Dear Editor, The novel coronavirus disease (COVID‐19) pandemic, caused by severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2), is a challenging situation globally due to its contagious nature. SARS‐CoV‐2 enters the host cell by the receptor‐binding domain of its spike protein interacting with the angiotensin‐converting enzyme 2 (ACE2) receptor present on the host cell surface.1 Host proteases, mainly transmembrane protease, serine 2 (TMPRSS2), play a vital role in cleaving the SARS‐CoV‐2 spike protein, thereby enabling the virus to enter the host cell by endocytosis. SARS‐CoV‐2 mainly affects the respiratory system of the infected host; however, its manifestation in other organs has also been reported.2 A functional ACE2 receptor and TMPRSS2 protease in a particular cell type in a tissue microenvironment are the major determinants in virus tissue tropism. No concrete evidence is available about skin tropism of SARS‐CoV‐2 and its implications in inflammatory dermatological conditions, but skin‐associated changes have been reported in patients with COVID‐19.3 However, it is inconclusive whether these skin‐specific changes are primarily due to SARS‐CoV‐2 infection or develop as a result of adverse reactions to drugs used in COVID‐19 management.
The status of SARS‐CoV‐2 determinants in inflammatory skin diseases like psoriasis is not known. Furthermore, interferons are considered the major antiviral host response, and a recent study has shown that ACE2 is the interferon‐stimulated gene.4 Interferons are the prominent proinflammatory cytokines and play a major role in psoriasis pathogenesis. Increased expression of interferon‐γ (IFN‐γ) is reported in psoriatic lesions.5 It may be possible that enhanced expression of interferons such as IFN‐γ in psoriatic lesions increases the ACE2 expression that may be exploited by SARS‐CoV‐2 towards skin manifestation. Therefore, to address this hypothesis, we determined the status of major determinants of SARS‐CoV‐2 infection (i.e. ACE2 and TMPRSS2) in the peripheral blood and skin of people with psoriasis.
We recruited 40 patients with psoriasis (30 male and 10 female) and 40 controls (30 male and 10 female), and blood samples were collected from both groups. Skin biopsy samples were collected from lesional skin of patients with psoriasis (n = 40) and the control group (n = 30; all male). Informed consent was obtained and the study was approved by the institutional ethics committee. Total RNA was isolated from peripheral blood mononuclear cells and skin homogenates using Trizol reagent. Transcripts levels of ACE2 and TMPRSS2 were determined by quantitative polymerase chain reaction using β‐actin as the endogenous control, as described previously,6 and data are represented as 2−ΔCt. Transcript levels of ACE2 were significantly increased in peripheral blood (P = 0.023) and lesional skin (P = 0.013) of patients with psoriasis compared with controls, but no significant difference was observed for TMPRSS2 (P > 0.05) (Figure 1a, b).
Figure 1.
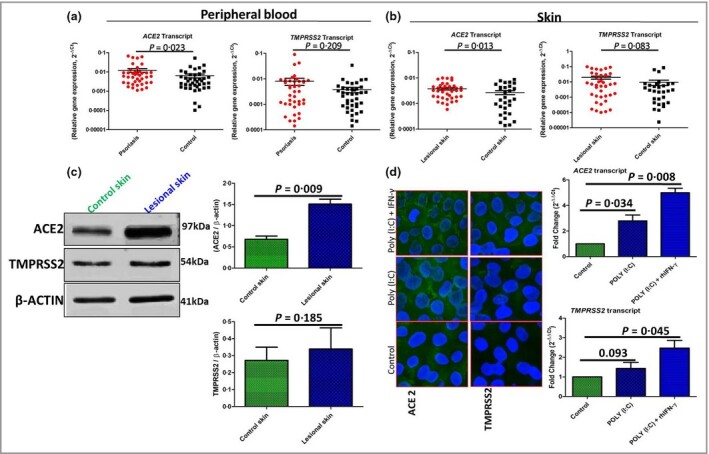
(a, b) ACE2 and TMPRSS2 transcript expression profiles in peripheral blood (a) and skin (b) of patients with psoriasis (n = 40) and healthy controls (n = 40 in blood, n = 30 in skin) (Mann–Whitney U‐test; mean and SEM). (c) Protein expression of angiotensin‐converting enzyme 2 (ACE2) and transmembrane protease, serine 2 (TMPRSS2) was determined in skin homogenates of patients with psoriasis (n = 40) and controls (n = 30) in triplicate (Student’s t‐test). (d) The effect of polyinosinic–polycytidylic acid [poly(I:C)] – alone or in combination with recombinant human interferon‐γ (rhIFN‐γ) – on ACE2 and TMPRSS2 expression. Left: immunofluorescence staining (original magnification × 40); right: transcript quantification (in triplicate) in adult human epidermal keratinocytes for 24 h (one‐way anova test). P‐values < 0.05 were deemed significant; bars and error bars indicated the mean and SEM.
Expression of ACE2 and TMPRSS2 proteins (Abcam, Cambridge, UK) was determined in tissue lysates of psoriatic and control skin by Western blotting using β‐actin as loading control, as described previously.7 Blot intensities quantified by densitometry analysis revealed significantly increased expression of ACE2 (P = 0.009) in lesional skin compared with control skin, but no significant difference was observed for TMPRSS2 (P = 0.19) (Figure 1c).
