24.7% of pil1Δ cells were Annexin V+/PI+ which was significantly reduced upon Pil1 overexpression (5%). Alternatively, we used trypan blue exclusion method. This dye penetrates the plasma membrane and stains the dead cells blue. The results that we obtained were consistent with the annexin V/PI staining assay (Fig. 6D). Thus, the absence of Pil1 triggers probably necrotic cell death. Taken together, our data suggest that absence of Pil1 leads to more cell death, but the pathway could be distinct from Fis1 and Dnm1-mediated cell death pathways.
Discussion
Mitochondrial homeostasis is known to be maintained in the cell with the help of various other organelles. Several studies have shown that mitochondrial dynamics and morphology is known to be sustained through communication with other cellular structures like endoplasmic reticulum (59), lysosomes (60), Golgi apparatus (61) and actin cytoskeleton (62, 63, 64, 65, 66, 67, 68). Mitochondrial fission in yeast is known to be executed by four major proteins—Dnm1, Fis1, Mdv1, and Caf4. Other accessory components that have also been shown to be involved in mitochondrial fission (69, 70). Given the complexity of the process, there is no denying the fact that unknown factors in the mitochondrial fission machinery are yet to be explored. Recent advances in the study of eisosome functions show that Pil1, a major eisosome complex protein, might be associated with cellular stress responses. There is also a possibility that Pil1 is dynamic in nature and does not remain confined to eisosomes (38). In this study, we have shown the evidence that Pil1 is present on mitochondrial outer membrane and maintains the mitochondrial shape. We have observed a drastic modification in mitochondrial morphology in both Pil1-deletion and Pil1-overexpression. pil1Δ cells have abnormally fused or spherical or aggregated mitochondria, while cells with Pil1 overexpression have fragmented mitochondria. However, change in mitochondrial morphology in Pil1 overexpression and deletion cannot be caused in the absence of Fis1 or Dnm1 which strengthens the fact that both these proteins are the major player in mitochondrial fission.
It is clear from emerging evidences that mitochondrial dynamics and mitophagy are integrated processes that ensure the quality control of mitochondria. However, the role of canonical fission machinery in mitophagy still remains ambiguous. Though mitochondrial dynamics was shown to modulate mitophagy in some previous reports (71, 72, 73), there are studies which show that mitophagy could be independent of these proteins (74, 75). According to our data, fis1Δ is defective in mitophagy while dnm1Δ is not, which contrasts with some of the previous findings. Though the exact reason behind these discrepancies are not known, there is a possibility that the change in phenotype that we observed could be due to differential expression of suppressors in the yeast deletion strains (76). Another possibility that we speculate is that the role of Fis1 and Dnm1 in mitophagy could be growth-phase dependent, hence there is an inconsistency in the phenotype in different studies. We found that absence of Pil1 reduces mitophagy significantly and overexpression enhances it irrespective of the presence or absence of Fis1/Dnm1. Our results also show that overexpression of Pil1 complements the defective mitophagy in fis1Δ cells. This study clearly shows that Pil1 regulates mitophagy pathway independent of Fis1 and Dnm1.
Mitochondrial dysfunction and defective autophagy are responsible for various irregularities in cell, mainly the accumulation of ROS and protein aggregates. Absence of Pil1 leads to accumulation of ROS and protein aggregates which are majorly cytosolic. However, we cannot completely rule out the possibility that mitochondria could be contributing to the elevated ROS levels that we observed in pil1Δ because the link between ROS and mitochondrial dysfunctions is a rather complex phenomenon. Mitochondrial dysfunctions need not necessarily increase ROS in the organelle itself but can trigger the ROS production in other organelles (77, 78). ROS release from mitochondria in order to maintain its health has also been reported (79). Though Pil1 interacts with Fis1 and Dnm1, the change in mitochondrial morphology caused by Pil1 are not seen in the absence of these proteins. On the other hand, Pil1 induces mitophagy independent of Fis1 and Dnm1. Also, there are irregularities in the shape of mitochondria when Pil1 is absent. It could be possible that aberrant mitophagy/autophagy leads to increased levels of cellular ROS and altered mitochondrial morphology.
More than nearly two decades ago, S. cerevisiae was shown to undergo apoptosis which opened the possibility to study this mode of cell death in a model organism that has a combined technical and logistic simplicity (80, 81). Our data show that cells undergo necrotic cell death in the absence of Pil1, even without any death stimuli which can be attributed to accumulated cellular ROS and protein aggregates. Also, the cell death pathway induced in the absence of Pil1 is clearly not dependent on already reported Fis1-mediated cell survival or Dnm1-mediated programmed cell death.
Several studies have established a functional link between mitochondrial fission, mitophagy, and cell death. However, there is a lack of sufficient study regarding the regulation of these processes, and our study depicts that Pil1 could be a potential regulator of these processes. Though in our study, we have reported the novel functions of eisosome protein Pil1 in mitophagy and cell death, further studies are required to explore how eisosome components interact with other cellular structures.
Eisosomes are present only in a few fungal species, and Pil1 is obviously not conserved beyond fungal kingdom. However, its ability to bind PI4,5P2 and promote membrane curvature are in common with those of mammalian BAR-domain family proteins (82, 83). Interestingly, recent studies have suggested potential similarities between MCC/eisosome domains and caveolae, a type of membrane invagination present in mammalian cells (33, 84). Role of BAR-domain proteins in modulating mitochondrial membrane dynamics is largely unknown. Presence of BAR-domain protein Pil1 on mitochondrial outer membrane and its potential role in mitophagy and maintaining mitochondrial shape opens up the possibility that counterpart of Pil1 is present in mammals with functional similarities.
