Summary
Trace metals are essential for various physiological processes, but their roles in innate immunity have not been fully explored. Here, we found that manganese (Mn) significantly enhanced DNA-mediated IFN-α, IFN-β, and IFN-λ1 production. Microarray analysis demonstrated Mn highly upregulated 351 genes, which were involved in multiple biological functions related to innate immune response. Moreover, we found that Mn2+ alone activates phosphorylation of TANK-binding kinase 1 (TBK1). Inhibiting ataxia telangiectasia mutated (ATM) kinase using ATM inhibitor or siRNA suppressed Mn-enhanced DNA-mediated immune response with decreasing phosphorylation of TBK-1, suggesting that ATM involves in Mn-dependent phosphorylation of TBK1. Given that TBK1 is an essential mediator in DNA- or RNA-mediated signaling pathways, we further demonstrated that Mn2+ suppressed infection of HSV-1 (DNA virus) or Sendai virus (RNA virus) into human macrophages by enhancing antiviral immunity. Our finding highlights a beneficial role of Mn in nucleic-acid-based preventive or therapeutic reagents against infectious diseases.
Subject areas: Biochemistry, Molecular physiology, Immunology
Graphical abstract
Highlights
-
•
Mn2+ enhances DNA- or RNA-mediated IFN-α, IFN-β and IFN-λ1 production
-
•
Mn2+ treatment activates phosphorylation of TBK1
-
•
Inhibiting ATM suppresses Mn2+-enhanced DNA-mediated innate immune response
-
•
Mn2+ suppresses DNA and RNA virus infection by enhancing antiviral immunity
Biochemistry; Molecular physiology; Immunology
Introduction
The innate immune response is an important process to initiate anti-pathogen immune activities against invaded pathogens in a host’s peripheral and lymphoid tissues (Thaiss et al., 2016). Before initiating innate immune responses, the pathogens must be detected by pattern recognition receptors (PRRs), which specifically recognize microbe-specific pathogen-associated molecular patterns (PAMPs) (Akira et al., 2006; Takeuchi and Akira, 2010). Each PRR activates specific signaling cascades to induce proinflammatory cytokines or antiviral interferons (IFNs). Both type I IFNs and type III IFNs play a critical role in antiviral innate immunity to eradicate replicating viruses (Ank et al., 2006; Donnelly and Kotenko, 2010; Kelly et al., 2011; Li et al., 2009; Syedbasha and Egli, 2017). Five PRR families have been identified so far: endosomal Toll-like receptors (TLRs) (Akira et al., 2001; Bowie and Haga, 2005), C-type lectin receptors (CLRs) (Bermejo-Jambrina et al., 2018), cytoplasmic retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) (Rehwinkel and Gack, 2020), nucleotide-binding oligomerization (NOD)-like receptors (NLRs) (Mason et al., 2012; Zhong et al., 2013), and cytosolic DNA sensors (CDS) (Jeffries and Marriott, 2020; Zahid et al., 2020). These PRRs commonly activate both type I and type III IFNs in addition to proinflammatory cytokines. Although the expression of these cytokine genes requires the activation of transcription factors, such as activating nuclear factor kappa B (NF-κB) and IFN regulatory factors (IRFs), each sensor molecule activates these transcription factors via different signaling pathways (Honda and Taniguchi, 2006; Iwanaszko and Kimmel, 2015; Sato et al., 2000).
Intracellular DNA and RNA are potent activators of innate immune responses in host cells (Dempsey and Bowie, 2015; Muruve, 2008; Paludan and Bowie, 2013; Sharma and Fitzgerald, 2011). The DNA-mediated innate immune response includes diverse signaling pathways leading to the production of type I or type III IFNs, chemokines, and pro-inflammatory cytokines (Gürtler and Bowie, 2013; Schoggins, 2019). For instance, the DNA-dependent activator of IFN-regulatory factors (DAI) is the first identified DNA sensor to recognize double-stranded DNA (dsDNA) and activate the STING (stimulator of IFN genes)-TBK1 (TANK-binding kinase)-IRF3 signaling pathway (Takaoka, 2007). Gamma-interferon-inducible protein 16 (IFI16) (Monroe et al., 2014; Thompson et al., 2014; Unterholzner et al., 2010) and DEAD box helicase 41 (DDX41) (Zhang et al., 2011b) were found to be cytosolic DNA sensors in diverse cellular processes to recognize DNA. More recently, cGAS (cyclic GMP-AMP [cGAMP] synthase) (Sun et al., 2013; Xia et al., 2016) was found to recognize cytosolic dsDNA and produce the second messenger cGAMP (Civril et al., 2013; Gao et al., 2013; Wu et al., 2015) to activate STING (Ishikawa and Barber, 2008; Sun et al., 2009; Zhong, 2008). In addition, we and other researchers previously identified that DNA repair proteins - Ku70 alone, a complex of Ku70 and Ku80, or DNA-dependent protein kinase (DNA-PK, composed of Ku70, Ku80, and DNA-PKcs) - serve as cytosolic DNA sensors to induce type I and type III IFNs (also called IFN-λs), the latter of which include IFN-λ1, IFN-λ2, IFN-λ3, and IFN-λ4 (Burleigh et al., 2020; Ferguson et al., 2012; Li et al., 2016; Sui et al., 2017, 2021a, 2021b, 2021b, 2017; Wang et al., 2017; Zhang et al., 2011a). Similar to type I IFNs, IFN-λs are important antiviral cytokines. During the early stage of viral infections, IFN-λs directly perform an antiviral immune response at epithelial surfaces and skew the balance of Th1 and Th2 cells to a Th1 phenotype (Zhou et al., 2018). RLRs are key sensors of RNA virus infection. This protein family encompasses three members: RIG-I, melanoma differentiation-associated protein 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2). RIG-I and MDA5 are the most well studied RLRs and provide key regulation of IFN pathways. Following post-translational modifications and activation, RIG-I and MDA5 translocate to the mitochondria, where they interact with the mitochondrial antiviral signaling (MAVS) to form a MAVS signalosome. This complex then activates TNF receptor-associated factor 3 (TRAF3), TBK1, and IκB kinase (IKK) to induce phosphorylation of IRF3 (Loo and Gale, 2011; Horner et al., 2011), which promotes its nuclear translocation and the transcription of type I and III IFNs (Diamond and Kanneganti, 2022).
DNA or RNA vaccination depends on nucleic acid recognition to initiate a powerful innate immune response. This innate response also assists the adaptive response. On the other hand, those nucleotides can also serve as a damage-associated molecular pattern (DAMP), accelerating inflammatory responses after their release from dying or damaged cells and directly contributing to the pathogenesis of various diseases, such as atherosclerosis (Oka et al., 2012) and deep vein thrombosis (Brill et al., 2012). Therefore, precisely regulating or balancing the innate immune response will help us enhance the vaccine efficacy or minimize side effects.
Trace metals play a role in many essential biological processes for sustaining life. Ionic forms of the metals act as cofactors in proteins and help to enable proteins’ biological function, regulate their activity, and stabilize their structure (Aisen et al., 2001; Andreini et al., 2008; Diaz-Ochoa et al., 2014; Waldron et al., 2009). Among trace metals, iron (Fe) is the most abundant metal in the human body. About 65–75% of total body Fe (3–5 g) in adults presents in erythrocytes and is utilized for the biosynthesis of heme, a tetrapyrrole molecule of hemoglobin, and then facilities the transportation of oxygen (Andrews, 2000). Fe is also important for other biochemical processes, such as DNA replication, protein synthesis, electron transport, and tissue cellular respiration (Lieu et al., 2001). Zinc (Zn) is a similarly essential mineral with a required daily intake of 15 mg for human adults (King, 2011). Approximately 95% of Zn in humans is intracellular, where it serves structural and functional roles for a large number of macromolecules and enzymes (Tapiero and Tew, 2003). Manganese (Mn) is one of the most common metals in the mammalian tissues, with a range of 0.3–2.9 μg/g wet tissue weight (Horning et al., 2015), and is indispensable for a variety of physiological processes, such as development, reproduction, immune regulation, and antioxidant defense (Roth et al., 2013). It is also a critical cofactor of some metalloenzymes, such as Mn superoxide dismutase, glutamine synthetase, and arginase (Taylor et al., 2006).
However, the role of trace metal ions in the innate immune system has not been fully investigated. Here, in the current study, we found that Mn2+ highly enhances DNA-mediated innate immune response, including the induction of IFN-α, IFN-β, and IFN-λ1. We further discovered that the enhancing effect by Mn2+ in the innate immune response was associated with an increase in the phosphorylation of TBK1. This enhanced phosphorylation was induced by Mn2+ alone via an ATM-TBK1 phosphorylation signaling pathway, suggesting that it primes the TBK1-dependent cell signaling. In addition, we validated that Mn2+ pretreatment inhibited not only herpes simplex virus type 1 (HSV-1, a DNA virus) infection but also Sendai virus (SeV, an RNA virus) infection in human primary macrophages. Such a protective effect was attributed to the Mn2+-enhanced induction of IFN-α, IFN-β, and IFN-λ1. These findings implicate the potential application value of Mn2+ in the development of DNA- and RNA-based preventive or therapeutic strategies to fight human infectious diseases, including the current COVID-19 pandemic.
Results
Pretreatment with manganese (Mn) enhances DNA-mediated innate immune response
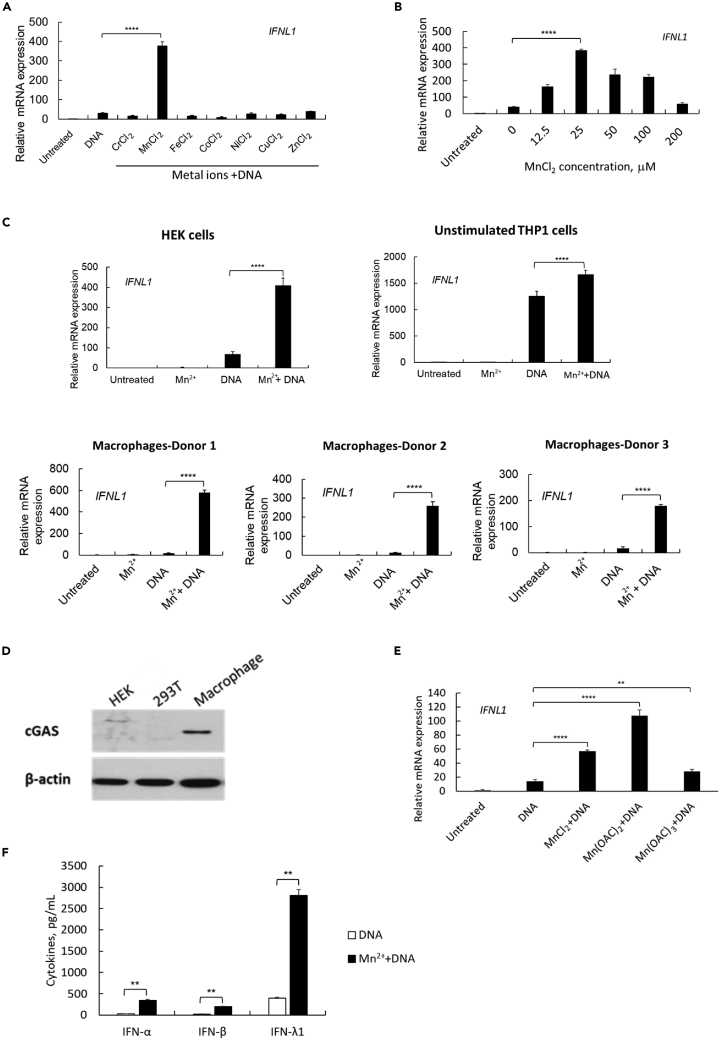
We previously reported that Ku70 is a cytosolic DNA sensor to initiate DNA (transfected DNA or DNA virus infection)-mediated type III IFN response in HEK cells, RD cells, and primary macrophages (Sui et al., 2017, 2021a, 2017; Zhang et al., 2011a). To assess the role of trace metals in the innate immune response, we pretreated HEK cells with different metal solutions for 24 h. The cells were then transfected by linearized DNA, and the total RNA was extracted at 24 h after transfection, or the cell culture supernatants were collected for IFN protein measurements at 48 h after DNA transfection. As shown in Figure 1A, among the trace metal ions (Cr2+, Mn2+, Fe2+, Co2+, Ni2+, Cu2+, or Zn2+), only pretreatment by MnCl2 demonstrated a significant enhancing effect on DNA-mediated IFN-λ1 induction. Further characterization indicated that the enhancing effect by Mn2+ showed a bell-shaped dose-dependent pattern. With Mn2+ at 12.5 μM, the DNA-mediated IFN-λ1 induction was significantly enhanced and at the concentration of 25 μM, Mn2+ exhibited the best enhancing effect (Figure 1B). This enhancing effect of Mn2+ occurred not only in HEK cells but also in THP-1 cells and primary human macrophages (Figure 1C). Consistent with our previous results (Sui et al., 2017), cGAS is not endogenously expressed in HEK or 293T cells (Figure 1D), so the cGAS-mediated cytosolic DNA sensing pathway is absent in HEK cells. However, Mn2+ enhanced the DNA-mediated innate immune response in HEK cells (Figures 1A–1C), indicating that the Mn-enhanced innate immune response is not only attributable to the cGAS-mediated signaling pathway (Wang et al., 2018). We also evaluated Mn ions in the form of Mn(OAc)2 and Mn(OAc)3, Mn2+ and Mn3+. HEK cells were pretreated by Mn(OAC)2 or Mn(OAC)3 at the same concentration of 25 μM, and IFN induction was assessed. The results showed that both forms of Mn ions enhanced the DNA-mediated IFN-λ1 induction, and that Mn(OAC)2 presented the best enhancing effect (Figure 1E); however, compared with the Mn(OAC)2 solution, MnCl2 shows much better stability and solubility, so we continued using MnCl2 in our subsequent studies. Furthermore, we confirmed that pretreatment with Mn2+ enhanced DNA-mediated induction of not only IFN-λ1 but also IFN-α and IFN-β (Figure 1F), suggesting that MnCl2 may enhance DNA-mediated innate immune cascades, leading to an enhanced induction of multiple IFNs and inflammatory cytokines. Of note, the dose of MnCl2 that we used for pretreatment did not induce any innate immune response by MnCl2 itself (Figure 1C). We further confirmed that such an enhancing effect had no impact on the kinetics of DNA-mediated IFN-λ1, IFN-α, and IFN-β induction. The measurement for gene expression level by real-time PCR and for protein expression level by ELISA consistently suggested that pretreatment with Mn2+ significantly enhanced DNA-mediated IFN induction at any early peak time points, 6 h or 18 h, or the late prolonged induction profile (Figure S1).
