Summary
Base pairs are fundamental building blocks of RNA. The base pairs of low stability are often critical in RNA functions. Here, we develop a solid-state NMR-based water-RNA exchange spectroscopy (WaterREXSY) to characterize RNA in solid. The approach uses different chemical exchange rates between iminos and water to evaluate base pair stability; the less stable ones would exchange more frequently, leading to stronger cross-peaks on WaterREXSY. Applied to the riboA71-adenine complex (the 71nt-aptamer domain of add adenine riboswitch from Vibrio vulnificus), the U47⋅U51 base pair, which is critical in ligand binding, was found to be less stable than other base pairs. The imino-water exchange rates of U47 at different temperatures are about 500–800 s−1, indeed indicative of low stability. This implies a highly complex and plastic triad involving U47⋅U51 and that the opening of the U47⋅U51 base pair may be the early stage of ligand release.
Subject areas: Biological sciences, Biochemistry, Structural biology
Graphical abstract

Highlights
-
•
A solid-state nuclear magnetic resonance (NMR) exchange spectroscopy to study RNA base pair stability
-
•
Low stable base pair exhibits strong cross-peaks on WaterREXSY spectra
-
•
The U47-U51 base pair of riboA71-adenine is less stable than other base pairs
Biological sciences; Biochemistry; Structural biology.
Introduction
Base pairs formed via inter-nucleotide hydrogen bonds are the fundamental building blocks of RNA. Each base pair involves a complex hierarchy of internal motions on various time scales (Al-Hashimi and Walter, 2008; Marusic et al., 2019; Nguyen and Qin, 2012; Rinnenthal et al., 2011a). The relative stability of an individual base pair determines the thermodynamics and kinetics of its open-close equilibrium, which depends on the nature of base pairs and the sequence context (Huang et al., 2009; Kierzek et al., 1999). Stable base pairs constituting the scaffold of RNA show slow opening kinetics. In contrast, base pairs of relatively low stability experience faster open-close dynamics that have seminal effects on RNA structures and functions, i.e., RNA folding (Manosas et al., 2008), local structural stability (Okada et al., 2006; Sheng et al., 2013), and site-specific catalysis in ribozymes (Juneja et al., 2014; Roy et al., 2008).
Site-specific characterization of the dynamics of the RNA base pairs with low stability is still challenging. Solution nuclear magnetic resonance (NMR) spectroscopy uses water-RNA magnetization transfer to indicate the thermodynamics and kinetics of base pair opening equilibria by measuring the apparent longitudinal relaxation rate constants of the imino protons with and without selectively inverting the water signal at the beginning of experiments (Huang et al., 2011; Kim et al., 2017; Lee et al., 2008; Lee and Pardi, 2007; Rinnenthal et al., 2010; Russu, 2004; Snoussi and Leroy, 2001; Wagner et al., 2015) or by phase-modulated CLEANEX approaches (Lee et al., 2009). However, solution NMR is often limited by molecular size and solubility of large RNA or RNA-protein complex (RNP), often not applicable in solid RNA (Bothe et al., 2011). It is also difficult to study RNA dynamics on the ns-μs timescale (Marusic et al., 2019). Other biophysical techniques can also be used to study RNA dynamics and thermodynamics, but they cannot provide site-specific information nor be applied in the solid state (Kortmann et al., 2011; Rinnenthal et al., 2011b). Recently, solid-state NMR (SSNMR) emerges as an important tool in studying large RNAs (Leppert et al., 2004; Riedel et al., 2006, 2005a, 2005b) and RNPs (Ahmed et al., 2020; Asami et al., 2013; Carlomagno, 2014; Marchanka and Carlomagno, 2019; Marchanka et al., 2015), and in the characterization of RNA dynamics over a wider range of timescales (Schanda and Ernst, 2016). To date, SSNMR-based deuterium line shape analysis has been used to obtain the ns-ms timescale overall dynamics of RNA (Emani et al., 2010; Huang et al., 2017; Olsen et al., 2008, 2009, 2010). However, SSNMR methods for the site-specific characterization of base pair dynamics in RNA have not been established. In relatively rigid biomolecules where strong proton-proton spin diffusion dominates the proton spin relaxation, the solution NMR-based strategies that measure proton relaxation rates for characterizing become invalid. Similarly, the solution NMR-based CLEANEX approaches (Hwang et al., 1997, 1998; Modig et al., 2004; Yuwen et al., 2018) cannot suppress the proton-proton dipolar couplings, thus not applicable in solid-state NMR. Thus, for RNA involved in the condensed phase, i.e., those involved in pathological-related RNA foci (Jain and Vale, 2017), a site-specific SSNMR-based method is urgently needed.
In this study, we developed an SSNMR-based two-dimensional (2D) 1H-15N water-RNA exchange spectroscopy (WaterREXSY) to detect the RNA base pairs dynamics, based on the one-dimensional (1D) SSNMR methodology for studying pure chemical exchange taking placed in membrane-bound proteins with the suppression of proton-proton spin diffusion (Fu et al., 2016). Our approach uses the chemical exchange between the imino protons involved in hydrogen bonds within base pairs and the water molecules to excite the imino proton signals, while the proton-proton spin diffusion is suppressed by a 1H Lee-Goldburg spin lock (LGSL) block (Zhang et al., 2021). The method can be used to characterize the imino-water chemical exchange with timescales of sub-millisecond. Because of their faster opening dynamics, the base pairs with low stability would experience more frequent exchanges between iminos and water, leading to a rapid build-up and strong cross-peaks on WaterREXSY spectra. In contrast, very stable base pairs are weakly or simply not detectable because of their slow exchange rates. Therefore, this approach detects selectively base pairs with low stability in RNA.
This strategy was used to characterize the adenine-bound form of a 71-nt aptamer domain of the add adenine riboswitch (riboA71). This purine riboswitch regulates add gene expression upon adenine binding (Reining et al., 2013; Serganov et al., 2004). The structure of the riboA71 complex with adenine has been determined (Zhang and Ferre-D'Amare, 2014) (Figure 1). Solution NMR indicated multiple conformational states exchanging with each other in solution (Ding et al., 2019; Reining et al., 2013). However, the relative stabilities of the base pairs in riboA71, particularly those around the ligand binding site, were still unclear. Using the WaterREXSY approach, we demonstrated that the imino proton of U47 within the U47·U51 base pair involved in ligand binding represents a less stable base pair with an imino-water exchange rate constant of about 500–800 s−1 at 15–35°C. The U47⋅U51 base pair in the minor groove of the riboA71 ligand binding pocket encompasses the complex hydrogen bond network with the U47⋅U51⋅(adenine-U74) base tetrad. The molecular dynamics simulation showed that U47⋅U51, but not other base pairs within this tetrad, experiences an equilibrium between the formation and breakage of the base pair. This result suggests that the opening of the U47·U51 base pair may be involved in the early stage of complex dissociation upon ligand release.
Figure 1.
Secondary and crystal structure of riboA71 in the adenine-bound form (PDB: 4TZX)
Stems P1, P2(L2), and P3(L3) are shown in green, orange, and blue. Junction-connecting segments J1-2, J2-3, and J3-1 are colored in cyan, hot pink, and violet. The symbol “A” presents the adenine ligand. The U47 and the U49 are colored in light blue. The dash lines represent the non-canonical base pairs around the ligand binding pocket.
Results
2D WaterREXSY experiments to identify base pairs with low stability in RNA
Differences in base pair stability within RNA can be determined by the chemical exchange between the imino protons involved in hydrogen bonds within base pairs and the water molecules (Figure 2A). According to the “open-close model”, base pairs in RNA experience equilibrium between the open state, wherein hydrogen bonds are broken, and the closed state, wherein hydrogen bonds are formed (Figure S2) (Snoussi and Leroy, 2001; Szulik et al., 2014). In the open state, imino protons are exchangeable with bulk water, while in the closed state, the water-imino exchange does not occur. Because the open kinetics (∼1–100 s−1) (Huang and Russu, 2017; Russu, 2004) is much slower than the imino-water exchange in the open state without catalyst (∼106 s−1) (Lee and Pardi, 2007), the base pair opening is the rate-limiting step of the water-imino exchange. Therefore, this process can be used as a “probe” to characterize the base pair stability in RNA. A stable base pair has strong hydrogen bonds, which lead to a relatively low population of the open state and a slow exchange between water and iminos. In contrast, the imino protons in less stable base pairs have much faster exchange rates because of the less stable hydrogen bonds. Empirically, the G-C and A-U canonical Watson-Crick RNA base pairs in double-stranded RNAs, are generally stable and have water-RNA exchange rates of 1–10 s−1 and 20–100 s−1, respectively (Lee et al., 2008; Maltseva et al., 1995; Mirau and Kearns, 1984; Snoussi and Leroy, 2001). The less stable base pairs have faster exchange rates (Lee et al., 2008; Varnai et al., 2004); earlier work has reported the exchange rate as fast as 200 s−1 between nucleic acids and water (Maltseva et al., 1995). Solution-state NMR can detect the RNA base pair opening with an exchange rate up to 400 s−1, beyond that it becomes not observable owing to exchange broadening (Lee et al., 2008). According to earlier works by Wagner, D. et al., imino protons of base pairs with kex rates of 100 s−1 or above are significantly less stable. In terms of the Gibbs energies to break up a base pair (ΔGdiss), the A-U and G-C base pairs were found to have a range from 10.1 to 26.5 kJ/mol and from 26.2 to 48.8 kJ/mol, respectively (Wagner et al., 2015). The base pairs with low stability would have ΔGdiss smaller than 10 kJ/mol.
Figure 2.