Experimental procedures
Strains and plasmids
The yeast strains used in this study are derivatives of BY4741 or BY4742 and are listed in Table 1. PCR-based targeted homologous recombination was used to replace complete open reading frame of PIL1 with KanMX6 or HIS3MX6 cassettes, as indicated (85). All plasmids and primers used in this work are listed in Tables 2 and 3, respectively. For the construction of p426TEF Pil1-FLAG, complete coding sequences of yeast PIL1 was amplified using yeast genomic DNA as a template and the primer pairs NB754/NB755. The PCR amplified product was digested with EcoRI/HindIII and ligated with pNB270 vector as described previously (86) to replace MXR1 gene with PIL1 to generate pNB441. Pil1-FLAG was subcloned from pNB441 in p425TEF vector at BamHI/XhoI sites to generate pNB656. For yeast two-hybrid vectors pGBD-Pil1 and pGAD-Pil1, the DNA fragment of PIL1 was PCR amplified from yeast genomic DNA using primers NB754/NB850 with EcoRI and SalI restriction sites. The double digested PCR product was ligated into the EcoRI and SalI sites of pGBD-C1 (pNB415) and pGAD-C1 (pNB422) vectors to generate pNB428 and pNB427, respectively. DNA fragments encoding Fis1 and Dnm1 were PCR amplified from yeast genomic DNA with primer pairs NB1149/NB1150 and NB1151/NB1152, respectively. PCR products were digested with BamHI/SalI and ligated into pGAD-C1 and pGBD-C1 to generate pGAD-Fis1 (pNB526) and pGBD-Dnm1 (pNB525), respectively. The same PCR products were also ligated into the BamHI and SalI sites of pUG35-GFP vector to generate pUG35 Fis1-GFP (pNB629) and pUG35 Dnm1-GFP (pNB630) constructs. For cloning of pGAD-C1 Mgm1 (pNB530), pGAD-C1 Fzo1 (pNB813), and pGAD-C1 Ugo1 (pNB531) vectors, MGM1, FZO1, and UGO1 were PCR amplified from yeast genomic DNA using primer pairs NB1195/NB1196, NB1197/NB1198, and NB1199/NB1200, respectively. PCR products of FZO1 and UGO1 were digested with BamHI/SalI and that of MGM1 was digested with XmaI/BglII. The double digested inserts were ligated into the respective sites of pGAD-C1.
Table 1.
Strains
| Strain | Genotype | Reference |
|---|---|---|
| YNB105 | BY4741; MATa his3Δ1; leu2Δ0; met15Δ0; ura3Δ | Euroscarf |
| YNB106 | BY4742; MATa his3Δ1; leu2Δ0; met15Δ0; ura3Δ | Euroscarf |
| YNB 339 | PJ469-A | (92) |
| YNB 263 | BY4741; PIL1-6XMYC::LEU2 | (41) |
| YNB 474 | BY4741; pil1Δ::KanMX | This study |
| YNB 245 | BY4742; pil1Δ::KanMX | Euroscarf |
| YNB 197 | BY4742; atg32Δ::KanMX | Euroscarf |
| YNB292 | BY4742; atg1Δ::KanMX | Euroscarf |
| YNB389 | BY4741; fis1Δ::KanMX | Euroscarf |
| YNB511 | BY4742; fis1Δ::KanMX | Euroscarf |
| YNB388 | BY4741; dnm1Δ::KanMX | Euroscarf |
| YNB512 | BY4742; dnm1Δ::KanMX | Euroscarf |
| YNB 475 | BY4741; pil1Δ::HIS3 | This study |
| YNB 454 | BY4741; fis1Δ::KanMX pil1Δ::HIS3 | This study |
| YNB 473 | BY4741; dnm1Δ::KanMX pil1Δ::HIS3 | This study |
| YNB437 | BY4741; HSP104-GFP::HIS3 | (93) |
| YNB438 | BY4741; HSP104-GFP::HIS3 pil1Δ::KanMX | This study |
Table 2.
Plasmids
| Plasmid | Source/Reference | |
|---|---|---|
| pNB356 | p425 TEF | (94) |
| pNB357 | p426 TEF | (94) |
| pNB656 | p425 TEF Pil1-flag | This study |
| pNB441 | p426 TEF Pil1-flag | This study |
| pNB478 | pFA6a-HisMX6 | (84) |
| pNB479 | pFA6a-KanMX6 | (84) |
| pNB812 | pHS12 Mito-mCherry | Addgene |
| pNB415 | pGBD-C1 | (92) |
| pNB422 | pGAD-C1 | (92) |
| pNB428 | pGBD-Pil1 | This study |
| pNB427 | pGAD-Pil1 | This study |
| pNB526 | pGAD-Fis1 | This study |
| pNB525 | pGBD-Dnm1 | This study |
| pNB530 | pGAD-C1 Mgm1 | This study |
| pNB531 | pGAD-C1 Ugo1 | This study |
| pNB813 | pGAD-C1 Fzo1 | This study |
| pNB636 | pUG35 | Gueldener U, Hegemann JH. Heinrich Heine University, Germany |
| pNB629 | pUG35 Fis1-GFP | This study |
| pNB630 | pUG35 Dnm1-GFP | This study |
| pNB362 | pRS423 GFP-Atg8 | Ravi Manjithaya, JNCASR Bangalore |
| pNB449 | pRS316 Su9-DHFR-GFP | (95) |
Table 3.
