Abstract
Background
Nirmatrelvir/ritonavir, the first severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) protease inhibitor, reduces the risk of hospitalization and death by coronavirus disease 2019 (COVID-19) but has been associated with symptomatic rebound after therapy completion.
Methods
Six individuals with relapse of COVID-19 symptoms after treatment with nirmatrelvir/ritonavir, 2 individuals with rebound symptoms without prior antiviral therapy and 7 patients with acute Omicron infection (controls) were studied. Soluble biomarkers and serum SARS-CoV-2 nucleocapsid protein were measured. Nasal swabs positive for SARS-CoV-2 underwent viral isolation and targeted viral sequencing. SARS-CoV-2 anti-spike, anti–receptor-binding domain, and anti-nucleocapsid antibodies were measured. Surrogate viral neutralization tests against wild-type and Omicron spike protein, as well as T-cell stimulation assays, were performed.
Results
High levels of SARS-CoV-2 anti-spike immunoglobulin G (IgG) antibodies were found in all participants. Anti-nucleocapsid IgG and Omicron-specific neutralizing antibodies increased in patients with rebound. Robust SARS-CoV-2–specific T-cell responses were observed, higher in rebound compared with early acute COVID-19 patients. Inflammatory markers mostly decreased during rebound. Two patients sampled longitudinally demonstrated an increase in activated cytokine-producing CD4+ T cells against viral proteins. No characteristic resistance mutations were identified. SARS-CoV-2 was isolated by culture from 1 of 8 rebound patients; Polybrene addition increased this to 5 of 8.
Conclusions
Nirmatrelvir/ritonavir treatment does not impede adaptive immune responses to SARS-CoV-2. Clinical rebound corresponds to development of a robust antibody and T-cell immune response, arguing against a high risk of disease progression. The presence of infectious virus supports the need for isolation and assessment of longer treatment courses.
Clinical trials registration. NCT04401436.
Keywords: COVID-19, nirmatrelvir/ritonavir, COVID-19 rebound, COVID-19 transmission, antiviral therapy
Clinical, virologic, and immune measurements were performed in 6 patients with coronavirus disease 2019 symptomatic rebound after nirmatrelvir/ritonavir treatment and 2 without previous treatment. There was no evidence of severe disease or impaired antibody and T-cell responses in people with rebound symptoms.
Nirmatrelvir-ritonavir (NMV-r) has been granted an emergency use authorization for treatment of early mild to moderate coronavirus disease 2019 (COVID-19) after demonstrating an 89% relative risk reduction of hospitalization or death in unvaccinated patients at high risk for severe disease, with an associated decrease in nasopharyngeal viral load at day 5 [1, 2]. Some patients who received NMV-r demonstrated a rise in viral load between day 10 and day 14 [1], and clinical rebound after completing NMV-r has now been reported [3]. The etiology of this phenomenon remains unknown, though immune evasion because of early viral suppression has been hypothesized [4]. Additionally, the risk of severe disease, viral transmissibility, and potential for NMV-r resistance remains unclear. To address these questions, we performed detailed virologic and immunologic evaluations of 8 patients with rebound COVID-19.
METHODS
Clinical Protocol
All participants were evaluated at the National Institutes of Health (NIH) between December 2021 and May 2022 and were enrolled in the clinical protocol COVID-19–Associated Lymphopenia Pathogenesis Study in Blood (NCT04401436) to evaluate adults (aged ≥18 years) with a diagnosis of COVID-19 by a molecular or other commercial assay or people who have recovered from COVID-19. Participation includes clinical evaluation, standard-of-care clinical management as indicated, laboratory studies, and research blood collection for storage of peripheral blood mononuclear cells (PBMCs), serum, and plasma. Additionally, plasma samples were obtained from 7 (5 women and 2 men) healthy volunteers with a median age of 59 years (range, 45–66) on a separate NIH institutional review board–approved protocol (NCT00001281) to provide a healthy control comparison for the biomarker analysis. All samples were collected 1–24 days from symptom onset. Samples collected during rebound symptoms after completing NMV-r were designated as the “rebound after NMV-r” group, the remainder of the samples collected within 4 days of symptom onset were designated as the “acute” group, and samples collected ≥8 days were referred to as the “late” time point group. Two patients were seen for acute visits and then later returned with rebound symptoms following NMV-r (Supplementary Figure 1A and 1B). All participants in both protocols provided written informed consent prior to any study procedures in accordance with the Declaration of Helsinki.
Viral Sequencing and Culture
Initial detection and estimation of viral load by cycle threshold (Ct values) were performed using the Hologic Panther Fusion severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) assay. Viral RNA sequencing was performed on SARS-CoV-2–positive nasal swab samples from acute and rebound patients as previously described [5] (see the Supplemental Methods). Two acute patients and 1 rebound patient had high Ct values, and virus could not be sequenced. Viral cultures were also performed on each sample by adding them to individual flasks containing Vero E6 cells expressing the transmembrane serine protease, TMPRSS2 (VeroE6/TMPRSS2), obtained from the Japanese Cell Culture Collection (Osaka, Japan). Inoculations were performed as previously described [6]. The infected cells were incubated at 37°C with 5% carbon dioxide in Dulbecco’s minimum essential medium (DMEM) supplemented with 2% heat-inactivated fetal bovine serum (HI-FBS) and observed daily for cytopathic effects (CPEs). Upon observation of CPEs, the virus supernatant was harvested and sequenced. This culture technique was subsequently repeated with the addition of 5 µg/mL of Polybrene (headimethrine bromide) to DMEM supplemented with 2% HI-FBS. Finally, SARS-CoV-2 viral nucleocapsid protein levels were measured in serum using a single-molecule immune bead assay (Quanterix; Billerica, MA) [7].
