Abstract
Estimating the effective reproduction number of Omicron subvariants is crucial for evaluating the effectiveness of control measures, and adjusting control measures promptly. We conducted a systematic review to synthesize the evidence from estimates of the reproduction numbers for Omicron subvariants, and estimated their effective reproduction number.
Omicron is circulating globally as the dominant variant of SARS-CoV-2 and posing a substantial threat in 2022.1 It is rapidly mutating into several descendent lineages and circulating recombinant forms.2 The effective reproduction number (Re) is a central concept in infectious disease epidemiology that refers to the average number of secondary infections generated by an infected case after considering the effects of population immunity and control measures. The relative reproduction number (relative Re) describes the ratio between the reproduction numbers for two different variants, such as the relative Re between Omicron subvariants and the Delta variant in this study. Accurate and timely estimation of the reproduction numbers is crucial for predicting disease development trends, evaluating the effectiveness of control measures and adjusting control measures promptly.
We identified 224 studies by searching PubMed, medRxiv and bioRxiv for articles that were published or pre-print from 1 January 2020 to 23 July 2022 (Supplementary Figure S1, Supplementary Methods). There were 153 studies removed during the process of title and abstract screening, leaving 71 studies for full-text assessment. Thirteen studies were eventually included in this review, providing 34 estimates of Re or relative Re. The reported Omicron substrains included BA.1, BA.2, BA.4, BA.5 and XG, where BA.1 and BA.2 were most studied (Supplementary Figures S2 and S3). These data involved 16 countries and mostly focused on Denmark, the United Kingdom, South Africa, the United States, South Korea and India (Supplementary Table S1).
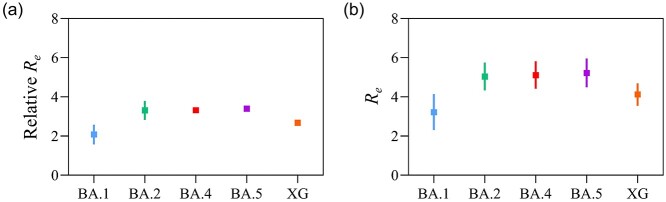
Transmissibility comparisons were made between Omicron subvariants and the Delta variant, and the relative Re estimates were summarized. Specifically, the pooled estimates of relative Re for BA.1 and BA.2 were 2.08 (95% confidence interval, 95% CI: 1.57–2.58) and 3.31 (95% CI: 2.83–3.79), respectively (Figure 1, Supplementary Figures S4 and S5). For studies where BA.1 and BA.2 were collectively reported, the median estimates of the relative Re ranged from 1.82 to 4.15. The other Omicron subvariants BA.4, BA.5, and XG were found to elicit 3.32 (95% credible interval, 95% CrI: 3.27–3.38), 3.39 (95% CrI: 3.30–3.49) and 2.67 (95% CrI: 2.62–2.73) times higher transmissibility than the Delta variant, respectively (Figure 1).3
Figure 1.
Reproduction number estimates for the Omicron subvariants. (a) Estimates of effective reproduction numbers for each Omicron subvariant (e.g. BA.1, BA.2, BA.4, BA.5 and XG). (b) Estimates of relative effective reproduction numbers to the Delta variant for each Omicron subvariant. The estimates of BA.1 and BA.2 were pooled values, whereas those of BA.4, BA.5 and XG were directly retrieved from Kimura et al.3. The dots and error bars show the estimated mean and 95% confidence interval, respectively
When not compared with the Delta variant, the pooled Re estimates of BA.1 and BA.2 were 3.22 (95% CI: 2.31–4.14) and 5.04 (95% CI: 4.33–5.75), respectively (Figure 1, Supplementary Figures S6 and S7). The median Re estimates ranged from 2.79 to 6.32 when BA.1 and BA.2 were collectively reported. BA.4, BA.5 and XG had higher Re estimates than the ancestral substrains, with median values of 5.11 (95% CrI: 4.56–5.66), 5.22 (95% CrI: 4.65–5.79) and 4.12 (95% CrI: 3.67–4.56), respectively (Figure 1).3
Meta-regression analyses were also performed for BA.1 and BA.2 to identify potential associations between the study regions and the estimated relative reproduction numbers. We found that the study region was associated with the reported relative ratio (Supplementary Figures S8 and S9) and the reproduction number (Supplementary Figures S10 and S11) of BA.1 and BA.2, by including the study region as a categorical variable.
The high transmission rate of Omicron has led to an epidemiological rebound in many countries, with the number of global BA.4 and BA.5 cases also rising and causing highly adverse impacts.4 Liu and Rocklöv found that the mean estimate of basic reproductive number of the Delta variant was 5.08, ranging from 3.2 to 8,5 and that of the Omicron variants, including BA.1 and BA.2, was 9.5 with a range from 5.5 to 24 (median: 10 and interquartile range, IQR: 7.25–11.88).6 We identified that the Omicron subvariants had higher transmission potential than that of the Delta variant, in which BA.5 has the highest reproduction number, followed by BA.4, BA.2, XG and BA.1. Liu and Rocklöv also stated an average Re value of 3.4 with a range from 0.88 to 9.4 (median: 2.8 and IQR: 2.03–3.85) for Omicron during outbreaks of BA.1 and BA.2,6 than which the Re estimates of Omicron subvariants are generally higher in our analyses.
