Abstract
Background
Iron overload is frequently observed in non-alcoholic fatty liver disease (NAFLD). Transferrin receptor 2 (TFR2) is an important key factor in iron regulation. We aimed to investigate whether TFR2 single nucleotide polymorphisms (SNPs) contribute to susceptibility to NAFLD in a Chinese Han population.
Methods
Five tag SNPs (rs10247962, rs4434553, rs2075672, rs1052897, and rs3757859) in the TFR2 gene were selected and genotyped in a case–control study on participants who visited two affiliated hospitals of Fujian Medical University between June 2011 and August 2017. Propensity score matching and inverse probability of treatment weighting analyses were used to verify the risk associated with TFR2 SNPs.
Results
Logistic regression analyses suggested that subjects with the rs4434553 GA or GG genotype had a lower risk of NAFLD than those carrying the AA genotype (odds ratio = 0.630, 95% confidence interval = 0.504–0.788). Moreover, the rs4434553 GA or GG genotype was negatively correlated with body mass index, hepatic steatosis index, and serum ferritin (b = −0.363, P = 0.008; b = −1.040, P = 0.009; b = −35.258, P = 0.015, respectively), and positively associated with serum hepcidin level (b = 35.308, P < 0.001). Moreover, rs10247962 and rs1052897 had multiplicative interactions with age in relation to the risk of NAFLD (P for interactions, 0.041 and 0.034, respectively). The cumulative effects of the rs10247962, rs1052897, and rs4434553 SNPs were positively associated with the risk of NAFLD (adjusted Ptrend = 0.012).
Conclusions
In this Chinese Han population, the rs4434553 polymorphism in TFR2 may be an independent influencing factor associated with the susceptibility to NAFLD. The ageing effect on the development of NAFLD may be inhibited by SNPs rs10247962 and rs1052897.
Keywords: NAFLD, transferrin receptor 2, single nucleotide polymorphism, gene–environment interaction
Introduction
Non-alcoholic fatty liver disease (NAFLD) is the leading chronic liver disease affecting the health of populations worldwide [1]. The prevalence of NAFLD is ∼25.2% globally [2] and ∼20.9% in China [3]. NAFLD can progress to non-alcoholic steatohepatitis, liver cirrhosis, and even hepatocellular carcinoma; however, its pathogenesis is complex and has not yet been clearly determined.
Multiple factors are involved in the occurrence of NAFLD, among which iron overload is a major concern [4]. The interaction between iron overload with insulin resistance and oxidative stress may drive towards the occurrence and development of NAFLD [5]. Our previous studies showed that high intake of heme-iron in the diet may be a risk factor for NAFLD [6], and the body iron load was also statistically correlated with the risk of NAFLD [7]. NAFLD is a genetic–environmental disease, and genetic variation relevant to iron metabolism might improve susceptibility to NAFLD. Thus, it is important to further explore the relationship between iron load and the risk of NAFLD at the genetic level.
Transferrin receptor 2 (TFR2) is an important factor in iron regulation [8]. It controls the secretion of hepcidin from the liver and improves the absorption of food iron to maintain systemic iron homeostasis. Disruption of TFR2 function may lead to excessive iron load, accumulation of iron in the liver, and the development of other diseases [9]. Research on TFR2 gene polymorphisms has mainly focused on the association of anaemia with type III hereditary hemochromatosis [10, 11]. However, the relationship between TFR2 gene polymorphisms and the risk of NAFLD remains unclear.
Given the strong association between iron overload and NAFLD, this study aimed to identify whether polymorphisms in the TFR2 gene are associated with increased risk of NAFLD in Chinese Han population. Subsequently, we examined the association of each SNP with certain metabolic phenotypes.
Methods
Participants and study design
This case–control study included new diagnostic NAFLD patients who underwent health-check examinations in two affiliated hospitals of Fujian Medical University (the Union Hospital and the Affiliated Nanping First Hospital, Fuzhou, China) between June 2011 and August 2017. Criteria for inclusion of patients were permanent residency in Fujian Province, China, and completion of ultrasonography examination and blood tests. Patients with a history of liver disease or alcohol misuse (weekly ethanol consumption of ≥140 g for men or ≥70 g for women) in the past year were excluded. Incident cases were diagnosed via liver ultrasonography using established criteria [12]. Controls were healthy participants without liver steatosis as determined by ultrasonography from the same centre during the same study period. Other inclusion and exclusion criteria for the controls were consistent with those of the cases. The study protocol was approved by the ethics committees of Fujian Medical University (ethics number 2014096) and written informed consent was obtained from all participants.
Anthropometric examination and blood sampling
All subjects underwent a complete physical examination in the morning after a 12-hour overnight fast and 48-hour alcohol abstinence. Health examination included a questionnaire on food intake, anthropometric measurements, laboratory data from blood samples, and abdominal ultrasonography. The patients’ weight and standing height were measured using a standardized protocol by a trained physician in order to calculate the body mass index (BMI), the ratio of weight (kg) to the square of the height (m2). Systolic and diastolic blood pressure readings were taken and hypertension was defined as systolic blood pressure of ≥140 mmHg and/or diastolic blood pressure of ≥90 mmHg. Venous blood samples were collected from an antecubital vein into vacutainer tubes containing ethylenediaminetetraacetic acid. Plasma was isolated using centrifugation (2,500 × g for 10 min at 4°C) and samples were aliquoted and frozen for subsequent analyses. The levels of fasting plasma glucose (FPG), total cholesterol (TC), total triglyceride (TG), high-density lipoprotein cholesterol (HDL-c), low-density lipoprotein cholesterol (LDL-c), apolipoprotein B, alanine aminotransferase (ALT), and aspartate aminotransferase (AST) were measured using standard clinical laboratory techniques, which have been described previously [13]. Diabetes was defined as serum FPG of ≥7.0 mmol/L. The hepatic steatosis index (HSI) was computed as 8 × (ALT/AST ratio) + BMI (+2, if female; +2, if diabetes mellitus). Overweight was defined as BMI of ≥24 kg/m2 [14].
