Abstract
Background
A cardiac resting phase is used when performing free-breathing cardiac magnetic resonance examinations.
Purpose
The purpose of this study was to test a cardiac resting phase detection system based on neural networks in clinical practice.
Material and Methods
Four chamber-view cine images were obtained from 32 patients and analyzed. The rest duration, start point, and end point were compared between that determined by the experts and general operators, and a similar comparison was done between that determined by the experts and neural networks: the normalized root-mean-square error (RMSE) was also calculated.
Results
Unlike manual detection, the neural network was able to determine the resting phase almost simultaneously as the image was obtained. The rest duration and start point were not significantly different between the neural network and expert (p = .30, .90, respectively), whereas the end point was significantly different between the two groups (p < .05). The start point was not significantly different between the general operator and expert (p = .09), whereas the rest duration and end point were significantly different between the two groups (p < .05). The normalized RMSEs of the rest duration, start point, and end point of the neural network were 0.88, 0.64, and 0.33 ms, respectively, which were lower than those of the general operator (normalized RMSE values were 0.98, 0.68, and 0.51 ms, respectively).
Conclusions
The neural network can determine the resting phase instantly with better accuracy than the manual detection of general operators.
Keywords: neural network, deep learning, cardiac resting phase, magnetic resonance image
Introduction
A cardiac resting phase is determined when performing free-breathing examinations, such as coronary magnetic resonance angiography (MRA), three-dimensional (3D) late gadolinium enhancement, and 3D non-contrast T1-weighted cardiovascular magnetic resonance image (MRI), that are usually performed on the entire heart with electrocardiogram gating.1,2,3 Particularly, in coronary MRA, the 2020 update of the standardized cardiovascular magnetic resonance imaging protocols recommends using the right coronary artery (RCA) to determine the resting phase.4 In previous studies, the resting phase was also determined based on the motion of the RCA on four chamber-view cine images.5 In addition, various reports found that the coronary artery motion is often minimal during the mid-diastole.6 Another study also reported that the longest rest period occurred during mid-diastole in 74% of the patients.7 In general practice, 3D volume images of the heart are obtained to determine these cardiac rest periods and the operators decide the resting phase visually. Hence, considerable experience is needed to accurately determine the cardiac resting phase. These operations make the examination of the cardiovascular MRI complicated. Moreover, determination of the resting phase can vary depending on the experience of the operators. If a fully automated neural network can immediately determine the resting phase similar to an expert, it can shorten the examination time and reduce the burden on the patients and the operators by replacing manual detection. This study aims to assess a neural network–based resting phase detection system from the viewpoint of accuracy and decision speed in clinical patients.
Material and methods
Study population
Forty patients who consented to participate in this study were selected between December 2020 and May 2021. Among these patients, those whose resting phase could not be determined manually by experts were excluded. Exclusion criteria were determined based on RCA with reference to the 2020 update of the standardized cardiovascular magnetic resonance imaging protocols.4 This study was approved by the ethical review board of our institution and the participants provided written informed consent.
Cine image protocol
All MRI examinations were performed using a clinical 3T MRI scanner (MAGNETOM Skyra, Siemens Healthcare, Erlangen, Germany). To evaluate the motion of the RCA, a set of four chamber-view cine images was acquired using a steady-state free precession sequence with breath-holding before contrast-medium injection. The imaging parameters were as follows: 3.2/1.4 ms (repetition time/echo time), 12 number of segments, 65° flip angle, 293 × 350 mm field of view, 157 × 208 acquisition matrix, 32 cardiac phases, three acceleration factor, and 6 mm slice thickness.
Deep learning algorithm
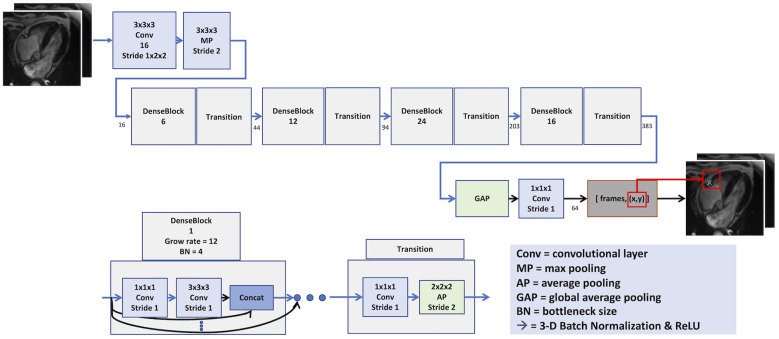
The deep learning algorithm consisted of a neural network to identify the RCA by landmark detection, followed by a quantification of the motion around the RCA through image registration. The training for the network used 1000 volunteers/patients of four chamber-view cine images that were split into the training set (70%), validation set (15%), and test set (15%). As a preprocessing step, all images were converted to a spatial resolution of 224 × 224 and 32 temporal frames. For the RCA localization network (Figure 1), a 3D-DenseNet8 was used for landmark detection. The ADAM optimizer was used to update the weight of network.9 The motion curve as shown in Figure 2 was quantitatively determined using elastic image registration.10 The period below the 0.15 threshold value was assigned as the resting phase. If the algorithm detected several resting periods, the longest period was adopted as the resting phase.
