Summary
DNA methylation is a key regulator of gene expression and a clinical therapeutic predictor. We examined global DNA methylation beyond the generally used promoter areas in human small cell lung cancer (SCLC) and find that gene body methylation is a robust positive predictor of gene expression. Combining promoter and gene body methylation better predicts gene expression than promoter methylation alone including genes involved in the neuroendocrine classification of SCLC and the expression of therapeutically relevant genes including MGMT, SLFN11, and DLL3. Importantly, for super-enhancer (SE) covered genes such as NEUROD1 or MYC, using H3K27ac and NEUROD1, ASCL1, and POU2F3 ChIP-seq data, we show that genic methylation is inversely proportional to expression, thus providing a new approach to identify potential SE regulated genes involved in SCLC pathogenesis. To advance SCLC transitional research, these data are integrated into our web portal (https://discover.nci.nih.gov/SclcCellMinerCDB/) for open and easy access to basic and clinical investigators.
Subject areas: Epigenetics, Cancer systems biology, Cancer
Graphical abstract

Highlights
-
•
Methylome signatures differentiate SCLC subtypes
-
•
Gene body methylation is highly predictive of gene expression
-
•
Genic Super-Enhancer regions are associated with local demethylation
-
•
The EPICmethylation array clinically available
Epigenetics; Cancer systems biology; Cancer.
Introduction
Small cell lung cancers (SCLCs) account for approximately 13% of all lung cancers (Govindan et al., 2006; Poirier et al., 2020), and are highly metastatic and rapidly chemoresistant (Gazdar et al., 2017; Sabari et al., 2017). Their genetic landscape is different from other lung cancers, with few activating driver mutations in oncogenes, near-universal loss of function-mutational prevalence in TP53 and RB1 tumor suppressor genes, and overexpression of cellular proliferation and replication pathways (Tlemsani et al., 2020). Many SCLCs also have increased copy-number changes in members of the MYC oncogene family, and/or disruptive mutations in chromatin remodeler and epigenetic genes such as histone lysine methyltransferases (KMT2D and KMT2C), and histone acetyltransferases (EP300, CREBBP) (George et al., 2015).
SCLC tumors and patient-derived cell lines and xenografts (PDXs) have recently been classified into four main subtypes based on the expression of 4 lineage-defining transcription factors (LTFs): NEUROD1, ASCL1, POU2F3, and YAP1 (briefly referred to as “NAPY”) (Rudin et al., 2019; Tlemsani et al., 2020). Further studies differentiate SCLC into ASCL1, NEUROD1, POU2F3 subtypes, and the inflammatory subtype, which benefits from combined chemotherapy and immunotherapy (Gay et al., 2021). The NEUROD1 and ASCL1 subtypes have a neuroendocrine (NE) phenotype, with elevated expression of NE markers including synaptophysin and chromogranin A. The POU2F3 and YAP1 subtypes are non-NE, with elevated NOTCH pathway expression and low expression of NE markers (Pozo et al., 2021; Tlemsani et al., 2020). Binding of the LTFs to many distinct sites in the genome has been proposed to act as enhancers and regulate different gene expression pathways that characterize each of the subtypes of SCLCs (Borromeo et al., 2016; Chakraborty et al., 2018; Pozo et al., 2021; Tlemsani et al., 2020).
The subtype classification of SCLC is not routinely implemented in clinical practice owing to limited access to well-preserved tumor tissues to perform immunohistochemistry analyses with many diagnostic biopsies coming from needle aspirations (Qu et al., 2022). RNA-seq analyses are also not routinely utilized to classify SCLCs based on the differential expression of the LTFs. Because of the challenges of RNA analyses and of the greater stability of DNA over RNA, DNA analyses are being effectively tested to classify subtypes of cancers, such as sarcomas (Koelsche et al., 2021). Taking advantage of the well-documented genomic databases of SCLC cell lines (Gay et al., 2021; George et al., 2015; Rudin et al., 2019; Tlemsani et al., 2020), we asked whether DNA methylation analyses based on the clinically used MethylationEPIC array could provide novel insights in the genetic regulatory pathways of SCLCs and be used as a classifier and therapeutic guide for SCLCs.
DNA methylation is used clinically to correlate survival (Gevaert et al., 2015; Hao et al., 2017), as well as drug response (Bacolod and Barany, 2021; Butler et al., 2020; Krushkal et al., 2020; Ortiz-Barahona et al., 2020). To that effect, O6-methylguanine (O6-MeG)-DNA methyltransferase (MGMT) promoter methylation is an established prognostic biomarker in glioblastomas treated with temozolomide (Butler et al., 2020), while Schlafen (SLFN11) expression inactivation through promoter methylation leads to resistance to PARP inhibitors and DNA damaging agents including etoposide, cisplatin, topotecan and lurbinectedin that are used as a standard of care for SCLCs (Kundu et al., 2021; Murai et al., 2016, 2019; Nogales et al., 2016; Tang et al., 2018). Although the DNA methylation of promoters is commonly used to predict the lack of gene expression (Butler et al., 2020; Esteller, 2021), in many cases it can give an incomplete picture when the promoters of non-expressed genes are not hypermethylated (Butler et al., 2020; Reinhold et al., 2017; Tang et al., 2018). Although many methylation probes in the 450k and 850k array platforms, widely used for detecting the methylation of specific gene regions, are outside of promoters, there have been limited analyses and use on their quantitative relationship with gene expression and on their potential translational value to predict gene expression (Lister et al., 2009; Spainhour et al., 2019; Su et al., 2018). Yet, gene body methylation has been shown to reflect gene activity (Bacolod and Barany, 2021; Yang et al., 2014), suppress alternative transcript isoforms by blocking promoters, and to regulate splicing (Huang et al., 2021; Lev Maor et al., 2015; Li et al., 2018; Neri et al., 2017; Teissandier and Bourc'his, 2017; Yang et al., 2014).
Studies have begun to explore the epigenetics of SCLCs by mapping the binding sites of the LTFs (Borromeo et al., 2016; Huang et al., 2018), the enhancers based on histone H3 lysine 27 acetylation (H3K27ac) (Pozo et al., 2021), and promoter DNA methylation (Krushkal et al., 2020; Tlemsani et al., 2020). To expand our understanding of how gene expression is regulated in SCLC and determine the potential translational value of the MethylationEPIC 850k array, we took advantage of the SCLC cell line databases (https://discover.nci.nih.gov/SclcCellMinerCDB/) and integrated whole genome DNA methylation (Krushkal et al., 2020; Tlemsani et al., 2020), enhancer H3K27ac ChIP-seq (Huang et al., 2018; Pozo et al., 2021) and ChIP-seq for the three main LTFs driving SCLC pathogenesis (ASCL1, NEUROD1, and POU2F3) (Borromeo et al., 2016; Huang et al., 2018). We developed algorithms to automatically quantify the levels of promoter and gene body methylation, as well as copy number and promoter acetylation for each gene. And for each epigenetic marker (DNA methylation, H3K27Ac, and ChIP-seq for ASCL1, NEUROD1, and POU2F3), we analyzed their genomic distribution and demonstrate how they mutually predict gene expression. The data are integrated in a public web-based genomic platform in our CellMinerCDB ecosystem (Luna et al., 2021), enabling individual researchers and clinicians to explore genomic and drug response correlations across the NCI, UT Southwestern, Broad-MIT (CCLE), and Welcome-Sanger databases (https://discover.nci.nih.gov/SclcCellMinerCDB/).
Results
General overview of the small cell lung cancer cell line methylome data and classification of small cell lung cancer cell lines based on their methylome
High-resolution DNA methylation (EPIC 850K array) was obtained for 68 different SCLC patient-derived cell lines (Krushkal et al., 2020; Tlemsani et al., 2020), H3K27ac data for 18 SCLC lines (Huang et al., 2018; Pozo et al., 2021) and ASCL1, NEUROD1, and POU2F3 ChIP-seq for 6 SCLC lines (Borromeo et al., 2016; Huang et al., 2018). We analyzed each dataset and studied their predictive value as classifiers of SCLCs and predictors of gene expression. The data included in the present study are summarized in Figure 1A and Table S1; they are publicly accessible in the updated version (v.1.1) of SCLC_Cellminer (https://discover.nci.nih.gov/SclcCellMinerCDB/).
Figure 1.
Overview of cell lines utilized in the study
(A) Data types available for each cell line (gene expression, DNA methylation, and H3K37ac) and their neuroendocrine status and NAPY classification.
(B) Pie chart of EPIC 850K array DNA methylation probe annotation distribution.
(C) Distribution of probe hypermethylation fraction in each cell stratifying by genomic regions using the EPICmethylation 850K array compared using Wilcoxon signed-rank test.