Figure 1.
Pretreatment of Mn enhances DNA-mediated innate immune response
(A) HEK cells were treated by different metal solutions at 100 μM, then transfected with DNA 24 h after metal treatment. The cells were collected for RNA extraction, relative IFNL1 expression was measured by real time RT-PCR. One representative experiment of at least three independent experiments is shown. And each was done as triplicate. Data are shown as mean ± SD (n = 3) and the gene expression level was compared to untreated cells. ∗∗∗∗p< 0.0001 (one-way ANOVA) was indicated in the figure where the IFN-λ1 expression level was compared between DNA-transfected and Mn2+/DNA-treated cells.
(B) HEK cells were treated by different concentrations of MnCl2, then transfected with DNA and collected to measure IFNL1 gene expression level. One representative experiment of at least three independent experiments is shown. And each was done as triplicate. Data are shown as mean ± SD (n = 3). ∗∗∗∗p< 0.0001 (one-way ANOVA) was indicated in the figure.
(C) HEK cells, unstimulated THP1 cells, and human primary macrophages (3 donors) were treated as indicated: untreated, MnCl2 treatment alone, DNA transfection alone, or MnCl2 pretreatment with DNA transfection. The cells were collected for real-time RT-PCR to measure IFNL1 gene expression level. For HEK and THP1 cells, one representative experiment of three independent experiments is shown. And each was done as triplicate. Data are shown as mean ± SD (n = 3). ∗∗∗∗p< 0.0001 (One-way ANOVA) were indicated in the figures where the IFN-λ1 expression level was compared between DNA-transfected and Mn2+/DNA-treated cells.
(D)Western blot was performed using the whole cell lysate of HEK, 293T and human primary macrophages. Anti-cGAS was probed to detect the endogenous expression of different cell lysate. The band of β-actin was included as a loading control for all the samples. One representative experiment of three independent experiments is shown.
(E) HEK cells were treated by MnCl2, Mn(OAC)2, or Mn(OAC)3 at 25 μM, then transfected with DNA 24 h later. The cells were collected to measure IFNL1 gene expression level 24 h after DNA transfection. One representative experiment of at least three independent experiments is shown. And each was done as triplicate. Data are shown as mean ± SD (n = 3). ∗∗p< 0.01, ∗∗∗∗p< 0.0001 (one-way ANOVA) was indicated in the figure.
(F) HEK cells were transfected with DNA with or without a pretreatment of MnCl2. The cell supernatants were collected 48 h after DNA transfection for ELISA to detect IFN-α, IFN-β, and IFN-λ1 protein expression levels. One representative experiment of at least three independent experiments is shown. And each was done as duplicate. Data are shown as mean ± SD (n = 2). ∗∗p < 0.01 (Student’s t-test) was indicated in the figures where the cytokine production was compared between DNA-transfected and Mn2+/DNA-treated cells.
Microarray analysis for the effect of Mn on DNA-transfected macrophages
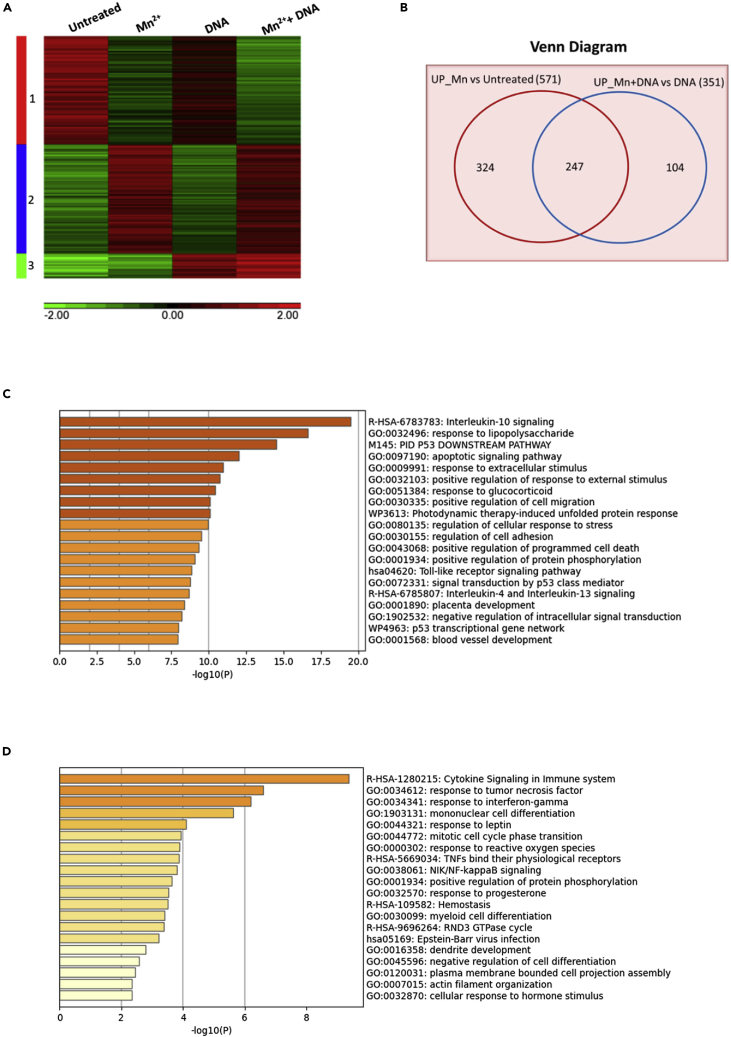
To fully illustrate the enhancing effect of Mn2+ on DNA-mediated innate immune response, we conducted a microarray analysis. Following the same experimental conditions as described above, macrophages were pretreated with MnCl2 at 25 μM, then transfected with DNA. The gene expression profiles in untreated, Mn2+-treated, DNA-transfected, and a combination of Mn2+- and DNA-treated cells were compared and as shown in a heat map (Figure 2A). A total of 1,302 genes with greater than two-fold increase or decrease and p value less than 0.05 in the gene expression were included in the map. Among those genes, 582 genes (cluster 1) were downregulated by Mn2+, and 587 genes (cluster 2) were upregulated by Mn2+. A total of 133 genes (cluster 3) were upregulated by DNA transfection alone. To specifically further define the genes that were upregulated by Mn2+ treatment, a Venn diagram, as shown in Figure 2B, was used to compare the Mn2+ upregulated-genes with the absence of DNA stimulation in Mn2+-treated macrophages versus untreated macrophages (as one group) against the Mn2+ upregulated-genes with the presence of DNA stimulation in DNA-transfected and Mn2+-treated macrophages vs. DNA-transfected macrophages (as another group). The Venn diagram showed that 351 genes are upregulated in DNA-mediated responses and 247 genes were upregulated by Mn2+ treatment. Of note, the expression of 104 genes were initiated by DNA stimulation alone and then enhanced by Mn2+ treatments. Using a functional enrichment analysis resource (Metascape (Zhou et al., 2019)), we found that the set of 247 genes (Mn2+-treatment-mediated upregulation) was significantly enriched for the genes involved in multiple biological processes, such as innate immune response and inflammatory cytokines, which include IL-10, IL-4, and IL-13 signaling and the Toll-like receptor signaling pathway. Meanwhile, we noticed that Mn2+ treatment significantly enhanced the responses of the host cells to external stimuli (Figure 2C). Similarly, the set of 104 genes was significantly enriched for cytokine signaling in the immune system and for responses to tumor necrosis factor (TNF) or IFN-γ (Figure 2D). More interestingly, we found that both functional enrichment analyses for the 247 and 104 genes shared a common category: 38 genes in the 247 genes and 20 genes of the 104 genes were involved in the positive regulation of protein phosphorylation.
Figure 2.
Microarray analysis for the impact of Mn treatment
Human primary macrophages (n = 3) were treated as indicated: untreated, MnCl2 treatment alone, DNA transfection alone, or MnCl2 pretreatment with DNA transfection. The cells were collected for RNA extraction, and the RNA samples were used as the input for the microarray platform. The microarray data was analyzed using 2-way ANOVA. The genes were selected by p-value <=0.05 and absolute fold change >=2.
(A) Heat map showing gene expression profiles among different treatments: untreated, MnCl2-treated, DNA-transfected, and Mn- and DNA-treated cells.
(B) Venn diagram showing overlap of differentially upregulated genes in macrophages on different treatments. (571 genes from Mn vs. Untreated and 351 genes from Mn + DNA vs DNA treated)
(C) Functional enrichment analysis by Metascape for 247 Mn-mediated and upregulated genes.
(D) Functional enrichment analysis by Metascape for 104 DNA-mediated and Mn-upregulated genes.
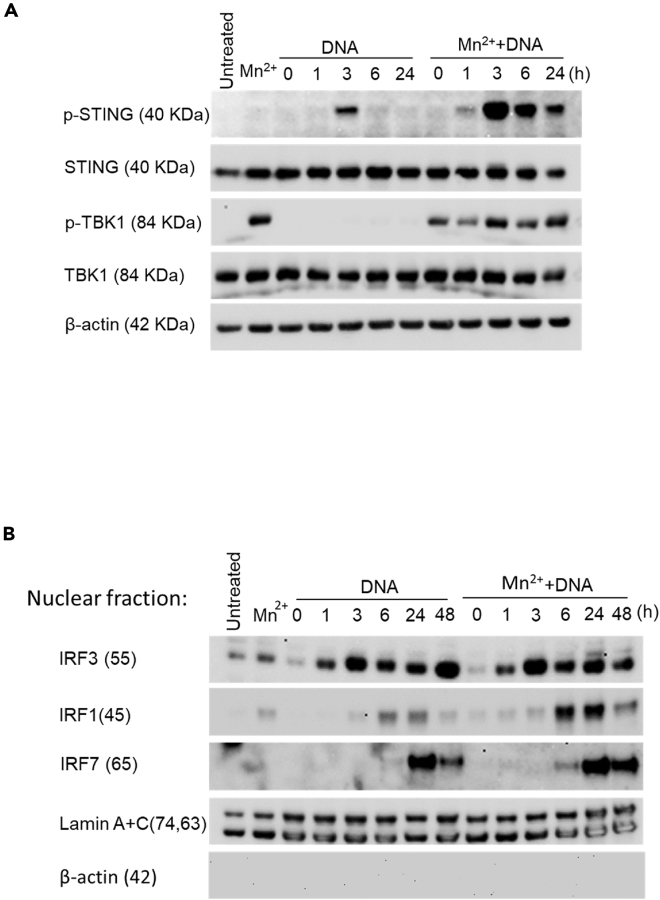
The enhancing effect of Mn treatment on the activity of the downstream STING-TBK1-IRFs axis
To further characterize how Mn2+ treatment enhances the DNA-mediated IFN induction, we compared the activity of major signaling mediators: STING, TBK1, IRF3, IRF1, and IRF7. The whole cell lysates at different time points after DNA stimulation were harvested from the cells with or without Mn2+ pretreatment. The lysates were then subjected to western blotting to detect phosphorylated STING and TBK1. Total STING, TBK1, and β-actin were also detected as input controls. Compared to the panel of DNA treatment alone, DNA transfection with Mn2+pretreatment yielded a robust and prolonged phosphorylation status of STING and TBK1. DNA transfection alone definitely activates phosphorylation of TBK1. However, compared with additional pretreatment by Mn, the signal of phosphorylation of TBK1 was much weaker, therefore, causing the band of p-TBK1 with DNA transfection was undetectable in the western blot analysis. The Mn-enhanced phosphorylation of STING was detected within 1 h, peaked at 3 h and persisted for 24 h after DNA transfection (Figure 3A). Interestingly, we noticed that Mn2+ pretreatment alone for 24 h already activated the phosphorylation of TBK1, which implicated the potential mechanism for the enhancing effect of Mn2+ on DNA-mediated innate immune responses: Mn2+ may activate TBK1 in a DNA-stimulation independent pathway.