Figure 1.
Right coronary artery localization network. The detected right coronary artery is marked by white X arrows. The motion quantification uses region of interest around the right coronary artery.
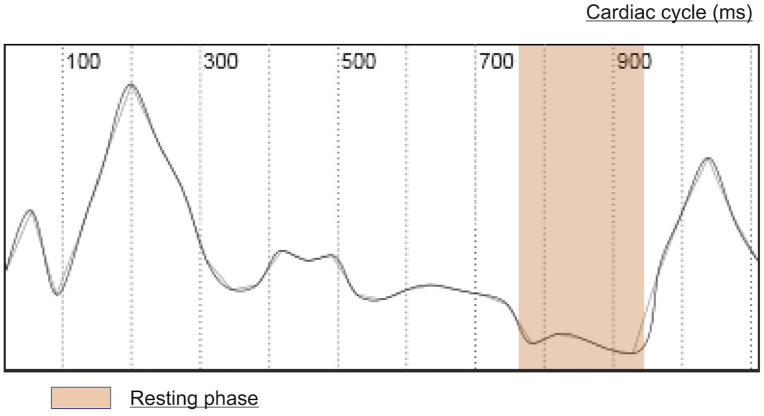
Figure 2.
Computed motion curve. The vertical axis shows the weight of the absolute mean value (mm) of the deformation vectors around the right coronary artery between the two frames. The horizontal axis represents the cardiac cycle.
Assessment of cardiac resting period
The resting phase was determined based on the motion of the RCA in the four chamber-view cine images. For resting phase detection, both the neural network–based resting phase and manual resting phase were determined. The analysis results of the neural network were checked on the MRI console. Manual detection was conducted by an experienced operator and general operator with 15 and 2 years of experience, respectively, in cardiovascular MRI examination. The resting phase was assessed based on the duration, start point, and end point. The time to decision of the neural network and manual detection were also compared.
Statistical analysis
The collected data are expressed as mean ± standard deviation or median (interquartile range). The rest duration, start time, and end time data sets collected from (1) the expert and general operator and (2) the expert and neural network were compared with each other using the Wilcoxon matched-pairs signed-rank test. Statistical significance was set at p < .05, and Dunnett’s correction was used for multiple comparisons. The normalized root-mean-square error (RMSE) between the expert and other groups was also calculated. Root-mean-square error is a common index for evaluating the performance of machine learning models because it can rigorously evaluate large errors. The normalized RMSE was calculated by dividing RMSE by the interquartile range. And additional analysis was performed by dividing the data into heart rate ≦60 (n = 16) and heart rate >60 (n = 16). The differences between the rest duration, start time, and end time data sets collected by the expert and other groups were assessed using the Bland–Altman method. All statistical analyses were conducted using the JMP software (version 11.2; SAS Institute, Inc., Cary, NC, USA).
Results
Summary of patients
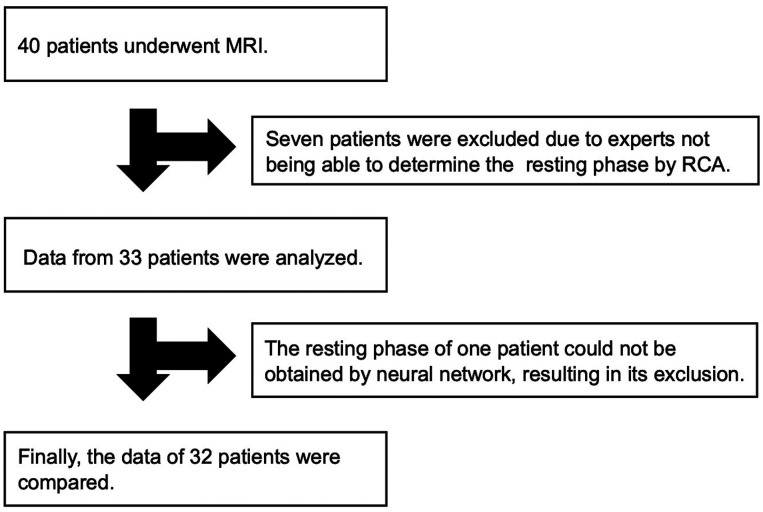
A total of 40 patients consented to participate in the study. Of these, seven patients whose RCA could not be confirmed and resting phase could not be manually determined by experts were excluded. The details of the seven patients were as follows: two had congenital heart disease, two had RCA hypoplasia, and three corresponded to poor image quality due to severe artifacts. Of 33 patients, the neural network could determine the resting phase in one patient. As a result, the final analysis was performed on 32 patients (Figure 3). Table 1 shows the detailed characteristics of the patients.