(D) UMAP clustering of cell lines using all methylation probes. YAP1 cell lines cluster with NSCLC cell lines.
(E) Correlation (Pearson) of MGMT expression and promoter methylation in the 66 SCLC cell lines from the NCI-DTP panel (snapshot from https://discover.nci.nih.gov/sclccellminercdb).
(F) Methylation probes at the MGMT promoter in 5 cell lines (2 with high and 3 with low MGMT expression).
The EPICmethylation 850K array probes are not only in promoters (40.6%), but also in genic areas (35.2%) and intergenic regions (24.2%) (Figure 1B). Compared to the 450K array, the higher resolution of the EPIC 850K methylation array has more than double the coverage in genic areas (Figure S1A). The comparison of the methylation levels of different genomic regions across all SCLC lines shows that DNA methylation probes overall tend to be hypermethylated, with approximately 58.4% (40.3–70%) of probes with beta value > 0.5 (Figure 1C, Table S2). This may be related to the fact that cell lines from SCLC express significantly higher levels of DNMT1 and DNMT3A (and DNMT3B) compared to other cancers (Figure S1B). Yet, as expected, CpG islands and promoters tend to be hypomethylated (median 22.9 and 48.4%, respectively) compared to gene body and intergenic regions (median 78%) (Figures 1C and S1C; Table S2).
DNA methylation has been previously used to cluster cell lines based on the tissue of origin (Ghandi et al., 2019). Thus, we assessed whether the genome-wide EPICmethylation array would differentiate SCLC subtypes. Although the non-neuroendocrine YAP1 and NSCLC cell lines showed distinct clustering, the SCLC cell lines showed limited clustering based on their NE status using UMAP clustering of all probes (Figure 1D, Table S3). As expected, some NEUROD1 cell lines with dual ASCL1-NEUROD1 expression (Figures S1D and S1E, Table S3) clustered with the ASCL1 subgroup (Figure S1D, green arrowheads) (Tlemsani et al., 2020). UMAP clustering using CpG, promoter, gene body, and intergenic probes yielded similar results. In case of gene body and promoter probes the NEUROD1 cell lines that did not express ASCL1 had a better separation from the ASCL1 cluster (Figure S1E). Overall, we conclude that the methylation array has limited value in clustering cells into subtypes.
Studies comparing gene expression and DNA methylation classically focus on the methylation of promoters, where hypermethylation is associated with low gene expression (Ortiz-Barahona et al., 2020; Reinhold et al., 2017). Yet, for many genes and across cell lines, we observed that low gene expression was observed despite low promoter methylation. This is illustrated in Figures S1F and S1G for the therapeutic response predictor Schlafen 11 (SLFN11) (Murai et al., 2019) and the mesenchymal Vimentin (VIM) genes. To extend this finding genome-wide, we selected the genes with highest SD bimodal gene expression distribution (Figure S1H), finding that more than 50% of the low-expressing cell lines showed hypomethylated promoters.
Among those genes where lack of expression fails to highly correlate with promoter hypermethylation is the methylguanine methyl transferase gene (MGMT), which is routinely analyzed in the context of temozolomide treatments (Butler et al., 2020; Farago et al., 2019; Ortiz-Barahona et al., 2020). MGMT showed a relatively low negative correlation (although significant) between promoter methylation and gene expression (Figures 1E and S1H). Detailed examination of the methylation probes in the promoter area of MGMT revealed that highly expressing cell lines showed consistent CpG hypomethylation, while cell lines with low expression displayed variable promoter methylation (Figure 1F). In addition, comparing the methylation (probe) distribution across the MGMT locus showed hypermethylation of genic probes in the highly expressing cells (Figure S1I). These observations indicate that promoter hypomethylation does not accurately predict gene expression and thus must be controlled by additional epigenetic regulatory mechanisms.
Relationship between gene body methylation and gene expression
To explore the relationship between DNA methylation in gene bodies and gene expression (Teissandier and Bourc'his, 2017), we focused on the distribution of DNA methylation by comparing methylation in promoter (−2.5kb–2.5kb area around transcription start sites), gene body (genic area excluding promoter regions) and upstream (5kb window upstream of promoter regions) and downstream (5kb window downstream of transcription end sites) regions of genes. Stratifying a large number of genes based on their expression showed a marked difference in overall DNA methylation patterns (Figure 2A). As expected, highly expressed genes showed hypomethylation centered in promoters (upper tracings in Figure 2A), while promoter hypermethylation was observed in lowly expressed genes (lower tracing in Figure 2A). Notably, the gene body of highly expressed genes was consistently hypermethylated (top tracings), while gene bodies were significantly less methylated and more variable in genes with low or no expression (bottom tracings in Figure 2A). These methylation profiles were similar in all cell lines in the SCLC database, and the methylation types were reproducible using reduced bisulfite sequencing data obtained from the CCLE (Ghandi et al., 2019) (Figures S2A–S2C). The bisulfite data are also available in CellMinerCDB (https://discover.nci.nih.gov/cellminercdb) (Luna et al., 2021).
Figure 2.
Relationship between gene body methylation, promoter DNA methylation, and gene expression
(A) Genic distribution of DNA methylation based on gene expression quintiles in NCI-H69 cells.
(B) Promoter and gene body methylation distribution in non-expressed, lowly expressed, and highly expressed genes.
(C) Heatmaps of promoter and gene body methylation in genes with low (left) or high range (right) of expression. For each gene, cell lines were sorted based on expression from low to high. The heatmaps on the left show promoter and gene body methylation for highly expressed genes (TOP2A and B2M) and non-expressed genes (DIRC1 and CRISP3). Heatmaps on the right demonstrate genes with high range of expression.
(D) Relationship of promoter and gene body methylation (Pearson correlation), and gene expression for YAP1 across the 66 NCI-DTP cell lines.
(E) Expression of YAP1 based on promoter and gene body methylation categories from panel D.
(F) Representative examples where gene body methylation predicts gene expression better than promoter methylation (Pearson correlation).
As gene body methylation was consistently associated with expression for many genes, we created a gene-level summarization algorithm of gene body methylation taking into account all probes instead of using a single-probe approach (Bacolod and Barany, 2021; Krushkal et al., 2020), and which can be used to assess independently promoter and gene body methylation. We included gene body probes outside of promoters, excluding CpG island overlapping probes, which tend to be hypomethylated (see Figure 1). In cases where a gene had multiple transcripts, we selected the transcript with the most positive correlation between gene body methylation level and gene expression. Only a very small number of very short genes such as ASCL1 could not be analyzed for gene body methylation because of insufficient probe coverage. Based on this approach, we were able to provide scores for both gene body and promoter methylation and integrated these features in SCLC-CellMinerCDB (https://discover.nci.nih.gov/sclccellminercdb).
Using the genome-wide methylome data from all 66 SCLC cell lines (see Figure 1A), we compared promoter and gene body methylation as a function of gene expression. The gene promoter methylation values and gene expression data were obtained from the SCLC-CellMinerCDB website (Tlemsani et al., 2020). Non-expressed genes across all SCLC cell lines (Figure 2B, left) clustered in 3 distinct groups: genes methylated both on the promoter and gene body (upper right quadrant), genes with no methylation at all (bottom left), and genes with both promoter hypomethylation and gene body methylation (upper left). By contrast, highly expressed genes (Figure 2B, right) were found to be hypomethylated on their promoters and consistently hypermethylated in their gene bodies. These analyses demonstrate that analyzing promoter methylation (bmt) is insufficient to accurately predict gene expression and that adding gene body methylation improves the gene expression correlation.
Figure 2C illustrates the relationship of gene expression and DNA methylation for specific genes exhibiting different expression ranges. Topoisomerase II-alpha (TOP2A) or beta-2-macroglobuin (B2M), which are housekeeping genes highly expressed across cell lines show the lack of promoter methylation and high gene body methylation, as expected (Figure 2C, upper left). By contrast, genes that are generally not expressed, such as DIRC1 and CRISP3 show concomitant hyper- or hypomethylation of both promoters and gene bodies (Figure 2C, bottom left). Additional examples demonstrating this concomitant hyper- and hypomethylation for low-expression genes are shown in Figure S2D for the GLT8D2, EYS and GJA8 genes. We also observed that the expression of genes with high range of expression such as MGMT, SLFN11, and NOTCH1 shows highest correlation with gene body methylation (Figure 2C, right) rather than promoter hypomethylation. Together these results stress the relevance of gene body methylation as a predictor of gene expression.