Serologic Assays
SARS-CoV-2 anti-spike (anti-S) and anti–receptor-binding domain (anti-RBD) antibody responses were analyzed with the enzyme-linked immunosorbent assay (ELISA) using a previously validated technique [8]. In this study, a threshold of 3 standard deviations above the mean of the negative controls for both the spike and RBD was determined to have a specificity of 100% for a positive result and therefore was considered the threshold for positivity. Levels of immunoglobulin G (IgG), IgM, and IgA were determined for anti-S and anti-RBD and compared between the rebound cases and controls. Anti-nucleocapsid antibodies were evaluated only for IgG and IgM.
The GenScript cPass assay, a surrogate viral neutralization test (sVNT) that has been shown to detect neutralizing antibodies [9], was also performed to evaluate for the presence of anti-S neutralizing antibodies against wild-type SARS-CoV-2 and the Omicron variant. This assay measures anti-S antibody-mediated inhibition of the interaction between the SARS-CoV-2 S–RBD and the angiotensin-converting enzyme 2 receptor. The result is reported as percent binding inhibition of this interaction (see the Supplementary Methods). The results were interpreted as positive if inhibition was ≥ 30%. For detection of neutralizing antibodies to the BA.1 Omicron variant, the wild-type S–RBD was replaced by the BA.1 S–RBD in the assay.
T-Cell Stimulation Assays
Frozen PBMCs were used to perform T-cell stimulation assays with SARS-CoV-2 wild-type and Omicron spike, nucleocapsid, and membrane proteins. Two peptide pools were used to measure spike-specific responses: spike pool A (peptides 1–160; residues 1–651) and spike pool B (peptides 161–316; residues 641–1273); responses from the 2 pools were summed to determine the response to full-length spike. See the Supplementary Methods for assay details and flow cytometry gating strategy in Supplementary Figure 2.
Soluble Biomarkers
Plasma samples were used to measure biomarkers according to the manufacturer’s instructions. Levels of interleukin 6 (IL-6), IL-8, IL-10, IL-18, tumor necrosis factor α (TNF-α), interferon γ (IFN-γ), and chemokine (C-X-C motif) ligand 10 (CXCL-10) were quantified by electrochemiluminescence assays from Meso Scale Discovery platforms (Gaithersburg, MD). Levels of CXCL-9, soluble CD25 (sCD25), IL-18 binding protein (BP), and sCD14 were quantified using ELISA kits from R&D Systems (Minneapolis, MN).
Statistical Analyses
All statistical analyses were conducted using GraphPad Prism (version 9) and R software (version 4.1.0).
RESULTS
Eight patients with rebound COVID-19 symptoms (6 following NMV-r, 2 without treatment) and 6 patients with Omicron-variant COVID-19 without rebound were evaluated. Two patients had samples collected during both their initial illness and when symptoms returned after completing NMV-r. Based on when samples were collected during the illness course, participants were subdivided into rebound after NMV-r (n = 6), acute (≤4 days, n = 7), and late (≥8 days, n = 3) groups (Table 1, Supplementary Figure 1, Supplementary Table 1). The 2 rebound patients who had no prior treatment were included in the late group.
Table 1.
Patient Characteristics
| Acute | Late | ||||
|---|---|---|---|---|---|
| Characteristic | Developed Rebound Symptoms (n = 2) |
No Rebound Symptom Development (n = 5) |
Rebound After NMV-r (n = 6) |
Rebound Without Treatment (n = 2) |
No Rebound Symptoms (n = 1) |
| Gender | |||||
| Male | 1 (50%) | 1 (20%) | 3 (50%) | 1 (50%) | 0 (0%) |
| Female | 1 (50%) | 4 (80%) | 3 (50%) | 1 (50%) | 1 (100%) |
| Age, years | |||||
| Median [min, max] | 39 [33, 45] | 50 [49, 65] | 42.5 [33, 74] | 44.5 [35, 54] | 23 |
| Comorbidity | |||||
| None | 0 (0%) | 1 (20%) | 0 (0%) | 2 (100%) | 1 (100%) |
| Pulmonary | 1 (50%) | 1 (20%) | 2 (33.3%) | 0 (0%) | 0 (0%) |
| Immunocompromised | 1 (50%)a | 1 (20%)b | 2 (33.3%)c | 0 (0%) | 0 (0%) |
| Cardiac | 0 (0%) | 0 (0%) | 2 (33.3%) | 0 (0%) | 0 (0%) |
| Other | 0 (0%) | 2 (40%)d | 0 (0%) | 0 (0%) | 0 (0%) |
| Days from initial symptom onset to visit | |||||
| Median [min, max] | 3.5 [3, 4]e | 3 [1, 3] | 16 [11, 17]f | 19 [14, 24] | 8 |
| Initial symptoms | |||||
| Upper respiratory only | 0 (0%) | 2 (40%) | 2 (33.3%) | 1 (50%) | 0 (0%) |
| Upper and lower respiratory | 1 (50%) | 1 (20%) | 1 (16.7%) | 0 (0%) | 1 (100%) |
| Upper respiratory and constitutional | 1 (50%) | 1 (20%) | 3 (50%) | 1 (50%) | 0 (0%) |
| Upper respiratory, lower respiratory, and constitutional | 0 (0%) | 1 (20%) | 0 (0%) | 0 (0%) | 0 (0%) |
| Receipt of NMV-r | |||||
| Recipients | 2 (100%) | 2 (40%) | 6 (100%) | 0 (0%) | 0 (0%) |
| Nonrecipients | 0 (0%) | 2 (40%) | 0 (0%) | 2 (100%) | 1 (100%) |
| Day of illness NMV-r started | |||||
| Median [min, max] | 2.5 [2, 3] | 2.5 [2, 3] | 2.5 [1, 4] | NA | NA |
| Day of illness symptoms returned | |||||
| Median [min, max] | 13 [11, 15] | NA | 12.5 [11, 15] | 14.5 [9, 20] | NA |
| Number of days symptoms returned after completing NMV-r | |||||
| Median [min, max] | 6 [4, 8] | NA | 6.5 [3, 9] | NA | NA |
Four rebound patients repeated rapid antigen tests after initial symptom resolution. Three became negative and the fourth became weakly positive. All 4 became positive again when rebound symptoms returned.