Figure 3.
Flow diagram of this study. Since this study is based on the resting phase manually determined by experts, we excluded seven cases wherein experts could not determine the resting phase by RCA. MRI, magnetic resonance image; RCA, right coronary artery.
Table 1.
Patient characteristics.
| Patients (n = 32) | |
|---|---|
| Male | n = 18 |
| Age (years) | 55.0 ± 20.7 |
| Heart rate (beats/min) | 60.5 (52.0–65.0) |
| Body mass index (kg/m2) | 21.7 (19.4–25.7) |
| Reasons for examination | |
| Suspect of ischemic heart disease | n = 4 |
| Suspect of cardiomyopathy or others | n = 21 |
| Congenital heart disease | n = 7 |
Presented as number, mean ± standard deviation, or median (range).
Time to decision
The neural network was able to determine the resting phase almost at the same instant as the four chamber-view cine image was obtained in all cases. By contrast, for manual detection of resting phase, the expert needed 50 (36–68) seconds and the general operator needed 61 (37–100) seconds.
Cardiac resting phase
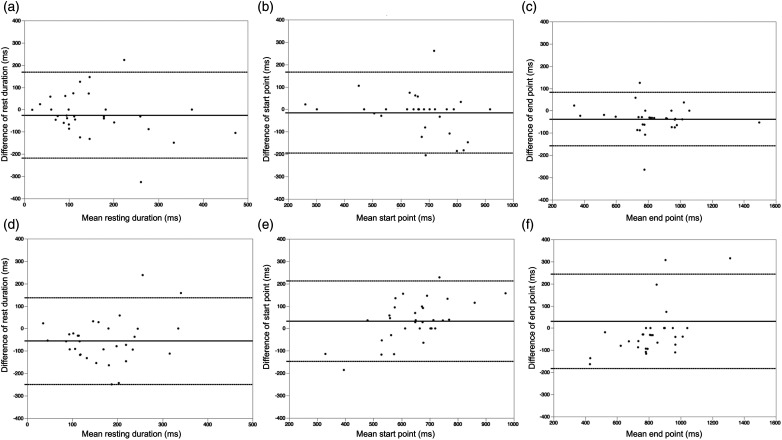
The rest duration, start point, and end point of the neural networks and manual detection are listed in Table 2. The rest duration and start point were not significantly different between the neural network and the expert (p = .30, .90). The end point was significantly different between the neural network and the expert (p < .05). The rest duration and end point were significantly different between the general operator and the expert (p < .05 in each). The start point was not significantly different between the two groups (p = .09). The normalized RMSEs between (1) the expert and general operator and (2) the expert and neural network are presented in Table 3. The normalized RMSE between the expert and neural network according to heart rate is shown in Table 4. The Bland–Altman plots of the neural network and general operator to experts are shown in Figure 4. Regarding the neural network, the mean differences (bias) were −23.0 ms [95% confidence interval (CI) −215.5 to 169.5 ms] for rest duration, −12.7 ms (95% CI −192.6 to 167.2 ms) for start points, and −36.3 ms (95% CI −156.6 to 84 ms) for end points. Regarding the general operator, mean differences (bias) were −54.0 ms (95% CI −247.8 to 139.8 ms) for rest duration, 33.9 ms (95% CI −146.1 to 213.9 ms) for start points, and 32.0 ms (95% CI −180.6 to 244.6 ms) for end points.
Table 2.
Rest duration, start point, and end point of manual detection and neural network detection.
| Expert | General operator | Neural network | |
|---|---|---|---|
| Rest duration (ms) | 99 (63–186) | 178 (132–261) | 123 (77–209) |
| Start point (ms) | 676 (583–730) | 628 (541–702) | 674 (567–791) |
| End point (ms) | 795 (709–942) | 829 (754–899) | 833 (759–987) |
Data are presented as median (first quartile, third quartile).