Primers
| Primer name | Sequence (5′-3′) | Restriction enzyme |
|---|---|---|
| NB754 PIL1F | GCCGAATTCACCATGCACAGAACTTACTCTTTAA | EcoRI |
| NB755 PIL1R | GCCAAGCTTAGCTGTTGTTTGTTGGGGAA | HindIII |
| NB850 PIL1R | GCCGTCGACAGCTGTTGTTTGTTGGGGAA | SalI |
| NB1149 FIS1F | GCCGGATCCACCATGACCAAAGTAGATTTTT | BamHI |
| NB1150 FIS1R | GCCGTCGACCCTTCTCTTGTTTCTTAAGA | SalI |
| NB1151 DNM1F | GCCGGATCCACCATGGCTAGTTTAGAAGATCTT | BamHI |
| NB1152 DNM1R | GCCGTCGACCAGAATATTACTAATAAGGGTTG | SalI |
| NB1195 Mgm1F | GCCCCCGGGATGAATGCGAGCCCAGTACGG | XmaI |
| NB1196 Mgm1R | GCCAGATCTTAAATTTTTGGAGACGCCCTT | BglII |
| NB1197 Fzo1F | GCCGGATCCATGTCTGAAGGAAAACAACAATT | BamHI |
| NB1198 Fzo1R | GCCGTCGACATCGATGTCTAAATTTATTTCTT | SalI |
| NB1199 Ugo1F | GCCGGATCCATGAACAACAATAATGTTACGGA | BamHI |
| NB1200 Ugo1R | GCCGTCGACGAACTTCTCTTGTTCCATGTTG | SalI |
Media and growth condition
Strains were grown to log phase (A600 = approximately 1) in synthetic complete dextrose media (0.8% yeast nitrogen base without amino acids [Becton Dickinson, 291940], 2% dextrose [Himedia, GRM077], pH 5.5) or synthetic minimal media (SMD; 0.67% yeast nitrogen base, 2% glucose, and auxotrophic amino acids and vitamins as needed) as described previously (87). For mitochondria proliferation, cells were grown in lactate medium (YPL; 1% yeast extract, 2% peptone, and 2% lactate) or synthetic minimal medium with lactate (0.67% yeast nitrogen base, 2% lactate, and auxotrophic amino acids and vitamins as needed). Autophagy/mitophagy was induced by shifting the cells to nitrogen starvation medium with glucose (0.17% yeast nitrogen base without ammonium sulfate or amino acids [Becton Dickinson, 233520], and 2% dextrose). Yeast cells were grown at 30 °C. Yeast transformations were performed using lithium acetate method as previously described (88).
Mitochondrial fractionation
Mitochondria isolation was done as described previously (89, 90, 91). In brief, yeast strains were grown in YPL medium at 30 °C, and cells were harvested when the cultures reached A600 of 1 or 2. Cultures were centrifuged at 5000 rpm for 5 min, and the cell pellets were washed with autoclaved distilled water. The cells were treated with 10 mM dithiothreitol (DTT; Sigma, D0632) in 0.1 M Tris-SO4 buffer, pH 9.4, for 15 min and centrifuged at 5000 rpm for 5 min. Cells were converted to spheroplasts by using Zymolyase (20T, USBiologicals, Z1000) in 1.2 M sorbitol/20 mM phosphate buffer, pH 7.0. After obtaining 50% lysis of cells (lysis correlated to a decrease in A600 of 100 μl of cells in 900 μl of water), the resulting spheroplasts were gently washed with 1.2 M sorbitol two or three times. The spheroplast pellets were resuspended in SEM buffer (250 mM sucrose [Amresco,0335], 1 mM EDTA [Himedia,GRM1195], 10 mM 3-(N-morpholino) propanesulfonic acid [MOPS; Sigma, M1254], 1 mM phenylmethylsulfonyl fluoride [Himedia, RM1592], 0.2% bovine serum albumin [BSA; Amresco, 0332], pH 7.0) and homogenized using a Dounce homogenizer (15 times). The homogenates were centrifuged at 3500 rpm for 5 min. The supernatants were collected and centrifuged at 10,000 rpm for 10 min. The resultant pellets were resuspended in SEM buffer, and the supernatant was collected (postmitochondrial supernatant) and centrifuged at 3500 rpm for 5 min. The supernatants were once again centrifuged at 10,000 rpm for 10 min, and pellets were washed three times in SEM buffer. The crude mitochondria were resuspended in SEM buffer (without bovine serum albumin) and were subjected to 10 to 15 strokes with a glass-Teflon potter and loaded onto a three-step sucrose gradient (60% 1.5 ml, 32% 4 ml, 23% 1.5 ml, 15% 1.5 ml sucrose in EM-buffer) and centrifuged for 1 h at 134,000g, yielding mitochondrial fraction at the 60 to 32% sucrose-interface. We repeated the above step to obtain a highly purified mitochondrial fraction.
Trypsin digestion and salt extraction
For trypsin digestion, 50 μg of intact mitochondria were taken in 50 μl of SEM buffer and treated with 1 μg, 2 μg, and 5 μg (final concentration) of trypsin (Amresco, 0785) on ice for 15 min. Protease treatment was stopped by adding 5 μg, 10 μg, and 25 μg (final concentration) soybean trypsin inhibitor (Amresco, K213), respectively, and incubated on ice for 15 min. Mitochondria were pelleted down at 14,000g for 10 min at 4 °C.
For high-salt treatment, 50 μg mitochondria in 50 μl of SEM buffer were incubated with 400 mM KCl on ice for 10 min. Supernatant and pellet were separated by centrifugation at 14,000 rpm for 15 min. For high pH treatment, mitochondria were incubated with 200 mM Na2CO3 (pH 11.5) for 15 min followed by centrifugation to separate the pellet and supernatant.