For the indicators related to iron, heme-iron intake was assessed using a semi-quantitative food frequency questionnaire and calculated using Chinese food composition tables. Serum hepcidin, ferritin, and TFR2 levels were analysed using commercially available enzyme-linked immunosorbent assay kits, as described previously [7].
Single nucleotide polymorphism selection and genotyping
The genomic region harbouring TFR2 was examined to select SNPs based on the linkage disequilibrium patterns of the Chinese Han population in Beijing from the International HapMap Project SNP database (HapMap Data Rel. 28 Phase II + III, 10 August, on National Centre for Biotechnology Information B36 assembly, dbSNP b126, accessed April 2013). We then used the pairwise tagging method in the Haploview software (Version 4.2, Broad institute, Cambridge, MA, USA) to capture SNPs with a minimum minor allele frequency of >0.02 and a minimum r2 of > 0.8. The SNPs were prioritized according to the following criteria: (i) known SNPs associated with iron metabolic traits from the literature; and (ii) SNPs within coding regions, the promoter region (2,500 bp before the start codon), 3′ untranslated region (500 bp after the stop codon), or 100 bp before exon–intron splicing boundaries. As a result, five SNPs (rs10247962, rs4434553, rs2075672, rs1052897, and rs3757859) were selected from unrelated Han Chinese individuals in Beijing. DNA was isolated from peripheral blood leukocytes following standard procedures. As previously described [6], genotyping was performed using iPLEX technology based on a MassARRAY (improved multiplex ligation detection reaction) platform in all participants. Primers for amplification and extension reactions were designed using the Mass Array Assay Design software (Version 3.1, Sequenom, San Diego, CA, USA), and SNP genotypes were determined according to the iPLEX protocol provided by the manufacturer. Genotyping assays were performed by laboratory technicians who were blinded to the NAFLD status of each subject. The genotyping quality was assessed using a detailed quality control procedure consisting of a >95% successful call rate, duplicate calling of genotypes, internal positive control samples, and Hardy–Weinberg equilibrium testing.
Genetic risk score
The association of heterozygosity for HFE2 (encoding hemojuvelin BMP co-receptor, also known as HJV) and NAFLD with iron overload might interfere with part of the current results, so we defined the polymorphisms of rs7551245 in HFE2 as confounders. A method of genetic risk score (GRS) determination, based on the average number of risk alleles (0, 1, or 2) in the SNPs genotyped, was used to analyse the cumulative effect of the SNPs showing significant association with NAFLD.
Statistical analysis
Propensity scores were calculated for each participant in the multivariate logistic regression model, with all potential confounding factors included as covariates (Table 1). Individuals were matched using a 1:1 nearest-neighbour propensity score matching (PSM) algorithm within a caliper of 0.2. Inverse probability of treatment weighting (IPTW) conducts data statistics without loss of sample size. Based on the IPTW method, we used weights of control subjects of the inverse of one minus the propensity score and weights of case subjects of the inverse of the propensity scores.
Table 1.
The demographic characteristics of the subjects
| Characteristic | All participants |
Propensity score matching |
Inverse probability of treatment weighting |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Controls (n = 766) | NAFLD cases (n = 772) | P-value | Controls (n = 679) | NAFLD cases (n = 679) | P-value | Controls (n = 1,537) | NAFLD cases (n = 1,539) | P-value | |
| Source | 0.964 | 0.126 | 0.998 | ||||||
| Nanping | 232 (30.3) | 233 (30.2) | 221 (32.5) | 195 (28.7) | 472 (30.7) | 473 (30.7) | |||
| Xiehe | 534 (69.7) | 539 (69.8) | 458 (67.5) | 484 (71.3) | 1,065 (69.3) | 1,066 (69.3) | |||
| Sex | 0.100 | 1.000 | 0.854 | ||||||
| Male | 579 (75.6) | 555 (71.9) | 496 (73.0) | 496 (73.0) | 1,140 (74.2) | 1,137 (73.9) | |||
| Female | 187 (24.4) | 217 (28.1) | 183 (27.0) | 183 (27.0) | 397 (25.8) | 402 (26.1) | |||
| Age (years) | 0.685 | 0.575 | 0.999 | ||||||
| <40 | 281 (36.7) | 269 (34.8) | 234 (34.5) | 252 (37.1) | 549 (35.7) | 549 (35.7) | |||
| 40–60 | 394 (51.4) | 414 (53.7) | 367 (54.0) | 355 (52.3) | 806 (52.5) | 809 (52.5) | |||
| ≥60 | 91 (11.9) | 89 (11.5) | 78 (11.5) | 72 (10.6) | 181 (11.8) | 181 (11.8) | |||
| Marital status | 0.058 | 0.523 | 0.968 | ||||||
| Married | 676 (88.3) | 704 (91.2) | 609 (89.7) | 616 (90.7) | 159 (10.4) | 160 (10.4) | |||
| Others | 90 (11.7) | 68 (8.8) | 70 (10.3) | 63 (9.3) | 1,377 (89.6) | 1,379 (89.6) | |||
| Education | 0.295 | 0.120 | 0.038 | ||||||
| Primary school or below | 484 (63.2) | 515 (66.7) | 427 (62.9) | 461 (67.9) | 977 (63.6) | 1,003 (65.2) | |||
| Middle school | 141 (18.4) | 122 (15.8) | 119 (17.5) | 96 (14.1) | 289 (18.8) | 238 (15.5) | |||
| University or above | 141 (18.4) | 135 (17.5) | 133 (19.6) | 122 (18.0) | 271 (17.6) | 298 (19.3) | |||
| Smoking | 0.002 | 0.233 | 0.923 | ||||||
| No | 573 (74.8) | 521 (67.5) | 490 (72.2) | 470 (69.2) | 1,096 (71.3) | 1,095 (71.2) | |||
| Yes | 193 (25.2) | 251 (32.5) | 189 (27.8) | 209 (30.8) | 441 (28.7) | 444 (28.8) | |||
| Drinking | 0.023 | 1.000 | 0.959 | ||||||
| No | 553 (72.2) | 516 (66.8) | 477 (70.3) | 477 (70.3) | 1,061 (69.0) | 1,063 (69.1) | |||
| Yes | 213 (27.8) | 256 (33.2) | 202 (29.7) | 202 (29.7) | 476 (31.0) | 475 (30.9) | |||
| Drinking tea | 0.209 | 0.162 | 0.919 | ||||||
| No | 529 (69.1) | 510 (66.1) | 451 (66.4) | 475 (70.0) | 1,032 (67.1) | 1,036 (67.3) | |||
| Yes | 237 (30.9) | 262 (33.9) | 228 (33.6) | 204 (30.0) | 505 (32.9) | 503 (32.7) | |||
NAFLD, non-alcoholic fatty liver disease.