Analyzing YAP1, a driver gene for a subset of non-NE SCLCs (Rudin et al., 2019; Tlemsani et al., 2020) reveals that YAP1-expressing cell lines have no hypomethylated promoter with a hypermethylated gene body (“Active” group) (Figures 2C and 2D). Many YAP1 non-expressing cell lines have either hypermethylation (“Hyper” group) or hypomethylation (“Hypo” group) of the YAP1 promoter and gene body. There are some cell lines that do not express YAP1 with a hypermethylated promoter and hypomethylated gene body (“Divergent” group). Thus, overall, the methylation categories based on both promoter and gene body methylation reveal that the “active” (low promoter methylation with high gene body methylation) group predicts the expression of YAP1 (Figure 2E).
Additional examples where gene body methylation predicts gene expression better than promoter methylation are provided in Figure 2F for MGMT, SLFN11 and the cell surface glycoprotein and cancer biomarker CEACAM5, which is selectively expressed in neuroendocrine SCLC cell lines (Tlemsani et al., 2020). Thus, scoring gene body methylation in addition to promoter methylation can be particularly helpful to predict gene expression for genes with a wide range of expression, and this feature has been implemented in our new SCLC-CellMinerCDB release (v.1.1) along with other features that are listed in Table S4 (https://discover.nci.nih.gov/SclcCellMinerCDB/).
Enhancer and small cell lung cancer-lineage-defining transcription factors distributions and their relationship with DNA methylation
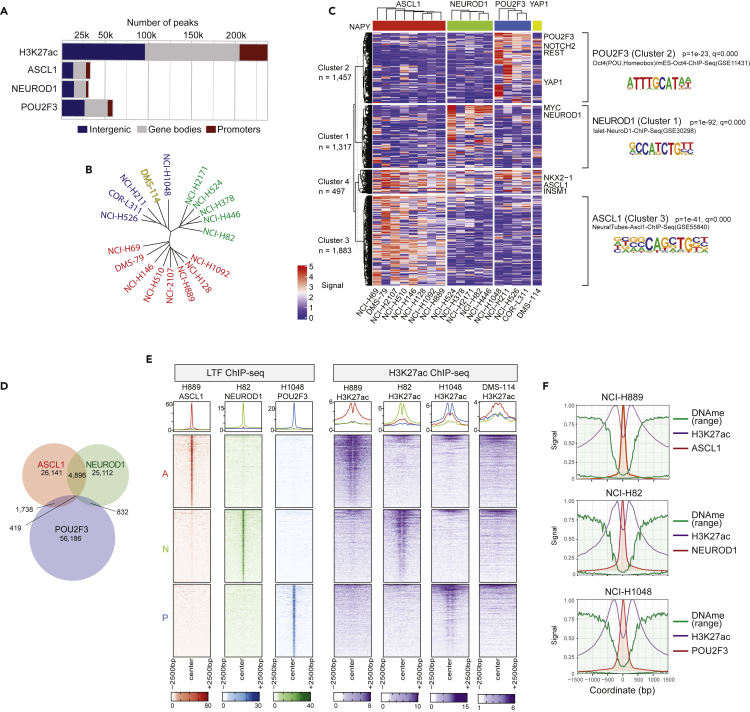
Because histone post-translational modifications are key epigenetic regulators, we integrated into our methylome analysis the recently available H3K27ac ChIP-seq data (Pozo et al., 2021) for eighteen of the SCLC cell lines (see Figure 1A). Our analysis identified ∼239,000 enhancers defined by H3K27ac peaks, which are mainly localized in intergenic and gene body areas, with a relatively small fraction in promoters (Figure 3A).
Figure 3.
Enhancer landscapes in SCLC cell lines
(A) Distribution of the binding sites of H3K27ac (enhancers) and the 3 LTFs ASCL1, NEUROD1, and POU2F3 identified from ChIP-seq data.
(B) Phylogenetic distance analysis of cell lines using the most variable enhancer sites.
(C) Heatmap of most variable enhancer sites (H3K27ac). HOMER motif analysis results are shown at right for each cluster.
(D) Limited overlap of the ASCL1, NEUROD1, and POU2F3 sites.
(E) Heatmap of ASCL1, NEUROD1, and POU2F3 specific binding sites (LTF ChIP-seq) overlayed with enhancer activity (H3K27ac ChIP-seq).
(F) DNA methylation and H3K27ac mean tracings centered to ASCL1 (NCI-H889), NEUROD1 (NCI-H82), and POU2F3 (NCI-H1048). Hypomethylation can be seen at the binding sites, with a valley of H3K27ac.
Phylogenic analysis of the H3K27ac data for the enhancers with highest range of signal intensity (to reduce background noise) across cell lines showed that the cell lines formed three main clusters, differentiating the ASCL1, NEUROD1, and POU2F3 subtypes, and confirming the distinct enhancer signatures of the SCLC subtypes, in agreement with the recently published super-enhancer analysis (Pozo et al., 2021). Cluster heatmap representation of the H3K27ac signals based on the 4 subtypes ASCL1, NEUROD1, POU2F3, and YAP1 produced 4 main clusters (Figure 3C; Table S5). HOMER sequence motif analysis revealed that clusters 1, 2 & 3 are significantly enriched for the DNA sequence motifs of ASCL1, POU2F3, and NEUROD1 (Figure 3C, right). The fourth enhancer cluster (cluster 4) mainly differentiates the NE and non-NE subtypes, with enrichment in promoter areas (Figure S3A). Differential GO analysis showed enrichment in neuronal pathways for the ASCL1 and NEUROD1-specific enhancers, and enrichment of the extracellular matrix organization and YAP1-TAZ pathways for the non-NE clusters (Figure S3B).
UMAP clustering of the enhancers with highest range of signal intensity across cell lines differentiated 2 main groups of enhancers: enhancers bound in promoter sites vs. non-promoter sites (Figure S3C, left, red points). Notably, enhancers matching the SCLC-LTF subtypes (ASCL1, NEUROD1, and POU2F3) were found enriched with non-promoter sites (Figure S3C, right). These data are consistent with the presence of multiple sites of specific enhancers and super-enhancer (SE) for each of the subtypes of SCLC cell lines located outside of promoters (Pozo et al., 2021).
To analyze the relationship between H3K27 acetylation (enhancers and SEs) and the SCLC-LTFs, we analyzed the ChIP-seq data obtained for ASCL1, NEUROD1, and POU2F3 in the 6 cell lines with overlapping data (see Figure 1A) (Borromeo et al., 2016; Huang et al., 2018). Like the H3K27ac histone marks, most sites were distributed in intergenic regions or within gene bodies. Yet, the binding sites of ASCL1, NEUROD1, and POU2F3 showed minor overlap (Figure 3D), which is consistent with the sequence specificity and specific distribution of each SCLC-LTF (Borromeo et al., 2016). Consistent with published results (Borromeo et al., 2016), the ASCL1, NEUROD1, and POU2F3 binding sites were found mainly localized outside of promoters (Figure 3A). Alignment of the genomic tracings for three representative cell lines corresponding to each subtype (NCI-H889 for ASCL1, H82 for NEUROD1 and H1048 for POU2F3) shows that the ASCL1, NEUROD1, and POU2F3 binding sites do not overlap with each other (Figure 3E). Furthermore, alignment with the H3K27ac data shows that the differential distribution of the LTF sites is concordant with enhancer signals in each of these regions (Figure 3E). For instance, regions bound by ASCL1 have matching active enhancers in NCI-H889 cells, but not in the other cell lines. Similar distribution could be seen in the case of the NEUROD1 sites in NCI-H82 cells and of the POU2F3 sites in NCI-H1048 cells (Figure 3E). Also, for each SCLC-LTF, the sites of LTF binding are within a short central segment with reduced H3K27ac signal corresponding to the LTF binding peaks (Figures 3E and 3F). The few overlapping binding sites across the three SCLC-LTFs show that the shared enhancers had the same lack of H3K27ac signal at the SCLC-LTF binding peaks (Figure S3E).
To establish the potential relationship between LTF-specific enhancers and DNA methylation, we overlaid the DNA methylation tracings with each of the SCLC-LTF enhancer regions. This showed that the summits of the ASCL1, NEUROD1, and POU2F3 ChIP-seq peaks are characterized by a dip in DNA methylation, consistent with the possibility that the SCLC-LTFs only bind unmethylated DNA sites outside of promoters and are flanked by hyperacetylated H3K27 regions (Figure 3F).
H3K27 promoter acetylation and gene expression
For the enhancers localized in gene promoters (∼40,000 sites corresponding to ∼30% of the total H3K27ac signals; see Figure 3A), enhancer signals (H3K27ac) were found positively correlated with gene expression and inversely correlated with DNA methylation (Figure 4A). Highly expressed genes displayed the highest levels of H3K27ac signal, and genes with intermediate expression across cell lines displayed intermediate H3K27ac signal (Figure 4A). Figure 4B (right panels) shows representative genomic tracings and quantitative analyses performed automatically on the CellMinerCDB web portal (https://discover.nci.nih.gov/) for MGMT and SLFN11. As expected, promoter acetylation is generally highly correlated with gene expression for effector genes.