Whole cell extraction and immunoblotting
Yeast cells (equivalent to A600 2-4) collected for the analysis were resuspended in 1 ml of water, spinned down at 13,000 rpm for 1 min, and the supernatant was discarded. Cells were resuspended in 160 μl of freshly prepared 1.85 M NaOH [Himedia, MB095], 7.4% β-mercaptoethanol [Sigma, M3148], mixed, and incubated on ice for 10 min. 160 μl of 50% TCA [Himedia,GRM6274] was added, mixed, and allowed to incubate on ice for 10 min. Cells were centrifuged at 13,000 rpm for 2 min, and the pellet was washed (not resuspended) with 500 μl of 1 M Tris base (not pH adjusted) (Sigma, T6066). The pellet was resuspended in 50 μl of SDS-PAGE loading buffer and heated at 95 °C for 5 min. After cooling down, cells were centrifuged at 13,000 rpm for 5 min, and the supernatant was collected. Ten microliter of the supernatant was loaded into SDS-PAGE gel (10–12%). After separation in SDS-PAGE, proteins were transferred onto nitrocellulose membranes (PALL, 66485) and blocked with 5% skimmed milk (Himedia, GRM1254) in Tris-buffered saline. The commercial monoclonal antibodies anti-GFP (ab183734, Abcam, 1:5000) and anti-Myc (ab9106, Abcam, 1:5000) were used to detect GFP or Myc epitope-fused protein on immunoblots. Anti-Dpm1 antibody (A-6429) was purchased from Invitrogen. Anti-Por1, Pgk1, Tom70, and Tom40 serums were kind gifts from Debkumar Pain (New Jersey Medical School, Rutgers University). Anti-Pdi1 antibody (ab4644, Abcam, 1:10,000) was a kind gift from Krishnaveni Mishra, University of Hyderabad. Anti-Pma1 antibody (sc-33735) was a kind gift from Rupinder Kaur, Centre for DNA Fingerprinting and Diagnostics, Hyderabad. Anti-Pil1 serum (Anti-rabbit, 1:3000) was generated in the laboratory using full-length Pil1-6XHis protein as immunogen. In the procedure of raising antibody, all the required guidelines were followed which were approved by the Institutional Animal Ethics Committee, University of Hyderabad (UH/IAEC/NS/2014-I/25). HRP-conjugated anti-rabbit (Jackson ImmunoResearch Laboratories, 111-035-144, 1:25,000) or anti-mouse (Jackson ImmunoResearch Laboratories, 115-035-146, 1:25,000) polyclonal secondary antibodies were used, followed by the detection of signal using ECL reagents (Advansta WesternBright, K-12045-D20) and imaging in Chemidoc Imaging System (Bio-Rad). Quantification of Western Blot images was done using Fiji software. Graphs and statistical analysis were done using GraphPad Prism8.
Co-immunoprecipitation
For Co-IP assays, cells were collected and lysed with glass beads (Sigma, G8772) in lysis buffer (20 mM Tris-HCL [pH 8.0], 100 mM NaCl [Himedia, GRM031], 1 mM EDTA and 0.5% Nonidet P-40[Sigma,I8896], protease inhibitor cocktail [Roche, 04693132001]). After bead-beating, cells were centrifuged at 12,000 rpm for 10 min. Supernatant was collected and incubated with anti-Pil1 serum (and preimmune serum as control) for 12 h at 4 °C in an end-over-end rotor. Thirty microliter of protein A/G PLUS-Agarose beads (SCBT, SC-2003)) were added and incubated for 2 h at 4 °C in an end-over-end rotor. Immunocomplexes were then washed three times with lysis buffer and eluted by SDS-PAGE loading buffer. The proteins were separated by SDS-PAGE and immunoblotted with anti-GFP antibody and anti-Pil1 serum.
Fluorescence microscopy
Cells grown till A600= approximately 1 were harvested by centrifugation, and the pellet was washed twice with fresh SMD media. Cells suspended in SMD media were added to slides coated with 0.1% Concanavalin A (Sigma, C2010) solution. The cells were allowed to settle for 2 to 3 min and mounted with coverslips. Images were acquired either on Leica TCS SP8 (HC PL APO CS2 63X/1.40 OIL objective) or Zeiss LSM NLO 710 (63×/1.5 OIL objective) confocal microscope. Fiji Software was used to process and analyze the microscopy data. Graphs and statistical analysis were done using GraphPad Prism8.
Measurement of ROS
For visualization of cellular ROS via fluorescence microscopy, cells growing in SMD medium were harvested by centrifugation (1 A600 cells), washed with PBS twice and then treated with 10 mM 5(6)-carboxy-2′7′-dichlorofluorescin diacetate (CM-H2DCFDA; Invitrogen, C6827) in 500 μl PBS for 30 min at 30 °C. Similarly, for measurement of mitochondrial ROS, cells (1 A600) were treated with 5 μM MitoSOX Red (Invitrogen, M36008) for 10 min at 30 °C in 500 μl of PBS.
Cell viability assays
Cells were grown in SMD media until A600 reached approximately 1. For colony-forming unit assays, cells were normalized to A600 = 0.5/ml, and 200 μl of each culture was collected into 96-well plates. Serial dilutions were done using multichannel pipettor, and 10 μl were plated onto SMD solid media. Plates were incubated for 2 days at 30 °C, and number of colonies were counted. Annexin V/PI co-staining assays using ApoAlert Annexin V-FITC Apoptosis kit (Takara, 630109) were performed as described previously (92). For trypan blue staining, 10 μl of cell suspension in PBS were mixed with 0.4% trypan blue (Gibco, 15250061) for 2 to 3 min at room temperature, and number of cells stained with the dye were counted.