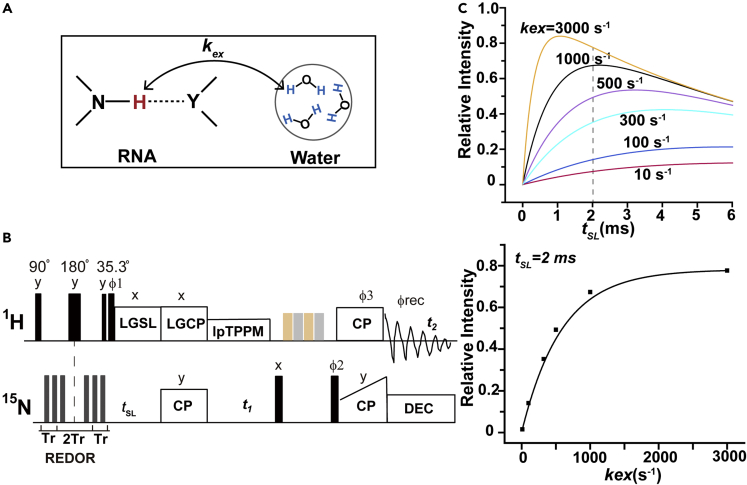
Theoretical considerations of the waterREXSY scheme
(A) Simplified water-RNA exchange model, where Y represents nitrogen or oxygen atom. The kex is the exchange rate constant for imino proton transfer with water.
(B) Pulse sequence of the 2D WaterREXSY.
(C) Numerical simulations of the time-dependent build-up in the 2D 1H-15N WaterREXSY experiments with T1ρ = 8 ms.
SSNMR spectroscopy edited by the water-RNA exchange rates could be used to characterize base pairs with relatively low stability. Here, we designed an SSNMR-based 2D 1H-15N WaterREXSY experiment to achieve this target. Figures 2B and S1 show the 2D WaterREXSY scheme. It includes two key steps: (i) a 1H-15N rotational echo double resonance (REDOR) period that dephases the initial imino proton magnetization but not the water, and (ii) a 1H LGSL, during which the water-RNA chemical exchange occurs and repolarizes the imino proton magnetization that is dephased during the REDOR period, while the proton-proton spin diffusion is effectively suppressed. Subsequently, the imino nitrogen resonances from the water-RNA exchange are recorded in the proton-detected 2D 1H-15N correlation spectra. Because the LGSL suppresses proton-proton spin-diffusion, the scheme avoids interference from their dipolar interactions and allows for the detection of pure chemical exchange (Zhang et al., 2021). In principle, this is the 2D version of the water-protein chemical exchange scheme developed from the previously described 1D version (Fu et al., 2016). Besides, the current work used the proton-detected experiments under a fast MAS rate (40 kHz), while the earlier work used the 15N-detected under a moderate MAS rate (10 kHz) (Fu et al., 2016). The proton detected 2D approach makes better sensitivity and chemical shift dispersion, with respect to the 15N-based 1D approach. Importantly, the 1H-15N 2D spectra combined with specific isotope labeling approaches could provide the site-specific information needed for RNA studies, which are not obtainable with the 1D 15N spectra.
The analytical solution of the Solomon equations (Fu et al., 2016) (Equation 1) shows that the build-up rate of the cross-peaks on 2D WaterREXSY is determined by the exchange rate constant (kex), and the proton relaxation time at the rotating frame (T1ρ) (Equation 1 in STAR methods). Figures 2C and S3 show the simulations of the time-dependent build-up in the 2D WaterREXSY experiments against tSL. The results indicate that as kex increases, a shorter tSL is needed to achieve equilibrium. For example, at a tSL of 2 ms, the intensity of an imino proton with kex of 500 s−1 (i.e. the base pairs with low stability) would be approximately 30-fold and 3-fold stronger than those of protons with kex of 10 s−1 and 100 s−1 (i.e., the stable G-C and A-U base pairs in double-stranded RNAs), respectively. It is also worth noting that the detection of very stable base pairs under this scheme would not be applicable even at a long tSL, because the relaxation loss would dramatically reduce efficiency. Therefore, the WaterREXSY scheme would obtain strong cross-peaks of iminos having kex with a time scale of sub-millisecond. Furthermore, fitting a build-up curve generated by a series of experiments with different tSL values would yield a quantitative kex for the observed hydrogen bonded imino proton in a less stable base pair.
The U47 is involved in less stable base pair within the riboA71-adenine complex in solid state
The 2D WaterREXSY scheme was applied to the riboA71-adenine complex in order to characterize the base pairs with low stability. In this work, the riboA71-adenine sample was prepared by ethanol-precipitation approach, according to earlier work (Zhao et al., 2019). A comparison of the spectral patterns of the precipitated riboA71-adenie complex in solid-state NMR and that in solution indicated the preservation of certain local structures as reported previously. However, This condition is different from the solution condition, thus possibly leading to different base pair dynamics.
The 2D hNH spectrum and several 2D WaterREXSY spectra at different tSL were collected to investigate the base pair dynamics in solid state. Figures 3A and S4 show the 2D hNH spectrum of the riboA71-adenine complex. The imino protons involved in the hydrogen bonds of base pairs are visible with characteristic chemical shifts, including imino groups in the uridines in both A-U Watson-Crick base pairs (15N and 1H chemical shifts of 162.7 ppm and 13–15 ppm, respectively) and non-canonical base pairs (G·U, A·U and U·U, with15N and 1H chemical shifts of 158.6 ppm and 10–12 ppm) and the guanines in G-C Watson-Crick base pairs (15N and 1H chemical shifts of 146.5 ppm and 11–14 ppm, respectively). The proton signals of imino of uridines in non-canonical base pairs, A-U base pairs and G-C base pairs were plotted against tSL. The 2D WaterREXSY spectra at tSL values of 0.5–1.2 ms include only the imino signals of uridines involved in non-canonical base pairs and reach the maximum at a tSL of 4 ms (Figures 3B–3G). In contrast, the imino peaks within the A-U and G-C Watson-Crick base pairs began to appear at a tSL of 2 and 3 ms (Figures 3C, 3D, 3I, and 3J), respectively. These iminos within the A-U and G-C regions of the 2D WaterREXSY spectra may represent less stable canonical base pairs in riboA71, i.e., those at the edges of stems.
Figure 3.
The WaterREXSY spectra of riboA71-adenine complex
(A) 2D hNH spectrum of the riboA71-adenine complex.
(B–E) 2D WaterREXSY spectra of riboA71-adenine complex at tSL of 2 ms (B), 3 ms (C), 4 ms (D) and 5 ms (E). The imino group regions are shown. The gray dash lines indicate the 15N chemical shifts of the 1D-1H extracted slices.
(F) The 1H signals against tSL of uridine imino protons depict non-canonical base pairs (blue), A-U base pairs (red), and guanines in G-C base pairs (gray).
(G, I, and J) 1D 1H slices with 15N chemical shifts of 158.6 ppm (G), 162.7 ppm (I), and 146.5 ppm (J). Non-canonical base pairs, A-U base pairs, and guanines in G-C base pairs were marked in blue, red, and gray, respectively.
(H) Structures of A-U, G-C Watson-Crick base pair, and U·U non-canonical base pair. The iminos are highlighted in red.
The above data suggest the presence of non-canonical base pairs with low stabilities involving uridines. Solution NMR revealed that the riboA71-adenine complex has five cross-peaks in this region, including U28, U31, and U39 in the P2 stem and U47 and U49 in J2-3 (Figure S6) (Ding et al., 2019; Noeske et al., 2007). To assign the less stable base pairs observed on WaterREXSY, we prepared two additional 15N,13C-uridine-labeled RNA samples, including a ligand-free wild-type riboA71 (Figure 4A) and a U31A/G42A mutant riboA71-adenine complex (Figure 4B). The NMR chemical shifts of the imino groups of the uridines in both solution and solid-state NMR were summarized in Table S2.
Figure 4.
SSNMR and solution NMR spectra of ligand-free wild-type riboA71 and U31A/G42A mutant riboA71-adenine complex
(A and B) Secondary structures of ligand-free wild-type riboA71 (A) and the U31A/G42A mutant riboA71-adenine complex (B).
(C) The intensity (I) of an imino proton of U47 at different tSL; the red line represents the fitting curve. The error bars in Figure 4C represent the noise level of the signals.
(D) 2D 1H-15N HSQC spectrum of ligand-free wild-type riboA71 in solution.
(E and F) 2D hNH (E) and WaterREXSY spectra (F) of ligand-free wild-type riboA71 in the solid state.
(G) 2D 1H-15N HSQC spectrum of the U31A/G42A mutant riboA71-adenine complex in solution.
(H and I) 2D hNH (H) and WaterREXSY spectra (I) of the U31A/G42A mutant riboA71-adenine complex in the solid state. tSL was set at 2 ms in both 2D WaterREXSY experiments. Only the regions of uridine imino groups in non-canonical base pairs are shown.
When we analyzed the ligand-free wild-type riboA71, we could not observe U47 and U49 resonances in the 1H-15N heteronuclear single-quantum coherence (HSQC) in solution, possibly owing to prominent dynamics around the binding site (Batey, 2012; Gilbert et al., 2006). Only three imino groups corresponding to U28, U31, and U39 in the stems were observed in the region representative of uridines within non-canonical base pairs (Figures 4D and S6). The cross-peaks of U28 were observed in the 2D hNH SSNMR spectrum of ligand-free wild-type riboA71 (Figure 4E), but no signals were detected in the 2D WaterREXSY spectrum at tSL of 2 ms (Figures 4F and S5). Increasing tSL to 5 ms, a weak peak of U28 started to appear, with only about 8% intensity with respect to that of hNH spectrum (Figure S5). This implies that the imino proton of U28 experiences a much slower water-imino exchange, not the base pair with low stability mentioned above. The U31 and U39 that are located at the edge of the P2 stem were not observed on the 2D hNH spectrum of ligand-free wild-type riboA71, possibly experiencing multiple local conformations in solid condition. Thus, we cannot conclude their characters.
