Abstract
Genes coding for homologs of the highly conserved cell division protein FtsZ were isolated from Bartonella henselae and Bartonella quintana, the causative agents of cat scratch disease and trench fever, respectively. DNA fragments coding for the ftsZ open reading frames (ORFs) were cloned into Escherichia coli following PCR amplification with primers based on the ftsZ sequence of the closely related species Bartonella bacilliformis. The amino acid sequences predicted from the cloned B. henselae and B. quintana ftsZ ORFs are 81 to 83% identical to the corresponding protein in B. bacilliformis. Like the FtsZ protein of B. bacilliformis, the B. henselae and B. quintana homologs are about twice as large as the FtsZ proteins reported in most other organisms. Localized sequence differences within the C-terminal coding regions of the Bartonella ftsZ genes were used as the basis for species-specific identification of these organisms at both the DNA and protein levels. Oligonucleotide primers which permit the amplification of an ftsZ fragment from each of the Bartonella species without amplifying DNA from the other two species were designed. Anti-FtsZ antisera raised in rabbits against synthetic peptides corresponding to the relatively divergent C-terminal regions were shown via Western blot analysis to react only with the FtsZ protein from the cognate Bartonella species. These observations raise the possibility that the differences in ftsZ sequences can be used as the basis for diagnostic tests to differentiate among these closely related pathogens.
The genus Bartonella consists of aerobic, fastidious, gram-negative bacilli belonging to the alpha-2 subgroup of the class Proteobacteria. Bartonella currently includes 11 species, 4 of which are recognized as human pathogens. Bartonella bacilliformis is the etiologic agent of bartonellosis (Carrion’s disease), a biphasic disease endemic to remote areas of South America. Bartonellosis is transmitted by the bite of certain nocturnal sandflies, whose short flight ranges and particular temperature and humidity requirements limit the spread of the disease to the areas of endemicity. Humans are the only known reservoir of B. bacilliformis. The primary, acute phase of bartonellosis is characterized by fever, hemolytic anemia, and bacteremia. In this phase, B. bacilliformis parasitizes almost 100% of the erythrocytes. The secondary, chronic phase is characterized by skin lesions, referred to as verruga peruana, in which B. bacilliformis invades endothelial cells, causing hemoangiomas. Research interest in this organism has increased in recent years due to a rural Peruvian epidemic in 1987 with a fatality rate of 88% in untreated cases (10).
Bartonella henselae, a close relative of B. bacilliformis, is the causative agent of cat scratch disease (CSD) and bacillary angiomatosis (BA). CSD is typically a benign and self-limited condition lasting 6 to 12 weeks in untreated individuals. Regional lymphadenopathy is the predominant clinical feature of CSD, with 25 to 60% of patients reporting a cutaneous lesion at the site of a cat scratch or bite (24). More serious symptoms, including encephalopathy and pulmonary disease, occur rarely.
BA is a newly recognized disease characterized by cutaneous and subcutaneous vascular lesions which bear distinct clinical and histological similarities to the lesions observed in the chronic secondary phase of bartonellosis. BA was first described in 1983, predominantly among patients infected with human immunodeficiency virus (31). Since then, the clinical spectrum has expanded to include patients with proliferative vascular lesions affecting virtually every organ system, including bone, liver, and spleen (24). Independently, an unknown gram-negative pathogen was isolated from human immunodeficiency virus patients with fever and bacteremia, the symptoms of CSD, but these patients lacked vascular lesions and were not recognized as having BA. Eventually it was shown that B. henselae is the causative agent of both CSD and BA and that the domestic cat is a reservoir for the organism. The cat flea (Ctenocephalides felis) was recently shown to be a vector for B. henselae, capable of transmitting the organism among felines (4).
Bartonella quintana has also been associated with BA and is the causative agent of trench fever, a disease that was prevalent among troops during World War I. The human body louse was shown to be the vector for B. quintana transmission, at least in the case of trench fever. Another focus of B. quintana infections (referred to as “urban trench fever”) has been identified among the homeless population of Seattle, Wash. (24). No other reservoir, including cats, has been demonstrated for B. quintana.
Recently, a 75-kDa antigen of B. bacilliformis was identified as a homolog of FtsZ, an essential cell division protein that is highly conserved among prokaryotes (23). The N-terminal half (321 amino acids) of the B. bacilliformis homolog, FtsZBb, exhibits 45 to 90% sequence identity with other FtsZ proteins and contains all of the domains known to be necessary for FtsZ cell division activities, including the GTP-binding domain. In addition, the extreme C terminus of FtsZBb contains a region of 14 amino acids which is conserved in almost all of the currently characterized FtsZ proteins. However, FtsZBb differs from most of the other FtsZ proteins reported to date in that it contains a unique C-terminal domain, resulting in a protein almost twice as large as the majority of the currently characterized FtsZ homologs. The C-terminal half of FtsZBb (271 amino acids) is very hydrophilic and is predicted to contain a high proportion of antigenic sites based on computer modeling (23).