Table 3.
Normalized root-mean-square error (1) between expert and general operator and (2) between expert and neural network.
| Expert and general operator | Expert and neural network | |
|---|---|---|
| Rest duration (ms) | 0.98 | 0.88 |
| Start point (ms) | 0.68 | 0.64 |
| End point (ms) | 0.51 | 0.33 |
Table 4.
Normalized root-mean-square error between expert and neural network according to heart rate.
| Heart rate ≦ 60 (n = 16) | Heart rate > 60 (n = 16) | |
|---|---|---|
| Rest duration (ms) | 0.82 | 1.16 |
| Start point (ms) | 0.81 | 0.44 |
| End point (ms) | 0.37 | 0.56 |
Figure 4.
Bland–Altman plot analysis. The Bland–Altman plots of the resting phase for neural networks and manual detection. The mean bias (solid line) and 95% confidence intervals (dotted lines) are shown. (A) Rest duration between the neural network and manual detection by experts. (B) Start point between the neural network and manual detection by the expert. (C) End point between the neural network and manual detection by the expert. (D) Rest duration between manual detection by the general operator and expert. (E) Start point between manual detection by the general operator and expert. (F) End point between manual detection by the general operator and expert.
Discussion
The main findings of this study are (1) the fully automated resting phase detection worked successfully in patients, (2) the time to decision for the neural network detection was shorter than that by manual detection, (3) the resting phase measured by the neural network was similar to that of the expert, and (4) the error was lower in the case of the neural network than in the case of the general operator. Therefore, the neural network achieved the objectives of this study, and it can determine the resting phase instantly with better accuracy than that of manual detection by general operators.
The automated RCA detection was sufficiently accurate to determine the RCA for motion detection. For RCA detection, a 3D-DenseNet-based network that uses separable spatiotemporal convolution and feature gating in an effective video classification system was applied to the proposed architecture. Manual detection required approximately 1 min to determine the resting phase, whereas the 3D DenseNet-based network could identify the RCA immediately because this system combines three key concepts: a top-heavy model design, temporally separable convolution, and spatiotemporal feature gating.11 A balance between speed and accuracy is important when building a neural network system and our network achieved both speed and accuracy. There was a significant difference only at the end points, but the mean bias was small (−36.3 ms), and the normalized RMSE (0.33 ms) was lower than that of the general operator (0.51 ms). It has been reported that the rest duration of the RCA ranges between 66 and 200 ms;6 the rest phase duration detected by the neural network and experts during our study is almost consistent with this finding. Moreover, the increase in speed of decision is substantial, which can be confirmed on-site in daily clinical practice.
A previous study demonstrated semi-automated systems for detection of rest period using methods other than neural networks,12,13 whereas our neural network system is a fully automated system. In another study of healthy volunteers, a fully automated system that adopted an alternative method, not a neural network, was verified to determine the cardiac rest period. By analyzing the high-speed motion of the heart in a set of cine images, Asou et al. specified the cardiac resting phase for coronary MRA in 10 healthy volunteers.14 Sato et al. reported a fully automated resting phase detection method based on a template-matching technique in 24 healthy volunteers.15 In contrast to these systems, we tested a novel artificial intelligence system, implemented it on the MRI scanner, and adopted it to clinical patients. It is important that the system works successfully in patients as well as in healthy volunteers. And operators could check results immediately on MRI console. This automated process is expected to result in faster examination times that will be beneficial to clinical workflow, operators, and patients. Moreover, this network can be extended to other anatomies, such as the atrioventricular system in the future. Therefore, it could pave the way for the development of similar solutions for other clinical evaluation procedures. If a fully automated examination is successful, the application of cardiovascular MRI examination can be more wide spread.
This study has some limitations. First, the study’s sample size is small. This study shows an early clinical experience; additional studies involving more patients should be carried out in the future.
Second, this study focused on the resting phase, and it did not obtain the coronary MRA. The resting phase of the neural network was similar to that of the expert, and the normalized RMSE of the neural network was lower than that of the general operator. There are various factors other than the resting phase, such as body movements, breathing, heart rate, acceptance rate, difference in time after injection of contrast medium, and artifacts, that affect the image quality of coronary MRA images. It is not possible to exclude these biases. Moreover, if the coronary MRA is performed two times in clinical practice, it will drastically increase the examination time and lead to a disadvantage for patients. Therefore, we assessed a neural network system from the viewpoint of accuracy.