Next, we performed in vitro studies using primary adult human epidermal keratinocytes (HEKa cells) maintained in keratinocyte growth medium‐2 (PromoCell, Heidelberg, Germany) supplemented with optimized growth factors (HiMedia Laboratories, Mumbai, India), antibiotics and antimycotics (Sigma‐Aldrich, St Louis, MO, USA) in 5% humidified CO2 at 37 °C. HEKa cells were treated with 0.1 μg mL−1 polyinosinic–polycytidylic acid [poly(I:C)] (Sigma‐Aldrich) alone, or a combination of poly(I:C) and recombinant human IFN‐γ (rhIFN‐γ, 1 ng mL−1; R&D Systems Inc., Minneapolis, MN, USA) for 24 h. Poly(I:C) mimics viral dsRNA and acts as a Toll‐like receptor 3 agonist. Significantly increased ACE2 expression (~2.5 fold) was observed in HEKa cells after poly(I:C) treatment compared with controls (P = 0.034) (Figure 1d). However, the combination poly(I:C) + rhIFN‐γ potentiated the ACE2 expression (~5 fold) (P = 0.008). No significant difference was observed in the expression of TMPRSS2 (P = 0.093) after treatment of HEKa cells with poly(I:C) alone, but the combination of poly(I:C) and rhIFN‐γ significantly enhanced the expression of TMPRSS2 (P = 0.045) (Figure 1d). Additional information will be made available on direct request.
Significantly increased expression of ACE2 in the peripheral blood and lesional skin of patients with psoriasis rendered them a risk group for SARS‐CoV‐2. Plaserico et al. reported no significant association between psoriasis and COVID‐19 risk, but the study was limited by the low number of patients with COVID‐19 (six out of 1830 people in the psoriasis cohort).8 Mediators of inflammation such as cytokines may mediate the upregulation of ACE2 receptor expression in skin keratinocytes, favouring the binding of SARS‐CoV‐2, and host cell proteases such as TMPRSS2 may facilitate entry of the virus, as proposed by Hoffmann et al.1 SARS‐CoV‐2 creates favourable conditions for skin tropism, particularly in patients with inflammatory skin diseases like psoriasis and may exacerbate the disease. However, further investigation is highly warranted in this direction.
Acknowledgments
This work was supported by intramural funding from AIIMS, New Delhi, and we extend our thanks to all the supporting medical healthcare workers and laboratory staff.
Author Contribution
MANOJ KUMAR TEMBHRE: Conceptualization (lead); Data curation (equal); Formal analysis (lead); Funding acquisition (lead); Investigation (lead); Methodology (lead); Project administration (lead); Resources (lead); Software (lead); Supervision (lead); Validation (lead); Visualization (lead); Writing‐original draft (lead); Writing‐review & editing (lead). ANITA SINGH PARIHAR: Data curation (equal); Formal analysis (equal); Methodology (equal); Project administration (equal); Resources (equal); Visualization (equal); Writing‐review & editing (equal). VINOD KUMAR SHARMA: Data curation (equal); Investigation (equal); Methodology (equal); Project administration (equal); Resources (equal); Writing‐review & editing (equal). Neetu Bhari: Funding acquisition (equal); Investigation (equal); Project administration (equal); Resources (equal). Shafaque Imran: Data curation (equal); Methodology (equal); Resources (equal). Lakshmy Ramakrishnan: Funding acquisition (equal); Investigation (equal); Project administration (equal); Resources (equal). Atul Bhalla: Methodology (equal); Resources (equal).
References
- Hoffmann M, Kleine‐Weber H, Schroeder S. et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020; 181:271–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puelles VG, Lütgehetmann M, Lindenmeyer MT. et al. Multiorgan and renal tropism of SARS‐CoV‐2. N Engl J Med 2020; 383:590–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisondi P, PIaserico S, Bordin C. et al. Cutaneous manifestations of SARS‐CoV‐2 infection: a clinical update. J Eur Acad Dermatol Venereol 2020; 10.1111/jdv.16774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler CGK, Allon SJ, Nyquist SK. et al. SARS‐CoV‐2 receptor ACE2 is an interferon‐stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell 2020; 181:1016–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K, Han L, Deng H. et al. The distinct role and regulatory mechanism of IL‐17 and IFN‐γ in the initiation and development of plaque vs guttate psoriasis. J Dermatol Sci 2018; 92:106–13. [DOI] [PubMed] [Google Scholar]
- Tembhre MK, Parihar AS, Sharma VK. et al. Alteration in regulatory T cells and programmed cell death 1‐expressing regulatory T cells in active generalized vitiligo and their clinical correlation. Br J Dermatol 2015; 172:940–50. [DOI] [PubMed] [Google Scholar]
- Tembhre MK, Parihar AS, Sharma A. et al. Participation of T cell immunoglobulin and mucin domain‐3 (TIM‐3) and its ligand (galectin‐9) in the pathogenesis of active generalized vitiligo. Immunol Res 2015; 62:23–34. [DOI] [PubMed] [Google Scholar]
- Piaserico S, Gisondi P, Cazzaniga S, Naldi L. Lack of evidence for an increased risk of severe COVID‐19 in psoriasis patients on biologics: a cohort study from northeast Italy. Am J Clin Dermatol 2020; 21:749–51. [DOI] [PMC free article] [PubMed] [Google Scholar]