Figure 3.
The enhancing effect of Mn treatment on the activity of the STING-TBK1-IRF signaling pathway
Macrophages were transfected with DNA with or without Mn2+ pretreatment.
(A) The cell lysate was collected at the indicated time points after DNA transfection, and Western blotting was performed using anti-p-STING, anti-STING, anti-p-TBK1, anti-TBK1 antibodies, and anti-β-actin antibody was used to probe β-actin as a loading control for all the samples. The untreated sample was collected at 48 h after cell seeding. One representative experiment of at least three independent experiments is shown.
(B) The nuclear fraction of the cell lysate was prepared at the indicated time points after DNA transfection, and Western blotting was performed using anti-IRF3, anti-IRF1, anti-IRF7, and anti-Lamin A + C antibody was probed as a loading control for all nuclear fractions. The untreated sample was collected at 48 h after cell seeding. One representative experiment of at least three independent experiments is shown.
To evaluate the activation of major transcription factors associated with IFN gene activation: IRF1, IRF3, and IRF7, we collected nuclear fractions from each time point. The western blot results consistently showed that DNA transfection highly enhanced the nuclear translocation of IRF1 and IRF7 on Mn2+ pretreatment (Figure 3B). The images in Figure 3B were further analyzed by Image J, the data was shown in Fig. S2. The analysis further clarified that Mn2+ highly enhanced the activity of IRF3, IRF1 and IRF7. Mn2+ has less enhancing effect on the nuclear accumulation of IRF3 between Mn2+-treated and untreated DNA-transfected cells, However, Mn2+ greatly enhances the activity of IRF1 and IRF7 at multiple time points after DNA transfection. The above data indicated that Mn2+ pretreatment at a concentration of 25 μM highly enhanced the DNA-mediated signaling pathway including the activity of STING, TBK1, IRF3, IRF1, and IRF7.
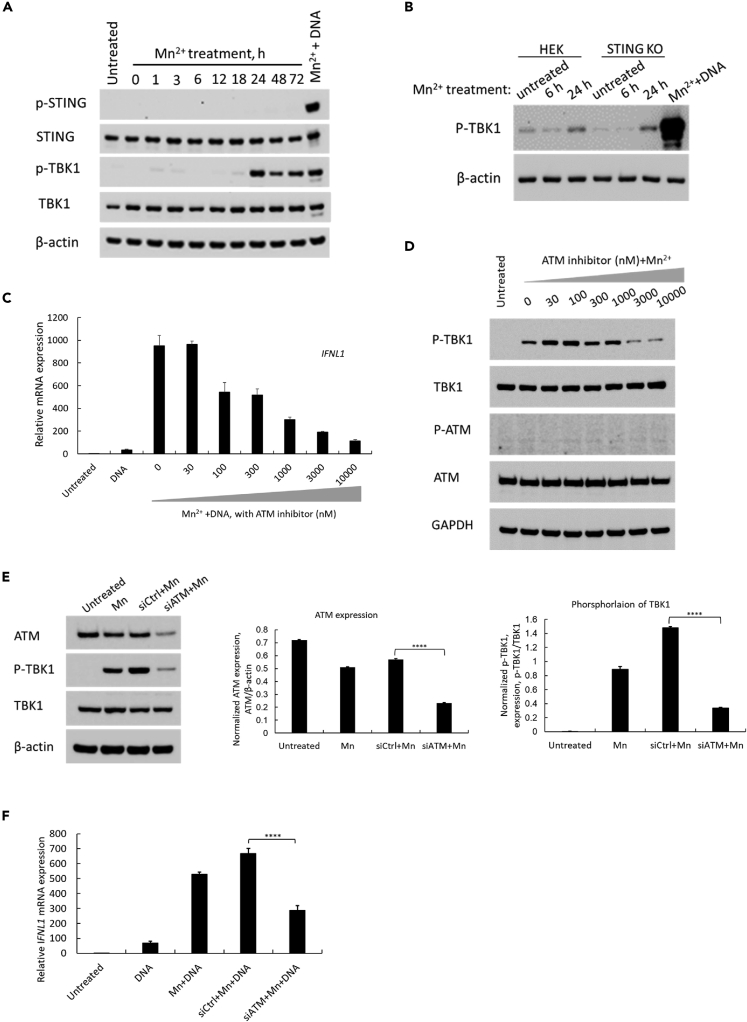
Pretreatment with Mn alone induces phosphorylation of TBK1 via an ATM-TBK1 phosphorylation signaling pathway
As mentioned in the above experiments and shown in the second lane of Figure 3A, we found that Mn2+ treatment alone induced the phosphorylation of TBK1 within 24 h. We further evaluated the time course of the phosphorylation of TBK1 by Mn2+. The result confirmed that Mn2+ treatment induced the phosphorylation of TBK1 without DNA transfection, which was abundantly induced at 24 h after treatment (Figure 4A). However, the phosphorylation of STING was not detected in the Mn2+-treated cells (Figure 4A). This result suggested that Mn2+ specifically induced the phosphorylation of TBK1. We also assessed whether the phosphorylation of TBK1 by Mn2+ is STING-dependent. HEK cells and STING-knockout HEK (STING KO) cells were treated with Mn2+ at 25 μM, and the cytosolic fractions were collected at 6 and 24 h after treatment. Phosphorylation of TBK1 was detected in both cells within 24 h after Mn2+ treatment (Figure 4B). The phosphorylation of TBK1 occurred in a STING-independent manner.
Figure 4.
Treatment with Mn alone induces phosphorylation of TBK1 via an ATM-TBK1 phosphorylation signaling pathway
(A) Macrophages were treated with MnCl2 at 25 μM. The cell lysate was collected at the indicated time points after MnCl2 treatment, and Western blotting was performed using anti-p-STING, anti-STING, anti-p-TBK1, anti-TBK1, and anti-β-actin antibodies. The untreated sample was collected at 24 h after cell seeding. One representative experiment of at least three independent experiments is shown.
(B) HEK cells or STING-knockout HEK (STING-KO) cells were treated with MnCl2 at 25 μM, and the cell lysate was collected at the indicated time points. Western blotting was performed using anti-p-TBK1 and anti-β-actin antibodies. The untreated sample was collected at 24 h after cell seeding. One representative experiment of at least three independent experiments is shown.
(C) Macrophages were treated with different concentration of ATM inhibitor for 30 min before Mn was added to the medium, and the cells were stimulated by DNA transfection at 24 h after Mn pretreatment. Total RNA was extracted at 24 h after DNA transfection. Relative IFNL1 mRNA expression level was measured using real-time RT-PCR assay. One representative experiment of three independent experiments is shown. And each was done as triplicate. Data are shown as mean ± SD (n = 3).
(D) Macrophages was treated first with different concentration of ATM inhibitor for 30 min. And then Mn2+ at the concentration of 25 μM was added to the cells. The whole cell lysate was collected after 24 h Mn pretreatment. WB was performed to detect P-TBK1, TBK1, P-ATM and ATM proteins. And anti-GAPDH antibody was also included as the loading control for all the samples. One representative experiment of at least three independent experiments is shown.
(E) Macrophages were transfected with siRNA targeting ATM (siATM) or control siRNA (siCtrl) at 25 nM, and then the cells were treated by Mn2+ at 25μMat 24 h after siRNA transfection. The whole cell lysate was collected at 24 h after Mn treatment. WB was performed to probe anti-ATM and anti-p-TBK1, anti-TBK1 antibodies, anti-β-actin is also included as a loading control for all the samples (the 1st panel). One representative experiment of at least three independent experiments is shown. The WB image was further analyzed using Image J to quantify the knocking-down efficiency of ATM expression (the 2nd panel), and the inhibition of phosphorylated TBK1 under the condition of ATM was knocked down (the 3rd panel). Data are shown as mean ± SD of three independent analyses (n = 3). ∗∗∗∗p< 0.0001 (One-way ANOVA) was indicated in the figures.
(F) Macrophages were treated with siATM for 24 h before Mn was added to the medium, and the cells were stimulated by DNA transfection at 24 h after Mn treatment. Total RNA was extracted at 24 h after DNA transfection. Relative IFNL1 mRNA expression level was measured using real-time RT-PCR assay. One representative experiment of at least three independent experiments is shown. Each was done as triplicate. Data are shown as mean ± SD (n = 3). ∗∗∗∗p< 0.0001 (One-way ANOVA) was indicated in the figure.
Manganese induces phosphorylation of 3 ATM targets, P53, CHK2 and H2AX, and ATM is an upstream kinase for Mn-dependent phosphorylation (Tidball et al., 2015). To define whether the kinase is associated with the phosphorylation of TBK1, we treated macrophages by different dose of specific ATM inhibitor (KU60019) for 30 min before Mn pre-treatment. And the data, as shown in Figure 4C, indicated that the ATM inhibitor dose dependently suppressed the Mn-enhanced, DNA-mediated IFNL1 gene expression in macrophages. At the concentration of 10,000 nM, the IFNL1 gene expression was inhibited up to 91.5% without any significant impact on cell viability. To further elucidate whether the ATM inhibitor blocks Mn-induced phosphorylation of TBK1, phosphorylation form of TBK1 was detected by WB using macrophages cell lysate treated by Mn with different dose of the ATM inhibitor. The result showed that ATM inhibitor also inhibited Mn-induced phosphorylation of TBK1, especially at the concentration of 10,000 nM, the band of phosphorylation of TBK1 was decreased by 70% (Figure 4D). This WB image was further analyzed by NIH Image J. The result suggested that ATM inhibitor does-dependently inhibits Mn-dependent phosphorylation of TBK1 (Figure S3). ATM is activated by phosphorylation at Ser1981 (Graham et al., 2017), thus we assessed the impact of phosphorylation of ATM by Mn using WB. The result demonstrated that Mn at the concentration of 25 μM was not able to induce phosphorylation of ATM at Ser1981 (Figure 4D).
To further confirm the involvement of ATM in the Mn-dependent phosphorylation of TBK1 pathway, we evaluated the impact of ATM knocking down on Mn-enhanced DNA-mediated innate immune response. A pooled siRNA targeting ATM (siATM) was used to knock down the expression of ATM. The data, as shown in Figure 4E, suggested that siATM successfully inhibited the expression of ATM in human macrophages by about 59% (the 1st and 2nd panel). In the same experimental condition, compared with Mn treatment alone, we found that phosphorylation of TBK1 is correlatively decreased by about 77% (the 3rd panel). This data further illustrated that the protein ATM is involved in Mn-dependent phosphorylation of TBK1. Based on those results, we further evaluated DNA transfection triggered IFNL1 induction in ATM-knocked down cells. The data (Figure 4F) indicated that the Mn-enhanced induction was suppressed about 57% by using siRNA to specifically knock down ATM, while compared with non-targeting siRNA(siCtrl)-transfected and Mn-enhanced DNA-mediated IFNL1 induction.
To further confirm the specific role of Mn2+ on the phosphorylation of TBK1, we also checked whether other metal ions have a similar role. Only treatment with Mn2+ induced the phosphorylation of TBK1; none of the other tested metal ions showed a similar effect (Figure S4A). As Mg2+ usually has a similar biological function as Mn2+, we further tested whether treatment with MgCl2 at a higher concentration induced phosphorylation of TBK1 similarly to MnCl2. Human primary macrophages were treated by MgCl2 at concentrations from 25 μM to 20 mM, and the whole-cell lysate was collected at 24 h after treatment. The result consistently suggested that the treatment with Mg2+, even at a higher concentration, still cannot induce the phosphorylation of TBK1 (Figure S4B). This confirms that treatment with Mn2+ specially and uniquely induced the phosphorylation of TBK1.
