Abstract
Background
Monoclonal antibodies (mAbs) that target severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are predominantly less effective against Omicron variants. Immunocompromised patients often experience prolonged viral shedding, resulting in an increased risk of viral escape.
Methods
In an observational, prospective cohort, 57 patients infected with Omicron variants who received sotrovimab alone or in combination with remdesivir were followed. The study end points were a decrease in SARS-CoV-2 RNA <106 copies/mL in nasopharyngeal swabs at day 21 and the emergence of escape mutations at days 7, 14, and 21 after sotrovimab administration. All SARS-CoV-2 samples were analyzed using whole-genome sequencing. Individual variants within the quasispecies were subsequently quantified and further characterized using a pseudovirus neutralization assay.
Results
The majority of patients (43 of 57, 75.4%) were immunodeficient, predominantly due to immunosuppression after organ transplantation or hematologic malignancies. Infections by Omicron/BA.1 comprised 82.5%, while 17.5% were infected by Omicron/BA.2. Twenty-one days after sotrovimab administration, 12 of 43 (27.9%) immunodeficient patients had prolonged viral shedding compared with 1 of 14 (7.1%) immunocompetent patients (P = .011). Viral spike protein mutations, some specific for Omicron (e.g., P337S and/or E340D/V), emerged in 14 of 43 (32.6%) immunodeficient patients, substantially reducing sensitivity to sotrovimab in a pseudovirus neutralization assay. Combination therapy with remdesivir significantly reduced emergence of escape variants.
Conclusions
Immunocompromised patients face a considerable risk of prolonged viral shedding and emergence of escape mutations after early therapy with sotrovimab. These findings underscore the importance of careful monitoring and the need for dedicated clinical trials in this patient population.
Keywords: immunodeficiency, sotrovimab, SARS-CoV-2, Omicron, escape
Immunodeficient patients had prolonged viral shedding of severe acute respiratory syndrome coronavirus 2 Omicron variants after treatment with sotrovimab. This was associated with rapid selection of escape mutations, which was confirmed in a pseudovirus neutralization assay but was significantly reduced by combination therapy with remdesivir.
During the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic, numerous studies showed that treatment options that directly target SARS-CoV-2 are most successful in the early phase of coronavirus disease 2019 (COVID-19), whereas in the late phases of COVID-19 with pneumonia and hyperinflammation, immunomodulation is the main therapeutic principle. Several monoclonal antibodies (mAbs) that target SARS-COV-2, such as bamlanivimab/etesevimab or casirivimab/imdevimab, became available starting in late 2020 and were successfully used in the early phase of COVID-19 to prevent disease progression in high-risk patients [1]. With the emergence of the currently dominating variant of concern Omicron in November 2021, a significant rise in infection rates was observed. This went along with a loss of in vitro activity of the mAb combination casirivimab/imdevimab, commonly used until then, because the target regions in the spike protein were altered through several mutations [2]. In January 2022, sotrovimab became available in Germany. It was one of the few mAbs found to be effective against the Omicron variant in vitro and thus represented a promising treatment option for early SARS-CoV-2 infection [3–5].
Sotrovimab was approved for use in children aged >12 years and in adults at high to moderate risk for developing severe infection [6]. To date, only 2 randomized, controlled trials have evaluated the efficacy of sotrovimab in preventing hospitalization and disease progression, but only the COVID-19 Monoclonal Antibody Efficacy Trial–Intent to Care Early (COMET-ICE) trial showed a benefit [3, 4, 7]. However, these trials did not include severely immunodeficient patients such as solid organ transplant (SOT) recipients. Case series as well as 2 cohort studies evaluated the efficacy and safety of sotrovimab in SOT patients in the context of Omicron and reported a reduction in disease severity [8, 9]. However, it was suggested that therapy of SARS-CoV-2 infections with a single mAb might promote the emergence of escape mutations in the spike protein, especially in immunocompromised patients [10]. Recently, mutations have been reported after sotrovimab therapy in patients infected with the Omicron variant, but the risk factors for the occurrence and the longitudinal development of resistance are still largely unclear [11, 12]. Therefore, we analyzed the outcome and risk factors for viral persistence after treatment with sotrovimab in our cohort of patients treated since January 2022, focusing specifically on the emergence of escape mutations.
METHODS
Study Design
We performed a prospective, observational cohort study in patients diagnosed with SARS-CoV-2 infection who received sotrovimab therapy between 20 January 2022 and 25 February 2022. Patients were either hospitalized or presented to the outpatient clinic at the University Hospital Düsseldorf. Inclusion criteria were polymerase chain reaction (PCR)–confirmed SARS-CoV-2 infection, age >12 years, weight >40 kg, and risk factors for developing a severe course of COVID-19. All patients provided informed consent. Patients were pseudonymized with an ID number. A single dose of 500 mg of sotrovimab was administered intravenously over a 1-hour period as part of routine clinical practice.
Baseline was defined as the day of sotrovimab administration. Nasopharyngeal swabs and clinical parameters were collected at baseline and during the follow-up period: every 7 days (±2 days) until viral clearance was achieved. The main study end points were percentage of patients with a decrease in SARS-CoV-2 RNA <106 copies/mL in nasopharyngeal swabs 21 days after sotrovimab administration and characterization of the viral variants including screening for escape mutations during the observation period of 28 days. Patients who did not attend their follow-up appointments and patients for whom viral genome sequencing was unsuccessful at any time during the study were excluded from the statistical analyses.