In the U31A/G42A mutant riboA71-adenine complex, G42 and U31 were mutated to adenine. Consequently, the U31·U39 and U28·G42 non-canonical base pairs were changed to the A31-U39 and U28-A42 Watson-Crick base pairs, respectively, which led to a shift in the corresponding cross-peaks of the uridine imino groups to the characteristic region corresponding to the A-U Watson-Crick base pairs. In this scenario, only two iminos, U47 and U49, remained in the region of uridines within non-canonical base pairs, and these had identical chemical shifts with respect to those observed in the wild-type riboA71-adenine complex (Figures 4G and S6). In SSNMR, two cross-peaks were detected in the region corresponding to non-canonical uridines (Figure 4H). One of the two peaks (with 1H and 15N chemical shifts of 10.6 and 159.5 ppm, respectively) was assigned to U49, owing to nearly identical 1H and 15N chemical shifts with respect to the U49 in solution NMR (Figures 4H and S6; Table S2). The other peak with 1H and 15N chemical shifts of 10.7 and 158.1 ppm was assigned to U47, according to the U47 cross peak in solution NMR (1H and 15N chemical shifts of 10.9 and 158.3 ppm, respectively).
The U47 signal, but not the U49, started to appear on the 2D WaterREXSY spectra at a tSL of 1 ms (about 18% signal strength compared with hNH spectrum). A comparison of the 15N chemical shift of this cross-peak with that in the 2D hNH SSNMR spectra allowed us to assign that peak as U47 (Figure 4I), thus demonstrating that U47 is involved in a less stable base pair. Figure 4C shows the cross-peak intensity of the imino group of U47 as a function of tSL at 15°C. The fitting of these peak intensities against tSL in accordance with Equation 1 yields a kex value of 546 ± 131 s−1 for U47, which is consistent with the expected exchange rate constants of base pairs with low stability. The U49 did not appear even with a tSL of 5 ms, showing that the imino-water exchange of U49 is too slow to be detected by WaterREXSY.
Furthermore, we measured the temperature-dependent kex values for the imino of U47, collected on the U31A/G42A riboA71-adenine complex sample at 15°C, 20°C, 25°C, 30°C, and 35°C. Fitting of these peak intensities against tSL in accordance with Equation 1 yields the kex values at different temperatures. At 15°C, 20°C, 25°C, 30 and 35°C, the kex values are 546 ± 131 s−1, 689 ± 126 s−1, 772 ± 132 s−1, 672 ± 137 s−1and 630 ± 126 s−1 respectively (Figure S7). All these values are higher than 100 s−1, indicative of low stability of this base pair.
The temperature-dependent stability of the base pairs of the U47 and the U79 in the solution
The solution NMR data were further collected to compare the stabilities of those non-canonical base pairs of riboA71-adenine complex in solution (Figure S8). The known solution NMR methods, e.g., those based on 1H-T1 relaxation rates, can only measure the imino-water exchange rates slower than 400 s−1 (Lee et al., 2008), incapable of studying kex in sub-millisecond timescale. Here, alternatively, the imino-water exchanges were determined by water pre-saturation coupled with HSQC, in which the imino signal intensities would be reduced owing to water-imino exchange. This method cannot derive quantitative results, but is capable of evaluating the relative stabilities of those non-canonical base pairs. At 5–15°C, the changing trend of signal intensities over saturation periods indicates that both U47 and U49 experience faster chemical exchange with water than other uridines, e.g., U28, U31, and U39 involved in non-canonical base pairs (Figures S8 and S9). It suggests that both U47 and U49 are involved in less stable base pairs in the temperature range of 5–15°C. Differently, at 20°C, the decay curves suggested that U47 has a similar exchange rate with U28, U31, and U39, but slower than U49 (Figure S8), demonstrating that U47 becomes more stable than U49 at 20°C. These results also showed that the stability of the U47 increased with an increase in the temperature range used in this work.
The imino-water exchanges in the solution were also measured on the U31A/G42A riboA71-adenine complex sample. Figure S10 compared the exchange of the U47 and the U49 to demonstrate the relative stability of the base pairs involving those nucleotides. Similar trends of relative stabilities were observed for the base pairs involving the U47 and the U49. At 5–15°C, the relative stability of these two base pairs is similar, while the base pair involving the U47 becomes more stable than that of the U49 at 20°C (Figure S10), showing that the mutations didn’t alter the relative stability of the base pairs involving the U47 and the U49.
The above solution and SSNMR data showed different dynamic behavior of the U47 and the U49. The solid sample of riboA71-adenine was prepared by ethanol-precipitation approach, the environment of which is different from the solution. It thus suggested that the base pair stability is dependent on sample conditions. The SSNMR method in providing complimentary dynamic messages of solid-state RNA is important.
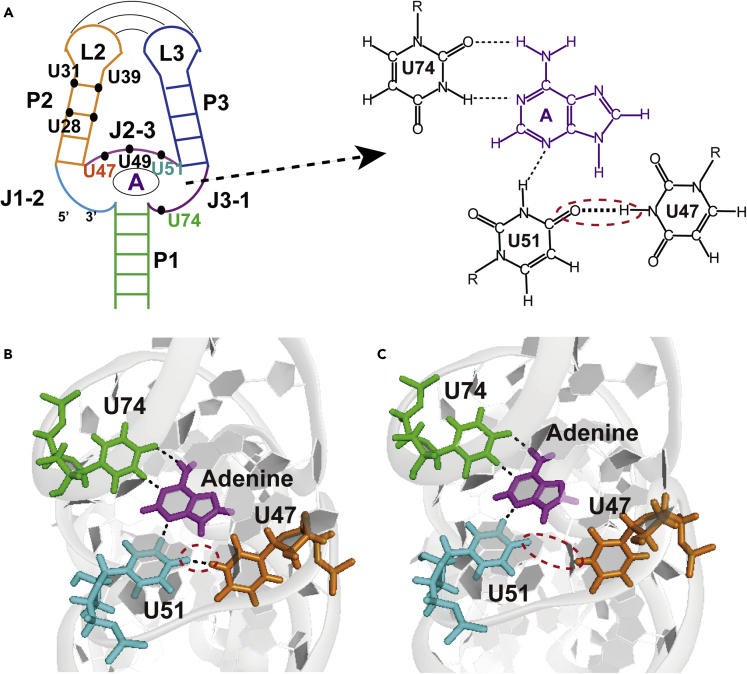
U47 is involved in a base pair with low stability within the base tetrad of the ligand-binding pocket
The crystal structure of the riboA71-adenine complex has been determined (PDB: 4TZX) (Zhang and Ferre-D'Amare, 2014). U47 is positioned in the minor groove, where it is involved in base pairing with U51. Adenine recognition occurs through the formation of a U47·U51·(adenine-U74) base tetrad, which is centered within a five-tiered triplex structure and surrounded by other nucleotides (Figure 5A). Two base triples, A23·(G46-C53) and A73·(A52-U22), are located above the adenine-binding site, while two other base triples, C50·(U75-A21) and U49·(A76-U20), are located below the adenine-binding site (Serganov et al., 2004). As the chemical shifts of the imino group of U47 on SSNMR spectra are almost identical to those in solution, the local structure is expected to remain solid.
Figure 5.
The topological view of riboA71-adenine complex
(A) The locations of the non-canonical base-pairs involving uridines are highlighted in the structure, e.g., U31, U39, U28 on stem P2 and U47, U49, U51 on J2-3. The structure of U47·U51·(adenine-U74) base tetrad is shown on the right.
(B and C) Closed (B) and open(C) state of U47·U51 base pair in U47·U51·(adenine-U74) base tetrad.
To visualize the dynamics of the U47·U51 base pair within the U47·U51·(adenine-U74) base tetrad, we further performed a 1 μs all-atom molecular dynamics (MD) simulation, using the crystal structure (PDB: 4TZX) as the starting point (Video S1). The hydrogen bonds in the U51·(adenine-U74) base triple, which is essential for the base tetrad stability, were retained throughout the simulation, indicating a very stable triple. In contrast, the open-close dynamics of the U47·U51 base pair were observed; here, the open state, wherein the hydrogen bond between U47 and U51 was broken (Figures 5B and 5C), had a population of approximately 4%. The hydrogen bond of U47·U51 is shown to be weaker than the other hydrogen bonds in the U51·(adenine-U74) base triple. The distances between the N3 of U47 and O4 of U51 range from 2.6 to 3.9 Å in the closed state and from 3.7 to 5.5 Å in the open state. Furthermore, the imino group of U47 in the open state flips to the outside of the binding pocket, away from the U51·(adenine-U74) base triple and more solvent-exposed (Figures 5C and S11C), whereas, in the closed state, the imino proton of U47 involves a hydrogen bond with the O4 of U51 and is more solvent-protected (Figures 5B and S11B; Videos S1 and S2). The probability of water molecules within 3 Å of imino proton of U47 in the open and closed state was also calculated using the MD simulation. The statistical results show that the probability of the existence of water molecules around U47 in the open and closed states is 95 and 12.5%, respectively, showing that the solvent accessibility of U47 imino proton in the open state is ∼8-fold higher than in closed state (Figure S11A).