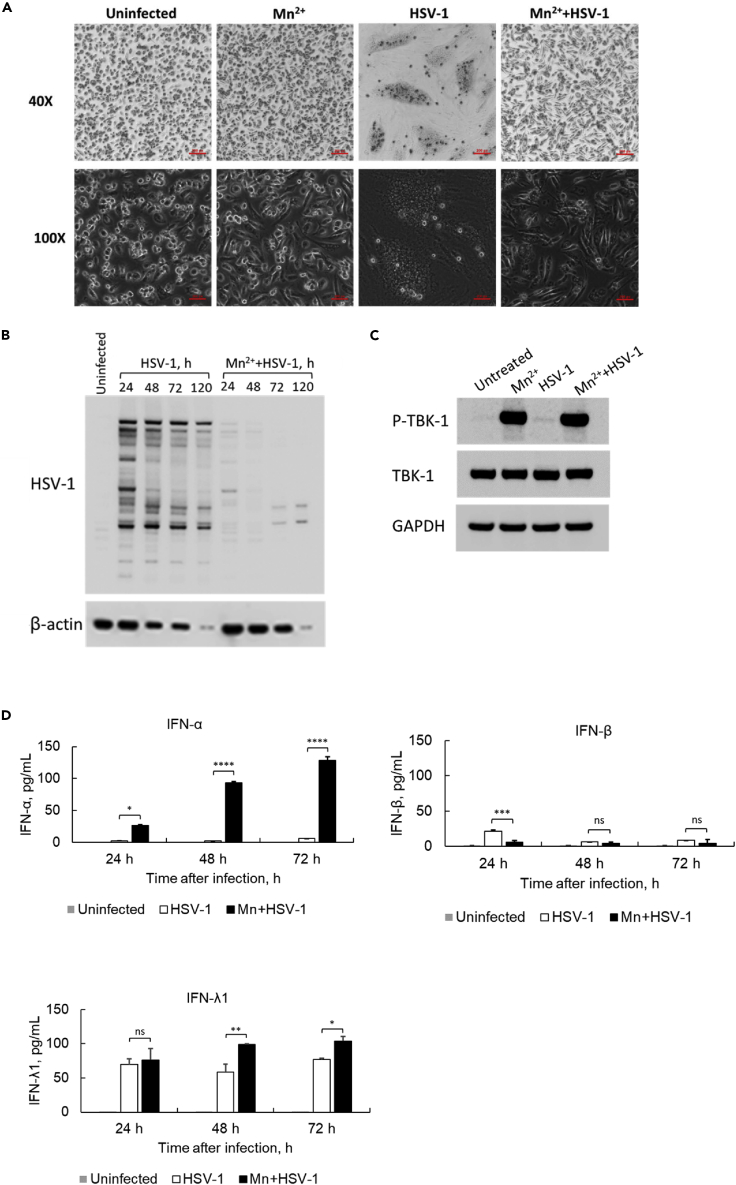
Mn treatment protects macrophages from HSV-1 McKrae infection
To further investigate the role of Mn2+ on virus infection stimulated innate immune system, we evaluated whether Mn2+ enhances DNA-virus-mediated innate immune response. Macrophages were infected with the DNA virus HSV-1 McKrae strain (Jiao et al., 2019; Watson et al., 2012) at an MOI of 1.0. Twenty-four hours later, the HSV-1-infected macrophages showed a dramatic change in morphology compared to uninfected cells (Figure 5A). However, those infection-induced morphological changes were completely diminished in the cells pretreated with Mn2+ before HSV-1 infection (Figure 5A). To further characterize the observation, the HSV-1-infected cells with and without Mn2+ treatment were collected at indicated time points, 24, 48, 72 and 120 h after infection (Figure 5B), and the HSV-1 protein expression was detected by Western blotting using anti-HSV-1 antibody. The results demonstrated that the HSV-1 viral proteins were acutely expressed in macrophages infected with the HSV-1 McKrae strain. Conversely, the viral proteins were either undetectable or detected at a much lower intensity in the Mn2+-pretreated macrophages (Figure 5B). To further confirm whether the pretreatment with Mn in HSV-1 infection cell culture model still activates phosphorylation of TBK1, a WB was performed to detect the phosphorylation of TBK1. The data in Figure 5C suggested a large amount of TBK1 was phosphorylated once the cells were treated with Mn for 24 h. In contrast, the band intensity of phosphorylation of TBK1 in HSV-1 infected cells was much weaker compared with the phosphorylation of TBK1 under Mn pretreatment. To further elucidate this protective effect, the culture supernatants of HSV-1-infected cells were collected for ELISA to quantify the protein expression level of the major antiviral cytokines IFN-α, IFN-β, and IFN-λ1. The results (Figure 5D) indicated that pretreatment with Mn2+ also potentially enhanced IFN-α and IFN-λ1 induction mediated by McKrae strain infection. A markedly lower level of IFN-β was detected, indicating that Mn2+ may has less enhancing effect on the production of IFN-β compared with the induction of IFN-α and IFN-λ1.
Figure 5.
Mn treatment protects macrophages from HSV-1 McKrae infection
(A) The effect of pretreatment with Mn2+ on morphological changes in human primary macrophages infected with the HSV-1 McKrae strain. Human macrophages were pretreated with Mn2+, then mock-infected (uninfected) or infected with the HSV-1 McKrae strain at an MOI of 1. Cells were observed under a phase-contrast microscope at 40× and 100× magnification 24 h after HSV-1 infection. Scale bar = 200 μM. One representative experiment of at least three independent experiments is shown.
(B) Human macrophages were treated with or without Mn2+, then infected with HSV-1 McKrae at an MOI of 1. The total cell lysate was collected at different time points after infection, and WB was performed using anti-HSV-1 and anti-β-actin antibodies. The untreated sample was collected at 48 h after cell seeding. One representative experiment of at least three independent experiments is shown.
(C) Macrophages were treated with or without Mn2+, then infected with HSV-1 McKrae at an MOI of 1. The total cell lysate was collected at 24 h after infection, and WB was performed using anti-p-TBK1, anti-TBK1 and anti-GAPDH antibodies. One representative experiment of at least three independent experiments is shown.
(D) Cell culture supernatants were collected at 24, 48, and 72 h after virus infection. The protein expression of IFN-α, IFN-β, and IFN-λ1 was detected by ELISA. One representative experiment of at least three independent experiments is shown. And each was done as duplicate. Data are shown as mean ± SD (n = 2). Not significant (ns) p> 0.05, ∗p< 0.05, ∗∗p< 0.01, ∗∗∗p< 0.001, ∗∗∗∗p< 0.0001 (One-way ANOVA) were indicated in the figures where the IFN protein expression level was compared between HSV-1 infected cells and Mn2+/HSV-1 infected cells.
Mn treatment also enhances innate immune signaling mediated by RNA agonists or RNA virus infection
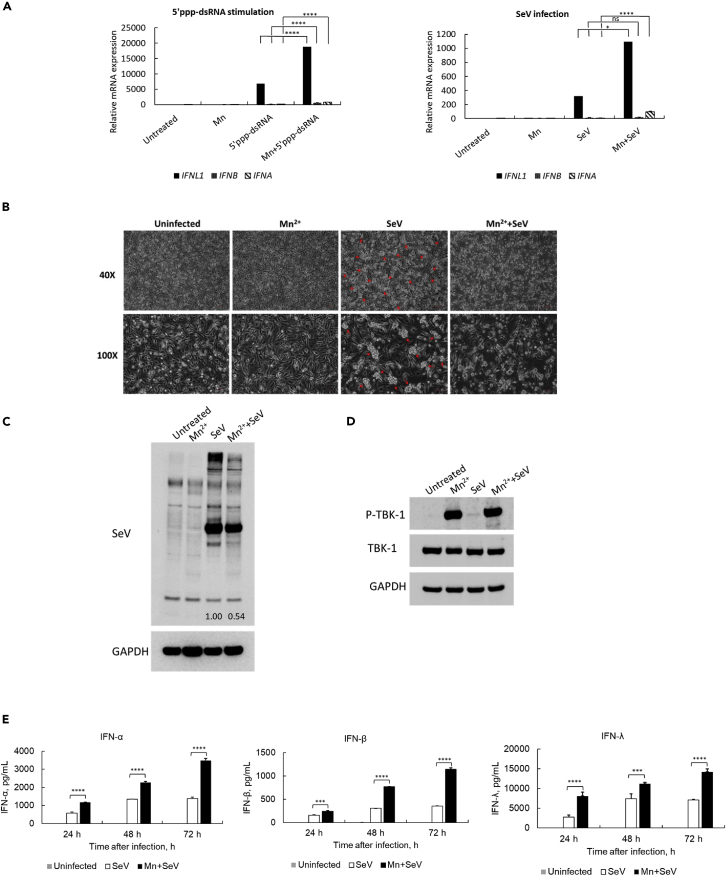
As we found above, Mn2+ treatment enhances DNA-mediated IFN-α, IFN-β, and IFN-λ1 induction by increasing the phosphorylation of TBK1. TBK1 is not only the downstream mediator of the DNA-mediated innate immune signaling pathway, but also an important mediator in the signaling pathway of RNA sensors, such as RIG-I and MDA5 (Rehwinkel and Gack, 2020; Yoneyama and Fujita, 2008,2009). Thus, we further investigated whether pretreatment with Mn2+ enhances RNA-mediated IFN induction in human macrophages. 5′ triphosphate double-stranded RNA (5′ppp-dsRNA, a synthetic ligand for RIG-I) and an RNA virus, Sendai virus (SeV), were tested to confirm the effect of Mn2+. The pretreatment enhanced 5′ppp-dsRNA-mediated IFNL1 induction by around four-fold (p< 0.001). The induction of IFNA and IFNB is much lower than that of IFNL1 but still enhanced by Mn2+ treatment (Figure 6A, upper panel). Consistently, the pretreatment with Mn2+ also enhanced SeV-infection-mediated IFNL1, IFNA, and IFNB induction (Figure 6A, lower panel). The impact of Mn2+ on the morphological changes in SeV-infected cells was also assessed via microscopy. Mn2+ treatment induced no apparent changes in macrophage morphology compared to uninfected cells (Figure 6B). At 48 h after SeV infection, the infection induced cell aggregation (indicated by the red arrow in Figure 6B). By contrast, the aggregates were not observed in Mn2+-pretreated and Sev-infected cells, indicating that Mn2+ treatment potentially protects cells from the viral infection. Finally, the whole-cell lysate was collected to measure SeV viral protein expression in the cells. The data further emphasized that the intensity of SeV proteins (multiple bands) was decreased in the cells with Mn2+ treatment. Image J was used to normalize SeV protein expression by the intensity of the GAPDH band. The results showed that Mn2+ inhibits SeV viral protein expression by 46%, indicating that Mn2+ induces partial inhibition of RNA virus replication in the cells (Figure 6C).
Figure 6.
Mn treatment enhances RNA-mediated or RNA-virus-infection-mediated innate immune responses
(A) Human macrophages were treated with or without Mn2+ at 25 μM, then transfected with 5′ppp-dsRNA (1 μg/mL) or infected with SeV at 40 HA units/mL. Total RNA was extracted 24 h after RNA stimulation. Real-time RT-PCR was performed to measure the gene expression level of IFNL1, IFNB, and IFNA. One representative experiment of at least three independent experiments is shown. And each was done as triplicate. Data are shown as mean ± SD (n = 3). Not significant (ns) p> 0.05, ∗p< 0.05, ∗∗∗∗p< 0.0001 (One-way ANOVA)) were indicated in the figures where the IFN mRNA expression level was compared between RNA-transfected/infected and Mn2+/RNA-treated cells.
(B) The effect of Mn2+ pretreatment on morphological changes of SeV-infected human primary macrophages. Human macrophages were pretreated with Mn2+, then mock-infected (uninfected) or infected with SeV at 40 HA units/mL. Cells were observed under a phase-contrast microscope at 40× and 100× magnification 48 h after SeV infection. Scale bar = 100 μM. One representative experiment of at least three independent experiments is shown.
(C) The total cell lysate was collected at 24 h after infection, and Western blotting was performed using anti-SeV and anti-GAPDH antibodies. One representative experiment of at least three independent experiments is shown. The band intensity of SeV proteins was normalized by the band intensity of GAPDH (Image J), the values are indicated in the image.
(D) Macrophages were treated with or without Mn2+, then infected with SeV at 40 HA units/mL. The total cell lysate was collected at 24 h after infection, and WB was performed using anti-p-TBK1, anti-TBK1 and anti-GAPDH antibodies. One representative experiment of at least three independent experiments is shown.
(E) Cell culture supernatants were collected at 24, 48, and 72 h after SeV virus infection. The protein expression of IFN-α, IFN-β, and IFN-λ1 was detected by ELISA. One representative experiment of at least three independent experiments is shown. Each was done as duplicate. Data are shown as mean ± SD (n = 2). ∗∗∗p < 0.001 and ∗∗∗p< 0.0001 (One-way ANOVA)) were indicated in the figures where the IFN protein expression level was compared between SeV-infected and Mn2+/SeV-infected cells.
Similarly, we also detected whether the pretreatment with Mn activates phosphorylation of TBK1 whereas the cells were infected by SeV virus. The WB result consistently indicated that Mn also highly activated phosphorylation of TBK1 compared with the phosphorylation of TBK1 in SeV infected cells (Figure 6D).
We further specifically measured the induction of IFN-α, IFN-β and IFN-λ1 in superannuants of SeV-infected macrophages. Mn also greatly enhanced SeV infection-induced IFN-α, IFN-β and more abundantly IFN-λ1 induction (Figure 6E). The result in the case of SeV infection (RNA virus) was consistent with that of DNA virus infection. Our results indicate that Mn enhances DNA and RNA virus infection induced innate immune response, and this enhanced antiviral interferons facilitates inhibiting virus infection or replication.