Figure 4.
Enhancer signals (H3K27ac) in promoters strongly predict gene expression
(A) Summary of genic H3K27ac and DNA methylation distribution based on gene expression. Highly expressed genes have highest promoter H3K27ac signal.
(B) Representative H3K27ac tracings at the MGMT and SLFN11 promoters for cell lines with different transcript levels. Plots at right are snapshots from SclcCellMinerCDB for the different NCI-DTP cell lines.
(C) Promoter enhancer signal intensity (derived from H3K27ac) strongly correlates with the expression of the SCLC-LTFs, ASCL1, NEUROD1, POU2F3, and YAP1.
(D and E) Enhancers and super-enhancers covering entire genes (NEUROD1 and NKX2-1 shown here) are associated with hypomethylation for the representative cell lines (tracings) and for the 66 cell lines in the database (plots are snapshots from SclcCellMinerCDB).
(F) Correlation summary of gene expression and H3K27ac, DNA promoter and gene body methylation and copy-number data using genes with the most variable expression.
Promoter enhancer signals for the SCLC-LTF genes were also closely correlated with the subtypes of SCLCs (Figure 4C), and selected SCLC-relevant genes including the MYC genes (MYC, MYCL, and MYCN), the NE SCLC markers (SYP, INSM1, and CHGA) and the NOTCH genes (NOTCH1, NOTCH2 and NOTCH3) all displayed tight correlation between promoter enhancer H3K27ac signals and gene expression (Figure S4A).
Additionally, the super enhancer (SE) signals for ASCL1, NEUROD1, and POU2F3 (Pozo et al., 2021) revealed overlap for each set of SE in the corresponding subtypes, correlating with the expression levels of each of the LTFs (Figure S4B). Exploring further the relationship between enhancers determined by H3K27ac and DNA methylation, we observed that DNA methylation probes overlapping enhancers tend to be hypomethylated compared to non-enhancer sites regardless of their localization (promoters, gene bodies, and intergenic regions as well) (Figure S4C). We also observed that in the case of strong enhancers (and SEs) covering entire genes, the gene body methylation probes tended to be hypomethylated. This can be seen in the case of NEUROD1 and NKX2-1, where cell lines with high genic H3K27ac signal show low gene body methylation, resulting in a negative correlation between gene expression and gene body methylation (Figures 4D and 4E). These observations demonstrate that gene body methylation, which for most expressed genes is closely associated with transcription, is low in genes that are covered by enhancers and/or super enhancers like NEUROD1.
To facilitate further analyses, we generated a gene-level H3K27ac summarization algorithm, which is implemented in SclcCellMinerCDB version 1.1 (https://discover.nci.nih.gov/cellminercdb). This feature was used to generate the plots and statistical analyses shown in the right panels of Figures 4B, 4D, and 4E. We also used this algorithm to compare promoter H3K27ac, promoter, and gene body methylation and copy number (CNV) with gene expression. Promoter enhancer signal shows the strongest correlation with gene expression (Figure 4F). Gene body methylation shows better predictive power over promoter methylation in the case of genes with high range of expression; yet promoter methylation has slightly better overall predictive power when including all genes (Figure S4D). Copy-number data showed the overall lowest correlation with gene expression when comparing genes with high range of expression.
Exploring the EPICmethylation array for precision medicine of small cell lung cancer
Based on the premise that two potential key elements of precision medicine are: 1) the classification of tumors in subgroups with well-defined pathways, and 2) Omics-driven therapies (Jo et al., 2021), we tested the potential of the EPIC 850k methylome array in the context of the SCLC cell lines as pilots for expansion studies in patient samples.
Regarding the classification of SCLCs, we used the “Multivariate Analyses” web tool of SclcCellMinerCDB to explore the predictive value of the EPIC array. Figure 5 displays snapshots that can be readily obtained by readers and users (https://discover.nci.nih.gov/sclccellminercdb). For NEUROD1, both promoter and gene body methylation give a highly significant prediction (p value = 1.3e-08) (Figure 5, upper left). As expected, promoter methylation (abbreviated “mth”) was negatively correlated with NEUROD1 expression. Gene body methylation (abbreviated “bmt”) is also highly correlated with NEUROD1 expression, as the NEUROD1 gene coincides with an SE (see section above and Figure 4D). Highly significant prediction is also observed for YAP1 and ASCL1 (Figure 5, middle left). In the case of YAP1, promoter methylation is highly predictive and gene body methylation is positively correlated with expression, as expected (see above) for genes not covered by an SE. For ASCL1, which is a short gene for which no gene body probes could be assigned, the predictive value of promoter methylation was very high (p value = 6.1e-10). Together, these analyses demonstrate the predictive value of the EPICmethylation array for SCLC classification based on their LTFs.
Figure 5.
Predictive value of gene body and promoter DNA methylation to classify SCLC and assess the expression of therapeutically relevant genes
The Multivariate Analyses tool of SclcCellMinerCDB (https://discover.nci.nih.gov/sclccellminercdb) was used to generate the analyses and panels (snapshots from the website). Queried genes are listed above each panel. The bottom panel shows the SLFN11 gene body and promoter methylation as predictors for topotecan response in the 66 SCLC cell lines from the NCI-DTP database. Insets below each plot show technical details and significance for gene body methylation (bmt) and promoter methylation (mth). Asterisks indicate statistical significance.
The predictive value of the EPIC array was tested for several therapeutically applicable genes (Figure 5, right and bottom panels). The expression of DLL3, which is specifically expressed in ASCL1-SCLC (Rudin et al., 2019; Tlemsani et al., 2020) and targeted by small molecules and radioligands (Morgensztern et al., 2019; Tully et al., 2022) can be predicted with high significance (p = 7.9e-10) by combining promoter (mth) and gene body (bmt) methylation (Figure 5, upper right). Lack of expression of MGMT, which predicts response to temozolomide (Butler et al., 2020; Farago et al., 2019; Thomas et al., 2017) is highly predicted by lack of promoter and gene body methylation (p = 2.8e-13) (Figure 5, right).
Finally, we tested the expression of SLFN11 as a sensitizer and biomarker for replication-targeted therapies commonly used in SCLC (etoposide, cisplatin, topotecan, lurbinectedin) (Jo et al., 2021; Kundu et al., 2021; Murai et al., 2019; Thomas and Pommier, 2016). Immunoblot analyses on the NCI-DTP SCLC cell lines (Polley et al., 2016) demonstrated high correlation between SLFN11 protein levels (“proSLFN11” in Figure 5, bottom right) and gene expression (Pearson correlation (r) = 0.93, p value = 1.5e-26), which is consistent with our initial studies performed in the NCI60 (Zoppoli et al., 2012). Next, we tested the predictive value of the EPIC array data for SLFN11 and found very high predictive value by combining promoter and gene body methylation (r = 0.82, p value = 5.9e-17) (Figure 5, bottom right). Both parameters obtained from the EPIC array combination were also predictive of the activity of topotecan (Figure 5, bottom) with predictive value primarily driven by gene body methylation (Figure S5C) rather than promoter methylation (Figure S5D).
Together these analyses demonstrate the predictive value of the EPICmethylation data analyzed using the CellMinerCDB algorithms (Luna et al., 2021) to classify SCLC and potentially predict therapies.
Discussion
The SCLC-CellMiner web portal (https://discover.nci.nih.gov/SclcCellMinerCDB/) provides a unique platform and data repository to examine multi-omics data for widely used patient-derived SCLC preclinical models with data originating from complementary sources (NCI, DepMap, UT Southwestern, Cold-spring Harbor) (Rudin et al., 2019; Tlemsani et al., 2020). SCLC-CellMiner enables researchers to directly compare and mine gene expression patterns and pathways, and determine how gene expression and mutations may be correlated and hence “predict” response to hundreds of drugs tested in the preclinical models (Polley et al., 2016; Tlemsani et al., 2020). Compared to our previously released version (Tlemsani et al., 2020), the updated version of SCLC-CellMiner integrates two new epigenetic markers: gene body methylation and promoter enhancer signals (H3K27ac). These epigenetic marks, in addition to promoter methylation and gene copy number (CNV), predict gene expression, classify SCLC cell lines, and show how omics analyses predict drug responses. These new features are summarized in Table S4.