Data availability
All data are contained within the manuscript.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Authors thank E.A. Craig (Washington University School of Medicine) for yeast-two-hybrid strains and plasmids, Victor J Cid (Complutense University of Madrid, Madrid) for Pil1-Myc strain, Krishnaveni Mishra (Department of Biochemistry, University of Hyderabad) for yeast deletion strains and anti-Pdi1 antibody, Shirisha Nagotu (IIT Guwahati) for providing pHS12 Mito-mCherry plasmid, Ravi Manjithaya (Jawaharlal Nehru Centre for Advanced Scientific Research, Bangalore) for pRS423 GFP-Atg8 plasmid, Liu Beidong (University of Gothenburg, Gotenborg, Sweden) for Hsp104-GFP strain, Jared Rutter (University of Utah School of Medicine, Salt Lake City) for pRS316 Su9-DHFR-GFP plasmid, Rupinder Kaur (Centre for DNA Fingerprinting and Diagnostics, Hyderabad) for anti-Pma1 antibody. Authors also thank Debkumar Pain (New Jersey Medical School, Rutgers University, Newark, NJ) for anti-Por1, Pgk1, Tom70, and Tom40 serums. We thank Prasad Tammineni for providing assistance in confocal microscopy.
Author contributions
A. P. and N. B. V. S. conceptualization; A. P. and N. B. V. S. methodology; A. P. and A. K. P. investigation; A. P. visualization; A. P. formal analysis; A. P. and N. B. V. S. writing-original draft; A. K. P., P. D., and N. B. V. S. writing-review and editing; P. D., A. C., P. R. P., and P. A. validation; N. B. V. S. supervision; N. B. V. S. funding acquisition.
Funding and additional information
This work was supported by funding from Department of Biotechnology (BUILDER-DBT-BT/INF/22/SP41176/2020), and Institute of Eminence, University of Hyderabad (UoH-IoE-RC3-21-010) to N. B. V. S. Lab. A. P., A. K. P., P. D., A. C., and P. A. acknowledge the Senior Research Fellowship (SRF) & Junior Research Fellowship (JRF) from Council of Scientific and Industrial Research (CSIR), India.
Edited by Ursula Jakob
References
- 1.Fischer F., Hamann A., Osiewacz H.D. Mitochondrial quality control: an integrated network of pathways. Trends Biochem. Sci. 2012;37:284–292. doi: 10.1016/j.tibs.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Gomes L.C., Di Benedetto G., Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackstone C., Chang C.R. Mitochondria unite to survive. Nat. Cell Biol. 2011;13:521–522. doi: 10.1038/ncb0511-521. [DOI] [PubMed] [Google Scholar]
- 4.Burman J.L., Pickles S., Wang C., Sekine S., Vargas J.N.S., Zhang Z., et al. Mitochondrial fission facilitates the selective mitophagy of protein aggregates. J. Cell Biol. 2017;216:3231–3247. doi: 10.1083/jcb.201612106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Youle R.J., van der Bliek A.M. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bockler S., Chelius X., Hock N., Klecker T., Wolter M., Weiss M., et al. Fusion, fission, and transport control asymmetric inheritance of mitochondria and protein aggregates. J. Cell Biol. 2017;216:2481–2498. doi: 10.1083/jcb.201611197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong E.D., Wagner J.A., Gorsich S.W., McCaffery J.M., Shaw J.M., Nunnari J. The dynamin-related GTPase, Mgm1p, is an intermembrane space protein required for maintenance of fusion competent mitochondria. J. Cell Biol. 2000;151:341–352. doi: 10.1083/jcb.151.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sesaki H., Jensen R.E. UGO1 encodes an outer membrane protein required for mitochondrial fusion. J. Cell Biol. 2001;152:1123–1134. doi: 10.1083/jcb.152.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hermann G.J., Thatcher J.W., Mills J.P., Hales K.G., Fuller M.T., Nunnari J., et al. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 1998;143:359–373. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bleazard W., McCaffery J.M., King E.J., Bale S., Mozdy A., Tieu Q., et al. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otsuga D., Keegan B.R., Brisch E., Thatcher J.W., Hermann G.J., Bleazard W., et al. The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. J. Cell Biol. 1998;143:333–349. doi: 10.1083/jcb.143.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mozdy A.D., McCaffery J.M., Shaw J.M. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 2000;151:367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tieu Q., Nunnari J. Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J. Cell Biol. 2000;151:353–366. doi: 10.1083/jcb.151.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffin E.E., Graumann J., Chan D.C. The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria. J. Cell Biol. 2005;170:237–248. doi: 10.1083/jcb.200503148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Legesse-Miller A., Massol R.H., Kirchhausen T. Constriction and Dnm1p recruitment are distinct processes in mitochondrial fission. Mol. Biol. Cell. 2003;14:1953–1963. doi: 10.1091/mbc.E02-10-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho D.H., Nakamura T., Fang J., Cieplak P., Godzik A., Gu Z., et al. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324:102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shirendeb U.P., Calkins M.J., Manczak M., Anekonda V., Dufour B., McBride J.L., et al. Mutant huntingtin’s interaction with mitochondrial protein Drp1 impairs mitochondrial biogenesis and causes defective axonal transport and synaptic degeneration in Huntington’s disease. Hum. Mol. Genet. 2012;21:406–420. doi: 10.1093/hmg/ddr475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narendra D., Tanaka A., Suen D.F., Youle R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding W.X., Yin X.M. Mitophagy: mechanisms, pathophysiological roles, and analysis. Biol. Chem. 2012;393:547–564. doi: 10.1515/hsz-2012-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanki T., Furukawa K., Yamashita S. Mitophagy in yeast: molecular mechanisms and physiological role. Biochim. Biophys. Acta. 2015;1853:2756–2765. doi: 10.1016/j.bbamcr.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 21.Liu L., Sakakibara K., Chen Q., Okamoto K. Receptor-mediated mitophagy in yeast and mammalian systems. Cell Res. 2014;24:787–795. doi: 10.1038/cr.2014.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malinsky J., Opekarova M., Tanner W. The lateral compartmentation of the yeast plasma membrane. Yeast. 2010;27:473–478. doi: 10.1002/yea.1772. [DOI] [PubMed] [Google Scholar]