Analyses were performed using SPSS version 19.0.0.1 (IBM Corp., Armonk, NY, USA), R software (version 3.6.1), and GraphPad Prism 9.0.0 (GraphPad Inc., San Diego, CA, USA). Student’s t-test, the nonparametric Mann–Whitney U test, and Pearson’s chi-squared test were used to compare the characteristics between the patients with NAFLD and the controls. Unconditional logistic regression models were used to evaluate the odds ratio (OR) and 95% confidence interval (CI) for the association between TFR2 polymorphism genotypes and NAFLD risk. For a protective SNP that was negatively associated with the risk of NAFLD, the risk alleles were the major alleles. A P-value of <0.05 was considered statistically significant.
Results
Characteristics of participants
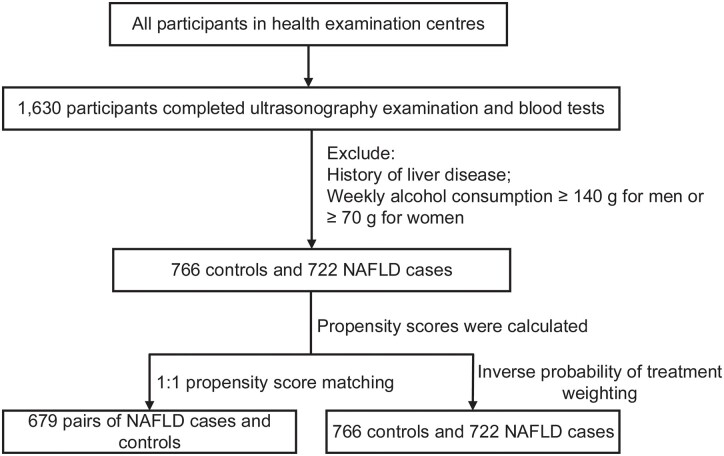
A total of 1,538 participants with quality-checked samples (766 patients with NAFLD and 772 healthy controls) were enrolled in this study. Individuals were matched by 1:1 PSM and 679 pairs of subjects were selected (Figure 1). The standardized mean difference of all variables was diminished after PSM and IPTW (Supplementary Figure 1).
Figure 1.
Flow chart of the study. NAFLD, non-alcoholic fatty liver disease.
As shown in Table 1, patients with NAFLD were more likely to smoke and drink alcohol than the controls. All demographic characteristics were similar between 679 NAFLD patients and 679 controls after PSM. After IPTW, the results showed that the patients with NAFLD had higher level of education than the controls. Furthermore, patients with NAFLD tended to have higher BMI and higher serum TG, TC, LDL-c, apolipoprotein B, ALT, and AST levels, but lower HDL-c levels (all P < 0.05), and were more likely to have hypertension and diabetes (both P < 0.001) than the control subjects (Table 2). The distribution of the polymorphic locus rs7551245 in HFE2 showed no significant difference between the patients with NAFLD and the controls (P = 0.804). Considering the variables related to iron metabolism, patients with NAFLD consumed more dietary heme-iron and had a lower level of serum TFR2 than the controls (P were 0.005 and 0.007, respectively).
Table 2.