We have confirmed that Mn enhanced the production of antiviral cytokines, IFN-α, IFN-β and IFN-λ1, in human primary macrophages and protected the host cells from infection of HSV-1 McKrae strain and SeV virus. To further evaluate the protection effect of Mn, we measured the induction of some ISGs at 24 h after virus infection. The data, however, indicated that among tested ISGs, OAS2, MX1, STAT1, IRF1, IFIT1, IRF7, MX2, EIF2AK2 and APOBEC3G, only IRF1 and IRF7 shows an enhanced induction by Mn at 24 h after virus infection, the induction of all other ISGs was at a low expression level or undetected (upper panel of Figures S5A and S5B). To confirm whether IFN genes were expressed when IRF1 and IRF7 were induced, we measured the mRNA expression of IFNA8, IFNB and IFNL1. Of interest, the results consistently indicated that Mn has enhanced the induction of IFNs at 24 h after infection (lower panel of Figures S5A and S5B). We assumed that the secreted protein levels of IFNs may not be sufficient to activate the induction of all ISGs, so only IRF1 and IRF7 may be early induced by a lower amount of IFNs, and the enhanced IRF1 and IRF7 may contribute to an abundant induction of IFN-α and IFN-λ1 at later time points. This observation is consistent with our previous finding about the kinetic induction of IFN-λ1 (Figure S1).
Discussion
Trace metals are essential components in all forms of life and are critically involved in the activities of many enzymes, facilitating various fundamental biological processes (Wang et al., 2020). Iron (Fe), zinc (Zn), manganese (Mn), nickel (Ni), copper (Cu), and cobalt (Co) are especially essential trace nutrients known to play key roles. They provide structural support, serve as enzymatic cofactors, and mediate electron transportation within metalloproteins (Wang et al., 2020). Recently, increasing attention has been drawn to the roles of metal ions in the immune system because accumulating evidence suggests that metals are critically involved in modulating both the innate immune sensing of and the host defense against invading pathogens.
Consequently, in the current study, we evaluated the role of metal divalent ions in the innate immune response mediated by DNA and RNA sensor proteins in the non-immune cell lines and human primary macrophages. We identified that among seven essential metal ions examined, only Mn2+ greatly enhances not only DNA-mediated but also RNA-mediated-induction of antiviral interferons, IFN-λ1, IFN-α, and IFN-β. However, the optimal concentration we used in the study is non-stimulatory by Mn2+ treatment itself. We further confirmed that the enhancing effect by Mn2+ was observed not only in HEK cells but also in unstimulated THP-1 cells and human primary macrophages. We have known that multiple DNA sensor proteins are present in both THP-1 cells and human macrophages, and we speculated that the enhancing effect by Mn2+ may present in multiple DNA-mediated signaling pathways, such as cGAS, Ku70, or IFI16-mediated innate immune response. Pretreatment with Mn2+ enhanced the production of antiviral interferons, enhanced the activation of the downstream signaling, phosphorylation of STING and TBK1, and increased the nuclear translocation of IRF3, IRF1 and IRF7. The collected data emphasize that pretreatment with Mn highly enhances the DNA-mediated innate immune response, regardless of what DNA sensor-mediated signaling pathway. Previously, Wang and colleagues reported that Mn is important for cGAS-STING-mediated cytosolic DNA sensing (Wang et al., 2018). They further discovered that Mn enhanced the sensitivity of cGAS to dsDNA and its enzymatic activity and that Mn enabled cGAS to produce secondary messenger cGAMP in the presence of low concentrations of dsDNA. Mn also enhanced STING activity by augmenting cGAMP-STING binding affinity. Our observation demonstrated that Mn’s effect is not restricted in the cGAS-STING-mediated signaling pathway because we demonstrated the enhancing effect of Mn also existed in HEK cells, in which cGAS expression is absent. In the current work, the DNA-mediated IFN-λ1 induction is highly upregulated by Mn2+ pretreatment. We therefore speculated that Mn2+ may have a role in the cytoplasmic translocation of Ku70, a cytosolic DNA sensor, to dominantly mediate type III IFN response (Sui et al., 2021a). The data, as shown in Fig. S6, did implicate a certain enhancing effect on the cytoplasmic accumulation of Ku70; however, the contribution to the enhanced IFN induction is limited.
The enhancing effect of Mn on multiple DNA-sensor-mediated signaling pathways was also underscored by microarray analysis and the mechanism we found in this study. Microarray analysis was used to further characterize the role of Mn on DNA-mediated innate immune response. The function enrichment analysis suggested that 351 genes are upregulated by Mn treatment. Those genes play roles in multiple biological processes, such as multiple cytokine responses in the immune system, enhanced response to external stimuli, and positive regulation of protein phosphorylation. The analysis implicated that the enhancing role of Mn in the innate immune system is not limited to one or two DNA-mediated signaling pathways. Treatment with Mn activated the host cells’ whole sensing system in response to intracellular stimulation. Interestingly, we further demonstrated that Mn treatment by itself activated phosphorylation of TBK1. TBK1 is a signaling mediator located downstream of multiple DNA sensors, such as Ku70, IFI16, DNA-PK, cGAS, IFIX, and DDX41 (Zhou et al., 2020). Such a mechanism is in line with our observations. In addition, we demonstrated that the phosphorylation of TBK1 by Mn treatment is STING-independent. This result further clarified that the phosphorylation of TBK1 by Mn is independent of DNA stimulation. The mechanism we found in the current study is partially in line with the proposed mechanism in another report (Sun et al., 2021), they also found that Mn-mediated potentiation of STING agonist is through STING-independent TBK1 and p65 phosphorylation and STING-dependent IRF3 phosphorylation.
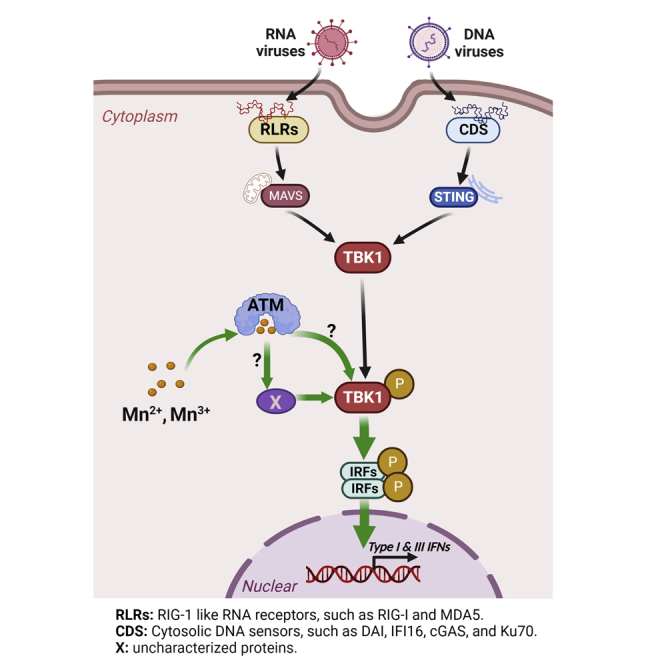
ATM is a serine/threonine kinase regulated by autophosphorylation. ATM is involved in multiple aspects of cellular metabolism, such as response to genotoxic stress, proliferation, and maintaining genetic stability in a Mn-dependent pattern (Graham et al., 2017). So, we speculated that whether this Mn related kinase plays a role in Mn-enhanced innate immunity. Using one of specific ATM inhibitor (KU60019)(Biddlestone-Thorpe et al., 2013; Golding et al., 2009; Nagane et al., 2015), we found that the ATM inhibitor does-dependently suppresses Mn-enhanced DNA-mediated IFN-λ1 induction. And this inhibition was correlated with the inhibition effect on phosphorylation of TBK1 by ATM inhibitor. This result was further hammered by the data obtained using siRNA to inhibit ATM activity. Those result suggested that ATM kinase may involve in Mn-dependent phosphorylation of TBK1. Based on the finding in our work, we proposed that Mn enhances DNA or RNA mediated innate immune response through Mn-dependent ATM-TBK1 phosphorylation signaling pathway. Autophosphorylation in regulating ATM kinase activity is very important (Kozlov et al., 2003). S1981 was identified as a major autophosphorylation site of ATM in vivo (Bakkenist and Kastan, 2003). Rapid intermolecular autophosphorylation in response to DNA damage caused dissociation of inactive ATM dimers to form active monomers. However, in our current study, the phosphorylation of ATM at S1981 was not detected with the concentration of Mn2+ at 25 μM, indicating that S1981 autophosphorylation may not be essential in the Mn-dependent phosphorylation of TBK1. Considering that the potential off-target effect may exist while using ATM inhibitor or siRNA, further detailed study is needed to clarify how TBK1 is phosphorylated by Mn through ATM and whether this process may involve any other cofactors or kinases (Graphical Abstract). A cell free kinase activity assay for TBK1 and ATM with Mn may help define a detailed mechanism.
To further validate the application relevance of our finding, we evaluated whether Mn enhances the antiviral innate immune response mediated by DNA virus infection. We used the HSV-1 McKrae strain to replace DNA transfection. Pretreatment with Mn appeared to protect macrophages from virus infection that would result in cell morphology damage and inhibited the expression of viral proteins. We confirmed that Mn treatment highly enhances the production of antiviral interferon cytokines IFN-α and IFN-λ1. We also confirmed that Mn enhances the DNA-transfection-mediated IFN-α, IFN-β, and IFN-λ1 induction without altering the initiation of the kinetics process of the induction. The kinetic curve of the IFN induction by DNA stimulation is consistent with Mn-enhanced DNA-mediated innate immune response. However, we noticed that Mn has a robust enhancing effect on the delayed induction stage of IFN-α and IFN-λ1, indicating that the enhancing effect by Mn affects the prolonged process. The time course study provided additional evidence that the induction of IFN-α and IFN-λ1 shares a similar mechanism; both depended on the activation of IRF1 and IRF7, suggesting Mn contributes to regulate a later prolonged innate immune response, because IRF1 and IRF7 are not endogenously expressed in the cells (Sui et al., 2017). By contrast, IFN-β induction was suggested to have an immediate induction process through the activation of IRF3. The signal transduction pathway in HSV-1-infection-mediated IFN induction was also consistent with that in the DNA-transfection-mediated IFN induction process. The significant enhancing effect of Mn on the induction of IFN-α and IFN-λ1 is the major contribution for the enhanced antiviral effects of Mn.
It is reported that TBK1 plays a role in the DNA-mediated signaling pathway and in the RNA-mediated innate immune response (Akira et al., 2006; Dempsey and Bowie, 2015; Gürtler and Bowie, 2013; Loo and Gale, 2011; Yoneyama and Fujita, 2009). Given that Mn2+ enhances DNA-mediated innate immune response by increasing the phosphorylation of TBK-1. Consequently, we further examined whether Mn enhances the RNA-mediated signaling pathway. Pretreatment with Mn also inhibited cell morphology changes induced by infection with the RNA virus SeV, as well as viral protein expression, although at less efficacy. In the current study, we also noticed that both DNA virus and RNA virus infection highly induced IFN-λ1, a type III interferon, in human primary macrophages compared with the induction of IFN-β (type I IFNs). The antiviral activity from Mn is mainly contributed by the enhanced induction of IFN-λ1. However, the IFN-λ1 gene in mice (ifnl1) lacks the entire exon 2 and contains a stop codon within exon 1 (Lasfar et al., 2006). Studies conducted in several labs have shown that this mutated mIFN-λ1 gene does not encode a functional IFN-λ1 protein (Bartlett et al., 2005; Donnelly and Kotenko, 2010). Therefore, the investigation of enhanced antiviral immunity from Mn2+ in a mice model was not feasible. Despite this fact, our findings provided further evidence to support applying Mn to the development of novel vaccines against both DNA and RNA viral infection.
In this study, we tested seven trace metal ions: chromium, manganese, iron, cobalt, nickel, copper, and zinc; however, only Mn, in the form of Mn2+ or Mn3+, enhanced the DNA-mediated innate immune response. Mn2+ is similar to Mg2+ in terms of physiochemical properties in many biological functions. Previous work demonstrated that Mn2+ is able to substitute for Mg2+in vitro in almost half of Mg2+-dependent enzymes and still maintain the enzymes’ catalytic activity (Wedler, 1993). Consequently, we also included Mg2+ in the screening and assessed whether Mg2+ by itself can activate TBK1. Mg2+ was not able to induce the phosphorylation of TBK1 at the same concentration as Mn2+ (25 μM) or even at a greater concentration. Similar results have been reported in the insulin-receptor-associated protein kinase, and this kinase was substantially activated by Mn2+ but not by Mg2+ (Wente et al., 1990). Multiple studies also suggested that Mn2+ replaces Mg2+ in cGAS activation and enhances its enzymatic activity and ligand sensitivity (Wang et al., 2018). They further found that in contrast to Mg2+, cGAS incubated with Mn2+ is inclined to precipitate; this observation implicated that Mn2+ may induce cGAS to shift into a compact conformation that is more easily activated (Zhao et al., 2020). Although the detailed mechanism by which Mn2+ activates TBK1 has not been fully elucidated in the current study, we hypothesize that the binding affinity of Mn2+ to TBK1 or other uncharacterized potential cofactors or kinase might be greater than that of Mg2+ and other trace metal ions; thus, Mn2+ could efficiently induce a certain conformational change in the signal mediator and activate the protein. Further study is required to provide more insight.