Definition of Prolonged Viral Shedding
Prolonged viral shedding was defined as a persistent SARS-CoV-2 RNA concentration above 106 copies/mL 21 days after sotrovimab administration. The threshold of 106 SARS-CoV-2 RNA copies/mL or a cycle threshold value >25 is considered a measure of infectivity based on in vitro cell culture data that show a correlation between viral load and viral cultivability and the associated probability of transmission [13]. This cutoff value as a correlate of contagiousness was also chosen following the German recommendations of the Robert Koch Institute for the isolation of SARS-CoV-2–infected hospitalized patients.
Laboratory SARS-CoV-2 Analyses
All detailed information on SARS-CoV-2 detection and quantification, SARS-CoV-2 whole-genome sequencing and resistance analysis, pseudovirus cloning, production, and neutralization assays is provided in the Supplementary Appendix.
Statistical Analyses
Detailed information on the statistical programs used and the statistical tests performed is provided in the Supplementary Appendix.
All investigations were performed in accordance with the Declaration of Helsinki. The study was approved via ethics vote of the local ethics committee of the medical faculty of Heinrich-Heine-University. All patients gave written informed consent.
RESULTS
Patients' Characteristics
A total of 57 patients (21 females; 36 males) were enrolled in the study, 47 (82.5%) of whom were infected with Omicron variant BA.1 and 10 (17.5%) with Omicron variant BA.2 (Table 1). No symptoms were present in 21 of 57 (36.8%) patients, while the rest had symptoms consistent with early COVID-19. The median time from onset of symptoms to administration of sotrovimab was 3 days (interquartile range, 1–3.3). All participants were in the early phase of COVID-19 when sotrovimab was administered; 2 of them required low levels of oxygen supplementation for reasons unrelated to COVID-19. Forty-two of 57 patients (73.7%) received at least 3 doses of SARS-CoV-2 vaccine in accordance with the recommendations of the Standing Committee on Vaccination (Supplementary Table 1). The median time span since the last vaccination was 3 months (range, 1–5). Two patients died from causes unrelated to COVID-19: 1 from stage IV malignant melanoma, the other from complications of acute lymphoblastic leukemia. In total, 5 patients could not be monitored because they either died (malignant melanoma) or did not present to follow-up (n = 4).
Table 1.
Baseline Characteristics of Patients Grouped by Immunodeficiency
| Variable | n |
|---|---|
| Total | 57 |
| Gender | |
| ȃMale | 36 |
| ȃFemale | 21 |
| Groups | |
| ȃImmunocompetent | 14 |
| ȃImmunodeficient | 43 |
| ȃȃSolid organ transplantation | |
| ȃȃȃȃKidney | 18 |
| ȃȃȃȃHeart | 2 |
| ȃȃȃȃHeart + kidney | 1 |
| ȃȃȃȃHeart + lung | 1 |
| ȃȃȃȃKidney + pancreas | 1 |
| ȃȃStem cell transplantation | |
| ȃȃȃȃAllogeneic | 5 |
| ȃȃȃȃAutologous | 2 |
| ȃȃLeukemia | |
| ȃȃȃȃAcute lymphoblastic leukemia | 2 |
| ȃȃȃȃAMLa | 2 |
| ȃȃȃȃAML + CMML | 1 |
| ȃȃȃȃCMML | 1 |
| ȃȃLymphoma | |
| ȃȃȃȃDiffuse large B-cell lymphoma | 1 |
| ȃȃȃȃT-cell lymphomaa | 1 |
| ȃȃAL amyloidosis/smoldering multiple myelomaa | 1 |
| ȃȃOther malignancies | |
| ȃȃȃȃStage IV malignant melanoma and stage IV non-small cell lung cancerb | 1 |
| ȃȃCommon variable immune deficiency | 1 |
| ȃȃAutoimmune diseases | |
| ȃȃȃȃCryoglobulinemic vasculitis | 1 |
| ȃȃȃȃp-ANCA vasculitis | 1 |
| ȃȃȃȃRheumatoid arthritis | 1 |
| ȃȃȃȃSystemic lupus erythematosus | 1 |
| ȃȃȃȃUlcerative colitis | 1 |
| ȃȃLiver cirrhosis Child–Pugh Ac | 1 |
| ȃȃLiver fibrosis with portal hypertensionc | 1 |
Abbreviations: ANCA, anti-neutrophil cytoplasmatic antibody; AML, acute myeloblastic leukemia; AL, amyloid; CMML, chronic myelomonocytic leukemia.
Patients with previous allogeneic (2) and autologous (1) stem cell transplantation and malignancy relapse.
Dexamethasone therapy for cerebral metastases.
Patients with liver fibrosis/cirrhosis had a concurrent autoimmune disease.
Patients were grouped into immunocompetent (n = 14) and immunodeficient (n = 43). Immunodeficiency mostly comprised SOT, stem cell transplantation (SCT), active hematologic malignancies, and autoimmune diseases. The full spectrum of diseases is presented in Table 1. Immunosuppressive medication was given to 39 of 43 patients (90%) classified as immunodeficient (Supplementary Table 1).