Abbreviations: NA, not applicable; NMV-r, nirmatrelvir-ritonavir.
Ankylosing spondylitis on golimumab.
Idiopathic thrombocytopenic purpura status post-splenectomy.
Multiple sclerosis on natalizumab; Ankylosing spondylitis on golimumab.
Primary biliary cirrhosis and obesity; epithelioid hemangioendothelioma and hypothyroidism.
Patients 1 and 2 were seen for acute visits 3 and 4 days from initial symptom onset, respectively.
Patients 1 and 2 were again seen 16 days and 11 days from initial symptom onset, respectively.
The SARS-CoV-2 polymerase chain reaction (PCR) test on nasal swabs from all patients was positive, and no additional pathogens were detected by BioFire FilmArray Respiratory Panel 2.1. All participants were vaccinated and boosted, and none received CYP3A4 inducers prior to NMV-r. None of the patients received any other COVID-19 directed therapy, including monoclonal antibody prophylaxis prior to sampling (except patient 2; Supplementary Figure 1B). All patients were managed in the outpatient setting for their entire disease course.
NMV-r was started between 1 and 4 days after initial symptom onset in those who later developed rebound symptoms. All rebound patients experienced significant symptomatic improvement prior to worsening. Five rebound patients repeated rapid antigen tests (RATs) after initial symptom resolution; 4 became negative and the fifth became weakly positive. Median time to symptom recurrence in the rebound after NMV-r group was 12.5 days after initial symptom onset and 6.5 days after completing NMV-r. During rebound after NMV-r, 4 patients reported milder symptoms than for their initial illness, 1 reported worse symptoms, and 1 reported similar symptom severity (Supplementary Table 2). No rebound patients required additional treatment or hospitalization.
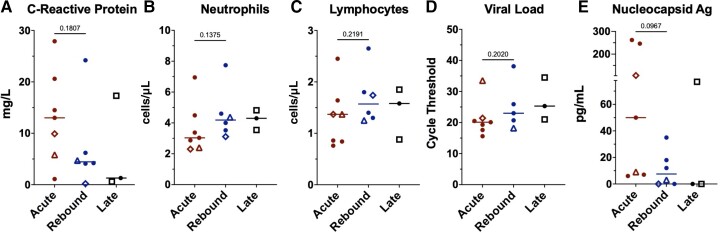
The median C-reactive protein (CRP) level was lower at time of rebound than during acute COVID-19, whereas neutrophil and lymphocyte counts and SARS-CoV-2 PCR Ct values were similar across groups (Figure 1A–D) with low or undetectable serum nucleocapsid antigen levels during rebound (Figure 1E).
Figure 1.
Comparison of clinical laboratory and virologic measurements across the groups. Lines represent median and points represent individual results. The 2 longitudinal patients are identified by an open triangle and open diamond, respectively. The open square represents the coronavirus disease 2019 (COVID-19) rebound patients who did not receive nirmatrelvir-ritonavir. Clinical values for C-reactive protein (A), absolute neutrophil count (B), and absolute lymphocyte count (C) across the acute, rebound, and late presenting COVID-19 cohorts. Severe acute respiratory syndrome coronavirus 2 cycle threshold from nasal swab samples (D) and serum nucleocapsid Ag (E). Cycle threshold was not available at rebound time point for longitudinal patient 1 (diamond) as it was run on a BioFire platform. Abbreviation: Ag, antigen.
All patients had Omicron BA.2 infections except for 1 acute and 1 rebound without NMV-r who were infected with the BA.5 subvariant. Infectious replication-competent SARS-CoV-2 was isolated from the nasal swabs of 6 of 7 acute controls with the only negative culture from longitudinal patient 2 (triangle data points in Figure 1, Figures 3–5, and Supplementary Figures 5–7) who had started NMV-r the day before. Comparatively, only 1 of 8 rebound patients (patient 1—diamond data points in Figure 1, Figures 3–5, and Supplementary Figures 5–7) was culture-positive. Positive culture was not always associated with low cycle threshold and/or high serum nucleocapsid antigen (Supplementary Figure 3). In a recent report, viral growth was observed in samples from 3 of 7 rebound patients after NMV-r [10]; however, viral culture media was supplemented with Polybrene, which can increase virus adsorption by target cells by approximately 10-fold [11]. We repeated viral cultures in our cohort using Polybrene and identified positive cultures in 5 of 8 rebound patients, including 4 of 6 who had rebound after NMV-r, supporting the greater sensitivity of this culture technique.
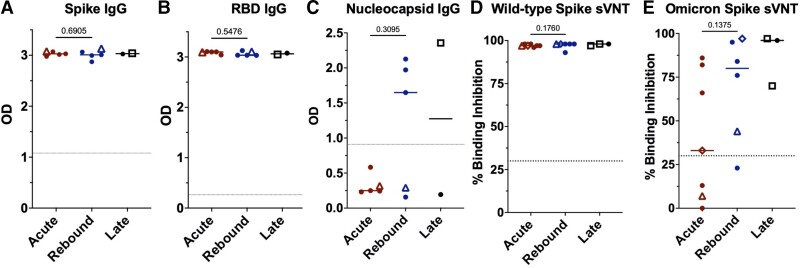
Figure 3.