It has been reported that Phosphatase PP4 negatively regulates type I IFN production and antiviral innate immunity by dephosphorylating and deactivating TBK1. TBK1 Ser172 phosphorylation is removed by the ser/thr phosphatase PP4 (Zhan et al., 2015). Among several trace metals, Zn was found to directly bind to and inhibit protein phosphatase 2A or other phosphatases (Tonks, 2013; Xiong et al., 2015). Given that have been investigated, even we did not find any effect from Zn in our study, however, Mn might act in a similar fashion on some phosphatase.
Mn is a nutritional inorganic trace element with diverse biological activities, an essential constituent of many metalloenzymes, and an enzyme activator. The concentration of Mn in the blood is within 0.029 μM and 1.2 μM, whereas the brain contains a higher level of Mn, from 20–53 μM (Bowman and Aschner, 2014). The physiological concentration of Mn in mammalian tissue is between 5 μM and 53 μM (Keen et al., 2000; Rehnberg et al., 1980). The distribution of Mn in the human body is a dynamic process. It has been reported that Mn is released from the mitochondrial area out to the cytoplasm of the cells on DNA stimulation and that cytoplasmic translocated Mn facilitates cGAS sensing (Wang et al., 2018). A certain level of Mn is indispensable to initiate DNA-induced innate immune response. Therefore, one can expect to observe that Mn-deficient mice are highly susceptible to DNA virus (Wang et al., 2018). We optimized the working concentration in our study and found that 25 μM is a lower concentration compared to other studies using Mn at 50–200 μM for stimulation. The current work demonstrated that Mn, even at a lower concentration, is still able to enhance DNA and RNA-mediated innate immune cascades, even it is non-stimulatory by itself. The finding also implicated Mn in various roles at different physiological concentrations under different biological contexts. A systemic evaluation is required to better understand the matter. Our study highlights the potential beneficial role of Mn as a vaccine adjuvant.
Given the enhancing role of Mn on innate immunity, extensive research has been carried out to demonstrate the hybrid or combination use of Mn to amplify antitumor immunotherapeutic efficacy. Hou et al. developed an Mn-based nano-activator for cancer immunotherapy, which demonstrated great efficiency in inducing the secretion of pro-inflammatory cytokines, such as TNF-α and IL-6, and exhibited superior tumor growth inhibition effects with improved survival (Hou et al., 2020). Only a minority of patients’ cancers respond to immunotherapies, evidently because of inadequate immunity. Antitumor immunity depends on the activation of the innate immune response pathway (Tang et al., 2018). A proof-of-concept study from Jiang’s group demonstrated that Mn is also essential in innate immune sensing of tumors and enhances adaptive immune responses against tumors. Mechanically, Mn promoted dendritic cell and macrophage maturation and tumor-specific antigen presentation, augmented CD8+T-cell differentiation and activation, augmented NK-cell activation, and increased memory CD8+T cells. Combining Mn with immune checkpoint inhibition synergistically boosted antitumor efficacies (Lv et al., 2020). In addition, Mn also could inhibit tumor development by enhancing CD8+T cell function via induction of type I IFN production (Song et al., 2021). Our current study, along with other studies we have discussed here, provides potential insight into the development of novel immunotherapies or vaccines to fight cancers or infectious diseases. Of note, our findings indicate Mn’s protective role against RNA virus infection, providing valuable information for the development of strategies to fight the current COVID-19 pandemic.
Limitations of the study
In the current study, we found that Mn enhances DNA- or RNA-mediated innate immune response through the increase in the phosphorylation of TBK1. Using ATM inhibitor and siRNA, we further discovered that ATM is involved in the Mn-dependent phosphorylation of TBK1. However, the underlying mechanism of the TBK1 phosphorylation pathway is still unclear. The effect might be caused by an off-target effect of the inhibitors. Considering this limitation, we need further study to precisely unveil the Mn-dependent phosphorylation of TBK1 pathway.
Data availability
All data generated or analyzed during this study are included in this article.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-cGAS | Cell Signaling Technology | Cat# 15102; RRID:AB_2732795 |
| Rabbit monoclonal anti-Phos-STING (Ser366) | Cell Signaling Technology | Cat# 19781; RRID:AB_2737062 |
| Rabbit monoclonal anti-STING | Cell Signaling Technology | Cat# 13647; RRID:AB_2732796 |
| Rabbit monoclonal anti-Phos-TBK1 (Ser172) | Cell Signaling Technology | Cat# 5483; RRID:AB_10693472 |
| Rabbit monoclonal anti-TBK1 | Cell Signaling Technology | Cat# 3504; RRID:AB_2255663 |
| Mouse monoclonal anti-Phos-ATM (Ser1981) | Cell Signaling Technology | Cat# 4526; RRID:AB_2062663 |
| Rabbit monoclonal anti-ATM | Cell Signaling Technology | Cat# 2873; RRID:AB_2062659 |
| Rabbit monoclonal anti-IRF1 | Cell Signaling Technology | Cat# 8478; RRID:AB_10949108 |
| Mouse monoclonal anti-IRF3 | Origene | Cat# TA500476; RRID:AB_2126996 |
| Rabbit polyclonal anti-IRF7 | Cell Signaling Technology | Cat# 4920; RRID:AB_2127551 |
| Rabbit monoclonal anti-Ku70 | Cell Signaling Technology | Cat# 4588; RRID:AB_11179211 |
| Rabbit polyclonal anti-Ku80 | Cell Signaling Technology | Cat# 2753; RRID:AB_2257526 |
| Rabbit polyclonal anti-HSV-1 | Abcam | Cat# ab9533; RRID:AB_307320 |
| Rabbit polyclonal anti-SeV | MBL | Cat# PD029; RRID:AB_10597564 |
| Mouse monoclonal anti-β-actin | Sigma Aldrich | Cat# A5316; RRID:AB_476743 |
| Rabbit polyclonal anti-GAPDH | Abcam | Cat# ab9485; RRID:AB_307275 |
| Rabbit monoclonal anti-Lamin A + C | Abcam | Cat# ab108922; RRID:AB_10860619 |
| Mouse lgG (HRP) | VWR | Cat# 95017–332 |
| Rabbit lgG (HRP) | VWR | Cat# 95017–556 |
| Bacterial and virus strains | ||
| HSV-1 McKrae strain | Dr. Jeffrey I. Cohen. Watson et al. (2012) | N/A |
| Sendai Cantell strain | Charles River Laboratories | Cat# VR-907 |
| Chemicals, peptides, and recombinant proteins | ||
| ATM inhibitor KU60019 | Sigma Aldrich | Cat# 5319780001 |
| Deposited data | ||
| Microarraydata | This paper | GEO: GSE214181 |
| Experimental models: Cell lines | ||
| Human embryonic kidney 293 (HEK) | ATCC | CRL-1573 |
| Human leukemia monocytic cell line (THP-1) | ATCC | TIB-202 |
| SV40 T-antigen transformed HEK293 (293T) | ATCC | CRL-11268 |
| STING KO HEK cell lines | Sui et al. (2017) | N/A |
| Oligonucleotides | ||
| Silencer™ Negative Control No. 1 siRNA | Thermo Fisher Scientific | AM4636 |
| ON-TARGETplus Human ATM (472) siRNA - SMARTpool | Horizon Discovery | L-003201-00-005 |
| GAPDH probe | Thermo Fisher Scientific | Hs02786624_g1 |
| IFNA6 probe | Thermo Fisher Scientific | Hs00819627_s1 |
| IFNA8 probe | Thermo Fisher Scientific | Hs00266883_s1 |
| IFNA13 probe | Thermo Fisher Scientific | Hs04190680_gh |
| IFNA14 probe | Thermo Fisher Scientific | Hs00353663_s1 |
| IFNA21 probe | Thermo Fisher Scientific | Hs00353738_s1 |
| IFNB probe | Thermo Fisher Scientific | Hs01077958_s1 |
| IFNL1 probe | Thermo Fisher Scientific | Hs01050642_gh |
| OAS2 probe | Thermo Fisher Scientific | Hs00942643_m1 |
| MX1 probe | Thermo Fisher Scientific | Hs00182073_m1 |
| MX2 probe | Thermo Fisher Scientific | Hs01550808_m1 |
| STAT1 probe | Thermo Fisher Scientific | Hs01014002_m1 |
| IRF1 probe | Thermo Fisher Scientific | Hs00233698_m1 |
| IRF7 probe | Thermo Fisher Scientific | Hs00185375_m1 |
| IFIT1 probe | Thermo Fisher Scientific | Hs01675197_m1 |
| EIF2AK2 probe | Thermo Fisher Scientific | Hs00169345_m1 |
| APOBEC3G probe | Thermo Fisher Scientific | Hs00222415_m1 |
| Recombinant DNA | ||
| pCR™2.1 Vector | Thermo fisher Scientific | Cat# K202020 |
| Software and algorithms | ||
| GraphPad Prism v9.3.1 | GraphPad Software | v9.3.1 |
| Image J 1.53s | National Institutes of Health | https://imagej.nih.gov/ij/ |
| Partek® Genomics Suite® 7.0 | Partek Inc. | https://www.partek.com/partek-genomics-suite/ |
| Metascape | Zhou et al. (2019) | https://metascape.org/gp/index.html#/main/step1 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Tomozumi Imamichi (timamichi@mail.nih.gov).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Ethics statement
Approval for these studies, including all sample materials, was granted by the National Institute of Allergy and Infectious Diseases Institutional Review Board, and participants gave written informed consent before blood being drawn. All experimental procedures in these studies were approved by the National Cancer Institute at Frederick (the protocol code number: 2016–19A6/11–30) and performed in accordance with the relevant guidelines and regulations.
Cell lines and viruses
Human embryonic kidney 293 (HEK), human leukemia monocytic cell line (THP-1), human rhabdomyosarcoma (RD), and SV40 T-antigen transformed HEK293 (293T) cells were obtained from American Type Culture Collection (ATCC, Manassas, VA) and maintained following the manufacturer’s instructions. To generate monocyte-derived macrophages (MDMs), isolated CD14+ monocytes were plated in 10 cm Petri dishes at 1.0 × 107 cells/dish. Monocytes were stimulated with 25 ng/mL M-CSF (R&D Systems, Minneapolis, MN) in macrophage serum-free medium (Thermo Fisher Scientific, Waltham, MA) for seven days. MDMs were then maintained in DMEM (Invitrogen, Thermo Fisher Scientific,) containing 10% FBS (HyClone Laboratories, Logan, UT), 25 mM HEPES (Quality Biology, Gaithersburg, MD), and 5 μg/mL Gentamicin (Thermo Fisher Scientific) before use in experiments. The HSV-1 McKrae strain was kindly provided by Dr. Jeffrey I. Cohen (National Institute of Allergy and Infectious Diseases/National Institutes of Health, Bethesda, MD). Virus stock was prepared using Vero cells (ATCC), and virus titer was determined by plaque-forming assay (Blaho et al., 2006). Sendai virus (SeV) was obtained from Advanced Biotechnologies, Inc. (Eldersburg, MD). HSV-1 was used at an MOI of 1, and SeV was used at a final concentration of 40 HA units/mL.
Method details
Plasmids
The pCR2.1 plasmid (Thermo Fisher Scientific) was digested with EcoRI, which was followed by purification using a PCR purification kit (QIAGEN, Germantown, MD). This digested plasmid was used as a noncoding DNA stimulant, as previously described (Sui et al., 2017; Zhang et al., 2011a).
DNA and siRNA transfections
DNA transfections were conducted with Lipofectamine 2000 (Thermo Fisher Scientific) for THP-1 cells, TransIT-293 (Mirus Bio, Madison, WI) for HEK cells, and Transit X2 (Mirus Bio) for human primary macrophages, all according to the manufacturers’ protocols. Lipofectamine RNAiMAX was used for siRNA transfection in human macrophages. The information about siRNA targeting ATM and corresponding negative control siRNA has been provided in key resources table.
RNA extraction and real-time RT-PCR
Total cellular RNA was isolated from cells using the RNeasy isolation kit (QIAGEN). The cDNA was synthesized from total RNA using TaqMan reverse transcription reagents (Thermo Fisher Scientific) with random hexamer primer according to the manufacturer’s instructions. IFN or inflammatory cytokine mRNA expression levels were measured using quantitative RT-PCR on a CFX96 real-time system (Bio-Rad, Hercules, CA); the two-temperature cycle of 95 °C for 15 s and 60 °C for one minute (repeated for 40 cycles) was used. Relative quantities of the transcript were calculated using the ΔΔCt method, with GAPDH as a reference. Normalized samples were expressed relative to the average ΔCt value for controls to obtain relative fold change in expression levels. The probes used for real time RT-PCR are listed in key resources table.