In this study, we report the isolation and characterization of ftsZ homologs in B. henselae and B. quintana. We also describe the development of strategies to identify and distinguish Bartonella species by using ftsZ DNA and protein sequences.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
B. bacilliformis KC584 (ATCC 35686) was grown on heart infusion agar plates supplemented with 5% defibrinated rabbit blood (BBL-Becton Dickinson, Cockeysville, Md.) at 28°C for 10 to 20 days. B. henselae Houston-1 (ATCC 49882) and B. quintana Oklahoma (OK 90-268) and Fuller (CIP 103739; ATCC VR-358) were grown on the same medium at 37°C for 10 to 20 days. Bacteria were harvested and resuspended in phosphate-buffered saline. Escherichia coli JM105 was grown in Luria broth (LB) or LB containing 50 μg ampicillin per ml when transformed with plasmids derived from pBluescript (Stratagene, La Jolla, Calif.) or pUC18 (Sigma, St. Louis, Mo.) vectors.
DNA extraction.
Total genomic DNA was extracted from Bartonella strains to serve as a template for PCR amplification by a protocol described previously (1). Plasmid DNA for sequencing was isolated with QIAGEN reagents, according to the manufacturer’s protocols (QIAGEN Inc., Chatsworth, Calif.).
PCR amplification.
PCR amplifications were performed with a kit from Boehringer Mannheim (Indianapolis, Ind.). The 100-μl reaction mixture (in 10 mM Tris-HCl, 1.5 mM MgCl2, 50 mM KCl [pH 8.3]) consisted of template (10 ng), forward and reverse primers (1 μg each), and a mixture of nucleotides (each at a final concentration of 200 μM). Both high- and low-stringency protocols were used. The high-stringency protocol consisted of 30 cycles of 94°C (1 min)–55°C (1 min)–72°C (3 min) followed by a 30-min extension period at 72°C. In the low-stringency protocol, 27 cycles under the conditions described above were preceded by three cycles of 94°C (1 min)–37°C (1 min)–72°C (3 min). PCR products were purified with the QIAQUICK PCR purification system (QIAGEN).
Molecular cloning of ORFs coding for ftsZ homologs.
Cloning of the gene coding for the FtsZ homolog of B. bacilliformis (ftsZBb) was described previously (23). Genes coding for the FtsZ homologs of B. henselae (ftsZBh) and B. quintana (ftsZBq) were cloned by insertion of PCR products into pUC18. ftsZBh and ftsZBq were amplified by a two-step strategy with primers derived from the published sequence of ftsZBb and preliminary sequence information on ftsZBh. All primers for PCR and sequencing were synthesized by a model 394 DNA synthesizer (Applied Biosystems, Foster City, Calif.). The forward and reverse primer sequences used for amplification of the 5′ portion of the gene were 5′ GAGGTAAGAATTCTCAACGTGTTGGTCAG 3′ (forward) and 5′ CGCATAGAAGTATCATCCAACAACGG 3′ (reverse). The forward primer sequence was derived from a region located approximately 90 bp upstream of the ftsZBb open reading frame (ORF) and was modified to contain an EcoRI site (underlined). The reverse primer corresponded to a sequence within the ftsZBh ORF, located about 750 bp from the initiation codon. For amplification of the 3′ portion of the gene, the sequence of the forward primer was 5′ CCGTTGTTGGATGATACTTCTATGCG 3′ while the sequences of the reverse primers were 5′ CTCTTTCGGATCCTATTCATTAATTCGCTTGGCGACG 3′ for B. henselae and 5′ CTCTTTCGGATCCTATTCATTAGTTCGCTTGGCGACG 3′ for B. quintana. In this case, the forward primer was derived from a sequence within the ftsZBh ORF overlapping that of the reverse primer for the 5′ amplification reaction. The reverse primers corresponded to a sequence consisting of the final 21 bp of the ftsZBh (or ftsZBq) ORF plus an additional 16 bp derived from sequence downstream of the ftsZBb ORF, modified to contain a BamHI site (underlined). The resulting amplification products were gel purified with the QIAQUICK kit and then subjected to another PCR containing only the flanking primers, resulting in a product consisting of the entire ORF plus flanking regions containing restriction enzyme sites. These PCR products were digested with EcoRI and BamHI and ligated into the appropriately digested and dephosphorylated pUC18 vector. The ligation reactions were transformed into competent E. coli JM105 cells which were then grown on LB containing 50 μg of ampicillin per ml to select cells harboring the recombinant plasmid (28). Incorporation of the PCR products into the vector was verified by gel electrophoresis of the recombinant plasmids. Plasmids containing PCR products derived from B. henselae and B. quintana were designated pUCBH-Z and pUCBQ-Z, respectively. Expression of FtsZBh and FtsZBq in E. coli was confirmed via Western blotting.
DNA sequencing.
The nucleotide sequences of both strands of pUCBH-Z and pUCBQ-Z were obtained by the methods of Sanger et al. (29) with an Applied Biosystems model 373 automated nucleic acid sequencer. These sequences were verified through sequence analysis of ftsZ amplification products from the chromosomes of B. henselae and B. quintana. Parallel DNA sequence analysis was also carried out on the previously cloned ftsZBb (23) sequence and on ftsZ amplification products from the chromosome of B. bacilliformis. The ftsZBb sequence derived from this analysis was found to differ from the previously published sequence for this ORF at 19 positions, resulting in 13 alterations of the amino acid sequence. Following verification, the corrected ftsZBb sequence was submitted to GenBank (accession no. AF007266) to reflect these changes.
DNA sequence analysis.