Prolonged Viral Shedding in Immunodeficient COVID-19 Patients Infected With an Omicron Variant and Treated With Sotrovimab
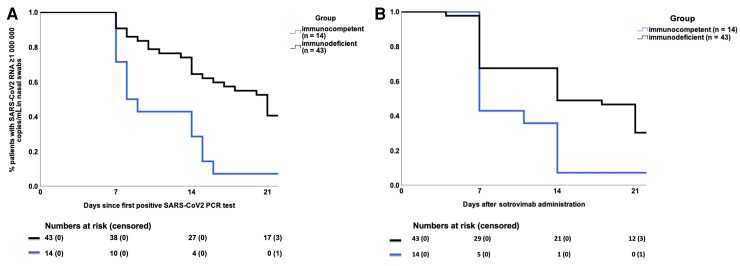
We analyzed the kinetics of viral clearance after the first positive SARS-CoV-2 PCR test and after sotrovimab administration in immunocompetent and immunodeficient patients (Figure 1). All but 1 of the immunocompetent patients had a viral load (VL) below 106 copies/mL at day 14, while 21 of 43 (48.8%) immunodeficient patients had prolonged viral shedding at this time point (P = .011). Moreover, even on day 21, 12 of 43 (27.9%) patients with immunodeficiency had not achieved a VL <106 copies/mL. The only immunocompetent patient who still had a VL >106 copies/mL on day 14 was lost to follow-up and therefore considered for analysis as having a VL >106 copies/mL on day 21 (patient 37; Supplementary Table 2). A higher proportion of patients who presented without COVID-related symptoms had prolonged viral shedding after days 14 and 21 (Supplementary Table 4).
Figure 1.
Patients with persistent viral replication (≥106 copies/mL) after sotrovimab administration. A, Prolonged viral shedding by day 21 after the first positive SARS-CoV-2 PCR test according to immunocompetence. B, Prolonged viral shedding by day 21 after sotrovimab administration in immunocompetent patients and patients with immunodeficiency. Numbers at risk are patients with a viral load ≥106 copies/mL; censored are patients lost to follow-up (1 patient was first lost to follow-up on day 28 and was included in numbers at risk). Abbreviations: PCR, polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Of note, 6 of 43 (13.9%) immunodeficient patients were infected with the BA.2 Omicron variant, characterized by higher levels of in vitro resistance of sotrovimab compared with Omicron BA.1. Twenty-nine of 43 (67.4%) immunodeficient patients received additional therapy with remdesivir at baseline (Supplementary Table 2). However, in the subgroup analysis, no significant association was found regarding the occurrence of prolonged viral shedding and the following factors: Omicron variant, remdesivir administration, number of vaccinations, and months since last vaccination or time between symptom onset and sotrovimab infusion (Table 2). The only risk factor identified for prolonged viral shedding was immunodeficiency (r = 0.329; P = .016).
Table 2.
Bivariate Correlation Among Clinical Parameters, Duration Until Viral Load <106 copies/mL, and Escape Mutations
| Parameter | VL <106 Copies/mL Since First Positive Polymerase Chain Reaction Test (d) | VL <106 Copies/mL Since Sotrovimab Administration (d) | Mutations Day 7 (0 = None, 1 = Mutation) | Mutations Day 14 (0 = None, 1 = Mutation) | Mutations overall (0 = None, 1 = Mutation) |
|---|---|---|---|---|---|
| Correlation Coefficient (r) P Value | |||||
| Remdesivir therapy at baseline (0 = 0, 1 = 3, and 2 = 5 d) | |||||
| ȃImmunocompetent | −0.355 | −0.224 | NA | NA | NA |
| 0.234 | 0.462 | NA | NA | NA | |
| ȃImmunodeficient | 0.057 | −0.036 | −0.372 | −0.261 | −0.392 |
| 0.726 | 0.827 | 0.015 | 0.099 | 0.009 | |
| Omicron variant (0 = BA.1,1 = BA.2) | |||||
| ȃImmunocompetent | 0.068 | 0.207 | NA | NA | NA |
| 0.824 | 0.498 | NA | NA | NA | |
| ȃImmunodeficient | 0.032 | 0.107 | −0.150 | −0.095 | −0.137 |
| 0.844 | 0.510 | 0.343 | 0.555 | 0.383 | |
| Time since last vaccination (mo) | |||||
| ȃImmunocompetent | 0.073 | 0.080 | NA | NA | NA |
| 0.822 | 0.805 | NA | NA | NA | |
| ȃImmunodeficient | 0.298 | 0.241 | 0.011 | 0.179 | 0.217 |
| 0.109 | 0.199 | 0.953 | 0.345 | 0.240 | |
| Number of vaccinations | |||||
| ȃImmunocompetent | 0.408 | 0.496 | NA | NA | NA |
| 0.167 | 0.085 | NA | NA | NA | |
| ȃImmunodeficient | 0.041 | 0.046 | 0.117 | −0.125 | −0.104 |
| 0.804 | 0.780 | 0.467 | 0.422 | 0.512 | |
| Immunodeficiency (0 = immunocompetent, 1 = immunodeficient) | 0.329 | 0.208 | 0.320 | 0.275 | 0.305 |
| 0.016 | 0.135 | 0.015 | 0.042 | 0.021 | |
| Viral clearance after sotrovimab administration (d) | NA | NA | 0.258 | 0.401 | 0.322 |
| NA | NA | 0.062 | 0.004 | 0.019 | |
| Tacrolimus levels at baseline (ng/mL) | 0.349 | 0.275 | 0.161 | 0.451 | 0.523 |
| 0.132 | 0.240 | 0.486 | 0.046 | 0.015 | |
| Days since first symptoms (number of pairs = 35a) | 0.075 | −0.251 | 0.090 | −0.144 | −0.10 |
| 0.669 | 0.146 | 0.600 | 0.417 | 0.955 | |
Significant correlations appear in bold.