The clinical characteristics of the subjects
| Characteristic | Controls (n = 764) | NAFLD cases (n = 772) | P-value for all participants | P-value for PSM | P-value for IPTW |
|---|---|---|---|---|---|
| BMI (kg/m2) | <0.001 | <0.001 | <0.001 | ||
| <24 | 605 (79.0) | 243 (31.5) | |||
| ≥24 | 161 (21.0) | 529 (68.5) | |||
| Hypertension | <0.001 | <0.001 | <0.001 | ||
| No | 670 (87.5) | 598 (77.5) | |||
| Yes | 96 (12.5) | 174 (22.5) | |||
| Diabetes | <0.001 | <0.001 | <0.001 | ||
| No | 740 (97.0) | 716 (92.7) | |||
| Yes | 23 (3.0) | 56 (7.3) | |||
| TG (mmol/L) | <0.001 | <0.001 | <0.001 | ||
| <1.7 | 617 (80.7) | 408 (52.8) | |||
| ≥1.7 | 148 (19.3) | 364 (47.2) | |||
| TC (mmol/L) | <0.001 | 0.001 | <0.001 | ||
| <5.2 | 446 (58.5) | 367 (47.5) | |||
| ≥5.2 | 317 (41.5) | 405 (52.5) | |||
| HDL-c (mmol/L) | <0.001 | <0.001 | <0.001 | ||
| <1.02 | 707 (92.4) | 616 (79.9) | |||
| ≥1.02 | 58 (7.6) | 155 (20.1) | |||
| LDL-c (mmol/L) | <0.001 | 0.002 | <0.001 | ||
| <3.12 | 387 (50.6) | 310 (40.2) | |||
| ≥3.12 | 378 (49.4) | 461 (59.8) | |||
| Apolipoprotein B (g/L) | <0.001 | <0.001 | <0.001 | ||
| <1.1 | 617 (80.7) | 510 (66.1) | |||
| ≥1.1 | 148 (19.3) | 262 (33.9) | |||
| ALT (U/L) | <0.001 | <0.001 | <0.001 | ||
| <40 | 702 (91.9) | 587 (76.0) | |||
| ≥40 | 62 (8.1) | 185 (24.0) | |||
| AST (U/L) | 0.041 | 0.015 | 0.003 | ||
| <40 | 734 (96.1) | 724 (93.8) | |||
| ≥40 | 30 (3.9) | 48 (6.2) | |||
| rs7551245 (HFE2 gene) | 0.804 | 0.599 | 0.697 | ||
| CC + TC | 196 (26.5) | 197 (25.9) | |||
| TT | 545 (73.5) | 564 (74.1) | |||
| Heme-iron (mg/d) | 4.36 (3.13–5.86) | 4.72 (3.65–6.30) | 0.005 | 0.002 | <0.001 |
| Hepcidin (ng/mL) | 90.39 (45.95–187.70) | 89.70 (39.96–179.73) | 0.388 | 0.645 | 0.141 |
| Ferritin (ng/mL) | 223.09 (84.49–359.34) | 250.15 (114.28–382.85) | 0.208 | 0.280 | 0.062 |
| TFR2 (ng/mL) | 15.14 (8.36–22.27) | 11.55 (6.00–19.96) | 0.007 | 0.008 | <0.001 |
NAFLD, non-alcoholic fatty liver disease; PSM, propensity score matching; IPTW, inverse probability of treatment weighting; BMI, body mass index; TG, total triglyceride; TC, total cholesterol; HDL-c, high-density lipoprotein cholesterol; LDL-c, low-density lipoprotein cholesterol; ALT, alanine aminotransferase; AST, aspartate aminotransferase.
The association between TFR2 genotypes and the risk of NAFLD
The genotype distribution frequencies of the five polymorphic loci in the TFR2 gene among the controls were assessed using the Hardy–Weinberg test, which indicated that the genotype frequencies of these SNPs were in accordance with genetic balance (all P > 0.05), suggesting that the controls could be representative of the target population (Supplementary Table 1).
The overall distribution of these five SNPs in all subjects and their associations with NAFLD risk are shown in Table 3. Logistic regression analyses suggested that the genotype of rs4434553 was statistically associated with the risk of NAFLD (P = 0.012). Patients with the GA or GG genotype of rs4434553 had a lower risk of NAFLD than those with the AA genotype; the OR (95% CI) was 0.702 (0.533–0.924) after matching for demographic characteristics. The results did not change markedly after further adjustment for age, sex, BMI, hypertension, diabetes, serum TG, TC, HDL-c, LDL-c, apolipoprotein B, ALT, and AST levels, and the polymorphisms in the HFE2 gene, and the OR (95% CI) was 0.625 (0.447–0.874). Among all participants after IPTW, the genotypes of rs10247962, rs1052897, and rs4434553 were all significantly associated with the risk of NAFLD; the ORs (95% CI) were 0.786 (0.650–0.950), 0.796 (0.657–0.964), and 0.701 (0.584–0.843), respectively. No statistical association was found between the other two SNPs and the risk of NAFLD (both P > 0.05).
Table 3.
The association between TFR2 genotypes and the risk of NAFLD
| Genotype | Controls [n (%)] | NAFLD cases [n (%)] | Propensity score matching |
Inverse probability of treatment weighting |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR1 (95% CI) | P 1 | OR2 (95% CI) | P 2 | OR1 (95% CI) | P 1 | OR2 (95% CI) | P 2 | |||
| rs10247962 | ||||||||||
| AA | 621 (81.7) | 652 (84.5) | 1 (reference) | – | 1 (reference) | – | 1 (reference) | – | 1 (reference) | – |
| GA + GG | 139 (18.3) | 120 (15.5) | 0.786 (0.590–1.046) | 0.098 | 0.859 (0.608–1.214) | 0.389 | 0.786 (0.650–0.950) | 0.013 | 0.839 (0.667–1.056) | 0.135 |
| rs1052897 | ||||||||||
| TT | 630 (82.2) | 655 (84.8) | 1 (reference) | – | 1 (reference) | – | 1 (reference) | – | 1 (reference) | – |
| TA + AA | 136 (17.8) | 117 (15.2) | 0.805 (0.603–1.075) | 0.142 | 0.862 (0.608–1.223) | 0.406 | 0.796 (0.657–0.964) | 0.019 | 0.840 (0.666–1.059) | 0.140 |
| rs2075672 | ||||||||||
| GG | 509 (66.4) | 501 (64.9) | 1 (reference) | – | 1 (reference) | – | 1 (reference) | – | 1 (reference) | – |
| GA + AA | 257 (33.6) | 271 (35.1) | 1.043 (0.834–1.305) | 0.713 | 0.939 (0.715–1.233) | 0.650 | 1.023 (0.882–1.187) | 0.763 | 0.939 (0.783–1.125) | 0.494 |
| rs3757859 | ||||||||||
| AA | 625 (81.7) | 643 (83.3) | 1 (reference) | – | 1 (reference) | – | 1 (reference) | – | 1 (reference) | – |
| CA + CC | 140 (18.3) | 129 (16.7) | 0.883 (0.666–1.170) | 0.385 | 0.954 (0.679–1.341) | 0.787 | 0.864 (0.717–1.041) | 0.124 | 0.918 (0.733–1.151) | 0.458 |
| rs4434553 | ||||||||||
| AA | 601 (78.9) | 649 (84.1) | 1 (reference) | – | 1 (reference) | – | 1 (reference) | – | 1 (reference) | – |
| GA + GG | 161 (21.1) | 123 (15.9) | 0.702 (0.533–0.924) | 0.012 | 0.625 (0.447–0.874) | 0.006 | 0.701 (0.584–0.843) | <0.001 | 0.630 (0.504–0.788) | <0.001 |
OR1 adjusted for age and sex.