Comparison of antibody level measurements across the groups. Antibody levels by enzyme-linked immunosorbent assay (ELISA) against the spike protein (A), spike–receptor binding domain (RBD) (B), and the nucleocapsid protein (C) presented as OD. ELISA data not available for longitudinal patient 1 (diamond), 1 acute patient, and 1 rebound without nirmatrelvir-ritonavir patient (square). sVNT to detect neutralizing antibodies against the wild-type (D) and Omicron (E) spike protein presented as percent binding inhibition. Dotted lines represent the cutoff for a positive result for the antibody tests (A–C). Mann–Whitney test was used to derive P values comparing the acute and rebound coronavirus disease 2019 cohorts. Abbreviations: Ig, immunoglobulin; OD, optical density; RBD, receptor-binding domain; sVNT, surrogate viral neutralization test.
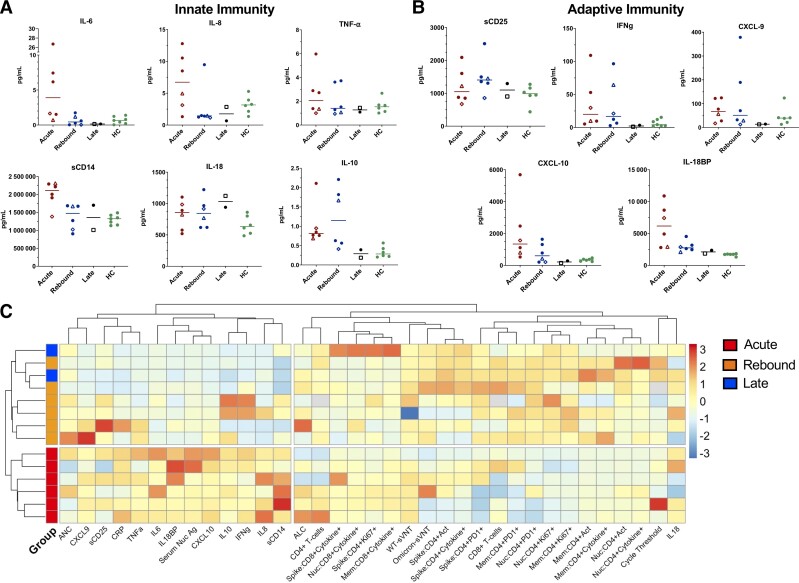
Figure 5.
Innate and adaptive biomarkers across study groups compared with healthy controls and heat map of data from study participants. Lines represent medians and points represent individual results across the acute, rebound, and late presentation coronavirus disease 2019 (COVID-19) clinical groups compared with healthy control population (HC). Healthy controls consisted of 5 women and 2 men with a median age of 59 years (range, 45–66). These samples were unmatched and are included to provide a baseline range for these biomarkers in an otherwise healthy population. The 2 longitudinal COVID-19 rebound patients are identified by an open diamond (patient 1) and open triangle (patient 2). The open square represents the COVID-19 rebound patient who did not receive nirmatrelvir-ritonavir (NMV-r). A, Innate biomarkers (IL-6, IL-8, TNF-α, CXCL-10, sCD14, and IL-18BP) classically increased in acute COVID-19 are downtrending at time of rebound. B, Adaptive biomarkers representing T-cell activation (IFN-γ, CXCL-9, sCD25) were stable or increasing at rebound consistent with a developing T-cell response. C, Comprehensive heat map with unsupervised clustering of variables including clinical laboratory tests, virologic measurements, biomarkers, and profiling of adaptive responses identified that all patients with rebound COVID-19 after NMV-r form a unique cluster distinct from those with acute infection. The late presenting patient and rebound patient without NMV-r cluster with the other rebound COVID-19 patients. Analysis performed in R using the pheatmap package. One patient with acute COVID-19 and 1 rebound patient without NMV-r (both with BA.5) were excluded from the heat map due to missing biomarker data. Abbreviations: CXCL-10, chemokine (C-X-C motif) ligand 10; HC, healthy controls; IFN-γ, interferon γ; IL, interleukin; IL-18BP, interleukin 18 binding protein; sCD25, soluble CD25; TNF-α, tumor necrosis factor α.
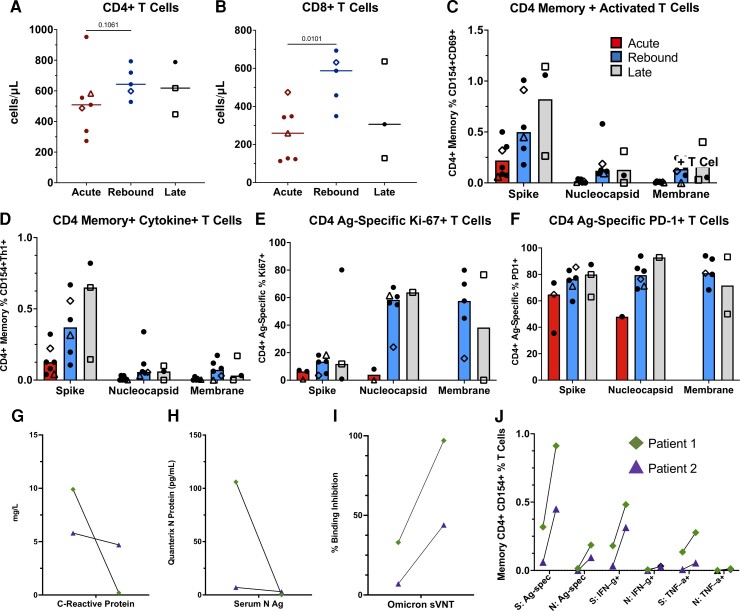
Figure 4.