Abbreviations: NA, not applicable; VL, viral load.
One patient was lost to follow-up and not included in this analysis.
Initial nonresponders, defined as patients whose symptoms either worsened despite sotrovimab administration and required hospitalization (patients 30, 34) or who experienced a viral rebound during the observation period (days 14–21: patients 8, 9, 10, 16; >21 days: patients 3, 26, 54), received further antiviral therapy. All patients who showed a slow but steady decline in SARS-CoV-2 viral load (VL) did not receive further antiviral therapy, and 5 patients were lost to follow-up. In all 10 patients re-treated with additional antiviral drugs, the virus was subsequently eliminated (Supplementary Table 2).
Taken together, these results show that immunocompromised patients have a substantial rate of prolonged viral shedding, even after administration of sotrovimab, which was the standard therapy for patients infected with SARS-CoV-2 at high risk for disease progression at the time of enrollment.
Emergence and Characterization of Escape Mutations in Omicron Variant of Concern After Use of Sotrovimab
Noting the prolonged viral shedding in immunocompromised patients after sotrovimab administration, we then performed whole-genome nanopore sequencing of all available viral samples with VL >106 copies/mL. Samples with detected resistance mutations were further analyzed with quantitative Illumina sequencing with spike amino acid coverage averaging 98.5% (range, 91.6%–100%; sample overview table on online data repository server, see Data availability section). This analysis revealed that mutations at spike protein residues associated with resistance to sotrovimab occurred in 14 of 57 patients (24.6%). No selection of escape mutations was observed in the immunocompetent patients, only in immunodeficient patients (14 of 43, 32.6%), most of whom had prolonged viral shedding. This group comprised 6 patients with SOT; 2 allogeneic SCT recipients; 2 patients with active hematologic malignancy who were receiving chemotherapy; 1 patient each with cryoglobulinemic vasculitis, systemic lupus erythematosus, and liver cirrhosis (Child–Pugh class A), each of whom received additional immunomodulatory therapies; and 1 patient with common variable immunodeficiency (Supplementary Table 2).
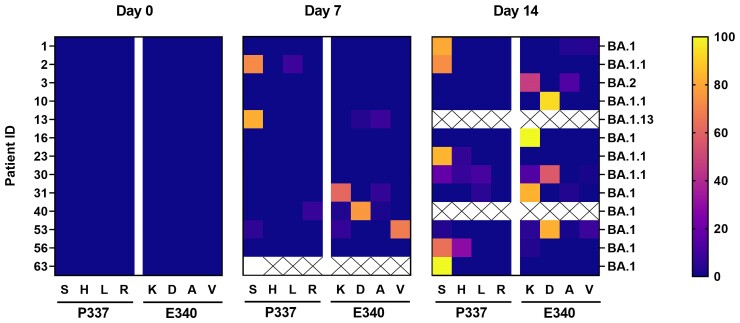
While no variants with reduced susceptibility to sotrovimab were detected at baseline confirmed by Illumina sequence analysis, 5 patients had sotrovimab-resistant variants by day 7, whereas most escape mutations occurred between day 7 and day 14. Details of the quantitative analysis of sotrovimab resistance mutations performed by Illumina sequencing on SARS-CoV-2 samples with evidence of immune escape in nanopore sequencing are shown in Figure 2. The resistance mutations that appeared first were detected exclusively at positions 337 or 340 in the spike protein, predominantly featuring the mutations P337S (n = 8), E340K (n = 9), and E340D (n = 5). In addition, amino acid substitutions P337H/L/R and E340A/V were found during our observation period of up to 28 days (Supplementary Table 3). During the observation period, not only an increase in escape variants (eg, patients 2, 31, 53) but also a change in the frequency of mutated variants were observed, for example, patient 10, E340D (d21) to E340K (d28) and patient 53, E340V (d7) to E340D (d14; Figure 2, Supplementary Table 3).
Figure 2.
Prevalence and evolution of escape mutations in the spike protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) after sotrovimab treatment. Detected amino acid exchanges in the spike protein at positions 337 and 340 on day 0, day 7, and day 14 after sotrovimab administration. The frequency of reads in % is indicated by the color scale. The determined patient-related SARS-CoV-2 variant is shown. Only patients with detected mutations after sotrovimab treatment are indicated. Patients selecting a spike protein mutation after day 14 are not included in this figure (patient 51).
