Abstract
The microtubule (MT) cytoskeleton provides the architecture that governs intracellular organization and the regulated motion of macromolecules through the crowded cytoplasm. The key to establishing a functioning cytoskeletal architecture is regulating when and where new MTs are nucleated. Within the spindle, the vast majority of MTs are generated through a pathway known as branching MT nucleation, which exponentially amplifies MT number in a polar manner. Whereas other MT nucleation pathways generally require a complex organelle such as the centrosome or Golgi apparatus to localize nucleation factors, the branching site is based solely on a simple, preformed MT, making it an ideal system to study MT nucleation. In this review, we address recent developments in characterizing branching factors, the branching reaction, and its regulation, as well as branching MT nucleation in systems beyond the spindle and within human disease.
Keywords: mitosis, meiosis, branching microtubule nucleation, γ-tubulin ring complex, γ-TuRC, augmin, TPX2
INTRODUCTION
The closest equivalent in the cell to the vertebrate skeleton is the microtubule (MT) cytoskeleton. It gives cells their shape and organizes their interior, generates diverse structures ranging from axons that transmit action potentials to cilia that generate force for movement, and forms spindles that segregate chromosomes. For many decades, centrosomes were thought to be the sole originators and organizers of the MT cytoskeleton and hence were dubbed the cell’s MT- organizing center (MTOC). Yet, it has become clear that a cell has, in fact, many other MTOCs, such as the Golgi apparatus, the nuclear envelope, and the plasma membrane, with new MTOCs being discovered on a regular basis. While many of these sites are complex organelles themselves, who would have guessed that MTs could also originate from MTs themselves? Consisting only of an MT seed and a few cytoplasmic factors, this possibly simplest MTOC is one of the most prevalent nucleation sites in the spindle: the site of MT-dependent MT nucleation, or branching MT nucleation.
Branching MT nucleation was first observed in Schizosaccharomyces pombe: Its small cell size and finite number of ~40 MTs allowed the observation of MTs that branch off existing MTs in an antiparallel fashion (at 180° angles) (Janson et al. 2005). Around the same time, MTs that branch off MTs were observed in plant cells at the cell cortex, where MT nucleation events could be clearly visualized (Chan et al. 2009, Murata et al. 2005). In plants, branching MT nucleation takes on quite a different morphology and produces daughter MTs roughly parallel relative to the mother MT (at 0 to 40° angles).
Inspired by these findings, MTs in animal spindles were proposed to originate not only from centrosomes but also within the spindle itself (Brugues et al. 2012, Mahoney et al. 2006). Intriguingly, the MT nucleator γ-tubulin was found to localize to the body of the spindle, further suggesting that MTs might be generated there locally. Eight genes that target γ-tubulin to spindle MTs but not centrosomes were identified—named dim γ-tubulin, or Dgt in Drosophila (Goshima et al. 2007)—and the complex was christened augmin, as it augments MTs in the spindle (Goshima et al. 2008, Hughes et al. 2008, Meireles et al. 2009). The augmin complex was also observed in human cells (Lawo et al. 2009, Uehara et al. 2009), where its subunits were coined HAUS1–8 (for homologous to augmin subunit). In its absence, Drosophila S2 spindles become elongated, lack MTs in their centers, and display only faint kinetochore fibers (Goshima et al. 2008, Lawo et al. 2009). This phenotype is more dramatic in human cells, where cells also arrest during cytokinesis (Uehara & Goshima 2010), and in acentrosomal meiotic spindles, where spindles become asymmetric and multipolar (Petry et al. 2011). Meanwhile, evidence both from in-cell MT scission experiments (Brugues et al. 2012) and theoretical modeling of spindle assembly (Clausen & Ribbeck 2007, Loughlin et al. 2010) pointed to the importance of spindle-derived MTs in animal cells. Yet, due to technical hurdles, how augmin and γ-tubulin were generating these MTs within the spindle body remained a mystery.
The very high MT density within the spindle body precludes resolving individual MT nucleation events via light microscopy. In contrast, electron microscopy (EM) methods provide high resolution, yet offer only a limited field of view at a single point in time, which is a poor approximation for the thousands of spindle MTs that turn over in seconds. These roadblocks were circumvented by using cell-free extracts of Xenopus eggs in conjunction with total internal fluorescence (TIRF) microscopy, which offered at least two advantages: (a) TIRF microscopy provides a high signal-to-noise ratio, and (b) the cell-free extract allowed the isolation of a single MT nucleation pathway, namely branching MT nucleation, without interference from MTs generated from other MTOCs. Thus, new MTs could clearly be seen nucleating from the sides of preexisting MTs in the mitotic extract, surprisingly at very shallow and mostly parallel branch angles (Petry et al. 2013). This was then directly observed within cells at anaphase close to the cell membrane (Verma & Maresca 2019) and inferred via light-sheet microscopy (David et al. 2019). Branching MT nucleation is now considered the major source of spindle MTs (Decker et al. 2018), with its newly discovered roles in interphase elevating its significance in biology (W.S. Chen et al. 2017, Cunha-Ferreira et al. 2018, Sanchez-Huertas et al. 2016) (Figure 1).
Figure 1.
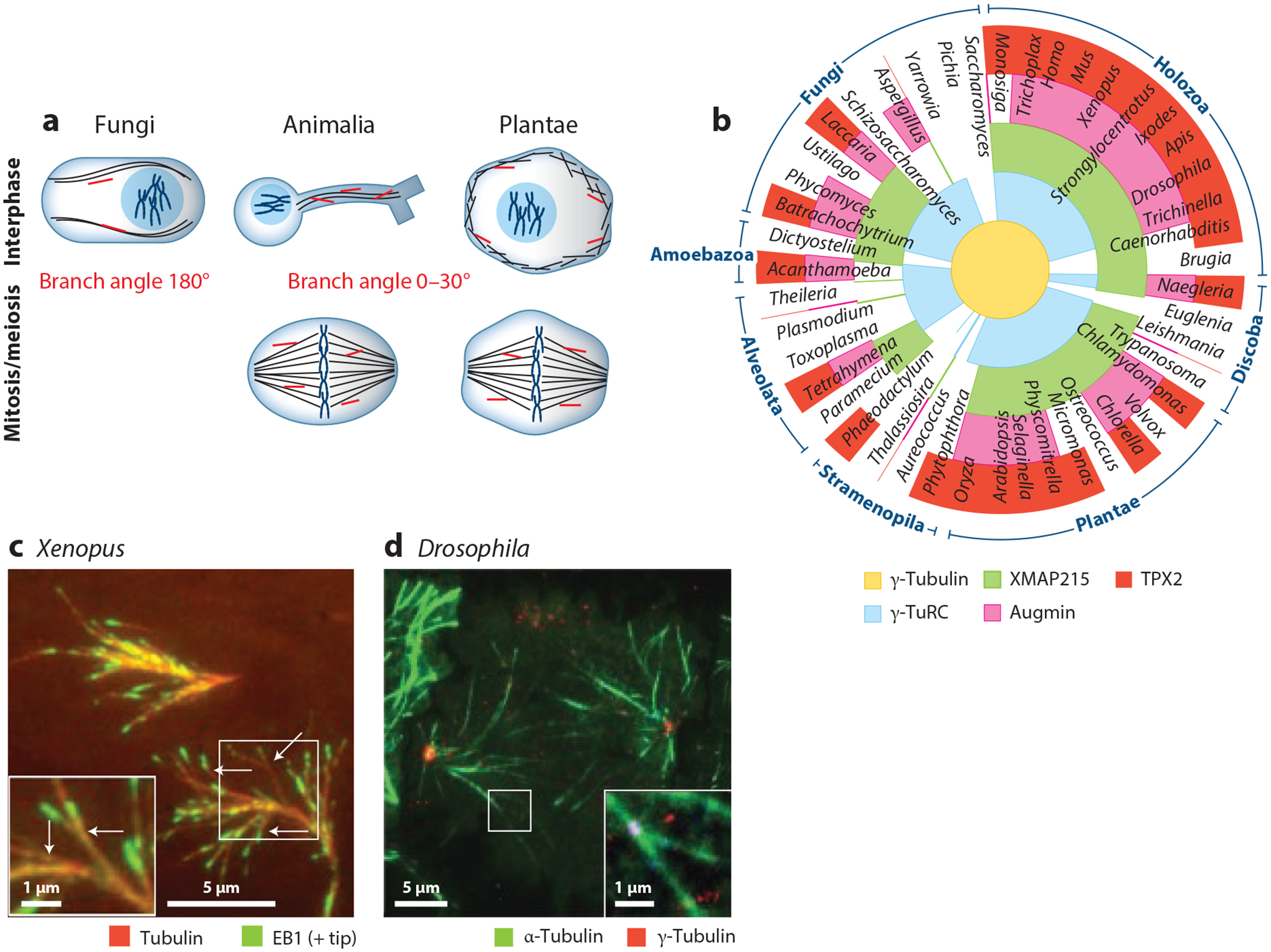
Branching microtubule (MT) nucleation is conserved across most eukaryotes and cell types. (a) Branching MT nucleation has been studied in both interphase and mitosis/meiosis across fungi, animals, and plants. MT branches are displayed in orange, and the ranges of branch angles relative to the parent MT are annotated at the center, where at 0° the two MTs are parallel and at 180° they are antiparallel. (b) Diverse eukaryotic species are represented as spokes in a circular eukaryotic phylogeny, in which phylum groupings are annotated on the exterior of the circle. The radial sections of each spoke are colored based on the presence or absence of nucleation and branching factors: from the center, γ-tubulin (yellow), γ-tubulin ring complex (γ-TuRC) subunit γ-tubulin complex protein 4 (GCP4) (blue), Xenopus MT-associated protein of 215 kDa (XMAP215) with a tumor overexpressed gene 6 (TOG6) domain to enable binding to γ-TuRC (green), augmin subunit Haus6 (pink), and targeting protein for Xklp2 (TPX2) (red). (c) Ex vivo visualization of branching MT nucleation in meiotic Xenopus laevis egg extract system. MTs are labeled in red via Alexa568 tubulin and growing plus ends via green fluorescent protein (GFP)-end-binding protein 1 (EB1). Arrows indicate branch sites. Panel c reproduced from Petry et al. (2013). (d) Visualization of MT branching in mitotic Drosophila melanogaster S2 cells during anaphase. Microtubules are labeled in green via GFP-α-tubulin and branch sites via γ-tubulin-red fluorescent protein (RFP). Panel d reproduced with permission from Verma & Maresca (2019).
Whereas other reviews have focused on branching MT nucleation in plants (Yi & Goshima 2018) and the augmin complex (Sanchez-Huertas & Luders 2015), we focus on branching MT nucleation during animal cell division. We first introduce and describe the major players in branching MT nucleation. Next, we describe how the MT branch site is established before a daughter MT is nucleated. This involves the deposition of the phase-separating targeting protein for Xklp2 (TPX2), followed by recruitment of the augmin complex, which in turn binds to the nucleators, the γ-tubulin ring complex (γ-TuRC) and XMAP215, to generate the daughter MT. Finally, we touch upon other MT branching systems and the link between this pathway and human health.
INDIVIDUAL FACTORS IN MICROTUBULE BRANCHING
γ-TuRC
γ-TuRC localizes to many cellular MTOCs and is essential for nucleating the majority of MTs in the cell (Roostalu & Surrey 2017, Tovey & Conduit 2018). γ-TuRC is composed of 14 copies of the tubulin paralog γ-tubulin assembled into a spiral shape (Kollman et al. 2010, Zheng et al. 1995). γ-Tubulin was first discovered in Aspergillus (Oakley & Oakley 1989) and later shown to be ubiquitous across eukaryotes (Findeisen et al. 2014). Within γ-TuRC, the γ-tubulin monomers approximate an MT end (Consolati et al. 2020, Liu et al. 2020, Wieczorek et al. 2020b), leaving the top of the spiral open to bind α-/β-tubulin dimers, which will form the 13-fold symmetric hollow helix of the growing MT (Oakley et al. 1990, Thawani et al. 2020).