Genomic DNA methylation is maintained by DNMT1, which recognizes and methylates hemi-methylated sites during replication. De novo DNA methylation is mainly deposited by DNMT3A and DNMT3B, which binds to histone H3 Lysine 4 (H3K4) before methylating DNA at inactive gene promoters. At actively transcribed genes, the promoters are marked by the trimethylation of H3K4, which inhibits the methyltransferase activity of DNMT3A/B, leading to decreased promoter methylation levels (Otani et al., 2009; Piunti and Shilatifard, 2016; Zhang et al., 2010). As genes are transcribed by POL2, SETD2 trimethylates the core histone H3 at lysine K36, which is recognized by DNMT3B, leading to hypermethylation in the gene body (Dhayalan et al., 2010; Greenberg and Bourc'his, 2019; Hervouet et al., 2018; Roadmap Epigenomics et al., 2015). Gene body methylation has been proposed to enhance transcription by suppressing the firing of alternate transcripts and by facilitating splicing (Bacolod and Barany, 2021; Huang et al., 2021; Neri et al., 2017; Teissandier and Bourc'his, 2017; Yang et al., 2014). Apart from transcription, DNA methylation has been shown to be mutually exclusive of the facultative heterochromatin marker H3K27me3 (Roadmap Epigenomics et al., 2015; Zhang et al., 2020). In addition, differential DNA methylation status has been seen in euchromatin (A) and heterochromatin (B) compartments, where A-compartments tended to be hypermethylated (Fortin and Hansen, 2015).
Our gene-level methylation profile analyses based on the high-resolution EPICmethylation 850K array demonstrate that genes expressed at high levels are characterized not only by promoter hypomethylation but also by gene body hypermethylation. The promoter hypomethylation valley is slightly narrower in lowly expressed genes paired with decreasing gene body methylation values. Because of this relationship, promoter methylation shows a negative correlation, while gene body methylation exhibits even higher positive correlation with gene expression levels. Looking at non-expressed genes, promoters and gene bodies are either both hypo- or hypermethylated. As transcription is lacking in these genes, methylation deposition may be coordinated by other epigenetic markers and chromatin states (Brinkman et al., 2012; Fortin and Hansen, 2015; Roadmap Epigenomics et al., 2015). From a translational viewpoint, our methylation algorithms (mth: promoter and bmt: gene body) demonstrate the value of the DNA EPICmethylation 850K array for predicting gene expression.
We also observed that DNA methylation levels are lower in enhancer areas, and that genic areas covered by super-enhancers (SEs) tend to be completely hypomethylated. This is probably related to the elevated promoter mark H3K4me3, which inhibits DNA methylation deposition that extends to wider genomic regions in super-enhancers (Cao et al., 2017; Khan et al., 2018). This results in a counterintuitively negative correlation between expression and gene body methylation and corresponds to a signature of genes covered by super-enhancers, such as NEUROD1, FOXA1/2 and NKX2-1 (see Table S6).
SCLC cell lines are classified into 4 main subtypes based on the four LTFs: ASCL1, NEUROD1, POU2F3, and YAP1 (Rudin et al., 2019), and recent attempts have been made to extend this classification to clinical samples (Qu et al., 2022). Basic studies recently showed that these subtypes can be clustered using their SE activity (Borromeo et al., 2016; Huang et al., 2018; Pozo et al., 2021). The clustering was also visible when we used variable enhancers, not limiting to SEs. This was not surprising, as ASCL1, NEUROD1, and POU2F3 binding sites have very little overlap, with elevated enhancer signal that is specific to each subtype. Additionally, DNA methylation levels are lower in the binding sites of ASCL1, NEUROD1, and POU2F3 (Yin et al., 2017). Finally, we demonstrate that promoter enhancer signal levels are strong predictors of gene expression, outperforming DNA methylation and DNA methylation-derived copy-number as well. Yet, in the absence of H3K27ac ChIP data, which is the case for clinical samples, our analyses demonstrate the potential of the DNA EPICmethylation 850K array to significantly predict gene expression and match tumors with the SCLC-LTF subgroups and with specific therapies such DLL3-based cytotoxins, temozolomide (based on lack of MGMT) and the DNA replication targeted chemotherapies (etoposide, cisplatin, camptothecin, and lurbinectedin) (based on the expression of SLFN11).
Limitations of the study
Our study has some limitations: 1) DNA methylation was measured with 850K arrays and RRBS (reduced representation bisulfite sequencing), which cover more positions than the 450k array, are still limited to predetermined areas; 2) as gene body methylation is usually deposited through transcription, in cases where a gene is expressed in all cell lines, the gene body methylation level may be less predictive of expression levels (which is similarly seen in case of promoter methylation); 3) Our data are based on cancer cell line models. Therefore, follow-up clinical studies including both RNA-seq and EPICmethylation 850K arrays are warranted to validate the predictive value of the high-resolution DNA methylation arrays in patient samples.
STAR★Methods
Key resources table
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Yves Pommier (yves.pommier@nih.gov).
Material availability
No new materials were generated for this study.
Experimental model and subject details
The cell lines used in this study were from human and include DMS 79 (M), NCI-H1092 (M), NCI-H146 (M), NCI-H1694 (M), NCI-H2029 (F), NCI-H2081 (F), NCI-H209 (M), NCI-H211 (F), NCI-H2141 (M), NCI-H2171 (M), NCI-H446 (M), NCI-H510 (M), NCI-H524 (M), NCI-H526 (M), NCI-H69 (M), NCI-H841 (M), SHP-77 (M), COLO 668 (F), COR L88 (M), COR-L279 (M), DMS 114 (M), DMS 273 (F), DMS 53 (M), NCI-H1105 (M), NCI-H1341 (F), NCI-H196 (M), SW 1271 (M), NCI-H1048 (F), NCI-H1436 (M), NCI-H1836 (M), NCI-H1876 (M), NCI-H1963 (M), NCI-H82 (M), NCI-H889 (F), COR-L47 (M), NCI-H1618 (F), NCI-H2066 (F), NCI-H128 (M), NCI-H1417 (F), NCI-H1522 (M), NCI-H1688 (M), NCI-H187 (M), NCI-H1930 (M), NCI-H2107 (M), NCI-H250 (M), NCI-H345 (M), NCI-H378 (F), NCI-H735 (F), NCI-H748 (M), NCI-H865 (F), COR-L32 (NA), NCI-H220 (M), NCI-H719 (F), NCI-H720 (M), NCI-H847 (M), NCI-H1672 (M), NCI-H1882 (M), NCI-H2198 (M), NCI-H711 (M), NCI-H774 (M), DMS 187 (NA), LXFS 605L (NA), LXFS 650L (NA), NCI-H2330 (F), NCI-H249 (M), NCI-H69/CPR (M), NCI-H69/LX10 (M), NCI-H69VCR/R (M) (F, female; M, male; NA, not available) (Tlemsani et al., 2020).
Method details
Bioinformatics analyses
Processing of raw methylation data
Raw methylation files (idat) format were processed in R using the minfi (v1.34.0) package (Aryee et al., 2014), normalized with the preprocessIllumina() function. Methylation data was then mapped to the hg19 genome version using the BSgenome.Hsapiens.UCSC.hg19 (v1.4.3) package as reference. Normalized methylation values for each cell line were converted to bigwig format using the rtracklayer (v1.48.0) package (Lawrence et al., 2009).
Comparison of methylation distribution in the genome
Methylation probes were annotated to CpG islands, promoters, genic regions and intergenic regions. CpG island annotation in BED format was obtained from the UCSC site for the hg19 genome version. For genic annotation the TxDb.Hsapiens.UCSC.hg19.knownGene (v3.2.2) package was used. Promoters were defined as 2.5kb area upstream and downstream from the transcription start site. Genic regions between the end of the promoter and transcription end site were defined as the gene body. Probes outside of CpGs, promoters and genes were categorized as intergenic probes. Probes were denoted as hypermethylated, if the beta value was above 0.5. The most variable methylation probes were used to perform UMAP clustering of cell lines in each category using the UMAP package (v0.2.7.0) (McInnes et al., 2018) setting the random state to 1, n_components to 2, n_neighbors to 6 and min_dist to 0.01.
DNA methylation profile of genes
The gene model was defined as promoter regions (2.5Kb area upstream and downstream of transcription start site), gene body (area between promoter region and transcription end site), gene upstream area (5kb upstream of promoter), and gene downstream area (5kb downstream of transcription end site). The promoter area was divided into 25 bins, the gene body area was divided into 50 bins, while the upstream and downstream areas were divided into 10 bins each. Genes were separated by expression quintiles. For each quintile, the average signal of each bin in each gene area was calculated, and plotted in R.