The U49, the A76, and the U20 form a base triple. The open-close dynamics of the U49·A76 were also observed by the MD simulations, while the hydrogen bonds of A76-U20 were retained during the simulations, showing the lower stability of U49·U76. The population of the open state of U49·A76 was 16%, higher than that of the U47·U51 base pair. The distance between the H1 of U49 and N3 of A76 range from 2.8 to 4.0 Å in the closed state and from 3.5 to 6.9 Å in the open state (Figure S11, Videos S1 and S2). Furthermore, the probability of the existence of water molecules was analyzed for both the open and closed states, showing 82.1 and 37%, respectively. Thus, our MD data showed that the U49 ·U76 is less stable than U47 ·U51 and prone to more frequently opening dynamics, which is consistent with solution NMR results.
Discussion
Here, we report an SSNMR approach to characterize the RNA base pairs with low stability. Although solution NMR methods have been established to characterize the individual base pair opening dynamics in solution (Huang et al., 2011; Lee et al., 2008; Lee and Pardi, 2007; Rinnenthal et al., 2010; Russu, 2004; Snoussi and Leroy, 2001; Wagner et al., 2015), the corresponding strategies for studying RNA in the solid state are under development. More importantly, the known solution NMR methods in studying the base pair opening cannot be transferred to SSNMR. This work established a novel SSNMR experimental design in applying the imino-water chemical exchange process to evaluate the base pair stability. It has unique advantages in evaluating the relative stability of an individual RNA base pair in the solid state. This SSNMR-based approach could measure the kex values of ∼1000 s−1. In contrast, the timescales of the kex that can be measured by corresponding solution NMR methods i.e., CLEANEX or water magnetization transfer experiments are in the range of 1–100 s−1 (Skinner et al., 2012) or up to 400 s−1 (Lee et al., 2008; Maltseva et al., 1995). Thus this method allows the detection of faster chemical exchange process in solids than in solution.
Through this approach, we identified a base pair with low stability in the riboA71-adenine complex, the base pair of U47·U51 within the U47·U51·(adenine-U74) base tetrad, which is involved in ligand binding pocket. The kex were measured for the imino of U47 at five different temperatures, all of which were 5–8 fold larger than 100 s−1. Compared with the other RNAs with known base pair opening kinetics, the obtained kex values for U47 supported the relatively low stable base pairs.
The MD simulation showed the stability of the other base pairs within the U47·U51·(adenine-U74) base tetrad. U47 and U51 are the key components in the ligand binding pocket of riboA71, and the relatively low stability of this base pair with respect to the others in the base tetrad implies that it might have a fundamental role in the ligand recognition and dissociation process. Interestingly, in solution the relative stabilities among the non-canonical base pairs are temperature dependent and have different trends with respect to them in solids. In solid, U47 is involved in less stable base pair in the investigated temperatures, and U49 is more stable than U47. Conversely, in the solution state, the relative stability of U47·U51 and U49·A76 base pairs are similar at 5–10°C, but U49·A76 is less stable than the U47·U51 base pair at 15–20°C. The data implied that the dynamics and the relative stability of base pairs could be dependent on condition and sample forms. Interestingly, both solution and SSNMR data showed increased stability of the U47·U51 base pair at higher temperatures. The U47 is involved in the tetrad of U47·U51·(adenine-U74). An increase in temperature may lead to subtle structure changes within the tetrad, i.e., bond lengths or angle of the H-bond of U47-U51, which elevates its stability. These also suggested the importance of new methods in providing complimentary dynamics pictures of RNA on different sample forms and conditions. Particularly, many RNAs are involved in forming pathological-related insoluble RNA foci (Jain and Vale, 2017). Comparison of the dynamics of RNA in the dilute state by solution NMR and in the condensed state by solid-state NMR would provide valuable clues in molecular mechanisms.
Previous structural studies suggested the complicated folding mechanism of the add adenine riboswitch upon ligand binding involving many distinct folding states (Neupane et al., 2011). Without the adenine ligand, two conformations are co-existed in solution, but with different secondary structures. One of the apo states is similar to ligand binding conformation, except for the structure of the ligand binding pocket and its proximity (Reining et al., 2013). In the presence of Mg2+, the ligand-free state has empty, but locally structured, binding pocket, and the nucleotides involving adenine binding form Watson-Crick base pairs (i.e., U47-A52) and wobble base pairs (i.e., U48 with U75 or U74). After ligand binding, these hydrogen bonds will break, and form different base pairs, including the base tetrad in adenine ligand recognition, i.e., (C53-G46) ·A23, (U22-A52) ·A73, (U74-adenine) ·U51·U47, (A21-U75) ·C50 and (U20-A76) ·U49, which lock P1 and J2-3 together (Stagno et al., 2017). During this process, some residues undergo large conformation changes. For example, the residue U51 can swing out from the binding pocket, which allows adenine to enter the binding pocket and forms stable interaction with adenine (Bao et al., 2019). These research provided conformation exchange information upon ligand binding. Our work provides new insight into the base pair stability of riboA71 folding upon ligand binding and ligand release. As the U47·U51 base pair is relatively less stable with respect to other base pairs of the tetrad, we suggest that upon ligand dissociation, breakage of the less stable base pair U47·U51 might be the early stage of ligand release, and U47 and U51 can subsequently switch their interaction patterns to shift the equilibrium toward a ligand-free state. In 2007, Janina Buck and her colleagues investigated the hypoxanthine-induced folding of the guanine-sensing riboswitch aptamer domain of the Bacillus subtilis xpt-pbuX operon (GSRapt) by measuring site-specific half-lives (t1/2) (Buck et al., 2007). They found that the residues close to hypoxanthine, such as U22, U47, U49, and U51, showed faster folding kinetics. Besides, U47 showed faster folding kinetics than U49 from the t1/2 data of U47 (21.2 ± 1.9 s) and U49 (23.5 ± 1.9 s). Compared to Buck’s work, we all found that the residues showed faster dynamic near the ligand-binding pocket. The difference is that the WaterREXSY experiment could provide a base pair open-close dynamic, and Buck’s work showed the overall folding dynamic in RNA. This mechanism may be general in the purine family riboswitches, in which such base tetrads have been proposed around ligand-binding pockets.
The example here proved the applicability of the WaterREXSY in studying RNA base pair stabilities. Moreover, high-resolution SSNMR spectra of RNA complex have been obtained under ultra-fast MAS spinning (with a rate >100 kHz) (Marchanka et al., 2018). The WaterREXSY is also applicable under ultra-fast MAS conditions. Especially, the 1H-T1ρ relaxation times under ultra-fast MAS spinning would be longer than that under fast or moderate MAS spinning (e.g. 40 kHz). Thus, the WaterREXSY under ultra-fast MAS spinning could observe base pairs with slower imino-proton exchange rates, possibly obtaining dynamics parameters of stable base pairs, e.g. A-U base pairs (Figure S3). Similarly, high-level deuteration on RNA would increase the T2 of imino protons, making it possible to observe more stable base pairs. Combining the sparse isotopic-labeling techniques and ultra-fast MAS spinning, it is anticipated that WaterREXSY can provide more site-specific information and be used in more challenging systems, i.e., the insoluble RNA aggregates relative to neurological diseases (Jain and Vale, 2017) and large RNAs and RNA-protein complexes that cannot be accessed by solution NMR.(Juen et al., 2016; LeBlanc et al., 2017; Liu et al., 2015, 2016a, 2016b; Longhini et al., 2016; Strebitzer et al., 2018).
Limitation of the study
The base pair dynamics of an RNA are crucial in understanding the RNA functions. Current studies provided an approach to measuring the base pair opening rates in solid-state. However, this study has some limitations. First, although the non-canonical uridine assignments were obtained through comparison between solution and solid-state NMR, the spectral resolution of an RNA under a MAS rate of 40 kHz is not good enough to achieve more site specific for the GC and the AU base pairs. The bottleneck of the resolution is expected to overcome by ultra-fast MAS spinning, which could be possible to obtain high resolution similar to solution NMR. Secondly, the current work is not applicable in obtaining the water-imino exchange rates in the presence of the catalyst, because the concentration of catalyst is difficult to quantify in solid state. Finally, the solid-state NMR sequential assignments of RNA as large as 71-nt have not been established, which requires further development of solid-state NMR methods on RNA in solid.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Oligonucleotides | ||
| 5ʹ-TTAATACGACTCACTATAGGGAAGATA TAATCCTAATGATATGGTTTGGGAGTTTC TACCAAGAGCCTTAAACTCTTGATTATC TTCCC-3ʹ and its complementary sequence |
GENEWIZ company | NA |
| 5ʹ-TTAATACGACTCACTATAGGGAAGATA TAATCCTAAAGATATGGTTTAGGAGTTTC TACCAAGAGCCTTAAACTCTTGATTATC TTCCC-3ʹ and its complementary sequence |
GENEWIZ company | NA |
| Software and algorithms | ||
| TOPSPIN | Bruker | Version 4.2 |
| Other | ||
| 15N,13C-labeled rNTP | Cambridge Isotope Laboratories | CNLM-7871-SL-4X10 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Shenlin Wang at wangshenlin@pku.edu.cn.
Materials availability
This study did not generate new unique reagents.
Method Details
Sample preparation
The uniform 15N,13C-labeled riboA71 was produced by an in vitro transcription reaction as previously described. (Milligan et al., 1987; Yang et al., 2017) Specifically, three 15N,13C isotope-labeled riboA71 samples were prepared, including a uniformly 15N,13C-labeled riboA71 complex with adenine, a 15N,13C-uridine-labeled riboA71 without adenine and a 15N,13C-uridine-labeled U31A/G42A mutant riboA71-adenine complex. The sequences of the wild-type and U31A/G42A mutant riboA71 are as follows.