In order to organize the γ-tubulin spiral, γ-TuRC contains γ-tubulin complex proteins (GCPs), paralogous adaptor subunits that directly bind and orient γ-tubulin (Guillet et al. 2011). Some eukaryotes, including Saccharomyces, in which γ-TuRC was first discovered, build γ-tubulin rings from seven copies of the γ-tubulin small complex (γ-TuSC), a heterotetramer composed of GCP2, GCP3, and two molecules of γ-tubulin (Knop et al. 1997, Zupa et al. 2020). However, in the last eukaryotic common ancestor, GCP2 and GCP3 appear to have duplicated and diverged to generate three additional paralogs that substitute for GCP2/3 at specific locations around the ring (Consolati et al. 2020, Liu et al. 2020, Wieczorek et al. 2020b) (Figure 2). The interfaces connecting adjacent GCPs are relatively small and thus presumably weakly associated, allowing the GCP spiral to breathe and reposition the attached γ-tubulins (Consolati et al. 2020, Liu et al. 2020, Wieczorek et al. 2020b). This conformational flexibility has been suggested to let γ-TuRC adopt a range of geometries that, depending on how closely the γ-tubulin helix mimics an MT, modulate γ-TuRC’s nucleation potential (Kollman et al. 2010).
Figure 2.
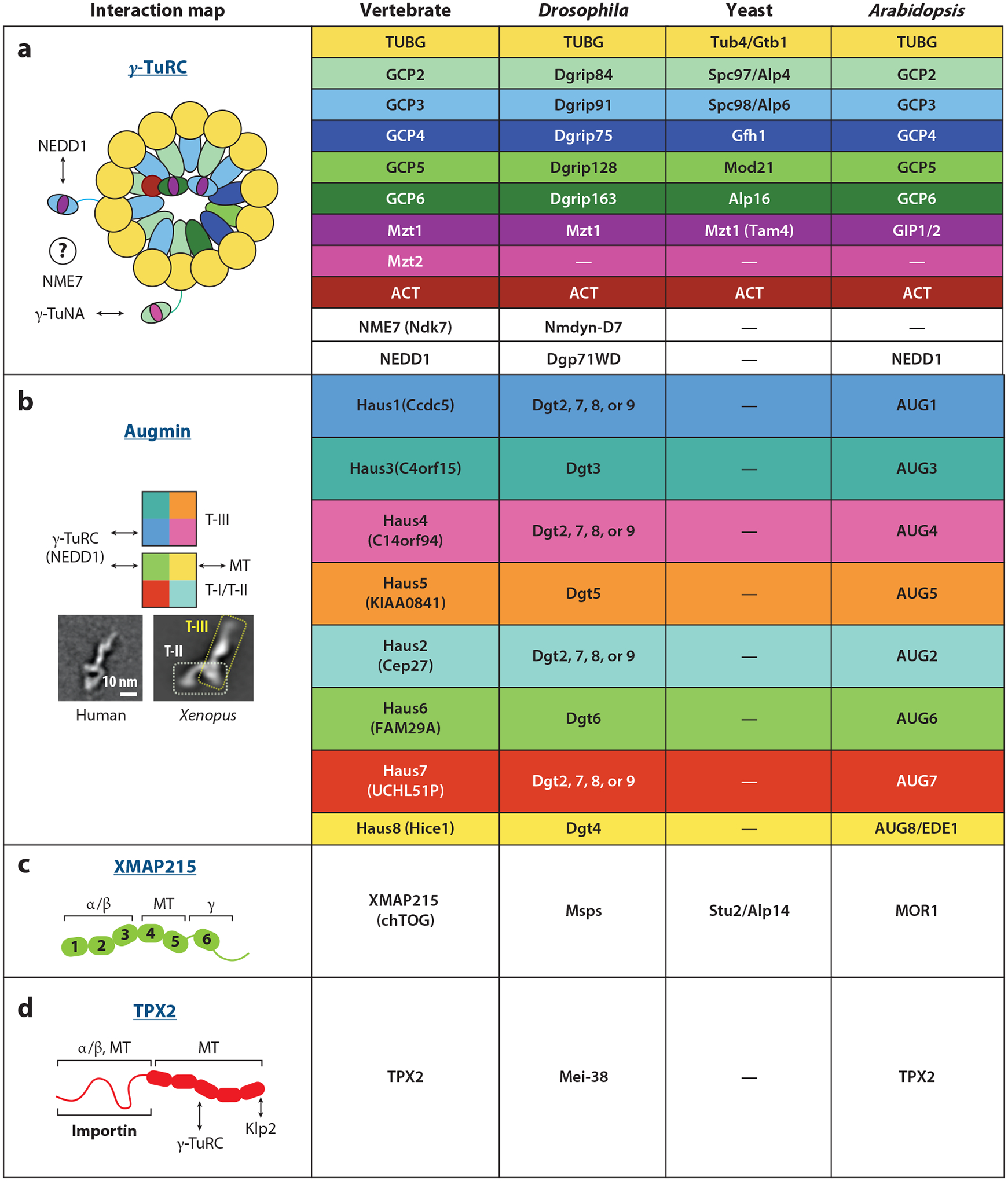
Orthologous branching factors across model organisms. (a) The nucleator γ-TuRC consists of a spiral of 14 γ-tubulins scaffolded by GCPs. Mzt/GCP complexes occupy diverse positions throughout the ring, binding the accessory subunit NEDD1 and the localizer/activator γ-TuNA. The cartoon of γ-TuRC is based on data from Consolati et al. (2020), Liu et al. (2020), and Wieczorek et al. (2020a,b). (b) The augmin complex is an “h” shaped hetero-octamer separable into the MT-binding tetramer I/II (T-I/II) and γ-TuRC-binding tetramer III (T-III). Augmin is lost in yeast, and low sequence homology prevents four of the eight Drosophila augmin subunits (Dgt2, Dgt7, Dgt8, and Dgt9) from being matched to their vertebrate and plant counterparts. The 2D classification of human augmin was reproduced with permission from Hsia et al. (2014); the 2D classification of Xenopus augmin was reproduced from Song et al. (2018). (c) The microtubule polymerase and nucleator XMAP215 consists of multiple tubulin-binding TOG domains, where TOG1–3 bind soluble tubulin, TOG4–5 bind polymerized tubulin, and TOG6 binds γ-tubulin. (d) TPX2, a large, poorly-ordered protein, binds tubulin in various forms, as well as itself, γ-TuRC, and importin regulatory proteins. The cartoon of TPX2 is based on data from Alfaro-Aco et al. (2017). Abbreviations: γ-TuNA, γ-tubulin nucleator activator; γ-TuRC, γ-tubulin ring complex; ACT, actin; Alp, altered polarity; AUG, augmin; CCDC, coiled-coil domain containing protein; Cep, centrosomal protein; chTOG, colonic and hepatic tumor overexpressed gene; Dgp, Drosophila γ-ring protein; Dgt, dim γ-tubulin; Dgrip, Drosophila γ-ring protein; EDE, endosperm defective; FAM, family with sequence similarity; GCP, γ-tubulin complex protein; Gfh, GCP4 homolog; GIP, GCP3-interacting protein; Haus, homologous to augmin subunit; Hice, Hec1-interacting and centrosome-associated; KIAA, Kazusa cDNA sequencing project gene; Klp, kinesin-like protein; Mei, meiotic mutant; Mod, morphology defective; MOR, microtubule organization; Msps, minispindles; MT, microtubule; Mzt, mitotic spindle organizing proteins associated with a ring of γ-tubulin; NEDD, neural precursor cell expressed, developmentally-downregulated; NME, non-metastatic cells; Spc, spindle pole component; Stu, suppressor of tubulin; Tam, transcript altered in meiosis; TPX, targeting protein for Xenopus kinesin-like protein; TUB, tubulin; UCHL5, ubiquitin carboxy-terminus hydrolase isozyme L5; XMAP215, Xenopus MT-associated protein of 215 kDa.
Along with GCPs, γ-TuRC contains accessory proteins that provide structural stability or allow attachment of recruitment factors. Tightly associated with the GCPs are the small paralogs Mzt1 and Mzt2, which intercalate into the N-terminal extensions (NTEs) of GCP3/5/6 and GCP2, respectively (Huang et al. 2020, Wieczorek et al. 2020a). These small Mzt/NTE modules are attached via long linkers to the core of γ-TuRC, modulating the dynamics of the ring (Wurtz et al. 2022) and binding γ-TuRC recruitment factors, such as the γ-tubulin nucleator activator (γ-TuNA, also called the CM1 domain) of the centrosomal anchor CDK5RAP2 (Wieczorek et al. 2020a). Further structural support is provided by a single nonpolymerizing actin molecule. Finally, two other subunits, NEDD1 (neural precursor cell expressed developmentally downregulated protein 1) and NME7, are present in at least one copy in the complex; however, their locations are unclear. NME7 is a poorly understood kinase of the nucleotide-diphosphate family [reviewed in Boissan et al. (2018)] that associates with γ-TuRC and promotes nucleation through unknown mechanisms likely involving phosphorylation (Liu et al. 2014). NEDD1 is a WD40 domain–containing protein (Gunawardane et al. 2003) integrated into γ-TuRC through binding of its disordered C-terminus to NTE/Mzt1 modules of GCP3 and/or GCP5 (Cota et al. 2017, Haren et al. 2006). NEDD1 is broadly required for γ-TuRC localization to both the centrosome (Haren et al. 2006) and to spindle MTs (Luders et al. 2006), suggesting a role in branching MT nucleation.
XMAP215
Best characterized as an MT polymerase, XMAP215 (Xenopus MT-associated protein of 215 kDa) (Gard & Kirschner 1987) is also essential in MT nucleation, including branching MT nucleation, in which it binds and acts in concert with γ-TuRC (Flor-Parra et al. 2018, Gunzelmann et al. 2018, Thawani et al. 2018). XMAP215 is comprised of a variable number of tubulin-binding TOG (tumor overexpressed gene) domains (Gard et al. 2004). Different TOG domains within XMAP215 have duplicated and specialized to bind different tubulins and/or tubulin conformers (Figure 2). TOG1–5 each bind to α-/β-tubulin: TOG1/2/3 preferentially bind to soluble tubulin (Howard et al. 2015, Nithianantham et al. 2018, Widlund et al. 2011), and TOG4/5 recognize the curved tubulin lattice (Byrnes & Slep 2017, Fox et al. 2014, Widlund et al. 2011). These TOG domains enable XMAP215 to concentrate and load new tubulin dimers at the growing MT plus end (Brouhard et al. 2008). In contrast, the C-terminal region of XMAP215, containing a sixth divergent TOG domain (Rostkova et al. 2018), binds γ-tubulin and allows XMAP215 to collaborate with γ-TuRC to promote MT nucleation (Thawani et al. 2018). Based on this interaction, multiple copies of XMAP215 could decorate the outside of the γ-tubulin spiral, locally concentrating α-/β-tubulin monomers and loading them onto the γ-tubulin ring.
Augmin
The augmin complex acts as a bridge between γ-TuRC and the template MT, thus recruiting the nucleation module to its correct location. Augmin has been characterized in Drosophila (Goshima et al. 2008), humans (Hutchins et al. 2010, Lawo et al. 2009), and Arabidopsis (Ho et al. 2011). During cell division, augmin localizes to the spindle, where it is essential for the recruitment of γ-tubulin (Goshima et al. 2008, Wainman et al. 2009). Augmin complexes are composed of eight subunits, named in vertebrates Haus1 through Haus8 (Figure 2). Haus3/5/6/8 are easiest to detect via sequence homology, likely because they comprise the important binding sites for γ-TuRC and MTs, whereas the remaining four subunits cannot be matched across species by their primary sequence. Vertebrate augmin has been structurally characterized at low resolution. Both human (Hsia et al. 2014) and Xenopus augmin (Song et al. 2018) form an “h” shape (Figure 3), where the stalk of the “h” is relatively rigid and extends ~40 nm. Augmin assembles from tetrameric subcomplexes. The first consists of Haus2/6/7/8—called T-I in human and T-II in Xenopus—and forms the arch at the base of the “h” (Hsia et al. 2014, Song et al. 2018). The other four subunits, Haus1/3/4/5, form T-III, the stalk of the “h” (Song et al. 2018). These two tetramers may connect via Haus1/4 and Haus6/8, because these form an additional tetramer in human augmin (Hsia et al. 2014); however, in Xenopus, this third tetrameric subcomplex could not be recapitulated (Song et al. 2018).