DNA and protein sequences were analyzed with the software package MacDNAsis (Hitachi Software Engineering America Ltd., South San Francisco, Calif.). Multiple protein sequence alignments were performed with the software package MacVector (Oxford Molecular Group, Beaverton, Oreg.).
Agarose gel electrophoresis.
For cloning of ftsZ homologs, purified PCR products were electrophoresed through 0.8% agarose gels in Tris-borate buffer (28) containing ethidium bromide. Appropriate fragments were extracted with the QIAQUICK gel purification kit (QIAGEN) for additional rounds of PCR or restriction enzyme digestion. For PCR-based diagnostics, purified PCR products were subjected to electrophoresis through 1.0% agarose gels in Tris-acetate buffer (28). Gels were stained with 10 μg of ethidium bromide per ml and photographed.
Peptide synthesis and antibody production.
Peptides were synthesized by a model 431A peptide synthesizer (Applied Biosystems). Peptides were dissolved in water at a concentration of 2 mg/ml and diluted 1:1 with Freund’s complete adjuvant (Sigma). The resulting 1-mg/ml inoculant was injected into New Zealand White rabbits (Myrtle’s Rabbitry Inc., Thompson Station, Tenn.) for antibody production. Animals were boosted after 2 weeks with an inoculant containing 1 mg of peptide/ml in Freund’s incomplete adjuvant (Sigma). Rabbits were bled at 3-week intervals, and serum was purified by centrifugation according to standard procedures (28).
SDS-polyacrylamide gel electrophoresis and Western blot analysis.
Proteins from E. coli and Bartonella strains were solubilized in 1× sample buffer (13) at 100°C for 5 min and subjected to electrophoresis on precast 12% Tris-glycine gels (Bio-Rad, Hercules, Calif.). Each gel included samples of protein extracts isolated from E. coli (pUC18) and from E. coli clones expressing cognate Bartonella FtsZ proteins. Gels were run in Tris-glycine-sodium dodecyl sulfate (SDS) running buffer (NOVEX, San Diego, Calif.) at 100 V. Separated proteins were electrotransferred to 0.45-μm-pore-size nitrocellulose membranes (NOVEX) according to the protocol of Towbin et al. (32). Transfer was performed in Tris-glycine buffer with 20% methanol for 1 h at 100 V with cooling. Membranes were blocked in Tris-buffered saline–Tween 20 containing 5% nonfat dry milk. Membranes were incubated with a primary antibody solution consisting of rabbit antipeptide antiserum diluted in blocking buffer. The secondary antibody was goat anti-rabbit immunoglobulin G conjugated to horseradish peroxidase (Kirkegaard and Perry Laboratories, Inc., Gaithersburg, Md.) diluted 1:5,000 in blocking buffer. Membranes were developed with a standard chromogenic substrate (TMB membrane peroxidase substrate system; Kirkegaard and Perry Laboratories, Inc.).
Nucleotide sequence accession numbers.
The nucleotide sequence of the 1,864-bp fragment of B. henselae DNA contained in pUCBH-Z has been deposited into the GenBank database and has been assigned accession no. AF061746. The nucleotide sequence of the 1,893-bp fragment of B. quintana DNA contained in pUCBQ-Z has been assigned accession no. AF061747.
RESULTS
DNA and protein sequence analysis.
Nucleotide sequence analysis of the 1,864-bp fragment of B. henselae DNA contained in pUCBH-Z revealed an ORF of 1,743 bp. This ORF encodes a protein of 581 amino acids (designated FtsZBh) with a predicted molecular mass of 62.3 kDa. Sequence analysis of the 1,893-bp fragment of B. quintana DNA contained in pUCBQ-Z revealed an ORF of 1,770 bp. This ORF encodes a protein of 590 amino acids (designated FtsZBq) with a predicted molecular mass of 63.8 kDa. About 8 nucleotides upstream of the start codon for each ORF is a sequence (AAGAGG) that is homologous to the consensus ribosome binding site of E. coli and other prokaryotes (30).