Analysis of RRBS based methylation data
The RRBS sequencing data for the CCLE SCLC cell lines were obtained from the SRA (https://www.ncbi.nlm.nih.gov/sra, accession number PRJNA523380). Reads were processed using the bismark (v0.22.3) pipeline (Krueger and Andrews, 2011), first aligning the reads with bismark to the hg19 genome version, followed by extracting the methylation values using the bismark_methylation_extractor with the “--comprehensive --cytosine_report --CX –bedGraph” flags to generate a comprehensive report.
Quantification of gene body methylation
Gene body methylation was obtained for every gene by calculating the mean methylation of probes overlapping gene bodies, excluding CpG probes, and probes that overlap promoter areas. In cases of genes with multiple transcripts, the transcript with the most positive correlation was selected, similar to the promoter methylation analysis (Tlemsani et al., 2020).
Alignment and analysis of ChIP-seq data
We obtained ChIP-seq data from GEO for H3K27ac (GSE151002 and GSE115124) and ASCL1 (GSE69398), NEUROD1 (GSE69398) and POU2F3 (GSE115124). Raw reads were trimmed using trimmomatic (v0.36) (Bolger et al., 2014), followed by read alignment using the bwa mem aligner (v 0.7.17) (Li, 2013). Reads were sorted using samtools (v1.8) (Li et al., 2009), followed by duplicate marking using picard-tools (v2.9.2). H3K27ac peaks were called using the MACS peak caller (Zhang et al., 2008) setting the “—nomodel” flag using the broad peak calling setting. ASCL1, NEUROD1 and POU2F3 peaks were called using the default settings of the MACS peak caller. Normalized sequencing depth coverages were prepared using the BAMscale (v0.06) tool (Pongor et al., 2020). Peaks were annotated using the ChIPseeker package (Yu et al., 2015). In case of the ASCL1, NEUROD1 and POU2F3 ChIP-seq experiments, we compared binding site BED files using the intervene venn program(Khan and Mathelier, 2017), which is also able to separate peaks based on the colocalization. The resulting BED files from the intervene package and bigwig files from BAMscale were used to create the colocalization heatmaps with the heatmapper script available in the BAMscale github repository (https://github.com/ncbi/BAMscale).
Enhancer H3K27ac assignment to gene promoters
When assigning peaks to genes, we first combined peaks that had 50% overlap with each other, and quantified the gene signal with the BAMscale cov function (Pongor et al., 2020), followed by normalization with the sva package (Leek et al., 2012) and TMM-FPKM normalized using the edgeR package (Robinson et al., 2010) followed by log2(TMM-FPKM + 1) transformation. Since multiple peaks can overlap the promoter of a single gene, we prioritized peaks based on distance and average signal. Peaks were prioritized using 2500bp, 5000bp, 10000bp and 15000bp distances intervals from the TSS site, where lower distance intervals had higher priority. To select the appropriate gene and peak pair, we selected peaks which were in the highest distance priority. In cases where multiple peaks were within a priority, the peak with highest mean signal in the cell lines was selected. Finally, we selected the cell replicates that had the best correlation with expression in the top 3000 genes with highest expression range based on RNA-seq data obtained from the SCLC-CellMinerCDB.
Enhancer H3K27ac signal analysis and clustering in cell lines
Enhancer peak calls from each cell line were combined into a single peak set using bedtools (Quinlan and Hall, 2010). Peak signals were quantified using BAMscale cov function (Pongor et al., 2020). Raw read counts were normalized using sva package (Leek et al., 2012) and TMM-FPKM normalized using edgeR (Robinson et al., 2010), and log2 (TMM-FPKM + 1) transformed. Since some cell lines had multiple replicates, we used the replicate that was assigned during the gene promoter assignment analysis. The most variable sites were used in the heatmap analysis, filtering for peaks with standard deviations above 1.25 in the cell lines. The heatmap was drawn using the ComplexHeatmap package (Gu et al., 2016) R package, clustering peaks into 4 groups, and separating cell lines based on their NAPY status. Peaks were annotated using the ChIPseeker (v1.28.3) R package (Yu et al., 2015). Differential pathway analysis of clusters was performed using the clusterProfiler (v4.0.5) package (Yu et al., 2012) and ReactomePA (Yu and He, 2016) in R. Peaks were also clustered using the UMAP package (v0.2.7.0) (McInnes et al., 2019) using default parameters and setting the metric to Euclidean distance. The unrooted phylogenetic tree was constructed using the APE R package (Paradis et al., 2004) using the hclust() hierarchical clustering of Z-scored peaks, setting the clustering to the “ward.D2” method.
Quantification and statistical details
Tests used to determine statistical significance include Wilcoxon signed-rank test (Figure 1C) and Pearson correlation coefficients (Figures 1E, 2D, 2F, 4B–4E, 5, S1B, S1F–S1G, S2C, and S5).
Acknowledgment
The studies performed by NCI members are supported by the Center for Cancer Research, the Intramural Program of the US National Cancer Institute, National Institutes of Health (Z01 BC 006150, ZIA BC010411). Grant support for JDM, JEJ. U01 CA213338, U24 CA213274, P50 CA070907. This work utilized the computational resources of the NIH HPC Biowulf cluster. (http://hpc.nih.gov).
Author contributions
Conceptualization, L.S.P., C.T., B.A.T, M.I.A., W.R., A.T., J.D.M, J.E.J, and Y.P.; Methodology, L.S.P., C.T., B.A.T, M.I.A., W.R., A.T., J.D.M, J.E.J, and Y.P.; Software, L.S.P., C.T., F.E., Y.A, U.J., J.M.G, S.M., S.V., R.K.K., N.R., B.A.T, M.I.A., W.R., A.T., J.D.M, J.E.J, and Y.P; Formal Analysis, L.S.P., C.T., F.E., Y.A, U.J., J.M.G, S.M., S.V., R.K.K., N.R., B.A.T, M.I.A., W.R., A.T., J.D.M, J.E.J, and Y.P.; Data Curation, L.S.P., C.T., F.E. S.V., M.I.A., W.R., A.T., J.D.M, J.E.J, and Y.P.; Writing – Original Draft, L.S.P., C.T., M.I.A., W.R., A.T., J.D.M, J.E.J, and Y.P; Writing – Review & Editing, L.S.P., C.T., F.E., Y.A., U.J., J.M.J, S.M., S.V., R.K.K., N.R., B.A.T., M.I.A, W.R., A.T., J.D.M., J.E.J., and Y.P.
Declaration of interests
The authors declare no competing interests. JDM receives royalties from the NCI and UT Southwestern Medical Center for the distribution of human tumor cell lines.
Published: November 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105338.
Supplemental information
Summary of cell lines with available data type annotated with NAPY status and NE status.
Fraction of hypermethylated probes is shown for each cell line and region type.
Table summarizes the UMAP clustering of each cell using methylation data for all probes, promoter probes, CpG island probes, gene body probes, and intergenic probes.
List of new features added in the new version of the website.
Annotation and quantification results of the most variable enhancers for each cell line used during analysis.
Correlation of gene body methylation, promoter methylation, and CNV used to predict potential super-enhancer covered genes.