- 23.Ziolkowska N.E., Christiano R., Walther T.C. Organized living: formation mechanisms and functions of plasma membrane domains in yeast. Trends Cell Biol. 2012;22:151–158. doi: 10.1016/j.tcb.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Malinska K., Malinsky J., Opekarova M., Tanner W. Visualization of protein compartmentation within the plasma membrane of living yeast cells. Mol. Biol. Cell. 2003;14:4427–4436. doi: 10.1091/mbc.E03-04-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malinska K., Malinsky J., Opekarova M., Tanner W. Distribution of Can1p into stable domains reflects lateral protein segregation within the plasma membrane of living S. cerevisiae cells. J. Cell Sci. 2004;117:6031–6041. doi: 10.1242/jcs.01493. [DOI] [PubMed] [Google Scholar]
- 26.Stradalova V., Stahlschmidt W., Grossmann G., Blazikova M., Rachel R., Tanner W., et al. Furrow-like invaginations of the yeast plasma membrane correspond to membrane compartment of Can1. J. Cell Sci. 2009;122:2887–2894. doi: 10.1242/jcs.051227. [DOI] [PubMed] [Google Scholar]
- 27.Moor H., Muhlethaler K. Fine structure in frozen-etched yeast cells. J. Cell Biol. 1963;17:609–628. doi: 10.1083/jcb.17.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walther T.C., Brickner J.H., Aguilar P.S., Bernales S., Pantoja C., Walter P. Eisosomes mark static sites of endocytosis. Nature. 2006;439:998–1003. doi: 10.1038/nature04472. [DOI] [PubMed] [Google Scholar]
- 29.Lee J.H., Heuser J.E., Roth R., Goodenough U. Eisosome ultrastructure and evolution in fungi, microalgae, and lichens. Eukaryot. Cell. 2015;14:1017–1042. doi: 10.1128/EC.00106-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karotki L., Huiskonen J.T., Stefan C.J., Ziolkowska N.E., Roth R., Surma M.A., et al. Eisosome proteins assemble into a membrane scaffold. J. Cell Biol. 2011;195:889–902. doi: 10.1083/jcb.201104040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Douglas L.M., Konopka J.B. Fungal membrane organization: the eisosome concept. Annu. Rev. Microbiol. 2014;68:377–393. doi: 10.1146/annurev-micro-091313-103507. [DOI] [PubMed] [Google Scholar]
- 32.Grossmann G., Opekarova M., Malinsky J., Weig-Meckl I., Tanner W. Membrane potential governs lateral segregation of plasma membrane proteins and lipids in yeast. EMBO J. 2007;26:1–8. doi: 10.1038/sj.emboj.7601466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabeche R., Howard L., Moseley J.B. Eisosomes provide membrane reservoirs for rapid expansion of the yeast plasma membrane. J. Cell Sci. 2015;128:4057–4062. doi: 10.1242/jcs.176867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grousl T., Opekarova M., Stradalova V., Hasek J., Malinsky J. Evolutionarily conserved 5’-3’ exoribonuclease Xrn1 accumulates at plasma membrane-associated eisosomes in post-diauxic yeast. PLoS One. 2015;10 doi: 10.1371/journal.pone.0122770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gournas C., Gkionis S., Carquin M., Twyffels L., Tyteca D., Andre B. Conformation-dependent partitioning of yeast nutrient transporters into starvation-protective membrane domains. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E3145–E3154. doi: 10.1073/pnas.1719462115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moharir A., Gay L., Appadurai D., Keener J., Babst M. Eisosomes are metabolically regulated storage compartments for APC-type nutrient transporters. Mol. Biol. Cell. 2018;29:2113–2127. doi: 10.1091/mbc.E17-11-0691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amen T., Kaganovich D. Stress granules sense metabolic stress at the plasma membrane and potentiate recovery by storing active Pkc1. Sci. Signal. 2020;13 doi: 10.1126/scisignal.aaz6339. [DOI] [PubMed] [Google Scholar]
- 38.Lacy M.M., Baddeley D., Berro J. Single-molecule imaging of the BAR-domain protein Pil1p reveals filament-end dynamics. Mol. Biol. Cell. 2017;28:2251–2259. doi: 10.1091/mbc.E17-04-0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walther T.C., Aguilar P.S., Frohlich F., Chu F., Moreira K., Burlingame A.L., et al. Pkh-kinases control eisosome assembly and organization. EMBO J. 2007;26:4946–4955. doi: 10.1038/sj.emboj.7601933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo G., Gruhler A., Liu Y., Jensen O.N., Dickson R.C. The sphingolipid long-chain base-Pkh1/2-Ypk1/2 signaling pathway regulates eisosome assembly and turnover. J. Biol. Chem. 2008;283:10433–10444. doi: 10.1074/jbc.M709972200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mascaraque V., Hernaez M.L., Jimenez-Sanchez M., Hansen R., Gil C., Martin H., et al. Phosphoproteomic analysis of protein kinase C signaling in Saccharomyces cerevisiae reveals Slt2 mitogen-activated protein kinase (MAPK)-dependent phosphorylation of eisosome core components. Mol. Cell. Proteomics. 2013;12:557–574. doi: 10.1074/mcp.M112.020438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohlmeier S., Hiltunen J.K., Bergmann U. Protein phosphorylation in mitochondria --a study on fermentative and respiratory growth of Saccharomyces cerevisiae. Electrophoresis. 2010;31:2869–2881. doi: 10.1002/elps.200900759. [DOI] [PubMed] [Google Scholar]