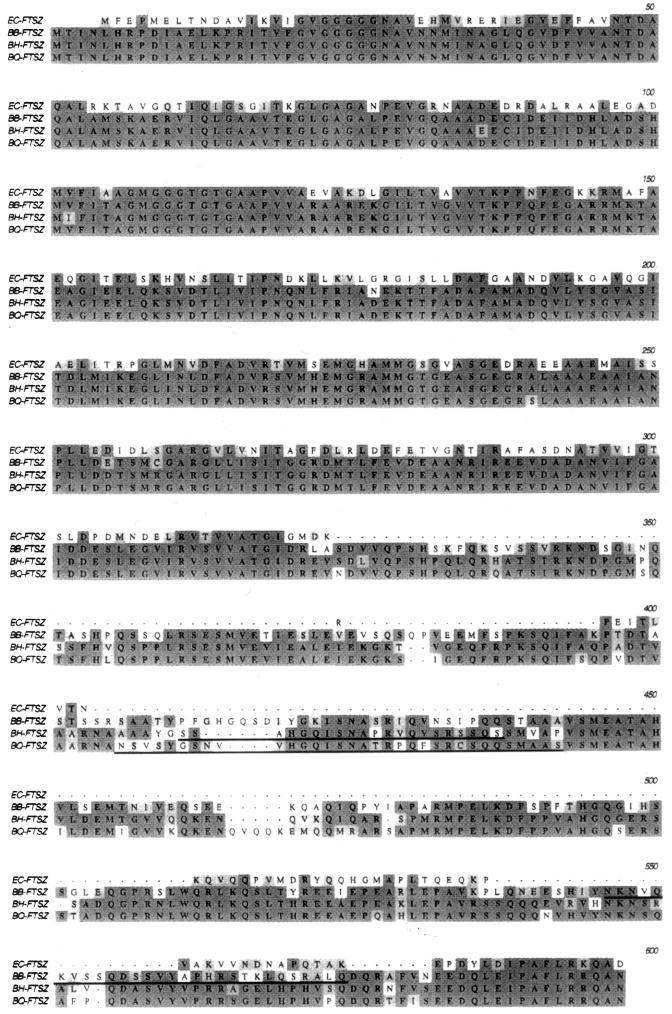
FtsZBh and FtsZBq exhibit 83 and 81% identity, respectively, to FtsZBb and are 91% identical to one another (Fig. 1). The degree of conservation for the three proteins is particularly striking within the N-terminal domains, which exhibit 98 to 99% identity over a length of 321 amino acids. The 321-amino-acid N-terminal halves of FtsZBh and FtsZBq exhibit 63 and 61% identity, respectively, to the N-terminal 321 amino acids of the FtsZ homolog from E. coli (FtsZEc). Like FtsZBb (23), FtsZBh and FtsZBq contain all of the functional domains identified in FtsZEc within their N-terminal domains, including the glycine-rich GTP-GDP binding pocket (residues 109 to 115). This region contains the amino acid sequence GGGTGTG, a sequence which is highly homologous to the tubulin signature motif (GGGTGSG) thought to be involved in GTP binding in eukaryotes (15).
FIG. 1.
Alignment of the amino acid sequences of the FtsZ homologs of E. coli (EC), B. bacilliformis (BB), B. henselae (BH), and B. quintana (BQ). Regions of identity are indicated by dark shading; regions of similarity are indicated by light shading. The peptides used to produce species-specific antisera are underlined in each Bartonella FtsZ homolog sequence.
The C-terminal domains of FtsZBh (260 amino acids) and FtsZBq (269 amino acids) are highly conserved, exhibiting 81% identity. However, the C-terminal domains of the B. henselae and B. quintana homologs both exhibit considerably less homology to the 271-amino-acid C-terminal domain of FtsZBb (57 and 55% identity, respectively).
PCR diagnostics.
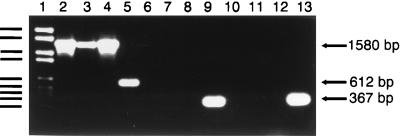
The differences in DNA sequence within the C-terminal regions of the ftsZBb, ftsZBh, and ftsZBq ORFs were exploited to design a species-specific PCR-based diagnostic test. Genomic DNA from B. bacilliformis, B. henselae, and B. quintana was incubated with primers designed to correspond either to sequences of the ftsZ gene that are highly conserved among prokaryotes or to regions that differ significantly among the three Bartonella species (Table 1 and Fig. 2). Primers corresponding to the conserved regions of ftsZ permitted amplification of a product of approximately 1,600 bp from all three Bartonella species under low-stringency conditions (Fig. 3).
TABLE 1.
Primers and peptides used in diagnostic studies
| Primer or peptide | Sequence (5′→3′) | Target organism | Nucleotide positionb | Amino acid positionc | Product size (bp) |
|---|---|---|---|---|---|
| Primers | |||||
| FTSZ-F-CONa | GCTTGGCGCAGCAGTGACA | Bartonella | 189–208 | 1,580 | |
| FTSZ-R-CON | GGCGACGTAAAAATGCTGGTATTTCC | Bartonella | 1768–1743 | ||
| BB-F | AAATCAGACAGCTTCTCATCC | B. bacilliformis | 1043–1063 | 612 | |
| BB-R | GAATCCTGAGAAGACACTTTG | B. bacilliformis | 1655–1634 | ||
| BH-F | GCCGCAAAGTTCTTTTCATG | B. henselae | 1044–1063 | 354 | |
| BH-R | AGGTGAACGCGCTTGTATTTG | B. henselae | 1398–1378 | ||
| BQ-F | GTCTCAGACTTCTTTTCATC | B. quintana | 1044–1063 | 367 | |
| BQ-R | GAGCCCGCATCTGTTGCATTTCCT | B. quintana | 1411–1388 | ||
| Peptides | |||||
| BB-1 | NKNVQKVSSQDSSVYAPHRSTKLQSRAL | B. bacilliformis | 541–568 | ||
| BH-3 | SSAHGQISNAPRVQVSRSSQS | B. henselae | 409–429 | ||
| BQ-5 | NSVSYGSNVVHGQISNATRPQFSRCSQQSMAAS | B. quintana | 404–436 |
FTSZ-F-CON and FTSZ-R-CON are derived from conserved Bartonella ftsZ sequences.
Position within the ftsZBb ORF (23) for conserved primers and within the ORF of the ftsZ homolog of the target organism for species-specific primers.
Position within the FtsZ homolog of the target organism.