Wild-type riboA71:
5ʹ-GGGAAGAUAUAAUCCUAAUGAUAUGGUUUGGGAGUUUCUACCAAGAGCCUUAAACUCUUG AUUAUCUUCCC-3ʹ.
U31A/G42A mutant riboA71:
5ʹ-GGGAAGAUAUAAUCCUAAAGAUAUGGUUUAGGAGUUUCUACCAAGAGCCUUAAACUCUUGA UUAUCUUCCC-3ʹ.
Wild-type riboA71 was prepared by in vitro transcription reactions according to a previously reported protocol. (Zhao et al., 2019) The double-stranded DNA templates were prepared by annealing single-stranded DNA with the sequence.
5ʹ-TTAATACGACTCACTATAGGGAAGATATAATCCTAATGATATGGTTTGGGAGTTTCTACCAAGA GCCTTAAACTCTTGATTATCTTCCC-3ʹ.
and its complementary sequence at 95°C for 10 min. The in vitro transcription reactions contained 5 mM 15N,13C-labeled rNTPs, 40 mM Tris-HCl (pH 7.0), 0.01% Triton X-100, 1 mM spermidine, 10 mM DTT, 45 mM MgCl2, 0.85 μM DNA template and 0.12 mg/mL T7 RNA polymerase and were incubated at 37°C for 24 h. To produce 15N,13C-uridine-labeled riboA71, the reactions were supplemented with 15N,13C-labeled rUTP and abundant natural rCTP, rGTP and rATP. The synthesized RNA was purified using 12% PAGE under denaturing conditions and eluted from the gel with buffer containing 20 mM Tris-HCl, 300 mM sodium acetate and 1 mM EDTA, pH 7.4. The final yield of the purified RNA sample was approximately 5 mg per 10 mL of reaction system. Purified riboA71 was buffer-exchanged into a buffer containing 10 mM KH2PO4, 30 mM KCl and 2 mM MgCl2, pH 6.8, and concentrated to a final concentration of 700 μM. To prepare the riboA71–adenine complex, an adenine stock solution was added to the riboA71 solution to a final concentration of 5 mM; the mixture was annealed at 95°C for 5 min and incubated at 0°C for 30 min.
U31A/G42A mutant riboA71-adenine complex was prepared using a similar in vitro transcription protocol. The 15N,13C-uridine-labeled U31A/G42A mutant riboA71 was obtained using 15N,13C-rUTP and natural abundant rATP, rGTP and rCTP. Purified riboA71 was buffer-exchanged into a deuterated buffer (75% D2O/25% H2O, pH 6.8) containing 10 mM KH2PO4, 30 mM KCl and 2 mM MgCl2, and concentrated to a final concentration of 700 μM. To prepare the riboA71–adenine complex, a stock adenine solution was added to the riboA71 solution to a final concentration of 5 mM; the mixture was annealed at 95°C for 5 min and incubated at 0°C for 30 min. Because of the high affinity between the riboA71 and the adenine (Reining et al., 2013; Serganov et al., 2004), the excess amount of adenine saturated the ligand binding sites of riboA71. The RNA solid-state samples were prepared using the ethanol-precipitated approach, following a previously described procedure. (Zhao et al., 2019) Details of the sample preparation are presented in the supplemental information.
Preparation of RNA samples with partial deuteration
The RNA solid-state samples were prepared using the ethanol-precipitated approach, following a previously described procedure. (Zhao et al., 2019). To produce the partially deuterated RNA samples, 15N,13C-labeled RNAs were annealed in a 75% D2O/25% H2O (v/v)-based buffer to achieve the desired level of H/D exchange of the imino and the amino protons. To obtain ethanol-precipitated, partially deuterated samples for SSNMR, the partially deuterated RNAs were mixed with a pre-chilled ethanol stock consisting of 75% C2D5OD/25% C2H5OH (v/v). The final concentration of ethanol in solution was 75%. The RNA pellets were collected by centrifugation at 9600 g for 3 min, followed by central packing into a 1.9 mm rotor for SSNMR studies.
Solid-state NMR spectroscopy
The solid-state NMR experiments were carried out on a 600 MHz Bruker Avance III spectrometer equipped with a 1.9 mm 1H-X-Y magic-angle spinning (MAS) probe. Approximately 4 mg of RNA were center-packed into a 1.9-mm SSNMR rotor. The experiments were performed under an effective temperature of 15–35°C and a MAS frequency of 40 kHz. The temperatures were calibrated using the T1 relaxation time of 79Br in KBr powder. The typical π/2 pulse lengths were 2.5 μs for 1H, 4.0 μs for 13C and 5.0 μs for 15N.
The 2D hNH experiments were performed using a previously reported pulse sequence. (Zhou and Rienstra, 2008) The diagram of the pulse sequence (Figure S1A) is described in the supplemental information. Both the 1H-15N and 15N-1H cross-polarization (CP) transfer were set at a constant field lock of 15N at 60 kHz and an experimentally optimized proton power around n = 1 Hartmann–Hahn (HH) conditions (10% linear ramp). The contact times for 1H-15N and 15N-1H CP were 4 ms and 300 μs, respectively. The long contact time of 4 ms of the forward 1H-15N CP transfer was set to use both the covalently bonded proton and the remote protons to polarize the 15N signals, while the short contact time of 300 μs was set to transfer the 15N magnetization back to the covalently bonded proton only, not the remote protons. The carrier frequencies were set to 10 ppm for 1H and 145 ppm for 15N. The total acquisition times for the 1H and 15N dimensions were 20 ms and 15 ms, respectively. The spectral width of the 15N dimension was set at 50 ppm. States–TPPI phase-sensitive detection was obtained in the indirect dimension (15N) by incrementing the first π/2 pulse of the 1H channel. MISSISSIPPI pulse trains were used to suppress the solvent signals.(Zhou and Rienstra, 2008) Low-power TPPM proton decoupling (nutation frequency ∼10 kHz) and WALTZ-16 15N decoupling (nutation frequency ∼10 kHz) were applied during the evolution of the chemical shifts of 15N and 1H, respectively. The recycle delay in the 2D hNH experiments was 2 s. The hNH experiments were recorded with 32 scans.
The 2D WaterREXSY experiments were performed using the pulse sequence described in Figure S2B. The source code of the pulse sequence was listed in Data S1. Compared to the 2D hNH experiments, the 2D WaterREXSY experiments included two additional blocks: a 1H-15N REDOR block and a 1H LGSL. 1H-15N REDOR dephasing was achieved by applying 16 rotor-synchronized π pulses to the 15N channel to dephase the 1H signals covalently attached to the 15N atoms. The total REDOR dephasing time was 0.4 ms. A π pulse in the middle of the REDOR dephasing block was applied to the 1H channel to refocus the 1H chemical shift evolution. During the LG spin-lock period, a 1H B1 field of 53.2 kHz was applied at an offset of +37.6 kHz, resulting in an effective spin-locking field of 65.2 kHz along the magic angle. After the LG spin-lock period, the 1H-15N LG-CP transfer was set with the LG spin-lock on 1H at the same parameters as those applied during the LG spin lock period, and on 15N with the power experimentally optimized to achieve an efficient transfer. The contact time of 1H-15N CP was 300 μs.
A series of 2D WaterREXSY experiments with different LG spin-lock times (tSL) were collected, and the 1D 1H slices with a 15N chemical shift of 158.6 ppm were used to create the plot of 1H signals versus tSL. The tSL were set to 0.2 ms, 0.5 ms, 0.8 ms, 1.2 ms, 2.0 ms, 3.0 ms, and 4.0 ms. According to Equation 1, the exchange rate constant could be obtained after fitting the curves of the 1H signal intensities as a function of tSL. The 1H-T1ρ times were measured under the 1H LGSL. The fitted 1H-T1ρ times of the imino protons of uridines were summarized in Table S1.
| (Equation 1) |
All NMR data were processed using the TOPSPIN 3.2 program. The spectra were analyzed using CARA. Chemical shifts were referenced to 2,2-dimethyl-2-silapentane-5-sulfonic acid (DSS), using adamantane as a secondary standard.(Morcombe and Zilm, 2003) The 1H and 15N chemical shifts were referenced indirectly using γ13C/γ1H = 0.25145020 and γ15N/γ13C = 0.40297994, respectively.
Solution NMR spectroscopy
2D HSQC experiments modified with water saturation during the relaxation delay were performed on riboA71-adenine complex at 5°C, 10°C, 15°C and 20°C on a Bruker Avance 600 MHz spectrometer equipped with a 5 mm triple-resonance TCI cryogenic probe. A series of spectra was collected under a weak B1 field of 50 Hz with different saturation times for the water presaturation. All spectra were processed and analyzed using NMRPipe (Delaglio et al., 1995) and autofit script to extract intensities.
Molecular dynamics (MD) simulations
The initial structure used in the 1 μs all-atom MD simulation was obtained from the crystal structure of riboA71-adenine complex (PDB ID: 4TZX) (Zhang and Ferre-D'Amare, 2014). Fifty-two negative changes were present in the crystal structure of riboA71-adenine complex (PDB ID: 4TZX) (Zhang and Ferre-D'Amare, 2014). Fifty-two potassium ions were added to neutralize the system. The effective ion concentration is zero. The riboA71-adenine complex was solvated in a truncated octahedral box using the SPCE water model and a margin distance of 10 Å. Hydrogen atoms were added to the adenine ligand by using Discovery Studio 3.5 (Biovia, 2012).The partial charges were calculated using the antechamber module of the Amber 18 package (Case. et al., 2018) and the AM1-BCC method. The force field used for RiboA71 was ff99bsc0XOL3. (Zgarbova et al., 2011) The general AMBER force-field (GAFF) parameter was used for the adenine ligand, and the other parameters required for the ligand were generated by parmchk. (Wang et al., 2004).