Figure 3.
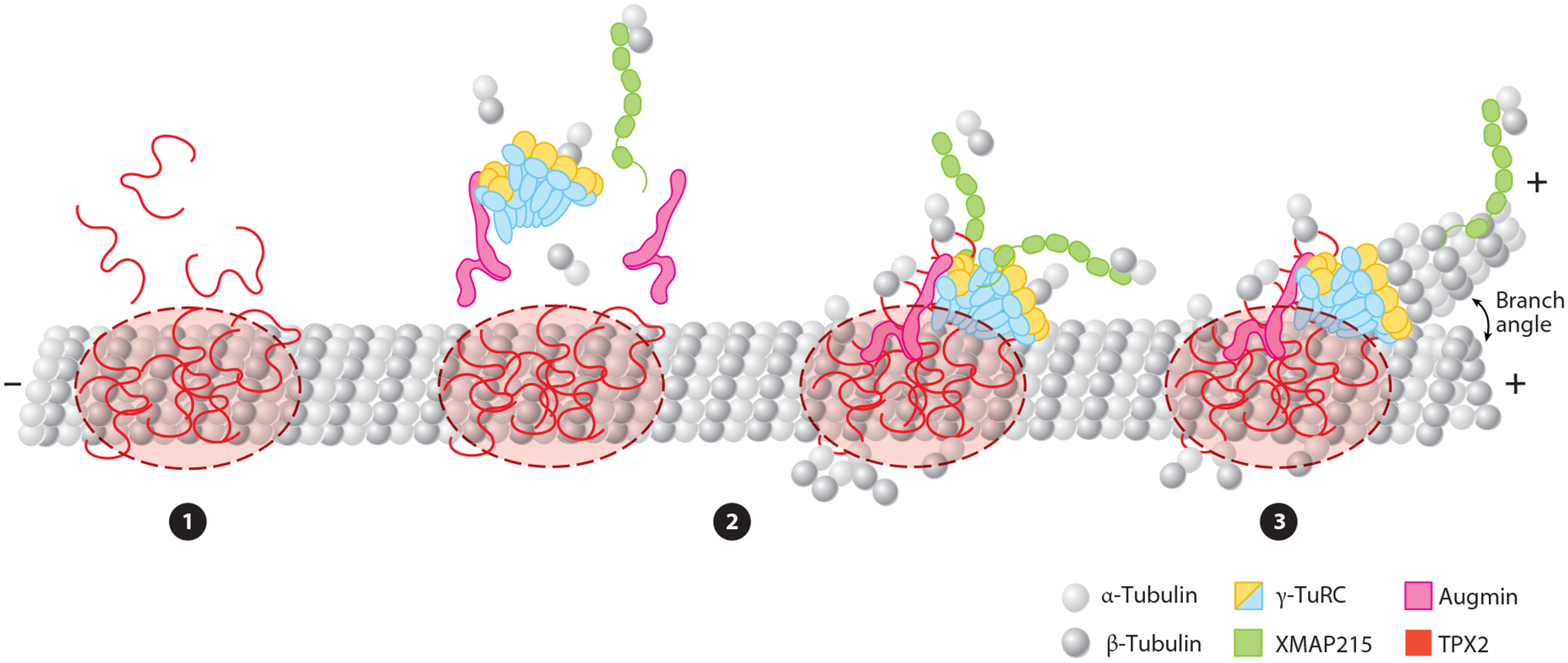
The temporal order of branching microtubule (MT) nucleation. ❶ TPX2 condenses into a liquid-like phase on the MT. ❷ Soluble tubulin and branching factors are recruited to the targeting protein for Xklp2 (TPX2) droplet. Augmin and Xenopus MT-associated protein of 215 kDa (XMAP215) may be recruited individually or in complex with γ-tubulin ring complex (γ-TuRC), and additional soluble tubulin may be recruited by XMAP215 and γ-TuRC. ❸ γ-TuRC with XMAP215 nucleates a new MT at an acute angle to the parent MT. After nucleation of the daughter MT, some molecules of XMAP215 track the plus tip of the growing MT.
Although these tetrameric subcomplexes of augmin do not appear to exist independently in the cell, they fulfill distinct functions by binding to γ-TuRC and to spindle MTs. The T-I/T-II arch binds to MTs via the disordered N-terminal polybasic region of Haus8 (Hsia et al. 2014, Wu et al. 2008). Because purified T-I/T-II has a tenfold higher affinity for MTs than does the N-terminus of Haus8 alone (Hsia et al. 2014), this tetramer is thought to contain a secondary binding site for MTs. The binding site or sites for γ-TuRC are less clear. Early reports suggested that the disordered C-terminus of human Haus6 might bind the γ-TuRC subunit NEDD1 (Bucciarelli et al. 2009, Uehara et al. 2009); however, further work with Xenopus and Drosophila augmins pointed to T-III (Song et al. 2018) and, more specifically, the N-termini of the Haus3/5 dimer (J.W.C. Chen et al. 2017) as the primary NEDD1/γ-TuRC binding site. The major outstanding questions are where augmin’s subunits are located, what they look like, and how precisely they bind γ-TuRC and ultimately orient the MT branch angle. Answering these will depend on a better structural characterization of the complex both on its own and while bound to γ-TuRC and the MT.
TPX2
Even though augmin is sufficient to target γ-TuRC to spindle MTs in Drosophila and in vitro, it came as a surprise that, in Xenopus, one more protein is required. TPX2 was first discovered for its role in bringing the mitotic kinesin Xklp2 (Kif15) to spindle poles (Wittmann et al. 1998), and it also binds the kinesin Eg5 (Kif11) (Eckerdt et al. 2008, Ma et al. 2010). However, TPX2 was soon demonstrated to enhance branching MT nucleation by interacting with a number of the key players. TPX2 binds to and stabilizes the MT lattice (Reid et al. 2016, Roostalu et al. 2015, Wieczorek et al. 2015). This effect can be explained by a recent cryoEM structure, which demonstrated that a small fragment of TPX2 (residues 300–339) bridges and stabilizes adjacent tubulin dimers, opposing the MT lattice conformational change that occurs as MTs age and hydrolyze GTP (Zhang et al. 2017). Intriguingly, TPX2 also undergoes a liquid–liquid phase separation, a state that co-condenses with soluble tubulin (King & Petry 2020), physically explaining the TPX2 so-called clusters previously reported to nucleate MTs in vitro (Brunet et al. 2004, Schatz et al. 2003). Unlike many droplets characterized in recent studies of biological condensates, TPX2 droplets prefer to form on the MT lattice itself (King & Petry 2020). Like macroscale droplet distribution—for example, dew on a spider web or a dripping faucet—the spacing of these TPX2 droplets is controlled by the Rayleigh–Plateau instability (Setru et al. 2021).
The disordered N-terminus of TPX2, by recruiting the nucleation substrate tubulin, forms a reaction crucible and enhances the rate of branching MT nucleation. In contrast, the C-terminal half of TPX2 is ordered and is required for MT branching to occur (Petry et al. 2013). It can also independently bind MTs (Alfaro-Aco et al. 2017) and harbors the Eg5 binding site (Figure 2). The C-terminus of TPX2 contains the Spc110/Pcp1 motif (SPM), which is present in the yeast spindle-pole body protein Spc110 and leads to the activation of γ-TuSC (Lin et al. 2014), as well as a split γ-TuNA sequence, the γ-TuRC binding/activation motif (Alfaro-Aco et al. 2017). However, it remains to be seen whether this split sequence from TPX2 is able to activate γ-TuRC to the same extent as the continuous γ-TuNA of CDK5RAP2 (Choi et al. 2010).
Other Factors
Three additional factors have been proposed to contribute to branching MT nucleation. The first is the kinesin Eg5, which will be discussed further in the section on branching and motors. In addition, a role in branching has been suggested for EML3 (Luo et al. 2019), a member of the MT-binding EML family (Fry et al. 2016). EML3 has been proposed to directly bind augmin and recruit it to the MT, regulated by Cdk1 phosphorylation (Luo et al. 2019). An additional recruitment factor for γ-TuRC has been proposed as well—the human receptor for hyaluronic acid–mediated motility (XRHAMM). Previous work (Groen et al. 2004, Scrofani et al. 2015) using a series of immunoprecipitation experiments coupled with MT nucleation assays in Xenopus egg extract suggests complex formation between XRHAMM, TPX2, and γ-TuRC. Further work will be needed to establish the roles of these factors in the branching MT nucleation reaction.
THE COMPLETE PATHWAY IN ANIMAL CELL DIVISION
The main branching factors were initially identified at the cellular level due to their localization or their role in determining spindle morphology. Shortly after γ-TuRC was found not only at centrosomes but within the spindle (Luders et al. 2006, Mahoney et al. 2006), augmin and TPX2 were found to be spindle-specific γ-TuRC localization factors. The knockdown of augmin or TPX2 led to the loss of spindle but not centrosomal γ-tubulin and reduced MT density within spindles, implicating the loss of MT nucleation within the spindle body (Goshima et al. 2008, Hughes et al. 2008, Lawo et al. 2009, Uehara et al. 2009, Wittmann et al. 2000). Later, ex vivo experiments demonstrated directly that γ-TuRC, augmin, TPX2, and XMAP215 were each required for branched MT networks to form (Petry et al. 2013, Thawani et al. 2018). At this point, the next tasks were to identify whether these factors were sufficient, and in what order they facilitated MT branching. In this section, we outline how the branching MT nucleation reaction occurs in Xenopus meiosis II and Drosophila mitosis. To a great extent, these models are based on ex vivo studies in Xenopus egg extract and in vitro reconstitutions using purified components.
The Branching Microtubule Nucleation Reaction in Meiosis II
Branching MT nucleation has been extensively characterized in Xenopus meiosis II. In this system, γ-TuRC, augmin, TPX2, and XMAP215 are all required for branching to occur. In addition, the MT branching reaction occurs in a strict temporal order: first, TPX2 binds; second, augmin, XMAP215, and γ-TuRC are recruited; and, third, nucleation occurs (Figure 3).
Step 1: Binding of TPX2 to microtubules to establish a reaction center
The first step in the branching MT nucleation reaction occurs when TPX2 binds to the mother MT (Figure 3), creating an environment that favors recruitment of branching factors and nucleation of a new MT. This has been shown in vitro, where prior incubation of MT seeds with TPX2 dramatically increases the quantity of augmin recruited; however, prior incubation with augmin has no effect on TPX2 recruitment (Alfaro-Aco et al. 2020). Similarly, immunodepletion/add-back experiments in egg extract show that the sequential addition of purified TPX2 followed by augmin yields robust nucleation, whereas the opposite order does not even allow branching MT nucleation to occur (Thawani et al. 2019). This result posed a conundrum, as, in the absence of TPX2, purified augmin can bind to MTs in vitro and recruit nucleation-competent γ-TuRC to generate MTs. The requirement for TPX2 in extract may highlight the importance of concentrating nucleation factors with tubulin in this more complex environment, or, alternatively, a role for TPX2 in displacing unrelated MT-binding proteins from the branch site and clearing space for augmin to bind. The TPX2 condensate may also recruit and concentrate so-far-unidentified factors that are needed to render the reaction robust in a physiological setting.