Comparison of CD4+ T-cell responses across the groups and longitudinal immune responses of the 2 patients with sampling at acute and rebound time points. Absolute T-cell counts compared across groups (A, B). Lines represent median values and points represent individual results. The 2 longitudinal patients are identified by an open triangle and open diamond. The empty square represents the coronavirus disease 2019 rebound patient who did not receive nirmatrelvir-ritonavir. T-cell subset flow cytometry data not available for longitudinal patient 2 (triangle) at rebound time point. T-cell stimulations were performed with peptide pools corresponding to spike, nucleocapsid, and membrane proteins as listed on the x-axis. Bars represent medians and groups are defined as acute, rebound, and late presentations. Severe acute respiratory syndrome coronavirus 2–specific CD4 T-cell responses are highlighted by memory (C), cytokine-producing (CD154 + IFN-γ+, CD154 + TNF-α+ or CD154 + IL-2+) (D), activated (CD154 + CD69+) (E), or Ag-specific proliferating (Ki-67+) and activated (PD-1+) T cells (F). For phenotyping of Ki-67 + and PD-1+ cells, a threshold of at least 20 events and a 2-fold increase over unstimulated cells was used, and samples were excluded if they did not meet these thresholds (E, F). Serum N Ag and C-reactive protein trends from the 2 longitudinal patients (G, H). T-cell responses and neutralizing antibodies from the acute and rebound presentation for 2 patients with longitudinal samples (I, J). T-cell responses are from S and N stimulations. Ag-specific CD4 T cells defined by (CD154 + CD69+, CD154 + IFN-γ+ and CD154 + TNF-α+), and neutralizing antibodies represented by percent binding inhibition on the sVNT. Abbreviations: Ag, antigen; S, spike; N, nucleocapsid; spec, specific; sVNT, surrogate virus neutralization test.
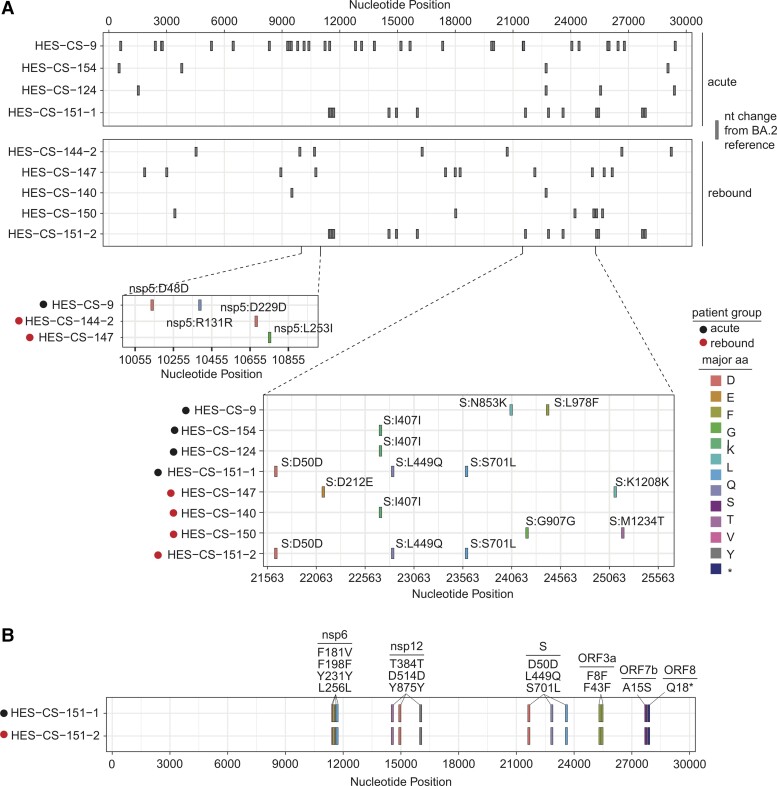
SARS-CoV-2 RNA sequencing was successfully performed on 5 patients with rebound COVID-19 after NMV-r and 4 acute controls. Due to low viral load, sequencing could not be performed for 1 rebound patient and the acute sample from longitudinal patient 2 (sampled while on NMV-r). No mutations associated with NMV resistance were detected. Sequences were compared to the Wuhan/Hu-1 strain (GenBank MN908947.3; Supplementary Figure 4) and the Omicron BA.2 strain (EPI_ISL_7190366; Figure 2A), but no specific mutations could differentiate the acute group from the rebound group. A full list of consensus changes and minority variants compared with the BA.2 reference is provided in Supplementary Table 3. Longitudinal evaluation of patient 1 from acute to rebound time point did not identify any changes to suggest within-host evolution (Figure 2B). Sequencing of cultured virus mapped to the same pangolin lineage as the clinical samples, suggesting that few or no notable mutations arose during the culture process.
Figure 2.
Viral sequencing of severe acute respiratory syndrome coronavirus 2 isolated from acute and rebound groups compared with Omicron BA.2 subvariant. A, Nucleotide mutations in the sequenced isolates from the acute and rebound groups compared with Omicron BA.2 subvariant. Vertical dashes for each isolate correspond to changes from the Omicron BA.2 subvariant. Zoomed images of the nsp5 region and the spike region are shown from isolates from the acute group (HES-CS-9, HES-CS-154, HES-CS-124, and HES-CS-151-1) and rebound group (HES-CS-144-2, HES-CS-147, HES-CS-140, HES-CS-150, and HES-CS-151-2) with specific mutations labeled. B, Longitudinal sequencing data for patient 1. Abbreviations: aa, amino acid; nt, nucleotide.