Western blot
Whole cell lysates and nuclear fraction were prepared using RIPA buffer (Boston BioProducts, Ashland, MA) and Nuclear Extract Kit (Active Motif, Carlsbad, CA), respectively in the presence of protease inhibitor cocktail (Millipore Sigma, St. Louis, MO) and Halt phosphatase inhibitor cocktail (Thermo Fisher Scientific). Protein concentrations of the cell lysates were quantified using a BCA protein assay (Thermo Fisher Scientific) to ensure equal amounts of total protein were loaded in each well of NuPAGE 4–12% Bis-Tris Gel (Thermo Fisher Scientific). Proteins were transferred onto a nitrocellulose membrane and blotted with the appropriate antibodies, followed by the HRP-conjugated secondary antibodies, and the detection was performed using ECL Plus Western blotting detection reagents (GE Healthcare, Chicago, IL). ECL signal was detected using the Azure 300 (Azure Biosystem, Dublin, CA). The antibodies used for WB are listed in key resources table.
Microarray
Total cellular RNA was isolated from cells using the RNeasy isolation kit (QIAGEN). RNA was quantitated and qualified with a Nanodrop 1000 (Thermo Fisher Scientific) and an Agilent Bioanalyzer RNA Nano 6000 chip (Agilent, Santa Clara, CA). cRNA synthesis, labeling, and hybridization to the Human GeneArray (Thermo Fisher Scientific) were performed according to the manufacturer’s protocol. Genechip data analysis was performed using Partek Genomics Suite (Partek, Inc., St Louis, MO). Briefly, raw expression values for each array were normalized using a quantile normalization procedure, and the significant differences in expression were determined by two-way ANOVA. Functional enrichment analysis was performed using Metascape (the database for annotation, visualization, and integrated discovery) (Zhou et al., 2019).
Quantification and statistical analysis
Results were representative of at least three independent experiments. The values were expressed as mean ± SD of individual samples. GraphPad Prism 9 was used to make statistical comparisons by Student’s t test or one-way analysis of variance (ANOVA) with multiple comparison. p-values less than 0.05 were considered statistically significance (∗p< 0.05, ∗∗p< 0.01, ∗∗∗p< 0.001, ∗∗∗∗p< 0.0001). p> 0.05 was considered not significant (ns).
Acknowledgments
We thank H. Clifford Lane for supporting this project and Jeffrey I. Cohen for providing the HSV-1 McKrae strain. We also thank Whitney Bruchey and Marjorie Bosche for running microarray assay. The graphical abstract is created by Biorender.com. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract number HHSN261200800001E. This research was supported, in part, by the National Institute of Allergy and Infectious Diseases, National Institutes of Health. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Author contributions
H.S. designed all experiments, performed assays, analyzed data and wrote the manuscript; Q.C. and S.S. performed assays; J.Y. analyzed data, and T.I. created the project, oversaw the data and wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: November 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105352.
Contributor Information
Hongyan Sui, Email: suih@mail.nih.gov.
Tomozumi Imamichi, Email: timamichi@mail.nih.gov.
Supplemental information
Data and code availability
-
•
Microscopy data reported in this article have been deposited at GEO and are publicly available as of the date of publication. Accession number is GSE214181 and has been listed in the key resources table.
-
•
This article does not report original code.
-
•
Any additional information required to reanalyze the data reported in this article is available from the lead contact on request.
References
- Aisen P., Enns C., Wessling-Resnick M. Chemistry and biology of eukaryotic iron metabolism. Int. J. Biochem. Cell Biol. 2001;33:940–959. doi: 10.1016/S1357-2725(01)00063-2. [DOI] [PubMed] [Google Scholar]
- Akira S., Takeda K., Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat. Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- Akira S., Uematsu S., Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Andreini C., Bertini I., Cavallaro G., Holliday G.L., Thornton J.M. Metal ions in biological catalysis: from enzyme databases to general principles. J. Biol. Inorg. Chem. 2008;13:1205–1218. doi: 10.1007/s00775-008-0404-5. [DOI] [PubMed] [Google Scholar]
- Andrews N.C. Iron homeostasis: insights from genetics and animal models. Nat. Rev. Genet. 2000;1:208–217. doi: 10.1038/35042073. [DOI] [PubMed] [Google Scholar]
- Ank N., West H., Bartholdy C., Eriksson K., Thomsen A.R., Paludan S.R. Lambda interferon (IFN-λ), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J. Virol. 2006;80:4501–4509. doi: 10.1128/jvi.80.9.4501-4509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakkenist C.J., Kastan M.B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Bartlett N.W., Buttigieg K., Kotenko S.V., Smith G.L. Murine interferon lambdas (type III interferons) exhibit potent antiviral activity in vivo in a poxvirus infection model. J. Gen. Virol. 2005;86:1589–1596. doi: 10.1099/vir.0.80904-0. [DOI] [PubMed] [Google Scholar]
- Bermejo-Jambrina M., Eder J., Helgers L.C., Hertoghs N., Nijmeijer B.M., Stunnenberg M., Geijtenbeek T.B.H. C-type lectin receptors in antiviral immunity and viral escape. Front. Immunol. 2018;9:590. doi: 10.3389/fimmu.2018.00590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddlestone-Thorpe L., Sajjad M., Rosenberg E., Beckta J.M., Valerie N.C.K., Tokarz M., Adams B.R., Wagner A.F., Khalil A., Gilfor D., et al. ATM kinase inhibition preferentially sensitizes p53-mutant glioma to ionizing radiation. Clin. Cancer Res. 2013;19:3189–3200. doi: 10.1158/1078-0432.Ccr-12-3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaho J.A., Morton E.R., Yedowitz J.C. Herpes simplex virus: propagation, quantification, and storage. Curr. Protoc. Microbiol. 2006 doi: 10.1002/9780471729259.mc14e01s00. [DOI] [PubMed] [Google Scholar]
- Bowie A.G., Haga I.R. The role of Toll-like receptors in the host response to viruses. Mol. Immunol. 2005;42:859–867. doi: 10.1016/j.molimm.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Bowman A.B., Aschner M. Considerations on manganese (Mn) treatments for in vitro studies. Neurotoxicology. 2014;41:141–142. doi: 10.1016/j.neuro.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill A., Fuchs T.A., Savchenko A.S., Thomas G.M., Martinod K., De Meyer S.F., Bhandari A.A., Wagner D.D. Neutrophil extracellular traps promote deep vein thrombosis in mice. J. Thromb. Haemost. 2012;10:136–144. doi: 10.1111/j.1538-7836.2011.04544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burleigh K., Maltbaek J.H., Cambier S., Green R., Gale M., James R.C., Stetson D.B. Human DNA-PK activates a STING-independent DNA sensing pathway. Science Immunology. 2020;5:eaba4219. doi: 10.1126/sciimmunol.aba4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civril F., Deimling T., de Oliveira Mann C.C., Ablasser A., Moldt M., Witte G., Hornung V., Hopfner K.P. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498:332–337. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey A., Bowie A.G. Innate immune recognition of DNA: a recent history. Virology. 2015;479–480:146–152. doi: 10.1016/j.virol.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond M.S., Kanneganti T.-D. Innate immunity: the first line of defense against SARS-CoV-2. Nat. Immunol. 2022;23:165–176. doi: 10.1038/s41590-021-01091-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Ochoa V.E., Jellbauer S., Klaus S., Raffatellu M. Transition metal ions at the crossroads of mucosal immunity and microbial pathogenesis. Front. Cell. Infect. Microbiol. 2014;4:2. doi: 10.3389/fcimb.2014.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly R.P., Kotenko S.V. Interferon-Lambda: a new addition to an old family. J. Interferon Cytokine Res. 2010;30:555–564. doi: 10.1089/jir.2010.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson B.J., Mansur D.S., Peters N.E., Ren H., Smith G.L. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. Elife. 2012;1:e00047. doi: 10.7554/eLife.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D., Wu J., Wu Y.-T., Du F., Aroh C., Yan N., Sun L., Chen Z.J. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science. 2013;341:903–906. doi: 10.1126/science.1240933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding S.E., Rosenberg E., Valerie N., Hussaini I., Frigerio M., Cockcroft X.F., Chong W.Y., Hummersone M., Rigoreau L., Menear K.A., et al. Improved ATM kinase inhibitor KU-60019 radiosensitizes glioma cells, compromises insulin, AKT and ERK prosurvival signaling, and inhibits migration and invasion. Mol. Cancer Ther. 2009;8:2894–2902. doi: 10.1158/1535-7163.Mct-09-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham M.E., Lavin M.F., Kozlov S.V. In: ATM Kinase: Methods and Protocols. Kozlov S.V., editor. Springer New York; 2017. Identification of ATM protein kinase phosphorylation sites by mass spectrometry; pp. 127–144. [DOI] [PubMed] [Google Scholar]
- Gürtler C., Bowie A.G. Innate immune detection of microbial nucleic acids. Trends Microbiol. 2013;21:413–420. doi: 10.1016/j.tim.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K., Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- Horner S.M., Liu H.M., Park H.S., Briley J., Gale M., Jr. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc. Natl. Acad. Sci. USA. 2011;108:14590–14595. doi: 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horning K.J., Caito S.W., Tipps K.G., Bowman A.B., Aschner M. Manganese is essential for neuronal health. Annu. Rev. Nutr. 2015;35:71–108. doi: 10.1146/annurev-nutr-071714-034419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L., Tian C., Yan Y., Zhang L., Zhang H., Zhang Z. Manganese-based nanoactivator optimizes cancer immunotherapy via enhancing innate immunity. ACS Nano. 2020;14:3927–3940. doi: 10.1021/acsnano.9b06111. [DOI] [PubMed] [Google Scholar]
- Ishikawa H., Barber G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. http://www.nature.com/nature/journal/v455/n7213/suppinfo/nature07317_S1.html [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwanaszko M., Kimmel M. NF-κB and IRF pathways: cross-regulation on target genes promoter level. BMC Genom. 2015;16:307. doi: 10.1186/s12864-015-1511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries A.M., Marriott I. Cytosolic DNA sensors and CNS responses to viral pathogens. Front. Cell. Infect. Microbiol. 2020;10:576263. doi: 10.3389/fcimb.2020.576263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X., Sui H., Lyons C., Tran B., Sherman B.T., Imamichi T. Complete genome sequence of herpes simplex virus 1 strain McKrae. Microbiology Resource Announcements. 2019;8:e00993. doi: 10.1128/MRA.00993-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen C.L., Ensunsa J.L., Clegg M.S. Manganese metabolism in animals and humans including the toxicity of manganese. Met. Ions Biol. Syst. 2000;23:137–170. [PubMed] [Google Scholar]
- Kelly C., Klenerman P., Barnes E. Interferon lambdas: the next cytokine storm. Gut. 2011;60:1284–1293. doi: 10.1136/gut.2010.222976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J.C. Zinc: an essential but elusive nutrient. Am. J. Clin. Nutr. 2011;94:679S–684S. doi: 10.3945/ajcn.110.005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov S., Gueven N., Keating K., Ramsay J., Lavin M.F. ATP activates ataxia-telangiectasia mutated (ATM) in vitro. J. Biol. Chem. 2003;278:9309–9317. doi: 10.1074/jbc.m300003200. [DOI] [PubMed] [Google Scholar]
- Lasfar A., Lewis-Antes A., Smirnov S.V., Anantha S., Abushahba W., Tian B., Reuhl K., Dickensheets H., Sheikh F., Donnelly R.P., et al. Characterization of the mouse IFN-λ ligand-receptor system: IFN-λs exhibit antitumor activity against B16 melanoma. Cancer Res. 2006;66:4468–4477. doi: 10.1158/0008-5472.can-05-3653. [DOI] [PubMed] [Google Scholar]
- Li M., Liu X., Zhou Y., Su S.B. Interferon-lambdas: the modulators of antivirus, antitumor, and immune responses. J. Leukoc. Biol. 2009;86:23–32. doi: 10.1189/jlb.1208761. [DOI] [PubMed] [Google Scholar]
- Li Y., Wu Y., Zheng X., Cong J., Liu Y., Li J., Sun R., Tian Z.G., Wei H.M. Cytoplasm-translocated Ku70/80 complex sensing of HBV DNA induces hepatitis-associated chemokine secretion. Front. Immunol. 2016;7:569. doi: 10.3389/fimmu.2016.00569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieu P.T., Heiskala M., Peterson P.A., Yang Y. The roles of iron in health and disease. Mol. Aspects Med. 2001;22:1–87. doi: 10.1016/S0098-2997(00)00006-6. [DOI] [PubMed] [Google Scholar]