- 43.Reinders J., Wagner K., Zahedi R.P., Stojanovski D., Eyrich B., van der Laan M., et al. Profiling phosphoproteins of yeast mitochondria reveals a role of phosphorylation in assembly of the ATP synthase. Mol. Cell. Proteomics. 2007;6:1896–1906. doi: 10.1074/mcp.M700098-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Twig G., Elorza A., Molina A.J., Mohamed H., Wikstrom J.D., Walzer G., et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanki T., Wang K., Cao Y., Baba M., Klionsky D.J. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okamoto K., Kondo-Okamoto N., Ohsumi Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell. 2009;17:87–97. doi: 10.1016/j.devcel.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 47.Shintani T., Klionsky D.J. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 2004;279:29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bhatti J.S., Bhatti G.K., Reddy P.H. Mitochondrial dysfunction and oxidative stress in metabolic disorders - a step towards mitochondria based therapeutic strategies. Biochim. Biophys. Acta Mol. Basis Dis. 2017;1863:1066–1077. doi: 10.1016/j.bbadis.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tal M.C., Sasai M., Lee H.K., Yordy B., Shadel G.S., Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc. Natl. Acad. Sci. U. S. A. 2009;106:2770–2775. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Filomeni G., De Zio D., Cecconi F. Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ. 2015;22:377–388. doi: 10.1038/cdd.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parsell D.A., Kowal A.S., Singer M.A., Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 52.Glover J.R., Lindquist S. Hsp104, Hsp70, and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 53.Bosl B., Grimminger V., Walter S. The molecular chaperone Hsp104--a molecular machine for protein disaggregation. J. Struct. Biol. 2006;156:139–148. doi: 10.1016/j.jsb.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 54.Doyle S.M., Wickner S. Hsp104 and ClpB: protein disaggregating machines. Trends Biochem. Sci. 2009;34:40–48. doi: 10.1016/j.tibs.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 55.Nillegoda N.B., Bukau B. Metazoan Hsp70-based protein disaggregases: emergence and mechanisms. Front. Mol. Biosci. 2015;2:57. doi: 10.3389/fmolb.2015.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shenouda S.M., Widlansky M.E., Chen K., Xu G., Holbrook M., Tabit C.E., et al. Altered mitochondrial dynamics contributes to endothelial dysfunction in diabetes mellitus. Circulation. 2011;124:444–453. doi: 10.1161/CIRCULATIONAHA.110.014506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu T., Robotham J.L., Yoon Y. Increased production of reactive oxygen species in hyperglycemic conditions requires dynamic change of mitochondrial morphology. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2653–2658. doi: 10.1073/pnas.0511154103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fannjiang Y., Cheng W.C., Lee S.J., Qi B., Pevsner J., McCaffery J.M., et al. Mitochondrial fission proteins regulate programmed cell death in yeast. Genes Dev. 2004;18:2785–2797. doi: 10.1101/gad.1247904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Friedman J.R., Lackner L.L., West M., DiBenedetto J.R., Nunnari J., Voeltz G.K. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wong Y.C., Ysselstein D., Krainc D. Mitochondria-lysosome contacts regulate mitochondrial fission via RAB7 GTP hydrolysis. Nature. 2018;554:382–386. doi: 10.1038/nature25486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nagashima S., Tabara L.C., Tilokani L., Paupe V., Anand H., Pogson J.H., et al. Golgi-derived PI(4)P-containing vesicles drive late steps of mitochondrial division. Science. 2020;367:1366–1371. doi: 10.1126/science.aax6089. [DOI] [PubMed] [Google Scholar]
- 62.Korobova F., Ramabhadran V., Higgs H.N. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 2013;339:464–467. doi: 10.1126/science.1228360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ji W.K., Hatch A.L., Merrill R.A., Strack S., Higgs H.N. Actin filaments target the oligomeric maturation of the dynamin GTPase Drp1 to mitochondrial fission sites. Elife. 2015;4 doi: 10.7554/eLife.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li S., Xu S., Roelofs B.A., Boyman L., Lederer W.J., Sesaki H., et al. Transient assembly of F-actin on the outer mitochondrial membrane contributes to mitochondrial fission. J. Cell Biol. 2015;208:109–123. doi: 10.1083/jcb.201404050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Manor U., Bartholomew S., Golani G., Christenson E., Kozlov M., Higgs H., et al. A mitochondria-anchored isoform of the actin-nucleating spire protein regulates mitochondrial division. Elife. 2015;4 doi: 10.7554/eLife.08828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moore A.S., Wong Y.C., Simpson C.L., Holzbaur E.L. Dynamic actin cycling through mitochondrial subpopulations locally regulates the fission-fusion balance within mitochondrial networks. Nat. Commun. 2016;7:12886. doi: 10.1038/ncomms12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee J.E., Westrate L.M., Wu H., Page C., Voeltz G.K. Multiple dynamin family members collaborate to drive mitochondrial division. Nature. 2016;540:139–143. doi: 10.1038/nature20555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen Y.C., Cheng T.H., Lin W.L., Chen C.L., Yang W.Y., Blackstone C., et al. Srv2 is a pro-fission factor that modulates yeast mitochondrial morphology and respiration by regulating actin assembly. iScience. 2019;11:305–317. doi: 10.1016/j.isci.