The energy minimization and MD simulation were carried out using the GPU accelerated version of pmemd in AMBER. According to the AMBER 18 manual (Case et al., 2018), the set of 8 Å of the non-bonded cut-off is commonly reasonable to simulation of bio-molecules. The solvated system was minimized for 1000 steps with harmonic restraints (force constant = 500 kcal mol−1·Å−2), followed by 1000 steps without restraints. The system was then heated for 20 ps from 0 to 288 K, and equilibrated for 1 ns at 288 K. The production stage of the MD simulation was conducted at 288 K using the NPT ensemble and a 2 fs integration step. During the simulations, all bonds involving hydrogen atoms were constrained using the SHAKE algorithm. Here, the non-bonded cut-off was set at 9 Å, in order to improve the accuracy of the simulations. DSSR was used to analyze the formation of hydrogen bonds in the MD trajectory at an interval of 1 ps (Lu et al., 2015).
Theoretical considerations
The formalism of proton exchange has been extensively described using the “open–close” model.(Choi et al., 2019; Kim et al., 2017; Snoussi and Leroy, 2001; Szulik et al., 2014) Equilibrium exists between the open state, in which the hydrogen-bond (H-bond) breaks, and the closed state, in which the H-bond forms. The exchangeable protons can exchange with water only in the open state. In this model (Figure S2), kex is the overall rate constant of the water–RNA chemical exchange, which can be described using Equation 1:
| (Equation 2) |
where kopen and kclose are the rate constants for the opening and closing of the base pair, respectively, and kex,open is the rate constant for proton exchange in the opening state.
Two kinetic regimes can characterize kex, depending on the relative magnitudes of the rate constants. In the regime where kex,open >> kclose, the exchange rate is limited by the formation of the exchange-accessible state (opening-limited exchange); thus, kex = kopen. This condition often occurs at high concentrations of a proton acceptor in the presence of a catalyst, such as NH3. Another regime occurs at low concentrations of a proton acceptor, wherein the base pairs open and close many times before the exchange (pre-equilibrium exchange); thus, the exchange rate kex can be described as follows:
| (Equation 3) |
where Kdiss (= kopen/kclose) is the equilibrium constant for base pair opening, which reflects the H-bond stability of base pairs. According to the earlier publication (Lee et al., 2008), the kex,open is fast in the range of 106 s−1. The base pairs having a kex of 10 s−1 and 500 s−1 can be estimated to have Kdiss of 10−5 and 5 × 10−4, respectively, which corresponds the ratios of the open state to be 0.0001 and 0.002%, respectively.
In the riboA71–adenine complex, the solution buffer pH was 6.8, and no other catalyst was added. Therefore, the RNA exchange regime is also referred to as the pre-equilibrium exchange regime (Equation 3). Here, the exchange rate between the hydrogen-bonded protons of the base pair and water is determined by Kdiss and the exchange rate constant between the protons in the open state and water. The latter rate is fast in the range of 106 s−1, whereas the former is slow and is the rate-limiting step, which is determined by the stability of the H-bond within the base pairs.
In the “open–close” model of the chemical exchange process between RNA and water (Figure S2.), the intermediate N-H bond state cannot stably exist. In our experimental design, exchangeable protons, namely those in hydrogen bonds (N-H…N or N-H…O=C), are initially suppressed by the 1H-15N REDOR cycles and then repolarized by water through a chemical exchange. Thus, we can use a simplified exchange model (Figure 2A in the manuscript) to describe our experiment: the proton in the H-bond is in exchange with a pool of nw water molecules at the exchange rate constant of kIM. Analogously (Fu et al., 2016), the chemical exchange between water (M) and the hydrogen-bonded proton (I), i.e., the proton in N-H…N and the proton in N-H…O=C, can be described by
| (Equation 4) |
where M(tSL) and I(tSL) are the M and I magnetizations at a given LG spin-lock time (tSL), respectively, and represents their relaxation times under the tSL. kIM represents the exchange rate constant between the hydrogen-bonded proton and the pool of nw water molecules. In these differential equations, the factor of 1/nw is taken into account to reflect that only one water molecule is in exchange with the I proton. Similarly, as detailed in the literature (Fu et al., 2016), the analytical solution can be obtained:
| (Equation 5) |
Therefore, the build-up of the 1H magnetization in the N-H…N or N-H…O=C H-bond in the experiments is depicted in the pulse sequence. Figure 2A represents the exchange process between the pool of water molecules and the N-H…Y H-bond at an exchange rate constant of , which should be equivalent to kex in the open–close model for the chemical exchange process between RNA and water. Thus, we obtain equation 1.
Here, I(0) is the initial magnetization before the exchange-induced signal build-up, such as the contribution from incomplete REDOR dephasing and the CP transfer from 1H to 15N along the magic angle, whereas is considered a constant coefficient that does not contribute to signal build-up. For the riboA71–adenine complex system, is 10−6.8 in a pH 6.8 solution. The T1ρ of the imino proton is 8.5 ms, as measured in separate experiments with variable tSL periods before 1H-15N CP. The kex was obtained by fitting the I(tSL) against the tSL.
Quantification and statistical analysis
Not applicable.
Acknowledgments
All of the solid-state NMR experiments were conducted at the Beijing NMR Center and the NMR Facility at the National Center for Protein Sciences at Peking University. The work was supported by the National Key Research and Development Program of the Ministry of Science and Technology of the People’s Republic of China (contract number 2016YFA0501203), the National Natural Science Foundation of China (22274050, 21874004, 91953104), the State Key Laboratory of Medicinal Chemical Biology (2020031), the Fundamental Research Funds for the Central Universities and the Beijing National Laboratory for Molecular Sciences. Dr. R. Fu thanks the support from the National High Magnetic Field Laboratory, which is supported by NSF Cooperative Agreement NSF/DMR-1644779 and the State of Florida. The authors thank Dr. Xiaogang Niu and Dr. Hongwei Li of Peking University for assistance in collecting solution NMR data. The authors thank Dr. Zhijun Liu at the National Center for Protein Science Shanghai for assistance with NMR data collection. Dr S. Wang thank Mr. Weidong Kong from WZ Biosciences Inc (Shandong, China) to provide access to the research facility during the COVID-19 lockdown.
Author contributions
R.F. and S.W. designed the project. S.Z. and Z.W. prepared the RNA samples. R.F. and S.W. developed the solid-state NMR pulse sequences. S.Z. collected the solution and the solid-state NMR spectra. X.L. and Y.X. performed the molecular dynamics simulation. S.Z., R.F., Y.X., L.Z., and S.W. prepared the article. All of the authors edited the article.
Declaration of interests
None declared.
Published: November 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105322.
Contributor Information
Yi Xue, Email: yixue@mail.tsinghua.edu.cn.
Riqiang Fu, Email: rfu@magnet.fsu.edu.
Shenlin Wang, Email: wangshenlin@pku.edu.cn.
Supplemental information
Data and code availability
-
•
The NMR data reported in the paper is available from the lead contact on request.