Data and code availability
-
•
Analysis scripts used in the study have been deposited to the github code repository and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
DNA methylation and H3K27ac ChIP-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Aryee M.J., Jaffe A.E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A.P., Hansen K.D., Irizarry R.A. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacolod M.D., Barany F. MGMT epigenetics: the influence of gene body methylation and other insights derived from integrated methylomic, transcriptomic, and chromatin analyses in various cancer types. Curr. Cancer Drug Targets. 2021;21:360–374. doi: 10.2174/1568009621666210203111620. [DOI] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borromeo M.D., Savage T.K., Kollipara R.K., He M., Augustyn A., Osborne J.K., Girard L., Minna J.D., Gazdar A.F., Cobb M.H., Johnson J.E. ASCL1 and NEUROD1 reveal heterogeneity in pulmonary neuroendocrine tumors and regulate distinct genetic programs. Cell Rep. 2016;16:1259–1272. doi: 10.1016/j.celrep.2016.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman A.B., Gu H., Bartels S.J.J., Zhang Y., Matarese F., Simmer F., Marks H., Bock C., Gnirke A., Meissner A., Stunnenberg H.G. Sequential ChIP-bisulfite sequencing enables direct genome-scale investigation of chromatin and DNA methylation cross-talk. Genome Res. 2012;22:1128–1138. doi: 10.1101/gr.133728.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler M., Pongor L., Su Y.T., Xi L., Raffeld M., Quezado M., Trepel J., Aldape K., Pommier Y., Wu J. MGMT status as a clinical biomarker in glioblastoma. Trends Cancer. 2020;6:380–391. doi: 10.1016/j.trecan.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao F., Fang Y., Tan H.K., Goh Y., Choy J.Y.H., Koh B.T.H., Hao Tan J., Bertin N., Ramadass A., Hunter E., et al. Super-enhancers and broad H3K4me3 domains form complex gene regulatory circuits involving chromatin interactions. Sci. Rep. 2017;7:2186. doi: 10.1038/s41598-017-02257-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty P., Huang J.T.J., Hiom K. DHX9 helicase promotes R-loop formation in cells with impaired RNA splicing. Nat. Commun. 2018;9:4346. doi: 10.1038/s41467-018-06677-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhayalan A., Rajavelu A., Rathert P., Tamas R., Jurkowska R.Z., Ragozin S., Jeltsch A. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J. Biol. Chem. 2010;285:26114–26120. doi: 10.1074/jbc.M109.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. DNA methylation in cancer: from mouse to human and back again. EBioMedicine. 2021;68:103393. doi: 10.1016/j.ebiom.2021.103393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farago A.F., Yeap B.Y., Stanzione M., Hung Y.P., Heist R.S., Marcoux J.P., Zhong J., Rangachari D., Barbie D.A., Phat S., et al. Combination olaparib and temozolomide in relapsed small cell lung cancer. Cancer Discov. 2019;9:1372–1387. doi: 10.1158/2159-8290.CD-19-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin J.P., Hansen K.D. Reconstructing A/B compartments as revealed by Hi-C using long-range correlations in epigenetic data. Genome Biol. 2015;16:180. doi: 10.1186/s13059-015-0741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay C.M., Stewart C.A., Park E.M., Diao L., Groves S.M., Heeke S., Nabet B.Y., Fujimoto J., Solis L.M., Lu W., et al. Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities. Cancer Cell. 2021;39:346–360.e7. doi: 10.1016/j.ccell.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdar A.F., Bunn P.A., Minna J.D. Small-cell lung cancer: what we know, what we need to know and the path forward. Nat. Rev. Cancer. 2017;17:725–737. doi: 10.1038/nrc.2017.87. [DOI] [PubMed] [Google Scholar]
- George J., Lim J.S., Jang S.J., Cun Y., Ozretić L., Kong G., Leenders F., Lu X., Fernández-Cuesta L., Bosco G., et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevaert O., Tibshirani R., Plevritis S.K. Pancancer analysis of DNA methylation-driven genes using MethylMix. Genome Biol. 2015;16:17. doi: 10.1186/s13059-014-0579-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandi M., Huang F.W., Jané-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R., 3rd, Barretina J., Gelfand E.T., Bielski C.M., Li H., et al. Next-generation characterization of the cancer cell line encyclopedia. Nature. 2019;569:503–508. doi: 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindan R., Page N., Morgensztern D., Read W., Tierney R., Vlahiotis A., Spitznagel E.L., Piccirillo J. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: analysis of the surveillance, epidemiologic, and end results database. J. Clin. Oncol. 2006;24:4539–4544. doi: 10.1200/JCO.2005.04.4859. [DOI] [PubMed] [Google Scholar]
- Greenberg M.V.C., Bourc'his D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019;20:590–607. doi: 10.1038/s41580-019-0159-6. [DOI] [PubMed] [Google Scholar]
- Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- Hao X., Luo H., Krawczyk M., Wei W., Wang W., Wang J., Flagg K., Hou J., Zhang H., Yi S., et al. DNA methylation markers for diagnosis and prognosis of common cancers. Proc. Natl. Acad. Sci. USA. 2017;114:7414–7419. doi: 10.1073/pnas.1703577114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervouet E., Peixoto P., Delage-Mourroux R., Boyer-Guittaut M., Cartron P.F. Specific or not specific recruitment of DNMTs for DNA methylation, an epigenetic dilemma. Clin. Epigenet. 2018;10:17. doi: 10.1186/s13148-018-0450-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Zhang X., Zong L., Gao Q., Zhang C., Wei R., Guan Y., Huang L., Zhang L., Lyu G., Tao W. Gene body methylation safeguards ribosomal DNA transcription by preventing PHF6-mediated enrichment of repressive histone mark H4K20me3. J. Biol. Chem. 2021;297:101195. doi: 10.1016/j.jbc.2021.101195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.H., Klingbeil O., He X.Y., Wu X.S., Arun G., Lu B., Somerville T.D.D., Milazzo J.P., Wilkinson J.E., Demerdash O.E., et al. POU2F3 is a master regulator of a tuft cell-like variant of small cell lung cancer. Genes Dev. 2018;32:915–928. doi: 10.1101/gad.314815.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo U., Murai Y., Takebe N., Thomas A., Pommier Y. Precision oncology with drugs targeting the replication stress, ATR, and schlafen 11. Cancers. 2021;13:4601. doi: 10.3390/cancers13184601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A., Mathelier A. Intervene: a tool for intersection and visualization of multiple gene or genomic region sets. BMC Bioinf. 2017;18:287. doi: 10.1186/s12859-017-1708-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A., Mathelier A., Zhang X. Super-enhancers are transcriptionally more active and cell type-specific than stretch enhancers. Epigenetics. 2018;13:910–922. doi: 10.1080/15592294.2018.1514231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelsche C., Schrimpf D., Stichel D., Sill M., Sahm F., Reuss D.E., Blattner M., Worst B., Heilig C.E., Beck K., et al. Sarcoma classification by DNA methylation profiling. Nat. Commun. 2021;12:498. doi: 10.1038/s41467-020-20603-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F., Andrews S.R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krushkal J., Silvers T., Reinhold W.C., Sonkin D., Vural S., Connelly J., Varma S., Meltzer P.S., Kunkel M., Rapisarda A., et al. Epigenome-wide DNA methylation analysis of small cell lung cancer cell lines suggests potential chemotherapy targets. Clin. Epigenet. 2020;12:93. doi: 10.1186/s13148-020-00876-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu K., Cardnell R.J., Zhang B., Shen L., Stewart C.A., Ramkumar K., Cargill K.R., Wang J., Gay C.M., Byers L.A. SLFN11 biomarker status predicts response to lurbinectedin as a single agent and in combination with ATR inhibition in small cell lung cancer. Transl. Lung Cancer Res. 2021;10:4095–4105. doi: 10.21037/tlcr-21-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M., Gentleman R., Carey V. rtracklayer: an R package for interfacing with genome browsers. Bioinformatics. 2009;25:1841–1842. doi: 10.1093/bioinformatics/btp328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev Maor G., Yearim A., Ast G. The alternative role of DNA methylation in splicing regulation. Trends Genet. 2015;31:274–280. doi: 10.1016/j.tig.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013 doi: 10.48550/arXiv.1303.3997. Preprint at. [DOI] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Zhang J., Huang S., He X. Genome-wide analysis reveals that exon methylation facilitates its selective usage in the human transcriptome. Briefings Bioinf. 2018;19:754–764. doi: 10.1093/bib/bbx019. [DOI] [PubMed] [Google Scholar]
- Lister R., Pelizzola M., Dowen R.H., Hawkins R.D., Hon G., Tonti-Filippini J., Nery J.R., Lee L., Ye Z., Ngo Q.M., et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna A., Elloumi F., Varma S., Wang Y., Rajapakse V.N., Aladjem M.I., Robert J., Sander C., Pommier Y., Reinhold W.C. CellMiner Cross-Database (CellMinerCDB) version 1.2: exploration of patient-derived cancer cell line pharmacogenomics. Nucleic Acids Res. 2021;49:D1083–D1093. doi: 10.1093/nar/gkaa968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L., Healy J., Saul N., Großberger L. UMAP: uniform manifold approximation and projection. J. Open Source Softw. 2018;3:861. doi: 10.21105/joss.00861. [DOI] [Google Scholar]