OR2 adjusted for age, sex, body mass index, hypertension, diabetes, serum total triglyceride, total cholesterol, high-density lipoprotein cholesterol, low-density lipoprotein cholesterol, apolipoprotein B, alanine aminotransferase, aspartate aminotransferase, and the polymorphisms in the HFE2 gene.
NAFLD, non-alcoholic fatty liver disease; OR, odds ratio; CI, confidence interval.
The association between TFR2 genotypes and the risk of NAFLD after stratification
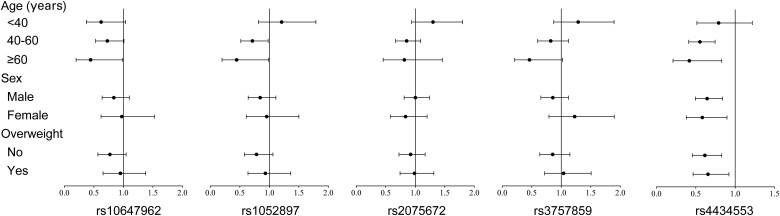
To further analyse the relationship between TFR2 polymorphisms and the risk of NAFLD, we stratified them by age, sex, and BMI after IPTW. As shown in Figure 2, for individuals ≥60 years old, subjects carrying the GA or AA genotype of rs10247962 had a lower risk of NAFLD than those with the GG genotype (P = 0.049). For subjects ≥40 years old (40–60 and ≥60 years old), subjects with the TA or TT genotype of rs1052897 and the GA or GG genotype of rs4434553 had a lower risk of NAFLD than those with the wild-type genotype (P = 0.042 and P = 0.049, respectively). In addition, we found that rs10247962 and rs1052897 had multiplicative interactions with age in relation to the risk of NAFLD (P-values for the interactions were 0.041 and 0.034, respectively). The risk of NAFLD was lower in patients carrying the GA or GG genotype of rs4434553 than in those carrying the AA genotype, whether or not the results were stratified by sex or BMI status. There was no significant association between rs207567 or rs3757859 and the risk of NAFLD.
Figure 2.
The association between TFR2 genotypes and the risk of NAFLD after stratification. NAFLD, non-alcoholic fatty liver disease.
The correlation between TFR2 genotypes and clinical parameters
After IPTW and adjustment for the polymorphisms of HFE2, multiple linear regression analyses revealed that the GA or GG genotype of rs4434553 was negatively correlated with BMI (b = −0.363, t = 2.655, P = 0.008), HSI (b = −1.040, t = 2.598, P = 0.009) and serum ferritin levels (b = −35.258, t = 2.447, P = 0.015). It was worth noting that the hepcidin level was significantly higher in NAFLD patients with the GA or GG genotype of rs4434553 than in those with the AA genotype (b = 35.308, t = 3.720, P < 0.001).
The association between GRS and the risk of NAFLD
The above findings suggested that the GA or AA genotype of rs10247962, the TA or TT genotype of rs1052897, and the GA or GG genotype of rs4434553 might be associated with lower risk of NAFLD. GRS was calculated to further analyse the impact of the cumulative effect of the three SNPs on NAFLD. Table 4 shows the distribution of NAFLD risk alleles in both cases and controls. Compared with the subjects with a GRS of 0, the risk ORs (95% CIs) of the subjects with GRS scores of 1 and ≥2 were 3.487 (1.624–7.485) and 3.778 (1.807–7.898), respectively, after IPTW. With the increased number of disease-associated genotypes, there was a stepwise increase in the risk of NAFLD (P for trend = 0.012).
Table 4.
The association between the genetic risk score and the risk of NAFLD
| GRS | Controls [n (%)] | NAFLD cases [n (%)] | Propensity score matching |
Inverse probability of treatment weighting |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR1 (95% CI) | P 1 | OR2 (95% CI) | P 2 | OR1 (95% CI) | P 1 | OR2 (95% CI) | P 2 | |||
| 0 | 17 (2.2) | 7 (0.9) | 1 (reference) | – | 1 (reference) | – | 1 (reference) | – | 1 (reference) | – |
| 1 | 122 (15.9) | 108 (14.0) | 2.329 (0.876–6.197) | 0.090 | 3.696 (1.144–11.943) | 0.029 | 2.113 (1.111–4.021) | 0.023 | 3.487 (1.624–7.485) | 0.001 |
| ≥2 | 627 (81.9) | 657 (85.1) | 2.822 (1.096–7.269) | 0.032 | 3.851 (1.241–11.953) | 0.020 | 2.611 (1.403–4.861) | 0.002 | 3.778 (1.807–7.898) | <0.001 |
| P for trend | 0.026 | 0.137 | 0.001 | 0.012 | ||||||
OR1 adjusted for age and sex.