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cerveny K.L., Studer S.L., Jensen R.E., Sesaki H. Yeast mitochondrial division and distribution require the cortical num1 protein. Dev. Cell. 2007;12:363–375. doi: 10.1016/j.devcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 70.Hammermeister M., Schodel K., Westermann B. Mdm36 is a mitochondrial fission-promoting protein in Saccharomyces cerevisiae. Mol. Biol. Cell. 2010;21:2443–2452. doi: 10.1091/mbc.E10-02-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Abeliovich H., Zarei M., Rigbolt K.T., Youle R.J., Dengjel J. Involvement of mitochondrial dynamics in the segregation of mitochondrial matrix proteins during stationary phase mitophagy. Nat. Commun. 2013;4:2789. doi: 10.1038/ncomms3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mao K., Wang K., Liu X., Klionsky D.J. The scaffold protein Atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy. Dev. Cell. 2013;26:9–18. doi: 10.1016/j.devcel.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bernhardt D., Muller M., Reichert A.S., Osiewacz H.D. Simultaneous impairment of mitochondrial fission and fusion reduces mitophagy and shortens replicative lifespan. Sci. Rep. 2015;5:7885. doi: 10.1038/srep07885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Graef M. A dividing matter: Drp1/Dnm1-independent mitophagy. J. Cell Biol. 2016;215:599–601. doi: 10.1083/jcb.201611079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mendl N., Occhipinti A., Muller M., Wild P., Dikic I., Reichert A.S. Mitophagy in yeast is independent of mitochondrial fission and requires the stress response gene WHI2. J. Cell Sci. 2011;124:1339–1350. doi: 10.1242/jcs.076406. [DOI] [PubMed] [Google Scholar]
- 76.Hawthorne D.C., Leupold U. Suppressors in yeast. Curr. Top. Microbiol. Immunol. 1974;64:1–47. doi: 10.1007/978-3-642-65848-8_1. [DOI] [PubMed] [Google Scholar]
- 77.Leadsham J.E., Sanders G., Giannaki S., Bastow E.L., Hutton R., Naeimi W.R., et al. Loss of cytochrome c oxidase promotes RAS-dependent ROS production from the ER resident NADPH oxidase, Yno1p, in yeast. Cell Metab. 2013;18:279–286. doi: 10.1016/j.cmet.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 78.Murphy M.P. Mitochondrial dysfunction indirectly elevates ROS production by the endoplasmic reticulum. Cell Metab. 2013;18:145–146. doi: 10.1016/j.cmet.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 79.Zorov D.B., Juhaszova M., Sollott S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014;94:909–950. doi: 10.1152/physrev.00026.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Madeo F., Frohlich E., Frohlich K.U. A yeast mutant showing diagnostic markers of early and late apoptosis. J. Cell Biol. 1997;139:729–734. doi: 10.1083/jcb.139.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Madeo F., Frohlich E., Ligr M., Grey M., Sigrist S.J., Wolf D.H., et al. Oxygen stress: a regulator of apoptosis in yeast. J. Cell Biol. 1999;145:757–767. doi: 10.1083/jcb.145.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Olivera-Couto A., Grana M., Harispe L., Aguilar P.S. The eisosome core is composed of BAR domain proteins. Mol. Biol. Cell. 2011;22:2360–2372. doi: 10.1091/mbc.E10-12-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zimmerberg J., McLaughlin S. Membrane curvature: how BAR domains bend bilayers. Curr. Biol. 2004;14:R250–R252. doi: 10.1016/j.cub.2004.02.060. [DOI] [PubMed] [Google Scholar]
- 84.Moreira K.E., Schuck S., Schrul B., Frohlich F., Moseley J.B., Walther T.C., et al. Seg1 controls eisosome assembly and shape. J. Cell Biol. 2012;198:405–420. doi: 10.1083/jcb.201202097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Longtine M.S., McKenzie A., 3rd, Demarini D.J., Shah N.G., Wach A., Brachat A., et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 86.Allu P.K., Marada A., Boggula Y., Karri S., Krishnamoorthy T., Sepuri N.B. Methionine sulfoxide reductase 2 reversibly regulates Mge1, a cochaperone of mitochondrial Hsp70, during oxidative stress. Mol. Biol. Cell. 2015;26:406–419. doi: 10.1091/mbc.E14-09-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 88.Gietz R.D., Woods R.A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 89.Sepuri N.B., Gorla M., King M.P. Mitochondrial lysyl-tRNA synthetase independent import of tRNA lysine into yeast mitochondria. PLoS One. 2012;7 doi: 10.1371/journal.pone.0035321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sepuri N.B., Yadav S., Anandatheerthavarada H.K., Avadhani N.G. Mitochondrial targeting of intact CYP2B1 and CYP2E1 and N-terminal truncated CYP1A1 proteins in Saccharomyces cerevisiae--role of protein kinase A in the mitochondrial targeting of CYP2E1. FEBS J. 2007;274:4615–4630. doi: 10.1111/j.1742-4658.2007.05990.x. [DOI] [PubMed] [Google Scholar]
- 91.Reinders J., Zahedi R.P., Pfanner N., Meisinger C., Sickmann A. Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J. Proteome Res. 2006;5:1543–1554. doi: 10.1021/pr050477f. [DOI] [PubMed] [Google Scholar]
- 92.Buttner S., Eisenberg T., Carmona-Gutierrez D., Ruli D., Knauer H., Ruckenstuhl C., et al. Endonuclease G regulates budding yeast life and death. Mol. Cell. 2007;25:233–246. doi: 10.1016/j.molcel.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 93.Song J., Yang Q., Yang J., Larsson L., Hao X., Zhu X., et al. Essential genetic interactors of SIR2 required for spatial sequestration and asymmetrical inheritance of protein aggregates. PLoS Genet 10. 2014:e1004539. doi: 10.1371/journal.pgen.1004539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mumberg D., Muller R., Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- 95.Heo J.-M. A stress-responsive system for mitochondrial protein degradation. Mol Cell. 2010;40:465–480. doi: 10.1016/j.molcel.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained within the manuscript.