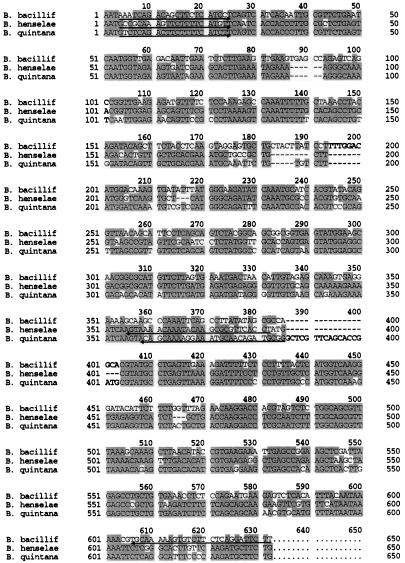
FIG. 2.
Alignment of the DNA sequences of the Bartonella ftsZ homologs within the region of PCR primer design. The sequences given correspond to nucleotides 1041 to 1657 of the ftsZBb ORF, nucleotides 1041 to 1627 of the ftsZBh ORF, and nucleotides 1041 to 1654 of the ftsZBq ORF. Sequences used to design species-specific primers for PCR diagnostic studies are underlined in each homolog sequence, with arrows indicating the 3′ end.
FIG. 3.
Identification of Bartonella species via PCR amplification of ftsZ. DNA Molecular Weight Marker VI (Boehringer Mannheim) in lane 1 consists of the following fragments: 2,176, 1,766, 1,230, 1,033, 653, 517, 453, 394, and 298 bp. Lanes 2 to 4, reaction mixtures containing conserved primers and chromosomal DNA from B. bacilliformis (lane 2), B. henselae (lane 3), and B. quintana Oklahoma (lane 4); lanes 5 to 7, reaction mixtures containing ftsZBb-specific primers and chromosomal DNA from B. bacilliformis (lane 5), B. henselae (lane 6), and B. quintana (lane 7); lanes 8 to 10, reaction mixtures containing ftsZBh-specific primers and chromosomal DNA from B. bacilliformis (lane 8), B. henselae (lane 9), and B. quintana (lane 10); lanes 11 to 13, reaction mixtures containing ftsZBq-specific primers and chromosomal DNA from B. bacilliformis (lane 11), B. henselae (lane 12), and B. quintana (lane 13).
In contrast, the use of primers corresponding to divergent regions of the ftsZ gene resulted in amplification only with those primers derived from the sequence of the cognate species (Fig. 3). All species-specific PCRs were performed under high-stringency conditions. PCR with the B. bacilliformis-specific primers resulted in a product of 612 bp in B. bacilliformis and no product in B. henselae or B. quintana. Incubation with the B. henselae-specific primers resulted in a product of 354 bp in B. henselae only. Finally, PCR including the B. quintana-specific primers resulted in a product of 367 bp in B. quintana only. From these observations, we conclude that a PCR-based diagnostic test utilizing the differences in ftsZ successfully distinguishes the genomes of B. henselae, B. quintana, and B. bacilliformis.
Serology.
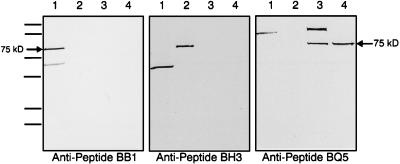
A comparison of the ORFs coding for FtsZBb, FtsZBh, and FtsZBq reveals that sequence divergence at the amino acid level, although limited, is predominantly localized within the C-terminal half of each protein. According to computer modeling predictions, the C-terminal regions also contain a high proportion of antigenic sites. Based on these considerations, synthetic peptides were designed for use in the development of a serological test to distinguish among B. bacilliformis, B. henselae, and B. quintana. Peptides corresponding to the regions of greatest divergence were synthesized and injected into rabbits to generate species-specific antisera (Table 1 and Fig. 1). Western blot analysis was then used to identify immunoreactive proteins extracted from the three Bartonella species. In all cases, a protein corresponding to the size of FtsZ (75 kDa) was observed only in extracts reacted with antisera from the cognate Bartonella species (Fig. 4). No immunoreactive protein of this size was detected when extracts were reacted with antisera generated in response to a heterologous peptide sequence. B. bacilliformis extracts were found to contain an additional immunoreactive protein of about 55 kDa that reacts with both B. bacilliformis- and B. henselae-specific antisera. A protein of about 150 kDa recognized by antibodies to B. quintana was also observed in B. bacilliformis extracts and in an extract derived from the Oklahoma strain of B. quintana. In both of these cases, however, the distinct migration patterns of these proteins make them clearly distinguishable from FtsZ.
FIG. 4.
Identification of Bartonella species via Western blotting with anti-FtsZ peptide antisera. Anti-peptide BB1 antiserum was diluted 1:5,000 in blocking buffer. Anti-peptide BH3 antiserum and anti-peptide BQ5 antiserum were diluted 1:1,000 in blocking buffer. Arrows indicate the positions of Bartonella FtsZ homologs expressed from ftsZ-encoding recombinant plasmids in E. coli JM105. The MultiMark molecular mass protein standard (NOVEX) consists of the following sizes: 250, 148, 60, 42, 30, and 22 kDa. Lanes: 1, B. bacilliformis cell lysate; 2, B. henselae cell lysate; 3, B. quintana Oklahoma cell lysate; 4, B. quintana Fuller cell lysate.