In addition to initiating nucleation, TPX2 appears to have further roles in the temporal control of MT branching. Interestingly, in extract and in vivo, TPX2 preferentially binds older MT lattice regions, i.e., those that have nucleated early in the reaction (Thawani et al. 2019, David et al. 2019). As TPX2 in vitro localizes predominantly to the GTP cap (Roostalu et al. 2015, Zhang et al. 2017), this surprising reversal suggests that, in extract, TPX2 localization is controlled by factors unrelated to sensing the state of the MT lattice. This can be explained by the slow on-rate of TPX2 binding to the MT lattice, which favors longer existing MT regions (Thawani et al. 2019). We speculate that the slow on-rate could be a consequence of TPX2’s preferred self-association into a condensate on the MT lattice.
Step 2: Recruitment of branching factors to mother microtubules
Upon depositing the base of the branch site, phase-separated TPX2 droplets on an MT specifically recruit free tubulin (King & Petry 2020, Setru et al. 2021), augmin, and to some extent γ-TuRC (Figure 3). It is unknown whether free augmin is recruited to TPX2 droplets, or if it first interacts with γ-TuRC in solution prior to recruitment. Augmin can interact with γ-TuRC in solution to some degree, supporting a simultaneous recruitment model (Song et al. 2018, Thawani et al. 2019). However, because γ-TuRC and augmin interact rather weakly in solution, and the branching reaction strictly requires the template MT as a substrate, we speculate that some change occurs when augmin binds the MT that promotes its interaction with γ-TuRC. Although it is at present unknown what could cause this shift, possibilities include a conformational change that augmin undergoes to enhance γ-TuRC affinity or an avidity effect due to the high local concentration of augmin and/or secondary weak binding sites provided by TPX2.
XMAP215 is required for MT nucleation to occur, but at which point during the reaction it associates with the branch site is unknown (Thawani et al. 2018, 2019). As the main binding partner at the branch site for XMAP215 is γ-TuRC (Thawani et al. 2018), XMAP215 likely arrives in complex with γ-TuRC and/or is recruited after γ-TuRC binds. Conflicting results have emerged as to whether XMAP215 nucleates MTs additively (i.e., independently) or synergistically (i.e., cooperatively within the same pathway) with γ-TuRC. When nucleation is assayed via single-molecule TIRF microscopy, both human (Consolati et al. 2020) and Xenopus XMAP215 (Thawani et al. 2018) increase the MT nucleation potential of γ-TuRC in a cooperative manner. However, when MT conucleation with γ-TuRC was measured using Xenopus XMAP215 in a bulk MT nucleation assay, only an additive effect could be observed, implying that XMAP215 nucleates MTs independently of γ-TuRC (King et al. 2020). Thus, further work will be needed to cement the timing and mechanistic role of XMAP215 in the branching MT nucleation reaction.
Step 3: Nucleation of daughter microtubules
Recent work has shown that the recruitment of branching factors to a mother MT is sufficient to initiate branching MT nucleation (Alfaro-Aco et al. 2020). Using an in vitro reconstitution system, the stepwise addition of TPX2, augmin, γ-TuRC, and finally unpolymerized tubulin causes new MTs to nucleate from GMPCPP-stabilized MT seeds in a uniform direction. Although not required for branching MT nucleation in vitro, when XMAP215 was added to this reaction, MT nucleation rates were significantly increased along with overall MT length. Surprisingly, and in contrast to branching MT nucleation in extract, nucleation in vitro, albeit to a lesser extent, can also occur in the absence of either TPX2 or augmin. Both in extract and in vitro, the polarity and branching angle of the resulting MTs are remarkably similar.
How an MT is physically nucleated from γ-TuRC once it is recruited to the mother MT during branching MT nucleation remains unclear. Early models suggested that γ-TuRC forms a perfect template for the growing MT (Zheng et al. 1995). However, recent cryo-EM structures of γ-TuRC found that the diameter of the complex was ~5 nm too wide to perfectly match a 13-protofilament MT (Liu et al. 2020, Wieczorek et al. 2020b). Thus, it was hypothesized that closure of the γ-tubulin ring would be necessary to activate γ-TuRC as a nucleation template (Kollman et al. 2010). Various binding partners have been proposed as γ-TuRC activators, including γ-TuNA-containing proteins and tubulin itself (Thawani et al. 2020). A centrosomally derived γ-TuNA has previously been shown to directly bind γ-TuRC and enhance MT nucleation (Choi et al. 2010). In addition, work in yeast has suggested that binding of γ-TuNA to γ-TuRC induces ring closure (Brilot et al. 2021), whereas this could not be observed in the cryo-EM structure of γ-TuNA-bound γ-TuRC (Wieczorek et al. 2020b).
TPX2 contains a γ-TuNA and an SPM motif, and it remains to be uncovered whether it indeed has an activation effect on γ-TuRC within the branch site, i.e., in association with the mother MT and augmin. TPX2 may also directly enhance the nucleation process by stabilizing the MT lattice (Reid et al. 2016, Roostalu et al. 2015, Wieczorek et al. 2015). Whether augmin acts as a pure bridge between the mother MT and γ-TuRC or whether it can also directly contribute to γ-TuRC’s nucleation activity remains an open question as well. Both factors together could also contribute to γ-TuRC’s nucleating ability within the branch site.
Branching Microtubule Nucleation in Mitosis
In addition to the pioneering work to image MT branching live in Drosophila S2 cells (Verma & Maresca 2019), endogenous mitotic augmin and γ-TuRC were recently purified from whole Drosophila embryos and used to reconstitute branching MT nucleation (Tariq et al. 2020). In Drosophila, TPX2 is not essential and not well conserved (Goshima 2011). Hence, it is not surprising that TPX2 was dispensable for branching MT nucleation in vitro. Yet, despite the absence of TPX2, the MT branching reaction proceeds in a similar order as described for Xenopus meiosis in the preceding section, beginning with the localization of augmin to the MT (Tariq et al. 2020). The average MT branch angle between mother and daughter MTs was much wider, from 30 to 60° (Tariq et al. 2020, Verma & Maresca 2019), compared to that in Xenopus. Although it is unclear whether there is any meaningful mechanistic consequence arising from this difference in branch angle, the species-specific difference of the branch angle offers an opportunity to understand how the branch angle is set.
Branching MT nucleation has also been visualized in fixed mitotic human cells using negative-stain electron tomography (Kamasaki et al. 2013). Not only could apparent MT branches be easily distinguished in these cells, but intriguing elongated connective density—approximately the length of augmin—was observed at the branch sites. Furthermore, the knockdown of Haus6 completely abolished this connecting density. These images, along with time-lapse microscopy in Drosophila S2 cells, suggested that augmin (and presumably γ-TuRC) continues to connect the daughter MT to the parent for some time after nucleation (Kamasaki et al. 2013, Verma & Maresca 2019).
Interestingly, even in the absence of TPX2, augmin and γ-TuRC appear to localize to spaced puncta distributed across the length of the MT (Alfaro-Aco et al. 2020, Tariq et al. 2020, Verma & Maresca 2019). It is currently unknown whether augmin (or γ-TuRC) has any intrinsic ability to form a condensed phase, whether in solution or, like TPX2, when bound to the MT lattice. However, note that the MT-binding subunit of augmin, Haus8, has the low complexity and predicted intrinsic disorder observed in many phase-separating proteins (Tiwary & Zheng 2019). In addition, purified Haus8 forms subtle puncta when bound to MTs (Wu et al. 2008). Further work will be needed to ascertain whether liquid–liquid phase separation is applicable to branching factors other than TPX2, but, together, these observations suggest the possibility that phase-separation may play a broader role in branching MT nucleation.
INTEGRATING THE PATHWAY INTO SPINDLE ASSEMBLY
The Ran Gradient
For 30 years, the Ran GTPase pathway has been recognized as a master regulator of spindle assembly via the spatially controlled release of spindle assembly factors (SAFs) from importins, as reviewed in Cavazza & Vernos (2015) and Forbes et al. (2015). The Ran pathway intersects with branching MT nucleation, because TPX2 is a SAF (Gruss et al. 2001). The details of how TPX2 is bound and inhibited by the importin dimer, composed of importin-α and importin-β, have been elucidated. TPX2 was first described as containing two noncanonical nuclear localization sequences (NLSs), and a crystal structure of both NLSs in complex with importin-α was determined (Giesecke & Stewart 2010). Recently, a third NLS, N-terminal to the previously described two, was identified, as well as an additional interaction between TPX2 and importin-β (Safari et al. 2021). Binding of TPX2 to importin-α or -β was shown to inhibit phase separation and prevents branching MT nucleation from occurring (Safari et al. 2021, Schatz et al. 2003).
The distribution of active—i.e., GTP-bound—Ran throughout the cell is not uniform. The activator of Ran, the RanGEF (Ran guanine nucleotide exchange factor) RCC1 (regulator of chromosome condensation 1), localizes to chromosomes and generates RanGTP, whereas RanGAPs (Ran GTPase-activating proteins) throughout the cytoplasm induce inactivating GTP hydrolysis. The combination of these two differentially localized regulators generates a gradient of active Ran that is highest near the chromosome (Figure 4). This gradient, acting through importin, presumably enables branching MT nucleation in the vicinity of chromosomes as well. In line with this hypothesis, branching MT nucleation factors are required for the nucleation of chromosome-associated MTs; enrich on MT bundles near chromosomes; and may be required for the polarized, kinetochore-directed growth of noncentrosomal MTs during kinetochore-fiber assembly (Bucciarelli et al. 2009, David et al. 2019, Goshima et al. 2008, Lawo et al. 2009, Zhu et al. 2008).
Figure 4.
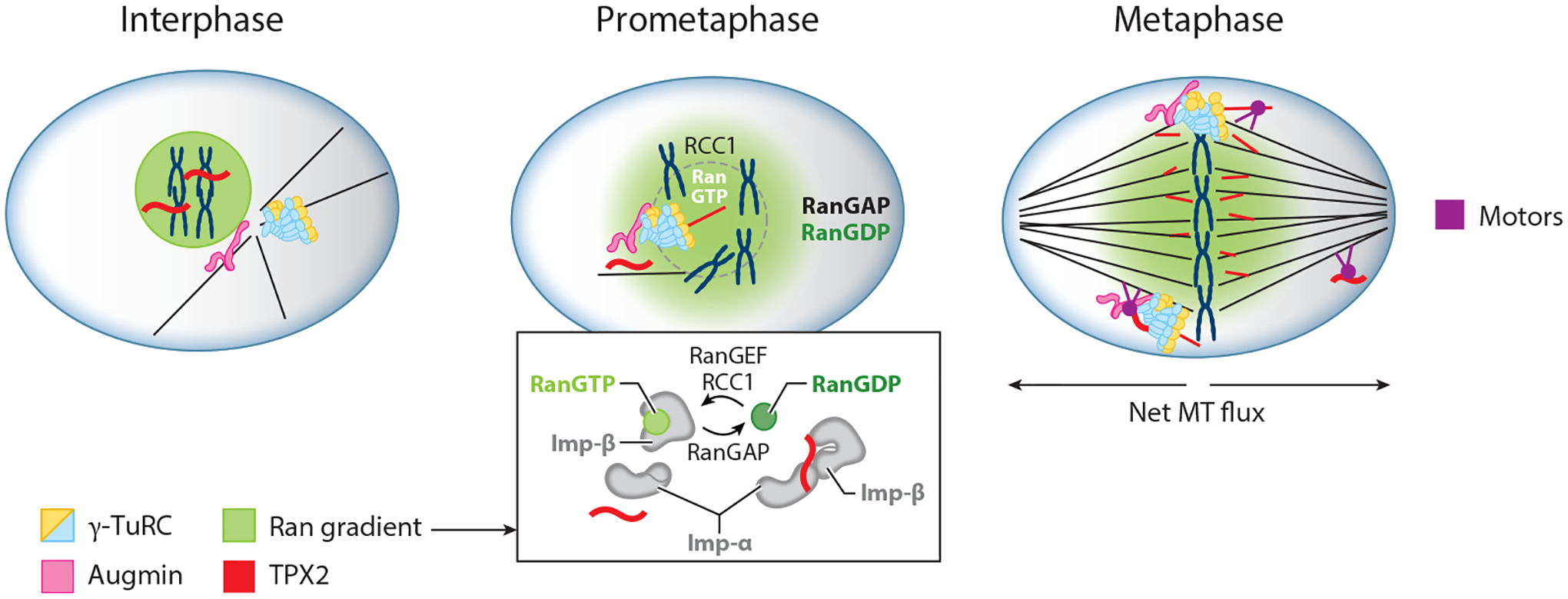
Branching MT nucleation is regulated within spindle assembly. In interphase, branching is inactive due to sequestration of TPX2 (red) within the nucleus by RanGTP. After nuclear envelope breakdown in prometaphase, RanGTP and active TPX2 are released to initiate branching near chromosomes due to the presence of RanGEF RCC1. The inset shows how RanGTP (green), acting through Imp-α and Imp-β, releases TPX2, and RanGDP (green-gray) causes TPX2 to be sequestered. During metaphase, the interaction of motors (purple) with TPX2 and MT branches causes both TPX2, new MTs, and perhaps other branching factors to relocate throughout the spindle. Abbreviations: γ-TuRC, γ-tubulin ring complex; Imp-α/β, importin α/β; MT, microtubule; RanGAP, Ran GTPase-activating protein; RanGEF, Ran guanine nucleotide exchange factor; RCC1, regulator of chromosome condensation 1; TPX2, targeting protein for Xklp2.