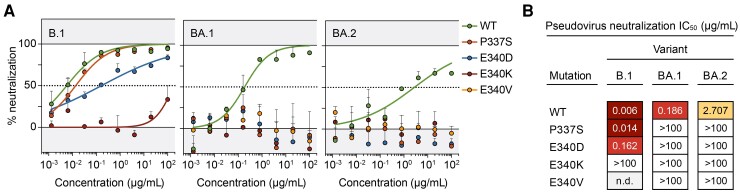
The sotrovimab-specific escape mutations (P337S, E340D/K/V) detected on day 7 were characterized in the BA.1 and BA.2 Omicron background using a pseudovirus neutralization assay (Figure 3). While in the B.1 background (a common lineage early in 2020 [14]), only E340K und E340D were associated with reduced neutralization by sotrovimab (IC50 (half maximal inhibitory concentration), >100 µg/mL and IC50, 0.162 µg/mL, respectively), all other detected mutations completely abrogated neutralization by sotrovimab in both the BA.1 and BA.2 backgrounds (IC50, >100 µg/mL).
Figure 3.
Neutralization of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike mutants by sotrovimab. A, SARS-CoV-2 variant-specific pseudoviruses harboring mutations that emerged after sotrovimab treatment were analyzed in sotrovimab neutralization assays. All samples were tested in duplicate. Symbols and bars indicate mean and standard deviation, respectively. The determined IC50 values are shown in (B). Abbreviation: IC50, half maximal inhibitory concentration.
To characterize the risk factors for the selection of escape mutations, correlation analysis was performed (Table 2). This analysis revealed that 2 factors correlated with the emergence of resistance mutations: immunodeficiency (r = 0.305, P = .021) and days until VL <106 SARS-CoV-2 RNA copies/mL achieved after sotrovimab administration (r = 0.322, P = .019). In detail, patients with emergence of mutations had significantly delayed time to viral clearance (mean, 28.2; standard deviation [SD], 16.2 days) compared with those without mutations (mean, 12.9; SD, 9.9 days; odds ratio, 5.04; 95% confidence interval, 1.29–18.3). In addition, for patients with tacrolimus therapy, higher tacrolimus levels at baseline positively correlated with the emergence of escape mutations (r = 0.523, P = .015). In immunodeficient patients, administration of remdesivir in combination with the corresponding duration correlated negatively with the occurrence of resistance mutations against sotrovimab (r = −0.392, P = .009). Most patients with selection of sotrovimab-specific escape mutations (13 of 14, 92.8%) were infected with the BA.1 variant; however, only 6 of 43 immunodeficient patients were infected with BA.2.
Together, these findings suggest that sotrovimab monotherapy in immunocompromised patients is associated with the risk of de novo development of specific mutations that lead to immune escape.
DISCUSSION
To our knowledge, this is one of the few studies to report the frequent emergence of escape mutations after sotrovimab treatment in a predominantly immunodeficient cohort of patients infected with Omicron variants.
Previous publications showed a decreased severity of SARS-CoV-2 disease with the Omicron variant [15]. Consistent with this, all patients in our high-risk cohort had uncomplicated disease throughout the follow-up period and there was no SARS-CoV-2–related mortality. Due to the observational nature of our study, it remains unclear whether the clinical course might have been less favorable in some patients without early antiviral therapy. When the BA.1 and BA.2 Omicron variants were compared in terms of prolonged viral shedding after sotrovimab administration, there was no significant difference found in our cohort. At this point, however, it must be emphasized that BA.2 was underrepresented in our study cohort compared with BA.1 (17.5% vs 82.5%, respectively). In our pseudovirus neutralization assays (Figure 3), as well as in other studies, a reduced neutralization activity of sotrovimab against BA.2 was described [16, 17]. These data led the US Food and Drug Administration to revoke the approval of sotrovimab for patients infected with BA.2 in April 2022 [18].
A unique feature of our cohort is the large number of immunodeficient patients, almost half of whom were patients with SOT, resulting in a higher risk of prolonged viral shedding, therefore potentially promoting the emergence of highly mutated viruses [19–21]. In this context, a higher baseline tacrolimus serum level was associated with the selection of escape mutations in our study, which highlights the importance of considering treatment adjustments of immunosuppressive medication during SARS-CoV-2 infection.
In our cohort, all but 1 of the immunocompetent patients (13 of 14, 92.9%) were below the defined viral threshold of 106 SARS-CoV-2 RNA copies/mL at day 14 and no selection of resistant variants to sotrovimab was detected. In contrast, sotrovimab escape mutations were detected in 32.6% of immunodeficient patients who predominantly experienced prolonged periods of viral replication. Similarly, treatment with other mAbs or antiviral agents (such as remdesivir) is also reported to promote the selection of viral mutations, particularly in immunosuppressed patients [10, 22–24].
Sotrovimab-specific resistance mutations were first described in an Australian cohort of patients infected with the Delta variant [25]. Genome sequencing of samples from the COMET-ICE trial detected sotrovimab escape mutations in 20 patients, of which P337L, E340A, and E340K showed reduced susceptibility to sotrovimab in pseudotyped viral-like particles (>100-fold change in EC50 (half maximal effective concentration) value) [3, 18]. In the study published by Rockett et al, 8 of 100 patients developed 1 of the following mutations, E340A/K/V or P337L, combined with the E340 mutation that occurred 6–13 days after sotrovimab administration [25]. While P337L and the E340A were selected primarily in the Delta variant of concern, other amino acids were selected in the Omicron variants at the same positions, predominantly P337S/R and E340D/K, as reported in other recent studies [11, 12].