The observed molecular mass of 75 kDa differs significantly from the predicted molecular mass of about 63 kDa. Similar discrepancies in the molecular weight of FtsZ have previously been reported for B. bacilliformis and Rhizobium meliloti (17, 23). In these cases, the discrepancies were attributed to aberrant migration of FtsZ through SDS-polyacrylamide gels.
DISCUSSION
The expression of ftsZ and the mechanism of action of its gene product in E. coli have been studied extensively. In E. coli, FtsZ initiates division by forming the cytokinetic ring at the division site, leading to circumferential invagination of the cytoplasmic membrane and cell wall followed by formation of the division septum (15). In nondividing cells, FtsZ is produced in amounts of 10,000 to 20,000 molecules per cell (6) and is recruited from the cytoplasm during division to form the cytokinetic ring on the inner surface of the cell membrane. Ring formation requires the self-assembly of FtsZ monomers, a process which is dependent on the GTP-binding activity of FtsZ (21). The importance of FtsZ in cell division is reflected in its strong conservation among the eubacteria. The identification of FtsZ homologs in both gram-positive and gram-negative bacteria (5) suggests that the role of this protein in cell division may be shared among all bacteria. FtsZ homologs have recently been reported among several members of the archaebacteria as well (33).
In this study, we report the sequences of FtsZBh and FtsZBq, two FtsZ homologs from the closely related species B. henselae and B. quintana. Within their N-terminal domains, FtsZBh and FtsZBq exhibit high degrees of similarity to the FtsZ family of proteins, features they share with the B. bacilliformis homolog, FtsZBb. Like FtsZBb, however, FtsZBh and FtsZBq each contain an additional C-terminal region of about 300 amino acids that is not present in most of the other FtsZ homologs. The level of amino acid sequence identity between the FtsZ homologs of B. henselae and B. quintana is 91% over the entire length of the proteins. FtsZBh and FtsZBq are slightly less homologous to FtsZBb, exhibiting sequence identities of 83 and 81%, respectively. The higher degree of conservation observed between the B. henselae and B. quintana homologs compared with that of B. bacilliformis is consistent with the pattern of relatedness in Bartonella based on comparisons of the citrate synthase gene sequence reported by Norman et al. (22).
Of the currently characterized FtsZ proteins, the Bartonella homologs appear to be most similar to FtsZRm1, a homolog from R. meliloti which also contains an extended C-terminal domain (17). The N-terminal domains of the Bartonella homologs and FtsZRm1 are 84 to 85% identical, with the degree of identity decreasing to approximately 35% in the C-terminal region. Overall, FtsZRm1 exhibits 68% identity with FtsZBh, 66% identity with FtsZBq, and 65% identity with FtsZBb. In addition to those from Bartonella and Rhizobium, a large FtsZ protein containing a unique C-terminal domain (FtsZAt) has been reported for Agrobacterium tumefaciens (5). The recently published sequence for the A. tumefaciens homolog (16) displays 67% identity with FtsZBh and 65% identity with FtsZBq and FtsZBb. The N-terminal domains of the Bartonella homologs and FtsZAt are 83 to 85% identical. Significantly, all of the bacteria reported to possess large FtsZ homologs belong to the alpha-2 subgroup of Proteobacteria, and they all interact closely with eukaryotic cells. The relationship with eukaryotic cells may be pathogenic, as in the case of Bartonella and A. tumefaciens, or endosymbiotic, as in the case of R. meliloti.
FtsZBb has been shown previously (23) to be recognized both by antiserum raised against E. coli FtsZ and by anti-Bartonella serum isolated from human patients. These antisera recognize different domains of the protein, however, with the E. coli antiserum recognizing the conserved N-terminal domain and the human anti-Bartonella antiserum exhibiting specificity for the unique C-terminal domain. These observations have led to the hypothesis that FtsZBb is associated with the cell membrane with the C-terminal domain exposed to the bacterial cell surface, allowing it to serve as an antigen during infection (23). Support for this theory comes from the observation that the 75-kDa antigen of B. bacilliformis is present in membrane fractions of lysed cells (12, 20).
The differences within the C-terminal regions of the Bartonella ftsZ ORFs, although few in number, were sufficient to allow us to develop strategies for the differentiation of B. henselae, B. quintana, and B. bacilliformis at both the DNA and protein levels. Since the diseases caused by these organisms can produce symptoms that are clinically and histologically similar, a rapid, reproducible method for identifying these pathogens is highly desirable. Because Bartonella species are notoriously difficult to culture by traditional means, many clinicians have relied on PCR-based and serological methods to diagnose these disorders.
PCR-based diagnostic procedures for differentiating Bartonella species have been available for several years. The first description of a PCR-based diagnostic test (26) utilized primers designed for the amplification of Bartonella 16S rRNA gene sequences, but these primers were not genus specific and resulted in amplification of rRNA gene fragments from many other Proteobacteria. Therefore, use of this test requires direct sequence analysis of amplified DNA for identification at the species level. More recently, protocols involving PCR amplification of an htrA-like fragment followed by hybridization of a species-specific probe (2), PCR-restriction fragment length polymorphism with gltA (22), and repetitive extragenic palindromic PCR (27) have been described for distinguishing among Bartonella species. The use of primers specific for unique sequences within the C-terminal region of ftsZ described here requires only one round of PCR and avoids some of the problems with specificity and consistency reported for other protocols. We are currently investigating the capability of the primers described here to amplify ftsZ fragments from multiple strains within the three Bartonella species. In addition, the utility of the ftsZ primers as tools for diagnosis of clinical specimens is currently being explored.