On a global scale, the Ran gradient has been proposed to regulate spindle size (Oh et al. 2016) and appears critical to shaping spindle morphology, as the addition of RCC1-coated beads to mitotic extract is sufficient to generate a bipolar spindle (Carazo-Salas et al. 1999, Ohba et al. 1999). Interestingly, the modulation of both Ran concentration and cell size alter the organization of MT nucleation and spindle morphology. This was proposed from recent work (Gai et al. 2021) using microfluidics to encapsulate Xenopus egg extract in emulsified monodisperse droplets. Using this approach, the architecture of Ran-induced MT networks was observed to be dependent on both droplet diameter and Ran concentration. It is currently unknown whether any other components of the branching MT nucleation pathway are also Ran-regulated SAFs.
Branching Microtubule Nucleation and Motion
In considering the regulation of MT branching by Ran, one seeming inconsistency appears: The Ran gradient should restrict branching MT nucleation to the vicinity of chromosomes, yet MTs formed by MT branching are distributed throughout the entire spindle body (David et al. 2019, Goshima et al. 2008, Lawo et al. 2009, Mahoney et al. 2006, Petry et al. 2011). This inconsistency can be resolved by considering the action of motors and other MT-motive forces. Although MT branches are stable in both in vitro reconstitutions and Xenopus extract assays (Alfaro-Aco et al. 2020, Petry et al. 2013, Tariq et al. 2020), this stability is due to the absence of motors and the inhibition of the dynein motor by vanadate or CC1, respectively. When dynein is not inhibited in Xenopus extract assays, MT branches are quickly dislodged and MTs disperse (Petry et al. 2013). In the spindle, MTs move en masse toward the poles through the combined action of kinesin-dependent antiparallel sliding and dynein-dependent poleward transport, as well as poleward flux (Steblyanko et al. 2020, Yang et al. 2008), which is defined as the net motion of MTs due to depolymerization at the pole-localized minus end and tubulin addition to the plus end. Both motor-based transport and poleward flux would distribute any MTs originating from branching MT nucleation. Intriguingly, binding between TPX2 and the kinesins Eg5 and Kif15 has been shown to be critical for spindle assembly (Ma et al. 2011, Mann et al. 2017, Tanenbaum et al. 2009). Eg5 and dynein are also required for the correct localization of TPX2 and its transport to the spindle poles (Ma et al. 2010). It remains to be determined whether and, if so, how factor-containing branching droplets remain close to the chromosome, where they continue to nucleate new MTs, and whether other MT branching factors are also distributed to the periphery of the Ran gradient, thereby activating MT branching throughout the spindle.
Phosphoregulation
At the onset of mitosis, 20% of the proteome undergoes regulation by kinases (Olsen et al. 2010), and the components of branching MT nucleation are no exception. Both augmin and γ-TuRC isolated from mitotic Drosophila embryos have been shown to have enhanced activity compared with those derived from interphase embryos (Tariq et al. 2020), likely as a result of phosphorylation (Tariq and Wakefield, unpublished observations). The phosphoregulation of branching MT nucleation mainly depends on the mitotic kinases Cdk, Plk1, and Aurora A. Cdk1 phosphorylates the C-terminal tail of the γ-TuRC subunit NEDD1, and this phosphorylation event is required for the recruitment of Plk1 to NEDD1 (Zhang et al. 2009). After further phosphorylating NEDD1, Plk1 also phosphorylates the augmin subunit Haus8 to promote MT binding (Johmura et al. 2011). Plk1 additionally phosphorylates TPX2 (Eckerdt et al. 2009), although this has not been shown to be NEDD1 dependent. The Plk1 phosphorylation of Tpx2 recruits Aurora A kinase (Eckerdt et al. 2009) and, when bound by TPX2, Aurora A autophosphorylates to become activated [as reviewed in Garrido & Vernos (2016)]. Active Aurora A then phosphorylates TPX2 to modulate MT flux through CLASP1 (Fu et al. 2015). Either dependent on or independent of TPX2, Aurora A also phosphorylates Haus8 to further modulate MT binding (Tsai et al. 2011) and NEDD1 to promote MT assembly at the chromosome (Courthéoux et al. 2019, Pinyol et al. 2013). Although the steps in this phosphorylation cascade have each been individually characterized and, in some cases, explicitly linked, the full order of the cascade has never been formally demonstrated. Furthermore, Nek9 has been shown to phosphorylate TPX2 to inhibit importin binding and modulate Eg5 localization (Eibes et al. 2018), and numerous additional mitotic and meiotic phosphorylation sites on NEDD1, TPX2, and augmin have been identified through high-throughput screening (Olsen et al. 2010, Peuchen et al. 2017), but their significance and the relevant kinases remain unknown. Further experiments will be needed to formally connect the inputs of this regulatory cascade to their direct consequences on the patterning of MT branching.
BRANCHING MICROTUBULE NUCLEATION IN OTHER SYSTEMS
Interphase
In one of the first screens identifying MT branching factors in Drosophila S2 cells, the knockdown of the poorly conserved augmin subunit Dgt2—but none of the other seven—was reported to alter morphology of the MT cytoskeleton during interphase (Hughes et al. 2008). In most vertebrate cells, however, branching MT nucleation is suppressed in interphase due to the sequestration of TPX2 within the nucleus (Figure 4), although augmin remains cytoplasmic or at the centrosome (Bucciarelli et al. 2009, Schweizer et al 2021, Zhu et al. 2008). One major exception has been recently characterized in vertebrates: postmitotic neurons (Chen et al. 2017b, Cunha-Ferreira et al. 2018, Sanchez-Huertas et al. 2016). Using super-resolution microscopy (Cunha-Ferreira et al. 2018) or in situ proximity ligation (Chen et al. 2017b), γ-TuRC, augmin, and TPX2 have all been shown to localize to MTs in mouse neurons, and, surprisingly, γ-TuRC and augmin still interact with one another despite downregulation and loss of the γ-TuRC subunit NEDD1 (Sanchez-Huertas et al. 2016). Due to the role of branching in controlling the orientation of new MTs, disruption of the augmin complex by the knockdown of Haus6 (Cunha-Ferreira et al. 2018) or Haus7 (Sanchez-Huertas et al. 2016) subsequently disrupts MT polarity in axons. Furthermore, depletion of TPX2, augmin, or γ-TuRC all decrease neuronal MT density, and depletion of augmin and γ-TuRC (although not TPX2) impairs dendritic development (Chen et al. 2017b, Cunha-Ferreira et al. 2018, Sanchez-Huertas et al. 2016). Either as a result of this defect in neuronal cytoskeletal architecture and/or due to cell division–dependent effects, the augmin complex is required in the mouse embryo. This is true not only in the blastocyst (Watanabe et al. 2016) but also, at later stages, during neuronal specification (Viais et al. 2021). Whether neuronal branching MT nucleation uses the same mechanism as the canonical mitotic one is unknown, because, surprisingly, the only putative augmin-binding subunit of γ-TuRC, NEDD1, is wholly absent (Chen et al. 2017b, Cunha-Ferreira et al. 2018, Sanchez-Huertas et al. 2016). The neuronal system may in fact represent evidence for a second, NEDD1-independent binding site on γ-TuRC for augmin.
Fungi
One of the first reports of branching MT nucleation described the process in the ascomycete S. pombe (Janson et al. 2005), which surprisingly lacks both augmin and TPX2 (Figure 1c). Rather, MT branching in S. pombe relies on the protein Mto2, a nonessential protein found only in Schizosaccharomyces. In conjunction with the γ-TuNA-containing protein Mto1, Mto2 recruits (Samejima et al. 2008) and activates (Lynch et al. 2014) γ-TuRC at the side of preexisting MTs during interphase. Intriguingly, MTs generated through this unique fungal pathway are antiparallel to the originating MT, and, unlike the animal and plant mitotic branching MT pathway, in S. pombe, branching is restricted to interphase, because the phosphorylation of Mto2 during cell division inactivates the pathway (Borek et al. 2015).
A more canonical MT branching pathway may exist in the filamentous fungus Aspergillus nidulans, which, despite also lacking TPX2, contains at least five conserved augmin subunits (Edzuka et al. 2014). Although the deletion of Aspergillus augmin has no effect on the mitotic spindle, further work will be necessary to determine whether this complex is required in meiosis or interphase.
Plants
Two excellent reviews (Tian & Kong 2019, Yi & Goshima 2018) have recently addressed branching MT nucleation in plants, to which we direct the reader for a more detailed treatment. In brief, Arabidopsis thaliana orthologs of γ-TuRC (Kong et al. 2010), augmin (Ho et al. 2011, Pignocchi et al. 2009), and TPX2 (Vos et al. 2008) are required during cell division and are essential for gametogenesis (Nakamura & Hashimoto 2009, Pignocchi et al. 2009, Vos et al. 2008). As in animal cells, during interphase, A. thaliana TPX2 is nuclear (Vos et al. 2008), yet, surprisingly, branching MT nucleation can still occur, relying on both γ-TuRC (Kong et al. 2010, Nakamura & Hashimoto 2009) and augmin (Ho et al. 2011). Interphase branching MT nucleation is required for the formation of cortical arrays, which contain branched MTs at an acute angle, as well as parallel MTs (Chan et al. 2009, Kirik et al. 2012). The ratio between these two geometries appears to be regulated by Ton2 phosphatase activity (Kirik et al. 2012), although the relevant kinase and substrate have not yet been identified. It remains unclear why plants can support branching MT nucleation during interphase whereas other organisms cannot, but recent work suggests that plants may be able to accomplish this due to possession of multiple Haus8 paralogs, which function at different points in the cell cycle (Lee et al. 2017).