Third, the neural network in this study did not support cases with hypoplastic RCA. To create a neural network that addresses these cases, we would have to increase the training data. Our group is also developing artificial intelligence that tracks the atrioventricular movement. If this artificial intelligence is used in combination with the neural network of this study, it would be possible to support cases with hypoplastic RCA in the future. And the information from this study will guide other researcher’s future work.
In conclusion, neural network algorithms can determine the cardiac resting phase instantly. It was observed that the resting phase is similar to that determined by an expert. The neural network worked successfully in patients and has the potential to improve the clinical workflow.
Acknowledgments
The authors are grateful to Yoshiaki Komori of Siemens Healthcare K.K. for the optimization of the sequence parameters.
Footnotes
The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: Seung Su Yoon receives a scholarship from Siemens Healthineers. Jens Wetzl and Michaela Schmidt are employed by Siemens Healthineers. All other authors declare that they do not have competing interests.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Research ethics and patient consent: This study was approved by the ethical review board of Ehime University (number: 2011015), and the participants provided written informed consent.
Authorship: We would like to request 8 authors. Please see attached Declaration-scheme.
ORCID iD
Ryo Ogawa https://orcid.org/0000-0002-3261-2752
References
- 1.Yang Q, Li K, Liu X, et al. Contrast-enhanced whole-heart coronary magnetic resonance angiography at 3.0T: a comparative study with X-ray angiography in a single center. J Am Coll Cardiol 2009; 54: 69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Munoz C, Bustin A, Neji R, et al. Motion-corrected 3D whole- heart water-fat high-resolution late gadolinium enhancement cardiovascular magnetic resonance imaging. J Cardiovasc Magn Reson 2020; 22: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hosoda H, Asaumi Y, Noguchi T, et al. Three-dimensional assessment of coronary high-intensity plaques with T1-weighted cardiovascular magnetic resonance imaging to predict periprocedural myocardial injury after elective percutaneous coronary intervention. J Cardiovasc Magn Reson 2020; 22: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kramer CM, Barkhausen J, Bucciarelli-Ducci C, et al. Standardized cardiovascular magnetic resonance imaging (CMR) protocols: 2020 update. J Cardiovasc Magn Reson 2020; 22: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weber OM, Martin AJ, Higgins CB. Whole-heart steady-state free precession coronary artery magnetic resonance angiography. Magn Reson Med 2003; 50: 1223–1228. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y, Vidan E, Bergman GW. Cardiac motion of coronary arteries: variability in the rest period and implications for coronary MR angiography. Radiology 1999; 213: 751–758. [DOI] [PubMed] [Google Scholar]
- 7.Jahnke C, Paetsch I, Achenbach S, et al. Coronary MR imaging: breath-hold capability and patterns, coronary artery rest periods, and beta-blocker use. Radiology 2006; 239: 71–78. [DOI] [PubMed] [Google Scholar]
- 8.Huang G, Liu Z, Van Der Maaten L, et al. Densely connected convolutional networks. arXiv:1608.06993.
- 9.Kingma DP, Ba J. Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980
- 10.Chefd'Hotel C, Hermosillo G, Faugeras O. Flows of diffeomorphisms for multimodal image registration. In: Proceedings IEEE International Symposium on Biomedical Imaging, Washington, DC, USA, 2002, pp. 753–756. [Google Scholar]
- 11.Xie S, Sun C, Huang J, et al. Rethinking spatiotemporal feature learning: speed-accuracy trade-offs in video classification. Lecture Notes in Computer Science. In: Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–4 September 2018: 318–335. [Google Scholar]
- 12.Ustun A, Desai M, Abd-Elmoniem KZ, et al. Automated identification of minimal myocardial motion for improved image quality on MR angiography at 3 T. Am J Roentgenol 2007; 188: W283–W290. [DOI] [PubMed] [Google Scholar]
- 13.Jahnke C, Paetsch I, Nehrke K, et al. A new approach for rapid assessment of the cardiac rest period for coronary MRA. J Cardiovasc Magn Reson 2005; 7: 395–399. [DOI] [PubMed] [Google Scholar]
- 14.Asou H, Imada N, Nishiyama Y, et al. Automated determination of cardiac rest period on whole-heart coronary magnetic resonance angiography by extracting high-speed motion of coronary arteries. Clin Imag 2018; 52: 183–188. [DOI] [PubMed] [Google Scholar]
- 15.Sato T, Okada T, Kuhara S, et al. Automatic identification of the cardiac rest period using template updating for magnetic resonance coronary angiography. Adv Biomed Eng 2016; 5: 26–31. [Google Scholar]