OR2 adjusted for age, sex, body mass index, hypertension, diabetes, serum total triglyceride, total cholesterol, high-density lipoprotein cholesterol, low-density lipoprotein cholesterol, apolipoprotein B, alanine aminotransferase, aspartate aminotransferase, and the polymorphisms in the HFE2 gene.
NAFLD, non-alcoholic fatty liver disease; OR, odds ratio; CI, confidence interval; GRS, genetic risk score.
Discussion
In this case–control study in a Chinese Han population, we adopted a genotype and GRS approach to explore the association between tag SNPs in the TFR2 gene and the risk of NAFLD. We found that certain genotypes of polymorphisms rs10247962, rs1052897, and rs443455 were significantly associated with decreased risk of NAFLD. Furthermore, the GA or GG genotype of rs4434553 was negatively correlated with BMI, HIS, and serum ferritin level, and positively correlated with the serum hepcidin level. Interaction effect analysis showed that the GA or GG genotype of rs10247962 and the TA or TT genotype of rs1052897 might be beneficial genotypes against the risk effect of age on NAFLD. The cumulative effect of the rs10247962, rs1052897, and rs4434553 SNPs was estimated, and a significantly increased trend in the risk of NAFLD with increasing GRS was observed (adjusted Ptrend = 0.012). This study investigated the association between gene polymorphisms in the promoter region of TFR2 and NAFLD, and demonstrated that TFR2 genetic polymorphisms might be influencing factors associating with the susceptibility to NAFLD independently or jointly in the Chinese Han population.
The TFR2 gene, which comprises 18 exons and maps to chromosome 7q22, encodes a protein that is involved in the regulation of iron metabolism in the body [15]. In 2000, Camaschella et al. [16] first discovered that the Y250X mutation in the TFR2 gene resulted in excessive deposition of iron and hereditary hemochromatosis. Subsequently, studies found that TFR2-mutant mice had decreased hepcidin expression and liver iron accumulation, and increased intestinal iron absorption [17, 18]. Hepcidin is a circulatory peptide that is synthesized in the liver and acts as the key regulator of iron metabolism by modulating iron absorption through the duodenum and the release of iron from macrophages [19]. A previous study suggested that Tfr2, combined with hepcidin, led to excessive iron and affected the process of NAFLD [20]. To date, the relationship between TFR2 gene polymorphisms and the risk of NAFLD remains unclear.
In the present study, we found that TFR2 SNPs rs10247962, rs1052897, and rs443455 were significantly associated with the risk of NAFLD. The effect of SNPs on disease mechanism is mainly achieved by the regulation of gene expression by polymorphic sites [21, 22]. These SNPs are located in the promoter region of TFR2. As the target site for transcription factors, promoters play a key role in the initiation and transcriptional regulation of gene expression. The SNPs in the TFR2 promoter region might regulate the TFR2 transcription, thereby participating in the pathogenesis of NAFLD. In addition, we found that the GA or GG genotype of rs4434553 was negatively correlated with the BMI, HSI, and serum ferritin level, and positively correlated with the serum hepcidin level. HSI is an index used to assess the severity of hepatic steatosis, which could be associated with sex, ALT, AST, BMI, and diabetic status [23]. The levels of BMI and HSI reflect events occurring in the liver and adipose tissue. Hepcidin acts as a key regulator of iron metabolism by binding to Tfr2. Our previous study found that hepcidin and central obesity have a multiplicative interaction in NAFLD [7]. Under the condition of obesity, inflammation and oxidative stress are caused by the stimulation of adipokines, and in turn results in iron metabolism being negatively regulated by hepcidin and TFR2, contributing to NAFLD development. Based on these findings, we hypothesize that the GA or GG genotype of rs4434553 might negatively regulate the expression of TFR2. However, we did not find such an association between rs4434553 and the level of serum TFR2, suggesting that there may be other factors associated with TFR2 expression. Auguet et al. [24] found that the expression of TFR2 mRNA in the liver of morbidly obese patients with NAFLD was higher than that in the liver of the healthy population. Furthermore, TFR2 mRNA levels were also elevated in non-alcoholic steatohepatitis [25]. By contrast, Mitsuyoshi et al. [26] reported that the TFR2 level decreased in advanced NAFLD. Thus, the association of TFR2 with the development of NAFLD remains controversial. In future research, the relationship between rs4434553 and the expression of TFR2 at both mRNA and protein levels should be verified, and the potential mechanism should be explored.
For other SNPs, we found that age influences the effects of the GA or GG genotype of rs10247962 and the TA or TT genotype of rs1052897 on the risk of NAFLD. The older the age, the stronger the protective effects of the GA or GG genotype of rs10247962 and the TA or TT genotype of rs1052897 against NAFLD. Ageing is one of the major environmental risk factors for complex diseases, including NAFLD. Many crucial biological functions significantly decline with age, such as DNA repair, wound healing, the immune response, and metabolism [27]. Systemic changes caused by age may affect complex phenotypes in a genotype-dependent manner [28]. Thus, we speculated that these three SNPs might exhibit age-related heterogeneity in the present study. In a meta-analysis, Simino et al. [29] reported nine age-related SNPs associated with blood pressure. In addition, the age effect may be influenced by genetic variance SNPs of TFR2 or other genes. As reported by Dongen et al. [30], the interaction between age and genetic influencing SNPs could affect the methylome in whole blood. To date, few studies have been conducted on the polymorphisms of rs10247962 or rs1052897, or the interaction between them and age. Therefore, the mechanisms by which age influences the effect of TFR2 SNPs on NAFLD require further study.