- Morgensztern D., Besse B., Greillier L., Santana-Davila R., Ready N., Hann C.L., Glisson B.S., Farago A.F., Dowlati A., Rudin C.M., et al. Efficacy and safety of rovalpituzumab tesirine in third-line and beyond patients with DLL3-expressing, relapsed/refractory small-cell lung cancer: results from the phase II TRINITY study. Clin. Cancer Res. 2019;25:6958–6966. doi: 10.1158/1078-0432.CCR-19-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai J., Feng Y., Yu G.K., Ru Y., Tang S.W., Shen Y., Pommier Y. Resistance to PARP inhibitors by SLFN11 inactivation can be overcome by ATR inhibition. Oncotarget. 2016;7:76534–76550. doi: 10.18632/oncotarget.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai J., Thomas A., Miettinen M., Pommier Y. Schlafen 11 (SLFN11), a restriction factor for replicative stress induced by DNA-targeting anti-cancer therapies. Pharmacol. Ther. 2019;201:94–102. doi: 10.1016/j.pharmthera.2019.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri F., Rapelli S., Krepelova A., Incarnato D., Parlato C., Basile G., Maldotti M., Anselmi F., Oliviero S. Intragenic DNA methylation prevents spurious transcription initiation. Nature. 2017;543:72–77. doi: 10.1038/nature21373. [DOI] [PubMed] [Google Scholar]
- Nogales V., Reinhold W.C., Varma S., Martinez-Cardus A., Moutinho C., Moran S., Heyn H., Sebio A., Barnadas A., Pommier Y., Esteller M. Epigenetic inactivation of the putative DNA/RNA helicase SLFN11 in human cancer confers resistance to platinum drugs. Oncotarget. 2016;7:3084–3097. doi: 10.18632/oncotarget.6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Barahona V., Joshi R.S., Esteller M. Use of DNA methylation profiling in translational oncology. Semin. Cancer Biol. 2020;83:523–535. doi: 10.1016/j.semcancer.2020.12.011. [DOI] [PubMed] [Google Scholar]
- Otani J., Nankumo T., Arita K., Inamoto S., Ariyoshi M., Shirakawa M. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep. 2009;10:1235–1241. doi: 10.1038/embor.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E., Claude J., Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Piunti A., Shilatifard A. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science. 2016;352:aad9780. doi: 10.1126/science.aad9780. [DOI] [PubMed] [Google Scholar]
- Poirier J.T., George J., Owonikoko T.K., Berns A., Brambilla E., Byers L.A., Carbone D., Chen H.J., Christensen C.L., Dive C., et al. New approaches to SCLC therapy: from the laboratory to the clinic. J. Thorac. Oncol. 2020;15:520–540. doi: 10.1016/j.jtho.2020.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polley E., Kunkel M., Evans D., Silvers T., Delosh R., Laudeman J., Ogle C., Reinhart R., Selby M., Connelly J., et al. Small cell lung cancer screen of oncology drugs, investigational agents, and gene and microRNA expression. J. Natl. Cancer Inst. 2016;108:djw122. doi: 10.1093/jnci/djw122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongor L.S., Gross J.M., Vera Alvarez R., Murai J., Jang S.M., Zhang H., Redon C., Fu H., Huang S.Y., Thakur B., et al. BAMscale: quantification of next-generation sequencing peaks and generation of scaled coverage tracks. Epigenet. Chromatin. 2020;13:21. doi: 10.1186/s13072-020-00343-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo K., Kollipara R.K., Kelenis D.P., Rodarte K.E., Ullrich M.S., Zhang X., Minna J.D., Johnson J.E. ASCL1, NKX2-1, and PROX1 co-regulate subtype-specific genes in small-cell lung cancer. iScience. 2021;24:102953. doi: 10.1016/j.isci.2021.102953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu S., Fetsch P., Thomas A., Pommier Y., Schrump D.S., Miettinen M.M., Chen H. Molecular subtypes of primary SCLC tumors and their associations with neuroendocrine and therapeutic markers. J. Thorac. Oncol. 2022;17:141–153. doi: 10.1016/j.jtho.2021.08.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajapakse V.N., Luna A., Yamade M., Loman L., Varma S., Sunshine M., Iorio F., Sousa F.G., Elloumi F., Aladjem M.I., et al. CellMinerCDB for integrative cross-database genomics and pharmacogenomics analyses of cancer cell lines. iScience. 2018;10:247–264. doi: 10.1016/j.isci.2018.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold W.C., Varma S., Sunshine M., Rajapakse V., Luna A., Kohn K.W., Stevenson H., Wang Y., Heyn H., Nogales V., et al. The NCI-60 methylome and its integration into CellMiner. Cancer Res. 2017;77:601–612. doi: 10.1158/0008-5472.CAN-16-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium. Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin C.M., Poirier J.T., Byers L.A., Dive C., Dowlati A., George J., Heymach J.V., Johnson J.E., Lehman J.M., MacPherson D., et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat. Rev. Cancer. 2019;19:289–297. doi: 10.1038/s41568-019-0133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari J.K., Lok B.H., Laird J.H., Poirier J.T., Rudin C.M. Unravelling the biology of SCLC: implications for therapy. Nat. Rev. Clin. Oncol. 2017;14:549–561. doi: 10.1038/nrclinonc.2017.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spainhour J.C., Lim H.S., Yi S.V., Qiu P. Correlation patterns between DNA methylation and gene expression in the cancer genome atlas. Cancer Inform. 2019;18 doi: 10.1177/1176935119828776. 1176935119828776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J., Huang Y.H., Cui X., Wang X., Zhang X., Lei Y., Xu J., Lin X., Chen K., Lv J., et al. Homeobox oncogene activation by pan-cancer DNA hypermethylation. Genome Biol. 2018;19:108. doi: 10.1186/s13059-018-1492-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S.W., Thomas A., Murai J., Trepel J.B., Bates S.E., Rajapakse V.N., Pommier Y. Overcoming resistance to DNA-targeted agents by epigenetic activation of schlafen 11 (SLFN11) expression with class I histone deacetylase inhibitors. Clin. Cancer Res. 2018;24:1944–1953. doi: 10.1158/1078-0432.CCR-17-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teissandier A., Bourc'his D. Gene body DNA methylation conspires with H3K36me3 to preclude aberrant transcription. EMBO J. 2017;36:1471–1473. doi: 10.15252/embj.201796812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A., Pommier Y. Small cell lung cancer: time to revisit DNA-damaging chemotherapy. Sci. Transl. Med. 2016;8:346fs12. doi: 10.1126/scitranslmed.aaf6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A., Tanaka M., Trepel J., Reinhold W.C., Rajapakse V.N., Pommier Y. Temozolomide in the era of precision medicine. Cancer Res. 2017;77:823–826. doi: 10.1158/0008-5472.CAN-16-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tlemsani C., Pongor L., Elloumi F., Girard L., Huffman K.E., Roper N., Varma S., Luna A., Rajapakse V.N., Sebastian R., et al. SCLC-CellMiner: a resource for small cell lung cancer cell line genomics and pharmacology based on genomic signatures. Cell Rep. 2020;33:108296. doi: 10.1016/j.celrep.2020.108296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully K.M., Tendler S., Carter L.M., Sharma S.K., Samuels Z.V., Mandleywala K., Korsen J.A., Delos Reyes A.M., Piersigilli A., Travis W.D., et al. Radioimmunotherapy targeting delta-like ligand 3 in small cell lung cancer exhibits antitumor efficacy with low toxicity. Clin. Cancer Res. 2022;28:1391–1401. doi: 10.1158/1078-0432.CCR-21-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Han H., De Carvalho D.D., Lay F.D., Jones P.A., Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26:577–590. doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., Morgunova E., Jolma A., Kaasinen E., Sahu B., Khund-Sayeed S., Das P.K., Kivioja T., Dave K., Zhong F., et al. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science. 2017;356:eaaj2239. doi: 10.1126/science.aaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., He Q.Y. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol. Biosyst. 2016;12:477–479. doi: 10.1039/c5mb00663e. [DOI] [PubMed] [Google Scholar]
- Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.G., He Q.Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- Zhang X., Jeong M., Huang X., Wang X.Q., Wang X., Zhou W., Shamim M.S., Gore H., Himadewi P., Liu Y., et al. Large DNA methylation nadirs anchor chromatin loops maintaining hematopoietic stem cell identity. Mol. Cell. 2020;78:506–521.e6. doi: 10.1016/j.molcel.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Jurkowska R., Soeroes S., Rajavelu A., Dhayalan A., Bock I., Rathert P., Brandt O., Reinhardt R., Fischle W., Jeltsch A. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res. 2010;38:4246–4253. doi: 10.1093/nar/gkq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoppoli G., Regairaz M., Leo E., Reinhold W.C., Varma S., Ballestrero A., Doroshow J.H., Pommier Y. Putative DNA/RNA helicase Schlafen-11 (SLFN11) sensitizes cancer cells to DNA-damaging agents. Proc. Natl. Acad. Sci. USA. 2012;109:15030–15035. doi: 10.1073/pnas.1205943109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of cell lines with available data type annotated with NAPY status and NE status.
Fraction of hypermethylated probes is shown for each cell line and region type.
Table summarizes the UMAP clustering of each cell using methylation data for all probes, promoter probes, CpG island probes, gene body probes, and intergenic probes.
List of new features added in the new version of the website.
Annotation and quantification results of the most variable enhancers for each cell line used during analysis.
Correlation of gene body methylation, promoter methylation, and CNV used to predict potential super-enhancer covered genes.
Data Availability Statement
-
•
Analysis scripts used in the study have been deposited to the github code repository and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
DNA methylation and H3K27ac ChIP-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.