In our longitudinal study, after detection of the sotrovimab-specific escape mutations P337S/L/R and E340A/D/K/V at day 7, an additional variant was detected during our observation period (P337H). Moreover, changes in frequency of different escape variants over time were observed, as described in infections with the Delta variant, presumably indicating ongoing viral evolution [25].
In the in vitro analyses carried out in our study, the pseudovirus neutralization assays confirmed that both sotrovimab mutations, E340K and E340V, which were also selected in Delta, and mutations P337S and E340D, newly described in the Omicron context, completely abrogated the neutralization activity of sotrovimab. In the B.1 background, on the other hand, a strongly reduced neutralization activity could only be observed for E340K, whereas the E340D mutation reduced the neutralization activity of sotrovimab to a much lesser extent. These data clearly show that not only the escape mutation itself but also the broader genetic background of the spike protein influence the impact of a specific escape mutation on mAb efficacy, as has been observed in several efficacy studies for mAbs [17, 26].
In a previous small cohort study conducted before the Omicron era, we found that the E484K mutation occurred with bamlanivimab monotherapy in 83% of patients and in a major portion of the viral population in the respective patients [10]. In contrast, in our study, the frequency of sotrovimab-resistant viral variants was lower in most patients and showed a very heterogeneous mutation spectrum [27, 28].
Our study has limitations that should be considered for future studies. First, the relatively small cohort made subgroup analysis difficult. Second, the quantitative analysis with Illumina sequencing was performed only in patients in whom spike protein mutations were detected in nanopore sequencing. Therefore, the diversity of viral quasispecies cannot be compared to patients without detection of mutations in nanopore sequencing. However, failure to account for possible viral minorities with spike protein mutations in this group seems unlikely, as these were not detected in the patients with emerging sotrovimab resistance mutations.
There is growing evidence to support the hypothesis that new SARS-CoV-2 variants preferentially occur in immunocompromised patients with persistent SARS-CoV-2 infection. Since some of these variants may be more transmissible or may have better immune escape, this potentially has significant implications for individual medical care and public health. In immunocompromised patients, prolonged viral shedding must therefore be considered with respect to infection control. Given the available data, administration of a single mAb or single antiviral drug should be avoided in immunocompromised patients because of the risk of emergent mutations. In our study, we demonstrated that the presence and length of remdesivir therapy at baseline was associated with a reduced emergence of escape mutations in immunodeficient patients. In addition, a second remdesivir administration over a longer period of 10 days and combination antiviral therapy resulted in a sustained decrease in viral load in the vast majority of patients with persistently high nasopharyngeal VL, and viral shedding was successfully terminated in those patients.
In summary, combination therapies with at least 2 mAbs or other antivirals, such as remdesivir, molnupiravir, and nirmatrelvir/ritonavir, should be considered when treating immunodeficient patients with SARS-CoV-2 infection. These results also highlight the importance of careful monitoring and the need to conduct dedicated clinical trials to establish the optimal treatment strategy for this patient population. This is especially true at this stage of the pandemic since, with the availability of vaccines that prevent severe disease courses for most patients, immunodeficient patients represent one of the most vulnerable and severely affected patient groups.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. S. G., N. L., A. K., T. L., and B.-E. O. J. were responsible for conceptualization and supervised the study. S. G., N. L., A. K., H. G., A. W., A. T. D., C. L., C. F., S. K., J. V., T. S., A. Z., T. F., J. B., H.-M. O., F. K., J. T., T. L., and B.-E. O. J. contributed to the investigation and data curation. S. G., N. L., A. K., B.-E. O. J., H. G., and J. T. conducted the formal analysis. S. G., B.-E. O. J., T. L., N. L., A. K., J. T., H. G., and F. K. were responsible for methodology, data validation, and visualization. S.G., N. L., A. K., H. G., J. T., T. L., and B.-E. O. J. contributed to the original draft. All authors critically revised the manuscript and approved the final version. Additionally, S. K., J. V. P., T. P. S., and A. Z. were instrumental in patient management, as well as the coordination of outpatient care for the study participants. A. T. and M. D. coordinated the Illumina sequencing and performed the bioinformatic analysis of the sequencing data.
Acknowledgments. The authors thank the patients and their relatives, the clinical staff involved in patient care, the laboratory staff involved in the virological analyses, and all others who contributed to the study, especially Kanika Vanshylla for support with pseudovirus neutralization assays.
Disclaimer. The funders had no role in study design, data collection, data analysis, interpretation, or writing of the report.
Financial support. This work was supported by COVIM (COVid IMmunity) (FKZ, 01KX2021), a joint project funded by the Federal Ministry of Education and Research (BMBF) reported by B.-E. O. J. and J. G. B.; the EuCARE project “European cohorts of patients and schools to advance response to epidemics,” which is funded by the European Commission as part of the HORIZON HLTH 2021 CORONA 01 (grant 101046016); and the joint project Beyond COVID-19 funded by The Ministry of Culture and Science of North Rhine-Westphalia reported by B.-E. O. J. In addition, this work was supported by the Ministry for Labor, Health and Social Affairs of the State of North Rhine-Westphalia (CPS-1-1A).
Data availability. Raw data are generated at the University Hospital Düsseldorf and in cooperation with the University Hospital Cologne. Derived or additional data that support the findings of this study can be provided by the corresponding author upon reasonable request. All Nanopore and Illumina sequence data can be found on the Open Science Framework server https://osf.io/q7js6/?view_only=59884b79343e449ea9f7103f99c118a9.