Accumulating evidence has indicated that the effect of single genetic variations might be dependent on environmental factors (gene–environment interactions) or other genetic variations (gene–gene interactions). The application of GRS has gained success in genetic research of complex diseases, providing improved predictability of genetic polymorphisms for diseases [31, 32]. In this study, three SNPs related to NAFLD (rs10247962, rs1052897, and rs4434553) were used as markers to calculate the GRS of each subject, and a positive association between GRS and the occurrence of NAFLD was observed (adjusted Ptrend = 0.012). GRS integrated the weak effects of these three SNPs, which improved the predictability of TFR2 gene polymorphisms for NAFLD.
There were several limitations in the current study. First, the studied patients were limited to the Han ethnicity in southern China; thus, the generalization of our findings to other ethnicities should be carried with caution and prospective studies with wider populations are necessary. Second, the biological functions of TFR2 SNPs in NAFLD have not been studied; therefore, the underlying mechanism remained unclear, and the expression of TFR2 mRNA and protein should be further investigated. Third, only one SNP related to iron metabolism was associated with the risk of NAFLD in this study, and the association between more SNPs and the risk of NAFLD needs to be further studied. Fourth, this study did not investigate the interaction between dietary iron and TFR2 SNPs on NAFLD risk. As some of the population in this study did not have dietary information, further studies will be conducted in the future. Finally, as an observational study, there were unmeasured confounders, though a comprehensive set of potential factors was used for minimizing the confounder effects. For instance, we could not exclude that there are other gene products that might interfere with iron metabolism besides HFE2.
Conclusions
The present study provides new evidence for the association between TFR2 SNPs and the risk of NAFLD in the Chinese Han population. Interactions between genes or between genes and the environment might play more important roles than a single locus. Our study provides important insights into the aetiology of NAFLD from both epidemiological and statistical viewpoints, enabling a better understanding of the complex pathogenic mechanisms. It would be worth validating our results in other ethnic populations, and prospective investigation should be conducted on in-depth mechanisms by which TFR2 gene SNPs influence susceptibility to NAFLD.
Supplementary Data
Supplementary data is available at Gastroenterology Report online.
Authors’ Contributions
P.X.E. designed the study. P.X.T., P.H.W., and Z.J.C. collected the data. P.X.T. and P.H.W. analysed the data. W.Y.L. and H.Z.J. contributed to the interpretation of results. P.X.T. and P.X.E. wrote and reviewed the manuscript. All authors have discussed the results and commented on the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China [grant number 81473047] and the Natural Science Foundation of Fujian Province [grant number 2019J01316].
Supplementary Material
Acknowledgements
We thank all the study participants for their cooperation. We also thank our staff for recruiting the subjects and for their technical assistance.
Contributor Information
Xinting Pan, Department of Epidemiology and Health Statistics, Fujian Provincial Key Laboratory of Environment Factors and Cancer, School of Public Health, Fujian Medical University, Fuzhou, P. R. China; The United Innovation of Mengchao Hepatobiliary Technology Key Laboratory of Fujian Province, Mengchao Hepatobiliary Hospital of Fujian Medical University, Fuzhou, P. R. China.
Hewei Peng, Department of Epidemiology and Health Statistics, Fujian Provincial Key Laboratory of Environment Factors and Cancer, School of Public Health, Fujian Medical University, Fuzhou, P. R. China.
Junchao Zhang, Department of Epidemiology and Health Statistics, Fujian Provincial Key Laboratory of Environment Factors and Cancer, School of Public Health, Fujian Medical University, Fuzhou, P. R. China.
Yunli Wu, Key Laboratory of Ministry of Education for Gastrointestinal Cancer, Fujian Medical University, Fuzhou, P. R. China.
Zhijian Hu, Department of Epidemiology and Health Statistics, Fujian Provincial Key Laboratory of Environment Factors and Cancer, School of Public Health, Fujian Medical University, Fuzhou, P. R. China.
Xian-E Peng, Department of Epidemiology and Health Statistics, Fujian Provincial Key Laboratory of Environment Factors and Cancer, School of Public Health, Fujian Medical University, Fuzhou, P. R. China; Key Laboratory of Ministry of Education for Gastrointestinal Cancer, Fujian Medical University, Fuzhou, P. R. China.
Conflict of Interest
All the authors declare that there are no financial or any other relationships that might lead to a conflict of interest of this article.