PCR-based tests commonly use lymph node biopsy specimens as sources of Bartonella DNA for clinical diagnosis. Many physicians prefer serological methods for diagnosis where possible because they do not require invasive procedures to acquire antigen. The serological assays currently used most frequently are the indirect fluorescence assay (IFA) and the enzyme-linked immunoassay, but the diagnostic value of these tests is questionable due to low sensitivity. The reported success in detecting B. henselae by IFA has ranged from 54 to 88% in patients with clinical CSD (9, 11, 24). A recent study specifically aimed at evaluating the diagnostic value of B. henselae-based IFA in patients with clinical CSD reported sensitivities ranging from 31.8 to 40.9% for anti-B. henselae immunoglobulin G, depending on the method of antigen preparation (3). In some cases, these variations may reflect differences in the definition of clinical CSD. Recently, a new serotype of B. henselae was described (8), and it now appears that B. henselae exists as a widely distributed group of serovariant strains (18). This antigenic variation among strains may help to explain the inconsistent IFA results.
A matter of significant concern in the use of IFA and other serological tests is the potential for misdiagnosis due to cross-reactivity. Serological cross-reactions have been observed between B. henselae and B. quintana antigens (7) and also between Bartonella species and other bacterial pathogens, such as Coxiella burnetii and Chlamydia species (14, 19). Such cross-reactivity represents a serious problem in the diagnosis of disorders caused by these agents. Endocarditis, for example, can be caused by B. henselae, B. quintana, C. burnetii, and at least two Chlamydia species. Since the antibiotic regimens used to treat infections caused by these pathogens differ significantly, proper diagnosis is critical. We are currently investigating the utility of the synthetic FtsZ peptides described here as species-specific antigens for serological diagnosis of Bartonella-related disorders.
In summary, we have described the use of the cell division gene ftsZ to differentiate among three closely related Bartonella species, each of which is a distinct human pathogen. These studies may provide a basis for reproducible molecular diagnosis of CSD and related disorders.
ACKNOWLEDGMENTS
We are grateful for the technical assistance provided by Judy Cooper in growing the B. henselae and B. quintana strains and to members of the Biology Core Facility at Georgia State University for DNA sequence analysis and synthesis of primers and peptides. We also thank Burt Anderson, University of South Florida, for providing B. quintana Oklahoma.
REFERENCES
- 1.Anderson B, Goldsmith C, Johnson A, Padmalayam I, Baumstark B. Bacteriophage-like particle of Rochalimaea henselae. Mol Microbiol. 1994;13:67–73. doi: 10.1111/j.1365-2958.1994.tb00402.x. [DOI] [PubMed] [Google Scholar]
- 2.Anderson B, Sims K, Regnery R, Robinson L, Schmidt M J, Goral S, Hager C, Edwards K. Detection of Rochalimaea henselae DNA in specimens from cat scratch disease patients by PCR. J Clin Microbiol. 1994;32:942–948. doi: 10.1128/jcm.32.4.942-948.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergmans A M C, Peeters M F, Schellekens J F P, Vos M C, Sabbe L J M, Ossewaarde J M, Verbakel H, Hooft H J, Schouls L M. Pitfalls and fallacies of cat scratch disease serology: evaluation of Bartonella henselae-based indirect fluorescence assay and enzyme-linked immunoassay. J Clin Microbiol. 1997;35:1931–1937. doi: 10.1128/jcm.35.8.1931-1937.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chomel B B, Kasten R W, Floyd-Hawkins K, Chi B, Yamamoto K, Roberts-Wilson J, Gurfield A N, Abbott R C, Pederson N C, Koehler J E. Experimental transmission of Bartonella henselae by the cat flea. J Clin Microbiol. 1996;34:1952–1956. doi: 10.1128/jcm.34.8.1952-1956.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corton J C, Ward J E, Jr, Lutkenhaus J. Analysis of cell division gene ftsZ (sulB) from gram-negative and gram-positive bacteria. J Bacteriol. 1987;169:1–7. doi: 10.1128/jb.169.1.1-7.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai K, Lutkenhaus J. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J Bacteriol. 1992;174:6145–6151. doi: 10.1128/jb.174.19.6145-6151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalton M J, Robinson L E, Cooper J, Regnery R L, Olsen J G, Childs J E. Use of Bartonella antigens for serologic diagnosis of cat-scratch disease at a national referral center. Arch Intern Med. 1995;155:1670–1676. [PubMed] [Google Scholar]
- 8.Drancourt M, Birtles R, Chaumentin G, Vandenesch F, Etienne J, Raoult D. New serotype of Bartonella henselae in endocarditis and cat-scratch disease. Lancet. 1996;347:441–443. doi: 10.1016/s0140-6736(96)90012-4. [DOI] [PubMed] [Google Scholar]