-
•
This article reports the NMR pulse sequence of WaterREXSY. The original code of WaterREXSY is available at the supplemental information. The code is also available from the lead contact on request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Ahmed M., Marchanka A., Carlomagno T. Structure of a protein-RNA complex by solid-state NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 2020;59:6866–6873. doi: 10.1002/anie.201915465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hashimi H.M., Walter N.G. RNA dynamics: it is about time. Curr. Opin. Struct. Biol. 2008;18:321–329. doi: 10.1016/j.sbi.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asami S., Rakwalska-Bange M., Carlomagno T., Reif B. Protein-RNA interfaces probed by 1H-detected MAS solid-state NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 2013;52:2345–2349. doi: 10.1002/anie.201208024. [DOI] [PubMed] [Google Scholar]
- Bao L., Wang J., Xiao Y. Molecular dynamics simulation of the binding process of ligands to the add adenine riboswitch aptamer. Phys. Rev. E. 2019;100:022412. doi: 10.1103/PhysRevE.100.022412. [DOI] [PubMed] [Google Scholar]
- Batey R.T. Structure and mechanism of purine-binding riboswitches. Q. Rev. Biophys. 2012;45:345–381. doi: 10.1017/S0033583512000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biovia D.S. Dassault Systèmes; 2012. Discovery Studio, 3.5. [Google Scholar]
- Bothe J.R., Nikolova E.N., Eichhorn C.D., Chugh J., Hansen A.L., Al-Hashimi H.M. Characterizing RNA dynamics at atomic resolution using solution-state NMR spectroscopy. Nat. Methods. 2011;8:919–931. doi: 10.1038/nmeth.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck J., Fürtig B., Noeske J., Wöhnert J., Schwalbe H. Time-resolved NMR methods resolving ligand-induced RNA folding at atomic resolution. Proc. Natl. Acad. Sci. USA. 2007;104:15699–15704. doi: 10.1073/pnas.0703182104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlomagno T. Present and future of NMR for RNA-protein complexes: a perspective of integrated structural biology. J. Magn. Reson. 2014;241:126–136. doi: 10.1016/j.jmr.2013.10.007. [DOI] [PubMed] [Google Scholar]
- Case D.A., Ben-Shalom I.Y., Brozell. S.R., Cerutti D.S., Cheatham T.E., III, Cruzeiro V.W.D., Darden. T.A. University of California; 2018. AMBER 2018. [Google Scholar]
- Choi S.R., Kim N.H., Jin H.S., Seo Y.J., Lee J., Lee J.H. Base-pair Opening Dynamics of Nucleic Acids in Relation to Their Biological Function. Comput. Struct. Biotechnol. J. 2019;17:797–798. doi: 10.1016/j.csbj.2019.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Ding J., Swain M., Yu P., Stagno J.R., Wang Y.X. Conformational flexibility of adenine riboswitch aptamer in apo and bound states using NMR and an X-ray free electron laser. J. Biomol. NMR. 2019;73:509–518. doi: 10.1007/s10858-019-00278-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emani P.S., Olsen G.L., Echodu D.C., Varani G., Drobny G.P. Slow exchange model of nonrigid rotational motion in RNA for combined solid-state and solution NMR studies. J. Phys. Chem. B. 2010;114:15991–16002. doi: 10.1021/jp107193z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu R., Miao Y., Qin H., Cross T.A. Probing hydronium ion histidine NH exchange rate constants in the M2 channel via indirect observation of dipolar-dephased (15)N signals in magic-angle-spinning NMR. J. Am. Chem. Soc. 2016;138:15801–15804. doi: 10.1021/jacs.6b08376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert S.D., Stoddard C.D., Wise S.J., Batey R.T. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain. J. Mol. Biol. 2006;359:754–768. doi: 10.1016/j.jmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Huang W., Emani P.S., Varani G., Drobny G.P. Ultraslow domain motions in HIV-1 TAR RNA revealed by solid-state deuterium NMR. J. Phys. Chem. B. 2017;121:110–117. doi: 10.1021/acs.jpcb.6b11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Chen C., Russu I.M. Dynamics and stability of individual base pairs in two homologous RNA-DNA hybrids. Biochemistry. 2009;48:3988–3997. doi: 10.1021/bi900070f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Russu I.M. Dynamic and energetic signatures of adenine tracts in a rA-dT RNA-DNA hybrid and in homologous RNA-DNA, RNA-RNA, and DNA-DNA double helices. Biochemistry. 2017;56:2446–2454. doi: 10.1021/acs.biochem.6b01122. [DOI] [PubMed] [Google Scholar]
- Huang Y., Weng X., Russu I.M. Enhanced base-pair opening in the adenine tract of a RNA double helix. Biochemistry. 2011;50:1857–1863. doi: 10.1021/bi1014997. [DOI] [PubMed] [Google Scholar]
- Hwang T.L., Mori S., Shaka A.J., vanZijl P.C.M. Application of phase-modulated CLEAN chemical EXchange spectroscopy (CLEANEX-PM) to detect water-protein proton exchange and intermolecular NOEs. J. Am. Chem. Soc. 1997;119:6203–6204. [Google Scholar]
- Hwang T.L., van Zijl P.C., Mori S. Accurate quantitation of water-amide proton exchange rates using the phase-modulated CLEAN chemical EXchange (CLEANEX-PM) approach with a Fast-HSQC (FHSQC) detection scheme. J. Biomol. NMR. 1998;11:221–226. doi: 10.1023/a:1008276004875. [DOI] [PubMed] [Google Scholar]
- Jain A., Vale R.D. RNA phase transitions in repeat expansion disorders. Nature. 2017;546:243–247. doi: 10.1038/nature22386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juen M.A., Wunderlich C.H., Nußbaumer F., Tollinger M., Kontaxis G., Konrat R., Hansen D.F., Kreutz C. Excited states of nucleic acids probed by proton relaxation dispersion NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 2016;55:12008–12012. doi: 10.1002/anie.201605870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneja A., Villa A., Nilsson L. Elucidating the relation between internal motions and dihedral angles in an RNA hairpin using molecular dynamics. J. Chem. Theor. Comput. 2014;10:3532–3540. doi: 10.1021/ct500203m. [DOI] [PubMed] [Google Scholar]
- Kierzek R., Burkard M.E., Turner D.H. Thermodynamics of single mismatches in RNA duplexes. Biochemistry. 1999;38:14214–14223. doi: 10.1021/bi991186l. [DOI] [PubMed] [Google Scholar]
- Kim W., Kim H.E., Lee A.R., Jun A.R., Jung M.G., Ahn J.H., Lee J.H. Base-pair opening dynamics of primary miR156a using NMR elucidates structural determinants important for its processing level and leaf number phenotype in Arabidopsis. Nucleic Acids Res. 2017;45:875–885. doi: 10.1093/nar/gkw747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortmann J., Sczodrok S., Rinnenthal J., Schwalbe H., Narberhaus F. Translation on demand by a simple RNA-based thermosensor. Nucleic Acids Res. 2011;39:2855–2868. doi: 10.1093/nar/gkq1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc R.M., Longhini A.P., Le Grice S.F.J., Johnson B.A., Dayie T.K. Combining asymmetric 13C-labeling and isotopic filter/edit NOESY: a novel strategy for rapid and logical RNA resonance assignment. Nucleic Acids Res. 2017;45:e146. doi: 10.1093/nar/gkx591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Jucker F., Pardi A. Imino proton exchange rates imply an induced-fit binding mechanism for the VEGF165-targeting aptamer, Macugen. FEBS Lett. 2008;582:1835–1839. doi: 10.1016/j.febslet.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Pardi A. Thermodynamics and kinetics for base-pair opening in the P1 duplex of the Tetrahymena group I ribozyme. Nucleic Acids Res. 2007;35:2965–2974. doi: 10.1093/nar/gkm184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.M., Lee E.H., Seo Y.J., Kang Y.M., Ha J.H., Kim H.E., Lee J.H. Measurement of hydrogen exchange times of the RNA imino protons using by phase-modulated CLEAN chemical exchange spectroscopy. Bull. Kor. Chem. Soc. 2009;30:2197–2198. [Google Scholar]
- Leppert J., Urbinati C.R., Häfner S., Ohlenschläger O., Swanson M.S., Görlach M., Ramachandran R. Identification of NH.N hydrogen bonds by magic angle spinning solid state NMR in a double-stranded RNA associated with myotonic dystrophy. Nucleic Acids Res. 2004;32:1177–1183. doi: 10.1093/nar/gkh288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Holmstrom E., Zhang J., Yu P., Wang J., Dyba M.A., Chen D., Ying J., Lockett S., Nesbitt D.J., et al. Synthesis and applications of RNAs with position-selective labelling and mosaic composition. Nature. 2015;522:368–372. doi: 10.1038/nature14352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Sousa R., Wang Y.X. Specific labeling: an effective tool to explore the RNA world. Bioessays. 2016;38:192–200. doi: 10.1002/bies.201500119. [DOI] [PubMed] [Google Scholar]
- Liu Y., Yu P., Dyba M., Sousa R., Stagno J.R., Wang Y.X. Applications of PLOR in labeling large RNAs at specific sites. Methods. 2016;103:4–10. doi: 10.1016/j.ymeth.2016.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhini A.P., LeBlanc R.M., Becette O., Salguero C., Wunderlich C.H., Johnson B.A., D'Souza V.M., Kreutz C., Dayie T.K. Chemo-enzymatic synthesis of site-specific isotopically labeled nucleotides for use in NMR resonance assignment, dynamics and structural characterizations. Nucleic Acids Res. 2016;44:e52. doi: 10.1093/nar/gkv1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X.J., Bussemaker H.J., Olson W.K. DSSR: an integrated software tool for dissecting the spatial structure of RNA. Nucleic Acids Res. 2015;43:e142. doi: 10.1093/nar/gkv716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltseva T.V., Zarytova V.F., Chattopadhyaya J. Base-pair exchange kinetics of the imino and amino protons of the 3'-phenazinium tethered DNA-RNA duplex, r(5'GAUUGAA3'):d(5'TCAATC3'-Pzn), and their comparison with those of B-DNA duplex. J. Biochem. Biophys. Methods. 1995;30:163–177. doi: 10.1016/0165-022x(94)00075-o. [DOI] [PubMed] [Google Scholar]
- Manosas M., Junier I., Ritort F. Force-induced misfolding in RNA. Phys. Rev. E - Stat. Nonlinear Soft Matter Phys. 2008;78:061925. doi: 10.1103/PhysRevE.78.061925. [DOI] [PubMed] [Google Scholar]
- Marchanka A., Carlomagno T. Solid-state NMR spectroscopy of RNA. Methods Enzymol. 2019;615:333–371. doi: 10.1016/bs.mie.2018.08.029. [DOI] [PubMed] [Google Scholar]