BA.4 and BA.5 lineages are replacing other Omicron subvariants and have been the dominant lineages.4,7 The government needs to take stricter public health measures and vaccinations before the outbreak is past the point of being contained. BA.4 and BA.5 have greatly increased neutralization resistance,8 and substantially escape neutralizing antibodies induced by infection and vaccination,9 thereby further compromising the efficacy of vaccines. There is a clear need for governments to accelerate the development of more efficient COVID-19 vaccines (e.g. universal coronavirus vaccines10).
However, estimates of reproduction numbers may vary widely due to various factors that were not included in this review, such as time and locations of studies, exposure patterns, vaccine promotion and distribution, and travel restrictions. In addition, we only studied the relative reproduction numbers to assess the transmission ability of Omicron and its subvariants and did not include studies of transmission advantage using other metrics. Besides, this review could be biased, given that our study contains some preprints that have not been certified by peer review.
In conclusion, multiple estimates of the effective reproduction numbers have been reported for the Omicron subvariant, and the study regions are considered to be associated with the relative reproduction numbers. COVID-19 is still posing serious threats, and it is necessary to strengthen public health measures while expanding vaccine coverage.
Supplementary Material
Contributor Information
Shuqi Wang, School of Cybersecurity, Northwestern Polytechnical University, Xian, China.
Fengdi Zhang, Mailman School of Public Health, Columbia University, New York, NY, USA.
Zhen Wang, School of Cybersecurity, Northwestern Polytechnical University, Xian, China; School of Artificial Intelligence, Optics, and Electronics (iOPEN), Northwestern Polytechnical University, Xian, China.
Zhanwei Du, WHO Collaborating Center for Infectious Disease Epidemiology and Control, School of Public Health, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong, SAR, China.
Chao Gao, School of Artificial Intelligence, Optics, and Electronics (iOPEN), Northwestern Polytechnical University, Xian, China.
Data availability
All data are collected from open source with detailed descriptions in the Methods section.
Author Contributions
S.W., F.Z. and Z.W. conceived the study, designed statistical and modelling methods, conducted analyses, interpreted results, and wrote and revised the manuscript; C.G., Z.D. and Z.W. interpreted results and revised the manuscript.
Code availability
Code used for data analysis is freely available upon request.
Funding
This work was supported by the Key Program for International Science and Technology Cooperation Projects of China (No. 2022YFE0112300), National Natural Science Foundation for Distinguished Young Scholars (No. 62025602), National Natural Science Foundation of China (No. 61976181), the Fundamental Research Funds for the Central Universities (Nos. D5000210738, D5000211001), Technological Innovation Team of Shaanxi Province (No. 2020TD-013), Natural Science Basic Research Plan in Shaanxi Province of China (No. 2022JM-325), the Tencent Foundation and XPLORER PRIZE and Tianjin Key Medical Discipline (Specialty) Construction Project (No. TJYXZDXK059B). The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. All code to perform the analyses and generate the figures in this study are available from the corresponding author upon reasonable request.
Competing interests
Not applicable.
References
- 1. Mohapatra RK, Sarangi AK, Kandi Vet al. Omicron (B.1.1.529 variant of SARS-CoV-2); an emerging threat: current global scenario. J Med Virol 2022; 94:1780–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. World Health Organization . Tracking SARS-CoV-2 variants. 11 Aug 2022. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/ (Accessed 15 Aug 2022, date last accessed).
- 3. Kimura I, Yamasoba D, Tamura Tet al. Virological characteristics of the novel SARS-CoV-2 Omicron variants including BA.2.12.1, BA.4 and BA.5 bioRxiv. 2022.
- 4. Mohapatra RK, Kandi V, Sarangi AKet al. The recently emerged BA.4 and BA.5 lineages of Omicron and their global health concerns amid the ongoing wave of COVID-19 pandemic - Correspondence. Int J Surg 2022; 103:106698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu Y, Rocklöv J. The reproductive number of the Delta variant of SARS-CoV-2 is far higher compared to the ancestral SARS-CoV-2 virus. J Travel Med 2021; 28:taab124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu Y, Rocklöv J. The effective reproductive number of the Omicron variant of SARS-CoV-2 is several times relative to Delta. J Travel Med 2022; 29:taac037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Desingu PA, Nagarajan K. The emergence of Omicron lineages BA.4 and BA.5, and the global spreading trend. J Med Virol 2022; 94:5077–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang Q, Guo Y, Iketani Set al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature 2022; 608:603–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shrestha LB, Foster C, Rawlinson Wet al. Evolution of the SARS-CoV-2 omicron variants BA.1 to BA.5: implications for immune escape and transmission. Rev Med Virol 2022; 32:e2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dolgin E. Pan-coronavirus vaccine pipeline takes form. Nat Rev Drug Discov 2022; 21:324–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are collected from open source with detailed descriptions in the Methods section.