Anti-S and anti-RBD IgG antibodies were at high levels in both groups (Figure 3A–E), consistent with prior vaccination. Anti-nucleocapsid IgG antibodies were absent in acute disease, detectable in 3 of 5 tested with rebound after NMV-r treatment and in 1 patient with rebound without prior treatment (Figure 1C). Significant variation was found in the IgM and IgA responses (Supplementary Figure 5A–E). sVNT [9] assays showed high levels of wild-type spike neutralizing antibodies in all patients; however, the percent binding inhibition was notably lower to Omicron spike protein, especially in the acute cohort (Figure 3D–E). More rebound patients had detectable neutralizing antibodies against Omicron spike that inversely correlated with serum antigen, although this relationship was less apparent in the 2 patients with the Omicron BA.5 subvariant (Figure 3E, Supplementary Figure 5F).
CD4 and CD8 T-cell counts increased in rebound cases compared with acute disease (Figure 4A–B). T-cell stimulation assays showed robust T-cell responses against the wild-type spike protein with higher levels of antigen-specific, cytokine-producing CD4+ T cells in those with rebound symptoms or late presentation (Figure 4C–D). Stimulations with Omicron spike protein produced similar results with a high correlation (r > 0.94) between the assays, which is consistent with a prior report [12] (Supplementary Figure 6). SARS-CoV-2–specific CD4+ T cells showed greater proliferation (Ki-67+) and activation (PD-1+) at rebound compared with acute presentations. These findings were even more prominent with nucleocapsid and membrane protein T-cell stimulations (Figure 4E–F). No notable difference was found across memory T-cell phenotypes (central, effector, terminal effector; Supplementary Figure 7A–C). Activated, cytokine-producing CD8+ T-cell responses were minimal across all groups, and there was no difference in CD4+ T-cell polyfunctional cytokine responses (Supplementary Figure 7D–E).
Two rebound patients were evaluated longitudinally (Supplementary Table 4). Both were evaluated and started NMV-r within 3 days of symptom onset, had symptom resolution and negative RATs after completing NMV-r, and had symptom return and positive RATs 8 and 5 days after completing NMV-r, respectively. During rebound, both exhibited significant drops in CRP and serum nucleocapsid antigen (Figure 4G–H) with concomitant increases in neutralizing antibodies and SARS-CoV-2–specific, cytokine-producing (IFN-γ, TNF-α) CD4+ T cells (Figure 4I–J).
We also assessed a spectrum of innate and adaptive immune markers. Innate markers such as IL-6, IL-8, and CXCL10 that are known to be associated with severe COVID-19 [13] were downtrending at rebound, whereas markers of adaptive immunity and T-cell activation such as IFN-γ and soluble CD25 were stable or increasing (Figure 5A–B). These biomarkers were incorporated into a comprehensive heat map summarizing clinical, virologic, and immunologic data. Unsupervised, hierarchical clustering identified the rebound COVID-19 patients clustered separately from the acute patients. This difference was driven primarily by the lower acute inflammatory markers and increased adaptive immune responses in the rebound cohort (Figure 5C).
DISCUSSION
NMV-r has been a long-awaited addition to the COVID-19 therapeutic armamentarium, providing an outpatient oral medication that can significantly improve disease prognosis in high-risk patients. Cases of clinical rebound after NMV-r reported recently have raised concerns about clinical deterioration and interference of early antiviral administration with the development of adaptive immune responses. The licensing trial did not identify significant differences in rebound incidence among NMV-r recipients vs placebo, although vaccinated patients were not included and the trial occurred during the Delta wave [2]. In contrast, NMV-r is now widely used for breakthrough infections by the Omicron variant, which may impact the incidence of clinical rebound. Although retrospective studies have suggested a low incidence [14], prospective epidemiologic studies will be required to more accurately measure the incidence and risk factors for rebound COVID-19 and compare them in those treated vs not treated with NMV-r.
In our case series of 6 patients with rebound COVID-19 symptoms after completing NMV-r and 2 patients with rebound symptoms without prior antiviral therapy, none of the patients developed severe symptoms or required additional therapy. High levels of SARS-CoV-2 anti-S IgG antibodies were found in all patients, consistent with prior vaccination. Anti-nucleocapsid IgG and Omicron-specific neutralizing antibodies were increased in patients with rebound symptoms. The development of sterilizing humoral immunity has been previously reported to occur between 2 and 3 weeks post-infection in a pre-vaccine and pre-Omicron era cohort [15]. As the median time from symptom onset to sample collection in our cohort was 16 days, we found no evidence of delayed development of sterilizing humoral immunity by this time line.
These findings are consistent with those from a report that showed higher virus neutralizing antibodies in a patient experiencing rebound symptoms after NMV-r compared with an uninfected, vaccinated, and boosted control [16]. Additionally, we detected robust cytokine-producing, proliferating, activated SARS-CoV-2–specific T-cell responses that were greater than those with acute COVID-19, along with rising T-cell counts in rebound patients. Two patients with longitudinal sampling demonstrated an increase in both antibody and cellular immune responses during rebound compared with their acute presentation. These findings argue against the hypothesis that impaired humoral and cellular immune responses lead to symptomatic rebound.