References
- 1. Loomba R, Friedman S, Shulman G.. Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell 2021;184:2537–64. [DOI] [PubMed] [Google Scholar]
- 2. Younossi ZM, Koenig AB, Abdelatif D. et al. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology (Baltimore, MD) 2016;64:73–84. [DOI] [PubMed] [Google Scholar]
- 3. Li Z, Xue J, Chen P. et al. Prevalence of nonalcoholic fatty liver disease in mainland of China: a meta-analysis of published studies. J Gastroenterol Hepatol 2014;29:42–51. [DOI] [PubMed] [Google Scholar]
- 4. Buzzetti E, Petta S, Manuguerra R. et al. Evaluating the association of serum ferritin and hepatic iron with disease severity in non-alcoholic fatty liver disease. Liver Int 2019;39:1325–34. [DOI] [PubMed] [Google Scholar]
- 5. Datz C, Müller E, Aigner E.. Iron overload and non-alcoholic fatty liver disease. Minerva Endocrinol 2017;42:173–83. [DOI] [PubMed] [Google Scholar]
- 6. Peng XE, Xu SH, Liu WJ. et al. Independent and combined effects of dietary iron composition and selected risk factors on the risk of NAFLD in a Chinese population. Sci Rep 2019;9:4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pan XT, Chen BB, Liu WJ. et al. Circulating iron levels interaction with central obesity on the risk of nonalcoholic fatty liver disease: a case-control study in Southeast China. Ann Nutr Metab 2019;74:207–14. [DOI] [PubMed] [Google Scholar]
- 8. Wortham A, Goldman D, Chen J. et al. Extrahepatic deficiency of transferrin receptor 2 is associated with increased erythropoiesis independent of iron overload. J Biol Chem 2020;295:3906–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Richard C, Verdier F.. Transferrin receptors in erythropoiesis. IJMS 2020;21:9713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tippairote T, Bjørklund G, Peana M. et al. The proteomics study of compounded HFE/TF/TfR2/HJV genetic variations in a Thai family with iron overload, chronic anemia, and motor neuron disorder. J Mol Neurosci 2021;71:545–55. [DOI] [PubMed] [Google Scholar]
- 11. Wang Y, Du Y, Liu G. et al. Identification of novel mutations in HFE, HFE2, TfR2, and SLC40A1 genes in Chinese patients affected by hereditary hemochromatosis. Int J Hematol 2017;105:521–5. [DOI] [PubMed] [Google Scholar]
- 12. Chalasani N, Younossi Z, Lavine JE. et al. The diagnosis and management of nonalcoholic fatty liver disease: practice guidance from the American Association for the Study of Liver Diseases. Hepatology 2018;67:328–57. [DOI] [PubMed] [Google Scholar]
- 13. Peng XE, Wu YL, Lu QQ. et al. Two genetic variants in FABP1 and susceptibility to non-alcohol fatty liver disease in a Chinese population. Gene 2012;500:54–8. [DOI] [PubMed] [Google Scholar]
- 14. Zhou B-F. Effect of body mass index on all-cause mortality and incidence of cardiovascular diseases: report for meta-analysis of prospective studies open optimal cut-off points of body mass index in Chinese adults. Biomed Environ Sci 2002;15:245–52. [PubMed] [Google Scholar]
- 15. Kawabata H, Yang R, Hirama T. et al. Molecular cloning of transferrin receptor 2: a new member of the transferrin receptor-like family. J Biol Chem 1999;274:20826–32. [DOI] [PubMed] [Google Scholar]
- 16. Camaschella C, Roetto A, Calì A. et al. The gene TFR2 is mutated in a new type of haemochromatosis mapping to 7q22. Nat Genet 2000;25:14–5. [DOI] [PubMed] [Google Scholar]
- 17. Guo S, Yang C, Jiang S. et al. Repeated restraint stress enhances hepatic TFR2 expression and induces hepatic iron accumulation in rats. Biol Trace Elem Res 2020;196:590–6. [DOI] [PubMed] [Google Scholar]
- 18. Drake SF, Morgan EH, Herbison CE. et al. Iron absorption and hepatic iron uptake are increased in a transferrin receptor 2 (Y245X) mutant mouse model of hemochromatosis type 3. Am J Physiol Gastrointest Liver Physiol 2007;292:G323–8. [DOI] [PubMed] [Google Scholar]
- 19. Prentice AM. Clinical implications of new insights into hepcidin-mediated regulation of iron absorption and metabolism. Ann Nutr Metab 2017;71:40–8. [DOI] [PubMed] [Google Scholar]
- 20. Roetto A, Mezzanotte M, Pellegrino RM.. The functional versatility of transferrin receptor 2 and its therapeutic value. Pharmaceuticals 2018;11:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Montesion M, Williams ZH, Subramanian RP. et al. Promoter expression of HERV-K (HML-2) provirus-derived sequences is related to LTR sequence variation and polymorphic transcription factor binding sites. Retrovirology 2018;15:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang X, Pittman GS, Bandele OJ. et al. Linking polymorphic p53 response elements with gene expression in airway epithelial cells of smokers and cancer risk. Hum Genet 2014;133:1467–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee JH, Kim D, Kim HJ. et al. Hepatic steatosis index: a simple screening tool reflecting nonalcoholic fatty liver disease. Dig Liver Dis 2010;42:503–8. [DOI] [PubMed] [Google Scholar]
- 24. Auguet T, Aragonès G, Berlanga A. et al. Hepcidin in morbidly obese women with non-alcoholic fatty liver disease. PLoS One 2017;12:e0187065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moya D, Baker SS, Liu W. et al. Novel pathway for iron deficiency in pediatric non-alcoholic steatohepatitis. Clin Nutr 2015;34:549–56. [DOI] [PubMed] [Google Scholar]
- 26. Mitsuyoshi H, Yasui K, Harano Y. et al. Analysis of hepatic genes involved in the metabolism of fatty acids and iron in nonalcoholic fatty liver disease. Hepatol Res 2009;39:366–73. [DOI] [PubMed] [Google Scholar]
- 27. Niccoli T, Partridge L.. Ageing as a risk factor for disease. Curr Biol 2012;22:R741–52. [DOI] [PubMed] [Google Scholar]
- 28. Wang K, Basu M, Malin J. et al. A transcription-centric model of SNP-age interaction. PLoS Genet 2021;17:e1009427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Simino J, Shi G, Bis JC. et al. ; LifeLines Cohort Study. Gene-age interactions in blood pressure regulation: a large-scale investigation with the CHARGE, Global BPgen, and ICBP Consortia. Am J Hum Genet 2014;95:24–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Van Dongen J, Nivard MG, Willemsen G. et al. ; BIOS Consortium. Genetic and environmental influences interact with age and sex in shaping the human methylome. Nat Commun 2016;7:11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lahelma M, Luukkonen PK, Qadri S. et al. Assessment of lifestyle factors helps to identify liver fibrosis due to non-alcoholic fatty liver disease in obesity. Nutrients 2021;13:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zusi C, Mantovani A, Olivieri F. et al. Contribution of a genetic risk score to clinical prediction of hepatic steatosis in obese children and adolescents. Dig Liver Dis 2019;51:1586–92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.