- Loo Y.-M., Gale M., Jr. Immune signaling by RIG-I-like receptors. Immunity. 2011;34:680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv M., Chen M., Zhang R., Zhang W., Wang C., Zhang Y., Wei X., Guan Y., Liu J., Feng K., et al. Manganese is critical for antitumor immune responses via cGAS-STING and improves the efficacy of clinical immunotherapy. Cell Res. 2020;30:966–979. doi: 10.1038/s41422-020-00395-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason D.R., Beck P.L., Muruve D.A. Nucleotide-binding oligomerization domain-like receptors and inflammasomes in the pathogenesis of non-microbial inflammation and diseases. J. Innate Immun. 2012;4:16–30. doi: 10.1159/000334247. [DOI] [PubMed] [Google Scholar]
- Monroe K.M., Yang Z., Johnson J.R., Geng X., Doitsh G., Krogan N.J., Greene W.C. IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science. 2014;343:428–432. doi: 10.1126/science.1243640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muruve D.A., Pétrilli V., Zaiss A.K., White L.R., Clark S.A., Ross P.J., Parks R.J., Tschopp J. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- Nagane M., Yasui H., Sakai Y., Yamamori T., Niwa K., Hattori Y., Kondo T., Inanami O. Activation of eNOS in endothelial cells exposed to ionizing radiation involves components of the DNA damage response pathway. Biochem. Biophys. Res. Commun. 2015;456:541–546. doi: 10.1016/j.bbrc.2014.12.002. [DOI] [PubMed] [Google Scholar]
- Oka T., Hikoso S., Yamaguchi O., Taneike M., Takeda T., Tamai T., Oyabu J., Murakawa T., Nakayama H., Nishida K., et al. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012;485:251–255. doi: 10.1038/nature10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paludan S.R., Bowie A.G. Immune sensing of DNA. Immunity. 2013;38:870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehnberg G.L., Hein J.F., Carter S.D., Laskey J.W. Chronic manganese oxide administration to preweanling rats: manganese accumulation and distribution. J. Toxicol. Environ. Health. 1980;6:217–226. doi: 10.1080/15287398009529844. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J., Gack M.U. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020;20:537–551. doi: 10.1038/s41577-020-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J., Ponzoni S., Aschner M. Manganese homeostasis and transport. Met. Ions Life Sci. 2013;12:169–201. doi: 10.1007/978-94-007-5561-1_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Suemori H., Hata N., Asagiri M., Ogasawara K., Nakao K., Nakaya T., Katsuki M., Noguchi S., Tanaka N., Taniguchi T. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- Schoggins J.W. Interferon-stimulated genes: what do they all do? Annu. Rev. Virol. 2019;6:567–584. doi: 10.1146/annurev-virology-092818-015756. [DOI] [PubMed] [Google Scholar]
- Sharma S., Fitzgerald K.A. Innate immune sensing of DNA. PLoS Pathog. 2011;7:e1001310. doi: 10.1371/journal.ppat.1001310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Liu Y., Teo H.Y., Hanafi Z.B., Mei Y., Zhu Y., Chua Y.L., Lv M., Jiang Z., Liu H. Manganese enhances the antitumor function of CD8+ T cells by inducing type I interferon production. Cell. Mol. Immunol. 2021;18:1571–1574. doi: 10.1038/s41423-020-00524-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui H., Chen Q., Imamichi T. Cytoplasmic-translocated Ku70 senses intracellular DNA and mediates interferon-lambda1 induction. Immunology. 2021;163:323–337. doi: 10.1111/imm.13318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui H., Hao M., Chang W., Imamichi T. The role of Ku70 as a cytosolic DNA sensor in innate immunity and beyond. Front. Cell. Infect. Microbiol. 2021;11:761983. doi: 10.3389/fcimb.2021.761983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui H., Zhou M., Imamichi H., Jiao X., Sherman B.T., Lane H.C., Imamichi T. STING is an essential mediator of the Ku70-mediated production of IFN-λ1 in response to exogenous DNA. Sci. Signal. 2017;10:eaah5054. doi: 10.1126/scisignal.aah5054. [DOI] [PubMed] [Google Scholar]
- Sun W., Li Y., Chen L., Chen H., You F., Zhou X., Zhou Y., Zhai Z., Chen D., Jiang Z. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. USA. 2009;106:8653–8658. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Wu J., Du F., Chen X., Chen Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Zhang Y., Li J., Park K.S., Han K., Zhou X., Xu Y., Nam J., Xu J., Shi X., et al. Amplifying STING activation by cyclic dinucleotide-manganese particles for local and systemic cancer metalloimmunotherapy. Nat. Nanotechnol. 2021;16:1260–1270. doi: 10.1038/s41565-021-00962-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syedbasha M., Egli A. Interferon lambda: modulating immunity in infectious diseases. Front. Immunol. 2017;8:119. doi: 10.3389/fimmu.2017.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka A., Wang Z., Choi M.K., Yanai H., Negishi H., Ban T., Lu Y., Miyagishi M., Kodama T., Honda K., et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Takeuchi O., Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Tang J., Yu J.X., Hubbard-Lucey V.M., Neftelinov S.T., Hodge J.P., Lin Y. The clinical trial landscape for PD1/PDL1 immune checkpoint inhibitors. Nat. Rev. Drug Discov. 2018;17:854–855. doi: 10.1038/nrd.2018.210. [DOI] [PubMed] [Google Scholar]
- Tapiero H., Tew K.D. Trace elements in human physiology and pathology: zinc and metallothioneins. Biomed. Pharmacother. 2003;57:399–411. doi: 10.1016/S0753-3322(03)00081-7. [DOI] [PubMed] [Google Scholar]
- Taylor M.D., Erikson K.M., Dobson A.W., Fitsanakis V.A., Dorman D.C., Aschner M. Effects of inhaled manganese on biomarkers of oxidative stress in the rat brain. Neurotoxicology. 2006;27:788–797. doi: 10.1016/j.neuro.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Thaiss C.A., Zmora N., Levy M., Elinav E. The microbiome and innate immunity. Nature. 2016;535:65–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- Thompson M.R., Sharma S., Atianand M., Jensen S.B., Carpenter S., Knipe D.M., Fitzgerald K.A., Kurt-Jones E.A. Interferon γ-inducible protein (IFI) 16 transcriptionally regulates type I interferons and other interferon-stimulated genes and controls the interferon response to both DNA and RNA viruses. J. Biol. Chem. 2014;289:23568–23581. doi: 10.1074/jbc.M114.554147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidball A.M., Bryan M.R., Uhouse M.A., Kumar K.K., Aboud A.A., Feist J.E., Ess K.C., Neely M.D., Aschner M., Bowman A.B. A novel manganese-dependent ATM-p53 signaling pathway is selectively impaired in patient-based neuroprogenitor and murine striatal models of Huntington's disease. Hum. Mol. Genet. 2015;24:1929–1944. doi: 10.1093/hmg/ddu609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonks N.K. Protein tyrosine phosphatases--from housekeeping enzymes to master regulators of signal transduction. FEBS J. 2013;280:346–378. doi: 10.1111/febs.12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L., Keating S.E., Baran M., Horan K.A., Jensen S.B., Sharma S., Sirois C.M., Jin T., Latz E., Xiao T.S., et al. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. http://www.nature.com/ni/journal/v11/n11/abs/ni.1932.html#supplementary-information [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron K.J., Rutherford J.C., Ford D., Robinson N.J. Metalloproteins and metal sensing. Nature. 2009;460:823–830. doi: 10.1038/nature08300. [DOI] [PubMed] [Google Scholar]
- Wang C., Guan Y., Lv M., Zhang R., Guo Z., Wei X., Du X., Yang J., Li T., Wan Y., et al. Manganese increases the sensitivity of the cGAS-STING pathway for double-stranded DNA and is required for the host defense against DNA viruses. Immunity. 2018;48:675–687.e7. doi: 10.1016/j.immuni.2018.03.017. [DOI] [PubMed] [Google Scholar]
- Wang C., Zhang R., Wei X., Lv M., Jiang Z. Metalloimmunology: the metal ion-controlled immunity. Adv. Immunol. 2020;145:187–241. doi: 10.1016/bs.ai.2019.11.007. [DOI] [PubMed] [Google Scholar]
- Wang J., Kang L., Song D., Liu L., Yang S., Ma L., Guo Z., Ding H., Wang H., Yang B. Ku70 senses HTLV-1 DNA and modulates HTLV-1 replication. J. Immunol. 2017;199:2475–2482. doi: 10.4049/jimmunol.1700111. [DOI] [PubMed] [Google Scholar]
- Watson G., Xu W., Reed A., Babra B., Putman T., Wick E., Wechsler S.L., Rohrmann G.F., Jin L. Sequence and comparative analysis of the genome of HSV-1 strain McKrae. Virology. 2012;433:528–537. doi: 10.1016/j.virol.2012.08.043. [DOI] [PubMed] [Google Scholar]
- Wedler F.C. In: Prog. Med. Chem. Ellis G.P., Luscombe D.K., editors. Elsevier; 1993. 3 biological significance of manganese in mammalian systems; pp. 89–133. [DOI] [PubMed] [Google Scholar]
- Wente S.R., Villalba M., Schramm V.L., Rosen O.M. Mn2(+)-binding properties of a recombinant protein-tyrosine kinase derived from the human insulin receptor. Proc. Natl. Acad. Sci. USA. 1990;87:2805–2809. doi: 10.1073/pnas.87.7.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.-j., Li W., Shao Y., Avey D., Fu B., Gillen J., Hand T., Ma S., Liu X., Miley W., et al. Inhibition of cGAS DNA sensing by a herpesvirus virion protein. Cell Host Microbe. 2015;18:333–344. doi: 10.1016/j.chom.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P., Wang S., Gao P., Gao G., Fan Z. DNA sensor cGAS-mediated immune recognition. Protein Cell. 2016;7:777–791. doi: 10.1007/s13238-016-0320-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Luo D.-J., Wang X.-L., Qiu M., Yang Y., Yan X., Wang J.-Z., Ye Q.-F., Liu R. Zinc binds to and directly inhibits protein phosphatase 2A in vitro. Neurosci. Bull. 2015;31:331–337. doi: 10.1007/s12264-014-1519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M., Fujita T. Structural mechanism of RNA recognition by the RIG-I-like receptors. Immunity. 2008;29:178–181. doi: 10.1016/j.immuni.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Yoneyama M., Fujita T. RNA recognition and signal transduction by RIG-I-like receptors. Immunol. Rev. 2009;227:54–65. doi: 10.1111/j.1600-065X.2008.00727.x. [DOI] [PubMed] [Google Scholar]
- Zahid A., Ismail H., Li B., Jin T. Molecular and structural basis of DNA sensors in antiviral innate immunity. Front. Immunol. 2020;11:613039. doi: 10.3389/fimmu.2020.613039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Z., Cao H., Xie X., Yang L., Zhang P., Chen Y., Fan H., Liu Z., Liu X. Phosphatase PP4 negatively regulates type I IFN production and antiviral innate immunity by dephosphorylating and deactivating TBK1. J. Immunol. 2015;195:3849–3857. doi: 10.4049/jimmunol.1403083. [DOI] [PubMed] [Google Scholar]
- Zhang X., Brann T.W., Zhou M., Yang J., Oguariri R.M., Lidie K.B., Imamichi H., Huang D.W., Lempicki R.A., Baseler M.W., et al. Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J. Immunol. 2011;186:4541–4545. doi: 10.4049/jimmunol.1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Yuan B., Bao M., Lu N., Kim T., Liu Y.J. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 2011;12:959–965. doi: 10.1038/ni.2091. http://www.nature.com/ni/journal/v12/n10/abs/ni.2091.html#supplementary-information [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Ma Z., Wang B., Guan Y., Su X.-D., Jiang Z. Mn2+ directly activates cGAS and structural analysis suggests Mn2+ induces a noncanonical catalytic synthesis of 2′3′-cGAMP. Cell Rep. 2020;32:108053. doi: 10.1016/j.celrep.2020.108053. [DOI] [PubMed] [Google Scholar]
- Zhong B. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Zhong Y., Kinio A., Saleh M. Functions of NOD-like receptors in human diseases. Front. Immunol. 2013;4:333. doi: 10.3389/fimmu.2013.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J.H., Wang Y.N., Chang Q.Y., Ma P., Hu Y., Cao X. Type III interferons in viral infection and antiviral immunity. Cell. Physiol. Biochem. 2018;51:173–185. doi: 10.1159/000495172. [DOI] [PubMed] [Google Scholar]
- Zhou R., Zhang Q., Xu P. TBK1, a central kinase in innate immune sensing of nucleic acids and beyond. Acta Biochim. Biophys. Sin. 2020;52:757–767. doi: 10.1093/abbs/gmaa051. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this article.
-
•
Microscopy data reported in this article have been deposited at GEO and are publicly available as of the date of publication. Accession number is GSE214181 and has been listed in the key resources table.
-
•
This article does not report original code.
-
•
Any additional information required to reanalyze the data reported in this article is available from the lead contact on request.