BRANCHING MICROTUBULE NUCLEATION IN CANCER
Given that branching MT nucleation is a key pathway in spindle assembly (David et al. 2019, Decker et al. 2018), it may not be surprising that it is implicated in uncontrolled cell division (Schvartzman et al. 2010). The overexpression of branching MT nucleation factors in several malignancies has been correlated with poor patient prognosis (Alvarado-Kristensson 2018, Ding et al. 2017, Dráberová et al. 2015, Neumayer et al. 2014, Zhang et al. 2019, Zou et al. 2018). TPX2 is upregulated in numerous cancers (Neumayer et al. 2014, Zou et al. 2018), which, besides its role in branching, could exert oncogenic effects by regulating Aurora A kinase activity and modulating the DNA damage response pathway. In addition, the overexpression of γ-TuRC subunits has been linked to various types of cancer, both through their role in MT nucleation and cell cycle progression, as well as in the regulation of the DNA damage response (Dráber & Dráberová 2021, Dráberová et al. 2015). Branching MT nucleation has been most clearly linked to cancer progression through the overexpression of augmin subunits. Recent evidence has shown that Haus3 and Haus5 are upregulated in aggressive forms of hepatocellular carcinoma and glioblastoma, respectively, and are considered prognostic markers (Ding et al. 2017, Zhang et al. 2019). Interestingly, the knockdown of Haus3 resulted in cells arrested in G2/M phase and the inhibition of hepatocellular carcinoma tumor growth both in vitro and in vivo (Zhang et al. 2019), but truncation of Haus3 has also been positively linked to progression of breast cancer (Shah et al 2009). In addition, as discussed in the section on phosphoregulation above, augmin is phosphorylated by the mitotic kinase Plk1, which is itself upregulated in many types of cancers (de Cárcer 2019, Gomez-Ferreria et al. 2012, Johmura et al. 2011). Thus, the hyperactivation of augmin and other branching factors, leading to an aberrant increase in branching MT nucleation, likely plays a significant role in oncogenesis.
The loss or knockdown of MT branching factors in cancer cells has been shown to dramatically decrease cancer cell viability and proliferation. In fact, the knockdown of MT branching factors strongly resembles the effect of treating cancer cells with the small-molecule MT stabilizer Taxol, a widely used and effective chemotherapeutic agent (Zasadil et al. 2014). For example, the loss of augmin or TPX2 in cells causes cell cycle arrest and reduces MT mass within spindles (Goshima et al. 2008, Gruss & Vernos 2004, van Gijn et al. 2019). Likewise, in vivo mouse models show a reduction in tumor size when augmin subunits are knocked down (Zhang et al. 2019). Similarly, the inhibition of γ-tubulin—and presumably γ-TuRC—drastically perturbs mitotic spindle assembly and induces cytotoxicity in cancer cells (Chinen et al. 2015, Ebisu et al. 2020, Traversi et al. 2019). In summary, understanding how branching MT nucleation can be targeted in cancer offers exciting opportunities (Zahreddine & Borden 2013).
CONCLUDING REMARKS
As a relatively simple MT nucleation pathway, originating from a preexisting MT, branching MT nucleation is an ideal model system to explore the underlying principles of how MTs are nucleated in space and time. Since the first reports of branching MT nucleation 17 years ago in yeast and plant cells (Janson et al. 2005, Murata et al. 2005), much has been determined about the primary molecular players, the order of the reaction, and how branching is regulated within the context of the spindle. In addition, branching has been firmly established as the dominant pathway generating spindle MTs during cell division.
Going forward, the major outstanding questions revolve around how the varied branching factors modulate one another’s activity and how the pathway integrates into global spindle assembly. In vitro reactions have established that TPX2, when present, concentrates and perhaps activates γ-TuRC and/or augmin, but what causes this cooperativity, as well as what establishes the angle of the resulting branch, remains an open question. In addition, the polymerase XMAP215 and other factors with less clearly defined roles, EML3 and XHRAMM, have not been placed within the timeline of the branching reaction, and their effects on the first three branching factors discovered are wholly unknown. Moreover, although the Ran gradient and mitotic phosphorylation both have documented effects on branching MT nucleation, it remains to be demonstrated how these regulatory pathways converge, along with other forces that act on the spindle, to connect these nanometer-scale recruitment centers to the micrometer-scale architecture of the spindle.
Finally, at the present date, branching MT nucleation has only been studied in a small subset of cell types and species, leaving many questions outstanding that could help explain the mechanism, regulation, and organism-level functions of the pathway. Branching out—pun intended—to new species or cell types may extend our understanding of the role of branching MT nucleation during interphase and the regulatory networks that modulate its activity throughout the cell cycle. Because branching MT nucleation creates polarized MT bundles, other roles in cellular structures and cell types are to be expected. In addition, studying organisms in which some branching factors are not represented in the genome may further clarify how the loss of different factors alters the shape of the branching reaction. In answering these questions about branching MT nucleation, we will continue to gain insight into MT nucleation more generally, leading to a better understanding of how the complex and dynamic MT cytoskeleton is organized and remodeled as a whole.
ACKNOWLEDGMENTS
We would like to thank all members of the Petry lab for their insightful comments and feedback, particularly Michael Rale and Venecia Valdez. This work was supported by National Institutes of Health grants F32GM142149 (SMT), New Innovator Award 1DP2GM123493 (SP) and R01 1R01GM141100-01A1 (SP), as well as the Pew Scholars Program in the Biomedical Sciences (SP), and the David and Lucile Packard Foundation (SP).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Alfaro-Aco R, Thawani A, Petry S. 2017. Structural analysis of the role of TPX2 in branching microtubule nucleation. J. Cell Biol 216:983–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfaro-Aco R, Thawani A, Petry S. 2020. Biochemical reconstitution of branching microtubule nucleation. eLife 9:e49797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarado-Kristensson M 2018. γ-tubulin as a signal-transducing molecule and meshwork with therapeutic potential. Signal Transduct. Target. Ther 3:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissan M, Schlattner U, Lacombe ML. 2018. The NDPK/NME superfamily: state of the art. Lab. Investig 98:164–74 [DOI] [PubMed] [Google Scholar]
- Borek WE, Groocock LM, Samejima I, Zou J, de Lima Alves F, et al. 2015. Mto2 multisite phosphorylation inactivates non-spindle microtubule nucleation complexes during mitosis. Nat. Commun 6:7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brilot AF, Lyon AS, Zelter A, Viswanath S, Maxwell A, et al. 2021. CM1-driven assembly and activation of yeast gamma-tubulin small complex underlies microtubule nucleation. eLife 10:e65168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouhard GJ, Stear JH, Noetzel TL, Al-Bassam J, Kinoshita K, et al. 2008. XMAP215 is a processive microtubule polymerase. Cell 132:79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugues J, Nuzzo V, Mazur E, Needleman DJ. 2012. Nucleation and transport organize microtubules in metaphase spindles. Cell 149:554–64 [DOI] [PubMed] [Google Scholar]
- Brunet S, Sardon T, Zimmerman T, Wittmann T, Pepperkok R, et al. 2004. Characterization of the TPX2 domains involved in microtubule nucleation and spindle assembly in Xenopus egg extracts. Mol. Biol. Cell 15:5318–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucciarelli E, Pellacani C, Naim V, Palena A, Gatti M, Somma MP. 2009. Drosophila Dgt6 interacts with Ndc80, Msps/XMAP215, and γ-tubulin to promote kinetochore-driven MT formation. Curr. Biol 19:1839–45 [DOI] [PubMed] [Google Scholar]
- Byrnes AE, Slep KC. 2017. TOG-tubulin binding specificity promotes microtubule dynamics and mitotic spindle formation. J. Cell Biol 216:1641–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carazo-Salas RE, Guarguaglini G, Gruss OJ, Segref A, Karsenti E, Mattaj IW. 1999. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature 400:178–81 [DOI] [PubMed] [Google Scholar]
- Cavazza T, Vernos I. 2015. The RanGTP pathway: from nucleo-cytoplasmic transport to spindle assembly and beyond. Front. Cell Dev. Biol 3:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J, Sambade A, Calder G, Lloyd C. 2009. Arabidopsis cortical microtubules are initiated along, as well as branching from, existing microtubules. Plant Cell 21:2298–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JWC, Chen ZA, Rogala KB, Metz J, Deane CM, et al. 2017a. Cross-linking mass spectrometry identifies new interfaces of Augmin required to localise the γ-tubulin ring complex to the mitotic spindle. Biol. Open 6:654–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W-S, Chen Y-J, Huang Y-A, Hsieh B-Y, Chiu H-C, et al. 2017b. Ran-dependent TPX2 activation promotes acentrosomal microtubule nucleation in neurons. Sci. Rep 7:42297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinen T, Liu P, Shioda S, Pagel J, Cerikan B, et al. 2015. The γ-tubulin-specific inhibitor gatastatin reveals temporal requirements of microtubule nucleation during the cell cycle. Nat. Commun 6:8722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y-K, Liu P, Sze SK, Dai C, Qi RZ. 2010. CDK5RAP2 stimulates microtubule nucleation by the γ-tubulin ring complex. J. Cell Biol 191:1089–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T, Ribbeck K. 2007. Self-organization of anastral spindles by synergy of dynamic instability, autocatalytic microtubule production, and a spatial signaling gradient. PLOS ONE 2:e244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consolati T, Locke J, Roostalu J, Chen ZA, Gannon J, et al. 2020. Microtubule nucleation properties of single human γTuRCs explained by their cryo-EM structure. Dev. Cell 53:603–17.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cota RR, Teixido-Travesa N, Ezquerra A, Eibes S, Lacasa C, et al. 2017. MZT1 regulates microtubule nucleation by linking γTuRC assembly to adapter-mediated targeting and activation. J. Cell Sci 130:406–19 [DOI] [PubMed] [Google Scholar]
- Courthéoux T, Reboutier D, Vazeille T, Cremet J-Y, Benaud C, et al. 2019. Microtubule nucleation during central spindle assembly requires NEDD1 phosphorylation on serine 405 by Aurora A. J. Cell Sci 132:jcs231118 [DOI] [PubMed] [Google Scholar]
- Cunha-Ferreira I, Chazeau A, Buijs RR, Stucchi R, Will L, et al. 2018. The HAUS complex is a key regulator of non-centrosomal microtubule organization during neuronal development. Cell Rep 24:791–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David AF, Roudot P, Legant WR, Betzig E, Danuser G, Gerlich DW. 2019. Augmin accumulation on long-lived microtubules drives amplification and kinetochore-directed growth. J. Cell Biol 218:2150–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cárcer G 2019. The mitotic cancer target Polo-like kinase 1: oncogene or tumor suppressor? Genes 10:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker F, Oriola D, Dalton B, Brugués J. 2018. Autocatalytic microtubule nucleation determines the size and mass of Xenopus laevis egg extract spindles. eLife 7:e31149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Herman JA, Toledo CM, Lang JM, Corrin P, et al. 2017. ZNF131 suppresses centrosome fragmentation in glioblastoma stem-like cells through regulation of HAUS5. Oncotarget 8:48545–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dráber P, Dráberová E. 2021. Dysregulation of microtubule nucleating proteins in cancer cells. Cancers 13:5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dráberová E, D’Agostino L, Caracciolo V, Sládková V, Sulimenko T, et al. 2015. Overexpression and nucleolar localization of γ-tubulin small complex proteins GCP2 and GCP3 in glioblastoma. J. Neuropathol. Exp. Neurol 74:723–42 [DOI] [PubMed] [Google Scholar]