- 9.Flexman J P, Chen S C A, Dickeson D J, Pearman J W, Gilbert G L. Detection of antibodies to Bartonella henselae in clinically diagnosed cat scratch disease. Med J Aust. 1997;166:532–535. doi: 10.5694/j.1326-5377.1997.tb123245.x. [DOI] [PubMed] [Google Scholar]
- 10.Gray G C, Johnson A A, Thornton S A, Smith W A, Knobloch J, Kelley P W, Escudero L O, Huayda M A, Wignall F S. An epidemic of Oroya Fever in the Peruvian Andes. Am J Trop Med Hyg. 1990;42:215–221. doi: 10.4269/ajtmh.1990.42.215. [DOI] [PubMed] [Google Scholar]
- 11.Hamilton D H, Zangwill K M, Hadler J L, Cartter M L. Cat-scratch disease—Connecticut, 1992–1993. J Infect Dis. 1995;172:570–573. doi: 10.1093/infdis/172.2.570. [DOI] [PubMed] [Google Scholar]
- 12.Knobloch J. Analysis and preparation of Bartonella bacilliformis antigens. Am J Trop Med Hyg. 1988;39:173–178. doi: 10.4269/ajtmh.1988.39.173. [DOI] [PubMed] [Google Scholar]
- 13.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 14.La Scola B, Raoult D. Serological cross-reactions between Bartonella quintana, Bartonella henselae, and Coxiella burnetii. J Clin Microbiol. 1996;34:2270–2274. doi: 10.1128/jcm.34.9.2270-2274.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lutkenhaus J. FtsZ ring in bacterial cytokinesis. Mol Microbiol. 1993;9:403–409. doi: 10.1111/j.1365-2958.1993.tb01701.x. [DOI] [PubMed] [Google Scholar]
- 16.Ma X, Sun Q, Wang R, Singh G, Jonietz E L, Margolin W. Interactions between heterologous FtsA and FtsZ proteins at the FtsZ ring. J Bacteriol. 1997;179:6788–6797. doi: 10.1128/jb.179.21.6788-6797.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Margolin W, Corbo J C, Long S R. Cloning and characterization of a Rhizobium meliloti homolog of the Escherichia coli cell division gene ftsZ. J Bacteriol. 1991;173:5822–5830. doi: 10.1128/jb.173.18.5822-5830.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maurin M, Birtles R, Raoult D. Current knowledge of Bartonella species. Eur J Clin Microbiol Infect Dis. 1997;16:487–506. doi: 10.1007/BF01708232. [DOI] [PubMed] [Google Scholar]
- 19.Maurin M, Eb F, Etienne J, Raoult D. Serological cross-reactions between Bartonella and Chlamydia species: implications for diagnosis. J Clin Microbiol. 1997;35:2283–2287. doi: 10.1128/jcm.35.9.2283-2287.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minnick M F. Identification of outer membrane proteins of Bartonella bacilliformis. Infect Immun. 1994;62:2644–2648. doi: 10.1128/iai.62.6.2644-2648.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mukherjee A, Lutkenhaus J. Guanine nucleotide-dependent assembly of FtsZ into filaments. J Bacteriol. 1994;176:2754–2758. doi: 10.1128/jb.176.9.2754-2758.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Norman A F, Regnery R, Jameson P, Greene C, Krause D C. Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J Clin Microbiol. 1995;33:1797–1803. doi: 10.1128/jcm.33.7.1797-1803.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Padmalayam I, Anderson B, Kron M, Kelly T, Baumstark B. The 75-kilodalton antigen of Bartonella bacilliformis is a structural homolog of the cell division protein FtsZ. J Bacteriol. 1997;179:4545–4552. doi: 10.1128/jb.179.14.4545-4552.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Regnery R, Tappero J. Unraveling mysteries associated with cat-scratch disease, bacillary angiomatosis, and related syndromes. Emerg Infect Dis. 1995;1:16–21. doi: 10.3201/eid0101.950103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Regnery R L, Olson J G, Perkins B A, Bibb W. Serological response to “Rochalimaea henselae” antigen in suspected cat-scratch disease. Lancet. 1992;339:1443–1445. doi: 10.1016/0140-6736(92)92032-b. [DOI] [PubMed] [Google Scholar]
- 26.Relman D A, Loutit J S, Schmidt T M, Falkow S, Tompkins L S. The agent of bacillary angiomatosis: an approach to the identification of uncultured pathogens. N Engl J Med. 1990;323:1573–1580. doi: 10.1056/NEJM199012063232301. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez-Barradas M C, Hamill R J, Houston E D, Georghiou P R, Clarridge J E, Regnery R L, Koehler J E. Genomic fingerprinting of Bartonella species by repetitive element PCR for distinguishing species and isolates. J Clin Microbiol. 1995;33:1089–1093. doi: 10.1128/jcm.33.5.1089-1093.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shine J, Dalgarno L. The 3′ terminal sequence of E. coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosomal binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stoler M H, Bonfiglio T A, Steigbigel R T, Pereira M. An atypical subcutaneous infection associated with acquired immune deficiency syndrome. Am J Clin Pathol. 1983;80:714–718. doi: 10.1093/ajcp/80.5.714. [DOI] [PubMed] [Google Scholar]
- 32.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Lutkenhaus J. FtsZ ring: the eubacterial division apparatus conserved in archaebacteria. Mol Microbiol. 1996;21:313–319. doi: 10.1046/j.1365-2958.1996.6421360.x. [DOI] [PubMed] [Google Scholar]