- Marchanka A., Simon B., Althoff-Ospelt G., Carlomagno T. RNA structure determination by solid-state NMR spectroscopy. Nat. Commun. 2015;6:7024. doi: 10.1038/ncomms8024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchanka A., Stanek J., Pintacuda G., Carlomagno T. Rapid access to RNA resonances by proton-detected solid-state NMR at >100 kHz MAS. Chem. Commun. 2018;54:8972–8975. doi: 10.1039/c8cc04437f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marušič M., Schlagnitweit J., Petzold K. RNA dynamics by NMR spectroscopy. Chembiochem. 2019;20:2685–2710. doi: 10.1002/cbic.201900072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan J.F., Groebe D.R., Witherell G.W., Uhlenbeck O.C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirau P.A., Kearns D.R. Effect of environment, conformation, sequence and base substituents on the imino proton exchange rates in guanine and inosine-containing DNA, RNA, and DNA-RNA duplexes. J. Mol. Biol. 1984;177:207–227. doi: 10.1016/0022-2836(84)90453-4. [DOI] [PubMed] [Google Scholar]
- Modig K., Liepinsh E., Otting G., Halle B. Dynamics of protein and peptide hydration. J. Am. Chem. Soc. 2004;126:102–114. doi: 10.1021/ja038325d. [DOI] [PubMed] [Google Scholar]
- Morcombe C.R., Zilm K.W. Chemical shift referencing in MAS solid state NMR. J. Magn. Reson. 2003;162:479–486. doi: 10.1016/s1090-7807(03)00082-x. [DOI] [PubMed] [Google Scholar]
- Neupane K., Yu H., Foster D.A.N., Wang F., Woodside M.T. Single-molecule force spectroscopy of the add adenine riboswitch relates folding to regulatory mechanism. Nucleic Acids Res. 2011;39:7677–7687. doi: 10.1093/nar/gkr305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen P., Qin P.Z. RNA dynamics: perspectives from spin labels. Wiley Interdiscip. Rev. RNA. 2012;3:62–72. doi: 10.1002/wrna.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noeske J., Schwalbe H., Wöhnert J. Metal-ion binding and metal-ion induced folding of the adenine-sensing riboswitch aptamer domain. Nucleic Acids Res. 2007;35:5262–5273. doi: 10.1093/nar/gkm565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K., Takahashi M., Sakamoto T., Kawai G., Nakamura K., Kanai A. Solution structure of a GAAG tetraloop in helix 6 of SRP RNA from Pyrococcus furiosus. Nucleos Nucleot. Nucleic Acids. 2006;25:383–395. doi: 10.1080/15257770600683979. [DOI] [PubMed] [Google Scholar]
- Olsen G.L., Bardaro M.F., Jr., Echodu D.C., Drobny G.P., Varani G. Hydration dependent dynamics in RNA. J. Biomol. NMR. 2009;45:133–142. doi: 10.1007/s10858-009-9355-6. [DOI] [PubMed] [Google Scholar]
- Olsen G.L., Bardaro M.F., Jr., Echodu D.C., Drobny G.P., Varani G. Intermediate rate atomic trajectories of RNA by solid-state NMR spectroscopy. J. Am. Chem. Soc. 2010;132:303–308. doi: 10.1021/ja907515s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen G.L., Echodu D.C., Shajani Z., Bardaro M.F., Jr., Varani G., Drobny G.P. Solid-state deuterium NMR studies reveal micros-ns motions in the HIV-1 transactivation response RNA recognition site. J. Am. Chem. Soc. 2008;130:2896–2897. doi: 10.1021/ja0778803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reining A., Nozinovic S., Schlepckow K., Buhr F., Fürtig B., Schwalbe H. Three-state mechanism couples ligand and temperature sensing in riboswitches. Nature. 2013;499:355–359. doi: 10.1038/nature12378. [DOI] [PubMed] [Google Scholar]
- Riedel K., Herbst C., Häfner S., Leppert J., Ohlenschläger O., Swanson M.S., Görlach M., Ramachandran R. Constraints on the structure of (CUG)97 RNA from magic-angle-spinning solid-state NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 2006;45:5620–5623. doi: 10.1002/anie.200600769. [DOI] [PubMed] [Google Scholar]
- Riedel K., Leppert J., Ohlenschläger O., Görlach M., Ramachandran R. Characterisation of hydrogen bonding networks in RNAs via magic angle spinning solid state NMR spectroscopy. J. Biomol. NMR. 2005;31:331–336. doi: 10.1007/s10858-005-1614-6. [DOI] [PubMed] [Google Scholar]
- Riedel K., Leppert J., Ohlenschläger O., Görlach M., Ramachandran R. TEDOR with adiabatic inversion pulses: resonance assignments of 13C/15N labelled RNAs. J. Biomol. NMR. 2005;31:49–57. doi: 10.1007/s10858-004-6066-x. [DOI] [PubMed] [Google Scholar]
- Rinnenthal J., Buck J., Ferner J., Wacker A., Fürtig B., Schwalbe H. Mapping the landscape of RNA dynamics with NMR spectroscopy. Acc. Chem. Res. 2011;44:1292–1301. doi: 10.1021/ar200137d. [DOI] [PubMed] [Google Scholar]
- Rinnenthal J., Klinkert B., Narberhaus F., Schwalbe H. Direct observation of the temperature-induced melting process of the Salmonella fourU RNA thermometer at base-pair resolution. Nucleic Acids Res. 2010;38:3834–3847. doi: 10.1093/nar/gkq124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinnenthal J., Klinkert B., Narberhaus F., Schwalbe H. Modulation of the stability of the Salmonella fourU-type RNA thermometer. Nucleic Acids Res. 2011;39:8258–8270. doi: 10.1093/nar/gkr314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A., Panigrahi S., Bhattacharyya M., Bhattacharyya D. Structure, stability, and dynamics of canonical and noncanonical base pairs: quantum chemical studies. J. Phys. Chem. B. 2008;112:3786–3796. doi: 10.1021/jp076921e. [DOI] [PubMed] [Google Scholar]
- Russu I.M. Probing site-specific energetics in proteins and nucleic acids by hydrogen exchange and nuclear magnetic resonance spectroscopy. Methods Enzymol. 2004;379:152–175. doi: 10.1016/S0076-6879(04)79009-6. [DOI] [PubMed] [Google Scholar]
- Schanda P., Ernst M. Studying dynamics by magic-angle spinning solid-state NMR spectroscopy: principles and applications to biomolecules. Prog. Nucl. Magn. Reson. Spectrosc. 2016;96:1–46. doi: 10.1016/j.pnmrs.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A., Yuan Y.R., Pikovskaya O., Polonskaia A., Malinina L., Phan A.T., Hobartner C., Micura R., Breaker R.R., Patel D.J. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem. Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng J., Gan J., Soares A.S., Salon J., Huang Z. Structural insights of non-canonical U∗U pair and Hoogsteen interaction probed with Se atom. Nucleic Acids Res. 2013;41:10476–10487. doi: 10.1093/nar/gkt799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner J.J., Lim W.K., Bédard S., Black B.E., Englander S.W. Protein hydrogen exchange: testing current models. Protein Sci. 2012;21:987–995. doi: 10.1002/pro.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoussi K., Leroy J.L. Imino proton exchange and base-pair kinetics in RNA duplexes. Biochemistry. 2001;40:8898–8904. doi: 10.1021/bi010385d. [DOI] [PubMed] [Google Scholar]
- Stagno J.R., Liu Y., Bhandari Y.R., Conrad C.E., Panja S., Swain M., Fan L., Nelson G., Li C., Wendel D.R., et al. Structures of riboswitch RNA reaction states by mix-and-inject XFEL serial crystallography. Nature. 2017;541:242–246. doi: 10.1038/nature20599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strebitzer E., Nußbaumer F., Kremser J., Tollinger M., Kreutz C. Studying sparsely populated conformational states in RNA combining chemical synthesis and solution NMR spectroscopy. Methods. 2018;148:39–47. doi: 10.1016/j.ymeth.2018.05.007. [DOI] [PubMed] [Google Scholar]
- Szulik M.W., Voehler M., Stone M.P. NMR analysis of base-pair opening kinetics in DNA. Curr. Protoc. Nucleic Acid Chem. 2014;59 doi: 10.1002/0471142700.nc0720s59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Várnai P., Canalia M., Leroy J.L. Opening mechanism of G.T/U pairs in DNA and RNA duplexes: a combined study of imino proton exchange and molecular dynamics simulation. J. Am. Chem. Soc. 2004;126:14659–14667. doi: 10.1021/ja0470721. [DOI] [PubMed] [Google Scholar]
- Wagner D., Rinnenthal J., Narberhaus F., Schwalbe H. Mechanistic insights into temperature-dependent regulation of the simple cyanobacterial hsp17 RNA thermometer at base-pair resolution. Nucleic Acids Res. 2015;43:5572–5585. doi: 10.1093/nar/gkv414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- Yang Y., Xiang S., Liu X., Pei X., Wu P., Gong Q., Li N., Baldus M., Wang S. Proton-detected solid-state NMR detects the inter-nucleotide correlations and architecture of dimeric RNA in microcrystals. Chem. Commun. 2017;53:12886–12889. doi: 10.1039/c7cc07483b. [DOI] [PubMed] [Google Scholar]
- Yuwen T., Bah A., Brady J.P., Ferrage F., Bouvignies G., Kay L.E. Measuring solvent hydrogen exchange rates by multifrequency excitation (15)N CEST: application to protein phase separation. J. Phys. Chem. B. 2018;122:11206–11217. doi: 10.1021/acs.jpcb.8b06820. [DOI] [PubMed] [Google Scholar]
- Zgarbová M., Otyepka M., Sponer J., Mládek A., Banáš P., Cheatham T.E., 3rd, Jurečka P. Refinement of the Cornell et al. Nucleic Acids Force Field Based on Reference Quantum Chemical Calculations of Glycosidic Torsion Profiles. J. Chem. Theor. Comput. 2011;7:2886–2902. doi: 10.1021/ct200162x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Ferré-D'Amaré A.R. Dramatic improvement of crystals of large RNAs by cation replacement and dehydration. Structure. 2014;22:1363–1371. doi: 10.1016/j.str.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Cross T.A., Fu R. Detecting water-protein chemical exchange in membrane-bound proteins/peptides by solid-state NMR spectroscopy. Magn. Reson. Lett. 2021;1:99–111. [Google Scholar]
- Zhao S., Yang Y., Zhao Y., Li X., Xue Y., Wang S. High-resolution solid-state NMR spectroscopy of hydrated non-crystallized RNA. Chem. Commun. 2019;55:13991–13994. doi: 10.1039/c9cc06552k. [DOI] [PubMed] [Google Scholar]
- Zhou D.H., Rienstra C.M. High-performance solvent suppression for proton detected solid-state NMR. J. Magn. Reson. 2008;192:167–172. doi: 10.1016/j.jmr.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The NMR data reported in the paper is available from the lead contact on request.
-
•
This article reports the NMR pulse sequence of WaterREXSY. The original code of WaterREXSY is available at the supplemental information. The code is also available from the lead contact on request.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.