Supplementary Material
Contributor Information
Smaranda Gliga, Department of Gastroenterology, Hepatology and Infectious Diseases, University Hospital Düsseldorf, Medical Faculty, Heinrich-Heine-University Düsseldorf, Düsseldorf, Germany; Institute of Virology, University Hospital Düsseldorf, Medical Faculty, Heinrich-Heine-University Düsseldorf, Düsseldorf, Germany.
Nadine Lübke, Institute of Virology, University Hospital Düsseldorf, Medical Faculty, Heinrich-Heine-University Düsseldorf, Düsseldorf, Germany.
Alexander Killer, Department of Gastroenterology, Hepatology and Infectious Diseases, University Hospital Düsseldorf, Medical Faculty, Heinrich-Heine-University Düsseldorf, Düsseldorf, Germany.
Henning Gruell, Institute of Virology, Faculty of Medicine and University Hospital Cologne, University of Cologne, Cologne, Germany.
Andreas Walker, Institute of Virology, University Hospital Düsseldorf, Medical Faculty, Heinrich-Heine-University Düsseldorf, Düsseldorf, Germany.
Alexander T Dilthey, Institute of Medical Microbiology and Hospital Hygiene, Medical Faculty, Heinrich-Heine-University Düsseldorf, Düsseldorf, Germany.
Alexander Thielen, Institute of Immunology and Genetics, Kaiserslautern, Germany.
Carolin Lohr, Department of Gastroenterology, Hepatology and Infectious Diseases, University Hospital Düsseldorf, Medical Faculty, Heinrich-Heine-University Düsseldorf, Düsseldorf, Germany.
Charlotte Flaßhove, Department of Gastroenterology, Hepatology and Infectious Diseases, University Hospital Düsseldorf, Medical Faculty, Heinrich-Heine-University Düsseldorf, Düsseldorf, Germany.
Sarah Krieg, Department of Gastroenterology, Hepatology and Infectious Diseases, University Hospital Düsseldorf, Medical Faculty, Heinrich-Heine-University Düsseldorf, Düsseldorf, Germany.
Joanna Ventura Pereira, Department of Gastroenterology, Hepatology and Infectious Diseases, University Hospital Düsseldorf, Medical Faculty, Heinrich-Heine-University Düsseldorf, Düsseldorf, Germany.
Tobias Paul Seraphin, Department of Gastroenterology, Hepatology and Infectious Diseases, University Hospital Düsseldorf, Medical Faculty, Heinrich-Heine-University Düsseldorf, Düsseldorf, Germany.
Alex Zaufel, Department of Gastroenterology, Hepatology and Infectious Diseases, University Hospital Düsseldorf, Medical Faculty, Heinrich-Heine-University Düsseldorf, Düsseldorf, Germany.
Martin Däumer, Institute of Immunology and Genetics, Kaiserslautern, Germany.
Hans-Martin Orth, Department of Gastroenterology, Hepatology and Infectious Diseases, University Hospital Düsseldorf, Medical Faculty, Heinrich-Heine-University Düsseldorf, Düsseldorf, Germany.
Torsten Feldt, Department of Gastroenterology, Hepatology and Infectious Diseases, University Hospital Düsseldorf, Medical Faculty, Heinrich-Heine-University Düsseldorf, Düsseldorf, Germany.
Johannes G Bode, Department of Gastroenterology, Hepatology and Infectious Diseases, University Hospital Düsseldorf, Medical Faculty, Heinrich-Heine-University Düsseldorf, Düsseldorf, Germany.
Florian Klein, Institute of Virology, Faculty of Medicine and University Hospital Cologne, University of Cologne, Cologne, Germany; Center for Molecular Medicine Cologne, University of Cologne, Cologne, Germany.
Jörg Timm, Institute of Virology, University Hospital Düsseldorf, Medical Faculty, Heinrich-Heine-University Düsseldorf, Düsseldorf, Germany.
Tom Luedde, Department of Gastroenterology, Hepatology and Infectious Diseases, University Hospital Düsseldorf, Medical Faculty, Heinrich-Heine-University Düsseldorf, Düsseldorf, Germany.
Björn-Erik Ole Jensen, Department of Gastroenterology, Hepatology and Infectious Diseases, University Hospital Düsseldorf, Medical Faculty, Heinrich-Heine-University Düsseldorf, Düsseldorf, Germany.
References
- 1. Kreuzberger N, Hirsch C, Chai KL, et al. SARS-CoV-2-neutralising monoclonal antibodies for treatment of COVID-19. Cochrane Database Syst Rev 2021; 9:CD013825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hoffmann M, Kruger N, Schulz S, et al. The Omicron variant is highly resistant against antibody-mediated neutralization: implications for control of the COVID-19 pandemic. Cell 2022; 185:447–56.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gupta A, Gonzalez-Rojas Y, Juarez E, et al. Early treatment for Covid-19 with SARS-CoV-2 neutralizing antibody sotrovimab. N Engl J Med 2021; 385:1941–50. [DOI] [PubMed] [Google Scholar]
- 4. Gupta A, Gonzalez-Rojas Y, Juarez E, et al. Effect of sotrovimab on hospitalization or death among high-risk patients with mild to moderate COVID-19: a randomized clinical trial. JAMA 2022; 327:1236–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yamasoba D, Kosugi Y, Kimura I, et al. Neutralisation sensitivity of SARS-CoV-2 Omicron subvariants to therapeutic monoclonal antibodies. Lancet Infect Dis 2022; 22:942–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. COVID-19: EMA recommends authorisation of antibody medicine Xevudy. Available at:https://www.ema.europa.eu/en/news/covid-19-ema-recommends-authorisation-antibody-medicine-xevudy. Accessed 14 May.