Resistance mutations were not identified at COVID-19 rebound, consistent with prior studies [3, 10, 16]. Interestingly, SARS-CoV-2 was isolated from culture in 1 of 8 patients with rebound compared with 6 of 7 acute controls. Although a recent report identified virus growth in samples from 3 of 7 rebound patients after NMV-r [10], Polybrene, which can increase virus adsorption to host cells, was used [11], and it is unclear if this is more reflective of clinical transmission potential of the virus. In our repeat virus cultures using Polybrene, 5 of 8 rebound patients were positive, highlighting this important difference in culture sensitivity. Further study is required to determine which technique most accurately predicts transmissibility as this is an important epidemiological point. Additionally, this raises the question of whether longer treatment could reduce the incidence of rebound symptoms. The rebound patient with culturable SARS-CoV-2 without Polybrene had underlying immunosuppression, making this issue particularly important for immunocompromised populations. The virus was isolated from the nasal swab while the serum nucleocapsid antigen was negative at rebound, and both antibodies and T-cell responses were higher than during acute infection, suggesting a more localized viral reactivation. However, this may not be the case in patients unable to mount a successful adaptive immune response.
Our findings of lower levels of serum nucleocapsid antigen and downtrending innate immune markers in rebound COVID-19 with an emerging adaptive immune response do not support a role of uncontrolled viral replication in driving inflammation or a significant risk for impending disease progression. Severe COVID-19 is characterized by myeloid cell activation and rises in innate biomarkers in the presence of mostly ineffective T-cell responses. In our cases, increases in both total and virus-specific T lymphocytes, biomarkers of T-cell activation, and rising antibodies suggest that the rebound symptoms may in fact be partially driven by the emerging immune response against residual viral antigens throughout the respiratory tract, which may be more clinically evident after use of potent antiviral treatment with quick clinical improvement [17]. Unsupervised clustering analysis of our comprehensive dataset revealed a distinct separation of the rebound COVID-19 patients from those with acute infection, highlighting the unique viro-immunologic profile of this rebound population. The time course of 2 weeks correlates with emergence of antibody-mediated cytotoxicity and with tissue repair, which can both cause release of virus from previously infected cells [17–19]. As dying cells shed antigen, this could lead to return of antigen positivity, and the developing adaptive immune response may drive the recurrence of symptoms. This could be more prominent in vaccinated people and could occur in both the presence and absence of NMV-r, though it may be more noticeable in patients taking NMV-r given the rapid clinical improvement many patients experience, whereas those not taking NMV-r may have prolonged symptoms that coalesce into this phase of the illness.
Our study has a few important limitations. Although we demonstrated differences in Omicron-specific neutralizing antibody levels between the groups that negatively correlated with SARS-CoV-2 antigenemia, we did not evaluate the functional activity of these antibodies. Recently, Fc-gamma receptor–mediated phagocytosis and activation of complement has been found to correlate with humoral immune protection [20], and further evaluation of these mechanisms in rebound COVID-19 should be pursued in future studies. Our sample size for each group was small, in particular, the late group, as patients who were 8–15 days from symptom onset and not exhibiting rebound symptoms were not actively recruited in this protocol. Moving forward, it will be important to recruit patients at this time point to evaluate immune responses of patients experiencing rebound after NMV-r and compare these to individuals with or without rebound symptoms in the absence of NMV-r. Additionally, longitudinal data were only available for 2 patients. Despite this limitation, it is worth noting that the within-person trends between their acute and rebound visits mirror the cross-sectional findings.
In conclusion, this case series provides important insights into the pathophysiology of rebound COVID-19 after NMV-r. None of our patients developed severe disease at rebound, and adaptive immunity against SARS-CoV-2 appeared intact. Our findings suggest that a more robust immune response rather than uncontrolled viral replication characterizes these clinical rebounds. Special consideration should be given for immunocompromised patients who cannot rely on adaptive immune responses and therefore may require prolonged or additional therapies. Further detailed evaluation in larger cohorts is required to assess the incidence, clinical, and, importantly, epidemiologic implications of rebound COVID-19.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Brian P Epling, Laboratory of Immunoregulation, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Joseph M Rocco, Laboratory of Immunoregulation, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Kristin L Boswell, Immunology Laboratory, Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Elizabeth Laidlaw, Laboratory of Immunoregulation, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Frances Galindo, Laboratory of Immunoregulation, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Anela Kellogg, Clinical Research Directorate, Frederick National Laboratory for Cancer Research, Leidos Biomedical Research, Frederick, Maryland, USA.
Sanchita Das, Department of Laboratory Medicine, Clinical Center, National Institutes of Health, Bethesda, Maryland, USA.
Allison Roder, Systems Genomics Section, Laboratory of Parasitic Diseases, Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Elodie Ghedin, Systems Genomics Section, Laboratory of Parasitic Diseases, Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Allie Kreitman, Systems Genomics Section, Laboratory of Parasitic Diseases, Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Robin L Dewar, Virus Isolation and Serology Laboratory, Frederick National Laboratory, Frederick, Maryland, USA.
Sophie E M Kelly, Trans-NIH Shared Resource on Biomedical Engineering and Physical Science, National Institute of Biomedical Imaging and Bioengineering, National Institutes of Health, Bethesda, Maryland, USA.
Heather Kalish, Trans-NIH Shared Resource on Biomedical Engineering and Physical Science, National Institute of Biomedical Imaging and Bioengineering, National Institutes of Health, Bethesda, Maryland, USA.
Tauseef Rehman, Virus Isolation and Serology Laboratory, Frederick National Laboratory, Frederick, Maryland, USA.
Jeroen Highbarger, Virus Isolation and Serology Laboratory, Frederick National Laboratory, Frederick, Maryland, USA.