- Ebisu H, Shintani K, Chinen T, Nagumo Y, Shioda S, et al. 2020. Dual inhibition of γ-tubulin and Plk1 induces mitotic cell death. Front. Pharmacol 11:620185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckerdt F, Eyers PA, Lewellyn AL, Prigent C, Maller JL. 2008. Spindle pole regulation by a discrete Eg5-interacting domain in TPX2. Curr. Biol 18:519–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckerdt F, Pascreau G, Phistry M, Lewellyn AL, DePaoli-Roach AA, Maller JL. 2009. Phosphorylation of TPX2 by Plx1 enhances activation of Aurora A. Cell Cycle 8:2413–19 [DOI] [PubMed] [Google Scholar]
- Edzuka T, Yamada L, Kanamaru K, Sawada H, Goshima G. 2014. Identification of the augmin complex in the filamentous fungus Aspergillus nidulans. PLOS ONE 9: e101471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eibes S, Gallisa-Sune N, Rosas-Salvans M, Martinez-Delgado P, Vernos I, Roig J. 2018. Nek9 phosphorylation defines a new role for TPX2 in Eg5-dependent centrosome separation before nuclear envelope breakdown. Curr. Biol 28:121–29.e4 [DOI] [PubMed] [Google Scholar]
- Findeisen P, Muhlhausen S, Dempewolf S, Hertzog J, Zietlow A, et al. 2014. Six subgroups and extensive recent duplications characterize the evolution of the eukaryotic tubulin protein family. Genome Biol. Evol 6:2274–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor-Parra I, Iglesias-Romero AB, Chang F. 2018. The XMAP215 ortholog Alp14 promotes microtubule nucleation in fission yeast. Curr. Biol 28:1681–91.e4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes DJ, Travesa A, Nord MS, Bernis C. 2015. Nuclear transport factors: global regulation of mitosis. Curr. Opin. Cell Biol 35:78–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JC, Howard AE, Currie JD, Rogers SL, Slep KC. 2014. The XMAP215 family drives microtubule polymerization using a structurally diverse TOG array. Mol. Biol. Cell 25:2375–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry AM, O’Regan L, Montgomery J, Adib R, Bayliss R. 2016. EML proteins in microtubule regulation and human disease. Biochem. Soc. Trans 44:1281–88 [DOI] [PubMed] [Google Scholar]
- Fu J, Bian M, Xin G, Deng Z, Luo J, et al. 2015. TPX2 phosphorylation maintains metaphase spindle length by regulating microtubule flux. J. Cell Biol 210:373–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai Y, Cook B, Setru S, Stone HA, Petry S. 2021. Confinement size determines the architecture of Ran-induced microtubule networks. Soft Matter 17:5921–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard DL, Becker BE, Romney SJ. 2004. MAPping the eukaryotic tree of life: structure, function, and evolution of the MAP215/Dis1 family of microtubule-associated proteins. Int. Rev. Cytol 239:179–272 [DOI] [PubMed] [Google Scholar]
- Gard DL, Kirschner MW. 1987. A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plus-end. J. Cell Biol 105:2203–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido G, Vernos I. 2016. Non-centrosomal TPX2-dependent regulation of the Aurora A kinase: functional implications for healthy and pathological cell division. Front. Oncol 6:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesecke A, Stewart M. 2010. Novel binding of the mitotic regulator TPX2 (target protein for Xenopus kinesin-like protein 2) to importin-α. J. Biol. Chem 285:17628–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Ferreria MA, Bashkurov M, Helbig AO, Larsen B, Pawson T, et al. 2012. Novel NEDD1 phosphorylation sites regulate γ-tubulin binding and mitotic spindle assembly. J. Cell Sci 125:3745–51 [DOI] [PubMed] [Google Scholar]
- Goshima G 2011. Identification of a TPX2-like microtubule-associated protein in Drosophila. PLOS ONE 6:e28120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Mayer M, Zhang N, Stuurman N, Vale RD. 2008. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J. Cell Biol 181:421–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, et al. 2007. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science 316:417–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen AC, Cameron LA, Coughlin M, Miyamoto DT, Mitchison TJ, Ohi R. 2004. XRHAMM functions in ran-dependent microtubule nucleation and pole formation during anastral spindle assembly. Curr. Biol 14:1801–11 [DOI] [PubMed] [Google Scholar]
- Gruss OJ, Carazo-Salas RE, Schatz CA, Guarguaglini G, Kast J, et al. 2001. Ran induces spindle assembly by reversing the inhibitory effect of importin α on TPX2 activity. Cell 104:83–93 [DOI] [PubMed] [Google Scholar]
- Gruss OJ, Vernos I. 2004. The mechanism of spindle assembly: functions of Ran and its target TPX2. J. Cell Biol 166:949–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillet V, Knibiehler M, Gregory-Pauron L, Remy MH, Chemin C, et al. 2011. Crystal structure of γ-tubulin complex protein GCP4 provides insight into microtubule nucleation. Nat. Struct. Mol. Biol 18:915–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane RN, Martin OC, Zheng Y. 2003. Characterization of a new γTuRC subunit with WD repeats. Mol. Biol. Cell 14:1017–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunzelmann J, Rüthnick D, Lin T-C, Zhang W, Neuner A, et al. 2018. The microtubule polymerase Stu2 promotes oligomerization of the γ-TuSC for cytoplasmic microtubule nucleation. eLife 7:e39932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren L, Remy MH, Bazin I, Callebaut I, Wright M, Merdes A. 2006. NEDD1-dependent recruitment of the γ-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. J. Cell Biol 172:505–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C-MK, Hotta T, Kong Z, Zeng CJT, Sun J, et al. 2011. Augmin plays a critical role in organizing the spindle and phragmoplast microtubule arrays in Arabidopsis. Plant Cell 23:2606–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard AE, Fox JC, Slep KC. 2015. Drosophila melanogaster mini spindles TOG3 utilizes unique structural elements to promote domain stability and maintain a TOG1- and TOG2-like tubulin-binding surface. J. Biol. Chem 290:10149–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia K-C, Wilson-Kubalek EM, Dottore A, Hao Q, Tsai K-L, et al. 2014. Reconstitution of the augmin complex provides insights into its architecture and function. Nat. Cell Biol 16:852–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T-L, Wang H-J, Chang Y-C, Wang S-W, Hsia K-C. 2020. Promiscuous binding of microprotein Mozart1 to γ-tubulin complex mediates specific subcellular targeting to control microtubule array formation. Cell Rep 31:107836 [DOI] [PubMed] [Google Scholar]
- Hughes JR, Meireles AM, Fisher KH, Garcia A, Antrobus PR, et al. 2008. A microtubule interactome: complexes with roles in cell cycle and mitosis. PLOS Biol 6:e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins JR, Toyoda Y, Hegemann B, Poser I, Heriche JK, et al. 2010. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 328:593–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson ME, Setty TG, Paoletti A, Tran PT. 2005. Efficient formation of bipolar microtubule bundles requires microtubule-bound γ-tubulin complexes. J. Cell Biol 169:297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johmura Y, Soung N-K, Park J-E, Yu LR, Zhou M, et al. 2011. Regulation of microtubule-based microtubule nucleation by mammalian polo-like kinase 1. PNAS 108:11446–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamasaki T, O’Toole E, Kita S, Osumi M, Usukura J, et al. 2013. Augmin-dependent microtubule nucleation at microtubule walls in the spindle. J. Cell Biol 202:25–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King BR, Moritz M, Kim H, Agard DA, Asbury CL, Davis TN. 2020. XMAP215 and γ-tubulin additively promote microtubule nucleation in purified solutions. Mol. Biol. Cell 31:2187–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MR, Petry S. 2020. Phase separation of TPX2 enhances and spatially coordinates microtubule nucleation. Nat. Commun 11:270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik A, Ehrhardt DW, Kirik V. 2012. TONNEAU2/FASS regulates the geometry of microtubule nucleation and cortical array organization in interphase Arabidopsis cells. Plant Cell 24:1158–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Pereira G, Geissler S, Grein K, Schiebel E. 1997. The spindle pole body component Spc97p interacts with the γ-tubulin of Saccharomyces cerevisiae and functions in microtubule organization and spindle pole body duplication. EMBO J 16:1550–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman JM, Polka JK, Zelter A, Davis TN, Agard DA. 2010. Microtubule nucleating γ-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature 466:879–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Z, Hotta T, Lee YR, Horio T, Liu B. 2010. The γ-tubulin complex protein GCP4 is required for organizing functional microtubule arrays in Arabidopsis thaliana. Plant Cell 22:191–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawo S, Bashkurov M, Mullin M, Ferreria MG, Kittler R, et al. 2009. HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr. Biol 19:816–26 [DOI] [PubMed] [Google Scholar]
- Lee YJ, Hiwatashi Y, Hotta T, Xie T, Doonan JH, Liu B. 2017. The mitotic function of Augmin is dependent on its microtubule-associated protein subunit EDE1 in Arabidopsis thaliana. Curr. Biol 27:3891–97.e4 [DOI] [PubMed] [Google Scholar]
- Lin T-C, Neuner A, Schlosser YT, Scharf AND, Weber L, Schiebel E. 2014. Cell-cycle dependent phosphorylation of yeast pericentrin regulates γ-TuSC-mediated microtubule nucleation. eLife 3:e02208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Choi YK, Qi RZ. 2014. NME7 is a functional component of the γ-tubulin ring complex. Mol. Biol. Cell 25:2017–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Zupa E, Neuner A, Bohler A, Loerke J, et al. 2020. Insights into the assembly and activation of the microtubule nucleator γ-TuRC. Nature 578:467–71 [DOI] [PubMed] [Google Scholar]
- Loughlin R, Heald R, Nedelec F. 2010. A computational model predicts Xenopus meiotic spindle organization. J. Cell Biol 191:1239–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luders J, Patel UK, Stearns T. 2006. GCP-WD is a γ-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol 8:137–47 [DOI] [PubMed] [Google Scholar]
- Luo J, Yang B, Xin G, Sun M, Zhang B, et al. 2019. The microtubule-associated protein EML3 regulates mitotic spindle assembly by recruiting the Augmin complex to spindle microtubules. J. Biol. Chem 294:5643–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch EM, Groocock LM, Borek WE, Sawin KE. 2014. Activation of the γ-tubulin complex by the Mto1/2 complex. Curr. Biol 24:896–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N, Titus J, Gable A, Ross JL, Wadsworth P. 2011. TPX2 regulates the localization and activity of Eg5 in the mammalian mitotic spindle. J. Cell Biol 195:87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N, Tulu US, Ferenz NP, Fagerstrom C, Wilde A, Wadsworth P. 2010. Poleward transport of TPX2 in the mammalian mitotic spindle requires dynein, Eg5, and microtubule flux. Mol. Biol. Cell 21:979–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney NM, Goshima G, Douglass AD, Vale RD. 2006. Making microtubules and mitotic spindles in cells without functional centrosomes. Curr. Biol 16:564–69 [DOI] [PubMed] [Google Scholar]
- Mann BJ, Balchand SK, Wadsworth P. 2017. Regulation of Kif15 localization and motility by the C-terminus of TPX2 and microtubule dynamics. Mol. Biol. Cell 28:65–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meireles AM, Fisher KH, Colombie N, Wakefield JG, Ohkura H. 2009. Wac: a new Augmin subunit required for chromosome alignment but not for acentrosomal microtubule assembly in female meiosis. J Cell Biol 184: 777–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T, Sonobe S, Baskin TI, Hyodo S, Hasezawa S, et al. 2005. Microtubule-dependent microtubule nucleation based on recruitment of γ-tubulin in higher plants. Nat. Cell Biol 7:961–68 [DOI] [PubMed] [Google Scholar]
- Nakamura M, Hashimoto T. 2009. A mutation in the Arabidopsis γ-tubulin-containing complex causes helical growth and abnormal microtubule branching. J. Cell Sci 122:2208–17 [DOI] [PubMed] [Google Scholar]
- Neumayer G, Belzil C, Gruss OJ, Nguyen MD. 2014. TPX2: of spindle assembly, DNA damage response, and cancer. Cell. Mol. Life Sci 71:3027–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nithianantham S, Cook BD, Beans M, Guo F, Chang F, Al-Bassam J. 2018. Structural basis of tubulin recruitment and assembly by microtubule polymerases with tumor overexpressed gene (TOG) domain arrays. eLife 7:e38922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley BR, Oakley CE, Yoon Y, Jung MK. 1990. γ-Tubulin is a component of the spindle pole body that is essential for microtubule function in Aspergillus nidulans. Cell 61:1289–301 [DOI] [PubMed] [Google Scholar]