- 7. Group AC-TfIwC-S. Efficacy and safety of two neutralising monoclonal antibody therapies, sotrovimab and BRII-196 plus BRII-198, for adults hospitalised with COVID-19 (TICO): a randomised controlled trial. Lancet Infect Dis 2022; 22:622–35. [DOI] [PMC free article] [PubMed]
- 8. Martin-Blondel G, Marcelin AG, Soulie C, et al. Outcome of very high-risk patients treated by sotrovimab for mild-to-moderate COVID-19 Omicron, a prospective cohort study (the ANRS 0003S COCOPREV study). J Infect 2022; 84:e101–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Solera JT, Arbol BG, Alshahrani A, et al. Impact of vaccination and early monoclonal antibody therapy on COVID-19 outcomes in organ transplant recipients during the Omicron wave. Clin Infect Dis 2022; 75:2193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jensen B, Luebke N, Feldt T, et al. Emergence of the E484K mutation in SARS-COV-2-infected immunocompromised patients treated with bamlanivimab in Germany. Lancet Reg Health Eur 2021; 8:100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Destras G, Bal A, Simon B, Lina B, Josset L. Sotrovimab drives SARS-CoV-2 Omicron variant evolution in immunocompromised patients. Lancet Microbe 2022; 3:e559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vellas C, Tremeaux P, Del Bello A, et al. Resistance mutations in SARS-CoV-2 Omicron variant in patients treated with sotrovimab. Clin Microbiol Infect 2022; 28:1297–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jefferson T, Spencer EA, Brassey J, Heneghan C. Viral cultures for coronavirus disease 2019 infectivity assessment: a systematic review. Clin Infect Dis 2021; 73:e3884–e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tallarita M, Giardina F, Novazzi F, et al. Spread of multiple SARS-CoV-2 lineages April-August 2020 anticipated the second pandemic wave in Lombardy (Italy). Pediatr Allergy Immunol 2022; 33(Suppl 27):89–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Abdullah F, Myers J, Basu D, et al. Decreased severity of disease during the first global Omicron variant covid-19 outbreak in a large hospital in Tshwane, South Africa. Int J Infect Dis 2022; 116:38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bruel T, Hadjadj J, Maes P, et al. Serum neutralization of SARS-CoV-2 Omicron sublineages BA.1 and BA.2 in patients receiving monoclonal antibodies. Nat Med 2022; 28:1297–302. [DOI] [PubMed] [Google Scholar]
- 17. Takashita E, Kinoshita N, Yamayoshi S, et al. Efficacy of antiviral agents against the SARS-CoV-2 Omicron subvariant BA.2. N Engl J Med 2022; 386:1475–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. FDA updates Sotrovimab emergency use authorization. Available at: https://www.fda.gov/drugs/drug-safety-and-availability/fda-updates-sotrovimab-emergency-use-authorization. Accessed 14 May.
- 19. Choi B, Choudhary MC, Regan J, et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N Engl J Med 2020; 383:2291–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Corey L, Beyrer C, Cohen MS, Michael NL, Bedford T, Rolland M. SARS-CoV-2 variants in patients with immunosuppression. N Engl J Med 2021; 385:562–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Truong TT, Ryutov A, Pandey U, et al. Increased viral variants in children and young adults with impaired humoral immunity and persistent SARS-CoV-2 infection: a consecutive case series. EBioMedicine 2021; 67:103355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gandhi S, Klein J, Robertson AJ, et al. De novo emergence of a remdesivir resistance mutation during treatment of persistent SARS-CoV-2 infection in an immunocompromised patient: a case report. Nat Commun 2022; 13:1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Leung WF, Chorlton S, Tyson J, et al. COVID-19 in an immunocompromised host: persistent shedding of viable SARS-CoV-2 and emergence of multiple mutations: a case report. Int J Infect Dis 2022; 114:178–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vellas C, Del Bello A, Debard A, et al. Influence of treatment with neutralizing monoclonal antibodies on the SARS-CoV-2 nasopharyngeal load and quasispecies. Clin Microbiol Infect 2022; 28:139.e5–e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rockett R, Basile K, Maddocks S, et al. Resistance mutations in SARS-CoV-2 Delta variant after sotrovimab use. N Engl J Med 2022; 386:1477–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Singh DD, Sharma A, Lee HJ, Yadav DK. SARS-CoV-2: recent variants and clinical efficacy of antibody-based therapy. Front Cell Infect Microbiol 2022; 12:839170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peiffer-Smadja N, Bridier-Nahmias A, Ferre VM, et al. Emergence of E484K mutation following bamlanivimab monotherapy among high-risk patients infected with the Alpha variant of SARS-CoV-2. Viruses 2021; 13:1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sanjuán R, Domingo-Calap P. Genetic diversity and evolution of viral populations. Encyclopedia Virol 2021:53–61. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.