Adam Rupert, AIDS Monitoring Laboratory, Frederick National Laboratory, Frederick, Maryland, USA.
Gregory Kocher, Integrated Research Facility at Fort Detrick, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Frederick, Maryland, USA.
Michael R Holbrook, Integrated Research Facility at Fort Detrick, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Frederick, Maryland, USA.
Andrea Lisco, Laboratory of Immunoregulation, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Maura Manion, Laboratory of Immunoregulation, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Richard A Koup, Immunology Laboratory, Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Irini Sereti, Laboratory of Immunoregulation, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA.
Notes
Disclaimer. The content of this publication does not necessarily reflect the views or policies of the US Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the US government.
Financial support. This work was supported in part by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH). R. L. D. and J. H. report support from the NIAID (HHSN261201500003I). A. Kellogg reports support from the National Cancer Institute, NIH (contract HHSN2612015000031 or 75N91019D00024). R. A. K. reports support from NIAID Intramural research funds.
Ethics approval. All patients evaluated at the NIH provided written informed consent and were enrolled on NCT04401436. This protocol was approved by the NIH Institutional Review Board.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Center for Drug Evaluation and Research (CDER) . Emergency Use Authorization (EUA) for Paxlovid (nirmatrelvir tablets co-packaged with ritonavir tablets) Center for Drug Evaluation and Research (CDER) Review. Silver Spring, MD: Food and Drug Administration, 2021. Available at: https://www.fda.gov/media/155194/download. Accessed 27 August 2022.
- 2. Hammond J, Leister-Tebbe H, Gardner A, et al. . Oral nirmatrelvir for high-risk, nonhospitalized adults with COVID-19. N Engl J Med 2022; 386:1397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Charness M, Gupta K, Stack G, et al. . Rapid relapse of symptomatic Omicron SARS-CoV-2 infection following early suppression with nirmatrelvir/ritonavir. Research Square [Preprint]. May 23, 2022. [cited 2022 Aug 9]. Available from: 10.21203/rs.3.rs-1588371/v3. [DOI] [Google Scholar]
- 4. Gupta K, Strymish J, Stack G, et al. . Rapid relapse of symptomatic SARS-CoV-2 infection following early suppression with nirmatrelvir/ritonavir. Research Square [Preprint, Version 1]. April 26, 2022. [cited 2022 Aug 9]. Available from: 10.21203/rs.3.rs-1588371/v1.. [DOI] [Google Scholar]
- 5. Nussenblatt V, Roder AE, Das S, et al. . Yearlong COVID-19 infection reveals within-host evolution of SARS-CoV-2 in a patient with B-cell depletion. J Infect Dis 2022; 225:1118–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bennett RS, Postnikova EN, Liang J, et al. . Scalable, micro-neutralization assay for assessment of SARS-CoV-2 (COVID-19) virus-neutralizing antibodies in human clinical samples. Viruses 2021; 13:893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. ACTIV-3/TICO Bamlanivimab Study Group, Lundgren JD, Grund B, et al. . Responses to a neutralizing monoclonal antibody for hospitalized patients with COVID-19 according to baseline antibody and antigen levels: a randomized controlled trial. Ann Intern Med 2022; 175:234–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Klumpp-Thomas C, Kalish H, Drew M, et al. . Standardization of ELISA protocols for serosurveys of the SARS-CoV-2 pandemic using clinical and at-home blood sampling. Nat Commun 2021; 12:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tan CW, Chia WN, Qin X, et al. . A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat Biotechnol 2020; 38:1073–8. [DOI] [PubMed] [Google Scholar]
- 10. Boucau J, Uddin R, Marino C, et al. . Characterization of virologic rebound following nirmatrelvir-ritonavir treatment for COVID-19. Clin Infect Dis 2022. doi: 10.1093/cid/ciac512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Davis HE, Morgan JR, Yarmush ML. Polybrene increases retrovirus gene transfer efficiency by enhancing receptor-independent virus adsorption on target cell membranes. Biophys Chem 2002; 97:159–72. [DOI] [PubMed] [Google Scholar]
- 12. Keeton R, Tincho MB, Ngomti A, et al. . T cell responses to SARS-CoV-2 spike cross-recognize Omicron. Nature 2022; 603:488–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lucas C, Wong P, Klein J, et al. . Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 2020; 584:463–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ranganath N, O'Horo JC, Challener DW, et al. . Rebound phenomenon after nirmatrelvir/ritonavir treatment of coronavirus disease-2019 in high-risk persons. Clin Infect Dis 2022. doi: 10.1093/cid/ciac481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Long QX, Liu BZ, Deng HJ, et al. . Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 2020; 26:845–8. [DOI] [PubMed] [Google Scholar]
- 16. Carlin AF, Clark AE, Chaillon A, et al. . Virologic and immunologic characterization of COVID-19 recrudescence after nirmatrelvir/ritonavir treatment. Clin Infect Dis 2022. doi: 10.1093/cid/ciac496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sefik E, Qu R, Junqueira C, et al. . Inflammasome activation in infected macrophages drives COVID-19 pathology. Nature 2022; 606:585–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lage SL, Amaral EP, Hilligan KL, et al. . Persistent oxidative stress and inflammasome activation in CD14(high)CD16(-) monocytes from COVID-19 patients. Front Immunol 2022; 12:799558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lage SL, Rocco JM, Laidlaw E, et al. . Activation of complement components on circulating blood monocytes from COVID-19 patients. Front Immunol 2022; 13:815833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang C, Li Y, Kaplonek P, et al. . The kinetics of SARS-CoV-2 antibody development is associated with clearance of RNAemia. mBio 2022; e0157722. doi: 10.1128/mbio.01577-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.