- Oakley CE, Oakley BR. 1989. Identification of γ-tubulin, a new member of the tubulin superfamily encoded by mipA gene of Aspergillus nidulans. Nature 338:662–64 [DOI] [PubMed] [Google Scholar]
- Oh D, Yu CH, Needleman DJ. 2016. Spatial organization of the Ran pathway by microtubules in mitosis. PNAS 113:8729–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T, Nakamura M, Nishitani H, Nishimoto T. 1999. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science 284:1356–58 [DOI] [PubMed] [Google Scholar]
- Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, et al. 2010. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal 3:ra3 [DOI] [PubMed] [Google Scholar]
- Petry S, Groen AC, Ishihara K, Mitchison TJ, Vale RD. 2013. Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2. Cell 152:768–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry S, Pugieux C, Nedelec FJ, Vale RD. 2011. Augmin promotes meiotic spindle formation and bipolarity in Xenopus egg extracts. PNAS 108:14473–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peuchen EH, Cox OF, Sun L, Hebert AS, Coon JJ, et al. 2017. Phosphorylation dynamics dominate the regulated proteome during early Xenopus development. Sci. Rep 7:15647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignocchi C, Minns GE, Nesi N, Koumproglou R, Kitsios G, et al. 2009. ENDOSPERM DEFECTIVE1 is a novel microtubule-associated protein essential for seed development in Arabidopsis. Plant Cell 21:90–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinyol R, Scrofani J, Vernos I. 2013. The role of NEDD1 phosphorylation by Aurora A in chromosomal microtubule nucleation and spindle function. Curr. Biol 23:143–49 [DOI] [PubMed] [Google Scholar]
- Reid TA, Schuster BM, Mann BJ, Balchand SK, Plooster M, et al. 2016. Suppression of microtubule assembly kinetics by the mitotic protein TPX2. J. Cell Sci 129:1319–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roostalu J, Cade NI, Surrey T. 2015. Complementary activities of TPX2 and chTOG constitute an efficient importin-regulated microtubule nucleation module. Nat. Cell Biol 17:1422–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roostalu J, Surrey T. 2017. Microtubule nucleation: beyond the template. Nat. Rev. Mol. Cell Biol 18:702–10 [DOI] [PubMed] [Google Scholar]
- Rostkova E, Burgess SG, Bayliss R, Pfuhl M. 2018. Solution NMR assignment of the C-terminal domain of human chTOG. Biomol. NMR Assign 12:221–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safari MS, King MR, Brangwynne CP, Petry S. 2021. Interaction of spindle assembly factor TPX2 with importins-α/β inhibits protein phase separation. J. Biol. Chem 297:100998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samejima I, Miller VJ, Groocock LM, Sawin KE. 2008. Two distinct regions of Mto1 are required for normal microtubule nucleation and efficient association with the γ-tubulin complex in vivo. J. Cell Sci 121:3971–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Huertas C, Freixo F, Viais R, Lacasa C, Soriano E, Luders J. 2016. Non-centrosomal nucleation mediated by augmin organizes microtubules in post-mitotic neurons and controls axonal microtubule polarity. Nat. Commun 7:12187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Huertas C, Luders J. 2015. The augmin connection in the geometry of microtubule networks. Curr. Biol 25: R294–99 [DOI] [PubMed] [Google Scholar]
- Schatz CA, Santarella R, Hoenger A, Karsenti E, Mattaj IW, et al. 2003. Importin α-regulated nucleation of microtubules by TPX2. EMBO J 22:2060–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvartzman JM, Sotillo R, Benezra R. 2010. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat. Rev. Cancer 10:102–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrofani J, Sardon T, Meunier S, Vernos I. 2015. Microtubule nucleation in mitosis by a RanGTP-dependent protein complex. Curr. Biol 25:131–40 [DOI] [PubMed] [Google Scholar]
- Setru SU, Gouveia B, Alfaro-Aco R, Shaevitz JW, Stone HA, Petry S. 2021. A hydrodynamic instability drives protein droplet formation on microtubules to nucleate branches. Nat. Phys 17:493–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer N, Haren L, Dutto I, Viais R, Lacasa C, et al. 2021. Sub-centrosomal mapping identifies augmin-gammaTuRC as part of a centriole-stabilizing scaffold. Nat Commun 12: 6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JG, King MR, Zhang R, Kadzik RS, Thawani A, Petry S. 2018. Mechanism of how augmin directly targets the γ-tubulin ring complex to microtubules. J. Cell Biol 217:2417–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steblyanko Y, Rajendraprasad G, Osswald M, Eibes S, Jacome A, et al. 2020. Microtubule poleward flux in human cells is driven by the coordinated action of four kinesins. EMBO J 39:e105432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Macurek L, Janssen A, Geers EF, Alvarez-Fernandez M, Medema RH. 2009. Kif15 cooperates with Eg5 to promote bipolar spindle assembly. Curr. Biol 19:1703–11 [DOI] [PubMed] [Google Scholar]
- Tariq A, Green L, Jeynes JCG, Soeller C, Wakefield JG. 2020. In vitro reconstitution of branching microtubule nucleation. eLife 9:e49769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thawani A, Kadzik RS, Petry S. 2018. XMAP215 is a microtubule nucleation factor that functions synergistically with the γ-tubulin ring complex. Nat. Cell Biol 20:575–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thawani A, Rale MJ, Coudray N, Bhabha G, Stone HA, et al. 2020. The transition state and regulation of γ-TuRC-mediated microtubule nucleation revealed by single molecule microscopy. eLife 9:e54253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thawani A, Stone HA, Shaevitz JW, Petry S. 2019. Spatiotemporal organization of branched microtubule networks. eLife 8:e43890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J, Kong Z. 2019. The role of the augmin complex in establishing microtubule arrays. J. Exp. Bot 70:3035–41 [DOI] [PubMed] [Google Scholar]
- Tiwary AK, Zheng Y. 2019. Protein phase separation in mitosis. Curr. Opin. Cell Biol 60:92–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovey CA, Conduit PT. 2018. Microtubule nucleation by γ-tubulin complexes and beyond. Essays Biochem 62:765–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traversi G, Staid DS, Fiore M, Percario Z, Trisciuoglio D, et al. 2019. A novel resveratrol derivative induces mitotic arrest, centrosome fragmentation and cancer cell death by inhibiting γ-tubulin. Cell Div 14:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CY, Ngo B, Tapadia A, Hsu P-H, Wu G, Lee W-H. 2011. Aurora-A phosphorylates Augmin complex component Hice1 protein at an N-terminal serine/threonine cluster to modulate its microtubule binding activity during spindle assembly. J. Biol. Chem 286:30097–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R, Goshima G. 2010. Functional central spindle assembly requires de novo microtubule generation in the interchromosomal region during anaphase. J. Cell Biol 191:259–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R, Nozawa RS, Tomioka A, Petry S, Vale RD, et al. 2009. The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. PNAS 106:6998–7003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gijn SE, Wierenga E, van den Tempel N, Kok YP, Heijink AM, et al. 2019. TPX2/Aurora kinase A signaling as a potential therapeutic target in genomically unstable cancer cells. Oncogene 38:852–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma V, Maresca TJ. 2019. Direct observation of branching MT nucleation in living animal cells. J. Cell Biol 218:2829–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viais R, Fariña-Mosquera M, Villamor-Payà M, Watanabe S, Palenzuela L, et al. 2021. Augmin deficiency in neural stem cells causes p53-dependent apoptosis and aborts brain development. eLife 10:e67989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos JW, Pieuchot L, Evrard JL, Janski N, Bergdoll M, et al. 2008. The plant TPX2 protein regulates prospindle assembly before nuclear envelope breakdown. Plant Cell 20:2783–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainman A, Buster DW, Duncan T, Metz J, Ma A, et al. 2009. A new Augmin subunit, Msd1, demonstrates the importance of mitotic spindle-templated microtubule nucleation in the absence of functioning centrosomes. Genes Dev 23:1876–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Shioi G, Furuta Y, Goshima G. 2016. Intra-spindle microtubule assembly regulates clustering of microtubule-organizing centers during early mouse development. Cell Rep 15:54–60 [DOI] [PubMed] [Google Scholar]
- Widlund PO, Stear JH, Pozniakovsky A, Zanic M, Reber S, et al. 2011. XMAP215 polymerase activity is built by combining multiple tubulin-binding TOG domains and a basic lattice-binding region. PNAS 108:2741–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek M, Bechstedt S, Chaaban S, Brouhard GJ. 2015. Microtubule-associated proteins control the kinetics of microtubule nucleation. Nat. Cell Biol 17:907–16 [DOI] [PubMed] [Google Scholar]
- Wieczorek M, Huang T-L, Urnavicius L, Hsia K-C, Kapoor TM. 2020a. MZT proteins form multi-faceted structural modules in the γ-tubulin ring complex. Cell Rep 31:107791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek M, Urnavicius L, Ti SC, Molloy KR, Chait BT, Kapoor TM. 2020b. Asymmetric molecular architecture of the human γ-tubulin ring complex. Cell 180:165–75.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T, Boleti H, Antony C, Karsenti E, Vernos I. 1998. Localization of the kinesin-like protein Xklp2 to spindle poles requires a leucine zipper, a microtubule-associated protein, and dynein. J. Cell Biol 143:673–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T, Wilm M, Karsenti E, Vernos I. 2000. TPX2, a novel Xenopus MAP involved in spindle pole organization. J. Cell Biol 149:1405–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Lin Y-T, Wei R, Chen Y, Shan Z, Lee W-H. 2008. Hice1, a novel microtubule-associated protein required for maintenance of spindle integrity and chromosomal stability in human cells. Mol. Cell. Biol 28:3652–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtz M, Zupa E, Atorino ES, Neuner A, Bohler A, et al. 2022. Modular assembly of the principal microtubule nucleator γ-TuRC. Nat. Commun 13:473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Cameron LA, Maddox PS, Salmon ED, Danuser G. 2008. Regional variation of microtubule flux reveals microtubule organization in the metaphase meiotic spindle. J. Cell Biol 182:631–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi P, Goshima G. 2018. Microtubule nucleation and organization without centrosomes. Curr. Opin. Plant Biol 46:1–7 [DOI] [PubMed] [Google Scholar]
- Zahreddine H, Borden KL. 2013. Mechanisms and insights into drug resistance in cancer. Front. Pharmacol 4:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasadil LM, Andersen KA, Yeum D, Rocque GB, Wilke LG, et al. 2014. Cytotoxicity of paclitaxel in breast cancer is due to chromosome missegregation of multipolar spindles. Sci. Transl. Med 6:3007965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Roostalu J, Surrey T, Nogales E. 2017. Structural insight into TPX2-stimulated microtubule assembly. eLife 6:e30959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chen Q, Feng J, Hou J, Yang F, et al. 2009. Sequential phosphorylation of Nedd1 by Cdk1 and Plk1 is required for targeting of the γTuRC to the centrosome. J. Cell Sci 122:2240–51 [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhuang R, Ye Q, Zhuo J, Chen K, et al. 2019. High expression of human AugminComplex Submit 3 indicates poor prognosis and associates with tumor progression in hepatocellular carcinoma. J. Cancer 10:1434–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Wong ML, Alberts B, Mitchison T. 1995. Nucleation of microtubule assembly by a γ-tubulin-containing ring complex. Nature 378:578–83 [DOI] [PubMed] [Google Scholar]
- Zhu H, Coppinger JA, Jang CY, Yates JR 3rd, Fang G. 2008. FAM29A promotes microtubule amplification via recruitment of the NEDD1-γ-tubulin complex to the mitotic spindle. J. Cell Biol 183:835–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Huang R-Y, Jiang F-N, Chen D-X, Wang C, et al. 2018. Overexpression of TPX2 is associated with progression and prognosis of prostate cancer. Oncol. Lett 16:2823–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupa E, Zheng A, Neuner A, Wurtz M, Liu P, et al. 2020. The cryo-EM structure of a γ-TuSC elucidates architecture and regulation of minimal microtubule nucleation systems. Nat. Commun 11:5705. [DOI] [PMC free article] [PubMed] [Google Scholar]


